Abstract
Crosslinking and immunoprecipitation (CLIP) followed by high-throughput sequencing identifies the binding sites of RNA binding proteins on RNAs. The covalent RNA-amino acid adducts produced by UV irradiation can cause premature reverse transcription termination and deletions (referred to as crosslink-induced mutation sites (CIMS)), which may decrease overall cDNA yield but are exploited in state-of-the-art CLIP methods to identify these crosslink sites at single-nucleotide resolution. Here, we show the ratio of both crosslinked base deletions and read-through versus termination are highly dependent on the identity of the reverse transcriptase enzyme as well as on buffer conditions used. AffinityScript and TGIRT showed a lack of deletion of the crosslinked base with other enzymes showing variable rates, indicating that utilization and interpretation of CIMS analysis requires knowledge of the reverse transcriptase enzyme used. Commonly used enzymes, including Superscript III and AffinityScript, show high termination rates in standard magnesium buffer conditions, but show a single base difference in the position of termination for TARDBP motifs. In contrast, manganese-containing buffer promoted read-through at the adduct site. These results validate the use of standard enzymes and also propose alternative enzyme and buffer choices for particularly challenging samples that contain extensive RNA adducts or other modifications that inhibit standard reverse transcription.
Keywords: CLIP-seq, eCLIP-seq, RNA binding protein
1.1 Introduction
RNA molecules play a variety of roles in cells, ranging from their well described responsibilities as messenger RNAs encoding instructions for the translation of proteins by the ribosome, to direct roles in modulating transcription, chromatin structure, RNA processing, and translation [1]. To achieve these varied tasks, each RNA undergoes numerous processing steps that are tightly controlled by the activity of RNA binding proteins [1, 2].
Early methods to discover RNA targets of RNA binding protein targets were limited to transcript-level resolution with techniques such as RIP-CHIP [3]. However, the development of crosslinking and immunoprecipitation (CLIP) methods enabled finer resolution mapping of binding sites through the incorporation of RNA fragmentation (typically by limited RNase treatment) [4]. Although CLIP methods initially identified binding sites with ~20–100nt resolution, analysis of early datasets revealed that reverse transcriptase (RT) enzymes often either terminated or created insertions and deletions (indels) at protein-RNA crosslink sites [5, 6].
To enable identification of binding sites and motifs at single-nucleotide resolution, iCLIP incorporated ligation of the second adapter after reverse transcription (which is maintained in the eCLIP method) [7]. In this way, the sequencing read identifies not only an RNA crosslinked to protein, but also maps the exact position of crosslinking if the RT terminates at the crosslink site [7]. This termination has been estimated to occur with up to 80% frequency, giving increased resolution to studies of RNA binding protein RNA targets [5, 8]. However, different RT enzymes have different levels of processivity and sensitivity to contaminants and RNA damage, which can be modified by altered reaction conditions. Of particular note, replacement of magnesium with manganese increases mis-incorporation of bases with many DNA polymerase enzymes and encourages the addition of non-templated nucleotides (particularly cytosines) by M-MLV reverse transcriptase enzymes [9, 10], and has been suggested to improve reverse transcription yield on targets that are highly crosslinked or contain RNA modifications.
To test the effectiveness of various reverse transcriptase enzymes and buffer formulations, we performed parallel eCLIP experiments for two RNA binding proteins previously shown to yield motifs at single-nucleotide resolution (RBFOX2 and TARDBP/TDP43). We observed that while overall cDNA yields were comparable across many enzymes, altering the choice of enzyme has a significant impact on read-through efficiency at crosslink sites. In particular, replacement of magnesium ions in standard reverse transcriptase buffer with manganese caused a dramatic decrease in termination, suggesting that this may present an alternative formulation that enables successful CLIP performed on samples with increased UV crosslinking. Further, we observed that some enzymes progress one base further than others through crosslinked uracil nucleotides, suggesting that computational analyses performed across multiple CLIP-seq datasets should be aware of the potential for differences due to reverse transcriptase enzyme choice.
1.2 Materials and methods
1.2.1 eCLIP experimental details
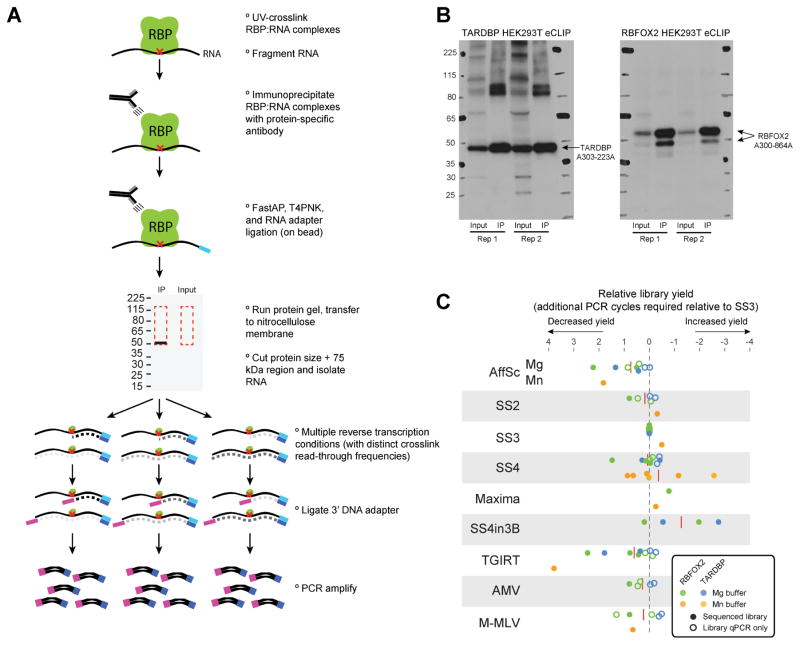
eCLIP experiments were performed largely as previously described in a detailed standard operating procedure, with modifications noted below [11]. As previously described, 107 cells were UV-crosslinked (254 nm, 400 mJ/cm2), lysed in 1 mL of 4°C eCLIP lysis buffer (50 mM TrisHCl pH 7.4, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 1:200 Protease Inhibitor Cocktail III (EMD Millipore)), incubated at 37°C for 5 min with 40 U of RNase I (Ambion) and 4 U Turbo DNase (Ambion), treated with RNase inhibitor (NEB), and clarified by centrifugation (4°C, 15kg for 15 min). Primary antibodies were pre-coupled with sheep anti-rabbit or anti-mouse IgG Dynabeads (ThermoFisher), and incubated 4°C overnight with rotation.
Following incubation, samples were magnetically separated and washed twice in high salt wash buffer (50 mM Tris-HCl pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, and 0.5% sodium deoxycholate) and twice in wash buffer (20 mM Tris HCl pH 7.4, 10 mM MgCl2, 0.2% Tween-20). Next, dephosphorylation of 5′ ends was performed with FastAP (ThermoFisher), followed by T4 PNK (NEB) treatment at low pH in the absence of ATP to remove 2′–3′ cyclic phosphates left by RNase I digestion. High efficiency ligation of 3′ adapters was then performed with T4 RNA Ligase I (NEB) with 18% PEG 8000 and 0.3% DMSO. After one additional high salt buffer wash and two additional wash buffer washes, samples were denatured in standard NuPAGE buffer with 0.1M DTT, and run at 150V on 4–12% NuPAGE Novel Bis-Tris protein gels (ThermoFisher). Replicate gels were transferred to both PVDF (for chemiluminescent imaging) and nitrocellulose (for RNA extraction) membranes. PVDF membranes were blocked with 5% milk, incubated with 1:5000 diluted primary antibody in 5% milk, washed three times in TBST, incubated with 1:2000 diluted TrueBlot anti-rabbit secondary antibody (Rockland Inc.), and imaged with standard enhanced chemiluminescence. To extract RNA samples from nitrocellulose membranes, a range from the observed protein size to 75 kDa above was isolated, finely fragmented, and treated with Proteinase K (NEB) in PK buffer (100 mM TrisHCl, pH 7.4, 50 mM NaCl, 10 mM EDTA) with 7M urea. RNA was then purified using phenol-chloroform extraction followed by RNA Clean & Concentrator column cleanup (Zymo).
At this stage, samples were divided based on the desired number of reverse transcription conditions to be assayed, and 2.5 μL of RNA (equivalent to approximately 3×106 cells) was used for each condition. Reaction conditions for reverse transcription are listed in section 1.2.2. After reverse transcription, excess oligonucleotides were removed with ExoSap-IT (Affymetrix), treated with EDTA to quench reactions, with NaOH to hydrolize remaining RNA, and pH balanced with HCl. Sample cleanups were performed with MyONE Silane beads as previously described except for the addition of 0.05% NP40 to all RLT buffer steps. 3′ DNA adapter ligation was then performed using T4 RNA Ligase I (NEB) in optimized reaction conditions including 22% PEG 8000. For the last batch of experiments (including Maxima, SSIII, and one replicate of all manganese buffer conditions), 0.3 μL of 5′ Deadenylase (NEB) was also added for increased ligation efficiency. PCR amplification was performed with Q5 master mix (NEB) for 6–18 cycles (chosen based on amplification Ct obtained from qPCR performed on the pre-amplified library). The library within a 175–300nt size range was size-selected by agarose gel electrophoresis and gel extracted (MinElute Gel Extraction, Qiagen). Libraries were quantitated and validated by Tapestation (Agilent), and sequenced on the Illumina HiSeq 4000 platform.
To estimate library yield, an extrapolated Ct (eCT) value was defined as the number of PCR cycles necessary to obtain 100 femtomoles of amplified library. This eCT value was calculated by taking the number of PCR cycles performed and subtracting the log2 ratio of final library yield divided by 100 femtomoles. To control for batch or biosample effects, eCT values were compared relative to Superscript III (which was included in all batches). For the subset of libraries that were only taken to the pre-amplified library stage, the Ct value obtained from qPCR of the pre-amplified library was used in place of the eCT value, and normalized against paired Superscript III experiments.
1.2.2 Reverse transcription reaction conditions
AffinityScript
To 2.5 μL RNA was added 1 μL 5 μM AR17 and 6.5 μL H2O, and samples were mixed, incubated at 65C for 2 minutes, and placed on ice. To each was added 4.4 μL H2O, 2 μL 10X AffinityScript buffer, 2 μL 0.1M DTT, 0.8 μL 25 mM dNTPs, 0.3 μL Murine RNase Inhibitor (NEB), and 0.5 μL AffinityScript enzyme. Samples were mixed and incubated at 55°C for 45 minutes.
Superscript II
To 2.5 μL RNA was added 1 μL 5 μM AR17, 1 μL 10 mM dNTPs, and 5.5 μL H2O, and samples were mixed, incubated at 65C for 2 minutes, and placed on ice. To each was added 4.2 μL H2O, 4 μL of the appropriate 5X Superscript buffer, 1 μL 0.1M DTT, 0.3 μL Murine RNase Inhibitor and 0.8 μL of Superscript II enzyme (ThermoFisher). Samples were mixed and incubated at 45°C for 45 minutes.
Superscript III, Superscript IV, and Superscript IV in III buffer
To 2.5 μL RNA was added 1 μL 5 μM AR17, 1 μL 10 mM dNTPs, and 5.5 μL H2O, and samples were mixed, incubated at 65C for 2 minutes, and placed on ice. To each was added 3 μL H2O, 4 μL 5X Superscript II buffer, 2 μL 0.1M DTT, 0.2 Murine RNase Inhibitor, and 0.5 μL of the appropriate Superscript enzyme. Samples were mixed and incubated at 55°C for 45 minutes.
Superscript IV in Mn buffer
To 2.5 μL RNA was added 1 μL 5 μM AR17, 1.4 μL 10 mM dNTPs, and 5.5 μL H2O, and samples were mixed, incubated at 65C for 2 minutes, and placed on ice. To each was added 2 μL H2O, 4 μL 5X Mn buffer (250 mM Tris pH 8.0, 375 mM KCl, 30 mM MnCl2), 2.8 μL 0.1M DTT, 0.3 μL Murine RNase Inhibitor, and 0.5 μL Superscript IV enzyme. Samples were mixed and incubated at 55°C for 45 minutes.
TGIRT
To 2.5 μL RNA was added 1 μL 5 μM AR17 and 5.5 μL H2O, and samples were mixed, incubated at 65C for 2 minutes, and placed on ice. To each was added 3 μL H2O, 4 μL of 5X TGIRT buffer (2.25M NaCl, 25 mM MgCl, 100 mM TrisHCl pH 7.5), 1 μL 0.1M DTT, 0.5 μL TGIRT-III enzyme (InGex), and 2.5 μL 10 mM dNTP mix. Samples were mixed and incubated at 55°C for 30 minutes followed by 60°C for 30 minutes.
AMV
To 2.5 μL RNA was added 1 μL 5 μM AR17, 1 μL 10 mM dNTPs, and 5.5 μL H2O, and samples were mixed, incubated at 65C for 2 minutes, and placed on ice. To each was added 7 μL H2O, 2 μL 10X AMV buffer (NEB), 0.2 μL RNase OUT RNase inhibitor (ThermoFisher), and 0.8 μL (8 units) of AMV enzyme (NEB AMV Reverse Transcriptase). Samples were mixed and incubated at 45°C for 45 minutes.
M-MLV
To 2.5 μL RNA was added 1 μL 5 μM AR17 and 5.5 μL H2O, and samples were mixed, incubated at 65C for 2 minutes, and placed on ice. To each was added 4 μL H2O, 4 μL 5X M-MLV buffer (Promega), 1 μL 10 mM dNTPs, 0.2 μL Murine RNase Inhibitor (NEB), and 0.8 μL of M-MLV enzyme (Promega M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant). Samples were mixed and incubated at 45°C for 45 minutes.
For all manganese buffer conditions, MgCl2 was replaced with the same concentration of MnCl2.
1.2.3 Primary antibodies used
RBFOX2 experiments were performed with 10 μg (10 μL) of A300-864A lot #002 (Bethyl) per 2×107 cells. TARDBP experiments were performed with 2 μg (10 μL) of A303-223A lot #001 (Bethyl) per 2×107 cells.
1.2.4 eCLIP data processing
Primary eCLIP data analysis, including read quality processing and adapter trimming (with Cutadapt), removal of repetitive element-mapping reads, mapping to the hg19 genome and transcriptome (with STAR), initial cluster identification (with CLIPper), and input normalization (with custom scripts) was performed as previously described [11, 12]. One set of paired size-matched inputs was generated for AffSc, SS3, SS4, SS4Mn, SS4in3B, and TGIRT conditions for each of RBFOX2 and TARDBP, which were used to normalize all IP datasets using each respective enzyme. Other enzymes (AMV, M-MLV, and SS2) were normalized against a previously published RBFOX2 HEK293XT size-matched input generated with AffinityScript [11]. Sequencing data has been deposited in the Gene Expression Omnibus (GSE101938).
1.2.5 Analysis of eCLIP data
To annotate peak overlaps with gene regions, each peak was compared against gene annotations from GENCODE (v.19). Overlaps with annotated regions were prioritized in the following order: exons (coding sequence (CDS), 3′ untranslated region (3′UTR) and 5′UTR), 5′ splice site (5′SS, defined as the 100 nt region beginning with the 5′ splice site) of coding or non-coding transcripts, 3′SS of coding or non-coding transcripts, proximal introns (the 400 nt intronic regions proximal to the 5′SS or 3′SS regions) of coding or non-coding transcripts, and distal introns (the remaining intronic sequence) of coding or non-coding transcripts.
To calculate correlation of peak fold-enrichment for a pair of eCLIP experiments, read density enrichment in IP versus size-matched input was calculated for each dataset using the set of input-enriched peaks (requiring ≥2-fold enrichment and p-value ≤ 0.1 in IP versus paired size-matched input) from the first dataset. Correlation (Pearson R) was then calculated comparing fold-enrichments in both datasets for all peaks. This process was repeated for all pairs of RBFOX2 and TARDBP datasets.
1.2.6 Single-nucleotide motif analysis at crosslink sites
To consider crosslink site termination or read-through, the start position for genomic mapping was obtained for each uniquely mapped, non-PCR duplicate read and defined as the “genomic 0 position” relative to the read start (using the second paired-end read). For positions ranging from −10 to +10nt around this start position, each k-mer (6nt for RBFOX2 analysis, 5nt for TARDBP analysis, or 1nt for uracil enrichment analyses) was counted for all reads to obtain the frequency of all motifs at positions relative to read starts, and normalized by the total number of reads. Frequency of uracil bases at positions within the read sequence were counted similarly, using the first base of the read as the “read 0 position”.
Analysis of crosslink-induced mutation sites (CIMS) was performed by first identifying deletions annotated by our standard STAR mapping. K-mers were then counted relative to the deletion, defining position 0 as last base of the deletion, and divided by the number of total reads to obtain the frequency of deletion-relative motifs.
1.3 Results
RBFOX2 and TARDBP are well-characterized RNA binding proteins with specific affinity for UGCAUG and GAAUG motifs respectively [13]. We previously observed that eCLIP performed on RBFOX2 and TARDBP not only identified these motifs as enriched, but also that these motifs were particularly enriched at specific locations relative to the start of eCLIP reads: the UGCAUG 6-mer was enriched in RBFOX2 eCLIP at the −2 and −6 positions relative to read starts, whereas the GAAUG 5-mer was enriched at the −4 position in TARDBP eCLIP [11].
1.3.1 Altering reverse transcription reactions does not dramatically effect eCLIP library yield
As the standard eCLIP method uses the AffinityScript reverse transcriptase enzyme [11], we set out to specifically test the effect on eCLIP binding maps and motif identification with the use of alternative reverse transcriptase enzymes. To do this, we performed standard eCLIP on RBFOX2 and TARDBP in HEK293T cells, validated successful immunoprecipitation by western blot, and partitioned the sample at the reverse transcription stage (Fig. 1A–B). We tested a core set of nine conditions (AffinityScript, Superscript II, Superscript III, Superscript IV, TGIRT, Superscript IV enzyme in Superscript III buffer, Maxima, AMV, and M-MLV enzymes), many in multiple biological replicates, as well as paired size-matched inputs (Fig. 1A,C). We observed generally minor differences across all nine conditions for both RBFOX2 and TARDBP, with the difference relative to Superscript III ranging from an average of 0.1 cycles (for Superscript IV) to 1.3 (Superscript IV enzyme with Superscript III buffer conditions)(Fig. 1C). Altered buffer conditions (particularly with manganese-containing buffer, which may not yet be optimized for all enzymes) showed more variable yields, with increases in some experiments but no changes in others (Fig. 1C). We note that unlike our results showing a decrease in library yield with TGIRT enzyme relative to Superscript III, other reports have suggested an increased library yield [14], suggesting that differences in CLIP protocols may yield different sensitivities for various reverse transcription enzymes or reaction conditions.
Figure 1. Testing reverse transcriptase conditions with eCLIP.
(A) eCLIP overall schematic. A single biological sample was lysed, immunoprecipitated, and taken through standard eCLIP library preparation until the reverse transcription stage, at which point it was split into multiple conditions. (B) Immunoprecipitation (IP) western blot images for (left) TARDBP and (right) RBFOX2 eCLIP performed in HEK293XT cells. (C) Library yield obtained in eCLIP experiments for RBFOX2 (filled circles) and TARDBP (empty circles), normalized to a Superscript III condition performed within that experiment batch. Average across all experiments is indicated by red dashed lines. For eCLIP experiments that were completed and sequenced (black), yield was calculated as the number of PCR cycles required to obtain 100 femtomoles of library (extrapolated from the library yield and number of PCR cycles performed). For additional experiments only taken to pre-amplified library stage (blue), library yield was determined as the Ct value obtained by qPCR of the pre-amplified library with standard library amplification primers. Reverse transcription conditions tested were AffinityScript (AffSc), Superscript II (SS2), Superscript III (SS3), Superscript IV (SS4), Superscript IV in manganese buffer (SS4Mn), Superscript IV in Superscript III buffer (SS4in3B), TGIRT-III enzyme (TGIRT), AMV, and M-MLV. Standard buffers and reaction conditions were used unless otherwise indicated (see Materials & Methods).
1.3.2 General properties of eCLIP profiles are insensitive to RT condition choice
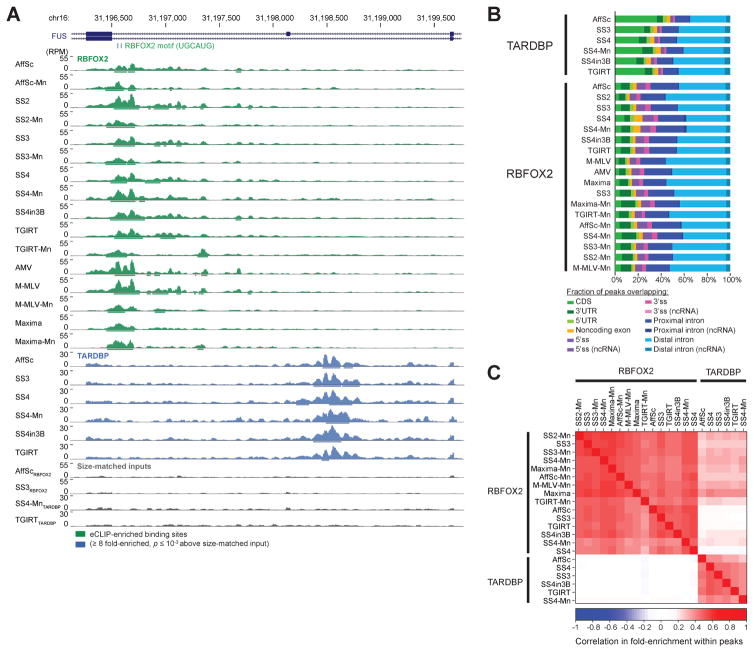
After high-throughput sequencing, reads were processed and mapped to the human genome using standard eCLIP analytical methods [11]. Next, we performed a variety of analyses to assay experimental success. Manual inspection of individual binding sites showed broadly similar results across all datasets, considering both peak locations and height (Fig. 2A). Considering the location of binding along transcripts, we observed that RBFOX2 showed particular enrichment for both proximal (within 500 nt of the splice site) and distal intronic regions whereas TARDBP showed relatively larger enrichment at distal intronic regions, matching previous results (Fig. 2B) [11, 15, 16].
Figure 2. eCLIP yields similar binding profiles with different reverse transcription conditions.
(A) Genome browser depiction of RBFOX2, TARDBP, and selected paired size-matched input eCLIP read density for exons 6–8 of FUS, with eCLIP performed with indicated reverse transcription conditions (abbreviated as in Fig. 1C). Read densities are shown as reads per million (RPM). Significantly enriched peaks are displayed as bars below read density tracks. (B) Bars indicate the cumulative fraction of significantly enriched peaks overlapping the indicated regions of annotated transcripts. (C) Color indicates Pearson correlation (R) between IP versus input fold-enrichment in in peaks for pair-wise comparison of eCLIP experiments, based on the peak list for the dataset indicated on the x-axis. Shown are all datasets with independent paired size-matched inputs.
To further validate these experiments, we calculated the pair-wise Pearson correlation in fold-enrichment (read density in IP versus size-matched input) from all peaks in each dataset. To maintain independence between datasets, we only considered the six conditions for which paired size-matched inputs were performed. We observed high correlation across reverse transcription reaction conditions for the same RBP, with average pairwise correlation of 0.49 for RBFOX2 and 0.42 for TARDBP (Fig. 2C). In contrast, we observed an average correlation of 0.06 between RBFOX2 and TARDBP experiments, with none higher than 0.23 (Fig. 2C). In summary, these results confirm that all tested reverse transcription conditions can be used to successfully perform eCLIP experiments and yield expected RBP-specific global binding profiles.
1.3.3 Reverse transcription termination frequency depends on reverse transcription conditions
Proteinase K treatment during CLIP procedures leaves a short peptide adduct on the RNA, which often causes reverse transcriptase enzymes to terminate extension of the cDNA (Fig. 1A). By performing the second adapter ligation after reverse transcription, iCLIP and subsequent CLIP procedures (including eCLIP) are thus able to use the 5′ end of the sequence read to identify enrichments characteristic of reverse transcription termination due to RBP crosslink sites [5]. In the standard eCLIP methodology, this site is positioned at the start of the second (paired-end) read (Fig. 1A) [11].
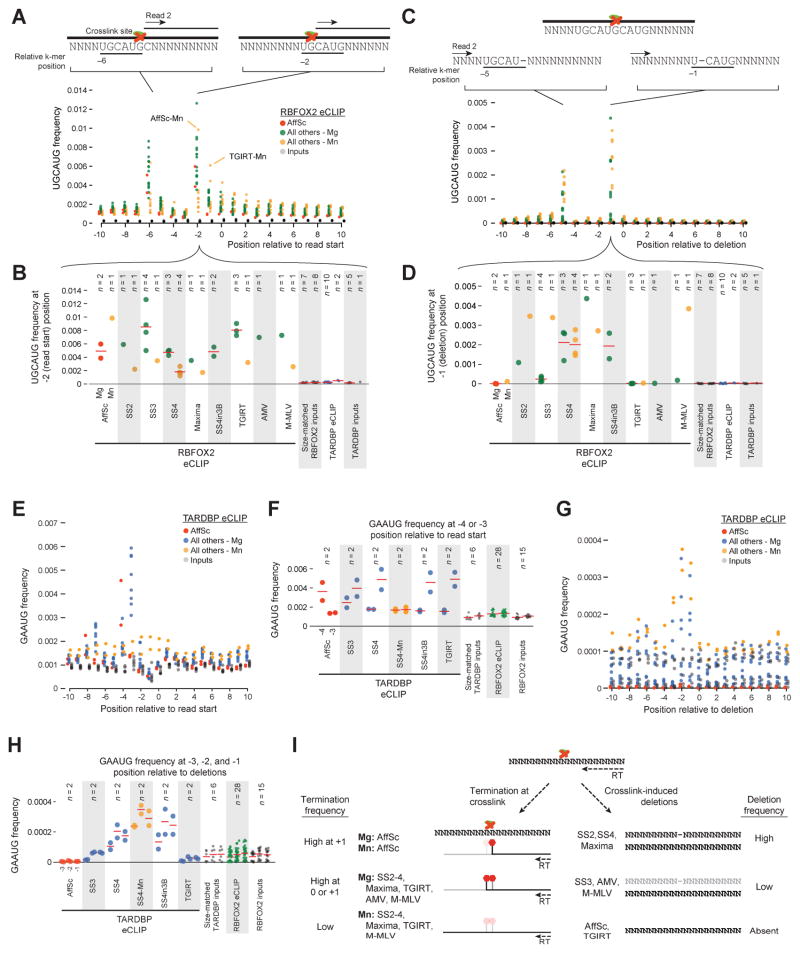
To consider whether this reverse transcription termination was altered by reverse transcriptase conditions, we counted the frequency of 5- and 6-nt sequences at positions around read start positions. For RBFOX2, we observed specific enrichments at the genomic −2 and −6 positions relative to the start of the second read, matching previous CLIP-seq observations (Fig. 3A) [11]. Further, previous structural studies of RBFOX2 confirm specific interaction with guanine residues in the RBFOX2 motif, suggesting that in RBFOX2 eCLIP reverse transcription termination is enriched for occurring at the base prior to the crosslink site (Fig. 3A) [17]. Comparing across conditions specifically at the genomic −2 position, we noted that the majority of conditions with standard magnesium-based buffers showed enriched motif frequency at these positions, although Maxima and SS4 had somewhat lower frequency (Fig. 3B). Conversely, manganese buffer conditions for a variety of enzymes showed a flat motif enrichment distribution, suggesting that reverse transcriptase is capable of processing through the amino acid adduct in this buffer (Fig. 3A–C). Notably, this pattern was not true for AffinityScript (which showed similar motif enrichment regardless of buffer ion) or TGIRT (which was instead shifted to a weaker −1 position peak in Mn conditions) (Fig. 3A–B).
Figure 3. Motif enrichment at read starts is altered by reverse transcription conditions.
(A) Circles indicate the frequency of the RBFOX2 motif (UGCAUG) (as a fraction of all reads) at indicated positions relative to the first mapped base of the second (paired-end) read in RBFOX2 eCLIP performed with (red) AffinityScript, (green) other reverse transcriptase (RT) enzymes in standard magnesium buffer, (yellow) RT enzymes in manganese buffer, and (grey) size-matched input datasets. (B) Circles indicate UGCAUG frequency at the −2 position relative to read starts from (B), with mean indicated with red lines. (C) Circles indicate frequency of UGCAUG as fraction of all reads at indicated positions relative to deletions within reads, with colors as in (A). (D) Circles indicate UGCAUG frequency at the −1 position relative to deletions from (C), with mean indicated with red lines. (E–H) Similar motif analysis for TARDBP motif (GAAUG) in TARDBP eCLIP, considered relative to read start positions (E–F) or deletions (G–H). Focused regions are (F) −4 and −3 for read start site analysis, and (H) −3, −2, and −1 relative to deletions. (I) Model of RT enzyme relative differences for (left) termination at crosslinked base and (right) deletion of crosslinked base.
If not leading to termination, the amino acid adduct can also lead to skipping of the crosslinked base, yielding reads with characteristic crosslinking induced mutation sites (CIMS) [6]. When we considered the frequency of UGCAUG motifs relative to deletions within reads, we observed enrichments at −1 and −5 positions, characteristic of deletion of the crosslinked G nucleotides (Fig. 3D). Surprisingly, however, these CIMS were highly variable across reverse transcriptase conditions: UGCAUG −1 position motifs were found in over 0.1% of reads in Superscript II and IV conditions, more than one hundred-fold higher than AffinityScript or TGIRT (Fig. 3D–E). CIMS were more common with Mn-based buffer for some conditions (Superscript 2 and 3, M-MLV) but not others (Superscript 4, Maxima), suggesting that the increased read-through does not always lead to increased CIMS frequency.
Considering TARDBP eCLIP, we again observed specific enrichment patterns for the canonical GAAUG TARDBP motif (Fig. 3D). A manganese buffer condition for SS4 again showed weaker position specificity, suggesting that increased read-through of the amino acid adduct may be a general property of performing reverse transcription with manganese-based buffer (Fig. 3D–F). Surprisingly, however, we observed that the specific position of GAAUG enrichment for TARDBP eCLIP was different between AffinityScript (enriched at genomic −4) and all Superscript or TGIRT (enriched at genomic −3) conditions (Fig. 3E). As reverse transcription occurs downstream of all crosslinking, fragmentation, and immunoprecipitation steps, this suggested a difference in processivity of these enzymes at the GAAUG motif. These results suggested a model where if the uracil base is crosslinked to protein, the AffinityScript reactions terminate before processing the uracil whereas other reactions terminate after (Fig. 3F). We note that a competing model in which it is the adenine that is crosslinked is disfavored both because previous CLIP analyses have indicated that UV crosslinking has a modest preference for uridine [5], and because such a model would require AffinityScript termination occur two bases prior to the adduct base.
Considering motif enrichment flanking deletions, we again observed that AffinityScript and TGIRT were not enriched for the expected motif (Fig. 3G–H), suggesting that CIMS analysis may not be fruitful for CLIP experiments performed with these reverse transcriptase enzymes. Surprisingly, we observed three positions of motif enrichment relative to deletion sites for TARDBP (although we note that two positions would not be surprising, due to mapping ambiguity if one of the two adenosines in the GAAUG motif is the deleted base). These results confirm that there is significant variability between both reverse transcriptase termination and base skipping at the crosslink site, with both enzyme and buffer ion choice playing roles in defining these properties (summarized in Fig. 3I). AffinityScript (regardless of buffer ion) terminates without including the crosslinked base, whereas other tested enzymes show variable inclusion of the crosslinked base between RBFOX2 and TARDBP in magnesium but have dramatically less termination in manganese buffer. AffinityScript and TGIRT show little to no crosslink-induced deletion of the crosslinked base, which are observed with all other enzymes (regardless of buffer ion) (Fig. 3I).
1.3.4 Differential uracil tailing and crosslinked base incorporation
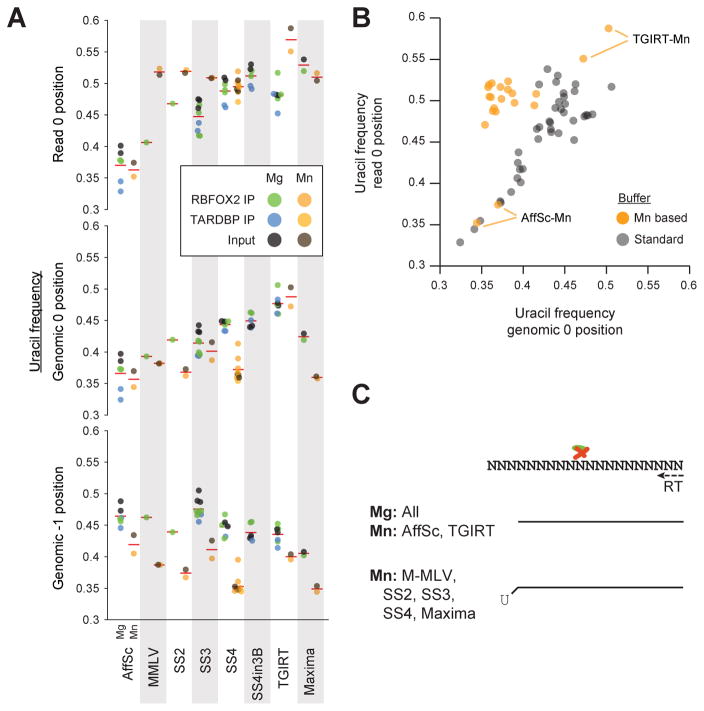
To further explore the observed difference between AffinityScript and other enzymes for TARDBP motifs, we calculated the uracil fraction for the genomic −1 and 0 positions relative to read starts as well as the first base contained within sequenced reads (read 0 position). We observed a significant depletion in 0 position uracil in AffinityScript conditions relative to all others (Fig. 4A). Thus, it appears that the observed result is not specific to TARDBP; rather, these results are consistent with uracil being more commonly crosslinked than other bases, leading to a uracil enrichment at the first base of read fragments for most enzymes. This enrichment is relatively decreased in AffinityScript conditions (with a corresponding increase at the genomic −1 position), suggesting that it more commonly terminates instead of incorporating the crosslinked uracil. In addition to AffinityScript, Superscript III experiments also had an increase in uracil at the genomic −1 position and decreased at the genomic 0 or read 0 position, suggesting an intermediate effect (Fig. 4A).
Figure 4. Differential U-tailing for reverse transcription conditions.
(A) Circles indicate the uracil fraction of (top) the first position of reads, (center) the genomic 0 position corresponding to the first position in the mapped read, and (bottom) the position one base 5′ of the genomic 0 position, with mean indicated by red lines. Significance was determined by Kolmogorov-Smirnov test. (B) Circles indicate the uracil fraction for genomic 0 (x-axis) versus read 0 (y-axis) positions for magnesium versus manganese buffer experiments. (C) Models summarizing observed reverse transcriptase termination properties relative to uracil residues.
When we directly compared uracil frequency at the first base of read sequences with the corresponding genomic 0 position, we observed that manganese buffer conditions for all enzymes except AffinityScript and TGIRT showed an increase in uracil frequency in the first base of reads relative to the genome-encoded sequence at that position (Fig. 4B). Although further work will be required to understand the mechanism of this uracil enrichment, we note that incorporation of non-templated nucleotides (typically cytosines) by M-MLV-based reverse transcriptase enzymes has been exploited for many high-throughput sequencing library methods [10]. Thus, it is possible that the manganese buffer conditions used here encourages tailing of the cDNA with non-templated adenine (Fig. 4C).
1.4 Conclusions
The ability to map binding sites and motifs with single-nucleotide precision is a major innovation developed in the iCLIP methodology and incorporated into eCLIP. Here we show that while altering reverse transcription conditions generally yields similar eCLIP read density profiles and peak enrichments, there are in fact significant differences at the single-nucleotide level. Notably, different reverse transcriptases show different patterns of termination at the amino acid adduct left after Proteinase K treatment of UV crosslinked protein-RNA complexes, with AffinityScript terminating at the base prior to the crosslinked base whereas other enzymes terminate at the crosslinked base. Surprisingly, we also observe a lack of crosslink-induced mutation sites with AffinityScript or TGIRT enzymes. These results have significant implications for comparisons of datasets generated with different CLIP methods, and suggests careful consideration of experimental conditions must be taken to ensure that identified differences are due to true biology and not artifacts of the experimental methodology used in the experiment.
Additionally, we show that the use of manganese-containing buffer can dramatically decrease reverse transcription termination frequencies. Although not generally recommended for standard CLIP analysis due to the loss of single-nucleotide resolution, we note that the ability to increase reverse transcription processivity may yield significant gains for transcripts that are highly damaged, crosslinked, or contain particular RNA modifications that are similarly challenging for standard reverse transcription.
Highlights.
Reverse transcription often terminates or creates deletions at protein-RNA crosslink sites
Altered enzyme and buffer choice can alter crosslink read-through frequency
Crosslink induced mutation sites (CIMS) are not observed with some enzymes
Acknowledgments
The authors would like to thank members of the Yeo lab for insightful discussions and critical reading of the manuscript, particularly S. Aigner and E. Wheeler. This work was supported by grants from the National Institute of Health [HG004659, HG007005 and NS075449] to G.W.Y. E.L.V.N. is a Merck Fellow of the Damon Runyon Cancer Research Foundati on [DRG-2172-13] and is supported by a K99 grant from the National Institute of Health [HG009530]. G.A.P. is supported by the National Science Foundation Graduate Research Fellowship.
Abbreviations
- RT
Reverse transcription
- eCLIP-seq
enhanced crosslinking and immunoprecipitation followed by high-throughput sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nature reviews Genetics. 2014;15(12):829–45. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nature reviews Genetics. 2013;14(4):275–87. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 3.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14085–90. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–5. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 5.Sugimoto Y, Konig J, Hussain S, Zupan B, Curk T, Frye M, et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome biology. 2012;13(8):R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nature biotechnology. 2011;29(7):607–14. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17(7):909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haberman N, Huppertz I, Attig J, Konig J, Wang Z, Hauer C, et al. Insights into the design and interpretation of iCLIP experiments. Genome biology. 2017;18(1):7. doi: 10.1186/s13059-016-1130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vartanian JP, Sala M, Henry M, Wain-Hobson S, Meyerhans A. Manganese cations increase the mutation rate of human immunodeficiency virus type 1 ex vivo. The Journal of general virology. 1999;80(Pt 8):1983–6. doi: 10.1099/0022-1317-80-8-1983. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt WM, Mueller MW. CapSelect: a highly sensitive method for 5′ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic acids research. 1999;27(21):e31. doi: 10.1093/nar/27.21.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nature methods. 2016;13(6):508–14. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Nostrand EL, Gelboin-Burkhart C, Wang R, Pratt GA, Blue SM, Yeo GW. CRISPR/Cas9-mediated integration enables TAG-eCLIP of endogenously tagged RNA binding proteins. Methods. 2017;118–119:50–9. doi: 10.1016/j.ymeth.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–7. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA. irCLIP platform for efficient characterization of protein-RNA interactions. Nature methods. 2016;13(6):489–92. doi: 10.1038/nmeth.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature neuroscience. 2011;14(4):459–68. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nature structural & molecular biology. 2013;20(12):1434–42. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell reports. 2014;6(6):1139–52. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]