Abstract
Language impairment is common in prodromal stages of Alzheimer’s disease (AD) and progresses over time. However, the genetic architecture underlying language performance is poorly understood. To identify novel genetic variants associated with language performance, we analyzed brain MRI and performed a genome-wide association study (GWAS) using a composite measure of language performance from the Alzheimer’s Disease Neuroimaging Initiative (ADNI; n=1,560). The language composite score was associated with brain atrophy on MRI in language and semantic areas. GWAS identified GLI3 (GLI family zinc finger 3) as significantly associated with language performance (p < 5 × 10−8). Enrichment of GWAS association was identified in pathways related to nervous system development and glutamate receptor function and trafficking. Our results, which warrant further investigation in independent and larger cohorts, implicate GLI3, a developmental transcription factor involved in patterning brain structures, as a putative gene associated with language dysfunction in AD.
Keywords: Alzheimer’s disease, language performance, GWAS, GLI3, neuroimaging
1. Introduction
Alzheimer’s disease (AD), the most common neurodegenerative disorder, is clinically characterized by progressive cognitive impairment primarily in memory in the earliest stages. However, as the disease progresses, deficits can also be observed in other cognitive domains such as judgment, orientation, executive function, visuospatial ability, and language. Language impairment is a prevalent clinical feature that often occurs early on – sometimes even before AD is diagnosed, and progresses during the course of the disease (Ahmed, Haigh, de Jager, & Garrard, 2013). Several language tests have been shown to discriminate between cognitively normal (CN) and those with AD (Bertola et al., 2014; Clark et al., 2014; Gomez & White, 2006; Henry, Crawford, & Phillips, 2004). Hence, language impairment is one of several areas of cognitive decline that can aid in the clinical diagnosis of AD dementia according to the most recent criteria (McKhann et al., 2011). Language deficits during prodromal AD include impairments in word finding (Bayles, Tomoeda, & Trosset, 1990), naming (Hodges, Salmon, & Butters, 1991), and verbal fluency (Monsch et al., 1992; Nutter-Upham et al., 2008).
A large portion of the neuropsychological tests used to assess language impairment, such as the Boston Naming Test (BNT) and Animal Fluency, are dependent on semantic memory (Verma & Howard, 2012). It has been suggested that the language impairment observed in AD is primarily the result of a decline in semantic processing due to structural and organizational deterioration of semantic memory (Chertkow & Bub, 1990). Other memory systems affected include episodic memory, one of the earliest and most severely affected systems in AD (Hodges & Graham, 2001). Language impairments in prodromal and clinical AD have been previously associated with cortical atrophy predominantly in left temporal and parietal lobe regions, in addition to several other brain regions (Ahn et al., 2011; Apostolova et al., 2008; Domoto-Reilly, Sapolsky, Brickhouse, Dickerson, & Alzheimer's Disease Neuroimaging, 2012; Dos Santos et al., 2011). Similarly, there are functional changes on semantic fMRI tasks manifest early stage AD (Saykin et al., 1999). The genetic factors that contribute to different modalities of language function have also been investigated in adolescents and young adults with and without language disorders. For example, genome-wide association studies (GWAS) have identified candidate genes related to reading and language in adolescents to young adults (Gialluisi et al., 2014; Luciano et al., 2013) and the morphology of Heschl’s gyrus in young adults (Cai et al., 2014). However, the genetic architecture underlying language performance in older adults with and at risk for AD has not been previously studied. To identify novel genetic variants specifically associated with language performance, we conducted a GWAS as well as a pathway-based analysis using the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort.
2. Material and methods
2.1 Alzheimer’s disease neuroimaging initiative (ADNI)
The ADNI was launched in 2003 to help researchers and clinicians develop new treatments for mild cognitive impairment (MCI) and early AD dementia, monitor their effectiveness, and decrease the time and cost of clinical trials. One of ADNI’s major goals is to test whether serial magnetic resonance imaging (MRI) (Grundman et al.), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and AD. This multi-year multi-site longitudinal study was started by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations as a $60 million, 5-year public-private partnership. It was then extended for an additional 7 years through two additional phases of funding. The ADNI participants consist of AD dementia, MCI, and healthy elderly individuals with and without significant memory concern (i.e., SMC and CN, respectively). They were aged 55–90 years and recruited from 59 sites across the U.S. and Canada including medical and academic institutions. Further information can be found at http://www.adni-info.org/ and see (Aisen et al., 2010). The current study involved 1,575 subjects with genotyping: 370 CN, 94 SMC, 283 early MCI (EMCI), 515 late MCI (LMCI), and 313 AD.
2.2 Phenotypes
A composite measure of language was generated as the target phenotype for this study using the following language measures from the ADNI neuropsychological test battery - the BNT, the Animal Fluency test, and the naming portion of AD Assessment Schedule-Cog (ADAS-COG). Given the invariable presence of episodic memory deficits in the cognitively impaired ADNI population, we sought to capture language performance independent of episodic memory performance (Supplementary Table 1). Therefore, each language test (BNT, Animal Fluency, and ADAS-COG naming) was individually adjusted with a linear regression model for concomitant episodic memory deficits using a composite score for episodic memory (Crane et al., 2012) in IBM SPSS (version 22, Armonk, NY). The residuals were then used to generate a composite score using principal component analysis taking the resulting first component, which accounted for 66% of the variance, with the following loadings: animal fluency = 0.62, BNT = 0.84, and ADAS-COG naming = 0.76 (Supplementary Fig. 1). For each diagnostic group, a positive linear relationship was observed between each language measure with respect to episodic memory (Supplementary Table 2). Thus, diagnostic status was not considered in the derivation of the language composite score. This summary measure of language performance was used as the primary phenotype for GWAS. A total of 1,560 subjects (365 CN, 94 SMC, 280 EMCI, 511 LMCI, 310 AD) had all three language test scores available for analyses. Age and sex were significantly associated (p<0.001; age, r = 0.15; sex, r = 0.11) with the generated language composite score pre-adjusted for episodic memory and were used as covariates in the following analyses.
2.3 Structural MRI scans
Baseline 1.5T (ADNI-1) and 3T (ADNI-GO/2) magnetization-prepared rapid gradient-echo (MPRAGE) images were downloaded from the ADNI LONI site for all participants (http://adni.loni.usc.edu/). Scan processing with voxel-based morphometry (VBM) in Statistical Parametric Mapping 8 (SPM8; Wellcome Trust Centre for Neuroimaging, http://www.fil.ion.ucl.ac.uk/spm/software/spm8/) and quality control were done as previously described (Risacher et al., 2013). Overall, 71 subjects (17 CN, 5 SMC, 12 EMCI, 21 LMCI, 16 AD) were excluded due to missing MRI scan data or failed processing, leaving 1,489 subjects available for analyses. Analyses were performed separately for each magnetic field strength.
2.4 Genotyping and imputation
The Illumina610-Quad BeadChip was used for genotyping all ADNI-1 participants and the Illumina HumanOmni Express BeadChip or the Illumina Omni 2.5M was used for participants enrolled in ADNI-GO or ADNI-2 (Saykin et al., 2015). Un-genotyped single nucleotide polymorphisms (SNPs) were imputed as previously described (Nho et al., 2015). Since population stratification is known to cause spurious association in disease studies, from the ADNI participants, we restricted our analyses to only non-Hispanic Caucasian subjects that clustered with CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) + TSI (Toscani in Italia) populations using HapMap 3 genotype data and the multidimensional scaling (MDS) analysis (www.hapmap.org). Standard sample and SNP quality control procedures were then implemented; SNPs were excluded if the genotyping call rate was < 95%, Hardy-Weinberg equilibrium test p < 1 × 10−6, sample call rate < 95%, or the minor allele frequency < 1%. In addition, sample sex marker mismatch check and DNA sample SNP fingerprint to microarray identity checks were also performed. MACH and 1000 Genomes Project data (build 37, hg19) as a reference panel were used for imputation. After quality control steps including filtering for MAF ≥ 5% on the imputed SNPs were applied, 1,575 of 1,716 ADNI participants and 5,574,300 SNPs remained for subsequent analyses.
2.5 Statistical analysis
A linear regression model in SPM8 using MRI scans was performed across voxels to evaluate the relationship of the language composite scores (unadjusted and pre-adjusted for episodic memory performance) with grey matter (GM) density. This enabled delineation of the regional associations between the language composite score and structural brain changes. Voxel-wise analysis was also used to determine the anatomical distribution of the association of the most significant SNP identified in the GWAS (see section below) with GM density. Voxel-wise analyses included age, sex, years of education, intracranial volume, and apolipoprotein E (APOE) ε4 status as covariates. Language abilities are typically lateralized to the left hemisphere, however some left handed individuals show right hemisphere or mixed dominance and thus handedness was also included as a covariate (Knecht et al., 2000). Significant results were displayed with a minimum cluster size (k;(Friston, Holmes, Worsley, Poline, & Frackowiak, 1995) of 100 contiguous voxels and a voxel-wise threshold of p < 0.001 (both uncorrected and corrected for multiple comparisons). Anatomical regions were defined using the x-y-z coordinates for the most significant voxel within each cluster. These coordinates were entered into the Talairach daemon (http://www.talairach.org/daemon.html) to receive the anatomical names for the GM regions closest to that coordinate (Lancaster et al., 2000).
A GWAS across the whole sample (n=1560) was performed using a linear regression under an additive genetic model in PLINK (Purcell et al., 2007) with the language composite pre-adjusted for episodic memory performance as the endophenotype to specifically target language-specific cognitive function in a population with known memory impairment (Rabin et al., 2009). Age, sex, handedness, years of education, APOE ε4 status, and genotyping platform were used as covariates. Bonferroni method was applied for correcting for multiple comparisons and SNPs with p < 5 × 10−8 were considered genome-wide significant (Pe'er, Yelensky, Altshuler, & Daly, 2008). Permutation testing (109 permutations) of the most significant SNP was implemented in PLINK to ensure that the language composite measure had did not deviate from normal distribution. Manhattan and Quantile-Quantile plots were generated in R using the ‘qqman’ package and regional association plots were generated with LocusZoom (Pruim et al., 2010). Linkage disequilibrium (LD) was estimated and visualized with D' using haploview (http://www.broad.mit.edu/mpg/haploview). The p-values obtained from the GWAS results and the GSA-SNP software (Nam, Kim, Kim, & Kim, 2010) were then utilized to identify pathways enriched with SNPs showing association to language performance (Ramanan, Kim, et al., 2012). We restricted downstream analysis to pathways containing 5–100 genes and used false discovery rate (FDR) to correct for pathway-level multiple comparisons. Pathway definitions were downloaded from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp; canonical pathways set, version 4.0).
3. Results
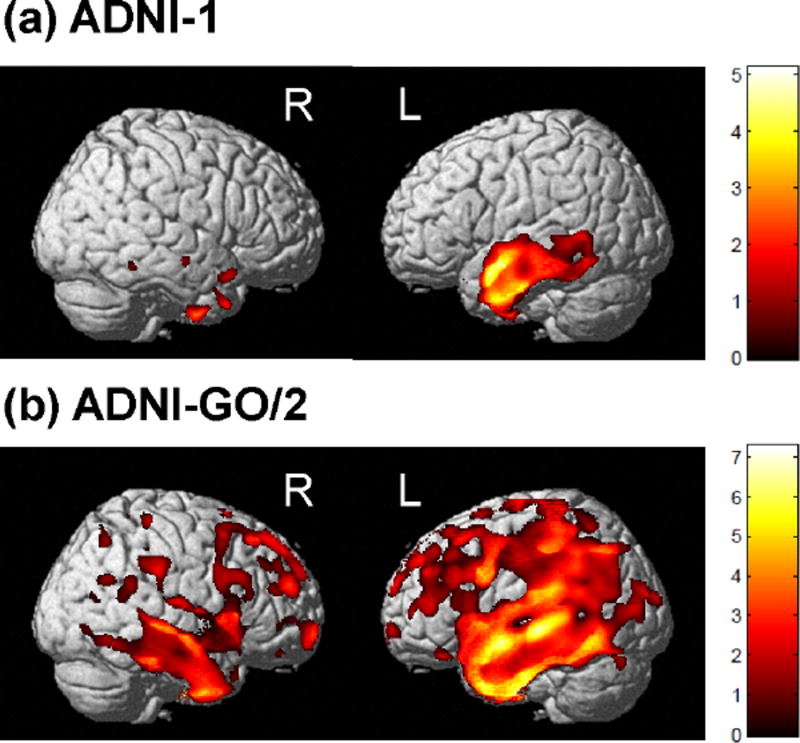
Sample characteristics of the participants used in this study are presented in Table 1. To assess brain regions in which grey matter (GM) density is associated with the generated language composite score pre-adjusted for episodic memory, we used VBM. A significant association between poorer language performance, both adjusted and unadjusted for episodic memory performance, and reduced GM density throughout the brain was observed (Fig. 1 and supplemental Fig. 2–3). Specifically, worse performance on the unadjusted score was associated with reduced GM density in nearly the entire cortex (Supplemental Fig. 2). However, performance on the language composite pre-adjusted for episodic memory performance showed a more localized pattern of association, with poorer performance associated with reduced GM density in the middle and superior temporal gyrus (Brodmann area (BA) 21 and BA22), predominantly lateralized to the left hemisphere (Fig. 1 and supplemental Fig. 3). We observed a larger region significantly associated with language performance in ADNI-GO/2, likely due to the increased sensitivity of the 3T scans to degeneration.
Table 1.
Baseline demographic characteristics.
| Baseline Diagnosis | CN | SMC | EMCI | LMCI | AD |
|---|---|---|---|---|---|
| No. of Subjects | 365 | 94 | 280 | 511 | 310 |
| Male/Female | 192/173 | 40/54 | 160/120 | 317/194 | 175/135 |
| Baseline Years of Age (SD) | 74.6 (5.6) | 71.8 (5.7) | 71.1 (7.4) | 73.5 (7.7) | 74.7 (7.8) |
| Baseline Years of Education (SD) | 16.3 (2.6) | 16.8 (2.6) | 16.1 (2.7) | 16 (2.9) | 15.2 (3) |
| Right/Left Handed | 338/27 | 82/12 | 251/29 | 457/54 | 286/24 |
| APOE (ε4−/ε4+) | 266/99 | 62/32 | 160/120 | 231/280 | 105/205 |
| Lang. Comp. Mean (SD)* | 0.6 (0.6) | 0.56 (0.5) | 0.32 (0.7) | −0.14 (0.9) | −0.94 (1.2) |
CN = cognitively normal; SMC = significant memory concern; EMCI = early mild cognitive impairment; LMCI = late mild cognitive impairment; AD = Alzheimer’s disease; SD = standard deviation
Not adjusted for memory composite score.
Figure 1.
Global regions associated with the language composite score pre-adjusted for episodic memory performance in ADNI participants. Grey matter density was positively correlated with the language composite score in the temporal lobe and language areas for both ADNI-1 (a) and ADNI-GO/2 (b). Covariates included age, sex, intracranial volume, APOE ε4 status, and handedness. Results are displayed at p<0.001 (uncorrected) and at a threshold (k) of 100 voxels.
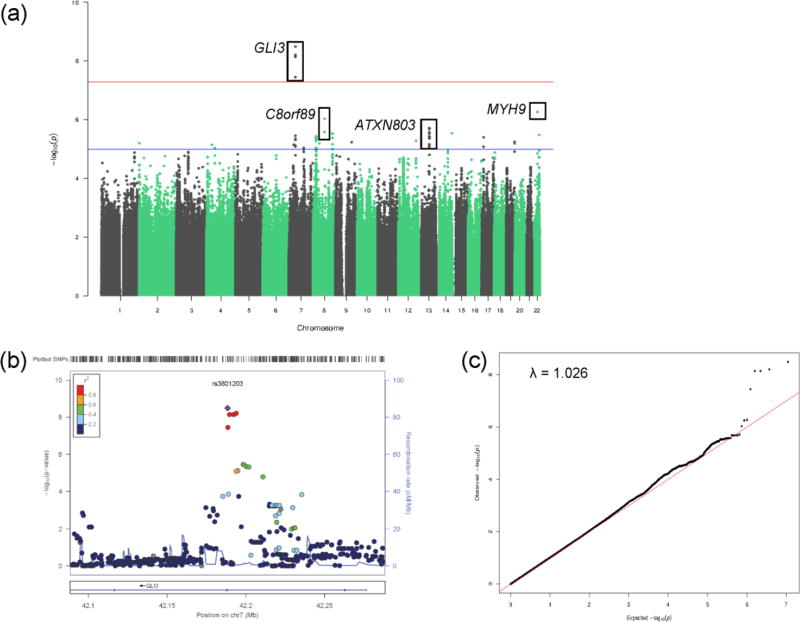
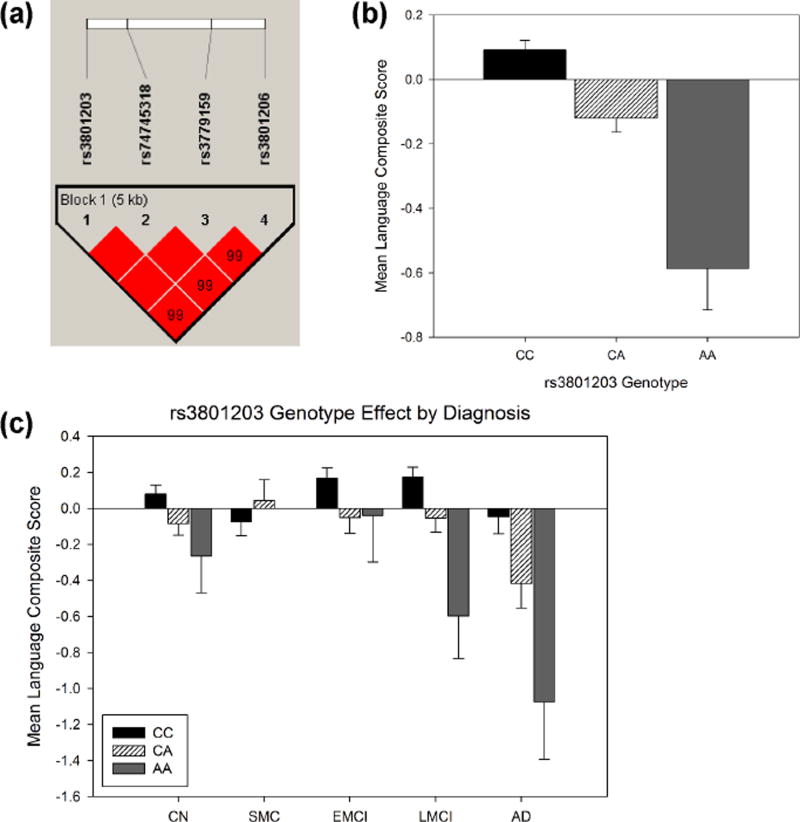
A GWAS was performed to identify genetic variants that might influence language performance using the language composite score pre-adjusted for episodic memory as the phenotype. GWAS identified four SNPs (rs3801203, p = 3.21 × 10−9; rs3779159, p = 7.12 × 10−9; rs3801206, p = 6.13 × 10−9; and rs74745318, p = 7.12 × 10−9; supplemental Table 1) in the region of GLI3 (GLI family zinc finger 3) that displayed genome-wide significant association with language function (Fig. 2a–b). The genomic inflation factor (λ) was 1.026 indicating no evidence for bias or inflation of our test statistics due to population stratification (Fig. 2c). These SNPs are intronic and demonstrated strong LD (Fig. 3a). To ensure our composite score had a normal distribution, permutation testing of the most significant SNP, rs3801203, resulted in the same p-value of 3.21 × 10−9 suggesting our phenotype had a normal distribution. Each SNP was also tested with BNT, ADAS-COG naming, and animal fluency, individually. All four GLI3 SNPs were significantly associated with BNT and ADAS-COG naming (p< 1 × 10−5). The minor allele of each SNP (data not shown) including rs3801203, was associated with worse language performance across all subjects (Fig. 3b) and diagnostic groups with the exception of the smallest group, SMC (Fig. 3c). An additional 31 SNPs in five additional genes displayed suggestive association (p < 5 × 10− 6) for language performance (Supplemental Table 3).
Figure 2.
Manhattan, regional association, and quantile-quantile plots for GWAS of language performance. (a) Observed –log10 p-values (y-axis) are displayed for all tested SNPs on each chromosome (x-axis). A SNP was considered genome-wide significant if p < 5 × 10−8 (above red line). Suggestive SNP associations were identified as those reaching genome-wide significance of p < 1 × 10−5 (above blue line). (b) Regional association plot showing the region around the most significant SNP in the GWAS. The SNPs within 500 kb of rs3801203 are plotted as their GWAS −log10 P-values against their NCBI 37 genomic position. The blue line indicates recombination rates estimated from the 1000 Genomes Project reference data. The color scale of r2 values is used to label SNPs based on their correlation with rs3801203. (c) The genomic inflation factor (λ) was 1.026 suggesting no population stratification effect.
Figure 3.
GLI3 SNPs are associated with lower language performance. (a) Linkage disequilibrium map of the four GLI3 SNPs significantly associated with language performance. The minor allele of rs3801203 (b-c, p = 3.21 × 10−9) was associated with lower language composite score in older adults at risk for or with AD, with the exception of the SMC group. Means adjusted for age, sex, years of education, genotyping platform, handedness, and APOE ε4 status. Standard errors are displayed.
CN = cognitively normal; SMC = significant memory concern; EMCI = early mild cognitive impairment; LMCI = late mild cognitive impairment; AD = Alzheimer’s disease
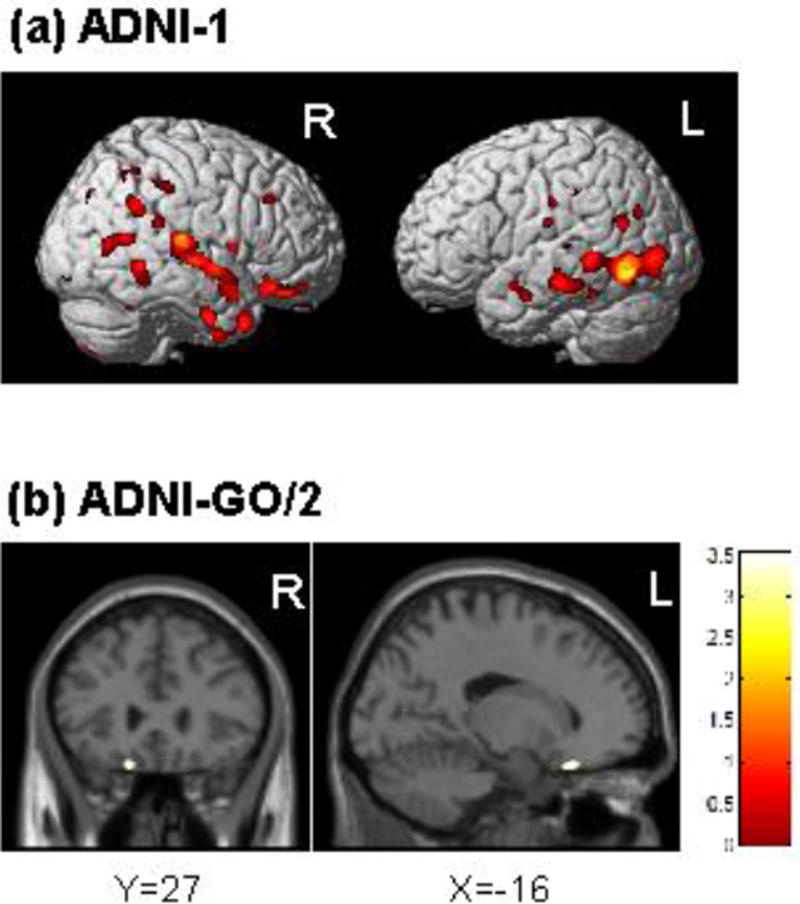
The four SNPs identified in the GWAS are in high LD thus we chose the most significant SNP, rs3801203, to determine if the genetic variants associated with language performance were also associated with GM density. GLI3 SNP rs3801203 was associated with lower GM density in both the ADNI-1 and ADNI-GO/2 cohorts (Fig. 4). Specifically, in the ADNI-1 cohort, the minor allele of rs3801203 was associated with atrophy in the middle and superior temporal gyrus (BA 21 and BA22, respectively; Fig. 4a), amongst other regions such as the fusiform, cingulate gyrus, and precuneus (Supplemental Table 4). However, in the ADNI-GO/2 cohort, the minor allele of rs3801203 was only found to be associated with atrophy in the inferior frontal gyrus (orbitofrontal cortex; Fig. 4b).
Figure 4.
The minor allele of GLI3 SNP rs3801203 is associated with lower grey matter density in the temporal cortex of ADNI-1 participants (a) and the orbitofrontal cortex for ADNI-GO/2 participants (b). Covariates included age, sex, intracranial volume, APOE ε4 status, and handedness. Results are displayed at p<0.001 (uncorrected) and at a threshold (k) of 100 voxels.
Pathway- enrichment analysis identified 24 pathways associated (FDR p < 0.01) with language performance. The top 20 pathways are presented in Table 2 and include pathways related to nervous system development, such as neuronal migration, axon guidance, and cell differentiation. Additional pathways included glutamate receptor trafficking and function, immune activation, apoptosis, and others.
Table 2.
Top 20 pathways associated with language performance.
| Pathway description (Source database) | Sizea | Uncorrected pb |
|---|---|---|
| Netrin-1 signaling (Reactome) | 36 | 2.38E-07 |
| L1CAM interactions (Reactome) | 76 | 9.39E-07 |
| Arrythmogenic right ventricular cardiomyopathy (KEGG) | 71 | 9.78E-07 |
| Ion channel transport (Reactome) | 48 | 5.33E-06 |
| SHH pathway (Biocarta) | 15 | 6.81E-06 |
| Phosphatidylinositol signaling system (KEGG) | 75 | 9.84E-06 |
| Trafficking of AMPA receptors (Reactome) | 26 | 1.17E-05 |
| NRAGE signals death through JNK (Reactome) | 37 | 1.39E-05 |
| Adherens junction (KEGG) | 71 | 1.39E-05 |
| Synthesis of PIPs at the plasma membrane (Reactome) | 28 | 1.59E-05 |
| Integrin cell surface reactions (Reactome) | 74 | 1.72E-05 |
| FC gamma R-mediated phagocytosis (KEGG) | 90 | 4.91E-05 |
| ECM-receptor interaction (KEGG) | 81 | 5.64E-05 |
| Cell death signaling via NGRAGE, NRIF, and NADE (Reactome) | 51 | 7.69E-05 |
| Semaphorin interactions (Reactome) | 61 | 9.89E-05 |
| Hypertrophic cardiomyopathy-HCM (KEGG) | 79 | 1.15E-04 |
| Unblocking of NMDA receptor glutamate binding and activation (Reactome) | 14 | 1.53E-04 |
| Interaction between L1 and Ankyrins (Reactome) | 20 | 1.57E-04 |
| PI metabolism (Reactome) | 44 | 1.59E-04 |
| Glioma (KEGG) | 63 | 1.73E-04 |
Number of genes in the pathway.
All pathway displayed are significant at false discovery rate (FDR)-corrected p < 0.01.
4. Discussion
We generated a language composite score using measures from animal fluency, BNT, and ADAS-COG naming. In the ADNI sample, MRI analysis identified significant GM atrophy across the whole-brain in relation to the language composite score unadjusted for episodic memory. However, GM atrophy in the left temporal, parietal, and frontal lobes were significantly associated with language performance pre-adjusted for episodic memory. These brain regions have previously been implicated in language and semantic memory processes (Apostolova et al., 2008; Domoto-Reilly et al., 2012; Joubert et al., 2010; McDonald, Bean, & Saykin, 2011; Saykin et al., 1999). We then investigated the effect of genetic variations on this language composite score using GWAS and pathway-enrichment analysis techniques in individuals at risk for and with AD from the ADNI cohort. GWAS identified variants in GLI3 significantly associated with language impairment. Genetic variation in GLI3 was also associated with lower GM density predominantly in the temporal lobes. Finally, neuronal development, glutamate receptor trafficking, immune function, and apoptotic pathways showed enriched associated with language performance in this sample. To our knowledge, this study is the first to identify GLI3 variation as associated with language performance.
GLI3 encodes one of three GLI zinc finger transcription factors expressed early in development that is normally involved in patterning brain structures (Blaess, Corrales, & Joyner, 2006; Ruppert, Vogelstein, Arheden, & Kinzler, 1990). The GLI3 protein is an important downstream mediator of the Sonic Hedgehog pathway and can act as an activator or repressor in the presence or absence of Sonic Hedgehog, respectively (Wang, Fallon, & Beachy, 2000). Mutations to this pathway are causative for developmental disorders which affect the limbs, head, and face (Kalff-Suske et al., 1999; Kang, Graham, Olney, & Biesecker, 1997).
GLI3 may also have a more direct role in development of the corpus callosum (Amaniti et al., 2013; Magnani et al., 2014) and hippocampus (Hasenpusch-Theil et al., 2012; Palma & Ruiz i Altaba, 2004). One study showed GLI3 expression was downregulated in the presence of Presenilin 1 (PSEN1), a protein which forms part of the γ-secretase complex involved in amyloid-beta production, ultimately leading to decreased neuronal differentiation (Paganelli et al., 2001). Notably, mutations in the PSEN1 gene are causative for some forms of early-onset autosomal dominant AD, including mutations with an aphasic phenotype (Denvir et al., 2015). Another study showed GLI3 expression is capable of repressing Pitrm1 (pitrilysin metallopeptidase) using limb tissue from GLI3 mutant mice (Town et al., 2009). The Pitrm1 protein is a metalloendopeptidase which is able to degrade amyloid-beta when it accumulates in mitochondria (Falkevall et al., 2006). Further Pitrm1 has been shown to have decreased expression in the temporal lobe of AD subjects (Alikhani et al., 2011), as well as decreased antisense expression in AD subjects compared to controls (Sekar et al., 2015). In light of these studies, our observed findings may suggest that this system could be dysregulated in older adults at risk for AD. Future studies exploring the relationships between GLI3, PSEN1, and Pitrm1 within the brain and in the context of AD would help to elucidate any potential role of this system in AD etiology.
To date, the four GLI3 SNPs identified in this study have not been linked to any other biological pathway or pathology to our knowledge. We found that these genetic variants were independently associated with BNT and ADAS-COG naming, but not with animal fluency, suggesting the language composite score was more reflective of naming rather than fluency. Furthermore, the minor allele of rs3801203 in the ADNI-1 cohort was associated with atrophy in brain regions associated with auditory processing. However, this finding did not replicate in the ADNI-GO/2 cohort which could be due to differences in scanner strength. This could also be due to the sample characteristics of these two cohorts. The ADNI-1 cohort includes mainly subjects who are later in the disease course and ADNI-GO/2 includes predominantly subjects that are earlier in the disease course. This could account for the larger variation in atrophy in relation to the language composite score seen in ADNI-GO/2. Moreover, the effect of rs3801203 may occur later in the disease course which could account for why more atrophy is observed in the ADNI-1 cohort.
Pathway-based analysis has provided useful insights into the underlying biological processes of complex genetic diseases by integrating and assessing the contribution of multiple genes and proteins across an intricate network of pathways and subnetworks (Ramanan, Shen, Moore, & Saykin, 2012). This pathway analysis identified several associations between nervous system development pathways and language performance. Specifically, one domain identified in the pathway analysis was glutamate receptor function and trafficking pathways. Glutamate is the primary excitatory neurotransmitter in the brain and is implicated in AD pathophysiology and neuronal cell death. Glutamate receptors have been under investigation as a therapeutic target for AD and other neurological disorders (Caraci et al., 2011; Morin et al., 2013; Olivares et al., 2012; Rosini, Simoni, Minarini, & Melchiorre, 2014). Memantine, a weak N-Methyl-D-aspartate channel blocker, has shown efficacy in reducing clinical decline in moderate AD and is widely used clinically (Reisberg et al., 2003; Tariot et al., 2004). Our results suggest that alterations in glutamate neurotransmission or receptor trafficking may play a critical role in language performance and functional semantic memory in AD.
One notable limitation of this study is the lack of a similar cohort to replicate our findings. Future studies investigating GLI3 in independent cohorts with similar neuroimaging and cognitive assessments should be done to confirm our results. In addition, further investigations about the functional impact of the observed significant GLI3 SNPs on a cellular or molecular level are warranted. Causal directions are yet to be determined but future studies may be able to capture the complicated relationship between the genetic, neuroimaging, and language domains using advanced statistical modeling such as mediation analysis or structural equation modeling. In view of the previously identified association of GLI3 with an amyloid-related protein (e.g.,PSEN1) as described above, future studies should also focus on evaluating the impact of GLI3 variation on amyloid phenotypes in older adults at risk for AD.
To our knowledge, this is the first reported GWAS of language performance in older adults at risk for or with AD. Our results identified novel associations of GLI3, a developmental transcription factor involved in patterning brain structures, with language dysfunction. Further, pathway analysis identified neuronal development and glutamate receptor pathways as enriched. Future studies will help to fully elucidate the underlying biology and importance of GLI3 and identified pathways in AD etiology and/or their potential as therapeutic targets.
Supplementary Material
Supplemental Figure 1: Distribution of language composite scores pre-adjusted for episodic memory in all subjects.
Supplemental Figure 2: Global regions associated with the language composite score unadjusted for episodic memory performance in ADNI participants with k = 100 voxels and p < 0.001 (uncorrected). Grey matter density was positively correlated with the unadjusted language composite score throughout the brain for both ADNI-1 (a) and ADNI-GO/2 (b).
Supplemental Figure 3: Global regions associated with the language composite score preadjusted for episodic memory performance in ADNI participants with k = 100 voxels and p < 0.05 (FWE corrected). Grey matter density was positively correlated with the pre-adjusted language composite score in temporal lobe and regions associated with language for both ADNI-1 (a) and ADNI-GO/2 (b).
Highlights.
We identified novel genetic variants associated with language performance.
Minor allele variants in GLI3 are associated with worse language performance.
Anatomical changes in language regions associated with language composite score.
Developmental and glutamate pathways were related to lower language performance.
Acknowledgments
Data collection and sharing was funded by the ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol- Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
The specific analyses reported here were supported in part by grants from the NIH including P30 AG10133, R01 AG19771, R01 LM011360, R01 AG040770, R01 AG042437, and R00 LM011384, K02 AG048240, as well the Alzheimer’s Association, Indiana CTSI (NIH grants U54 RR025761, RR027710-01, and RR020128), NIA grants R36 AG053445 and KO1 AG049050, and NSF grant IIS-1117335.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors declare no conflict of interest.
References
- Ahmed S, Haigh AM, de Jager CA, Garrard P. Connected speech as a marker of disease progression in autopsy-proven Alzheimer's disease. Brain. 2013;136(Pt 12):3727–3737. doi: 10.1093/brain/awt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn HJ, Seo SW, Chin J, Suh MK, Lee BH, Kim ST, Na DL. The cortical neuroanatomy of neuropsychological deficits in mild cognitive impairment and Alzheimer's disease: a surface-based morphometric analysis. Neuropsychologia. 2011;49(14):3931–3945. doi: 10.1016/j.neuropsychologia.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG Alzheimer's Disease Neuroimaging, I. Clinical Core of the Alzheimer's Disease Neuroimaging Initiative: progress and plans. Alzheimers Dement. 2010;6(3):239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhani N, Guo L, Yan S, Du H, Pinho CM, Chen JX, Yan SS. Decreased proteolytic activity of the mitochondrial amyloid-beta degrading enzyme, PreP peptidasome, in Alzheimer's disease brain mitochondria. J Alzheimers Dis. 2011;27(1):75–87. doi: 10.3233/JAD-2011-101716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaniti EM, Hasenpusch-Theil K, Li Z, Magnani D, Kessaris N, Mason JO, Theil T. Gli3 is required in Emx1+ progenitors for the development of the corpus callosum. Dev Biol. 2013;376(2):113–124. doi: 10.1016/j.ydbio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Lu P, Rogers S, Dutton RA, Hayashi KM, Toga AW, Thompson PM. 3D mapping of language networks in clinical and pre-clinical Alzheimer's disease. Brain Lang. 2008;104(1):33–41. doi: 10.1016/j.bandl.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles KA, Tomoeda CK, Trosset MW. Naming and categorical knowledge in Alzheimer's disease: the process of semantic memory deterioration. Brain Lang. 1990;39(4):498–510. doi: 10.1016/0093-934x(90)90158-d. [DOI] [PubMed] [Google Scholar]
- Bertola L, Mota NB, Copelli M, Rivero T, Diniz BS, Romano-Silva MA, Malloy-Diniz LF. Graph analysis of verbal fluency test discriminate between patients with Alzheimer's disease, mild cognitive impairment and normal elderly controls. Front Aging Neurosci. 2014;6:185. doi: 10.3389/fnagi.2014.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133(9):1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Cai DC, Fonteijn H, Guadalupe T, Zwiers M, Wittfeld K, Teumer A, Hagoort P. A genome-wide search for quantitative trait loci affecting the cortical surface area and thickness of Heschl's gyrus. Genes Brain Behav. 2014;13(7):675–685. doi: 10.1111/gbb.12157. [DOI] [PubMed] [Google Scholar]
- Caraci F, Molinaro G, Battaglia G, Giuffrida ML, Riozzi B, Traficante A, Nicoletti F. Targeting group II metabotropic glutamate (mGlu) receptors for the treatment of psychosis associated with Alzheimer's disease: selective activation of mGlu2 receptors amplifies beta-amyloid toxicity in cultured neurons, whereas dual activation of mGlu2 and mGlu3 receptors is neuroprotective. Mol Pharmacol. 2011;79(3):618–626. doi: 10.1124/mol.110.067488. [DOI] [PubMed] [Google Scholar]
- Chertkow H, Bub D. Semantic memory loss in dementia of Alzheimer's type. What do various measures measure? Brain. 1990;113(Pt 2):397–417. doi: 10.1093/brain/113.2.397. [DOI] [PubMed] [Google Scholar]
- Clark DG, Kapur P, Geldmacher DS, Brockington JC, Harrell L, DeRamus TP, Marson DC. Latent information in fluency lists predicts functional decline in persons at risk for Alzheimer disease. Cortex. 2014;55:202–218. doi: 10.1016/j.cortex.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A Alzheimer's Disease Neuroimaging, I. Development and assessment of a composite score for memory in the Alzheimer's Disease Neuroimaging Initiative (ADNI) Brain Imaging Behav. 2012;6(4):502–516. doi: 10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denvir J, Neitch S, Fan J, Niles RM, Boskovic G, Schreurs BG, Alkon DL. Identification of the PS1 Thr147Ile Variant in a Family with Very Early Onset Dementia and Expressive Aphasia. J Alzheimers Dis. 2015 doi: 10.3233/JAD-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domoto-Reilly K, Sapolsky D, Brickhouse M, Dickerson BC Alzheimer's Disease Neuroimaging, I. Naming impairment in Alzheimer's disease is associated with left anterior temporal lobe atrophy. Neuroimage. 2012;63(1):348–355. doi: 10.1016/j.neuroimage.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos V, Thomann PA, Wustenberg T, Seidl U, Essig M, Schroder J. Morphological cerebral correlates of CERAD test performance in mild cognitive impairment and Alzheimer's disease. J Alzheimers Dis. 2011;23(3):411–420. doi: 10.3233/JAD-2010-100156. [DOI] [PubMed] [Google Scholar]
- Falkevall A, Alikhani N, Bhushan S, Pavlov PF, Busch K, Johnson KA, Glaser E. Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J Biol Chem. 2006;281(39):29096–29104. doi: 10.1074/jbc.M602532200. [DOI] [PubMed] [Google Scholar]
- Friston K, Holmes A, Worsley K, Poline J-B, Frackowiak R. Statistical parametric maps in functional imaging: a general linear approach. Hum Brain Mapp. 1995;2:189–210. [Google Scholar]
- Gialluisi A, Newbury DF, Wilcutt EG, Olson RK, DeFries JC, Brandler WM, Fisher SE. Genome-wide screening for DNA variants associated with reading and language traits. Genes Brain Behav. 2014;13(7):686–701. doi: 10.1111/gbb.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez RG, White DA. Using verbal fluency to detect very mild dementia of the Alzheimer type. Arch Clin Neuropsychol. 2006;21(8):771–775. doi: 10.1016/j.acn.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Grundman M, Petersen RC, Ferris SH, Thomas RG, Aisen PS, Bennett DA Alzheimer's Disease Cooperative, S. Mild cognitive impairment can be distinguished from Alzheimer disease and normal aging for clinical trials. Arch Neurol. 2004;61(1):59–66. doi: 10.1001/archneur.61.1.59. [DOI] [PubMed] [Google Scholar]
- Hasenpusch-Theil K, Magnani D, Amaniti EM, Han L, Armstrong D, Theil T. Transcriptional analysis of Gli3 mutants identifies Wnt target genes in the developing hippocampus. Cereb Cortex. 2012;22(12):2878–2893. doi: 10.1093/cercor/bhr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JD, Crawford JR, Phillips LH. Verbal fluency performance in dementia of the Alzheimer's type: a meta-analysis. Neuropsychologia. 2004;42(9):1212–1222. doi: 10.1016/j.neuropsychologia.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Hodges JR, Graham KS. Episodic memory: insights from semantic dementia. Philos Trans R Soc Lond B Biol Sci. 2001;356(1413):1423–1434. doi: 10.1098/rstb.2001.0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges JR, Salmon DP, Butters N. The nature of the naming deficit in Alzheimer's and Huntington's disease. Brain. 1991;114(Pt 4):1547–1558. doi: 10.1093/brain/114.4.1547. [DOI] [PubMed] [Google Scholar]
- Joubert S, Brambati SM, Ansado J, Barbeau EJ, Felician O, Didic M, Kergoat MJ. The cognitive and neural expression of semantic memory impairment in mild cognitive impairment and early Alzheimer's disease. Neuropsychologia. 2010;48(4):978–988. doi: 10.1016/j.neuropsychologia.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Kalff-Suske M, Wild A, Topp J, Wessling M, Jacobsen EM, Bornholdt D, Grzeschik KH. Point mutations throughout the GLI3 gene cause Greig cephalopolysyndactyly syndrome. Hum Mol Genet. 1999;8(9):1769–1777. doi: 10.1093/hmg/8.9.1769. [DOI] [PubMed] [Google Scholar]
- Kang S, Graham JM, Jr, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15(3):266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- Knecht S, Drager B, Deppe M, Bobe L, Lohmann H, Floel A, Henningsen H. Handedness and hemispheric language dominance in healthy humans. Brain. 2000;123(Pt 12):2512–2518. doi: 10.1093/brain/123.12.2512. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Fox PT. Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp. 2000;10(3):120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Evans DM, Hansell NK, Medland SE, Montgomery GW, Martin NG, Bates TC. A genome-wide association study for reading and language abilities in two population cohorts. Genes Brain Behav. 2013;12(6):645–652. doi: 10.1111/gbb.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani D, Hasenpusch-Theil K, Benadiba C, Yu T, Basson MA, Price DJ, Theil T. Gli3 controls corpus callosum formation by positioning midline guideposts during telencephalic patterning. Cereb Cortex. 2014;24(1):186–198. doi: 10.1093/cercor/bhs303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald BC, Bean J, Saykin AJ. fMRI Wada test: Prospects for presurgical mapping of language and memory. In: Faro SH, Mohamed FB, editors. Functional Neuroradiology: Principles and Clinical Applications. Springer; 2011. [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, Phelps CH. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsch AU, Bondi MW, Butters N, Salmon DP, Katzman R, Thal LJ. Comparisons of verbal fluency tasks in the detection of dementia of the Alzheimer type. Arch Neurol. 1992;49(12):1253–1258. doi: 10.1001/archneur.1992.00530360051017. [DOI] [PubMed] [Google Scholar]
- Morin N, Gregoire L, Morissette M, Desrayaud S, Gomez-Mancilla B, Gasparini F, Di Paolo T. MPEP, an mGlu5 receptor antagonist, reduces the development of L-DOPA-induced motor complications in de novo parkinsonian monkeys: biochemical correlates. Neuropharmacology. 2013;66:355–364. doi: 10.1016/j.neuropharm.2012.07.036. [DOI] [PubMed] [Google Scholar]
- Nam D, Kim J, Kim SY, Kim S. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic Acids Res. 2010;38(Web Server issue):W749–754. doi: 10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho K, Ramanan VK, Horgusluoglu E, Kim S, Inlow MH, Risacher SL, Saykin AJ. Comprehensive Gene- and Pathway-Based Analysis of Depressive Symptoms in Older Adults. J Alzheimers Dis. 2015 doi: 10.3233/JAD-148009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutter-Upham KE, Saykin AJ, Rabin LA, Roth RM, Wishart HA, Pare N, Flashman LA. Verbal fluency performance in amnestic MCI and older adults with cognitive complaints. Arch Clin Neuropsychol. 2008;23(3):229–241. doi: 10.1016/j.acn.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares D, Deshpande VK, Shi Y, Lahiri DK, Greig NH, Rogers JT, Huang X. N-methyl D-aspartate (NMDA) receptor antagonists and memantine treatment for Alzheimer's disease, vascular dementia and Parkinson's disease. Curr Alzheimer Res. 2012;9(6):746–758. doi: 10.2174/156720512801322564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganelli AR, Ocana OH, Prat MI, Franco PG, Lopez SL, Morelli L, Carrasco AE. The Alzheimer-related gene presenilin-1 facilitates sonic hedgehog expression in Xenopus primary neurogenesis. Mech Dev. 2001;107(1-2):119–131. doi: 10.1016/s0925-4773(01)00458-0. [DOI] [PubMed] [Google Scholar]
- Palma V, Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131(2):337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin LA, Saykin AJ, West JD, Borgos MJ, Wishart HA, Nutter-Upham KE, Santulli RB. Judgment in Older Adults with Normal Cognition, Cognitive Complaints, MCI, and Mild AD: Relation to Regional Frontal Gray Matter. Brain Imaging Behav. 2009;3(2):212–219. doi: 10.1007/s11682-009-9063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK, Kim S, Holohan K, Shen L, Nho K, Risacher SL Alzheimer's Disease Neuroimaging, I. Genome-wide pathway analysis of memory impairment in the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain Imaging Behav. 2012;6(4):634–648. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012;28(7):323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisberg B, Doody R, Stoffler A, Schmitt F, Ferris S, Mobius HJ, Memantine Study G. Memantine in moderate-to-severe Alzheimer's disease. N Engl J Med. 2003;348(14):1333–1341. doi: 10.1056/NEJMoa013128. [DOI] [PubMed] [Google Scholar]
- Risacher SL, Kim S, Shen L, Nho K, Foroud T, Green RC Alzheimer's Disease Neuroimaging Initiative, d. The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI) Front Aging Neurosci. 2013;5:11. doi: 10.3389/fnagi.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosini M, Simoni E, Minarini A, Melchiorre C. Multi-target design strategies in the context of Alzheimer's disease: acetylcholinesterase inhibition and NMDA receptor antagonism as the driving forces. Neurochem Res. 2014;39(10):1914–1923. doi: 10.1007/s11064-014-1250-1. [DOI] [PubMed] [Google Scholar]
- Ruppert JM, Vogelstein B, Arheden K, Kinzler KW. GLI3 encodes a 190-kilodalton protein with multiple regions of GLI similarity. Mol Cell Biol. 1990;10(10):5408–5415. doi: 10.1128/mcb.10.10.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Flashman LA, Frutiger SA, Johnson SC, Mamourian AC, Moritz CH, Weaver JB. Neuroanatomic substrates of semantic memory impairment in Alzheimer's disease: patterns of functional MRI activation. J Int Neuropsychol Soc. 1999;5(5):377–392. doi: 10.1017/s135561779955501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL Alzheimer's Disease Neuroimaging, I. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 2015;11(7):792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar S, McDonald J, Cuyugan L, Aldrich J, Kurdoglu A, Adkins J, Liang WS. Alzheimer's disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging. 2015;36(2):583–591. doi: 10.1016/j.neurobiolaging.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariot PN, Farlow MR, Grossberg GT, Graham SM, McDonald S, Gergel I, Memantine Study G. Memantine treatment in patients with moderate to severe Alzheimer disease already receiving donepezil: a randomized controlled trial. JAMA. 2004;291(3):317–324. doi: 10.1001/jama.291.3.317. [DOI] [PubMed] [Google Scholar]
- Town L, McGlinn E, Fiorenza S, Metzis V, Butterfield NC, Richman JM, Wicking C. The metalloendopeptidase gene Pitrm1 is regulated by hedgehog signaling in the developing mouse limb and is expressed in muscle progenitors. Dev Dyn. 2009;238(12):3175–3184. doi: 10.1002/dvdy.22126. [DOI] [PubMed] [Google Scholar]
- Verma M, Howard RJ. Semantic memory and language dysfunction in early Alzheimer's disease: a review. Int J Geriatr Psychiatry. 2012;27(12):1209–1217. doi: 10.1002/gps.3766. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100(4):423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Distribution of language composite scores pre-adjusted for episodic memory in all subjects.
Supplemental Figure 2: Global regions associated with the language composite score unadjusted for episodic memory performance in ADNI participants with k = 100 voxels and p < 0.001 (uncorrected). Grey matter density was positively correlated with the unadjusted language composite score throughout the brain for both ADNI-1 (a) and ADNI-GO/2 (b).
Supplemental Figure 3: Global regions associated with the language composite score preadjusted for episodic memory performance in ADNI participants with k = 100 voxels and p < 0.05 (FWE corrected). Grey matter density was positively correlated with the pre-adjusted language composite score in temporal lobe and regions associated with language for both ADNI-1 (a) and ADNI-GO/2 (b).