Abstract
With the completion of the human genome project at the beginning of the 21st century, the biological sciences entered an unprecedented age of data generation, and made its first steps towards an era of personalized medicine. This abundance of sequence data has led to the proliferation of numerous sequence-based techniques for associating variation with disease, such as Genome-Wide Association Studies (GWAS) and Candidate Gene Association Studies (CGAS). However, these statistical methods do not provide an understanding of the functional effects of variation. Structure-based drug discovery and design is increasingly incorporating structural bioinformatics techniques to model and analyze protein targets, perform large scale virtual screening to identify hit to lead compounds, and simulate molecular interactions. These techniques are fast, cost-effective, and complement existing experimental techniques such as High Throughput Sequencing (HTS).
In this paper, we will discuss the contributions of structural bioinformatics to drug discovery, focusing particularly on the analysis of non-synonymous Single Nucleotide Polymorphisms (SNPs). We conclude by suggesting a protocol for future analyses of the structural effects of non-synonymous SNPs on proteins and protein complexes.
Introduction
With the completion of the human genome project in 2003, biological science entered the genomic era. Since then, the rate of data generation has been increasing at an unprecedented rate. Improved technologies have given rise to Next Generation Sequencing (NGS) capabilities that are able to sequence genomes faster and at a fraction of the cost of technologies that came before. These advances have made previously unfeasible undertakings, such as the 1000 Genomes Project [1] and the International HapMap Project [2], possible.
More recently, the Human Heredity and Health in Africa (H3Africa) Initiative was founded to facilitate genomic studies and to build research capacity on the African continent [3]. As part of this project, thousands of genomes from various populations around Africa are being sequenced and massive amounts of new data are being generated. One of the goals of the project is to identify and understand Single Nucleotide Polymorphisms (SNPs) linked to disease. In order to identify SNPs associated with disease, various sequence-level techniques can be employed, including Genome Wide Association Studies (GWAS) and Candidate Gene Association Studies (CGAS). These techniques associate SNPs with diseases by comparing the genomes/genes of healthy individuals with those of unhealthy individuals to determine which SNPs mostly occur in disease-affected patients. SNPs that occur at a statistically significant higher rate in the unhealthy individuals are said to be associated with disease.
Where techniques such as GWAS and CGAS are used to analyze variation at the DNA level, structural bioinformatics techniques provide a means for the downstream analysis of variation i.e. the analysis of variation at the protein level. These techniques include methods such as homology modeling, molecular docking, molecular dynamics, and Residue Interaction Network (RIN) analysis, and let researchers form hypotheses on what effects SNPs have on protein structure, stability, and inter- and intra-protein interactions. Unfortunately, structural bioinformatics techniques can be extremely computationally expensive. As such, even the filtered data sets provided by GWAS and CGAS can be too large. In this paper, we discuss the importance of structural bioinformatics in SNP analysis and drug discovery, and provide a suggested approach for analyzing variation at the protein level.
Retrieving and filtering SNPs for use in structural studies
There are roughly 100 million validated human variants in dbSNP build 147 [4]. It is simply not feasible to study each and every on of these variants in detail. Techniques such as GWAS and CGAS are applied at the sequence level and provide a quick means of filtering out SNPs that are likely not important for a disease. Additionally, tools that predict the effects of SNPs on protein function and stability can be used to further filter these datasets. This does not mean that the remaining SNPs are important, however. Further studies are required to confirm their importance as well as to understand their role, if any, in the disease. It is at this point that structural bioinformatics techniques can be employed.
Variation databases
One of the challenges of bioinformatics is storing the enormous amounts of data being generated by NGS projects. In line with this, various databases have been developed to store variation identified via these projects (Table 1). The most well-known of these databases is probably dbSNP [5], a database created and managed by the National Center for Biotechnology Information (NCBI) as a central repository for all known short variation. The dbSNP database incorporates data from projects such as 1000 Genomes and HapMap, as well many others.
Table 1.
Variation databases
| Database | Description | Link | Reference |
|---|---|---|---|
| COSMIC | Cancer-associated mutations | http://cancer.sanger.ac.uk/cosmic | [6] |
| ClinVar | Clinical significance of variation | http://www.ncbi.nlm.nih.gov/clinvar/ | [7] |
| dbGaP | Database of genotypes and phenotypes | http://www.ncbi.nlm.nih.gov/gap/ | [8] |
| dbNSFP | Functional predictions and annotations of non-synonymous SNPs | https://sites.google.com/site/jpopgen/dbNSFP | [9–11] |
| dbSNP | Short variation | http://www.ncbi.nlm.nih.gov/projects/SNP/ | [5] |
| dbVAR | Structural variation | http://www.ncbi.nlm.nih.gov/dbvar/ | [12] |
| DGVa | Structural variation | http://www.ebi.ac.uk/dgva | [12] |
| EGA | Private variation archive | https://www.ebi.ac.uk/ega/home | [13] |
| EVA | Public variation archive | http://www.ebi.ac.uk/eva/ | n/a |
| Ensembl | Comprehensive biological database including variation | http://www.ensembl.org/ | [14] |
| HGMD | Disease related gene lesions | http://www.hgmd.cf.ac.uk/ | [15] |
| HGVD | Japanese genetic variation | http://www.genome.med.kyoto-u.ac.jp/SnpDB/ | [16] |
| HUMA | Comprehensive biological database including variation | https://huma.rubi.ru.ac.za | n/a |
| LS-SNP/PDB | Non-synonymous SNPs likely to affect biological function | http://ls-snp.icm.jhu.edu/ls-snp-pdb/ | [17] |
| NHGRI-EBI Catalog | Manually-curated database of published genome-wide association studies | http://www.ebi.ac.uk/gwas/home | [18] |
| OMIM | Human genes and genetic disorders | http://www.omim.org/ | [19] |
| PinSnps | Protein-protein interaction networks | http://fraternalilab.kcl.ac.uk/PinSnps/ | [20] |
| SNPeffect | Characterization and annotation of SNPs | http://snpeffect.switchlab.org/ | [21] |
| SNPs3D | Functional effects of non-synonymous SNPs | http://www.snps3d.org/ | [22] |
| TCGA | Cancer-associated mutations | http://cancergenome.nih.gov/ | [23] |
| Uniprot | Protein database including non-synonymous SNPs | http://www.uniprot.org/ | [24] |
| VnD | Variation and drugs | http://vnd.kobic.re.kr/ | [25] |
The NCBI also has various other variation databases including dbVAR [12], dbGaP [8], and ClinVar [7]. Where dbSNP focuses on short variation, dbVAR stores structural variation such as insertions and deletions. On the other hand, dbGaP and ClinVar are focused on the relationship between genotype and phenotype and the clinical significance of variation, respectively.
The European Bioinformatics Institute (EBI) also hosts various variation databases including the European Variation Archive (EVA), the Database of Genomic Variants archive (DGVa) [12], and the European Genome-phenome Archive (EGA) [13]. EVA is a public variation archive, which stores all types of variation. DGVa, on the other hand, is EBI’s version of dbVAR i.e. a database for structural variation. Variation in EVA, DGVa, dbVAR, and dbSNP is exchanged on a regular basis, meaning that these databases generally mirror each other. EVA also stores data from ClinVar, making it a rich source for variation data.
The European Genome-phenome Archive (EGA) stores complete data sets from genomic studies, allowing users to browse various aspects of the data. Unlike EVA, EGA is not a public data archive. Data sets are stored privately and researchers must be granted access by the specified Data Access Committee in order to view the data.
The EBI, along with the National Human Genome Research Institute (NHGRI), have also produced the NHGRI-EBI GWAS Catalog [18], a high-quality, manually-curated collection of published genome-wide association studies. The GWAS Catalog stores SNP and SNP-trait associations for over 11 000 SNPs and from over 1700 publications.
Some variation databases focus of variation related to a disease or group of diseases. Examples of this include COSMIC [6] and The Cancer Genome Atlas (TCGA) [23], which focus on variation related to cancer. Other databases, such as the Online Mendelian In Man (OMIM) [19] database link variation to phenotypes. Uniprot [24], a database focused on proteins, maps non-synonymous SNPs to these proteins.
One of the most comprehensive biological databases is hosted by Ensembl [14]. The Ensembl database stores various biological data including genes, transcripts, proteins, exons, and more. To this data, it links phenotypes and variation. Ensembl incorporates variation from numerous sources including dbSNP, ClinVar, COSMIC, dbGaP, DGVa, EGA, OMIM, and Uniprot. All this data is stored within a single, relational database and can be queried using BioMart [26], a powerful tool that provides simple and uniform access to various data sources.
The above-mentioned databases all focus on the analysis of SNPs at the sequence level. PinSnps [20] is one database where variation is mapped to protein structures. Variation data is collected from various sources including OMIM and COSMIC. Users of the PinSnps web server are then able to select their SNPs of interest and visualize them in the protein structure. PinSnps also links SNPs to protein interaction networks.
LS-SNP/PDB [17] is another variation database where SNPs are pre-mapped to protein structures. As with PinSnps, users can query the database for a protein or SNP of interest and then visualize SNPs in the structure of the protein.
Tools and databases, such as PinSnps and LS-SNP/PDB, that focus on the structural impacts of variation are, unfortunately, few and far between. Additionally, these databases tend to neglect the sequence level data. We have developed the Human Mutation Analysis (HUMA) web server and database, which focuses on the analysis of variation in humans both at the sequence and structural level. The HUMA database stores genes, proteins, proteins structures, diseases and variants.
Variation is pre-mapped to gene and protein sequences based on chromosome co-ordinates. Variants are also mapped to protein structures based on alignments between the protein sequences and sequences extracted from the PDB files for the respective proteins. Additional information about the protein structures, such as the ligands that were solved with the structure, as well as drugs that are known to target the structure are also stored. Proteins, genes, and variation are all linked to disease via data obtained from ClinVar and Uniprot. As part of the pipeline for mapping variation to protein sequences, HUMA also stores the coding sequences (CDS), coding DNA (cDNA) and exons for proteins. As such, HUMA provides a resource for querying variation both at the sequence and structural level.
Predicting disease associated/deleterious mutations
The main challenge of computational SNP analysis at the sequence level is determining whether a SNP is associated with, or likely to be associated with, disease. As previously discussed, GWAS and CGAS are useful techniques for associating variants with disease. Association via these techniques is no guarantee that mutation is disease-related, however. Additionally, these techniques can miss variation that is important. As such, other methods are still required to further analyze the effects of variation.
At the protein level, numerous tools have been developed which predict the impact of non-synonymous SNPs on protein function (Table 2). These tools usually fall into one of two categories. The first category is made up of tools that make predictions based solely on the sequence of a protein, while the second is made up of tools that incorporate structural information when making predictions [27].
Table 2.
Tools for predicting the functional effects of non-synonymous SNPs
Tools such as SIFT [39], PROVEAN [38], and PANTHER-PSEP [34] fall into the first category. These tools look at sequence conservation to determine whether mutations at a particular position will be deleterious. This is based on the theory that highly conserved regions of a sequence must be important to protein function. Mutations in these regions will therefore have detrimental effects. SIFT and PROVEAN look at the conservation of amino acids across homologs. While SIFT can predict the effects of SNPs, PROVEAN has the added advantage of being able to predict the effects of in-frame insertions and deletions. PANTHER-PSEP, on the other hand, looks at evolutionary conservation i.e. the time since the last mutation occurred at a particular position in an amino acid sequence.
FATHMM [29] is sequence-based SNP analysis tool. As with the above tools, the FATHMM makes conservation-based predictions. However, FATHMM also includes a second, weighted algorithm. This algorithm essentially allows predictions that were made via the first method, to be adjusted based on the tolerance of the region of the protein to mutations.
Machine learning techniques have also been used to predict the functional effects of variation. PhD-SNP [42] and Parepro [35] are sequence-based Support Vector Machine (SVM) methods for predicting the functional effects of SNPs. SVM methods are popular for handling biological data due to their ability to work with large data sets and to handle noise effectively.
PolyPhen-2 [36], Auto-Mute 2.0 [28], and SNAP [40] incorporate structural information when making predictions on the functional effects of mutations. As such, they fall into the second category of SNP analysis tools. PolyPhen-2 uses three structure-based predictive features as well as eight sequence-based predictive features to classify variation. Predictions are made via a naïve Bayes classifier. Similarly, Auto-Mute 2.0 combines structural features with trained, machine-learning methods. SNAP, on the other hand, only requires sequence information as input, but structural and functional annotations help to improve predictions.
There are various other methods for predicting the functional effects of SNPs, which have not been discussed here. None of these methods are perfect, however. As such, it is a good idea to get a consensus from several different tools before deciding, which SNPs to select for further analysis. With this in mind, classifiers such as PredictSNP [37] and Meta-SNP [31] combine the predictions of various existing tools to gain a consensus on which SNPs are deleterious to protein function.
We have developed the Variant Analysis Portal (VAPOR), which has been incorporated into the HUMA web server. VAPOR is a workflow, which accepts either a protein sequence or protein structure as input along with a list of SNPs. From here, it gets predictions from PROVEAN, PolyPhen-2, PhD-SNP, PANTHER-PSEP, and FATHMM and merges the results into a single table. Unlike PredictSNP and Meta-SNP, VAPOR does not generate a consensus score from these results. It remains as a useful tool for quickly getting results from multiple SNP analysis methods, however.
Predicting changes in protein stability due to mutations
Predicting the impact of SNPs on protein stability is another important area of SNP analysis. Non-synonymous SNPs can result in changes of the internal energy of a protein as well as lead to changes in the structure of the protein. Calculating the change in Gibbs free energy between a wild type protein and the mutated form is a common measure of how much a mutation affects protein stability [43]. One thing to note when analyzing changes in protein stability is that increase and decreases in protein stability do not necessarily correspond to deleterious and beneficial effects, as increases in protein stability can also hamper protein function.
Various tools have been developed to predict changes in protein stability due to non-synonymous SNPs (Table 3). The Auto-Mute 2.0 suite discussed earlier includes functionality for predicting stability changes. Additionally, I-Mutant2.0 [44] and MuPro [45] provide SVM based methods for predicting changes instability. Both tools can be used, either to simply predict the sign of the change in stability, or to predict the actual size of the change. Both tools can also incorporate structural information when making predictions, but MuPro can achieve nearly the same accuracy when only the primary sequence is considered, making it a useful option when the tertiary structure of the protein is unknown.
Table 3.
Tools for predicting changes in stability due to non-synonymous SNPs
| Tool | Description | Link | Reference |
|---|---|---|---|
| Auto-Mute 2.0 | Sequence-and structure-based | http://binf2.gmu.edu/automute/ | [28] |
| CUPSAT | Structure-based | http://cupsat.tu-bs.de/ | [46] |
| Eris | Structure-based | http://troll.med.unc.edu/eris/login.php | [47] |
| I-Mutant2.0 | Sequence-and structure-based | http://folding.biofold.org/i-mutant/imutant2.0.html | [44] |
| MuPro | Sequence-and structure-based | http://mupro.proteomics.ics.uci.edu/ | [45] |
| NeEMO | Residue interaction networks | http://protein.bio.unipd.it/neemo/help.html | [48] |
| PoPMuSiC 2.1 | Structure-based | https://soft.dezyme.com/query/create/pop | [49] |
NeEMO [48] is a machine learning method based on Residue Interaction Networks (RINs). It incorporates information from RINs in a non-linear neural network to improve prediction accuracy. RINs provide useful information regarding changes in residue interactions when a mutation is introduced as they implicitly incorporate detailed maps of chemical interactions within proteins.
The Vapor workflow makes use of I-Mutant 2.0 and MuPro predictions to complement the functional predictions described in the previous section. Unfortunately, NeEMO is not available for download and, as such, could not be included as part of Vapor. Including stability prediction tools in Vapor, however, adds an additional dimension to the workflow and differentiates it from similar tools.
Role of structural bioinformatics – SNP analysis in drug discovery
Structural bioinformatics is an area of bioinformatics focused on the structure, movement and interaction of biological macromolecules in three-dimensional space. Structural bioinformatics techniques play an important role in drug discovery and can be used at every stage of the drug design process [50–54], where they can be used to complement, and sometimes replace more costly experimental techniques [55–57]. For example, protein structure prediction software provides alternatives to X-ray crystallography and NMR techniques, while virtual screening and molecular dynamics simulations can complement High-Throughput Screening (HTS).
The use of computational techniques in drug discovery and design is often referred to as Computer-Aided Drug Design (CADD) [54]. In this section, we will discuss the uses of structural bioinformatics as part of CADD, specifically in the context of non-synonymous SNP analysis.
Mutations have been associated with drug resistance in numerous diseases such as influenza, tuberculosis, HIV and cancer [58–62]. Similarly, mutations can be linked to drug sensitivity in patients [63]. This opens the door to personalized medicines, where knowledge of drug resistant and drug sensitive SNPs allow treatments to be tailored to individual patients [64,65]. Understanding structural changes caused by non-synonymous SNPs will enable the design of novel drugs to target these mutations and, thus, be key in advancing personalized medicine [25].
Protein structure prediction
In the post-genomic era, there is an abundance of available protein sequences. Unfortunately, solving the structures of these proteins is a slow an expensive process. As such, the gap between known protein sequences and solved protein structures is growing. To illustrate this, as of September 2016, the Protein Data Bank [66] contained a little over 120 thousand protein structures, which pales in comparison to the 65 million sequences available in the Uniprot protein sequence database. Having the protein structure available lets researchers gain insight into the molecular function of the protein. An understanding of the structural and functional aspects of proteins opens up the door to drug design and discovery [53,67] and, as such, is of great interest to chemists as well as biologists. To counter the growing sequence-structure gap, various computational structure prediction methods have been developed. These methods can be categorized into two distinct groups, namely, template-based modeling, and ab initio (or de novo) techniques.
Ab initio modeling attempts to construct a model of a protein based on solely on its amino acid sequence. This is a computationally intensive task that, despite ever increasing computational power, is currently only practical for small systems [68]. Additionally, according to the latest CASP results [69], ab initio methods have not yet to catch up to template-based modeling techniques in terms of accuracy.
Template-based modeling is currently the most reliable method for protein structure prediction, producing decent quality models for roughly two-thirds of proteins with unsolved structures [69–71]. Template-based modeling can be divided into homology modeling and protein threading techniques.
Homology modeling is a structure prediction technique that relies on the observation that the structural conformation of a protein is more conserved than its amino acid sequence. As such, solved protein structures can be used as ‘templates’ for predicting the tertiary structure of a ‘target’ sequence, provided the sequence identity between the ‘target’ and ‘template’ sequences is high enough (roughly >30%) [53,72].
Protein threading is similar to homology modeling in that it uses the structures of previously solved proteins to predict the structure of a target sequence. Where homology modeling uses the structures of homologous proteins as templates, however, threading uses the structures of proteins, which are predicted to have the same folds. Threading is useful when there are no homologous proteins available that have solved structures [73].
Protein structure prediction can be used to introduce SNPs into a structure and determine the effects that these SNPs might have on the protein’s function and stability. Once modeled, the wild type structure can be compared to the mutant structure in several ways. For example, the Residue Interaction Networks (RINs) of the structures can be compared to see if introducing SNPs influences intra-protein communication. The structures can also be compared to see if new bonds have been introduced or existing bonds have been broken. In addition, the models can be further analyzed using molecular docking and molecular dynamics simulations, two important techniques for drug discovery.
Homology modeling has been used in various stages of drug discovery including the study of protein function and mechanisms [74], analysis of the effects of mutations in binding sites of receptor proteins [75], identification of druggable pockets [76], and various virtual screening studies [77–80].
Molecular docking and virtual screening
Molecular docking is a technique for predicting the bound conformations of a protein-ligand complex, and is used in structure-based drug design to study biomolecular interactions [81]. Docking is fast enough to allow libraries containing thousands of compounds to be docked against a receptor protein in a process called virtual screening. Virtual screening is used to scan a compound library for potential drug candidates [82–84]. As compounds are docked against the receptor, a score is calculated to determine the binding affinity of each compound to the receptor. Compounds with the highest binding affinity scores are selected for further study. Binding affinity scores are not infallible, and rankings based on these scores are, therefore, not necessarily reliable. Nevertheless, these binding affinity scores can distinguish likely from unlikely compounds, and can be used as potential hit compounds in the drug design process [82].
Molecular docking can also be used to assess the impact of SNPs on drug response. Mutations in the binding sites of receptor proteins can affect the binding affinity of drugs. This can lead to drug resistance or drug susceptibility. Molecular docking can be used in conjunction with protein structure prediction to predict the effect these mutations will have on drug response [75].
Virtual screening has become a routine procedure in drug discovery and can be used as a cheaper alternative to HTS [85]. Having access to a comprehensive compound library is an important part of virtual screening. As such, numerous compound libraries have been made available via online databases and portals such as ZINC [86], ChemSpider [87], the Traditional Chinese Medicine (TCM) Database@Taiwan [88] and SANCDB [89].
Molecular dynamics simulations
Protein structure prediction and molecular docking provide a snapshot in time of a protein structure and protein-ligand complex, respectively. Molecular dynamics, on the other hand, simulates the movements and trajectories of all the atoms in these structures over a period time. It can be used to check if a protein structure remains stable after the introduction of one or more SNPs. Similarly, it can be used to determine the stability of protein-ligand complexes after docking [90]. While molecular docking predicts how well a compound docks to a receptor, molecular dynamics can predict how stably bound the compound is and whether it will stay bound over a specified period.
Molecular dynamics results are usually analyzed via plots of their Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF). There first measurement, RMSD, measures the average movement in the structure’s backbone over the course of the simulation. If, by the end of the simulation, it appears that the plot of the RMSD has leveled out, it can be assumed that the structure has stabilized.
Where RMSD measures the global movement of the protein, RMSF, measures local movement i.e. how much individual residues fluctuate over the course of the simulation. Spikes in this plot indicate residues which move a lot over the course of the simulation, while low values indicate residues that remain relatively fixed throughout.
Molecular docking simulations are often used in combination with homology modeling and virtual screening [90,91]. In terms of computational SNP analysis, molecular dynamics can be used to determine whether introducing a SNP will destabilize a protein or perhaps cause the protein to move or fold in a different way [92].
Inter- and intra-protein interactions
Inter- and intra-protein interactions play important roles in protein folding as well as in the stability and function of proteins and protein complexes. Due to protein folding, residues that are far apart in a protein’s sequence can be right next to one another in three-dimensional space. Interactions between these residues help the protein to adopt the correct structural conformation [93]. As such, disruptions to these interactions (e.g. residue substitutions) could cause instability and loss of protein function. It is, therefore, useful to understand, which residues are important in the structure and function of a protein. This can be done by analyzing the types of bonds (e.g. hydrogen bonds, di-sulphide bonds, etc.) that occur between residues.
RINs provide another means of analyzing protein structures. RINs have been analyzed using a branch of Mathematics known as graph theory. In a RIN, each residue in the protein is a node in the network. An edge (or connection) between two nodes exists if there is an interaction between the two residues that they represent [94]. In RINs, an interactions between residues exist if the residues are within a user-defined cut-off (usually around 6.5 – 7.5 Å) of each other [95].
Various network measures have been used to analyze RINs. Previously, the change in the average shortest path to each residue (ΔL) and the change in betweenness centrality of each residue (ΔBC) has been used to perform Alanine scanning, where each residue is mutated to Alanine to see its effect on the overall network[96].
The shortest path (L) between two nodes is the minimum number of edges that must be traversed to travel from one node to another. The average shortest path to a residue is calculated by summing the shortest path between a given residue and all other residues in the structure and dividing the result by N-1, where N is the number of residues in the structure. The result of this calculation is the average accessibility of the given residue from any other residue in the structure i.e. selecting any other residue at random, what are the average number of edges that will need to be traversed to reach the given residue. When comparing a wild type protein to a mutant, ΔL can be calculated for each residue by subtracting the average shortest path to each residue in the mutant from the average shortest path to each respective residue in the wild type. The result describes whether the residue is more or less accessible in the mutated structure [96].
The betweenness centrality (BC) of a given node is a measurement of how often a shortest path between two nodes passes through the given node. As such, it measures the importance of the given node to efficient navigation of the network. A high BC means that the node occupies a central position in the network. When using this measure to perform an Alanine scan, ΔBC for a residue is calculated by getting the difference between the BC for a residue in the mutant and wild type [96].
Network analysis techniques such as those describe above can be applied to both experimental and predicted PDB structures. In addition, network analysis can be carried out over the trajectory of a molecular dynamics simulation to monitor how the network changes over time [97]. Although L and BC have previously only been used to perform Alanine scanning, we propose that these same techniques could be applied to SNP analysis.
Protocol for analyzing SNPs using structural bioinformatics
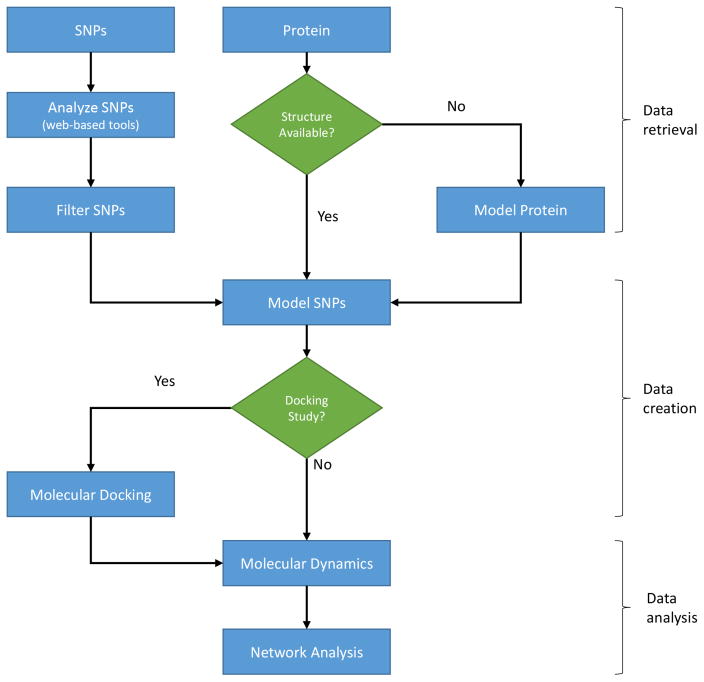
Structural bioinformatics is an important part of the drug discovery process. As discussed in previous sections, it can contribute to every stage of the drug design process. Here we propose a protocol for determining the effects of non-synonymous SNPs on protein structure, function, and stability using structural bioinformatics techniques (Figure. 1).
Figure 1. Protocol for analyzing non-synonymous SNPs.
A flowchart depicting the steps required to analyze the effects of non-synonymous SNPs using structural bioinformatics. The process can be divided into 3 phases: 1) data retrieval; 2) data creation; and 3) data analysis.
The first requirement of any type of analysis is data. In our case, the required data to perform the analysis is the protein sequence and structure and the non-synonymous SNPs that occur in the protein. As previously discussed, there are various public databases available that provide access to variation data (Table 1). For our purposes, the most useful of these databases are arguably Ensembl and HUMA. Both databases allow the user to search for their protein of interest and make both the sequence and all the known variation in that sequence available for download. Mutation data from these databases is linked to phenotypes, where possible. If there are experimentally determined structures available for the protein, these structures are also linked to. As such, Ensembl and HUMA provide convenient locations to access all of our required data.
If no protein structures are available, or if there are important missing residues in available structures, the structure of the protein must be modeled. Fortunately, various online structure prediction pipelines exist. Commonly used tools include HHPred [98], SWISS-MODEL [99], I-TASSER [100], and Phyre2 [101]. We have also developed PRIMO (https://primo.rubi.ru.ac.za), an interactive homology modeling platform that assists users through the modeling process.
As structural bioinformatics techniques tend to be computationally intensive, it is not possible to analyze every SNP in the protein in detail using these methods. As such, the SNP data set must be filtered before we move on to more computationally expensive techniques. Tools that predict the effects of SNPs on function (Table 2) and stability (Table 3) can be used to quickly analyze large SNP data sets. The results of this analysis, although not infallible, can be used to filter the data set to contain only SNPs that are likely to negatively affect function or stability. As a general rule of thumb, at least four or five of these tools should be run to gain a consensus as to the effect of the SNP.
To complement this analysis, the SNPs should be checked for known disease-associations in literature. Ensembl and HUMA link diseases to SNPs and, as such, provide useful resources for this purpose.
If a structure is available for the protein, or once the structure of the protein has been modeled, it may be useful to check, which residues in the structure are interacting. Interacting residues are likely to be important for protein function and stability and, as such, SNPs occurring at these locations may be important. Thus, protein intra- and inter-actions can be used to further filter the SNP data set. Various tools have been developed to calculate these interactions by determining the bonds, such as hydrogen bonds and di-sulphide bonds, that form between residues. These include web servers such as PIC [102], COCOMAPS [103], InterProSurf [104], PDBParam [105], and PDBSum [106].
Once the SNP data set has been filtered to a low enough level (dependent on available computational resources), the SNPs can be introduced into the protein structure via homology modeling. A model should be produced for every SNP i.e. if there are 20 SNPs in the data set, 20 models should be produced, each containing one of the SNPs. Combinations of SNPs can also be modeled into the structure if, for example, it is known that the SNPs co-occur.
If the goal of the research is to determine whether SNPs will affect the binding affinity of a drug, it is at this point that molecular docking runs should be performed, both on the wild type structure and the mutants. Analyzing changes in the binding affinity of the drug between the wild type and the mutants will give an idea of whether drug responses may be affected in the mutants.
To improve the reliability of the docking results, or to analyze the stability of the wild type and protein models, molecular dynamics simulations should be run. Currently, the most popular molecular dynamics software available are arguably GROMACS [107] and NAMD [108]. These simulations will give insight into whether the docked drug will remain bound to the mutant proteins over a period of time. If the protein has been destabilized, this may not be the case. A destabilized protein may also have impaired function, which could indicate the involvement of the respective SNP in a disease phenotype.
Residue Interaction Network analysis can be performed after modeling or docking to determine how these methods have affected the network. Previous methods have minimized the protein structure before performing network analysis [96]. Another interesting option is to perform network analysis over the trajectory of the molecular dynamics simulation [97].
To predict whether a given SNP is associated with a disease, the networks of mutant models containing SNPs that are associated with the disease in literature (or in Ensembl and HUMA) can be compared with the network of the mutant model containing the given SNP. Similar changes in the network may indicate similar effects on protein function and stability.
Conclusions
Structural bioinformatics techniques such as protein structure prediction, molecular docking, and molecular dynamics provide low cost alternatives to experimental techniques such as X-ray crystallography, NMR, and HTS. In this paper, we have discussed the use of these techniques in drug discovery, with a focus on the analysis of non-synonymous SNPs. Mutations, such as SNPs, contribute to differences in drug response between individuals. Gaining further understanding of the reasons behind these differences will gives us insight into how we can take advantage of them and, thereby, usher in the age of personalized medicine.
Acknowledgments
H3ABioNet is supported by the National Institutes of Health Common Fund [grant number U41HG006941]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- BC
Betweenness Centrality
- H3Africa
Human Heredity and Health in Africa
- HTS
High Throughput Sequencing
- HUMA
Human Mutation Analysis web server
- L
Shortest path
- PRIMO
Protein Interactive Modeling web server
- RIN
Residue Interaction Network
- RMSD
Root Mean Square Deviation
- RMSF
Root Mean Square Fluctuation
- VAPOR
Variant Analysis Portal
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International T, Consortium H. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 3.The H3Africa Consortium. Research capacity. Enabling the genomic revolution in Africa. Science. 2014;344:1346–8. doi: 10.1126/science.1251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.dbSNP 147 Data Summary. n.d https://www.ncbi.nlm.nih.gov/projects/SNP/snp_summary.cgi?view+summary=view+summary&build_id=147.
- 5.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014:42. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Jian X, Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–9. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: A database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013:34. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat. 2016;37:235–41. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lappalainen I, Lopez J, Skipper L, Hefferon T, Spalding JD, Garner J, et al. DbVar and DGVa: Public archives for genomic structural variation. Nucleic Acids Res. 2013:41. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lappalainen I, Almeida-King J, Kumanduri V, Senf A, Spalding JD, Ur-Rehman S, et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat Genet. 2015;47:692–5. doi: 10.1038/ng.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stenson PD, Mort M, Ball EV, Shaw K, Phillips AD, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higasa K, Miyake N, Yoshimura J, Okamura K, Niihori T, Saitsu H, et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J Hum Genet. 2016;61:547–53. doi: 10.1038/jhg.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: Annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25:1431–2. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM) Hum Mutat. 2000;15:57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 20.Lu H-C, Braga JH, Fraternali F. PinSnps: structural and functional analysis of SNPs in the context of protein interaction networks. Bioinformatics. 2016:btw153. doi: 10.1093/bioinformatics/btw153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reumers J, Schymkowitz J, Ferkinghoff-Borg J, Stricher F, Serrano L, Rousseau F. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005;33:D527–32. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer T, Atlas G. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leinonen R, Garcia Diez F, Binns D, Fleischmann W, Lopez R, Apweiler R. UniProt archive. Bioinformatics. 2004;20:3236–7. doi: 10.1093/bioinformatics/bth191. [DOI] [PubMed] [Google Scholar]
- 25.Yang JO, Oh S, Ko G, Park S-J, Kim W-Y, Lee B, et al. VnD: a structure-centric database of disease-related SNPs and drugs. Nucleic Acids Res. 2011;39:D939–44. doi: 10.1093/nar/gkq957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, et al. BioMart--biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mah JTL, Low ESH, Lee E. In silico SNP analysis and bioinformatics tools: A review of the state of the art to aid drug discovery. Drug Discov Today. 2011;16:800–9. doi: 10.1016/j.drudis.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Masso M, Vaisman II. AUTO-MUTE 2.0: A portable framework with enhanced capabilities for predicting protein functional consequences upon mutation. Adv Bioinformatics. 2014 doi: 10.1155/2014/278385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GLA, Edwards KJ, et al. Predicting the Functional, Molecular, and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–86. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capriotti E, Altman RB, Bromberg Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics. 2013;14(Suppl 3):S2. doi: 10.1186/1471-2164-14-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wainreb G, Ashkenazy H, Bromberg Y, Starovolsky-Shitrit A, Haliloglu T, Ruppin E, et al. MuD: an interactive web server for the prediction of non-neutral substitutions using protein structural data. Nucleic Acids Res. 2010;38:W523–8. doi: 10.1093/nar/gkq528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–50. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H, Thomas PD. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016:1–3. doi: 10.1093/bioinformatics/btw222. [DOI] [PubMed] [Google Scholar]
- 35.Tian J, Wu N, Guo X, Guo J, Zhang J, Fan Y. Predicting the phenotypic effects of non-synonymous single nucleotide polymorphisms based on support vector machines. BMC Bioinformatics. 2007;8:450. doi: 10.1186/1471-2105-8-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendl J, Stourac J, Salanda O, Pavelka A, Wieben ED, Zendulka J, et al. PredictSNP : Robust and Accurate Consensus Classifier for Prediction of Disease-Related Mutations. 2014;10:1–11. doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS One. 2012:7. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bromberg Y, Rost B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–35. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–44. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- 42.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–34. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 43.Thiltgen G, Goldstein RA. Assessing Predictors of Changes in Protein Stability upon Mutation Using Self-Consistency. PLoS One. 2012:7. doi: 10.1371/journal.pone.0046084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005:33. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–32. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 46.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–42. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods. 2007;4:466–7. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 48.Giollo M, Martin AJ, Walsh I, Ferrari C, Tosatto SC. NeEMO: a method using residue interaction networks to improve prediction of protein stability upon mutation. BMC Genomics. 2014;15:S7. doi: 10.1186/1471-2164-15-S4-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics. 2011;12:151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou K-C. Impacts of bioinformatics to medicinal chemistry. Med Chem (Los Angeles) 2015;11:218–34. doi: 10.2174/1573406411666141229162834. [DOI] [PubMed] [Google Scholar]
- 51.Blundell TL, Sibanda BL, Montalvão RW, Brewerton S, Chelliah V, Worth CL, et al. Structural biology and bioinformatics in drug design: opportunities and challenges for target identification and lead discovery. Philos Trans R Soc Lond B Biol Sci. 2006;361:413–23. doi: 10.1098/rstb.2005.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taboureau O, Baell JB, Fernández-Recio J, Villoutreix BO. Established and emerging trends in computational drug discovery in the structural genomics era. Chem Biol. 2012;19:29–41. doi: 10.1016/j.chembiol.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 53.Cavasotto CN, Phatak SS. Homology modeling in drug discovery: current trends and applications. Drug Discov Today. 2009;14:676–83. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 54.Kapetanovic IM. Computer-aided drug discovery and development (CADDD): In silico-chemico-biological approach. Chem Biol Interact. 2008;171:165–76. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scapin G. Structural biology and drug discovery. Curr Pharm Des. 2006;12:2087–97. doi: 10.2174/138161206777585201. [DOI] [PubMed] [Google Scholar]
- 56.Congreve M, Murray CW, Blundell TL. Structural biology and drug discovery. Drug Discov Today. 2005;10:895–907. doi: 10.1016/S1359-6446(05)03484-7. [DOI] [PubMed] [Google Scholar]
- 57.Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9:71. doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sim S, Kacevska M, Ingelman-Sundberg M. Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects. Pharmacogenomics J. 2012;13:1–11. doi: 10.1038/tpj.2012.45. [DOI] [PubMed] [Google Scholar]
- 59.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–86. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gottesman MM. Mechanisms of Cancer Drug Resistance. Annu Rev Med. 2002;53:615–27. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 61.LI J, Linley L, Kline R, Ziebell R, Heneine W, Johnson JA. Sensitive sentinel mutation screening reveals differential underestimation of transmitted HIV drug resistance among demographic groups. Aids. 2016:1. doi: 10.1097/QAD.0000000000001099. [DOI] [PubMed] [Google Scholar]
- 62.Pielak RM, Schnell JR, Chou JJ. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc Natl Acad Sci U S A. 2009;106:7379–84. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar RD, Chang LW, Ellis MJ, Bose R. Prioritizing Potentially Druggable Mutations with dGene: An Annotation Tool for Cancer Genome Sequencing Data. PLoS One. 2013:8. doi: 10.1371/journal.pone.0067980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niu B, Scott AD, Sengupta S, Bailey MH, Batra P, Ning J, et al. Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat Genet. 2016;48:827–37. doi: 10.1038/ng.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kantardjieff K, Rupp B. Structural bioinformatic approaches to the discovery of new antimycobacterial drugs. Curr Pharm Des. 2004;10:3195–211. doi: 10.2174/1381612043383205. [DOI] [PubMed] [Google Scholar]
- 68.Chen M, Lin X, Zheng W, Onuchic JN, Wolynes PG. Protein Folding and Structure Prediction from the Ground Up: The Atomistic Associative Memory, Water Mediated, Structure and Energy Model. J Phys Chem B. 2016;120:8557–65. doi: 10.1021/acs.jpcb.6b02451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction: Progress and new directions in round XI. Proteins Struct Funct Bioinforma. 2016 doi: 10.1002/prot.25064. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacobson M, Sali A. Comparative Protein Structure Modeling and its Applications to Drug Discovery. Annu Rep Med Chem. 2004;39:259–76. doi: 10.1016/S0065-7743(04)39020-2. [DOI] [Google Scholar]
- 71.Ma J, Wang S, Zhao F, Xu J. Protein threading using context-specific alignment potential. Bioinformatics. 2013;29 doi: 10.1093/bioinformatics/btt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 73.Peng J, Xu J. Low-homology protein threading. Bioinforma. 2010;26:i294–300. doi: 10.1093/bioinformatics/btq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petrey D, Chen TS, Deng L, Garzon JI, Hwang H, Lasso G, et al. Template-based prediction of protein function. Curr Opin Struct Biol. 2015;32:33–8. doi: 10.1016/j.sbi.2015.01.007. http://dx.doi.org/10.1016/j.sbi.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blair JMA, Bavro VN, Ricci V, Modi N, Cacciotto P, Kleinekathöfer U, et al. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc Natl Acad Sci. 2015;112:3511–6. doi: 10.1073/pnas.1419939112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vyas VK, Ghate M, Patel K, Qureshi G, Shah S. Homology modeling, binding site identification and docking study of human angiotensin II type I (Ang II-AT1) receptor. Biomed Pharmacother. 2015;74:42–8. doi: 10.1016/j.biopha.2015.07.008. http://dx.doi.org/10.1016/j.biopha.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 77.Messaoudi A, Belguith H, Ben Hamida J. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 β-lactamase. Theor Biol Med Model. 2013;10:22. doi: 10.1186/1742-4682-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ung PM-U, Song W, Cheng L, Zhao X, Hu H, Chen L, et al. Inhibitor Discovery for the Human GLUT1 from Homology Modeling and Virtual Screening. ACS Chem Biol. 2016;11:1908–16. doi: 10.1021/acschembio.6b00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morya VK, Dung NH, Singh BK, Lee H-B, Kim E. Homology modelling and virtual screening of P-protein in a quest for novel antimelanogenic agent and In vitro assessments. Exp Dermatol. 2014;23:838–42. doi: 10.1111/exd.12549. [DOI] [PubMed] [Google Scholar]
- 80.Fazi R, Tintori C, Brai A, Botta L, Selvaraj M, Garbelli A, et al. Homology Model-Based Virtual Screening for the Identification of Human Helicase DDX3 Inhibitors. J Chem Inf Model. 2015;55:2443–54. doi: 10.1021/acs.jcim.5b00419. [DOI] [PubMed] [Google Scholar]
- 81.Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11:905–19. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Irwin JJ, Shoichet BK. Docking Screens for Novel Ligands Conferring New Biology. J Med Chem. 2016;59:4103–20. doi: 10.1021/acs.jmedchem.5b02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pyzer-Knapp EO, Suh C, Gómez-Bombarelli R, Aguilera-Iparraguirre J, Aspuru-Guzik A. What Is High-Throughput Virtual Screening? A Perspective from Organic Materials Discovery. Annu Rev Mater Res. 2015;45:195–216. doi: 10.1146/annurev-matsci-070214-020823. [DOI] [Google Scholar]
- 84.Kumar V, Krishna S, Siddiqi MI. Virtual screening strategies: Recent advances in the identification and design of anti-cancer agents. Methods. 2015;71:64–70. doi: 10.1016/j.ymeth.2014.08.010. http://dx.doi.org/10.1016/j.ymeth.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 85.Lyne PD. Structure-based virtual screening: an overview. Drug Discov Today. 2002;7:1047–55. doi: 10.1016/s1359-6446(02)02483-2. http://dx.doi.org/10.1016/S1359-6446(02)02483-2. [DOI] [PubMed] [Google Scholar]
- 86.Irwin JJ, Shoichet BK. ZINC - A free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–82. doi: 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pence HE, Williams A. ChemSpider: An Online Chemical Information Resource. J Chem Educ. 2010;87:1123–4. doi: 10.1021/ed100697w. [DOI] [Google Scholar]
- 88.Chen CY-C. TCM Database@Taiwan: The World’s Largest Traditional Chinese Medicine Database for Drug Screening In Silico. PLoS One. 2011;6:e15939. doi: 10.1371/journal.pone.0015939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hatherley R, Brown DK, Musyoka TM, Penkler DL, Faya N, Lobb KA, et al. SANCDB: A South African Natural Compound Database. J Cheminform. 2015:7. doi: 10.1186/s13321-015-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Musyoka TM, Kanzi AM, Lobb KA, Tastan Bishop Ö. Analysis of non-peptidic compounds as potential malarial inhibitors against Plasmodial cysteine proteases via integrated virtual screening workflow. J Biomol Struct Dyn. 2016;34:2084–101. doi: 10.1080/07391102.2015.1108231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Musyoka TM, Kanzi AM, Lobb KA, Tastan Bishop Ö. Structure Based Docking and Molecular Dynamic Studies of Plasmodial Cysteine Proteases against a South African Natural Compound and its Analogs. Sci Rep. 2016;6:23690. doi: 10.1038/srep23690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kumar A, Purohit R. Use of Long Term Molecular Dynamics Simulation in Predicting Cancer Associated SNPs. PLoS Comput Biol. 2014:10. doi: 10.1371/journal.pcbi.1003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gromiha MM, Selvaraj S. Inter-residue interactions in protein folding and stability. Prog Biophys Mol Biol. 2004;86:235–77. doi: 10.1016/j.pbiomolbio.2003.09.003. http://dx.doi.org/10.1016/j.pbiomolbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 94.Grewal RK, Roy S. Modeling Proteins as Residue Interaction Networks. Protein Pept Lett. 2015;22:923–33. doi: 10.2174/0929866522666150728115552. [DOI] [PubMed] [Google Scholar]
- 95.Atilgan AR, Turgut D, Atilgan C. Screened Nonbonded Interactions in Native Proteins Manipulate Optimal Paths for Robust Residue Communication. Biophys J. 2007;92:3052–62. doi: 10.1529/biophysj.106.099440. http://dx.doi.org/10.1529/biophysj.106.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ozbaykal G, Rana Atilgan A, Atilgan C. In silico mutational studies of Hsp70 disclose sites with distinct functional attributes. Proteins Struct Funct Bioinforma. 2015;83:2077–90. doi: 10.1002/prot.24925. [DOI] [PubMed] [Google Scholar]
- 97.Doshi U, Holliday MJ, Eisenmesser EZ, Hamelberg D. Dynamical network of residue–residue contacts reveals coupled allosteric effects in recognition, catalysis, and mutation. Proc Natl Acad Sci. 2016;113:4735–40. doi: 10.1073/pnas.1523573113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005:33. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tina KG, Bhadra R, Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007:35. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vangone A, Spinelli R, Scarano V, Cavallo L, Oliva R. COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinforma. 2011;27:2915–6. doi: 10.1093/bioinformatics/btr484. [DOI] [PubMed] [Google Scholar]
- 104.Negi SS, Schein CH, Oezguen N, Power TD, Braun W. InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatics. 2007;23:3397–9. doi: 10.1093/bioinformatics/btm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nagarajan R, Archana A, Thangakani AM, Jemimah S, Velmurugan D, Gromiha MM. PDBparam: Online Resource for Computing Structural Parameters of Proteins. Bioinform Biol Insights. 2016;10:73–80. doi: 10.4137/BBI.S38423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–2. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 108.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]