Abstract
The tribbles (trbl) pseudokinases play important roles in signaling and physiology in multiple contexts, ranging from innate immunity to cancer, suggesting fundamental cellular functions for the trbls’ gene products. Despite expression of the trbl pseudokinases in the nervous systems of invertebrate and vertebrate animals, and evidence that they have a function within mouse and human dopamine neurons, there is no clear case for a function of a Trbl protein that influences behavior. Indeed, the first and only evidence for this type of function comes from Drosophila melanogaster, where a mutation of the single trbl gene was identified in a genetic screen for short-term memory mutant flies. The current study tested flies containing multiple trbl mutant alleles and potential transgenic rescue in both operant place memory and classical olfactory memory paradigms. Genetic complementation tests and transgenic rescue of memory phenotypes in both paradigms show that the D. melanogaster trbl pseudokinase is essential for proper memory formation. Expression analysis with a polyclonal antiserum against Trbl shows that the protein is expressed widely in the fly brain, with higher expression in the cellular rind than the neuropil. Rescue of the behavioral phenotype with transgenic expression indicates the trbl function can be localized to a subset of the nervous system. Thus, we provide the first compelling case for the function of a trbl pseudokinase in the regulation of behavior.
Keywords: tribbles, pseudokinase, learning, memory, behavior
1 INTRODUCTION
The tribbles (trbl) family of pseudokinases play multiple critical roles in physiology and disease (Eyers, Keeshan, & Kannan, 2016). These pseudokinases are thought to both link substrate binding to specific protein stability by recruiting ubiquitin ligases and as regulators of MAPK and AKT/FOXO - signaling (Eyers et al., 2016). There is one version of the trbl pseudokinase in Drosophila melanogaster and Caenorhabditis elegans (Kim, Thakur, Piggott, Omi, Polanowska, Jin, & Pujol, 2016; Mata, Curado, Ephrussi, & Rorth, 2000; Pujol, Cypowyj, Ziegler, Millet, Astrain, Goncharov, Jin, Chisholm, & Ewbank, 2008). Mouse and man have three, named trbl 1, 2, and 3 (Boudeau, Miranda-Saavedra, Barton, & Alessi, 2006; Eyers et al., 2016). There is wide-spread expression of the three mammalian trbl products in brain and the D. melanogaster trbl gene is expressed in the developing nervous system (Aime, Sun, Zareen, Rao, Berman, Volpicelli-Daley, Bernd, Crary, Levy, & Greene, 2015; Fisher, Li, Hammonds, Brown, Pfeiffer, Weiszmann, MacArthur, Thomas, Stamatoyannopoulos, Eisen, Bickel, Biggin, & Celniker, 2012; Ord, Innos, Lillevali, Tekko, Sutt, Ord, Koks, Vasar, & Ord, 2014). Nevertheless, there is little known about how trbl influences brain function. The vertebrate trbl 3 has been implicated in Parkinson’s disease as a cell death promoter in dopaminergic neurons (Aime et al., 2015). However, knock-out of trbl 3 in the mouse has no effect on feeding behavior, or learning and memory in three different paradigms (Ord et al., 2014). Behavioral tests on trbl 1 or 2 have not been reported (Lin, Yang-Yen, Lien, Liao, Huang, Lin, Li, & Yen, 2016; Satoh, Kidoya, Naito, Yamamoto, Takemura, Nakagawa, Yoshioka, Morii, Takakura, Takeuchi, & Akira, 2013). Thus far the only evidence for a function of trbl in regulating behavior is from a genetic screen to identify short-term memory fly mutants (LaFerriere, Guarnieri, Sitaraman, Diegelmann, Heberlein, & Zars, 2008; Masoner, Das, Pence, Anand, LaFerriere, Zars, Bouyain, & Dobens, 2013). Whether the Drosophila trbl gene, as a key example of trbl pseudokinase function, plays a definitive role in memory is the focus of this study.
Memory is readily examined in D. melanogaster. Operant place learning tests individual flies for the ability to avoid part of a simple chamber associated with aversive high temperatures (Ostrowski & Zars, 2014; Wustmann, Rein, Wolf, & Heisenberg, 1996; Zars, 2010). That is, flies are conditioned to avoid one of two halves of a narrow chamber. Flies can be trained in minutes, memory lasts for up to 2 hours (Diegelmann, Zars, & Zars, 2006; Ostrowski, Kahsai, Kramer, Knutson, & Zars, 2015; Putz & Heisenberg, 2002). Flies show a memory by persistent avoidance of the chamber half associated with high temperature. A second well established learning paradigm is olfactory classical conditioning (Guven-Ozkan & Davis, 2014; Tully & Quinn, 1985; Zars, Fischer, Schulz, & Heisenberg, 2000a). In this case, flies are presented with an odor that is paired in time with electric shocks or sugar reward. A second odor is also presented to flies but not associated with the shock or sugar. Flies can again be trained in minutes, and memory can be readily tested for hours to days after training. Flies show a memory in a T-maze choice point by avoiding an odor previously associated with electric shock, or approaching an odor previously associated with sugar.
The role of the trbl gene in memory formation was investigated here. First, complementation tests using flies with multiple mutant alleles were tested with both operant place conditioning and aversive olfactory conditioning. Second, to gain insights into where in the nervous system trbl is expressed, the expression pattern of the Trbl protein was examined. Finally, spatially restricted expression of the wild-type trbl transgene in an otherwise mutant trbl fly was used to rescue the mutant phenotype in both the operant place conditioning paradigm and the olfactory aversive conditioning paradigm. The results show that the D. melanogaster trbl gene can influence nervous system function in behavior and is required in a specific subset of the nervous system for normal memory formation.
2 MATERIALS AND METHODS
2.1 Fly rearing
D. melanogaster were raised on cornmeal-based media in a light, temperature, and humidity controlled chamber. Flies were kept on a 12-hour light-dark cycle and held at 24 °C and 60% relative humidity. Flies used for behavioral experiments were between three and six days old and were never anesthetized.
2.2 Fly stocks and crosses
To control for genetic background, all potential mutant lines were out-crossed to a white-mutant (w1118) wild-type Canton S (CS) background for six generations. The X-chromosomes were then replaced using balancer chromosomes that were themselves in a wild-type CS background. Mutant trbl alleles that were tested were trbl3-54, trbl1119 and trbl3519 (LaFerriere et al., 2008; Rorth, Szabo, & Texido, 2000). The trbl1119 and trbl3519 are hypomorphic alleles (Masoner et al., 2013). Based on complementation tests trbl3-54 is also likely a loss-of-function allele (below). The UAS-trbl transgene was provided by Pernille Rorth (Rorth et al., 2000). The c155Gal4 driver is an enhancer trap in the elav gene, Df(3L)ri-79c is a deficiency at the trbl locus; both were provided by the Bloomington Drosophila Stock Center (Juergens, Wieschaus, Nuesslein-Volhard, & Kluding, 1984; Lin & Goodman, 1994).
2.3 Operant place conditioning in the heat box
Operant place conditioning was performed in the heat-box. In this apparatus, single flies are allowed to walk in a dark narrow chamber (34 × 1 × 3 mm) that is lined both top and bottom with Peltier elements (Wustmann et al., 1996). There is no light source with which the flies might see. The position of the fly was monitored at 10 Hz, temperature within the box was controlled by the Peltier elements (Zars, Wolf, Davis, & Heisenberg, 2000b). One half of the experiments associate high temperatures with the front half of the chamber. The other half of the experiments associates high temperatures with the back half. The temperature of 24 °C was used for the non-punished temperature as flies have a strong preference for this temperature over both higher and lower temperatures (Sayeed & Benzer, 1996; Zars, 2001) and 41 °C was used as the high temperature aversive reinforcement as this is a temperature they avoid. Conditioning consists of three phases. First flies were allowed to walk in the chamber for 30 s during a pre-test phase where the chamber is held at 24 °C. Conditioning immediately follows the pre-test. Here flies are trained for twenty minutes to avoid the punished half of the chamber. When a fly enters the punished half of the chamber by crossing an invisible midline the whole chamber heats to 41 °C and when it enters the unpunished half the whole chamber cools to 24 °C. In this case, place memory was measured directly after training for three minutes. This test provides a single measure of a memory (Putz & Heisenberg, 2002; Sitaraman, Zars, & Zars, 2007; Zars & Zars, 2006). During the memory test the chamber temperature is held at 24 °C.
2.4 Thermosensitivity
Control experiments tested the ability of wild-type and potentially mutant flies to sense and avoid a high temperature source (Zars, 2001). In this test, one half of the chamber was heated to the same temperature as that used for conditioning while the other half of the chamber is kept at 24 °C, a temperature that flies normally prefer. This provides a step-gradient for the flies, in which one half of the chamber is at a higher temperature than the other. This is in contrast to the conditioning experiments, in which the temperature of the whole chamber rises and falls depending on whether the fly moves to the front or rear of the chamber. An equal number of experiments paired the front or back chamber-half with the higher temperature.
2.5 Aversive olfactory conditioning
Aversive olfactory conditioning was performed by pairing one of two odorants (4-methylcyclohexanol or octanol) with 100 volts of electric shock (Tully & Quinn, 1985). This was done under dim red light at between 85–95% relative humidity. 100–150 flies are trapped in a copper wire-lined tube where they are presented with either of the two odors. The first odor presentation was paired with electric shock every five seconds for 1.2 seconds over one minute, followed by a one-minute rest period with a clean airstream. The second odor was then presented with no shock. Memory tests were performed 3 min after training. Altered olfactory preferences were tested in a T-maze. Flies were allowed one minute to choose between two arms one containing the odor associated with shock and the other containing the non-shock associated odor. This choice was made in complete darkness.
2.6 Shock sensitivity and odor avoidance
Control experiments for olfactory conditioning measured flies’ sensitivity to the odors and shock used in the conditioning experiment. For odor sensitivity tests, naïve flies were given a choice at the T-maze choice point between entering an arm containing an odor at the same concentration used in the conditioning experiments and entering the other arm which had air from the room. In the shock test, two shock tubes were placed at the T-maze choice point and one of these was pulsed with 100 volt electric shocks every five seconds for 1.2 seconds over one minute as in the conditioning experiment. In both odor and shock control experiments, flies were allowed to choose for 1 minute (the amount of time used in the conditioning experiments). The number of flies in both tubes were counted.
2.7 Performance index
A performance index is used to quantify fly behavior and memory in each paradigm and is calculated the same way for conditioning and control experiments.
A performance index (PI) for operant place memory is calculated as the time in the punishment-associated chamber half subtracted from the time in the non-punishment-associated chamber half, all divided by the total time in a training session (Wustmann et al., 1996). The maximum performance index is 1.0 and indicates perfect avoidance of the chamber-half associated with high temperature. A performance index of zero indicates preference for neither chamber half.
For olfactory conditioning a performance index is quantified as the number of flies that approach the non-shock associated odor minus the number of flies that approach the shock associated odor all divided by the total number of flies in a half-test. A complete olfactory experiment PI is calculated as the average from a pair of half-test PIs, where each half came from using one of the two odors as the punished odor in one conditioning experiment. Memory or avoidance performance is represented on a scale from −1 to 1, with 0 indicating no memory.
2.8 Behavior Statistics
Place memory and thermosensitivity scores were tested using non-parametric statistics (Putz, Bertolucci, Raabe, Zars, & Heisenberg, 2004; Putz & Heisenberg, 2002). That is, when multiple groups were compared a non-parametric Kruskal Wallis ANOVA was used. Tests for significant differences in olfactory conditioning and control experiments used a parametric ANOVA with Neuwmann-Keuls post-hoc tests (Zars et al., 2000a). Statistica 8.0 software was used for all tests.
2.9 Immunohistochemical analysis of the fly brain
Drosophila brains were extracted from 3–5 day old adults (Rein, Zockler, Mader, Grubel, & Heisenberg, 2002). First the proboscis and then the eyes were removed with fine forceps while the whole fly was pinned at the thorax and abdomen in Ringer’s solution (130 mM NaCl, 0.7 mM KH2PO4, 0.35 mM Na2HPO4, 18 mM MgCl2 and 4.7 mM KCl). The brains were then fixed in 2% formaldehyde in PAT (100 ml 1XPBS, 1% BSA and 0.5% Triton X-100) solution overnight at 4 °C. Brains used to visualize GFP expression were blocked for two hours with 3% normal goat serum in PAT solution at room temperature followed by overnight incubation with the two primary antibodies, anti-5HT (1:10 in PAT; Biomeda) and anti-GFP (1:20 in PAT; Sigma) at 4 °C. Secondary antibodies Alexa 488 goat anti rabbit (1:100 in PAT; Invitrogen) and Alexa 647 goat anti-mouse (1:50 in PAT; Invitrogen) were added together and incubated overnight at 4 °C.
Brains immunolabeled with the Trbl antiserum: Fixed brains were blocked using Hen-Block (1:10 in PAT; Aves Lab) at room temperature for one and a half hours followed by overnight incubation at 4 °C in anti-Trbl (1:1000 in PAT). The affinity-purified chicken polyclonal anti-Trbl antibody was raised against a peptide corresponding to amino acids CZDKHEYEDIGVEPLDYTR of the Drosophila Trbl protein (Aves Lab, Tigard, OR). The brains were also incubated with anti-Bruchpilot (Brp) (Rein et al., 2002), a synaptic active zone marker, (1:100 in PAT) for 2 hours at room temperature followed by incubation with secondary antibodies FITC anti-chick (1:500) and Alexa 647 goat anti-mouse (1:100; Invitrogen) both in PAT. All antibody incubations were followed by three 10 minute washes with PAT.
Brains were mounted in Vectashield reagent (Vector Laboratories) (1:3 Ringers: Vectashield) in a narrow well made from four coverslips and fingernail polish. The whole mount brains were visualized using an LSM 510 NLO confocal microscope with 20X objective. Images were examined using the LSM examiner software.
3 RESULTS
Complementation tests were used to determine whether the mutant phenotype observed in the insertion line trbl3-54 is indeed due to a defect in the trbl gene (LaFerriere et al., 2008). Multiple insertion alleles of trbl were used. These included trbl3-54 containing a P[GawB] P-element insertion ~250 bp 5′ of exon 1 of the trbl gene, and two additional alleles trbl1119 and trbl3519 (Mata et al., 2000; Rorth et al., 2000), which each contain an EP P-element. The trbl1119 insertion is ~185 bp downstream of the transcription start site and trbl3519 is ~8 bp upstream of the transcription start site. Few progeny can be obtained that are homozygous for either trbl1119 and trbl3519, so all tests performed were trans-heterozygous with the trbl3-54 allele or with a wild type allele.
3.1 Complementation tests using multiple trbl alleles indicate that the trbl gene is necessary for operant place memory
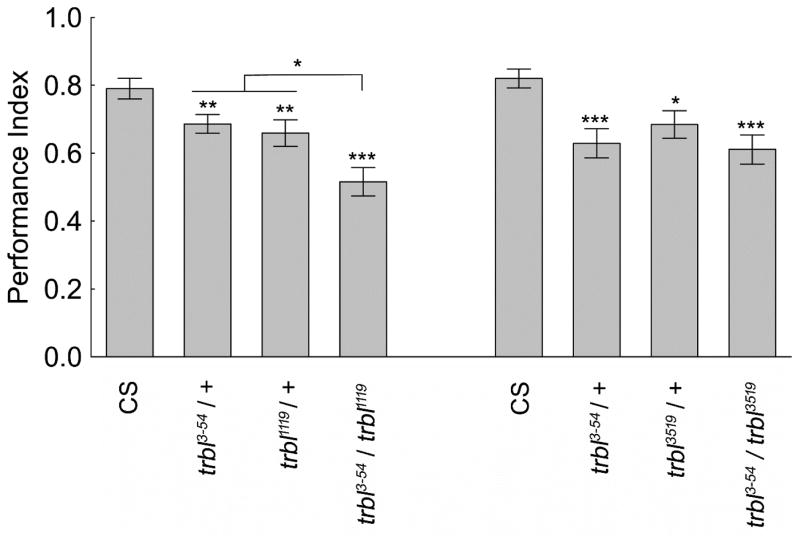
trbl3-54 homozygous flies perform significantly worse than wild-type flies in the operant place conditioning paradigm (LaFerriere et al., 2008). Tests were performed in the heat-box to determine whether two additional alleles might fail to complement the trbl3-54 phenotype. Following twenty minutes of conditioning using a 41 °C punishment temperature, place memory levels were reduced in all flies that have at least one copy of any of the mutant trbl alleles compared to wild type CS flies (Figure 1). Flies with only one mutant allele of the trbl gene had reductions of 13–25% compared to wild-type CS flies’ performance. Flies that were trans-heterozygous for trbl3519/trbl3-54 did not have a significant decrease in their performance relative to those carrying only one mutant allele. In contrast, flies of the genotype trbl1119/trbl3-54 performed significantly lower than wild-type as well as the flies heterozygous for other combinations of the mutant trbl alleles. The memory of flies of the genotype trbl1119/trbl3-54 relative to wild-type CS was ~ 30% lower. With the similarity in phenotypes between the flies with the three different insertion alleles of the trbl gene and the reduction in performance of one trans-heterozygous combination, these results argue that the trbl gene is necessary for operant place memory in the heat-box.
Figure 1. Complementation test of place memory performance for the trbl gene.
The memory performance level of each of the fly lines carrying a mutant trbl allele was significantly reduced compared to wild type CS. Flies that were trans-heterozygous for the trbl gene trbl3-54/trbl1119 performed significantly lower than CS and flies with only one copy of a mutant allele (left-most comparisons H(3, N=910) = 53.32, p < 0.001, Kruskal-Wallis ANOVA test with multiple comparisons (p<0.05 = *, p<0.01 = **, and p<0.001 = ***)). The reduction in memory performance found with flies carrying only a single mutant trbl allele was not increased with the trans-heterozygote trbl3-54/trbl3519 (right-most comparisons H(3,N=750) = 32.67, p < 0.001 Kruskal-Wallis ANOVA test with multiple comparisons (p<0.01 = ** and p<0.001 = ***)). The values represent means and the error bars are SEMs.
Flies of each genotype were tested for the ability to sense and avoid the high temperatures used in the heat box in a thermosensitivity assay. Flies were allowed to choose between chamber halves that were set at either 24 °C or 41 °C. The only significant difference found was between wild-type CS flies and those trans-heterozygous for trbl3519/trbl3-54 (Table 1). Thus, for the majority of the allelic combinations that were tested there was no significant change in the ability of the flies to sense and avoid the high temperature. There was only one case where a change in the ability to sense temperature could influence place memory. Overall, this indicates that the reductions in memory performance levels with mutation of the trbl gene is largely independent from an inability to sense and avoid the high temperature.
Table 1.
Control behaviors in wild-type CS and trbl mutant flies.
| Genotype | Shock avoidance | MCH acuity | Oct acuity | 41 °C avoidance |
|---|---|---|---|---|
| N = 48 | N = 50 | N = 54 | N = 799 | |
| CS | 73.8±3.1 | 25.7±4.5 | 29.4±5.4 | 0.64±0.03 |
| trbl1119/+ | 73.8±6.7 | 15.9±4.2 | 23.5±5.6 | 0.64±0.02 |
| trbl3-54/trbl1119 | 68.9±7.3 | 15.0±4.5 | 8.7±3.9* | 0.58±0.02 |
| trbl3-54/+ | 77.2±4.8 | 12.3±6.8 | 17.5±4.5 | 0.63±0.02 |
| trbl3519/+ | 77.6±3.3 | 29.4±3.1 | 10.4±5.5 | 0.58±0.02 |
| trbl3-54/trbl3519 | 77.6±4.2 | 16.3±3.0 | 12.5±7.1 | 0.54±0.02** |
Few significant differences were found in the control behaviors of wild-type and trbl mutant flies. Shock avoidance: ANOVA F(5, 42)=0.40, p=0.85; MCH acuity: F(5, 44)=1.90 p=0.11; Oct acuity: F(5, 48)=2.37, p=0.05, trbl3-54/trbl1119 was significantly different from CS after Multiple Comparisons, * = p < 0.05; 41 °C avoidance: Kruskal-Wallis Test H(5, N=799) =18.47, p =.002, the only significant difference between genotypes after Multiple Comparisons was between CS and trbl3-54/trbl3519 **= p < 0.01. The values represent means ± the SEMs.
3.2 Complementation tests using multiple trbl alleles indicates that the trbl gene is important for aversive olfactory memory
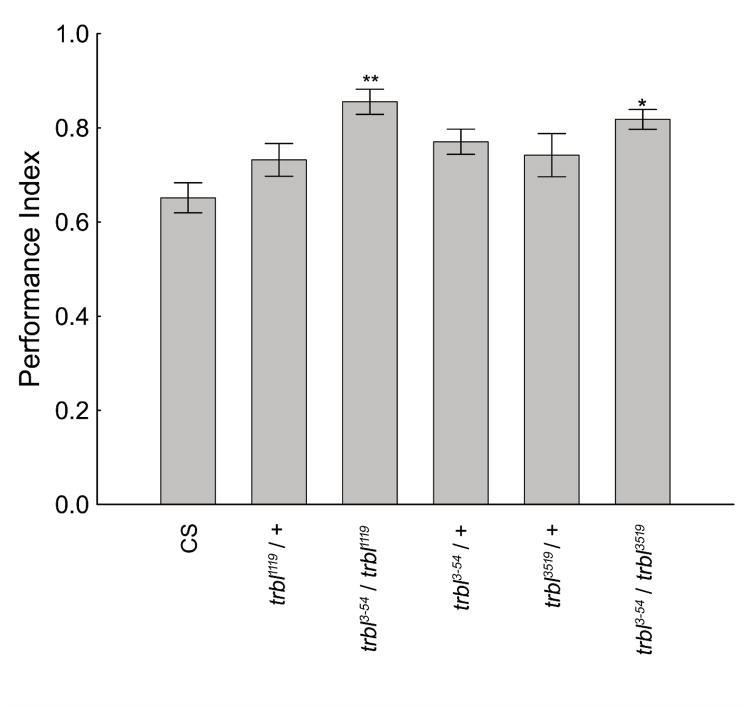
The role of trbl in olfactory learning was examined by testing flies using three different mutant trbl alleles. trbl3-54 homozygous flies perform significantly higher than wild-type flies in this paradigm (LaFerriere et al., 2008). Only flies that were trans-heterozygous with two mutant trbl alleles performed significantly higher than wild-type CS flies, at ~125% of wild-type levels (Figure 2). Flies with only a single mutant trbl allele performed slightly better than wild-type, but never reached statistically significant levels. Thus the trbl gene is necessary for the altered phenotypes in olfactory memory tests.
Figure 2. Complementation test in the olfactory classical conditioning paradigm for the trbl gene.
Three trbl insertion alleles were tested. Complementation tests reveal significantly better memory compared to wild-type CS in the olfactory paradigm for the trans-heterozygotes, trb1119/trbl3-54 and trbl3519/trbl3-54. Flies with a single mutant copy of the trbl have increase performance compared to wild-type, but this does not reach significant levels ANOVA: F(5,58) = 5.04, p < 0.001 Newman-Keuls post-hoc test (p< 0.05 = * and p<0.01 = **) N=8 for each genotype. The values represent means and error bars are SEMs.
Flies from each genotype were next tested for their ability to sense and avoid the odors and electric shock used for the olfactory aversive conditioning experiment. Although there was variation in the odor avoidance and shock avoidance scores between genotypes, there was only one significant difference found between trbl3-54/trbl1119 and CS flies in octanol avoidance (Table 1). A significant difference was not found, however, between flies of other mutant alleles and wild-type, suggesting that there is not a consistent relationship between altered octanol avoidance and olfactory memory in mutant flies. Moreover, changes in olfactory memory levels was not a consequence of changes in the ability to sense and avoid MCH or the shocks used for conditioning.
3.3 The Trbl protein is expressed throughout the adult fly brain
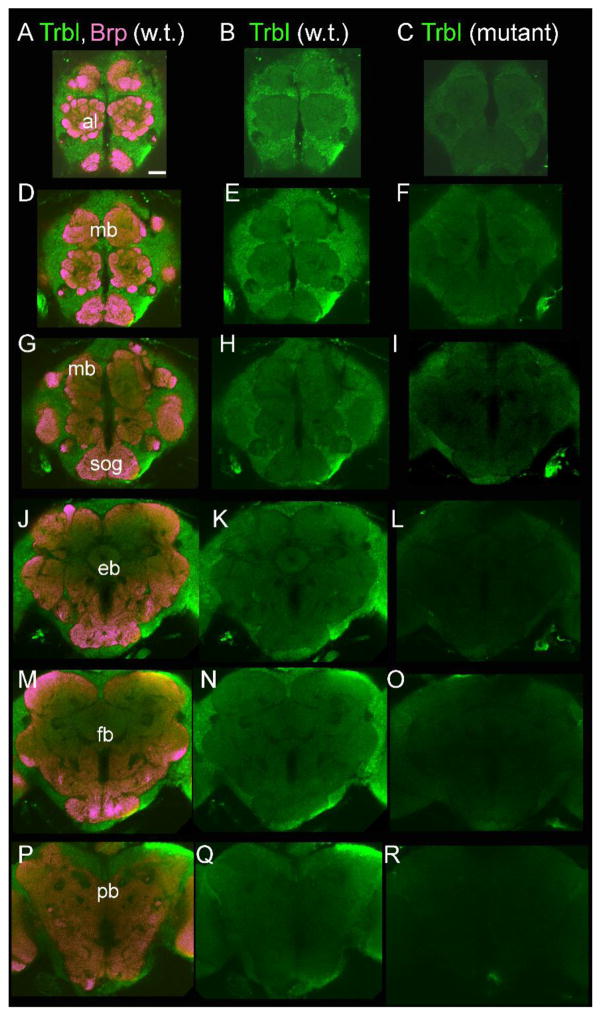
The expression pattern of the Trbl protein was examined using a polyclonal antiserum. Anti-Trbl was used to visualize expression of Trbl in the adult fly brain of both wild-type CS flies and mutant trbl flies (Df(3L)ri-79c/trbl1119) (Figure 3). In the CS fly brain, Trbl expression is visible in cell bodies throughout the brain. There is also detectable expression in the neuropil. There is a marked decrease in Trbl expression levels in trbl mutant flies. This reduction in anti-Trbl signal between CS flies and trbl mutant flies indicates that it is indeed the Trbl protein that is visualized.
Figure 3. Tribbles immunostaining in the Drosophila brain.
Trbl is expressed in cell bodies and the neuropil throughout the fly brain visualized in green in both wild-type CS brains (left and center columns) and trbl mutant fly brains (right column). Mutant flies have reduced Tribbles expression compared to wild-type CS levels, comparing center and right most panels. Brains were co-labeled with the bruchpilot (brp) monoclonal antibody marking the synapses in purple on the left most panels. Labeled structures: antennal lobes (al), mushroom bodies (mb), subesophogial ganglia (sog), ellipsoid body (eb), fan shaped body (fb), protocerebral bridge (pb). Scale bar = 50 μm.
3.4 The trbl mutant phenotype can be rescued in operant place memory
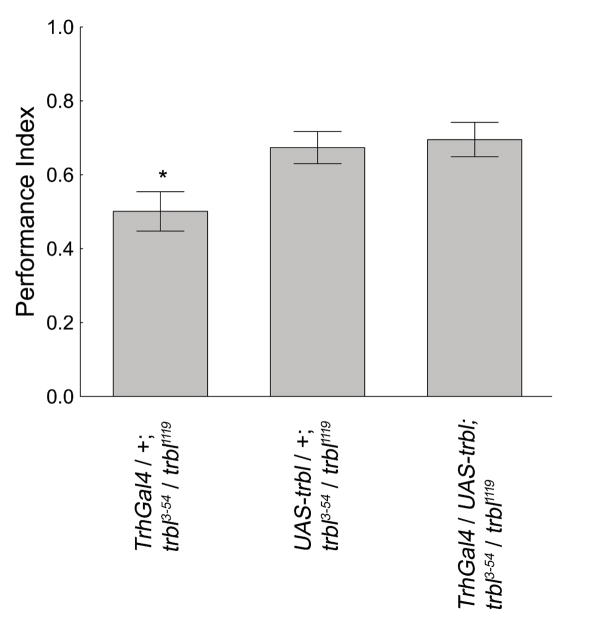
The reduced memory phenotype of trbl mutant flies in the place-conditioning paradigm was rescued with expression of a wild-type version of trbl as a transgene. Since the trans-heterozygote trbl1119/trbl3-54 gave the strongest mutant phenotype, these flies along with the appropriate genetic controls were used in the rest of the conditioning experiments. trbl3-54 Gal4 driven expression of UAS-trbl (Mata et al., 2000) is sufficient to rescue the reduced memory phenotype of trbl mutant flies in the heat-box (Figure 4). We further tested whether the addition of a serotonin Gal4 driver, Trh-Gal4 (Park, Lee, Lee, Kim, Song, Kim, Bae, Kim, Shong, Kim, & Chung, 2006), could further increase memory levels, since serotonin is the key biogenic amine for operant place memory (Sitaraman, Zars, Laferriere, Chen, Sable-Smith, Kitamoto, Rottinghaus, & Zars, 2008). The addition of the Trh-Gal4 driver does not significantly increase the performance of trbl3-54-rescued flies (Figure 4). These results indicate that trbl expression is sufficient in cells expressing the trbl3-54 Gal4 for normal memory formation.
Figure 4. Rescue of the trbl mutant phenotype in the operant place-learning paradigm.
trbl3-54 driven expression of UAS-trbl, in otherwise mutant flies, was able to rescue the trbl mutant phenotype in the heat-box. The addition of Trh Gal4 did not increase performance further H(2,N=476) = 16.18, p < 0.001 Kruskal-Wallis ANOVA. (p<0.05 = *). The values represent means and error bars are SEMs.
Flies of the same genotypes used in the rescue experiments were tested for their ability to sense and avoid the high temperature in a thermosensitivity assay. There are no significant differences in thermosensitivity between Trh-Gal4/UAS-trbl;trbl1119/trbl3-54 and Trh-Gal4/+;trbl1119/trbl3-54 or between Trh-Gal4/UAS-trbl;trbl1119/trbl3-54 and UAS-trbl/+;trbl1119/trbl3-54 (Table 2). A significant difference was found between UAS-trbl/+;trbl1119/trbl3-54 and Trh-Gal4/+;trbl1119/trbl3-54. The flies from one of the genotypes that were rescued for place memory was different from control flies in the ability to avoid high temperatures, but the other rescuing line was not different from the genotypic control for this behavior. These mixed results show that the memory rescue can be dissociated from changes in high temperature avoidance behavior.
Table 2.
Control behaviors for trbl mutant rescue in the heat box paradigm
| Genotype | 41 °C avoidance |
|---|---|
| N = 357 | |
| TrhGal4/+; trbl3-54/trbl1119 | 0.67±0.03 |
| UAS-trbl/+; trbl3-54/trbl1119 | 0.76±0.03 |
| TrhGal4/UAS trbl; trbl3-54/trbl1119 | 0.71±0.03 |
Kruskal-Wallis Test; H(2, N=357) = 15.43 p =0.0004. A significant difference was found between UAS-trbl/+; trbl3-54/trbl1119 and TrhGal4/+; trbl3-54/trbl1119 p<0.001 after Multiple Comparisons for 41 °C avoidance. No other significant differences were found.
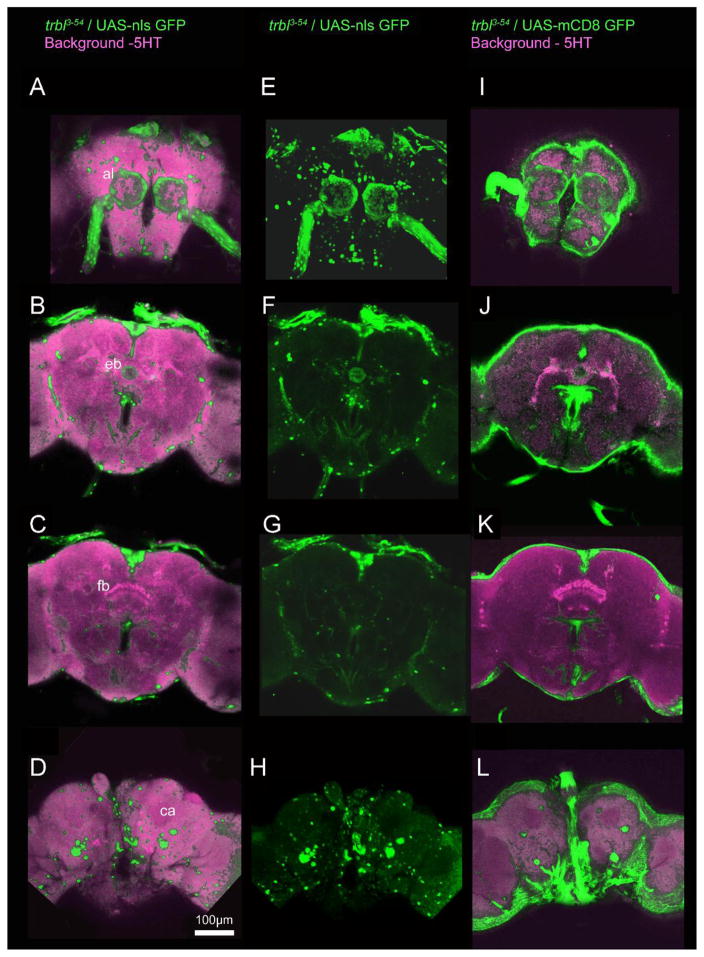
3.5 The expression pattern of the trbl3-54 Gal4 driver examined with UAS-GFP
The expression pattern of the trbl3-54 Gal4 driver was examined using UAS-GFP driven expression. This was done to find where trbl expression is sufficient to rescue the reduced memory phenotype in the heat-box. Two different UAS-GFP fly lines were used, UAS-nls-GFP (targeting GFP expression to the nucleus) and UAS-mCD8-GFP (targeting GFP expression to the membranes) (Lee & Luo, 1999; Neufeld, de la Cruz, Johnston, & Edgar, 1998). Expression is visualized throughout the brain (Figure 5). Expression in the antennal lobes and the antennal nerve are visible in the anterior most sections using either UAS-GFP (Figure 5). More posterior, expression in the median bundle and the ellipsoid body is visible along with a cluster of cells on the dorsal surface of the brain in the pars intercerebralis (Figure 5). The pars intercerebralis cluster of cell bodies becomes more noticeable moving farther posterior to the level of the fan shaped body (Figure 5). The most posterior sections have several cell bodies present (Figure 5). There is also expression around the periphery of the whole brain in all sections with UAS-mCD8-GFP, which may correspond to glial cells. Although unlikely, we cannot rule out the possibility that expression outside of the nervous system could be important for the trbl-dependent function in behavior.
Figure 5. trbl3-54 Gal4 driven expression of two different UAS-GFPs in the adult brain.
GFP expression is visualized from anterior to posterior A–D with UAS-nls-GFP and anti-5HT, E–H with UAS-nls-GFP alone, and I–L with UAS-mCD8-GFP and anti-5HT. Notable structures are labeled: antennal lobes (al), median bundle (meb), ellipsoid body (eb), fan shaped body (fb) and calyces (ca). Expression is visualized throughout the brain including in the antennal lobes, median bundle, ellipsoid body, and around the periphery of the brain. Scale bar = 100 μm.
3.6 The trbl mutant phenotype can be rescued for aversive olfactory memory
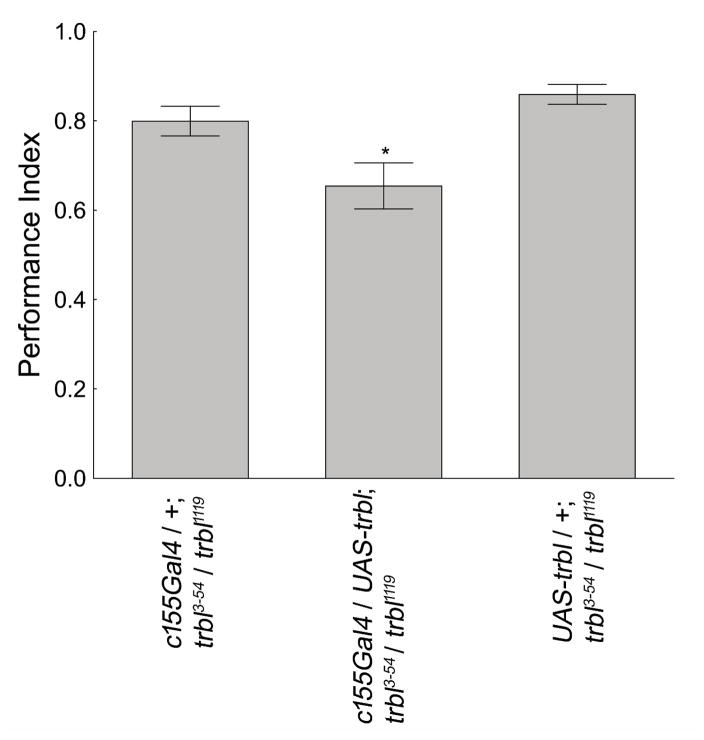
Experiments to attempt a genetic rescue of the increased memory phenotype of trbl mutant flies in olfactory aversive conditioning paradigm were performed. The trans-heterozygous trbl1119/trbl3-54 flies gave the strongest mutant phenotype, so they were used as the trbl mutant flies in the rest of the olfactory conditioning experiments. The trbl3-54 driven expression of UAS-trbl was not sufficient to rescue the olfactory memory changes (Figure 6). Addition of the c155Gal4 driver, by contrast, did rescue the elevated olfactory memory scores of trbl mutant flies.
Figure 6. Rescue of the trbl mutant phenotype in olfactory aversive conditioning.
c155Gal4 driven expression of UAS-trbl, in otherwise mutant flies was able to rescue the trbl mutant phenotype in the olfactory conditioning paradigm, causing these flies to have significantly lower memory scores comparted to the control genotypes. ANOVA: F(2,18) = 7.83, p = 0.004 Newman-Keuls post-hoc test (p<0.05 = *). N=7 for each group. The values represent means and error bars are SEMs.
The same genotypes of flies used for the olfactory aversive rescue experiments were examined for their ability to sense and avoid the odors and electric shock used during the experiment (Table 3). The only significant difference found was between c155Gal4/+; UAS-trbl/+;trbl1119/trbl3-54 and UAS-trbl/+;trbl1119/trbl3-54 for MCH avoidance. This difference is unlikely to cause the memory phenotypes observed, since this difference in MCH acuity does not seem to be related with the altered memory performance index.
Table 3.
Control behaviors for trbl mutant rescue in the aversive olfactory paradigm
| Genotype | Shock acuity | MCH acuity | Oct acuity |
|---|---|---|---|
| N = 18 | N = 18 | N = 12 | |
| c155Gal4/+; trbl3-54/trbl1119 | 72.6±5.8 | −4.4±2.2 | 11.1±9.2 |
| UAS-trbl/+; trbl3-54/trbl1119 | 65.4±5.7 | 9.6±4.3 | 24.7±8.3 |
| c155Gal4/+;UAS-trbl/+; trbl3-54/trbl1119 | 52.4±7.4 | −4.1±6.7 | 12.8±13.2 |
Only a single significant difference was found for the aversive olfactory rescue experiment control behaviors. Shock avoidance: ANOVA F(2,15)=2.6, p=0.11; MCH acuity: F(2,12)=4.4, p=0.038; Oct acuity: F(2,9)= 0.5, p=0.62. MCH acuity is significantly different between c155Gal4/+;UAS-trbl/+; trbl3-54/trbl1119 and UAS-trbl/+; trbl3-54/trbl1119, p=0.04 following Newman-Keuls post-hoc test. The values represent means ± the SEMs.
4 DISCUSSION
Although the trbl pseudokinase in Drosophila, and the vertebrate trbl 1–3, are expressed in the brain (Figure 3) (Aime et al., 2015; Fisher et al., 2012; Ord et al., 2014), the results here provide the first definitive evidence for the function of this protein in regulating behavior. We show that the trbl gene is critical for normal memory formation. That is, the trbl gene provides a function that permits memories to be formed normally. The results here cannot discriminate a function for trbl in different components or stages of memory formation. Mutation of the trbl gene causes opposite effects in the two conditioning paradigms tested. Nevertheless, when multiple alleles are tested each results in abnormal phenotypes in the two paradigms. A decrease in memory performance in operant place conditioning was identified in flies heterozygous for three different trbl alleles. In one case, this defect was increased in flies trans-heterozygous for two different mutant trbl alleles. Conversely, an increase in memory performance in classical olfactory aversive conditioning was observed only for flies that were trans-heterozygous for two different mutant alleles. The memory phenotypes from flies with multiple mutant alleles argues that the trbl gene is necessary for normal memory formation. Moreover, the trbl mutant phenotype is rescued by targeted gene expression in both of the conditioning paradigms tested. Using transgenic expression of a UAS-trbl gene, we are able to rescue place learning using trbl3-54 Gal4 driven expression. The addition of a Trh-Gal4 did not increase the memory performance further relative to expression driven by trbl3-54 Gal4 alone. We were also able to rescue aversive olfactory memory using c155Gal4 driven expression. c155Gal4 is expressed throughout the nervous system (Lin & Goodman, 1994). Since expression of UAS-trbl in neurons is sufficient to normalize aversive olfactory memory, we conclude that trbl expression in neurons of the brain is important for aversive olfactory memory formation. Thus, using multiple allelic combinations and transgenic rescue, the Drosophila trbl gene is clearly important for memory formation.
That trbl mutation causes opposite phenotypes when tested in these two paradigms is rare but not unique. Behavioral changes such as these have been observed in flies mutant for the white ABC transporter (Diegelmann et al., 2006). The mutant white gene causes defects in both place memory and aversive olfactory memory. For place memory, flies’ mutant for white perform significantly worse than wild type, but when tested using different shock intensities for olfactory aversive memory, mutant flies perform significantly better than wild type. Both serotonin and dopamine levels are reduced by mutation of the white ABC transporter and these biogenic amines play a role in place and olfactory memory (Sitaraman et al., 2008; Waddell, 2010). Perhaps the trbl gene alters the biogenic amine systems or its effects to alter memory formation. Future experiments testing the role trbl in biogenic amine systems will address this possibility.
There are two primary open questions about the function of trbl in memory formation. Where in the fly brain trbl is acting to regulate memory formation is not clear. The expression pattern of Trbl is throughout the adult brain (Figure 3). The Trbl signal is strongest in the cell body rind. Weaker but clear expression is detected throughout the neuropil. There was no region with relatively high or low expression that would provide a clue to where Trbl might be acting in memory formation. At a minimum one can conclude that Trbl is expressed in brain regions that have been previously shown as important for both place and olfactory memory, including the mushroom bodies and the median bundle (Guven-Ozkan & Davis, 2014; Heisenberg, 2003; Zars, 2000; Zars et al., 2000a; Zars et al., 2000b). The rescue of the place memory phenotype with trbl3-54 suggests regions that might be important for this type of conditioning. Gene expression driven by trbl3-54 GAL4 was detected in the antennal lobe, ellipsoid body, and median bundle (Figure 5). These are the same regions that have been shown to be important for rutabaga-dependent place memory (Zars et al., 2000b). That the olfactory memory phenotype could only be rescued with the addition wide-spread expression in the nervous system suggests that trbl expression is required in places in addition to the antennal lobe, ellipsoid body, and median bundle. Or, alternatively, that there is more expression of UAS-trbl in the trbl3-54 GAL4 positive cells with the addition of the c155GAL4 driver. In any event, a good candidate structure for local rescue of trbl would be in the mushroom bodies, as these have been repeatedly shown as a site critical for olfactory memory (Guven-Ozkan & Davis, 2014; Heisenberg, 2003; Zars, 2000; Zars et al., 2000a). When in the life cycle trbl is acting to support normal memory is also not clear. The trbl gene product has multiple cellular functions and a role in early development (Eyers et al., 2016; Masoner et al., 2013). One cannot differentiate with the current results whether or not trbl acts through development or in the adult fly for memory formation. That there is widespread expression of the Trbl protein in the fly brain suggests that there will be adult-specific functions of this protein. Future investigation of trbl will be needed to address where and when this gene is sufficient for proper memory formation.
The finding that trbl is critical for place memory and olfactory memory suggests a new set of signaling mechanisms that could be important for memory formation. The trbl gene encodes a pseudokinase. The conserved serine/threonine kinase domain is missing key residues necessary for normal kinase activity including an ATP binding region. Observed interactions indicate that trbl acts as a signal transducer to target specific proteins to a ubiquitin ligase followed by protein degradation (Eyers et al., 2016; Mata et al., 2000; Qi, Heredia, Altarejos, Screaton, Goebel, Niessen, Macleod, Liew, Kulkarni, Bain, Newgard, Nelson, Evans, Yates, & Montminy, 2006). In Drosophila, trbl is a known regulator of both string/CDC25 (a cell cycle regulator) and slow boarders (a homolog to the C/EBP family of transcription factors) by targeting each to the proteasome pathway (Masoner et al., 2013; Mata et al., 2000; Rorth et al., 2000). In mammals, trbl homologous genes products regulate the activity of a number of proteins. Mammalian Trbl proteins are implicated in regulating AKT/FOXO and MAPK signaling in various cancer models (Du, Herzig, Kulkarni, & Montminy, 2003; Erazo, Lorente, Lopez-Plana, Munoz-Guardiola, Fernandez-Nogueira, Garcia-Martinez, Bragado, Fuster, Salazar, Espadaler, Hernandez-Losa, Bayascas, Cortal, Vidal, Gascon, Gomez-Ferreria, Alfon, Velasco, Domenech, & Lizcano, 2016; Salazar, Lorente, Garcia-Taboada, Perez Gomez, Davila, Zuniga-Garcia, Maria Flores, Rodriguez, Hegedus, Mosen-Ansorena, Aransay, Hernandez-Tiedra, Lopez-Valero, Quintanilla, Sanchez, Iovanna, Dusetti, Guzman, Francis, Carracedo, Kiss-Toth, & Velasco, 2015; Sung, Guan, Czibula, King, Eder, Heath, Suvarna, Dower, Wilson, Francis, Crossman, & Kiss-Toth, 2007; Yokoyama, Kanno, Yamazaki, Takahara, Miyata, & Nakamura, 2010; Zareen, Biswas, & Greene, 2013). They also play a role in regulation of transcription factors like ATF4 and C/EBPalpha (Bowers, Scully, & Boylan, 2003; Dedhia, Keeshan, Uljon, Xu, Vega, Shestova, Zaks-Zilberman, Romany, Blacklow, & Pear, 2010) and interact with a number of additional proteins including p65 (regulating NF-kappaB-dependent transcription), CtIP (cell cycle regulation), and MAPKK (Hegedus, Czibula, & Kiss-Toth, 2007; Kiss-Toth, Bagstaff, Sung, Jozsa, Dempsey, Caunt, Oxley, Wyllie, Polgar, Harte, O’Neill L, Qwarnstrom, & Dower, 2004; Wu, Xu, Zhai, & Shu, 2003; Xu, Lv, Qin, Shu, Xu, Chen, Xu, Sun, & Wu, 2007). The trbl-dependent signaling functions from other systems provide a rich target for novel memory formation mechanisms in Drosophila and across species.
To summarize, flies mutant for multiple trbl alleles show abnormalities in two forms of memory. Transgenic rescue with a wild-type version of the trbl gene reverses the mutant phenotype to wild-type levels. Expression analysis shows widespread expression of Trbl in the fly brain. Thus, the trbl pseudokinase can be critical for regulating complex behavior.
Highlights.
The tribbles class of pseudokinases is critical for signaling and physiology
The Drosophila tribbles gene is expressed widely in the brain
The Drosophila tribbles gene is essential for two forms of memory
Acknowledgments
The authors thank Pernille Rorth and Leonard Dobens for kindly providing fly strains. The Bloomington Drosophila Stock also provided fly strains.
Funding Resources: Research in the laboratory of TZ is funded by the National Sciences Foundation [Grants 1535790 and 1654866] and National Institutes of Health [Grants DK107900 and NS076980].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aime P, Sun X, Zareen N, Rao A, Berman Z, Volpicelli-Daley L, Bernd P, Crary JF, Levy OA, Greene LA. Trib3 Is Elevated in Parkinson’s Disease and Mediates Death in Parkinson’s Disease Models. J Neurosci. 2015;35:10731–10749. doi: 10.1523/JNEUROSCI.0614-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Bowers AJ, Scully S, Boylan JF. SKIP3, a novel Drosophila tribbles ortholog, is overexpressed in human tumors and is regulated by hypoxia. Oncogene. 2003;22:2823–2835. doi: 10.1038/sj.onc.1206367. [DOI] [PubMed] [Google Scholar]
- Dedhia PH, Keeshan K, Uljon S, Xu L, Vega ME, Shestova O, Zaks-Zilberman M, Romany C, Blacklow SC, Pear WS. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood. 2010;116:1321–1328. doi: 10.1182/blood-2009-07-229450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diegelmann S, Zars M, Zars T. Genetic dissociation of acquisition and memory strength in the heat-box spatial learning paradigm in Drosophila. Learn Mem. 2006;13:72–83. doi: 10.1101/lm.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- Erazo T, Lorente M, Lopez-Plana A, Munoz-Guardiola P, Fernandez-Nogueira P, Garcia-Martinez JA, Bragado P, Fuster G, Salazar M, Espadaler J, Hernandez-Losa J, Bayascas JR, Cortal M, Vidal L, Gascon P, Gomez-Ferreria M, Alfon J, Velasco G, Domenech C, Lizcano JM. The New Antitumor Drug ABTL0812 Inhibits the Akt/mTORC1 Axis by Upregulating Tribbles-3 Pseudokinase. Clin Cancer Res. 2016;22:2508–2519. doi: 10.1158/1078-0432.CCR-15-1808. [DOI] [PubMed] [Google Scholar]
- Eyers PA, Keeshan K, Kannan N. Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol. 2016 doi: 10.1016/j.tcb.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher WW, Li JJ, Hammonds AS, Brown JB, Pfeiffer BD, Weiszmann R, MacArthur S, Thomas S, Stamatoyannopoulos JA, Eisen MB, Bickel PJ, Biggin MD, Celniker SE. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci U S A. 2012;109:21330–21335. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Davis RL. Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem. 2014;21:519–526. doi: 10.1101/lm.034363.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus Z, Czibula A, Kiss-Toth E. Tribbles: a family of kinase-like proteins with potent signalling regulatory function. Cell Signal. 2007;19:238–250. doi: 10.1016/j.cellsig.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Juergens G, Wieschaus E, Nuesslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Wilhelm Roux’s Archives of Developmental Biology. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Kim KW, Thakur N, Piggott CA, Omi S, Polanowska J, Jin Y, Pujol N. Coordinated inhibition of C/EBP by Tribbles in multiple tissues is essential for Caenorhabditis elegans development. BMC Biol. 2016;14:104. doi: 10.1186/s12915-016-0320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Toth E, Bagstaff SM, Sung HY, Jozsa V, Dempsey C, Caunt JC, Oxley KM, Wyllie DH, Polgar T, Harte M, O’Neill LA, Qwarnstrom EE, Dower SK. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J Biol Chem. 2004;279:42703–42708. doi: 10.1074/jbc.M407732200. [DOI] [PubMed] [Google Scholar]
- LaFerriere H, Guarnieri DJ, Sitaraman D, Diegelmann S, Heberlein U, Zars T. Genetic dissociation of ethanol sensitivity and memory formation in Drosophila melanogaster. Genetics. 2008;178:1895–1902. doi: 10.1534/genetics.107.084582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Lin KR, Yang-Yen HF, Lien HW, Liao WH, Huang CJ, Lin LI, Li CL, Yen JJ. Murine tribbles homolog 2 deficiency affects erythroid progenitor development and confers macrocytic anemia on mice. Sci Rep. 2016;6:31444. doi: 10.1038/srep31444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoner V, Das R, Pence L, Anand G, LaFerriere H, Zars T, Bouyain S, Dobens LL. The kinase domain of Drosophila Tribbles is required for turnover of fly C/EBP during cell migration. Dev Biol. 2013;375:33–44. doi: 10.1016/j.ydbio.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi A, Rorth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Ord T, Innos J, Lillevali K, Tekko T, Sutt S, Ord D, Koks S, Vasar E, Ord T. Trib3 is developmentally and nutritionally regulated in the brain but is dispensable for spatial memory, fear conditioning and sensing of amino acid-imbalanced diet. PLoS ONE. 2014;9:e94691. doi: 10.1371/journal.pone.0094691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski D, Kahsai L, Kramer EF, Knutson P, Zars T. Place memory retention in Drosophila. Neurobiol Learn Mem. 2015;123:217–224. doi: 10.1016/j.nlm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- Ostrowski D, Zars T. Place Memory. In: Dubnau J, editor. Handbook of Behavior Genetics of Drosophila melanogaster: Behavioral Phenotypes and Models of Neurobehavioral Disorders. Cambridge: Cambridge University Press; 2014. pp. 125–134. [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz G, Bertolucci F, Raabe T, Zars T, Heisenberg M. The S6KII (rsk) gene of Drosophila melanogaster differentially affects an operant and a classical learning task. J Neurosci. 2004;24:9745–9751. doi: 10.1523/JNEUROSCI.3211-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz G, Heisenberg M. Memories in Drosophila heat-box learning. Learn Mem. 2002;9:349–359. doi: 10.1101/lm.50402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Heredia JE, Altarejos JY, Screaton R, Goebel N, Niessen S, Macleod IX, Liew CW, Kulkarni RN, Bain J, Newgard C, Nelson M, Evans RM, Yates J, Montminy M. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312:1763–1766. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- Rein K, Zockler M, Mader MT, Grubel C, Heisenberg M. The Drosophila standard brain. Curr Biol. 2002;12:227–231. doi: 10.1016/s0960-9822(02)00656-5. [DOI] [PubMed] [Google Scholar]
- Rorth P, Szabo K, Texido G. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol Cell. 2000;6:23–30. doi: 10.1016/s1097-2765(05)00008-0. [DOI] [PubMed] [Google Scholar]
- Salazar M, Lorente M, Garcia-Taboada E, Perez Gomez E, Davila D, Zuniga-Garcia P, Maria Flores J, Rodriguez A, Hegedus Z, Mosen-Ansorena D, Aransay AM, Hernandez-Tiedra S, Lopez-Valero I, Quintanilla M, Sanchez C, Iovanna JL, Dusetti N, Guzman M, Francis SE, Carracedo A, Kiss-Toth E, Velasco G. Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ. 2015;22:131–144. doi: 10.1038/cdd.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kidoya H, Naito H, Yamamoto M, Takemura N, Nakagawa K, Yoshioka Y, Morii E, Takakura N, Takeuchi O, Akira S. Critical role of Trib1 in differentiation of tissue-resident M2-like macrophages. Nature. 2013;495:524–528. doi: 10.1038/nature11930. [DOI] [PubMed] [Google Scholar]
- Sayeed O, Benzer S. Behavioral genetics of thermosensation and hygrosensation in Drosophila. Proc Natl Acad Sci U S A. 1996;93:6079–6084. doi: 10.1073/pnas.93.12.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D, Zars M, Laferriere H, Chen YC, Sable-Smith A, Kitamoto T, Rottinghaus GE, Zars T. Serotonin is necessary for place memory in Drosophila. Proc Natl Acad Sci U S A. 2008;105:5579–5584. doi: 10.1073/pnas.0710168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D, Zars M, Zars T. Reinforcement pre-exposure enhances spatial memory formation in Drosophila. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2007;193:903–908. doi: 10.1007/s00359-007-0243-9. [DOI] [PubMed] [Google Scholar]
- Sung HY, Guan H, Czibula A, King AR, Eder K, Heath E, Suvarna SK, Dower SK, Wilson AG, Francis SE, Crossman DC, Kiss-Toth E. Human tribbles-1 controls proliferation and chemotaxis of smooth muscle cells via MAPK signaling pathways. J Biol Chem. 2007;282:18379–18387. doi: 10.1074/jbc.M610792200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol [A] 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Waddell S. Dopamine reveals neural circuit mechanisms of fly memory. Trends Neurosci. 2010;33:457–464. doi: 10.1016/j.tins.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Xu LG, Zhai Z, Shu HB. SINK is a p65-interacting negative regulator of NF-kappaB-dependent transcription. J Biol Chem. 2003;278:27072–27079. doi: 10.1074/jbc.M209814200. [DOI] [PubMed] [Google Scholar]
- Wustmann G, Rein K, Wolf R, Heisenberg M. A new paradigm for operant conditioning of Drosophila melanogaster. J Comp Physiol [A] 1996;179:429–436. doi: 10.1007/BF00194996. [DOI] [PubMed] [Google Scholar]
- Xu J, Lv S, Qin Y, Shu F, Xu Y, Chen J, Xu BE, Sun X, Wu J. TRB3 interacts with CtIP and is overexpressed in certain cancers. Biochim Biophys Acta. 2007;1770:273–278. doi: 10.1016/j.bbagen.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Kanno Y, Yamazaki Y, Takahara T, Miyata S, Nakamura T. Trib1 links the MEK1/ERK pathway in myeloid leukemogenesis. Blood. 2010;116:2768–2775. doi: 10.1182/blood-2009-10-246264. [DOI] [PubMed] [Google Scholar]
- Zareen N, Biswas SC, Greene LA. A feed-forward loop involving Trib3, Akt and FoxO mediates death of NGF-deprived neurons. Cell Death Differ. 2013;20:1719–1730. doi: 10.1038/cdd.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zars M, Zars T. High and low temperatures have unequal reinforcing properties in Drosophila spatial learning. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2006;192:727–735. doi: 10.1007/s00359-006-0109-6. [DOI] [PubMed] [Google Scholar]
- Zars T. Behavioral functions of the insect mushroom bodies. Curr Opin Neurobiol. 2000;10:790–795. doi: 10.1016/s0959-4388(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Zars T. Two thermosensors in Drosophila have different behavioral functions. J Comp Physiol [A] 2001;187:235–242. doi: 10.1007/s003590100194. [DOI] [PubMed] [Google Scholar]
- Zars T. Short-term memories in Drosophila are governed by general and specific genetic systems. Learn Mem. 2010;17:246–251. doi: 10.1101/lm.1706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000a;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- Zars T, Wolf R, Davis R, Heisenberg M. Tissue-specific expression of a type I adenylyl cyclase rescues the rutabaga mutant memory defect: In search of the engram. Learn Mem. 2000b;7:18–31. doi: 10.1101/lm.7.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]