Abstract
Objectives
The practice of evidence-based medicine involves integrating the latest best available evidence into patient care decisions. Yet, critical barriers exist for clinicians’ retrieval of evidence that is relevant for a particular patient from primary sources such as randomized controlled trials and meta-analyses. To help address those barriers, we investigated machine learning algorithms that find clinical studies with high clinical impact from PubMed®.
Methods
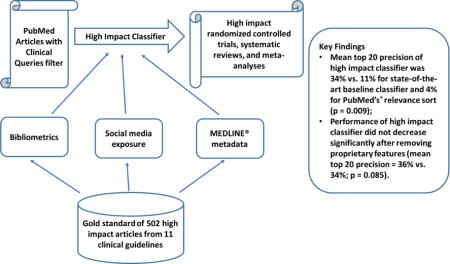
Our machine learning algorithms use a variety of features including bibliometric features (e.g., citation count), social media attention, journal impact factors, and citation metadata. The algorithms were developed and evaluated with a gold standard composed of 502 high impact clinical studies that are referenced in 11 clinical evidence-based guidelines on the treatment of various diseases. We tested the following hypotheses: 1) our high impact classifier outperforms a state-of-the-art classifier based on citation metadata and citation terms, and PubMed’s® relevance sort algorithm; and 2) the performance of our high impact classifier does not decrease significantly after removing proprietary features such as citation count.
Results
The mean top 20 precision of our high impact classifier was 34% versus 11% for the state-of-the-art classifier and 4% for PubMed’s® relevance sort (p = 0.009); and 2) the performance of our high impact classifier did not decrease significantly after removing proprietary features (mean top 20 precision = 34% vs. 36%; p = 0.085).
Conclusion
The high impact classifier, using features such as bibliometrics, social media attention and MEDLINE® metadata, outperformed previous approaches and is a promising alternative to identifying high impact studies for clinical decision support.
Graphical abstract
1. Introduction
On average, clinicians raise more than one clinical question for every two patients seen, and the majority of these questions are left unanswered [1,2]. Unmet information needs can lead to suboptimal patient care decisions and lower patient care quality [3]. “Lack of time” and “doubt that a useful answer exists” are two major reasons that prevent clinicians from pursuing clinical questions at the point of care [2]. Yet, online knowledge resources, such as primary literature resources (e.g., PubMed®) and evidence summaries (e.g. clinical guidelines, UpToDate®), can provide answers to most clinical questions [4].
Evidence based medicine (EBM) practice advocates clinicians to integrate individual clinical expertise and the best available evidence, ideally from methodologically sound randomized controlled trials (RCTs), systematic reviews (SRs), and meta-analyses (MAs) [5,6]. In the past twenty years, the publication of RCTs, SRs, and MAs has experienced steady growth [6]. Despite recommendations for clinicians to integrate high quality evidence in patient care decisions, the use of primary literature resources in patient care is still low [7]. Challenges include: 1) only a very small fraction of the studies indexed in PubMed® warrant changes in clinical practice - the findings of most studies are false due to weaknesses such as small sample size, small effect size, biases, unstandardized study design, flaws in statistical analysis, and conflicts of interest [8]; and 2) finding and interpreting high quality studies that have an impact on the care of a specific patient is very time-consuming and unfeasible in busy care settings [9].
To promote clinical use of PubMed®, several promising approaches have been investigated to retrieve high quality (i.e., scientifically rigorous) studies, mainly using MeSH terms/keywords or bibliometric information [10–15]. However, previous approaches focused primarily on retrieving studies with scientifically sound methodology. In the present study, we investigate approaches to retrieve articles that have a high clinical impact, and are likely to influence clinicians’ patient care decisions. Our method is built over the following previous approaches: 1) the Clinical Query filters [10]; 2) citation count [13]; and 3) the high quality study classifier by Kilicoglu et al. [11]. We combined the approaches above and explored several novel features as surrogates for an article’s clinical impact. We hypothesize that 1) our high impact classifier outperforms Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s® relevance sort in terms of top 20 precision; and 2) the performance of our high impact classifier does not decrease significantly after removing proprietary features.
2. Background
Multiple research efforts have investigated algorithms to retrieve scientifically rigorous clinical studies. Overall, they can be divided into three categories: search filters, citation-based algorithms, and supervised machine learning algorithms.
2.1 Search filters
Clinical Queries, a PubMed® built-in feature, have been designed to help clinicians find citations of scientifically sound clinical studies [10,12]. Clinical Query filters are Boolean-based search strategies that include MeSH terms and keywords that are often found in the abstracts of scientifically sound clinical studies. The approach has been developed and validated through a rigorous systematic process [10,12]. Filters for five topics have been developed (i.e., therapy, diagnosis, etiology, prognosis, clinical prediction guides), with the option to maximize precision or recall. Since Clinical Queries are openly available through PubMed®, they are often used as a baseline for evaluating the efficacy of novel approaches aimed at retrieving high quality clinical studies from PubMed®.
2.2 Citation-based algorithms
Citation-based algorithms, such as the approach proposed by Bernstam et al. [13], are based on approaches that are widely used on the Web, such as citation count and PageRank [16]. Both citation count and PageRank are based on linkage analysis between the nodes (i.e., citations/websites) of a graph. The difference is that citation count considers only one layer of linkage (i.e., only the documents that directly cite the document of interest), whereas PageRank looks at multiple layers (i.e., all documents that recursively cite the document of interest). Using the Society of Surgical Oncology’s Annotated Bibliography (SSOAB) as the gold standard, Bernstam et al. found that both citation count and PageRank outperformed MeSH and keyword-based algorithms, such as Clinical Queries and machine learning classifiers (top precision = 6% versus 0.85%) [13]. Limitations of citation-based algorithms include 1) not considering the scientific quality of a study; 2) citation count relies on proprietary data; and 3) citation count is time-dependent and does not work for very recent studies.
2.3 Supervised machine learning
Examples of the supervised machine learning approach include studies conducted by Aphinyanaphongs et al. and Kilicoglu et al. [11,14,15]. Aphinyanaphongs et al. found that a polynomial support vector machine (Poly SVM) classifier outperformed Clinical Queries’ specificity filter for retrieving internal medicine articles included in the American College of Physicians Journal Club (ACPJ) on treatment (recall = 0.80 versus 0.40) and etiology (recall = 0.76 versus 0.28) tasks [14]. The features included MeSH terms, publication type, and words in the title and abstract. Next, they proposed that each PubMed® article retrieval system should be built upon a particular task and an appropriate gold standard for the task [15]. They compared the performance of different machine learning methods using three gold standards (i.e., the SSOAB for surgical oncology, ACPJ-treatment for internal medicine treatment, and ACPJ-etiology for internal medicine etiology) [15]. The study found that gold-standard-specific machine learning classifiers (e.g., applying the model built on ACPJ-treatment to find internal medicine treatment articles) outperformed non-specific machine learning (e.g., applying the model built on SSOAB to find internal medicine treatment articles) in terms of the area under the curve (0.966 versus 0.770). The main limitation of this study is the generalizability of the classifier (i.e., only explored the internal medicine domain). We may need to develop new classifiers for every different domain.
More recently, Kilicoglu et al. employed a stacking ensemble method that combined the features used in Aphinyanaphongs et al. with Unified Medical Language System (UMLS) concepts, UMLS semantic relations, and semantic predications [11]. Classifiers were built and evaluated with a large gold standard developed by McMaster University and consisting of 49,028 high quality clinical studies selected through a rigorous manual process from 161 clinical journals [11,17]. The stacking classifier had 73.7% precision and 61.5% recall for scientifically rigorous studies, and 82.5% precision and 84.3% recall for treatment/prevention studies. The main strength of this study is good generalizability as it covered multiple clinical domains. The main limitation of this study is still focusing on scientifically rigorous studies, but not high clinical impact ones.
3. Methods
Our overall method is based on machine learning algorithms with a variety of features, including bibliometrics, MEDLINE® metadata, and social media exposure. The method was developed according to the following steps (Figure 1): 1) development of a gold standard of high impact articles cited in 11 clinical guidelines; 2) retrieval of candidate PubMed® citations covering the main topic of each guideline using a search strategy based on PubMed’s® Clinical Queries filter [17]; 3) preparation of bibliometrics, MEDLINE® metadata, and social media exposure features; 4) ranking of features; 5) training and optimization of classifiers to identify high impact clinical studies; and 6) testing of a set of hypotheses regarding the performance of the classifiers.
Figure 1.
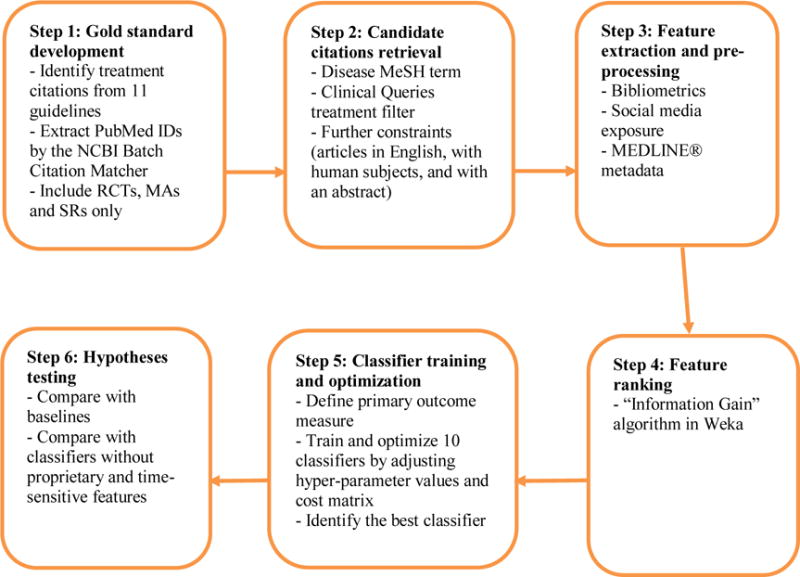
Method Steps
3.1 Gold standard development
We used studies cited in clinical practice guidelines as a surrogate for high impact studies in a clinical topic. Clinical guidelines contain evidence-based recommendations on the diagnosis and treatment of specific conditions. Through rigorous systematic review development methodology, domain experts identify all studies relevant to the topic of the clinical guideline, screen out studies that do not meet minimum quality criteria (e.g., randomized controlled trials), and derive guideline recommendations from the included studies [18,19]. We focused on treatment citations since most clinical questions are related to the treatment of patient conditions [2]. In our study, we 1) manually extracted those citations (i.e., RCTs, MAs and SRs) from each guideline (Table 1); and 2) automatically mapped each extracted citation to PubMed® IDs using the NCBI Batch Citation Matcher tool [18]. We manually mapped citations that could not be automatically mapped.
Table 1.
Clinical guidelines used in the gold standard and number of citations per guideline.
| Disease Topic | Guideline Title | Medical Society | Number of Citations |
|---|---|---|---|
| Rheumatoid Arthritis (RA) | 2012 update of the 2008 American College of Rheumatology recommendations for the use of disease-modifying antirheumatic drugs and biologic agents in the treatment of rheumatoid arthritis. [20] | ACR | 66 |
| Asthma in Children and Adults | VA/DoD Clinical Practice Guideline For Management of Asthma in Children and Adults. [21] | VA/DoD | 31 |
| Major Depressive Disorder (MDD) | VA/DoD Clinical Practice Guideline For Management of Major Depressive Disorder (MDD). [22] | VA/DoD | 65 |
| Outpatient Chronic Obstructive Pulmonary Disease(COPD) 2007 | VA/DoD Clinical Practice Guideline For Management of Outpatient COPD. [23] | VA/DoD | 95 |
| Outpatient Chronic Obstructive Pulmonary Disease(COPD) 2014 | VA/DoD Clinical Practice Guideline For the Management of Chronic Obstructive Pulmonary Disease. [24] | VA/DoD | 58 |
| Extracranial Carotid and Vertebral Artery Disease | 2011ASA/ACCF/AHA/AANN/AANS/ACR/ASNR/CNS/SAIP/SCAI/SIR/SNIS/SVM/SVS Guideline on the Management of Patients With Extracranial Carotid and Vertebral Artery Disease. [25] | ACC | 22 |
| Stable Ischemic Heart Disease | 2012ACCF/AHA/ACP/AATS/PCNA/SCAI/STS Guideline for the Diagnosis and Management of Patients With Stable Ischemic Heart Disease. [26] | ACC | 66 |
| ST- Elevation Myocardial Infarction | 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. [27] | ACC | 69 |
| Heart Failure | 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. [28] | ACC | 88 |
| Valvular Heart Disease | 2014 AHA/ACC Guideline for the Management of Patients With Valvular Heart Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. [29] | ACC | 32 |
| Atrial Fibrillation (AFib) | 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. [30] | ACC | 56 |
ACC: the American College of Cardiology;
ACR: the American College of Rheumatology;
VA/DoD: the US Veterans Administration/Department of Defense Clinical Practice Guidelines;
To find these 11 guidelines, the overall approach was to search for recent guidelines on the treatment of a range of common complex chronic conditions. We also included guidelines based on the following criteria: 1) articles cited in the guideline must have been selected through a systematic search, screening, and appraisal process; 2) eligible guidelines must have provided explicit treatment recommendations, along with citations to the original studies that supported each recommendation. For guidelines with multiple versions, we selected the latest version available at the time of our search.
3.2 Candidate citations retrieval
Candidate citations were retrieved using a search strategy specifically designed for RCTs, MAs and SRs (Box 1). The strategy included three components. First, a suitable disease MeSH term was manually selected based on the main condition covered in each guideline. Second, filters were applied to retrieve high quality treatment studies. RCTs were retrieved by using the Clinical Queries narrow therapy filter [17], which is designed to retrieve high quality therapy studies and is optimized for precision. MAs and SRs were retrieved with a combination of PubMed’s® systematic review filter and studies whose titles contained “systematic review” or “meta-analysis” or were published in the Cochrane Database of Systematic Reviews. Third, a date range constraint was applied matching the same time period of the systematic search conducted to support the development of the corresponding guideline. Further constraints included articles written in English, studies with human subjects, and articles with an abstract [31].
Box 1: Search strategy pattern for retrieving candidate PubMed® citations.
“Disease”[MeSH Terms] AND (Therapy/Narrow[filter] OR (“therapy” [Subheading] AND systematic[sb] AND (“systematic review”[ti] OR “meta-analysis” [ti] OR “Cochrane Database Syst Rev”[journal])))AND (Guideline Coverage Start Date[PDAT] : Guideline Coverage End Date[PDAT])AND “humans”[MeSH Terms] AND “english”[language] AND hasabstract[text]
3.3 Feature extraction and pre-processing
Features of retrieved citations were extracted through a set of automated scripts, and stored in a relational database. The features are as follows:
3.3.1 Journal Impact Factor (JIF)
JIF measures how often articles published in a particular journal are cited by other articles. Specifically, JIF is calculated by dividing the number of citations to publications in the journal of interest in the previous two years by the number of original research articles and reviews published in that journal in the previous two years [32,33]. JIF eliminates the bias of higher citation counts from large journals, frequently published journals and old journals. We used JIF as a surrogate for the reputation of a journal and consequently for the impact of articles published in the journal. We obtained the JIFs from the Journal Citation Reports® (JCR®), published by Thomson Reuters [34].
3.3.2 Registration in ClinicalTrials.gov
This feature indicates whether the study is registered in the ClinicalTrials.gov registry. National regulations and most reputable journals require registration of clinical trials in national registries such as ClinicalTrials.gov before the trial is initiated. Our assumption is that registration in ClinicalTrials.gov is a predictor of the study quality and impact. This feature is determined by the presence of a ClinicalTrial.gov ID in the citations’ PubMed® metadata.
3.3.3 Publication in PubMed Central®
This feature indicates whether the article is available in the PubMed Central® database. All studies funded by the US National Institutes of Health (NIH) are published in PubMed Central® and available open access. Since these studies are not funded by commercial entities, they tend to be more balanced and potentially have a stronger clinical impact [35–37]. This feature is determined by the presence of a PubMed Central® ID in the PubMed® metadata.
3.3.4 Article Age
This feature represents the number of months since the article was published. More recent articles may have a stronger clinical impact. Article age was determined based on the number of months elapsed between the date the citation was added to PubMed® (the Entrez Date in the PubMed® metadata) and the month when the Article Age feature was processed (i.e., August 2016).
3.3.5 Study Sample Size
This feature represents the number of participants in the study according to the study record in ClinicalTrials.gov. A large sample size might be a predictor of high impact studies [8].
3.3.6 Comparative Study
This feature indicates whether the study compared two or more treatment alternatives as opposed to a treatment versus placebo. Comparative studies generally provide more useful information to support clinical decisions than intervention versus placebo trials [38]. This feature was extracted from the publication type field in the PubMed® metadata.
3.3.7 Study Quality
This feature represents the probability that a given citation is a high quality article according to the classifier developed by Kilicoglu et al. [11]. The probability score for each retrieved citation was generated using a model based on a Naïve Bayes classifier with two types of features (i.e., MeSH indexing terms and publication type). The rationale behind this classifier is similar to the rationale of PubMed’s® Clinical Query filters, i.e. that attributes of strong study designs are indexed as MeSH terms and publication type in the citation metadata. Examples include MeSH terms such as “random allocation” and “clinical trials” and publication types such as “randomized controlled trials”. Other publication types may serve as negative predictors, such as “case-control study” or “case report”.
3.3.8 Number of comments on PubMed®
This feature indicates the number of editorial comments on a given citation. Articles that receive editorial comments might be more important. The number of editorial comments was extracted from the CommentsCorrectionsList field in the PubMed® metadata.
3.3.9 Citation Count
This feature indicates how many times an article has been cited according to the Scopus system. As a rough adjustment for the time elapsed since the publication date, we also calculated the rate of citations per month. We obtained the citation counts in August 2016 using a Web service API provided by Scopus [39].
3.3.10 Altmetric® score
Altmetric® tracks the online exposure of scientific work based on social media (e.g., Facebook, Twitter), traditional media (e.g., New York Times) and online reference managers (e.g., Mendeley). A different weight is assigned to each specific source. The score is calculated based on both the quantity and quality of posts [40,41]. We also calculated a monthly-adjusted score. We obtained Altmetric® scores in August 2016 using an Altmetric® API that is freely available for research purposes [42].
3.3.11 High Impact Journal
This feature indicates whether the study was published in a journal included in a list of high impact clinical journals. The list was compiled by combining the MEDLINE® Abridged Index Medicus (AIM or “Core Clinical") journals [43] and the McMaster Plus (Premium LiteratUre Service) journals [44]. The quality and relevance of these journals are rigorously and periodically evaluated by a group of experts [45–47].
3.4 Feature ranking
To evaluate the contribution of each individual feature, we employed the Information Gain evaluator in the Weka data mining package [48]. This evaluator is one of the best feature ranking methods according to Hall and Holmes’s benchmarking study [49]. We selected citations from an average-size guideline (heart failure dataset) among the 11 guidelines for feature ranking.
3.5 Classification Method
To identify an optimal classifier, we chose the heart failure dataset as the training dataset and the major depressive disorder dataset as the validation dataset based on our primary outcome (top 20 precision). We chose these two datasets because their sizes are closest to the average size of all datasets, their positive sample rates are close to the average positive sample rate across datasets, and they are focused on different medical domains.
We evaluated 12 classification algorithms with their parameter settings (Table 2). Since our dataset is very unbalanced (3.2% positive vs. 96.8% negative cases), we also employed cost-sensitive data mining with meta cost algorithm where all mentioned classifiers were trained based on different costs for false positive and false negative errors determined by various cost matrices [50]. Our experimental setting is aligned with similar studies on performance comparison among classifiers [51,52].
Table 2.
Classification algorithms and their parameter settings.
| Algorithm | Parameter Setting |
|---|---|
| K-Nearest Neighbors | number of neighbors and instance weighting methods |
| Naïve Bayes | kernel density estimator |
| Bayes Net | search algorithm and estimator algorithm |
| Naïve Bayes Multinomial | default parameter setting in Weka |
| Logistic | kernel type and the corresponding parameters of each kernel type |
| Multilayer Perceptron | number of hidden layers, number of nodes in each layer, learning rate, and momentum |
| Simple Logistic | default parameter setting in Weka |
| Stochastic Gradient Descent | learning rate, lambda and loss function |
| Decision Table | attribute search method |
| J48 | minimum number of instances per leaf, reduced error pruning and confidence threshold for pruning |
| Random Forest | number of trees, maximum depth of the trees, and number of attributes |
| Support Vector Machine | kernel type and the corresponding parameters of each kernel type |
We selected the best classifier based on our primary outcome (top 20 precision). If the performance of two or more classifiers was similar, we selected the one that is easiest to implement and interpret. After finalizing the optimal parameter setting for the best classifier, we applied it to the remaining nine disease datasets for hypothesis testing.
3.6 Hypotheses testing
Hypothesis 1. The high impact classifier outperforms Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s® relevance sort in terms of top 20 precision. For the Kilicoglu baseline, we ranked the citations according to the probability output of the Naïve Bayes classifier. PubMed’s® relevance sort is a relevance-based ranking algorithm. The ranking is determined by the frequency and the location of search terms in the retrieved citation, and the age of the retrieved citation [53].
Hypothesis 2. The performance of the high impact classifier does not decrease significantly after removing citation count and social media exposure features. This experiment assessed the contribution of the Scopus citation count and the Altmetric® score. These two features are less desirable since both are based on proprietary information, and there is a time lag between the time of publication and the presence of the first citation to an article for the Scopus citation count.
3.6.1 Study outcomes
By default, the PubMed® search results page displays 20 items per page. Since busy clinicians are less likely to look past the top 20 citations [54], we adopted top 20 precision a priori as the primary outcome for all the hypotheses. We also measured top 20 mean average precision, and top 20 mean reciprocal rank [55]. The main difference between top 20 precision and top 20 mean average precision is that top 20 precision only measures the percentage of true positive citations among the first 20 retrieved citations, whereas top 20 mean average precision measures the average ranking position of the true positive citations; the closer the true positive citations to the top of the ranking, the better the retrieval system is. Top 20 mean average precision is computed as follows by: 1) for each true positive citation in the top 20 retrieved citations, divide its position among true positive citations by its position in the overall retrieved results; 2) calculate the average of the values obtained in the previous step. Since our overall dataset is unbalanced, we also measured precision, recall, and F-measure of positive samples in all experiments.
3.6.2 Statistical analysis
The goal of the statistical analyses included three aspects: 1) in Hypothesis 1, determining if our classifier was superior to Kilicoglu et al.’s high quality Naïve Bayes classifier; 2) in Hypothesis 1, determining if our classifier was superior to PubMed’s® relevance sort classifier; 3) in Hypothesis 2, determining if our classifier was equivalent to the classifier without the citation count and social media exposure features. Since these were separate study questions, rather than the more general question of whether our classifier is better than other classifiers without being specific, the statistical issue of multiple comparisons did not arise in our study [56,57]. Besides multiple classifiers (multiple groups), another way that multiplicity, or the multiple comparison problem can arise is from having multiple outcome measures. To address that, we selected top 20 precision a priori as our primary outcome measure. The other five measures were secondary measures. The hypothesis test, then, for answering the research question was limited to the single primary measure. The secondary measures are simply exploratory, or descriptive, and have been included as others in the field may be interested in seeing them. This approach to multiplicity is called the primary-secondary approach to multiplicity [58], which is the most commonly used approach in randomized controlled trials reported in The New England Journal of Medicine [59]. To compare our classifier with any of the other three classifiers, we used a paired sample Wilcoxon signed rank test. We employed the Wilcoxon test in place of a paired sample t-test so that no data value could overly influence the result in an outlier fashion. We performed all the statistical analyses using Stata IC 14.
4. Results
A total of 15,845 citations were retrieved with the PubMed® search strategy for the diseases represented in the 11 guidelines. Among these citations, 502 (recall of 77.5% for the total 648 guideline citations (Table 1)) were high impact clinical studies. Feature ranking results are shown in Table 3. We found that Scopus citation count and journal impact factor were the top two features followed by number of comments on PubMed®, high impact journal, Altmetric® score and other PubMed® metadata.
Table 3.
Feature ranking results
| Rank | Feature | Information Gain |
|---|---|---|
| 1 | Citation count | 0.05154 |
| 2 | Citation count (monthly) | 0.04851 |
| 3 | Journal impact factor | 0.03784 |
| 4 | Number of comments on PubMed® | 0.03563 |
| 5 | High impact journal | 0.01887 |
| 6 | Altmetric® score | 0.01771 |
| 7 | Altmetric® score (monthly) | 0.01275 |
| 8 | Study sample size | 0.01242 |
| 9 | Registration in ClinicalTrials.gov | 0.00763 |
| 10 | Article age | 0.00584 |
| 11 | Comparative study | 0 |
| 12 | Study quality | 0 |
| 13 | Publication in PubMed Central® | 0 |
We found that hyper-parameter optimization with cost matrix improved the performance of some but not all of the classifiers (see online supplement Table s1 for details). The performance of the Naïve Bayes classifier with default parameter settings was similar to the performance of several other classifiers (e.g., Bayesian network, Naïve Bayes Multinomial). As the Naïve Bayes classifier is easiest to implement and understand, we chose it as the final classifier for hypotheses testing.
Experiment #1: The high impact classifier outperforms Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s® relevance sort. Figure 2 summarizes the results. The high impact classifier with all features performed significantly better than Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s® relevance sort in terms of top 20 precision (mean = 34% vs. 11% and 4% respectively; both p = 0.009). Similar results were found for the secondary outcomes top 20 mean average precision (mean = 23% vs. 6% and 1% respectively; both p = 0.008), top 20 mean reciprocal rank (mean = 0.78 vs. 0.30 and 0.05 respectively; p = 0.012 and p = 0.007), precision (mean = 33% vs. 5% and 4% respectively; both p = 0.008) and F-measure (mean = 21% vs. 9% and 8% respectively; p = 0.015 and p = 0.008). The high impact classifier performed significantly worse than Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s® relevance sort in terms of recall (mean = 23% vs. 55% and 65% respectively; p = 0.009 and p = 0.008) (see online supplement Table s2 for details).
Experiment #2: The performance of the high impact classifier does not decrease significantly after removing Scopus citation count and social media exposure features. Figure 3 summarizes the results. The high impact classifier with all features had an equivalent performance to the classifier without Scopus citation count and social media exposure in terms of top 20 precision (mean = 34% vs. 36%; p = 0.085). Similar results were found for the secondary measures top 20 mean average precision (mean = 23% vs. 24%; p = 0.441), top 20 mean reciprocal rank (mean = 0.78 vs. 0.60; p = 0.187), precision (mean = 33% vs. 29%; p = 0.406), and recall (mean = 23% vs. 20%; p = 0.094). In terms of F-measure, the high impact classifier performed better than the classifier without Scopus citation count and social media exposure (mean = 21% vs. 18%; p = 0.044) (see online supplement Table s3 for details).
Figure 2.
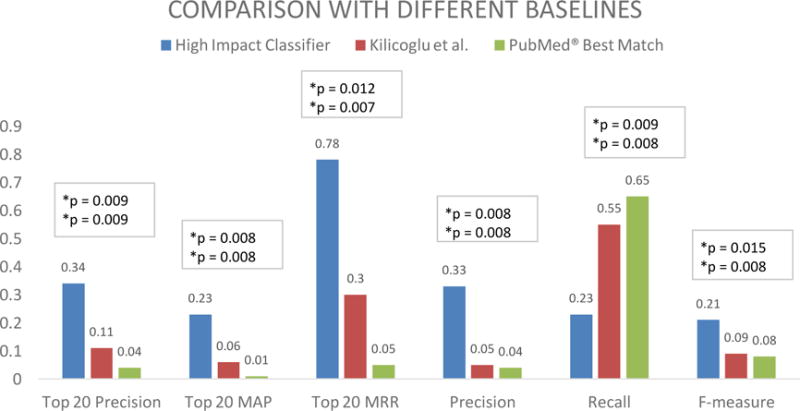
Average top 20 precision, top 20 mean average precision (MAP), top 20 mean reciprocal rank (MRR), precision, recall and F-measure, of the high impact classifier, Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s® relevance sort (Experiment #1).
Figure 3.
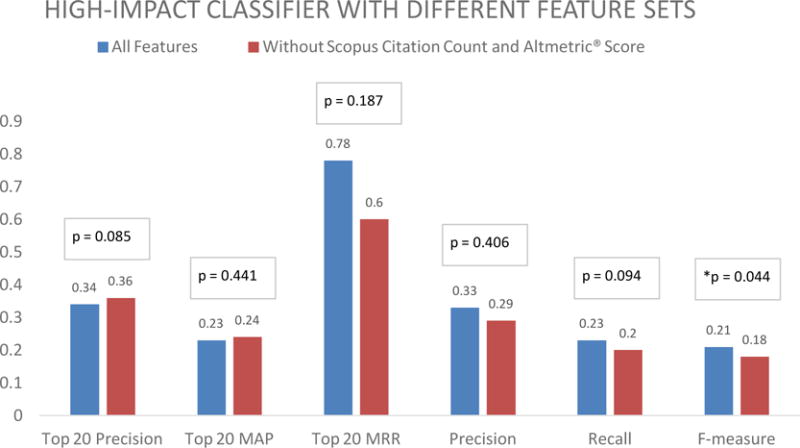
Average top 20 precision, top 20 mean average precision (MAP), top 20 mean reciprocal rank (MRR), precision, recall and F-measure of the high impact classifier, and the all feature without Scopus citation count and Altmetric® score classifier (Experiment #2).
5. Discussion
In this study, we investigated machine learning methods to automatically identify high impact clinical studies in PubMed® for supporting clinical decision making. Our approach builds over previous research that relied on MeSH metadata, abstract terms, and bibliometrics to retrieve scientifically sound studies from PubMed®[10–15]. Our approach is innovative because we combined features and classifiers used in previous studies with new features, such as social media attention. Our high impact classifier outperformed Kilicoglu et al.’s high study quality Naïve Bayes classifier and PubMed’s® relevance sort ranking algorithm. In addition, the level of performance does not change significantly after removing time-sensitive and proprietary features (i.e., citation count and social media exposure features). Our method can be used to support patient care by helping time-constrained clinicians meet their information needs with the latest available evidence. For example, the algorithm could be integrated with existing tools, such as an additional filter within PubMed®, or integrated with new clinical decision support tools, such as the Clinical Knowledge Summary [59]. The method can also be used to support physicians in training, who can incorporate evidence searching in their learning routine.
Strengths of our study include: 1) generalizability to multiple domains, since the 11 diseases included in our study cover multiple medical domains such as autoimmune (e.g., rheumatoid arthritis), cardiac (e.g., heart failure), and respiratory diseases (e.g., Asthma); 2) retrieval of high impact clinical citations that influence clinicians’ patient care decisions; 3) less dependency on time-sensitive and proprietary data of our automatic classifier.
5.1 Experiment 1
Experiment 1 confirmed the hypothesis that the high impact classifier outperforms Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s® relevance sort ranking in terms of top 20 precision. The top 20 precision of our classifier was on average 34%, which means that roughly 6 to 7 out of the top 20 retrieved PubMed® articles are high impact articles. Compared with PubMed’s® relevance sort algorithm (roughly 1 out of 20 retrieved articles) and with a state-of-the-art algorithm (roughly 2 out of 20 articles), our classifier provides a significantly higher chance for clinicians to find high impact articles among the top ranked ones. For information retrieval systems, it is very imperative to retrieve the best results in the first page. According to a classic study, more than 75% of users using a general search engine do not view the results beyond the first 20 hits [60]. This issue is even more pronounced in busy clinical settings [61]. On average, clinicians are not willing to spend more than 2 minutes seeking information during patient care [62], and more than 91% of clinicians do not view PubMed® citations beyond the first 20 [54]. Therefore, to effectively support clinical decision making, it is critical to retrieve the best results on the first page.
5.2 Experiment 2
Experiment 2 confirmed the hypothesis that the performance of the high impact classifier does not decrease significantly after removing citation count and social media exposure features. Citation count has been a well-established surrogate for measuring the quality of PubMed® articles [63,64], and it was the strongest feature in our study according to feature selection, confirming the finding of Bernstam et al. [13]. Journal Impact Factor (JIF) is another proprietary metric, based on citation counts of articles published within that journal. Although JIF changes each year, we kept it in Experiment 2 because, unlike citation count, JIF 1) is easier to obtain than article-level citation counts, 2) changes at a slower rate, and 3) does not have a time lag. Altmetric® score is a non-traditional surrogate for article quality and impact. No strong correlation was found between citation count and Altmetric® score, suggesting that the two features are complementary [65,66]. Although both citation count and Altmetric® score are strong predictors for article quality, their utility is compromised by their time-sensitive and proprietary nature. In our study, it is possible that other features served as surrogates for citation count and social media attention and, when combined, compensated for the absence of these features. For example, journal impact factor is calculated based on the number of citations to each of the articles published in a journal. Thus, journal impact factor may serve as a proxy for an article’s citation count. Therefore, our finding that other features combined compensate for the absence of citation count and social media attention is important for the feasibility of integrating our high impact classifier into a production system.
5.3 Limitations
This first limitation of our approach is that we only employed one guideline (heart failure) dataset for feature ranking and selecting an optimal classifier, which could potentially bring some bias into this study. We have 11 guideline datasets, for the purpose of boosting statistical power, we employed maximum number of datasets (i.e., 9) for the statistical analyses regarding the performance of our classifiers. In the future, we will include more guideline datasets so that number of the guidelines used for feature selection and optimal classifier identification and number of the guidelines used for statistical analyses could be well balanced.
The second limitation of our approach is that it does not account for concept drift [67,68] and several features used in our high impact classifier change their values over time, which are likely to affect the performance of a classifier in a production system. Ideally, we should have extracted data for time-sensitive features reflecting the values of those features at the time when articles were searched by the guideline authors. However, historical data for the Scopus citation count and Altmetric® score are not available. In addition, our approach depends on citation metadata, such as MeSH terms and publication type, but those features are not available immediately after a citation becomes available in PubMed®. The time-to-indexing of an article in PubMed® varies from less than a month to eight months, depending on multiple factors such as journal impact factor, focus area, and discipline [69]. This poses a challenge upon using our classifier for very recent articles, which may be quite desirable for clinicians who are experts in a domain and are mostly interested in keeping up with very recent evidence. In future studies, we plan to investigate approaches to overcome this limitation, such as relying on off-the-shelf auto-indexing tools (e.g., MTI indexer [70]) and leveraging other citation metadata (e.g., journal impact factor, author and affiliation, and references) that are available at the first time the article appears in PubMed®.
6. Conclusion
This study shows that a high impact Naïve Bayes classifier, using features such as bibliometrics, social media attention and MEDLINE® metadata, is a promising approach to identifying high impact studies for clinical decision support. Our current classifier is optimal for classifying PubMed® articles that have been published after a certain period of time, roughly 6 to 9 months. Further research is warranted to investigate time-sensitive approaches that address concept drift.
Supplementary Material
Highlights.
High impact clinical studies provide evidence influencing clinicians’ patient care.
An automated approach is proposed to classify high impact studies from PubMed®.
Our approach identified 6–7 high impact studies out of top 20 articles from PubMed®.
Our approach outperformed a state-of-the-art classifier and PubMed’s® relevance sort.
The high impact classifier performed similarly without proprietary features.
Acknowledgments
This project was supported by National Library of Medicine grants: 1R01LM011416-01, R00LM011389 and T15LM007124. We would like to thank the Richard A. Fay and Carol M. Fay Endowed Graduate Fellowship for funding support. We also would like to thank Dr. Duy Bui for extracting PMIDs for the guideline citations used as a gold standard, and Dr. Liqin Wang for extracting Journal Citation Reports® (JCR®) data from Thomson Reuters.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None declared.
References
- 1.Covell DG, Uman GC, Manning PR. Information needs in office practice: are they being met? Ann Intern Med. 1985;103:596–9. doi: 10.7326/0003-4819-103-4-596. http://www.ncbi.nlm.nih.gov/pubmed/4037559 (accessed July 18, 2012) [DOI] [PubMed] [Google Scholar]
- 2.Del Fiol G, Workman TE, Gorman PN. Clinical questions raised by clinicians at the point of care: a systematic review. JAMA Intern Med. 2014;174:710–8. doi: 10.1001/jamainternmed.2014.368. [DOI] [PubMed] [Google Scholar]
- 3.Leape LL, Bates DW, Cullen DJ, Cooper J, Demonaco HJ, Gallivan T, Hallisey R, Ives J, Laird N, Laffel G. Systems analysis of adverse drug events. ADE Prevention Study Group. JAMA. 1995;274:35–43. http://www.ncbi.nlm.nih.gov/pubmed/7791256 (accessed May 13, 2015) [PubMed] [Google Scholar]
- 4.Maviglia SM, Yoon CS, Bates DW, Kuperman G. KnowledgeLink: impact of context-sensitive information retrieval on clinicians’ information needs. J Am Med Inform Assoc. 2006;13:67–73. doi: 10.1197/jamia.M1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sackett DL, Rosenberg WMC, Gray JAM, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn’t. BMJ. 1996;312:71–72. doi: 10.1136/bmj.312.7023.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung BT, Long NP, Hung LP, Luan NT, Anh NH, Nghi TD, Van Hieu M, Trang NTH, Rafidinarivo HF, Anh NK, Hawkes D, Huy NT, Hirayama K. Research Trends in Evidence-Based Medicine: A Joinpoint Regression Analysis of More than 50 Years of Publication Data. PLoS One. 2015;10:e0121054. doi: 10.1371/journal.pone.0121054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoogendam A, Stalenhoef AFH, de V Robbé PF, Overbeke AJPM. Answers to questions posed during daily patient care are more likely to be answered by UpToDate than PubMed. J Med Internet Res. 2008;10:e29. doi: 10.2196/jmir.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ioannidis JPA. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook DA, Sorensen KJ, Hersh W, Berger RA, Wilkinson JM. Features of effective medical knowledge resources to support point of care learning: a focus group study. PLoS One. 2013;8:e80318. doi: 10.1371/journal.pone.0080318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haynes RB, Wilczynski N, McKibbon KA, Walker CJ, Sinclair JC. Developing optimal search strategies for detecting clinically sound studies in MEDLINE. J Am Med Inform Assoc. 1994;1:447–58. doi: 10.1136/jamia.1994.95153434. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=116228&tool=pmcentrez&rendertype=abstract (accessed November 28, 2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilicoglu H, Demner-Fushman D, Rindflesch TC, Wilczynski NL, Haynes RB. Towards automatic recognition of scientifically rigorous clinical research evidence. J Am Med Inform Assoc. 2009;16:25–31. doi: 10.1197/jamia.M2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilczynski NL, McKibbon KA, Walter SD, Garg AX, Haynes RB. MEDLINE clinical queries are robust when searching in recent publishing years. J Am Med Informatics Assoc. 2013;20:363–368. doi: 10.1136/amiajnl-2012-001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstam EV, Herskovic JR, Aphinyanaphongs Y, Aliferis CF, Sriram MG, Hersh WR. Using citation data to improve retrieval from MEDLINE. J Am Med Inform Assoc. 2006;13:96–105. doi: 10.1197/jamia.M1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, Hardin D, Aliferis CF. Text categorization models for high-quality article retrieval in internal medicine. J Am Med Inform Assoc. 2005;12:207–16. doi: 10.1197/jamia.M1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aphinyanaphongs Y, Statnikov A, Aliferis CF. A comparison of citation metrics to machine learning filters for the identification of high quality MEDLINE documents. J Am Med Inform Assoc. 2006;13:446–55. doi: 10.1197/jamia.M2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brin S, Page L, Brin S, Page L. The anatomy of a large-scale hypertextual Web search engine, Comput. Networks ISDN Syst. 1998;30:107–117. doi: 10.1016/S0169-7552(98)00110-X. [DOI] [Google Scholar]
- 17.Wilczynski NL, Morgan D, Haynes RB. An overview of the design and methods for retrieving high-quality studies for clinical care. BMC Med Inform Decis Mak. 2005;5:20. doi: 10.1186/1472-6947-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moher D, Liberati A, Tetzlaff J, Altman DG, Altman D. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.JPT Higgins S. Cochrane handbook for systematic reviews of interventions. Wiley-Blackwell; 2008. (Sally E. Green, Cochrane Collaboration., Wiley InterScience (Online service) https://books.google.com/books?hl=en&lr=&id=NKMg9sMM6GUC&oi=fnd&pg=PT13&dq=systematic+review+methodology+cochrane&ots=LIRAR-HGC6&sig=qAntasDRxidNqJG_KFz4wOHD3HU#v=onepage&q=systematic_review_methodology_cochrane&f=false (accessed July 20, 2017) [Google Scholar]
- 20.Singh JA, Furst DE, Bharat A, Curtis JR, Kavanaugh AF, Kremer JM, Moreland LW, O’Dell J, Winthrop KL, Beukelman T, Bridges SL, Chatham WW, Paulus HE, Suarez-Almazor M, Bombardier C, Dougados M, Khanna D, King CM, Leong AL, Matteson EL, Schousboe JT, Moynihan E, Kolba KS, Jain A, Volkmann ER, Agrawal H, Bae S, Mudano AS, Patkar NM, Saag KG. 2012 update of the 2008 American College of Rheumatology recommendations for the use of disease-modifying antirheumatic drugs and biologic agents in the treatment of rheumatoid arthritis. Arthritis Care Res (Hoboken) 2012;64:625–39. doi: 10.1002/acr.21641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curran D, Blain A, Orio K, Daughaday C, Bray D, Potter L, Khachikian D, Colluraburke C, Quinn J, Littner M, Dodson D, Radebaugh N, Sees G, Davenport T, Robson K, Hsu D, Roach C, Lee R, Schriner W, Liss J, Tucker M, Manning J, Wojtczak H, Morris M, Jacknewitz-Woolard J, Markusfeld J, Cassidy C, Degenhardt E, Klar A, Susskind O, Lux L, Viswanathan M, Lohr K, Fishman R, Marko J. VA/DoD Clinical Practice Guideline For Management of Asthma in Children and Adults. 2009:127. [Google Scholar]
- 22.Cassidy C, Carchedi L, Craig T, Engel C, Gerrity M, Knittel D, Labbate L, Lowry P, McQuaid J, Manos G, Muller A, McLaughlin E, Oslin D, Ramos M, Ray J, KTrice S, Semla T, Wilson R, Williams J, Zeiss A, Susskind O, Coats V, Erinoff E, Schoelles K, Snyder D, D’Erasmo M, Fishman R, Marko J. VA/DoD Clinical Practice Guideline For Management of Major Depressive Disorder (MDD) 2009:203. [Google Scholar]
- 23.Almenoff P, Cote C, Doman S, Habib M, Doherty D, Khachikian D, Littner M, Mahutte K, McGlashan P, Sethi S, Sharafkhaneh A, Carnahan D, Kallish M, Kang C, Klar A, Mitchell J, Musket M, Stephens M, Susskind O, Coats V, Erinoff E, Snyder D, Turkelson C, D’Erasmo M, Fishman R, Marko J. VA/DoD Clinical Practice Guideline For Management of Outpatient COPD. 2007:138. [Google Scholar]
- 24.Render M, Sherner J, Rice K, Anekwe T, Sharafkhaneh A, Feinberg J, Ellis J, Sall J, Fritz A, Sapp MJ, Hintz C, Selvester RM, Khachikian D, Shaw S, Staropoli C, Tennant M, Wallinger J, Rodgers ME, Degenhardt E, Sutton RM, Sall J, Goodman C, Murphy F, Beam E, Stettler N, Uhl S, Kleiner H, D’Anci K, Jones C, Akinyede O, Rishar R, Tsou A, Laws K, Ramanathan A. VA/DoD CLINICAL PRACTICE GUIDELINE FOR THE MANAGEMENT OF CHRONIC OBSTRUCTIVE PULMONARY DISEASE. 2014:94. [Google Scholar]
- 25.Brott TG, Halperin JL, Abbara S, Bacharach JM, Barr JD, Bush RL, Cates CU, Creager MA, Fowler SB, Friday G, Hertzberg VS, McIff EB, Moore WS, Panagos PD, Riles TS, Rosenwasser RH, Taylor AJ. 2011 ASA/ACCF/AHA/AANN/AANS/ACR/ASNR/CNS/SAIP/SCAI/SIR/SNIS/SVM/SVS Guideline on the Management of Patients With Extracranial Carotid and Vertebral Artery Disease. J Am Coll Cardiol. 2011;57:e16–e94. doi: 10.1016/j.jacc.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Fihn SD, Gardin JM, Abrams J, Berra K, Blankenship JC, Dallas AP, Douglas PS, Foody JM, Gerber TC, Hinderliter AL, King SB, Kligfield PD, Krumholz HM, Kwong RYK, Lim MJ, Linderbaum JA, Mack MJ, Munger MA, Prager RL, Sabik JF, Shaw LJ, Sikkema JD, Smith CR, Smith SC, Spertus JA, Williams SV. 2012 ACCF/AHA/ACP/AATS/PCNA/SCAI/STS Guideline for the Diagnosis and Management of Patients With Stable Ischemic Heart Disease: Executive Summary. J Am Coll Cardiol. 2012;60:2564–2603. doi: 10.1016/j.jacc.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 27.O’Gara PT, Kushner FG, Ascheim DD, Casey DE, Chung MK, de Lemos JA, Ettinger SM, Fang JC, Fesmire FM, Franklin BA, Granger CB, Krumholz HM, Linderbaum JA, Morrow DA, Newby LK, Ornato JP, Ou N, Radford MJ, Tamis-Holland JE, Tommaso CL, Tracy CM, Woo YJ, Zhao DX, Anderson JL, Jacobs AK, Halperin JL, Albert NM, Brindis RG, Creager MA, DeMets D, Guyton RA, Hochman JS, Kovacs RJ, Ohman EM, Stevenson WG, Yancy CW. 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;61:e78–140. doi: 10.1016/j.jacc.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Drazner MH, Fonarow GC, Geraci SA, Horwich T, Januzzi JL, Johnson MR, Kasper EK, Levy WC, Masoudi FA, McBride PE, McMurray JJV, Mitchell JE, Peterson PN, Riegel B, Sam F, Stevenson LW, Tang WHW, Tsai EJ, Wilkoff BL. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;62:e147–239. doi: 10.1016/j.jacc.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura RA, Otto CM, Bonow RO, Carabello BA, Erwin JP, Guyton RA, O’Gara PT, Ruiz CE, Skubas NJ, Sorajja P, Sundt TM, Thomas JD. 2014 AHA/ACC Guideline for the Management of Patients With Valvular Heart Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2014;63:e57–e185. doi: 10.1016/j.jacc.2014.02.536. [DOI] [PubMed] [Google Scholar]
- 30.January CT, Wann LS, Alpert JS, Calkins H, Cleveland JC, Cigarroa JE, Conti JB, Ellinor PT, Ezekowitz MD, Field ME, Murray KT, Sacco RL, Stevenson WG, Tchou PJ, Tracy CM, Yancy CW. 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. J Am Coll Cardiol. 2014;64:e1–e76. doi: 10.1016/j.jacc.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 31.Demner-Fushman D, Lin J. Answering Clinical Questions with Knowledge-Based and Statistical Techniques. Comput Linguist. 2007;33:63–103. doi: 10.1162/coli.2007.33.1.63. [DOI] [Google Scholar]
- 32.Garfield E. The history and meaning of the journal impact factor. JAMA. 2006;295:90–3. doi: 10.1001/jama.295.1.90. [DOI] [PubMed] [Google Scholar]
- 33.McVeigh ME, Mann SJ. The Journal Impact Factor Denominator. JAMA. 2009;302:1107. doi: 10.1001/jama.2009.1301. [DOI] [PubMed] [Google Scholar]
- 34.Journal Citation Reports® (JCR®) by Thomson Reuters, (nd) https://jcr.incites.thomsonreuters.com/ (accessed April 4, 2017).
- 35.Lundh A, Sismondo S, Lexchin J, Busuioc OA, Bero L. Industry sponsorship and research outcome. Cochrane Database Syst Rev. 2012;12:MR000033. doi: 10.1002/14651858.MR000033.pub2. [DOI] [PubMed] [Google Scholar]
- 36.Sismondo S. Pharmaceutical company funding and its consequences: a qualitative systematic review. Contemp Clin Trials. 2008;29:109–13. doi: 10.1016/j.cct.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Lexchin J, Bero LA, Djulbegovic B, Clark O. Pharmaceutical industry sponsorship and research outcome and quality: systematic review. BMJ. 2003;326:1167–70. doi: 10.1136/bmj.326.7400.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumock GT, Pickard AS. Comparative effectiveness research: Relevance and applications to pharmacy. Am J Health Syst Pharm. 2009;66:1278–86. doi: 10.2146/ajhp090150. [DOI] [PubMed] [Google Scholar]
- 39.Scopus Abstract Citations Count API, (nd) http://api.elsevier.com/documentation/AbstractCitationCountAPI.wadl (accessed April 4, 2017)
- 40.About Altmetric and the Altmetric score, (nd) https://help.altmetric.com/support/solutions/articles/6000059309-about-altmetric-and-the-altmetric-score (accessed April 4, 2017)
- 41.How is the Altmetric score calculated?, (nd) https://help.altmetric.com/support/solutions/articles/6000060969-how-is-the-altmetric-score-calculated- (accessed April 4, 2017)
- 42.Altmetric API, (nd) http://api.altmetric.com/ (accessed April 4, 2017)
- 43.MEDLINE Abridged Index Medicus (AIM or “Core Clinical”) Journal Titles, (nd) https://www.nlm.nih.gov/bsd/aim.html (accessed April 4, 2017)
- 44.McMaster Plus (Premium LiteratUre Service) journals, (nd) http://hiru.mcmaster.ca/hiru/journalslist.asp (accessed April 4, 2017)
- 45.Journal Selection for Index Medicus®/Medline®, J Can Chiropr Assoc. 1996;40:47. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2485146/ (accessed March 14, 2016) [Google Scholar]
- 46.Committed selection: abridged index medicus. N Engl J Med. 1970;282:220–1. doi: 10.1056/NEJM197001222820410. [DOI] [PubMed] [Google Scholar]
- 47.Hemens BJ, Haynes RB. McMaster Premium LiteratUre Service (PLUS) performed well for identifying new studies for updated Cochrane reviews. J Clin Epidemiol. 2012;65:62–72.e1. doi: 10.1016/j.jclinepi.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software. ACM SIGKDD Explor Newsl. 2009;11:10. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- 49.Hall MA, Holmes G. Benchmarking attribute selection techniques for discrete class data mining, IEEE Trans. Knowl Data Eng. 2003;15:1437–1447. doi: 10.1109/TKDE.2003.1245283. [DOI] [Google Scholar]
- 50.Charles L, Sheng V. Encycl Mach Learn. Springer US; Boston, MA: 2011. Cost-Sensitive Learning; pp. 231–235. [DOI] [Google Scholar]
- 51.Soni J, Ansari U, Sharma D, Soni S. Predictive Data Mining for Medical Diagnosis: An Overview of Heart Disease Prediction. Int J Comput Appl. 2011;17:43–48. doi: 10.5120/2237-2860. [DOI] [Google Scholar]
- 52.Seera M, Lim CP. A hybrid intelligent system for medical data classification. Expert Syst Appl. 2014;41:2239–2249. doi: 10.1016/j.eswa.2013.09.022. [DOI] [Google Scholar]
- 53.Canese K. PubMed Relevance Sort. NLM Tech Bull. 2013 https://www.nlm.nih.gov/pubs/techbull/so13/so13_pm_relevance.html accessed July 20, 2017.
- 54.Hoogendam A, Stalenhoef AFH, de V Robbé PF, Overbeke AJPM. Analysis of queries sent to PubMed at the point of care: Observation of search behaviour in a medical teaching hospital. BMC Med Inform Decis Mak. 2008;8:42. doi: 10.1186/1472-6947-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manning CD, Raghavan P, Schütze H. Introduction to Information Retrieval. Cambridge University Press; 2008. [Google Scholar]
- 56.Charles D, Charles G. Statistics in the Pharmaceutical Industry. In: Buncher R, Tsay J-Y, editors. Stat Pharm Ind. 3rd. Chapman & Hall/CRC; New York, New York, USA: 2006. pp. 421–452. [Google Scholar]
- 57.Dmitrienko A, Tamhane A, Bretz F. Multiple Testing Problems in Pharmaceutical Statistics, New York, New York, USA. 2009 [Google Scholar]
- 58.International Conference on Harmonisation E9 Expert Working Group. ICH harmonised tripartite guideline: statistical principles for clinical trials. Stat Med. 1999;18:1905–42. [PubMed] [Google Scholar]
- 59.Van Cutsem E, Köhne C-H, Hitre E, Zaluski J, Chang Chien C-R, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P. Cetuximab and Chemotherapy as Initial Treatment for Metastatic Colorectal Cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 60.Jansen BJ, Spink A, Saracevic T. Real life, real users, and real needs: a study and analysis of user queries on the web. Inf Process Manag. 2000;36:207–227. doi: 10.1016/S0306-4573(99)00056-4. [DOI] [Google Scholar]
- 61.Ely JW, Osheroff JA, Chambliss ML, Ebell MH, Rosenbaum ME. Answering physicians’ clinical questions: obstacles and potential solutions. J Am Med Inform Assoc. 2005;12:217–24. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ely JW, Osheroff JA, Ebell MH, Bergus GR, Levy BT, Chambliss ML, Evans ER. Analysis of questions asked by family doctors regarding patient care. BMJ. 1999;319:358–61. doi: 10.1136/bmj.319.7206.358. http://www.ncbi.nlm.nih.gov/pubmed/10435959 (accessed March 14, 2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Margolis J. Citation indexing and evaluation of scientific papers. Science. 1967;155:1213–9. doi: 10.1126/science.155.3767.1213. http://www.ncbi.nlm.nih.gov/pubmed/5335153 (accessed March 5, 2017) [DOI] [PubMed] [Google Scholar]
- 64.Bornmann L, Daniel H. What do citation counts measure? A review of studies on citing behavior. J Doc. 2008;64:45–80. doi: 10.1108/00220410810844150. [DOI] [Google Scholar]
- 65.Barbic D, Tubman M, Lam H, Barbic S. An Analysis of Altmetrics in Emergency Medicine. Acad Emerg Med. 2016;23:251–268. doi: 10.1111/acem.12898. [DOI] [PubMed] [Google Scholar]
- 66.Rosenkrantz AB, Ayoola A, Singh K, Duszak R. Alternative Metrics (“Altmetrics”) for Assessing Article Impact in Popular General Radiology. Journals Acad Radiol. 2017 doi: 10.1016/j.acra.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 67.Widmer G, Kubat M. Learning in the presence of concept drift and hidden contexts. Mach Learn. 1996;23:69–101. doi: 10.1007/BF00116900. [DOI] [Google Scholar]
- 68.Tsymbal A. The Problem of Concept Drift: Definitions and Related Work. 2004 citeulike-article-id:2350391. [Google Scholar]
- 69.Irwin AN, Rackham D. Comparison of the time-to-indexing in PubMed between biomedical journals according to impact factor, discipline, and focus. Res Soc Adm Pharm. 2017;13:389–393. doi: 10.1016/j.sapharm.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 70.Mork J, Aronson A, Demner-Fushman D. 12 years on – Is the NLM medical text indexer still useful and relevant? J Biomed Semantics. 2017;8:8. doi: 10.1186/s13326-017-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


