Abstract
Microtubule-actin crosslinking factor 1 (MACF1), also known as actin crosslinking factor 7 (ACF7), is essential for proper modulation of actin and microtubule cytoskeletal networks. Most MACF1 isoforms are expressed broadly in the body, but some are exclusively found in the nervous system. Consequentially, MACF1 is integrally involved in multiple neural processes during development and in adulthood, including neurite outgrowth and neuronal migration. Furthermore, MACF1 participates in several signaling pathways, including the Wnt/β-catenin and GSK-3 signaling pathways, which regulate key cellular processes, such as proliferation and cell migration. Genetic mutation or dysregulation of the MACF1 gene has been associated with neurodevelopmental and neurodegenerative diseases, specifically schizophrenia and Parkinson’s disease. MACF1 may also play a part in neuromuscular disorders and have a neuroprotective role in the optic nerve. In this review, the authors seek to synthesize recent findings relating to the roles of MACF1 within the nervous system and explore potential novel functions of MACF1 not yet examined.
Keywords: MACF1, ACF7, neuron, brain, nervous system, development, dendrite, axon, neurite, neuron migration, proliferation
1. Introduction
Microtubule-actin crosslinking factor 1 (MACF1), also widely known as actin crosslinking factor 7 (ACF7), is a member of the spectraplakin family of cytoskeletal crosslinking proteins. Spectraplakins are large proteins distinguished by their ability to bind to different cytoskeletal networks. There are only two known mammalian spectraplakins, MACF1/ACF7 and bullous pemphigoid antigen 1 (BPAG1)/dystonin, and this family of proteins is evolutionarily conserved in most multicellular organisms [1]. MACF1 was originally identified as an actin-crosslinking protein in 1995 [2]. MACF1 belongs to a subset of microtubule plus-end tracking proteins (+TIPs), functioning at the microtubule plus-end to coordinate microtubule and F-actin interactions at the plasma membrane [3]. The most widely researched function of MACF1 is in regulation of cytoskeletal proteins, specifically F-actin and microtubules [4]. Microtubules, the actin cytoskeleton and their interacting components are involved in many polarized cellular processes including cell shape, cell division, intracellular transport, adhesion, and cell migration [5–8]. MACF1 interacts with microtubules and F-actin via distinct microtubule and actin-binding domains to regulate the polarization of cells and coordination of cellular movements [1, 4, 9]. MACF1 stabilizes the downstream cytoskeleton structure by either directly binding to microtubules or forming links between microtubules and F-actin [10], and plays an important role in cell migration via its regulation of Golgi polarization [11, 12]. This large and complex protein, however, is involved in a wide range of cellular signaling networks and processes, including Wnt/β-catenin signaling, cell migration, proliferation, survival and autophagy [13–18]. MACF1 has recently received increased attention due to its broad expression in the nervous system, more specifically, in the brain [15, 19, 20]. MACF1 mutations have been linked to neurological diseases including Parkinson’s disease (PD), autism spectrum disorder (ASD), and schizophrenia [21–23]. On a related note, several contemporary studies from our group and others have found that MACF1 is essential for proper neural progenitor proliferation, neuronal migration and neurite development [15, 16, 20, 24, 25].
In this review, we provide a brief overview of the MACF1 protein and its known functions and interactions, followed by an in-depth analysis of the roles of MACF1 in nervous system development and function. We also seek to highlight current research questions and potential explanations relating to MACF1 and its neuronal activities and related disorders.
2. Isotype structure and expression
MACF1 is expressed in multiple tissues throughout the body and has various isoforms with distinctive structures. MACF1 is a large protein of ~600 kD [2] and its primary function is cross-linking microtubules and F-actin microfilaments. MACF1 is encoded by the MACF1 gene, which is located on the human chromosome 1p32 and on chromosome 4 in mice [2, 26, 27], and is a unique hybrid of dystrophin/spectrin and plakin genetic domains [4, 27]. The MACF1 actin-binding domain (ABD) is located at the N-terminus and is composed of either one or two calponin homology domains, CH1 and CH2, respectively [4, 28–31]. Furthermore, the MACF1 ABD is conserved within the spectrin superfamily [4, 30]. Adjacent to the ABD in the N-terminus, all MACF1 isoforms possess a plakin domain stemming from spectrin repeats [4, 9, 28, 32], which can be observed throughout the plakin superfamily [33]. Separating the functionally distinct N- and C-terminal domains, each MACF1 protein exhibits 23 flexible, α-helical spectrin repeats in one domain [1, 4, 33–35]. At the C-terminus of MACF1, two calcium-binding EF-hand motifs can be found [9], followed by a spectraplakin-specific Gas2-related protein (GAR) domain responsible for microtubule binding and stabilization (Figure 1A) [4, 9, 33].
Figure 1. MACF1 structure and role in the Wnt/β-catenin signaling.
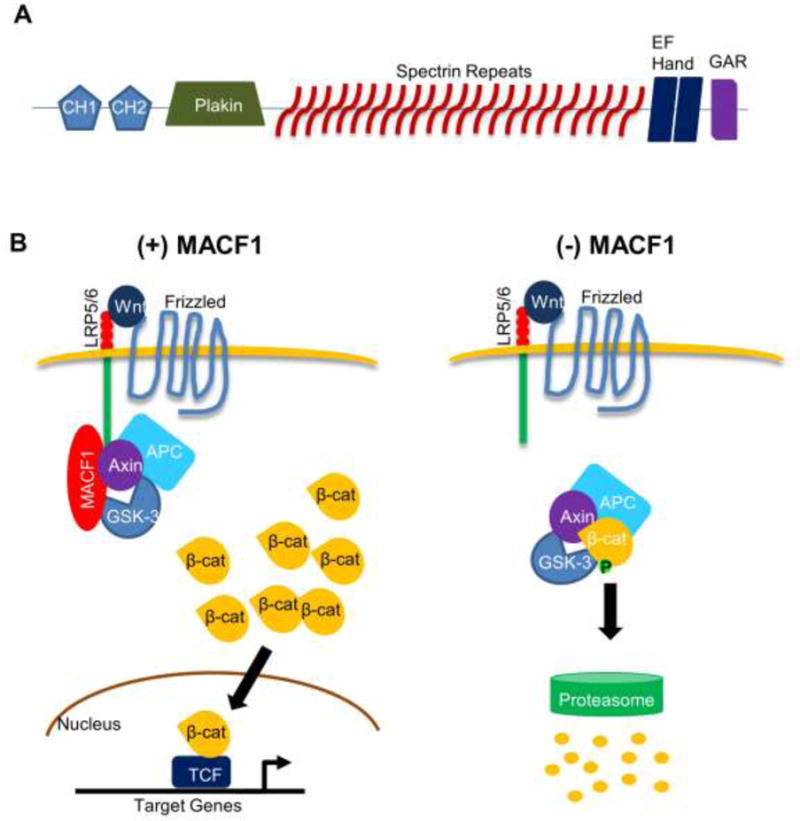
(A) General protein structure of MACF1. The five functional domains found in most MACF1 isotypes are shown: the actin-binding domain (ABD) comprised of CH1 and CH2 fragments, a plakin domain, 23 α-helical spectrin repeats, two EF hand motifs, and a GAR domain at the C-terminus. CH1: calponin homology domain 1. CH2: calponin homology domain 2. GAR: Gas2-related domain. (B) MACF1 knockdown inhibits Wnt/β-catenin signaling. Upon Wnt binding to the receptor, MACF1 translocates axin and associated molecules to the cell membrane, allowing accumulation of β-catenin in the cytosol. Some β-catenin proteins enter the nucleus to turn on target gene expression. In the absence of MACF1, Axin is unable to translocate to the cell membrane and facilitate formation of the destruction complex containing β-catenin, resulting in proteasome-mediated β-catenin degradation. LRP5/6: low-density lipoprotein receptor-related protein 5/6. GSK-3: glycogen synthase kinase-3. APC: adenomatous polyposis coli.
There are six identified murine MACF1 isoforms [36]. The first three isoforms to be discovered are currently known as MACF1a1, MACF1a2 and MACF1a3 [26, 37]. They possess identical 3′ RNA sequences, but display significant variation in the 5′ region leading to distinct protein N-termini [26]. MACF1a1 and MACF1a2 are both broadly expressed, although MACF1a1 is more predominantly found in the kidney, stomach and skin [19, 26, 37]. MACF1a2 is detected at higher levels in the lung and central nervous system [19, 26, 37, 38]. MACF1a3 expression is mainly restricted to the brain and spinal cord [19, 26]. In 2001, a fourth MACF1 isoform, MACF1-4, was discovered, with heightened expression levels in the placenta, pituitary gland, heart and lung. MACF1-4 is unique in that it lacks an ABD and instead expresses a series of plectin repeats at its N terminus [27]. Successively, a further, exceptionally-large MACF1 isoform, MACF1b, was found to be expressed throughout the body. It contains additional plakin repeats between its N-terminal plakin domain and its spectrin repeat domain [37]. The most recent MACF1 isoform to be isolated, MACF1c, is thought to only be expressed in the nervous system. It is structurally similar to the MACF1a isoforms, but lacks an ABD at its N-terminus [15]. A recent, brief review from Hu et al. provides a summary of all MACF1 isotypes and their functions [36].
In mice, MACF1 is broadly expressed throughout the developing brain. MACF1 protein can be detected in somas and neurites of cortical neurons [20]. During early brain development, MACF1 levels are highest in the ventricular zone and upper cortical areas near the marginal zone of the developing cerebral cortex [20]. As neurodevelopment progresses, MACF1 expression in the ventricular zone gradually decreases while MACF1 levels in the cortical plate steadily increase, following the established pattern of radial neuronal migration [20]. Additionally, MACF1 expression in postmitotic neurons is mainly restricted to the marginal zone at early stages of brain development, but transitions into the cortical plate by embryonic day 15.5 (E15.5) [20], indicating that MACF1 may participate in neuronal migration and differentiation.
3. Cellular signaling associated with MACF1
Beyond its role crosslinking cytoskeletal proteins, MACF1 is actively involved in multiple signaling cascades. In 2006, Chen and colleagues published that Macf1 knockout (Macf1−/−) mice do not survive beyond gastrulation, as evidenced by a failure to develop a primitive streak, node or axial mesoderm. Interestingly, they also found that knockout of BPAG1, a closely related plakin protein, had strikingly different effects (mice survived until weaning), indicating a unique role for MACF1 in regulation of embryonic development [13]. They further noted that the developmental defects present in Macf1−/− embryos mirror those seen in Wnt3−/− and LRP5/6 double-knockout mice [13, 39, 40], indicating a potential role for MACF1 in the Wnt/β-catenin signaling pathway. Consequently, they demonstrated that MACF1 interacts with the β-catenin destruction complex in the cell, binding directly to Axin using the MACF1-spectrin repeat 0 (SR0) domain. The SR0 domain is defined as the region between the MACF1 plakin domain and the first spectrin repeat [41]. They also illustrated that either knockdown of MACF1 or overexpression of the MACF1 deletion fragment of SR0 successfully inhibits Wnt/β-catenin signaling by preventing Axin translocation to the cell membrane (Figure 1B) [13]. It was further shown that MACF1 interacts directly with Wnt co-receptors LRP5/6 at the cell membrane via its SR0 domain.
Interestingly, it was later shown that MACF1 is directly phosphorylated by GSK-3 at its C-terminal microtubule-binding domain in skin stem cells, effectively preventing MACF1-microtubule interactions and nullifying microtubule polarization along actin focal-adhesion networks [42]. We have demonstrated that MACF1 and GSK-3 physically bind to one another and that MACF1 is phosphorylated in a GSK-3-dependent manner in the developing brain [20], similar to what was seen in skin stem cells [42]. It is unclear, however, whether the GSK-3 and MACF1 interaction is part of the Wnt destruction complex or downstream of growth factor signaling. While Wnt signaling utilizes a protein-protein interaction mechanism to control GSK-3, growth factors regulate a different pool of GSK-3 in the cell by phosphorylation at serine 21 (α) and 9 (β). Both signaling pathways are thought to be insulated. Wu and colleagues have shown that phosphorylation-refractile constructs of GSK-3 modulate MACF1 phosphorylation and activity in skin cells [42]. Thus, at least a part of MACF1 regulation by GSK-3 appears to be induced by growth factor signaling. In a breast carcinoma model, it was found that MACF1 is involved in microtubule stabilization via an ErbB2 receptor tyrosine kinase signaling pathway. Heregulin β activates ErbB2, which leads to the phosphorylation and inhibition of GSK-3 through the Memo-RhoA-mDial pathway. Inhibition of GSK-3 kinase activity blocks the phosphorylation of two other cytoskeletal regulators, adenomatous polyposis coli (APC) and cytoplasmic linker-associated protein 2 (CLASP2), and their subsequent translocation to the cell membrane. MACF1 is recruited to the membrane by APC, but not CLASP2, where it regulates microtubule dynamics [43].
Additionally, MACF1 plays a role in DOCK 180-ELMO-Rac signaling in cell protrusion/lamellipodium extension during cell migration. In this system, ELMO recruits MACF1 to sites of emerging protrusions, where MACF1 orchestrates microtubule capture and stabilization [44]. Following stimulation by integrin, Elmo and MACF1 colocalize at the cell membrane [36, 44]. MACF1 then organizes the cytoskeleton to extend stable membrane protrusions [44].
MACF1 is also integrally involved in some forms of vesicular trafficking, specifically relating to axonal vesicle transport [45] and autophagy [14]. MACF1 can act as a Rab21 effector. The complex of Rab21, Kif5A, GolginA4, and MACF1 acts together to transport TI-VAMP from the Golgi to neurite tips along microtubule [45]. In autophagy, MACF1 and its binding partner, the trans-Golgi protein p230, are responsible for trafficking of mAtg9 from the trans-Golgi network to the cell surface, a necessary step in phagophore formation. MACF1 knockdown impairs mAtg9 transport and blocks early steps in autophagy in a state of amino acid starvation [14].
4. MACF1 in cell proliferation
Cell proliferation is the process that results in an increased number of cells. During cell division, microtubule and actin interactions regulate spindle positioning and cytokinesis. Abnormal microtubules and actin cytoskeleton dynamics cause cytokinesis defects, thus altering cell proliferation [46–48]. In osteoblast cells, MACF1 knockdown inhibits cell proliferation and induces S phase cell cycle arrest [49]. Additionally, the microtubule organizing center (MTOC) fails to form in proximity to the condensed α-tubulin fibers surrounding the nucleus in osteoblasts [49]. These observations indicate dysregulated cytokinesis following MACF1 knockdown [46–48, 50]. Wu and colleagues, however, have observed no significant deficit in cell proliferation in epidermal or endodermal cells in the absence of MACF1 expression [42, 51]. This cell type-specific function of MACF1 could be explained by unknown unique MACF1 protein-protein interactions in osteoblast cells or by additional proteins fulfilling the same functional role as MACF1 in epidermal and endodermal cells during cytokinesis [49, 50]. Taken together, these findings may provide insight into the role of MACF1 in the proliferation of neural progenitor cells.
In dividing neural progenitor cells, proper positioning of the centrosome, the main MTOC, is necessary for cell proliferation [52]. Neurons originate from a limited number of neural progenitor cells during embryonic development [53]. Neural progenitors can either self-renew (symmetric division) or undergo the process of neurogenesis, in which one daughter remains a neural progenitor cell and the other undergoes sequential differentiations toward becoming a neuron (asymmetric division) [53–58]. This process takes place in the ventricular zone (VZ) or subventricular zone (SVZ) of the developing cerebral cortex for most excitatory pyramidal neural progenitors and in the VZ or SVZ of the medial ganglionic eminence (MGE) for most inhibitory interneuron progenitors [53–55, 57, 58]. Throughout the process of neurogenesis, a significant reorganization of cellular components is required before mitosis can take place. Following the completion of S phase, the nucleus must migrate before apical mitosis can be undergone in a process known as interkinetic nuclear migration, which requires the interplay of the actin and microtubule cytoskeletons [59–61]. Initially, neural progenitors and/or stem cells divide symmetrically along a vertical axis before asymmetrical division along a horizontal axis can begin [55, 61, 62]. The plane of neural progenitor division is highly regulated by the cytoskeleton, specifically the orientation of mitotic spindles [61, 63, 64], thus microtubules and their regulatory proteins are crucial to proper proliferation and cell division throughout neural progenitor proliferation [53, 61, 64, 65]. During mitosis, microtubule assembly and disassembly at the plus- and minus-ends is required for proper separation of chromosomes and cytokinesis [66–69]. +TIPs crucially regulate microtubule dynamics during cell division and must be maintained at proper levels [66, 70], as abnormal microtubule stabilization can suppress microtubule dynamics, preventing cell division and resulting in apoptosis [70, 71]. Like other +TIPs, MACF1 is localized to microtubule plus-ends and physically interacts with several other +TIPs, including EB1, APC and CLASP [13, 72], and regulates centrosome movement [20]. All of this circumstantial evidence suggests the importance of MACF1 in neural cell proliferation. However, its function in this process is still unclear.
Examining a Macf1 conditional knockout mouse model (Macf1 cko), in which Macf1 expression is eliminated in the developing nervous system, Goryunov and colleagues observed extensive heterotopia, or distinct disorganization of neural layers, in the cortex and hippocampus of Macf1 cko mice [15]. The majority of early-born cortical neurons were found in their proper, deep layers, whereas neurons with a late-born phenotype appeared to be mixed in with the deep-layer neurons and not in their typical outer cortical layers [15]. Heterotopia is often attributed to neuronal migration impediments but can also be caused by defective neuronal proliferation. It is unclear whether the layer positioning defects in Macf1 cko mice are due to a reduced neuronal migration rate, aberrant migratory guidance, or defective neuronal proliferation [15].
5. MACF1 in neuronal and non-neuronal cell migration
Cell migration is a fundamental cellular process and is essential for embryonic development, tissue repair and regeneration, and tumor metastasis [73]. Cell migration begins with various extracellular cues such as chemokines and signals from the extracellular matrix that lead to the polarization and the extension of protrusions in the direction of movement [74]. Migrating cells must acquire a polarized, asymmetric morphology and develop a single leading edge with one filopodia [75]. During the migration process, cells actively reorganize the actin cytoskeleton and microtubules [73]. MACF1 directly interacts with other +TIPs, such as the EB1 protein, to recruit cell polarity and signaling molecules to microtubule tips [76]. MACF1 also interacts with CLASP2, another +TIP protein, and regulates CLASP localization. CLASP2 is involved in microtubule stabilization and is required for efficient, persistent motility [77]. It was recently discovered that MACF1 also directly interacts with the ELMO protein (engulfment and cell motility protein), as mentioned above. ELMO1 recruits MACF1 to the cell membrane, where MACF1 regulates microtubule capture and stabilization of cellular protrusions [44].
In MACF1 null cells, many microtubules exhibit irregular trajectories and are more sensitive to depolymerizing agents. Moreover, loss of MACF1 causes defective polarization of stable microtubules in epidermal cells, and a lack of coordinated migration in response to wounding [10]. In migrating skin stem cells, GSK-3β phosphorylates the microtubule-binding domain of MACF1, resulting in the dissociation of MACF1 from microtubules. Thus, phosphorylation of MACF1 is necessary for microtubule growth and for skin stem cell migration [42]. Moreover, it was recently suggested that the FAK/Src kinase phosphorylation of MACF1 is essential for its binding to F-actin and coordination of cytoskeletal dynamics at focal adhesions. The effects of MACF1 phosphorylation in focal adhesion dynamics and cell motility have been clearly observed in epithelial cells [78]. In motile fibroblasts, MACF1 regulates cortical CLASP localization, allowing microtubule stabilization and promoting directionally persistent motility [77]. In breast carcinoma cells, the ErbB2 receptor controls microtubule capture by recruiting MACF1 to the plasma membrane, where MACF1 contributes to microtubule guidance and capture in migrating cells [43]. miR-34a regulates cytoskeletal proteins such as MACF1, LMNA, GFAP, ALDH2 and LOC100129335, and transfection of miR-34a into carcinoma cells causes inhibition of cell migration and invasion [79].
During brain development, neurons migrate from their place of birth to a specified final destination (Figure 2). Excitatory pyramidal neurons migrate from the cortical ventricular zone into the cortical plate along radial glial processes in a pattern known as radial migration (Figure 2A) [80–83]. Later-born pyramidal neurons migrate outwards beyond earlier-born neurons, resulting in more superficially-positioned late-born neurons in mature brains. Inhibitory interneurons (GABAergic neurons) begin migration in the medial ganglionic eminence of the ventral telencephalon and tangentially migrate into the dorsal telencephalon before changing course and radially entering the cortical plate (Figure 2B) [84–86]. Neuronal migration and positioning are critical steps for establishing functional neural circuitry in the developing brain. Therefore, abnormal neuronal migration during development causes brain malformations, which have been linked to a variety of neurodevelopmental and neuropsychiatric diseases such as ASD, attention deficit hyperactivity disorder (ADHD), intellectual disability, and schizophrenia [87–90]. Neuronal migration is a dynamic process, which requires persistent reconstruction of the cytoskeleton. In this context microtubules and microtubule-related proteins, including MACF1, play important roles in the regulation of neuronal migration during brain development [91–93]. We and others have reported that MACF1 is highly expressed in the nervous system and developing brain [4, 20]. Macf1 conditional knockout brains using a nestin-cre driver display partially-mixed upper- and deeper-layer neurons in the cerebral cortex [15]. The expression pattern of MACF1 and the heterotopic cortical phenotype in Macf1/nestin-cre conditional knockout mice strongly suggest a role for MACF1 in neuronal migration. Indeed, our study shows that neuron-specific Macf1 deletion using a Nex-cre driver or in utero electroporation of the Dcx-cre-iGFP construct suppresses the radial migration of cortical pyramidal neurons, resulting in aberrant positioning of excitatory pyramidal neurons in the cortical layers [20]. During radial neuron migration, MACF1 regulates leading process morphogenesis and dynamics. Macf1-deleted neurons develop short and unstable leading processes resulting in unidirectional and slow radial neuron migration. Also, Macf1-deleted pyramidal neurons exhibit microtubule destabilization and static centrosomes [20]. Centrosomes show dynamic back-and-forth movements along the leading process to pull the soma during normal neuron migration. However, centrosomes in Macf1-deleted neurons have little movement and remain close to the cell body, resulting in the creation of insufficient tension for somal translocation. Thus, MACF1-mediated regulation of microtubule stability and centrosome movement contributes to radial neuron migration in the developing brain. Consistent with migrating skin stem cells, GSK-3-mediated phosphorylation is an important mechanism for the MACF1 function in neuronal migration [20, 42]. In addition to the role of MACF1 in radial migration, we have recently shown that MACF1 is also a key molecule in tangential neuron migration [16]. MACF1 is highly expressed in the tangential migratory stream [16]. Macf1 deletion in interneuron progenitors and progeny using Nkx2.1-cre or Dlx5/6-cre lines results in abnormal migration and defective positioning of GABAergic inhibitory interneurons in the mouse cerebral cortex and hippocampus [16]. Macf1-deleted GABAergic interneurons show slower speed and aberrant orientation of movement during migration. Importantly, MACF1 regulates the transition of migration direction from a tangential to a radial route during cortical development [16]. Macf1-deleted interneurons develop abnormal leading processes and disrupted microtubule stability and severing [16]. Overall, MACF1 is an essential regulator of cell migration via its management of microtubule and actin cytoskeleton dynamics.
Figure 2. Modes of neuronal migration in the developing brain.
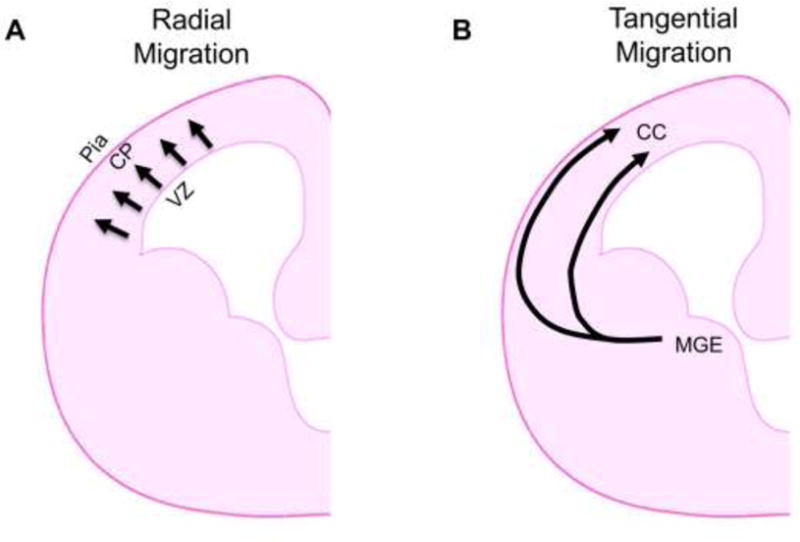
(A) Radial migration. Excitatory pyramidal projection neurons migrate outward from the ventricular zone of the cerebral cortex toward the cortical plate during brain development. CP: cortical plate. VZ: ventricular zone. (B) Tangential migration. Inhibitory interneurons migrate tangentially from the medial ganglionic eminence (MGE) in the ventral brain to the cerebral cortex, where they undergo further movements until they reach their final cortical destinations. CC: cerebral cortex.
6. MACF1 in neurite development
Neurite outgrowth is an essential event in neural development, which involves coordinated changes between the actin cytoskeleton and the microtubule network [94, 95]. This process is regulated by various proteins that manipulate the cytoskeletal network by various means [96, 97]. Recent studies indicate that MACF1 plays a vital role in neurite outgrowth. MACF1 controls the extension and differentiation of neurites in Drosophila neurons [25]. Knockdown of MACF1 decreases axon outgrowth, a process dependent on its F-actin- and microtubule-binding domains in Drosophila neuronal cultures [25]. We have recently provided evidence that supports the MACF1 function in mouse neurite growth in vivo [24]. Using an in utero electroporation method and conditional knockout mouse lines to generate temporal and spatial Macf1 deletion, we have knocked out or down Macf1 in developing cortical and hippocampal pyramidal neurons. We have found that MACF1 deletion decreases dendrite growth and branching in mouse pyramidal neurons. Accordingly, Macf1-deleted neurons show reduced density and abnormal morphology of dendritic spines. Macf1-deleted spines appear long and thin with short spine heads and necks [24]. The cellular cytoskeletal network is critical in dendritic spine morphogenesis, a process which is regulated by a complex network of signaling molecules [98–100]. Dendritic spine morphology is dependent on the amount and structure of F-actin within neurons [101, 102]. MACF1 interacts with F-actin to regulate cell polarization [4, 9]. Loss of MACF1 also impairs the elongation of callosal axons in the brain. MACF1 is thought to regulate neurite development via GSK-3 signaling in the brain (Figure 3). As described above, knockdown of the MACF1 protein inhibits Wnt signaling, which is mediated by GSK-3 [13]. GSK-3 is a master-regulator of the cellular cytoskeletal network, neural progenitor regulation and neurite growth [103–106]. Over-expression of a constitutively-active GSK-3β (ca-GSK-3β) construct reduces the number and length of dendrites. However, co-expression of MACF1 S:A (phosphorylation-refractile form) rescues the inhibitory effects of ca-GSK-3β [24], suggesting that GSK-3-mediated phosphorylation is an important mechanism for the MACF1 function in neurite development. Future studies will be needed to expand our understanding of MACF1 as to regulatory mechanisms and cellular signaling pathways in neurite development.
Figure 3. MACF1 in neurite outgrowth.
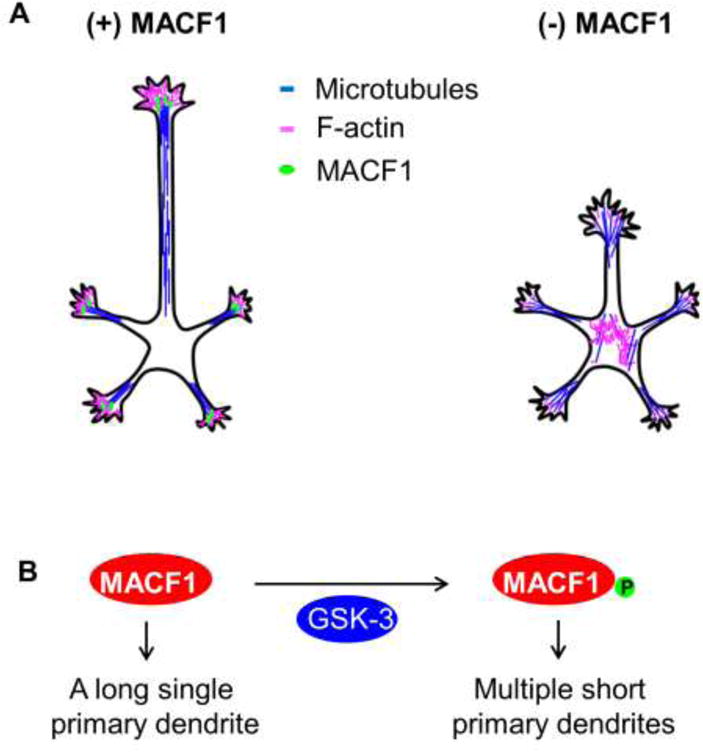
(A) MACF1 regulation of neurite outgrowth. MACF1 localizes near the distal ends of growing axons and dendrites, where it stabilizes microtubule bundles and leads to the assembly of polymerized actin. In the absence of MACF1, microtubules and actin are disorganized and unstable at the neurite tip and scattered in the cytosol. (B) GSK-3 regulates the activity of MACF1 in dendrite outgrowth and arborization. When MACF1 is phosphorylated by GSK-3, it results in multiple short primary dendrites. When MACF1 remains unphosphorylated, neurons extend a single long primary dendrite.
7. Neural diseases and MACF1
MACF1 gene mutations have been associated with neuromuscular diseases. Mutations in cytoskeletal genes, such as dystonin, dystrophin, and plectin result in myopathic consequences, thus suggesting MACF1 may have similar muscular phenotypes [17]. In a family with novel neuromuscular conditions including diminished motor skills, lax muscles, and occasional hypotonia, the Macf1 gene product is found at low levels due to a chromosome modification in one gene locus. This novel myopathy is termed “spectraplakinopathy type I,” based on MACF1 belonging to the spectraplakin protein family [17]. Ultrastructural changes and altered motility are accompanied in muscle tissues of affected individuals [17].
MACF1 mutations have been shown to contribute to psychological disorders. Two schizophrenia risk genes, disrupted in schizophrenia 1 (DISC1) and dysbindin (DTNBP1), are associated with cognitive deficits in schizophrenics [107]. DISC1 and DTNBP1 are important molecules in many aspects of neural development including neural progenitor proliferation and neurogenesis, neurite outgrowth, neuronal migration, and synaptic differentiation [108–112]. Several instances of synaptic pathology have been reported in individuals diagnosed with schizophrenia [113]. Both proteins form a similar network of protein-protein interactions, and the profiles of proteins that they interact with suggest similar functions in cytoskeletal stability and organization, intracellular transport, and cell cycle progression [114]. Camargo and colleagues have shown that MACF1 is one of the proteins involved in critical interactions with both DISC1 and DTNBP1. They suggest that DISC1 and DTNBP1 may play a converging role in affecting synapse structure and function by disrupting intracellular transport and cytoskeletal stability via interactions with MACF1, contributing to cognitive deficits in schizophrenics [114]. Furthermore, schizophrenia and ASD may share underlying pathology, as suggested by shared risk genes [22]. For example, rare mutations in genes that are functional in the synapse have been identified in ASD and schizophrenia cases [23]. Several novel loss-of-function variants overlap in both cases, including those coding for proteins involved in protein-protein interactions with DISC1, such as MACF1 [23]. These results suggest that mutations in multiple genes involved in synapse formation, including Macf1, are a risk factor for both ASD and schizophrenia.
Several neurodegenerative disorders show evidence of cytoskeletal collapse. In particular, a hallmark of Parkinson’s Disease (PD) is degeneration of dopaminergic (DA) neurons in the substantia nigra pars compacta [21]. It has been observed that both genetic and neurotoxic causes of PD may target the cytoskeleton, and resulting cytoskeletal disorganization and dysregulation may be a mechanistic cause of PD [115]. Two lines of evidence suggest that MACF1 is involved in the pathogenesis of PD. First, Macf1 knockout inhibits Wnt signaling, which is important in the development of dopamine neurons. Second, MACF1 mRNA levels in DA neurons of PD patients are significantly lower than in controls [116]. Thus, reduced MACF1 levels leading to dysregulation of the cytoskeleton may cause vulnerability of DA neurons to neurodegeneration. More directly, MACF1 has been shown to be a risk gene for PD [21]. MACF1 is a downstream target of PD biochemical pathways, and has been found to be significantly associated with PD in 713 families studied. In addition, knockdown of the Macf1 orthologue Vab-10 in C. elegans results in selective loss of DA neurons [21]. These results suggest that MACF1 may contribute to genetic etiology of PD, and may be a mechanistic cause.
Optic nerve injury is another neurological condition in which MACF1 has been implicated. Retinal ganglion cells (RGCs) are the final neuronal output of the retina, receiving visual signals from amacrine and bipolar cells and transmitting them to the brain via the optic nerve [18]. Degeneration of RGCs and their axons in the optic nerve leads to vision loss in multiple optic neuropathies, including glaucoma most commonly. The work of Munemasa and colleagues shows strong Nell2 and MACF1 expression in RGCs [18]. Nell2 has a strong neuroprotective function, increasing survival of neurons in the hippocampus and cerebral cortex. After an optic nerve injury, Nell2 interacts with MACF1 to promote survival of RGCs [18].
8. Concluding remarks
In this review, we have summarized and synthesized much of the current research regarding MACF1 in nervous system development and maintenance. MACF1 is a large protein with multiple distinct isoforms, expressed at varying levels throughout the body. The MACF1 protein is involved in important signaling pathways and plays roles in many cellular processes. Genetic mutations of Macf1 or deficits in MACF1 protein function have far reaching effects in nervous system development and activity. Specifically, MACF1’s interactions with cytoskeletal proteins and other cytoskeletal regulators are required for proper polarity, proliferation, migration and neurite outgrowth in neurons and neural progenitors. Beyond these established roles, MACF1 has also been shown to participate in the Wnt/β-catenin signaling pathway, as well as several other pathways related to cellular processes such as vesicular transport and autophagy. Despite increased understanding of MACF1 and its functions in the nervous system, there are still many questions to be answered and opportunities for further research. For example, although several studies have implicated MACF1 mutations as a risk factor for neural disorders including psychiatric and neurodegenerative diseases, relatively few studies have examined how MACF1 genetic abnormalities relate to the pathology of these conditions on a mechanistic or disease-specific level. Additionally, due to MACF1’s broad roles in nervous system regulation, is it possible to modulate MACF1 expression or activity in a specific setting related to neurological symptoms? On a more basic level, although recent studies have offered a role of MACF1 in neurite outgrowth and the formation and/or pruning of dendritic spines during development, detailed regulatory mechanisms of MACF1 in these neural processes remain to be studied.
Acknowledgments
Funding
This work was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS091220 to W.K.
List of Abbreviations
- MACF1
microtubule-actin crosslinking factor 1
- ACF7
actin crosslinking factor 7
- BPAG1
bullous pemphigoid antigen 1
- PD
Parkinson’s disease
- ASD
autism spectrum disorder
- ABD
actin-binding domain
- CH1
calponin homology domain 1
- CH2
calponin homology domain 2
- GAR
Gas2-related protein
- LRP5/6
low-density lipoprotein receptor-related protein 5/6
- GSK3
glycogen synthase kinase 3
- APC
adenomatous polyposis coli
- CLASP2
cytoplasmic linker-associated protein 2
- MTOC
microtubule organizing center
- VZ
ventricular zone
- SVZ
subventricular zone
- MGE
medial ganglionic eminence
- ELMO
engulfment and cell motility protein
- VAMP
vesicle-associated membrane protein
- LMNA
Lamin A/C
- GFAP
glial fibrillary acidic protein
- ALDH2
aldehyde dehydrogenase 2
- ADHD
attention deficit hyperactivity disorder
- DA
dopaminergic
- Vab-10
variable abnormal morphology 10
- DISC1
disrupted in schizophrenia 1
- DTNBP1
dysbindin
- RGC
retinal ganglion cell
- Nell2
neural EGFL-like 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
None
Authors’ contributions
J.J.M., M.K., A.L.S., E.J, and W.K. analyzed the published studies and wrote the paper. W.K. conceived the study.
References
- 1.Suozzi KC, Wu X, Fuchs E. Spectraplakins: master orchestrators of cytoskeletal dynamics. J Cell Biol. 2012;197(4):465–75. doi: 10.1083/jcb.201112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byers TJ, Beggs AH, McNally EM, Kunkel LM. Novel actin crosslinker superfamily member identified by a two step degenerate PCR procedure. FEBS Lett. 1995;368(3):500–4. doi: 10.1016/0014-5793(95)00722-l. [DOI] [PubMed] [Google Scholar]
- 3.Gupta T, Marlow FL, Ferriola D, Mackiewicz K, Dapprich J, Monos D, Mullins MC. Microtubule actin crosslinking factor 1 regulates the Balbiani body and animal-vegetal polarity of the zebrafish oocyte. PLoS Genet. 2010;6(8):e1001073. doi: 10.1371/journal.pgen.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung CL, Sun D, Zheng M, Knowles DR, Liem RK. Microtubule actin cross-linking factor (MACF): a hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J Cell Biol. 1999;147(6):1275–86. doi: 10.1083/jcb.147.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goode BL, Drubin DG, Barnes G. Functional cooperation between the microtubule and actin cytoskeletons. Curr Opin Cell Biol. 2000;12(1):63–71. doi: 10.1016/s0955-0674(99)00058-7. [DOI] [PubMed] [Google Scholar]
- 6.Palazzo AF, Gundersen GG. Microtubule-actin cross-talk at focal adhesions. Sci STKE. 2002;2002(139):pe31. doi: 10.1126/stke.2002.139.pe31. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nature cell biology. 2003;5(7):599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- 8.Yarm F, Sagot I, Pellman D. The social life of actin and microtubules: interaction versus cooperation. Curr Opin Microbiol. 2001;4(6):696–702. doi: 10.1016/s1369-5274(01)00271-5. [DOI] [PubMed] [Google Scholar]
- 9.Sun D, Leung CL, Liem RK. Characterization of the microtubule binding domain of microtubule actin crosslinking factor (MACF): identification of a novel group of microtubule associated proteins. J Cell Sci. 2001;114(Pt 1):161–172. doi: 10.1242/jcs.114.1.161. [DOI] [PubMed] [Google Scholar]
- 10.Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115(3):343–54. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 11.Etienne-Manneville S. Actin and microtubules in cell motility: which one is in control? Traffic. 2004;5(7):470–7. doi: 10.1111/j.1600-0854.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 12.Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes & development. 2007;21(5):483–96. doi: 10.1101/gad.1511207. [DOI] [PubMed] [Google Scholar]
- 13.Chen HJ, Lin CM, Lin CS, Perez-Olle R, Leung CL, Liem RK. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes & development. 2006;20(14):1933–45. doi: 10.1101/gad.1411206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohda M, Misumi Y, Ogata S, Sakisaka S, Hirose S, Ikehara Y, Oda K. Trans-Golgi protein p230/golgin-245 is involved in phagophore formation. Biochem Biophys Res Commun. 2015;456(1):275–81. doi: 10.1016/j.bbrc.2014.11.071. [DOI] [PubMed] [Google Scholar]
- 15.Goryunov D, He CZ, Lin CS, Leung CL, Liem RK. Nervous-tissue-specific elimination of microtubule-actin crosslinking factor 1a results in multiple developmental defects in the mouse brain. Mol Cell Neurosci. 2010;44(1):1–14. doi: 10.1016/j.mcn.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ka M, Moffat JJ, Kim WY. MACF1 Controls Migration and Positioning of Cortical GABAergic Interneurons in Mice. Cerebral cortex. 2016 doi: 10.1093/cercor/bhw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorgensen LH, Mosbech MB, Faergeman NJ, Graakjaer J, Jacobsen SV, Schroder HD. Duplication in the microtubule-actin cross-linking factor 1 gene causes a novel neuromuscular condition. Sci Rep. 2014;4:5180. doi: 10.1038/srep05180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munemasa Y, Chang CS, Kwong JM, Kyung H, Kitaoka Y, Caprioli J, Piri N. The neuronal EGF-related gene Nell2 interacts with Macf1 and supports survival of retinal ganglion cells after optic nerve injury. PloS one. 2012;7(4):e34810. doi: 10.1371/journal.pone.0034810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernier G, Pool M, Kilcup M, Alfoldi J, De Repentigny Y, Kothary R. Acf7 (MACF) is an actin and microtubule linker protein whose expression predominates in neural, muscle, and lung development. Dev Dyn. 2000;219(2):216–25. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1041>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 20.Ka M, Jung EM, Mueller U, Kim WY. MACF1 regulates the migration of pyramidal neurons via microtubule dynamics and GSK-3 signaling. Dev Biol. 2014;395(1):4–18. doi: 10.1016/j.ydbio.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Li N, Xiong N, You Q, Li J, Yu J, Qing H, Wang T, Cordell HJ, Isacson O, Vance JM, Martin ER, Zhao Y, Cohen BM, Buttner EA, Lin Z. Genetic Variants of Microtubule Actin Cross-linking Factor 1 (MACF1) Confer Risk for Parkinson’s Disease. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-9861-y. [DOI] [PubMed] [Google Scholar]
- 22.Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, Zhang N, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Kendler KS, Freedman R, Dudbridge F, Pe’er I, Hakonarson H, Bergen SE, Fanous AH, Holmans PA, Gejman PV. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168(3):302–16. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenny EM, Cormican P, Furlong S, Heron E, Kenny G, Fahey C, Kelleher E, Ennis S, Tropea D, Anney R, Corvin AP, Donohoe G, Gallagher L, Gill M, Morris DW. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Molecular psychiatry. 2014;19(8):872–9. doi: 10.1038/mp.2013.127. [DOI] [PubMed] [Google Scholar]
- 24.Ka M, Kim WY. Microtubule-Actin Crosslinking Factor 1 Is Required for Dendritic Arborization and Axon Outgrowth in the Developing Brain. Mol Neurobiol. 2016;53(9):6018–6032. doi: 10.1007/s12035-015-9508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Soriano N, Travis M, Dajas-Bailador F, Goncalves-Pimentel C, Whitmarsh AJ, Prokop A. Mouse ACF7 and drosophila short stop modulate filopodia formation and microtubule organisation during neuronal growth. J Cell Sci. 2009;122(Pt 14):2534–42. doi: 10.1242/jcs.046268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernier G, Mathieu M, De Repentigny Y, Vidal SM, Kothary R. Cloning and characterization of mouse ACF7, a novel member of the dystonin subfamily of actin binding proteins. Genomics. 1996;38(1):19–29. doi: 10.1006/geno.1996.0587. [DOI] [PubMed] [Google Scholar]
- 27.Gong TW, Besirli CG, Lomax MI. MACF1 gene structure: a hybrid of plectin and dystrophin. Mamm Genome. 2001;12(11):852–61. doi: 10.1007/s00335-001-3037-3. [DOI] [PubMed] [Google Scholar]
- 28.Karakesisoglou I, Yang Y, Fuchs E. An epidermal plakin that integrates actin and microtubule networks at cellular junctions. J Cell Biol. 2000;149(1):195–208. doi: 10.1083/jcb.149.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Way M, Pope B, Weeds AG. Evidence for functional homology in the F-actin binding domains of gelsolin and alpha-actinin: implications for the requirements of severing and capping. J Cell Biol. 1992;119(4):835–42. doi: 10.1083/jcb.119.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winder SJ, Hemmings L, Maciver SK, Bolton SJ, Tinsley JM, Davies KE, Critchley DR, Kendrick-Jones J. Utrophin actin binding domain: analysis of actin binding and cellular targeting. J Cell Sci. 1995;108(Pt 1):63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- 31.Bandi S, Singh SM, Mallela KM. Interdomain Linker Determines Primarily the Structural Stability of Dystrophin and Utrophin Tandem Calponin-Homology Domains Rather than Their Actin-Binding Affinity. Biochemistry. 2015;54(35):5480–8. doi: 10.1021/acs.biochem.5b00741. [DOI] [PubMed] [Google Scholar]
- 32.Jefferson JJ, Ciatto C, Shapiro L, Liem RK. Structural analysis of the plakin domain of bullous pemphigoid antigen1 (BPAG1) suggests that plakins are members of the spectrin superfamily. J Mol Biol. 2007;366(1):244–57. doi: 10.1016/j.jmb.2006.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roper K, Gregory SL, Brown NH. The ‘spectraplakins’: cytoskeletal giants with characteristics of both spectrin and plakin families. J Cell Sci. 2002;115(Pt 22):4215–25. doi: 10.1242/jcs.00157. [DOI] [PubMed] [Google Scholar]
- 34.Yan Y, Winograd E, Viel A, Cronin T, Harrison SC, Branton D. Crystal structure of the repetitive segments of spectrin. Science. 1993;262(5142):2027–30. doi: 10.1126/science.8266097. [DOI] [PubMed] [Google Scholar]
- 35.Pascual J, Pfuhl M, Walther D, Saraste M, Nilges M. Solution structure of the spectrin repeat: a left-handed antiparallel triple-helical coiled-coil. J Mol Biol. 1997;273(3):740–51. doi: 10.1006/jmbi.1997.1344. [DOI] [PubMed] [Google Scholar]
- 36.Hu L, Su P, Li R, Yin C, Zhang Y, Shang P, Yang T, Qian A. Isoforms, structures, and functions of versatile spectraplakin MACF1. BMB Rep. 2016;49(1):37–44. doi: 10.5483/BMBRep.2016.49.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin CM, Chen HJ, Leung CL, Parry DA, Liem RK. Microtubule actin crosslinking factor 1b: a novel plakin that localizes to the Golgi complex. J Cell Sci. 2005;118(Pt 16):3727–38. doi: 10.1242/jcs.02510. [DOI] [PubMed] [Google Scholar]
- 38.Okuda T, Matsuda S, Nakatsugawa S, Ichigotani Y, Iwahashi N, Takahashi M, Ishigaki T, Hamaguchi M. Molecular cloning of macrophin, a human homologue of Drosophila kakapo with a close structural similarity to plectin and dystrophin. Biochem Biophys Res Commun. 1999;264(2):568–74. doi: 10.1006/bbrc.1999.1538. [DOI] [PubMed] [Google Scholar]
- 39.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. 1999;22(4):361–5. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 40.Kelly OG, Pinson KI, Skarnes WC. The Wnt co-receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development. 2004;131(12):2803–15. doi: 10.1242/dev.01137. [DOI] [PubMed] [Google Scholar]
- 41.Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM. The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape. The Journal of biological chemistry. 2016;291(36):18643–62. doi: 10.1074/jbc.M116.732909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Shen QT, Oristian DS, Lu CP, Zheng Q, Wang HW, Fuchs E. Skin stem cells orchestrate directional migration by regulating microtubule-ACF7 connections through GSK3beta. Cell. 2011;144(3):341–52. doi: 10.1016/j.cell.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaoui K, Benseddik K, Daou P, Salaun D, Badache A. ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc Natl Acad Sci USA. 2010;107(43):18517–22. doi: 10.1073/pnas.1000975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margaron Y, Fradet N, Cote JF. ELMO recruits actin cross-linking family 7 (ACF7) at the cell membrane for microtubule capture and stabilization of cellular protrusions. The Journal of biological chemistry. 2013;288(2):1184–99. doi: 10.1074/jbc.M112.431825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgo A, Proux-Gillardeaux V, Sotirakis E, Bun P, Casano A, Verraes A, Liem RK, Formstecher E, Coppey-Moisan M, Galli T. A molecular network for the transport of the TI-VAMP/VAMP7 vesicles from cell center to periphery. Dev Cell. 2012;23(1):166–80. doi: 10.1016/j.devcel.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 46.Moulding DA, Blundell MP, Spiller DG, White MR, Cory GO, Calle Y, Kempski H, Sinclair J, Ancliff PJ, Kinnon C, Jones GE, Thrasher AJ. Unregulated actin polymerization by WASp causes defects of mitosis and cytokinesis in X-linked neutropenia. J Exp Med. 2007;204(9):2213–24. doi: 10.1084/jem.20062324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hossain MM, Hwang DY, Huang QQ, Sasaki Y, Jin JP. Developmentally regulated expression of calponin isoforms and the effect of h2-calponin on cell proliferation. Am J Physiol Cell Physiol. 2003;284(1):C156–67. doi: 10.1152/ajpcell.00233.2002. [DOI] [PubMed] [Google Scholar]
- 48.Zhu J, Beattie EC, Yang Y, Wang HJ, Seo JY, Yang LX. Centrosome impairments and consequent cytokinesis defects are possible mechanisms of taxane drugs. Anticancer Res. 2005;25(3B):1919–25. [PubMed] [Google Scholar]
- 49.Hu L, Su P, Li R, Yan K, Chen Z, Shang P, Qian A. Knockdown of microtubule actin crosslinking factor 1 inhibits cell proliferation in MC3T3-E1 osteoblastic cells. BMB Rep. 2015;48(10):583–8. doi: 10.5483/BMBRep.2015.48.10.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D’Avino PP, Giansanti MG, Petronczki M. Cytokinesis in animal cells. Cold Spring Harb Perspect Biol. 2015;7(4):a015834. doi: 10.1101/cshperspect.a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135(1):137–48. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30(6):276–83. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Homem CC, Repic M, Knoblich JA. Proliferation control in neural stem and progenitor cells. Nat Rev Neurosci. 2015;16(11):647–59. doi: 10.1038/nrn4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huttner WB, Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr Opin Cell Biol. 2005;17(6):648–57. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6(10):777–88. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 56.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10(10):724–35. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14(11):755–69. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franco SJ, Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77(1):19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67(6):906–14. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 60.Miyata T, Okamoto M, Shinoda T, Kawaguchi A. Interkinetic nuclear migration generates and opposes ventricular-zone crowding: insight into tissue mechanics. Front Cell Neurosci. 2014;8:473. doi: 10.3389/fncel.2014.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mora-Bermudez F, Huttner WB. Novel insights into mammalian embryonic neural stem cell division: focus on microtubules. Mol Biol Cell. 2015;26(24):4302–6. doi: 10.1091/mbc.E15-03-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taverna E, Gotz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- 63.Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nature cell biology. 2008;10(1):93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 64.Mora-Bermudez F, Matsuzaki F, Huttner WB. Specific polar subpopulations of astral microtubules control spindle orientation and symmetric neural stem cell division. Elife. 2014;3 doi: 10.7554/eLife.02875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zigman M, Laumann-Lipp N, Titus T, Postlethwait J, Moens CB. Hoxb1b controls oriented cell division, cell shape and microtubule dynamics in neural tube morphogenesis. Development. 2014;141(3):639–49. doi: 10.1242/dev.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferreira JG, Pereira AL, Maiato H. Microtubule plus-end tracking proteins and their roles in cell division. Int Rev Cell Mol Biol. 2014;309:59–140. doi: 10.1016/B978-0-12-800255-1.00002-8. [DOI] [PubMed] [Google Scholar]
- 67.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19(1):31–5. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 68.Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17(6):488–98. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 69.Tanenbaum ME, Galjart N, van Vugt MA, Medema RH. CLIP-170 facilitates the formation of kinetochore-microtubule attachments. EMBO J. 2006;25(1):45–57. doi: 10.1038/sj.emboj.7600916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wade RH. On and around microtubules: an overview. Mol Biotechnol. 2009;43(2):177–91. doi: 10.1007/s12033-009-9193-5. [DOI] [PubMed] [Google Scholar]
- 71.Wood KW, Cornwell WD, Jackson JR. Past and future of the mitotic spindle as an oncology target. Curr Opin Pharmacol. 2001;1(4):370–7. doi: 10.1016/s1471-4892(01)00064-9. [DOI] [PubMed] [Google Scholar]
- 72.Scheffler K, Tran PT. Motor proteins: kinesin can replace Myosin. Curr Biol. 2012;22(2):R52–4. doi: 10.1016/j.cub.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 73.Watanabe T, Noritake J, Kaibuchi K. Regulation of microtubules in cell migration. Trends Cell Biol. 2005;15(2):76–83. doi: 10.1016/j.tcb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 74.Horwitz R, Webb D. Cell migration. Curr Biol. 2003;13(19):R756–9. doi: 10.1016/j.cub.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 75.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84(3):359–69. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 76.Slep KC, Rogers SL, Elliott SL, Ohkura H, Kolodziej PA, Vale RD. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J Cell Biol. 2005;168(4):587–98. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Drabek K, van Ham M, Stepanova T, Draegestein K, van Horssen R, Sayas CL, Akhmanova A, Ten Hagen T, Smits R, Fodde R, Grosveld F, Galjart N. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol. 2006;16(22):2259–64. doi: 10.1016/j.cub.2006.09.065. [DOI] [PubMed] [Google Scholar]
- 78.Yue J, Zhang Y, Liang WG, Gou X, Lee P, Liu H, Lyu W, Tang WJ, Chen SY, Yang F, Liang H, Wu X. In vivo epidermal migration requires focal adhesion targeting of ACF7. Nat Commun. 2016;7:11692. doi: 10.1038/ncomms11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng J, Zhou L, Xie QF, Xie HY, Wei XY, Gao F, Xing CY, Xu X, Li LJ, Zheng SS. The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells. Proteomics. 2010;10(8):1557–72. doi: 10.1002/pmic.200900646. [DOI] [PubMed] [Google Scholar]
- 80.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145(1):61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 81.Moffat JJ, Ka M, Jung EM, Kim WY. Genes and brain malformations associated with abnormal neuron positioning. Molecular brain. 2015;8(1):72. doi: 10.1186/s13041-015-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan SS, Kalloniatis M, Sturm K, Tam PP, Reese BE, Faulkner-Jones B. Separate progenitors for radial and tangential cell dispersion during development of the cerebral neocortex. Neuron. 1998;21(2):295–304. doi: 10.1016/s0896-6273(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 83.Evsyukova I, Plestant C, Anton ES. Integrative mechanisms of oriented neuronal migration in the developing brain. Annu Rev Cell Dev Biol. 2013;29:299–353. doi: 10.1146/annurev-cellbio-101512-122400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sultan KT, Brown KN, Shi SH. Production and organization of neocortical interneurons. Front Cell Neurosci. 2013;7:221. doi: 10.3389/fncel.2013.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson SA, Marin O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128(3):353–63. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- 86.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8(6):427–37. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 87.Gleeson JG, Walsh CA. Neuronal migration disorders: from genetic diseases to developmental mechanisms. Trends Neurosci. 2000;23(8):352–9. doi: 10.1016/s0166-2236(00)01607-6. [DOI] [PubMed] [Google Scholar]
- 88.Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11(5):316–28. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cerebral cortex. 2000;10(10):981–91. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 90.Wegiel J, Kuchna I, Nowicki K, Imaki H, Wegiel J, Marchi E, Ma SY, Chauhan A, Chauhan V, Bobrowicz TW, de Leon M, Louis LA, Cohen IL, London E, Brown WT, Wisniewski T. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119(6):755–70. doi: 10.1007/s00401-010-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feng Y, Walsh CA. Protein-protein interactions, cytoskeletal regulation and neuronal migration. Nat Rev Neurosci. 2001;2(6):408–16. doi: 10.1038/35077559. [DOI] [PubMed] [Google Scholar]
- 92.Bielas SL, Gleeson JG. Cytoskeletal-associated proteins in the migration of cortical neurons. J Neurobiol. 2004;58(1):149–59. doi: 10.1002/neu.10280. [DOI] [PubMed] [Google Scholar]
- 93.Xie Z, Samuels BA, Tsai LH. Cyclin-dependent kinase 5 permits efficient cytoskeletal remodeling–a hypothesis on neuronal migration. Cerebral cortex. 2006;16(Suppl 1):i64–8. doi: 10.1093/cercor/bhj170. [DOI] [PubMed] [Google Scholar]
- 94.da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3(9):694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- 95.Tsaneva-Atanasova K, Burgo A, Galli T, Holcman D. Quantifying neurite growth mediated by interactions among secretory vesicles, microtubules, and actin networks. Biophys J. 2009;96(3):840–57. doi: 10.1016/j.bpj.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tian D, Diao M, Jiang Y, Sun L, Zhang Y, Chen Z, Huang S, Ou G. Anillin Regulates Neuronal Migration and Neurite Growth by Linking RhoG to the Actin Cytoskeleton. Curr Biol. 2015;25(9):1135–45. doi: 10.1016/j.cub.2015.02.072. [DOI] [PubMed] [Google Scholar]
- 97.Belliveau DJ, Bani-Yaghoub M, McGirr B, Naus CC, Rushlow WJ. Enhanced neurite outgrowth in PC12 cells mediated by connexin hemichannels and ATP. The Journal of biological chemistry. 2006;281(30):20920–31. doi: 10.1074/jbc.M600026200. [DOI] [PubMed] [Google Scholar]
- 98.Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21(1):165–76. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Penzes P, Rafalovich I. Regulation of the actin cytoskeleton in dendritic spines. Adv Exp Med Biol. 2012;970:81–95. doi: 10.1007/978-3-7091-0932-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shirao T, Gonzalez-Billault C. Actin filaments and microtubules in dendritic spines. J Neurochem. 2013;126(2):155–64. doi: 10.1111/jnc.12313. [DOI] [PubMed] [Google Scholar]
- 101.Okamoto K, Nagai T, Miyawaki A, Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nature neuroscience. 2004;7(10):1104–12. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- 102.Koleske AJ. Molecular mechanisms of dendrite stability. Nat Rev Neurosci. 2013;14(8):536–50. doi: 10.1038/nrn3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim WY, Snider WD. Functions of GSK-3 Signaling in Development of the Nervous System. Frontiers in molecular neuroscience. 2011;4:44. doi: 10.3389/fnmol.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim WY, Wang X, Wu Y, Doble BW, Patel S, Woodgett JR, Snider WD. GSK-3 is a master regulator of neural progenitor homeostasis. Nature neuroscience. 2009;12(11):1390–7. doi: 10.1038/nn.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou FQ, Zhou J, Dedhar S, Wu YH, Snider WD. NGF-induced axon growth is mediated by localized inactivation of GSK-3beta and functions of the microtubule plus end binding protein APC. Neuron. 2004;42(6):897–912. doi: 10.1016/j.neuron.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 106.Kim WY, Zhou FQ, Zhou J, Yokota Y, Wang YM, Yoshimura T, Kaibuchi K, Woodgett JR, Anton ES, Snider WD. Essential roles for GSK-3s and GSK-3-primed substrates in neurotrophin-induced and hippocampal axon growth. Neuron. 2006;52(6):981–96. doi: 10.1016/j.neuron.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tamminga CA, Holcomb HH. Phenotype of schizophrenia: a review and formulation. Molecular psychiatry. 2005;10(1):27–39. doi: 10.1038/sj.mp.4001563. [DOI] [PubMed] [Google Scholar]
- 108.Steinecke A, Gampe C, Valkova C, Kaether C, Bolz J. Disrupted-in-Schizophrenia 1 (DISC1) is necessary for the correct migration of cortical interneurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32(2):738–45. doi: 10.1523/JNEUROSCI.5036-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nature cell biology. 2005;7(12):1167–78. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 110.Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF, Murdoch H, Dunlop AJ, Kubo K, Furukori K, Huang B, Zeledon M, Hayashi-Takagi A, Okano H, Nakajima K, Houslay MD, Katsanis N, Sawa A. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473(7345):92–6. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, Liu CY, Ganesan S, Cheng HJ, Ming GL, Lu B, Song H. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130(6):1146–58. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, Sawa A. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nature neuroscience. 2010;13(3):327–32. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Molecular psychiatry. 2005;10(1):40–68. doi: 10.1038/sj.mp.4001558. image 5. [DOI] [PubMed] [Google Scholar]
- 114.Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, Bonnert TP, Whiting PJ, Brandon NJ. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Molecular psychiatry. 2007;12(1):74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 115.Feng J. Microtubule: a common target for parkin and Parkinson’s disease toxins. Neuroscientist. 2006;12(6):469–76. doi: 10.1177/1073858406293853. [DOI] [PubMed] [Google Scholar]
- 116.Simunovic F, Yi M, Wang Y, Macey L, Brown LT, Krichevsky AM, Andersen SL, Stephens RM, Benes FM, Sonntag KC. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain. 2009;132(Pt 7):1795–809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]


