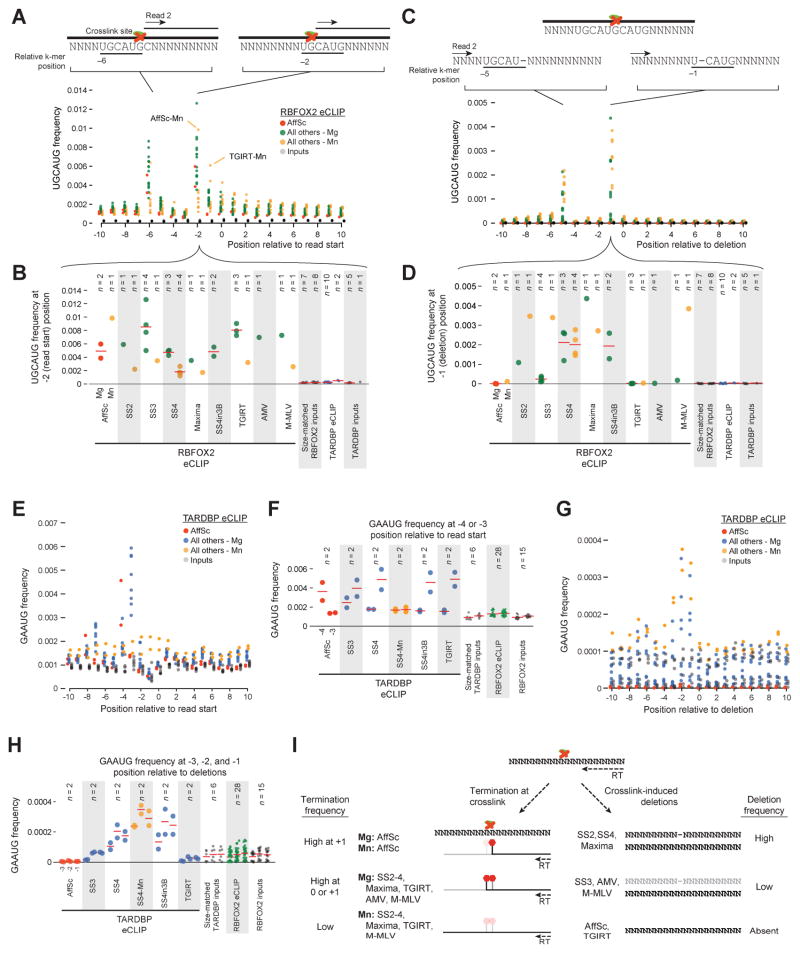
Figure 3. Motif enrichment at read starts is altered by reverse transcription conditions.
(A) Circles indicate the frequency of the RBFOX2 motif (UGCAUG) (as a fraction of all reads) at indicated positions relative to the first mapped base of the second (paired-end) read in RBFOX2 eCLIP performed with (red) AffinityScript, (green) other reverse transcriptase (RT) enzymes in standard magnesium buffer, (yellow) RT enzymes in manganese buffer, and (grey) size-matched input datasets. (B) Circles indicate UGCAUG frequency at the −2 position relative to read starts from (B), with mean indicated with red lines. (C) Circles indicate frequency of UGCAUG as fraction of all reads at indicated positions relative to deletions within reads, with colors as in (A). (D) Circles indicate UGCAUG frequency at the −1 position relative to deletions from (C), with mean indicated with red lines. (E–H) Similar motif analysis for TARDBP motif (GAAUG) in TARDBP eCLIP, considered relative to read start positions (E–F) or deletions (G–H). Focused regions are (F) −4 and −3 for read start site analysis, and (H) −3, −2, and −1 relative to deletions. (I) Model of RT enzyme relative differences for (left) termination at crosslinked base and (right) deletion of crosslinked base.