Abstract
Since blood borne circulating tumour cells (CTCs) initially shed from the primary tumour can seed and initiate metastasis at distant sites a better understanding of the biology of CTCs and their dissemination could provide valuable information that could guide therapeutic intervention and real time monitoring of disease progression. Although CTC enumeration has provided a reliable prognostic readout for a number of cancers, including lung cancer, the precise clinical utility of CTCs remains to be established. The rarity of CTCs together with the vanishingly small amounts of nucleic acids present in a single cell as well as cell to cell heterogeneity has stimulated the development of a wide range of powerful cellular and molecular methodologies applied to CTCs. These technical developments are now enabling researchers to focus on understanding the biology of CTCs and their clinical utility as a predictive and pharmacodynamics markers. This review summarises recent advances in the field of CTC research with focus on technical and biological challenges as well the progress made towards clinical utility of characterisation of CTCs with emphasis on studies in lung cancer.
Keywords: Circulating tumour cells (CTCs), lung cancer, liquid biopsy, biomarkers, metastasis
Introduction
Circulating tumour cells (CTCs) were identified and first proposed as part of the fundamental process of cancer dissemination in the mid-19th century by the Australian pathologist, Thomas Ashworth (1). Since more than 90% of cancer- related deaths are thought to be due to metastatic disease (2,3), it is not surprising that the presence of CTCs indicates metastatic disease is generally linked with poor prognosis (4,5) and dissemination of tumour cells starts at early stages of disease progression (5,6). However, the mechanism of dissemination and the accompanying biology of CTCs are still poorly understood.
In this review, we will present our view of the technical challenges of working with CTCs as well as some of the key biological and clinical findings including the potential use of CTCs in the clinical management of lung cancer including their use as prognostic, pharmacodynamic and predictive biomarkers. We will reference CTC studies across all cancer types but will highlight studies addressing lung cancer CTCs. Given the broad scope of CTC research and the large numbers of CTC publications we cannot provide an exhaustive coverage of the subject so for those requiring more detail we would recommend recent excellent CTC reviews (7-9).
Technical challenges
The biggest challenge for CTC research is the rarity of CTCs which is estimated at a level of one CTC per 106–107 white blood cells (WBCs). Detecting and isolating rare CTCs from the bloodstream requires sensitive technologies that can efficiently pick out these cells amongst the millions of other blood cells. Several of the key technical challenges that face CTC researchers and potential solutions are addressed in the following section.
Identification and isolation of CTCs
There are three broad basic strategies for CTC enrichment, identification and analysis from whole blood: (I) negative selection through removal of blood cells thereby enriching the remaining CTCs; (II) positive selection of CTCs; and (III) analyse all cells accompanied by CTC identification through intensive image analysis. Each of these strategies has their own strengths and weaknesses so the choice of approach should be guided primarily by the clinical/biological aim of the study as well as pragmatic considerations such as cost and time available.
Many negative selection approaches are based on the availability of antibodies recognising blood specific cell surface epitopes and the fact that these epitopes are not widely expressed in cancer cells. The attraction of this approach is that by targeting the unwanted cells, in this case red blood cells (RBCs) and WBCs, the CTCs are left untouched and are less likely to be influenced or damaged by the enrichment approach. The commercial RosetteSep™ kit (StemCell Technologies) provides a simple method to remove the bulk of mature blood cells including both RBCs and WBCs and has been shown to leave behind viable CTCs that can be cultured (10,11) or implanted into mice and grown as tumours (12-14). Recently, a new enhanced negative depletion strategy named Multi-marker Immuno-magnetic Negative Depletion Enrichment of CTCs (MINDEC) has been developed where blood cell fraction is isolated by density gradient centrifugation followed by the use of superparamagnetic beads coupled with a cocktail of antibodies against different blood classes (15). Although negative selection approaches have proven useful, due to the overwhelming numbers of WBCs compared to CTCs, the output from negative selection is often heavily contaminated with WBCs which may interfere with downstream CTC based assays.
For positive selection of CTCs there are two major approaches based either on physical properties such as size, density and deformability or biological properties such as cell surface epitopes. The most successful and widely used CTC technology is the FDA recognised CellSearch® system (Jannsen Diagnostics, Raritan, NJ, USA) which is a semi-automated platform that employs ferromagnetic beads coated with the epithelial cell surface marker, EpCAM to “pull out” or enrich epithelial CTCs (4). Following the initial CTC enrichment CellSearch® utilises automated fluorescence staining to identify and enumerate CTCs based on their cell/nuclear morphology along with detectable expression of epithelial cytokeratins and low expression of the WBC marker CD45 (4). Since CellSearch® has been shown to be both reliable and reproducible it has been widely used in multi-center trials and CellSearch® CTC counts have been shown to be prognostic for metastatic breast (4), lung (16), colorectal (CRC) (17) and prostate cancer (18). A key feature of the CellSearch® platform which led to its widespread uptake has been the use of the CellSave® preservative blood collection tube (Jannsen Diagnostics, Raritan, NJ, USA) which allows whole blood to be stored or shipped at room temperature for up to 96 h prior to analysis (4,17). Other systems have been described which also employ an EpCAM-based enrichment technologies including the CTC-Chip (19), MagSweeper (Stanford University, USA) (20), iCHIP (Massachussets General Hospital Center, USA) (21) and GILUPI CellCollector® (GILUPI Nanomedizin, Germany) (22) (Table 1) Since EpCAM-based enrichment technologies are limited to epithelial CTC detection alternative strategies are required for detection of CTCs which are not epithelial. For example, CellSearch® has developed a melanoma tailored enrichment and staining kit, where melanoma CTCs are captured via the CD146 cell surface marker (also known as melanoma cell adhesion molecule or MCAM) and positively stained for the high molecular weight melanoma associated antigen (HMW-MAA) while CD45 and CD34 are used as WBC markers (23).
Table 1. Technologies for enrichment and isolation of CTCs from a blood sample.
| CTC isolation technologies | Approach | Advantages | Disadvantages |
|---|---|---|---|
| Antibody-dependent enrichment | |||
| CellSearch® | EpCAM-coated ferromagnetic beads | FDA clearance and clinical relevance in many cancer types; semi-automated; visual confirmation of CTCs; highly reproducible and automated | EpCAM positivity dependent; expensive; operator dependent visual confirmation; sample volume analysable (7.5 mL) |
| MagSweeper | Flow through EpCAM based immunomagnetic capture | Automated; high purity; can process whole blood, possibility of live cell capture | EpCAM positivity dependent; expensive; sample volume analysable (7.5 mL) |
| GILPUI CellCollector® | EpCAM-coated Seldinger guide wire into the cubital vein | Allows for analysis of all peripheral blood in an individual | EpCAM positivity dependent; invasive (minimally) |
| IsoFlux | EpCAM-coated magnetic beads combined with microfluidic processing | Automated; Enriched cells can be recovered in low volume (3 µL) and hence suitable for downstream molecular analysis | Sample volume analysable (7.5 mL) |
| Herringbone chip | Microfluidic device consisting of EpCAM-coated microposts | Visual confirmation of CTCs; potential for further staining using immunofluorescence-conjugated antibodies; high detection rate; potential for detection of CTMs | EpCAM positivity dependent; operator dependent visual confirmation; sample volume analysable (7.5 mL) |
| HD-SCA | Peripheral blood mononuclear cells (PBMCs) are applied onto a slide and subsequently stained using markers of choice | Potential for tailoring a selection of antibodies; visual confirmation of CTCs with automated digital microscopy; potential for downstream molecular analysis | Time consuming due to the number of slides required to be scanned; requires additional instrumentation for cell collection |
| RareCyte | PBMCs are applied onto a slide and subsequently stained using markers of choice | Potential for tailoring a selection of antibodies; visual confirmation of CTCs; potential for downstream molecular analysis; plasma fraction of the blood sample is removed and replaced with buffer, hence potential for ctDNA analysis | Time consuming due to the number of slides required to be scanned; requires additional instrumentation for cell collection |
| Physical properties based enrichment | |||
| Parsortix™ | Cell size and deformability based enrichment | Label free approach; avoids staining protocols; broad population of CTCs enriched; plasma fraction of the blood sample is removed and replaced with buffer, hence potential for ctDNA analysis | WBC contamination as enrichment is purely size based; sample volume analysable (7.5 mL) |
| ISET® | Filtration based on cell size based | Label free approach; avoids staining protocols; broad population of CTCs enriched | WBC contamination as enrichment is purely size based; pre-RBC lysis required |
| ClearCell® | Microfluidic approach based on cell size, deformability and inertia | Label free approach; avoids staining protocols; broad population of CTCs enriched | WBC contamination as enrichment is purely size based; pre-RBC lysis required |
| CTC iChip | Size-based separation followed by negative and positive selection using magnetic beads | Enrichment of broad population of CTCs by positive selection by EpCAM or negative selection by CD45; | Immuno-affinity based selection; require a clean room and manual labor, and expensive deep reactive ion etching procedures; current manufacturing process not compatible for clinical studies; sample volume analysable (7.5 mL) |
In this table we summarise technologies used for isolation of CTCs using antibody-dependent and physical property-dependent enrichment technologies. CTCs, circulating tumour cells; ctDNA, circulating tumour DNA.
Although positive selection utilising cell surface markers has been very successful it is also clear that there are CTCs which change or lack these markers (see further discussion regarding EMT below) and this has prompted the development of marker-independent CTC enrichment platforms. Marker-independent CTC enrichment platforms are generally devices which retain CTCs on the basis of physical properties such as size (ISET®, Rarecells Diagnostics, France) (24), cell size and deformability (Parsortix™ Cell Separation, ANGLE plc, UK) (25,26) (Table 1) and flow properties (ClearCell®, Clearbridge Biomedics, Singapore) (27) (Table 1). Captured CTCs can be then recovered for further molecular analysis, however CTCs may not be always larger and stiffer than WBCs, thus can be washed away through the filters (28).
More recently, imaging and scanning technologies have been developed to a point where CTCs can be visualised, identified and isolated without any prior enrichment (8). A number of integrated systems have been described for the interrogation of whole blood including high definition single cell analysis (HD-SCA) (29) and the RareCyte system (RareCyte, Seattle, WA, USA) (30) (Table 1). Both are based on the principle of “no cell left behind” where all nucleated blood cells are deposited on microscope slides and stained with markers allowing discrimination of blood and non-blood cells thereby increasing the possibility to detect rare CTCs subpopulations (8). An additional feature for both the HD-SCA and RareCyte® platforms is that, once candidate CTCs have been identified, they have an integrated capacity to isolate target cells directly from the slide in a format that is compatible with single cell molecular analysis. In contrast, most CTC enrichment technologies require an additional step for CTC visualisation and isolation. This has been achieved in a variety of ways including fluorescence-activated cell sorting (FACS) (31), CTC visualisation followed by isolation via micromanipulation (21,32-34), and the DEPArray™ automated system which incorporates cell manipulation via electrostatic charge (35,36) (Table 1).
CTC molecular analysis
The striking increase in the sensitivity of DNA and RNA technologies have been pivotal for CTC research since they have enabled researchers to confirm the tumour identity of cells identified by morphology or tissue specific surface markers by establishing that they harbour tumour specific molecular signatures. Early observations generally using targeted sequencing or PCR demonstrated the presence of tumour specific mRNAs or DNA changes in either whole blood or samples enriched for CTCs. Stathopoulou et al. developed an assay for detection of cytokeratin-19 (CK-19) mRNA in peripheral blood of patients with breast cancer (37-39). Similarly, in CRC a panel of genes including carcinoembryonic antigen (CEA), CK-19, CK-20 and CD133 has been used to verify the presence of colorectal tumour cells in enriched blood samples (40). However, any study carried out on enriched CTC populations will inevitably suffer from reduced sensitivity through the effective dilution caused by varying levels of contaminating WBCs.
Over the last decade, advances in both CTC enrichment protocols and molecular methodologies has culminated in a range of successful strategies for genome wide amplification and analysis of single CTCs which have extended many of the observations made with enriched CTCs. There are several advantages of single CTC analysis including: (I) single CTC analysis is 100% pure (assuming a CTC was isolated) and avoids any complications or dilution of signal due to contaminating cells; (II) since a cell has on average only two DNA copies of each gene and around 100,000 mRNA molecules (41) the degree of next generation sequencing (NGS) required for generating a representative single cell snapshot is relatively low; (III) single cell genomic analysis can unambiguously identify co-existing genetic changes; (IV) the integration of RNA and DNA analysis from the same cell can establish linkage between the genetic alterations and alterations in RNA pathways (42); (V) single cell data provides an ideal means of establishing the degree of heterogeneity and estimating tumour evolution (43,44). For single CTC molecular analysis the main problems encountered are largely technical, financial and logistical. Since the amplification process necessary for single CTC analysis is extremely sensitive it is critical that steps are taken to avoid or identify potential contamination such as defining separate clean areas and use of negative controls.
Given the vanishingly small amounts of DNA (6.6 pg) present in a single cell, whole genome amplification (WGA) is a pre-requisite for extensive genomic analysis of CTCs. The widespread use of CTC WGA was pioneered by Christoph Klein and co-workers through the establishment of a simple single tube amplification system (available as the AMPLI1™ WGA kit, Silicon Biosystems) based on restriction enzyme digestion, ligation of primer and subsequent PCR amplification (45). There are now many commercially available single cell WGA systems to choose from generally based on PCR (46,47) or linear amplification (48-50). There are obvious pros and cons for each WGA system (51) and the choice of which WGA system to use needs to be based on the clinical/scientific aim of the study, whether the WGA system is compatible with the CTC enrichment process used and the resources available.
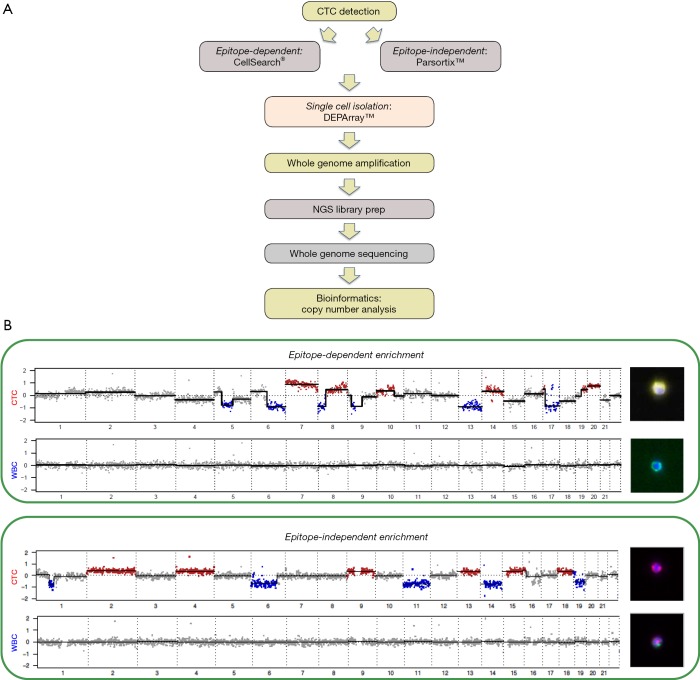
Following WGA of CTCs one of the most widely used approach for CTC molecular analysis has been establishing genome copy number alterations (CNA) which provides a simple snapshot of each CTC which will allow comparisons between CTCs and, when available, the matching tumour [Figure 1 provides examples of non-small cell lung cancer (NSCLC) CTC CNA profiles]. Initial single cell studies compared targeted mutation profiles and copy number profiles of CTCs with the primary tumours and metastases of the corresponding patients. One such study of circulating melanoma cells using single cell comparative genomic hybridisation (single-cell CGH) revealed multiple CNA consistent with melanoma which was in sharp contrast to the normal cells that revealed balanced CGH profiles (52). In addition, the use of hierarchical clustering provided a means of establishing linkages between each cell and suggested a clonal origin of the melanoma cells examined (52).
Figure 1.
Determining CTC copy number alterations (CNA). (A) Shows a schematic outline of the CTC NGS analysis workflow used. (B) Presents genome wide CNA profiles and accompanying images of single CTCs and WBCs isolated by epitope dependent and independent technologies from two patients with NSCLC. For all four cells the CNA pattern is presented in a linear format with marked copy number increases shown in red and losses in blue alongside the fluorescent stained images of the starting cells. The top panel presents the CNA patterns and images of a NSCLC cytokeratin (CK) positive, CD45 negative CTC and a CK negative, CD45 positive WBC obtained following CellSearch® EpCAM enrichment and subsequent CK and CD45 staining with CK staining depicted as yellow, CD45 as green and nuclear staining (DAPI) as blue. The bottom panel presents the CNA patterns and images of a NSCLC vimentin (VIM) positive, CD45/CD31 negative CTC and a VIM positive, CD45/CD31 positive WBC obtained following Parsortix™ size enrichment and subsequent VIM plus combined CD45/CD31 staining with VIM staining depicted as red, CD45/CD31 as green and nuclear staining (DAPI) as blue. CTC, circulating tumour cell; NGS, next generation sequencing; NSCLC, non-small cell lung cancer.
For de novo detection of variants in single cells there is a profound challenge of how to distinguish the artefactual “variants” that accrue during the million fold or more amplification during WGA from the actual mutations present in the starting cell. Although it is difficult to be completely certain of true biological origin of variants called in a single cell or CTC, the use of suitable controls, technical replicates, biological replicates and bioinformatics can be used to establish the extent of amplification errors (53). It may be possible to reduce the impact of amplification errors by incorporating molecular bar codes (54) or using low error amplification processes (53,55) but it is not yet clear how much benefit these strategies will bring. For the moment the most effective means of identifying true variants in CTCs is the use of biological replicates, i.e., identify variants seen in two or more independent CTCs (56).
In addition to the advances in genomic analysis of CTCs there have been similar exciting advances in CTC RNA profiling and particularly in the plethora of single cell RNA-Seq methods that have emerged in the last 5 years (57). The attraction of genome-wide RNA profiling of CTCs is that it can give a broad picture of the biological activity of the cell including pathway activation and the presence of expressed drug targets. Transcriptomic profiles of CTCs can be viewed as an emergent property of all of the genetic and epigenetic changes within the cell and as such may be easier to characterise than the often complex genomic changes seen in CTCs. The ability to carry out effective single CTC RNA profiling is largely driven by CTC enrichment protocols and the impact they have on the RNA within the final enriched cell. Since RNA is chemically more labile than DNA and subject to enzymatic degradation it is clear that CTC enrichment and identification strategies that work well for DNA isolation and genomic analysis may not be suitable for RNA analysis. For example, the CellSearch® platform that utilises the CellSave® preservative blood tube works well for DNA but it is clear that both the preservative and the downstream cell processing steps have a negative impact on RNA quality. For this reason most genome wide CTC RNA profiling methods have avoided the use of preservative and have isolated viable cells suitable for direct lysis and processing. A number of groups have now described a broad range of effective genome wide and targeted RNA approaches applied to either enriched pools or single CTCs (36,58,59). Transcriptomic profiling of CTCs is now providing valuable insights into CTC biology (see discussion below) and further advances in methodologies such as combined RNA/DNA SCA (42) and droplet based analysis of many 1000s of cells (60) will provide an even greater depth of understanding.
Blood sampling considerations
Since CTCs are generally very rare it is also important to consider the volume of blood sampled and the implications this has for effective CTC identification and isolation (61). Fischer et al. in 2008 demonstrated the possibility of combining cell detection approach such as CellSearch® along with diagnostic leukapheresis (DLA) to increase the possibility of detecting CTCs (62). In this study, they increased the CTC detection rate in patient samples from a range of cancer types by 44% by using DLA followed by CellSearch® as compared to using 7.5 mL of blood on CellSearch®. DLA is a clinically safe approach that enables detection of rare CTCs from larger volume of blood. The other approach that can increase effective blood sampling volumes is the GILUPI CellCollector® which introduces a EpCAM-coated Seldinger guide wire into the cubital vein allowing CTC enumeration and isolation from the entire circulating blood in an individual patient (22).
Understanding CTC biology
Having established an impressive tool kit for isolating and analysing CTCs recent research is now beginning to unravel the complex biology of CTCs. In addition to examining the relationship between CTCs, primary tumour and metastasis it is important to establish that CTCs are themselves tumourigenic and if properties of CTCs can shed light on the process of metastatic spread. Two key hallmarks of the malignant tumour cells responsible for the initiation of the metastatic process are motility and invasiveness (63) and it is likely that there will be features in CTCs that reflect these characteristics. The dissemination of cells from the primary tumour and resultant metastatic seeding may well start early even at stages where primary tumour is thought to be localised with no overt metastases (5,64-66). Tumour cells that enter the circulatory system to become CTCs do so through a process known as intravasation and the rate of intravasation from solid tumours in patients is largely unknown although several clinical and animal model studies suggest that millions of tumour cells are shed even from small tumours (67). Although most cells that intravasate die (68,69), a small proportion of cells exit the vasculature to enter organs, a process known as extravasation and colonise distant organs. CTCs as wells as their pre-cursors and descendants must overcome many obstacles such as immune surveillance and physical/cellular barriers in order to survive in the circulatory system and to colonise the host organ (70,71). The biological processes associated with propagation, survival and subsequent colonisation of organs distant to the primary tumour are discussed in the following section.
Tumour and CTC heterogeneity
An early model of clonal evolution linked to the generation of genetic diversity in tumours was proposed in 1976 (72) and the emergence of intra-tumour heterogeneity was confirmed experimentally in murine models (73). More recently, NGS technologies have been used to understand the breadth and depth of this overwhelming genetic diversity in primary and the metastatic tumours (74-79).
NGS and bioinformatic strategies have been developed and applied to tumour tissue, which have allowed both a measure of heterogeneity and the generation of phylogenetic models which provide a picture of the inferred development and branched evolution of the tumour (78,80-82). Advances in NGS technologies now allow routine analysis of single cells or nuclei and this has been fruitfully applied to dissociated tumour cells to provide unambiguous detection of tumour sub-clones (81,83-86). The picture that has emerged from both whole tissue and single cell tumour analysis is one of considerable intra and inter patient tumour heterogeneity that is most likely contributing to metastatic spread and the emergence of therapy resistance. Studies in animal models have provided evidence that this cellular and molecular heterogeneity is matched by functional heterogeneity whereby only a subset of cells, often known as tumour initiating cells or tumour stem cells, are capable of initiating tumours in solid tumours such as colon (87,88), pancreas (89), melanoma (90), liver (91) and brain (92).
Disseminated tumour cells (DTCs) provide another fascinating view of cellular heterogeneity since in some malignancies, notably in breast cancer, DTCs are often detectable in apparently healthy organs such as bone marrow and can reside there for many years after surgical removal of the primary tumour until signs of overt malignant growth (93,94). Bone marrow can function as a reservoir for DTCs (71,95) not only for breast cancer but also for other malignancies such as lung cancer and CRC which do not usually metastasise in the bone (65,96). It has been proposed that in the vast majority of cancer patients, DTCs can enter state of proliferative quiescence, process known as cellular dormancy which can represent the main reason why DTCs evade systemic therapy persisting as minimal residual disease (71,97,98). Genetic and transcriptomic characterisation of DTCs could help understand the nature, origin and biology of this cancer cell type in the individual patients and could subsequently provide extremely valuable information of the future course of the patients’ disease. In breast and prostate cancer, DTCs were found with fewer copy number changes compared to the primary tumour, suggesting that the dissemination may happen at an early tumour stage (99). In contrast, Demeulemeester et al. demonstrated by single cell sequencing of DTCs isolated from the bone marrow of breast cancer patients that they originate from either the main primary tumour clone, primary tumour subclones or subclones in an axillary lymph node metastasis, indicating that breast cancer dissemination may arise relatively late in tumour evolution (64).
Although several groups have also reported the presence of DTCs in the bone marrow of patients with localised NSCLC, and shown a correlation with worse clinical outcome (65,100,101) these studies included relatively small numbers of patients (particularly in comparison to breast cancer) making it unclear to what extent NSCLC is driven by DTCs. DTCs were also detected in the bone marrow of 18% of patients with SCLC using a RT-PCR approach to detect levels of gastrin-releasing peptide, which is preferentially expressed in SCLC (102). However, given the rapid aggressive nature of SCLC and the high numbers of CTCs observed in this disease it is not clear if there is a role for a long term DTCs located in the bone marrow.
Although it is likely that the first CTCs arise from the primary tumour and play a role in metastasis, once metastasis are established they can also be a source of CTCs. In one SCLC study CNA and mutational profiles were established from CTCs, primary tumor as well metastasis and the comparison indicated that there was more overlap between CTCs and metastasis than between CTCs and the primary tumor (103). Given the aggressive nature of SCLC it is not surprising that metastasis can also contribute to CTC based spread and further studies are required to establish if CTC contribution from developing metastasis is a general feature of all cancers. A CRC study revealed similar results with CNA and mutational profiles of CTCs presenting with more overlap with liver metastases than the primary tumour (43).
CTC-derived tumour xenografts (CDX)
One of the major challenges for CTC research is to establish if there is any functional capacity of CTCs and trace this back to molecular changes that are shared by CTCs and tumour. Recently it has been shown that CTCs isolated from patient blood can form tumours when introduced into immune compromised mice firmly establishing that CTCs are viable and capable of forming tumours (12-14). In a study investigating breast cancer, CTCs isolated from 110 patients were transplanted into the femoral medullar cavity of immunocompromised mice and three patient-derived CTCs were able to form multiple metastases (104). In a SCLC CDX study it was also established that CTCs from both patients who responded to chemotherapy (chemosensitive) and CTCs from patients who did not (chemorefractory) were capable of forming tumours in immunocompromised mice and that when mice harbouring CDXs were treated with platinum and etoposide the response seen mirrored the response observed in the corresponding patient (12). In the same SCLC study, genomic analyses revealed that CellSearch® CTCs and the matching CTC-derived CDX are highly related in terms of both copy number and TP53 and RB1 mutations (12). CTCs were also shown to be tumorigenic in melanoma, where CDXs were established with a success rate of 13% and they had similar metastatic tropism as the corresponding patient (14).
Epithelial-to-mesenchymal transition (EMT)
The key feature of EMT relevant to CTC biology is the switch from relatively restricted or anchored epithelial cell to a more motile flexible mesenchymal cell type (105). EMT was first coined by embryologists and describes cell migration from the ectoderm during the gastrulation process to produce the mesoderm and is now also referred to as type I EMT (106,107). A similar process was also observed in wound healing and fibrosis and this is referred to as type II EMT (108). For CTCs it has been proposed that EMT facilitates intravasation from primary epithelial tumour through the switch to a mesenchymal phenotype associated with increased cell motility and migration (105,109). However, histopathological examination of metastatic tumours reveals a predominantly epithelial tumour and relatively fewer mesenchymal characteristics (110) indicating that, for CTCs, there is a reciprocal mesenchymal-to-epithelial transition (MET) associated with extravasation and reestablishment of an epithelial metastasis (5,109,111,112).
During EMT there is a loss or redistribution of the epithelial adhesion molecule, E-cadherin, a trans-membrane glycoprotein that is responsible for the tight junction between epithelial cells and integral to apical-basal polarity (111). Along with the modulation of cell-cell contact markers, EMT is also associated with remodelling of the cytoskeleton with an increase in vimentin-rich intermediate filaments providing a more flexible mesenchymal cellular structure as opposed to the more rigid cytokeratin-rich starting point (113). The process of EMT can be triggered by paracrine signalling of several factors such as TGF-beta, WNT or interleukin-6 (105,114,115) and these triggers in turn activate transcription factors such as Snail, Twist and Zeb that maintain the mesenchymal phenotype by autocrine signalling (115). These EMT associated alterations have also been observed in CTC sub-populations in breast cancer by single cell RNA-seq analysis (116). Determining the biological relevance of EMT for CTC mediated metastatic spread is confounded by the fact that EMT and MET are processes that occur over time (it is not clear how long either process will be) and that metastatic spread may occur either continually or sporadically. In this context, several studies have reported identification of CTCs with mesenchymal markers in patients with advanced stage of breast cancer as compared to patients with localised breast cancer (117). To get a clearer picture of the relevance of CTC biology including EMT it is possible to take advantage of the simplicity of blood sampling and establish multiple longitudinal blood samples to build a picture of CTC biology during the course of disease progression. Yu et al. applied this strategy to monitor breast cancer CTCs and identified emergence of a mesenchymal CTC phenotype in samples from patients undergoing chemotherapy when compared to samples taken at baseline (116). This study utilised RNA in situ hybridization to detect epithelial and mesenchymal genes and highlighted the role of TGF-beta activation along with the transcription factor FOXC1 in EMT in breast cancer CTCs. EMT and MET switching in CTCs can also poses a challenge for detecting the resultant mesenchymal CTCs since they are in a hematopoietic cell environment consisting of many mesenchymal cells this will be expanded on in the section summarising technical challenges.
One of the earliest reports of EMT in CTCs from patients with NSCLC compared CTC identification and enumeration using epithelial epitopes to enrich and stain CTCs (CellSearch®) to an epitope independent CTC identification system based on size selection (ISET®) and showed both increased CTC numbers as well as heterogeneous expression of EMT markers detected using the ISET® approach (112). Subsequently, other studies in SCLC (112), NSCLC (118), head and neck (119), breast (117,120) and prostate (120) cancer supported the observation of heterogeneous expression of mesenchymal and epithelial markers in CTCs. Although, in patients with NSCLC the CellSearch®-ISET® comparison revealed more NSCLC CTCs detected by ISET® than CellSearch® both systems provided CTC numerical readouts that were prognostic suggesting both EpCAM positive and negative CTCs may play a role in tumour spread (121).
A recent examination of blood samples from a patient with NSCLC revealed the presence of both cytokeratin positive CTCs expressing or not expressing the epithelial marker EpCAM suggesting that epithelial markers alone may not be sufficient for complete CTC enumeration (122). Interestingly, the authors went on to show only the EpCAM positive CTCs were associated with clinical outcome suggesting that, for this patient cohort, the EpCAM negative CTCs may not play a role in propagating the disease (122). Also, a study in a mouse model of SCLC identified a population of long term tumour propagating cells and this population presented with marked high expression of EpCAM and CD24 which is also prevalent in human primary SCLC tumours (123). Moreover, in the 47 attempts to generate CDX models (12), the 15 human blood samples that generated a mouse model also presented with more than 50 EpCAM positive CTCs in 7.5 mL suggesting that EpCAM positive CTC subpopulation that has tumour-initiating capacity (123).
There is an unmet requirement of comprehensive evaluation of markers associated with mesenchymal and epithelial characteristics that will allow assess the degree of EMT in different solid tumour types. This type of study has the potential to explain the low yield of EpCAM positive NSCLC CTCs on CellSearch® (5) and further highlights the need to combine both CTC enrichment technologies and analytical methods to more clearly understand the underlying CTC biology.
Vascular mimicry (VM)
In addition to CTCs adopting a mesenchymal phenotype which may facilitate intravasation it is also been recently reported that CTCs may also take on a vascular phenotype through a process known as VM (124). The term VM was first coined by Mary Hendrix and described the formation of tumour-derived blood vessels and it was hypothesised that this may provide an easy access for tumour cells to the bloodstream (125). VM was first described in human melanoma where it was shown that tumour cells shown to co-express endothelial and tumour markers and formed blood vessels (125) and further studies have given insights in VM induction across a variety of cancer types including breast, ovary, lung, prostate, bladder cancer (126). VM has been associated with tumour dissemination and metastasis (127) and in SCLC it has been shown that a subpopulation of CTCs co-express VE-cadherin (a VM marker) as well as epithelial cytokeratins indicating VM is involved in CTCs dissemination (124).
Circulating tumour microemboli (CTM)
Once CTCs arrive in the blood stream the next problem they face is to survive in the circulation. Reports of tumour cell clusters or CTM in the blood stream of patients with lung, colon, kidney and prostate cancer (16,128-130) have indicated that inclusion within a CTM may provide protection for CTCs particularly since the presence of CTMs is linked with poor prognosis (16,131). Circulating tumour emboli have been defined as groups of tumour cells either alone or associated with fibroblasts (132), leukocytes (133,134), endothelial cells (135-137) or platelets (138-140). There is growing evidence suggesting the co-operative behaviour exhibited by cells within CTMs can promote mutual survival and increased tumorigenic potential (16,33,141-144). It has also been suggested that interactions between blood cell and platelets can benefit both CTMs and single CTCs by protecting them from the host immune system (134,145-147).
Cellular cooperation has been elegantly demonstrated using a model derived from hamster oral keratinocytes of both mesenchymal and epithelial cells (148). At the subcutaneous site, injection of either pure population of cells formed tumours. However, only the mesenchymal cells entered the blood stream but neither cell type alone was capable of forming lung metastases. If pure populations of cells were injected intravenously, only the epithelial cell population formed lung metastases. When mixed population of cells were subcutaneously injected, although both cell types entered the blood stream the epithelial cells formed the lung metastases. This study shed light on the cooperation between cell types in transit which could be crucial in the metastatic cascade. In SCLC, a mouse model was used to examine the relevance and potential co-operation between neuroendocrine and mesenchymal tumour cells (144). The introduction of either cell type individually into immunocompromised mice resulted in the formation of local tumours without any liver metastasis. However, both cell types administered together resulted in formation of liver metastases along with the local tumour (144) and preformed CTMs injected into mice have a higher propensity to initiate metastases when compared to single cells suggesting a survival advantage of the CTMs (16,133,149).
Given the growing body of evidence it seems likely that CTMs play a pivotal role in tumour dissemination. However a number of questions remain unanswered regarding CTMs such as what is the role of cell heterogeneity (including blood cells) within CTMs and how do CTMs traverse narrow capillaries? Intriguingly, recent modelling using microscale devices, computational simulations and animal studies provides evidence that clusters can transit through capillaries by unfolding into single-file conga lines providing a potential explanation for how CTMs can travel throughout a patient’s body and seed distant sites (150).
CTCs and clinical utility
Along with helping us to understand the biology of primary and metastatic cancer, an examination of CTCs may also provide clinical benefit in the areas of: early detection; prognostic/predictive biomarkers; drug target identification; monitoring disease response/progression and identifying mechanisms of drug resistance. In this section, we address the potential clinical utility of CTCs.
Early detection
Early detection of disease is crucial for effective treatment of cancer. For example, SCLC is generally diagnosed when the disease is well advanced and treatment does not provide long term benefit. In general, the numbers of CTCs present in a 7.5 mL blood sample correlates with clinical staging; with the highest numbers seen in patients with metastases and lower numbers in patients without overt metastases (151,152). However, a recent study has demonstrated the predictive value of CTCs in early NSCLC where patients with chronic obstructive pulmonary disease (COPD) harboured detectable CTCs and then developed lung nodules 1–4 years later with four patients diagnosed with invasive adenocarcinoma and a fifth diagnosed with squamous cell carcinoma (153). However, in the same study there was a clear false positive rate since three patients who harboured detectable CTCs did not develop overt cancer (153). Clearly the potential for using CTCs in early detection needs to be further explored particularly using broader CTC detection systems in large screening programs.
Prognostic biomarkers
Prognostic biomarkers are indicators of the patient’s overall clinical outcome including progression free survival (PFS) and overall survival (OS) (154). With a number of emerging therapeutic options for the treatment of oligometastatic disease including local radiotherapy and immunotherapy that could potentially have an impact on the PFS in patients, the potential of CTCs as prognostic markers has been explored by a multitude of groups. The FDA have recognised the prognostic utility of the CellSearch® platform in many solid cancers such as breast (4), prostate (18) and CRC (17). The presence of more than 5 CTCs in breast (4) and prostate (18) and 3 CTCs in CRC (17) per 7.5 mL of blood correlates positively with shorter OS. This prognostic utility of CellSearch® CTCs has also been demonstrated for NSCLC where the CTC count cut-off is 5 CTCs per 7.5 mL of peripheral blood (5). However, SCLC is one of the solid tumours where the CTCs are more abundant than any other solid tumour thus far reported with a range of 0–44,896 cells per 7.5 mL blood and hence the corresponding CellSearch® CTC count cut-off for SCLC is set at a higher value of 50 CTCs per 7.5 mL blood (16). A NSCLC study using the ISET® CTC technology (CTCs enriched based on cell size) examined bloods from 208 patients with stages I–IV identified CTCs in 50% of patients (155). Although, this study found no correlation between the numbers of CTCs and the staging of the disease, it concluded that a threshold of >50 CTCs corresponded to shorter PFS and OS (155). However, a similar ISET® CTC study of 101 stage III/IV NSCLC patients showed a threshold of 5 CTCs in 7.5 mL blood corresponded with poor prognosis (5) indicating potential differences in the patients recruited to each study and also highlighting the need for standardisation of CTC calling.
The prognostic relevance of CTC clusters or CTMs has also been discussed in many studies. For example, CTMs were detected by ISET® technology in 43% of patients with stage IIIB/IV NSCLC (16) and in an independent study using HD-CTC assays CTMs were seen in 50% of patients with stage I–IV NSCLC (156) Both studies highlighted the prognostic value of CTMs. Furthermore, analysis of pulmonary vein blood in patients with NSCLC also revealed the existence of CTCs and CTMs in 33% of patients which predicted tumour recurrence and poorer disease-free survival (33,66).
Predictive biomarkers
Predictive biomarkers provide information on the likelihood of response to therapy and, in the clinic, can be used to support the selection of suitable personalised therapies (154). For example, over 64% of lung adenocarcinomas have an underlying driver mutation (157) and around 30% of these mutations are linked to an approved targeted therapy with the most common genetic alterations being EGFR mutations and translocations of ALK or ROS1 (158). Another potential for CTCs in personalised therapy is the longitudinal monitoring of therapy linked mutations as exemplified in a NSCLC study where an examination of 12 patients with NSCLC with EGFR mutated tumour revealed the same detectable EGFR mutations in CTCs in 11 patients (159). Similarly, a study by Marchetti et al. detected EGFR mutation in CTCs of 84% (31 out of 37 patients) EGFR mutant primary tumour (160). Several similar studies were able to reproduce similar results in detecting mutations in CTCs that matched the primary tumour and in cases where the mutations were not detected was likely due to the tumour heterogeneity between primary tumour and the metastatic tumours.
A number of studies have examined the feasibility of identifying rearrangements or translocations of ALK and ROS1 genes in CTCs as a means of guiding therapy (161,162). In one study fluorescence in-situ hybridisation (FISH) has been used to generate the filter-adapted FISH (FA-FISH) technique which was able to identify unique ALK rearrangement patterns in mesenchymal CTCs (105). It was suggested that the mesenchymal CTCs detected by FA-FISH could represent a highly invasive subpopulation with the potential to generate metastatic lesions (105). A similar FA-FISH approach was also used to detect both ROS1 translocations and CNA in NSCLC CTCs which correlated with poor response to crizotinib treatment (163).
In contrast to NSCLC, molecular analysis of SCLC has shown that the overall genomic landscape is chaotic with many mutations and widespread CNA but, despite the near ubiquitous inactivation of RB1 and TP53 frequent “actionable druggable” mutations have not been found (164,165). However, in a recent SCLC study a biomarker was developed based on CNA patterns detectable in single and pooled CTCs from 31 pre-treatment patients and was able to classify patients as either chemosensitive or chemorefractory to standard cisplatin etoposide with an accuracy of 83.3% (44). Interestingly, in the same study five patients who responded to treatment and relapsed with chemorefractory disease showed unaltered CTC CNA patterns at relapse indicating that mechanism of acquired chemoresistance differs from de novo chemoresistance (44).
Treatment response and mechanisms of resistance
Besides use of CTCs as prognostic or predictive markers, they have also been used as a pharmacodynamic marker where a change in the numbers of CTCs or a particular subset of CTCs following therapy indicates response to therapy. For example, in SCLC the total numbers of CTCs were significantly decreased following a single dose of chemotherapy (166,167). Similar results were observed in patients with NSCLC (5,168-170). In contrast, an increase in CTC numbers is indicative of failure to respond to therapy. Changes in CTC numbers were also examined in patients with localised NSCLC undergoing radiation treatment the change in CTC numbers post treatment was evaluated as a surrogate for disease response (171). Additional studies have investigated subsets of CTCs linked to the therapy used. For example, CTCs expressing insulin-like growth factor-1 receptor (IGF-1R) were enumerated in patients with prostate cancer treated with a monoclonal human antibody, CP-751,871, targeting IGF-IR (172) and HER2 expressing CTCs were examined in patients with breast cancer treated in the neoadjuvant GeparQuattro trial (173).
Two clinical studies evaluated targeting the Notch ligand, delta-like ligand 3 (DLL3), and the T-cell checkpoint receptors programmed cell-death protein 1 (PD-1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) for SCLC. Rovalpituzumab tesirine, DLL3 targeting agent has shown encouraging results in phase I clinical trials and is now moving on to phase II studies. Several other agents are in clinical trials for treatment of lung cancer such as new generation EGFR and ALK inhibitors (174-176).
In the recent past, immune checkpoint inhibitors have emerged as promising therapeutic targets in both NSCLC and SCLC yielding unprecedented improvements in OS and quality of life. More recently, expression of PDL-1 in CTCs has been examined in trials with immune checkpoint inhibitors and has linked with patients who experienced progressive disease (177).
New drug target identification via CTC analysis
Since CTCs have been shown to be tumourigenic and can be obtained from a simple blood sample taken at pre-treatment, on-treatment or post-treatment CTCs also represent an attractive source for drug target identification. Although as yet there has been no systematic use of CTCs for de novo drug target identification the recent advances in single cell profiling methodologies which now enable genetic and epigenetic analysis of CTCs (178) now provide this opportunity. For example, putative drug targets could be identified by examining genes encompassed in the CNA patterns seen in single and pooled CTCs which are linked to treatment response (44).
Conclusions
The study of CTCs represents an important field of research in cancer biology and also holds great promise as diagnostic, prognostic and predictive biomarkers. Currently the use of a blood samples as a “liquid biopsy” in oncology has been dominated by the striking advances in examining the cell-free component of blood and particularly circulating tumour DNA (ctDNA) (179,180). However, there are several features of CTCs that are not shared by ctDNA that justify continued CTC research including the generation of CDX and CTC culture models that can provide a direct means of testing new drugs/drug combinations (181). In addition, RNA analysis of CTCs can provide insights into their biological status and serial CTC RNA profiling can be used to identify underlying mechanisms of resistance. Furthermore, DNA analysis of individual CTCs provides an unambiguous means of understanding the mutational burden of the tumour as well as providing additional means for establishing the degree of tumour heterogeneity. Since a single blood sample encompasses both cell-free and cellular components, CTC and ctDNA can be readily combined to provide an enhanced understanding of the patient’s disease status (26,180,182).
In the field of oncology CTCs remain a key focus of biological and clinical research. With the recent improvements in CTC enrichment researchers are now able to explore key ongoing biological questions in CTC research such as: how many CTCs are required for a representative snapshot of the donor; can CTCs can be detected reliably in early disease and can CTCs be used to routinely guide cancer patient care.
Acknowledgements
We would like to thank Sarah Hilton, Dawar Pasha and Jackie Pierce for developing the staining protocol for identification of CTCs post enrichment from an epitope independent system (Parsortix™), Debbie Burt for isolation of single cells via the DEPArray and Sakshi Gulati for bioinformatic analysis and generation of CNA plots.
Funding: We would like to acknowledge the help of the CR-UK Clinical and Experimental Pharmacology Group (C5759/A20971) for their support of this study, as well as the financial support from Medical Research Council Manchester Single Cell Research Centre (MR/M008908/1). F Chemi is funded by Cancer-ID (115749-Cancer-ID) and S Mohan is funded by AstraZeneca (D1330N00013). Additional research funding was also provided by the companies Menarini Silicon Biosysytems (provider of DEPArray) and ANGLE plc UK (provider of Parsortix).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aus Med J 1869;14:146-9. [Google Scholar]
- 2.Weiss L. Metastasis of cancer: a conceptual history from antiquity to the 1990s. Cancer Metastasis Rev 2000;19:I-XI, 193-383. [PubMed] [Google Scholar]
- 3.Wittekind C, Neid M. Cancer invasion and metastasis. Oncology 2005;69 Suppl 1:14-6. 10.1159/000086626 [DOI] [PubMed] [Google Scholar]
- 4.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 5.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. 10.1200/JCO.2010.28.7045 [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa T, Martinez SR, Goto Y, et al. Detection of Circulating Tumor Cells in Early-Stage Breast Cancer Metastasis to Axillary Lymph Nodes. Clin Cancer Res 2007;13:4105. 10.1158/1078-0432.CCR-07-0419 [DOI] [PubMed] [Google Scholar]
- 7.Pantel K, Alix-Panabieres C. Liquid biopsy in 2016: Circulating tumour cells and cell-free DNA in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol 2017;14:73-4. 10.1038/nrgastro.2016.198 [DOI] [PubMed] [Google Scholar]
- 8.Thiele JA, Bethel K, Kralickova M, et al. Circulating Tumor Cells: Fluid Surrogates of Solid Tumors. Annu Rev Pathol 2017;12:419-47. 10.1146/annurev-pathol-052016-100256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawal S, Yang YP, Cote RJ, et al. Identification and Quantitation of Circulating Tumor Cells. Annu Rev Anal Chem (Palo Alto Calif) 2017;10:321-43. 10.1146/annurev-anchem-061516-045405 [DOI] [PubMed] [Google Scholar]
- 10.Kulasinghe A, Perry C, Warkiani ME, et al. Short term ex-vivo expansion of circulating head and neck tumour cells. Oncotarget 2016;7:60101-9. 10.18632/oncotarget.11159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao D, Vela I, Sboner A, et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014;159:176-87. 10.1016/j.cell.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkinson C, Morrow C, Tugwood J, et al. Circulating tumour cells from small cell lung cancer patients are tumourigenic. Lung Cancer 2015;87:S1. 10.1016/S0169-5002(15)50001-925644283 [DOI] [Google Scholar]
- 13.Morrow CJ, Trapani F, Metcalf RL, et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Ann Oncol 2016;27:1155-60. 10.1093/annonc/mdw122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girotti MR, Gremel G, Lee R, et al. Application of Sequencing, Liquid Biopsies, and Patient-Derived Xenografts for Personalized Medicine in Melanoma. Cancer Discov 2016;6:286-99. 10.1158/2159-8290.CD-15-1336 [DOI] [PubMed] [Google Scholar]
- 15.Lapin M, Tjensvoll K, Oltedal S, et al. MINDEC-An Enhanced Negative Depletion Strategy for Circulating Tumour Cell Enrichment. Sci Rep 2016;6:28929. 10.1038/srep28929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. 10.1200/JCO.2010.33.3716 [DOI] [PubMed] [Google Scholar]
- 17.Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 18.de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- 19.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9. 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talasaz AH, Powell AA, Huber DE, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A 2009;106:3970-5. 10.1073/pnas.0813188106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 2013;5:179ra47. 10.1126/scitranslmed.3005616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saucedo-Zeni N, Mewes S, Niestroj R, et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol 2012;41:1241-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao C, Bui T, Connelly M, et al. Circulating melanoma cells and survival in metastatic melanoma. Int J Oncol 2011;38:755-60. [DOI] [PubMed] [Google Scholar]
- 24.Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer 2011;105:847-53. 10.1038/bjc.2011.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu L, Mao X, Imrali A, et al. Optimization and Evaluation of a Novel Size Based Circulating Tumor Cell Isolation System. PLOS ONE 2015;10:e0138032. 10.1371/journal.pone.0138032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chudziak J, Burt DJ, Mohan S, et al. Clinical evaluation of a novel microfluidic device for epitope-independent enrichment of circulating tumour cells in patients with small cell lung cancer. Analyst 2016;141:669-78. 10.1039/C5AN02156A [DOI] [PubMed] [Google Scholar]
- 27.Hou HW, Warkiani ME, Khoo BL, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Scientific Reports 2013;3:1259. 10.1038/srep01259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. Embo Mol Med 2015;7:1. 10.15252/emmm.201303698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieva J, Wendel M, Luttgen M, et al. High definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients; a longitudinal analysis. Phys Biol 2012;9:016004. 10.1088/1478-3975/9/1/016004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campton DE, Ramirez AB, Nordberg JJ, et al. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer 2015;15:360. 10.1186/s12885-015-1383-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swennenhuis JF, Reumers J, Thys K, et al. Efficiency of whole genome amplification of single circulating tumor cells enriched by CellSearch and sorted by FACS. Genome Med 2013;5:106. 10.1186/gm510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CL, Mahalingam D, Osmulski P, et al. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate 2013;73:813-26. 10.1002/pros.22625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158:1110-22. 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yusa A, Toneri M, Masuda T, et al. Development of a New Rapid Isolation Device for Circulating Tumor Cells (CTCs) Using 3D Palladium Filter and Its Application for Genetic Analysis. PLOS ONE 2014;9:e88821. 10.1371/journal.pone.0088821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabbri F, Carloni S, Zoli W, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Letters 2013;335:225-31. 10.1016/j.canlet.2013.02.015 [DOI] [PubMed] [Google Scholar]
- 36.Peeters DJ, De Laere B, Van den Eynden GG, et al. Semiautomated isolation and molecular characterisation of single or highly purified tumour cells from CellSearch enriched blood samples using dielectrophoretic cell sorting. Br J Cancer 2013;108:1358-67. 10.1038/bjc.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stathopoulou A, Gizi A, Perraki M, et al. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the lightcycler system. Clin Cancer Res 2003;9:5145-51. [PubMed] [Google Scholar]
- 38.Xenidis N, Ignatiadis M, Apostolaki S, et al. Cytokeratin-19 mRNA-Positive Circulating Tumor Cells After Adjuvant Chemotherapy in Patients With Early Breast Cancer. J Clin Oncol 2009;27:2177-84. 10.1200/JCO.2008.18.0497 [DOI] [PubMed] [Google Scholar]
- 39.Chimonidou M, Kallergi G, Georgoulias V, et al. Breast cancer metastasis suppressor-1 promoter methylation in primary breast tumors and corresponding circulating tumor cells. Mol Cancer Res 2013;11:1248-57. 10.1158/1541-7786.MCR-13-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iinuma H, Watanabe T, Mimori K, et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes' stage B and C colorectal cancer. J Clin Oncol 2011;29:1547-55. 10.1200/JCO.2010.30.5151 [DOI] [PubMed] [Google Scholar]
- 41.Marinov GK, Williams BA, McCue K, et al. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res 2014;24:496-510. 10.1101/gr.161034.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein CA, Seidl S, Petat-Dutter K, et al. Combined transcriptome and genome analysis of single micrometastatic cells. Nat Biotechnol 2002;20:387-92. 10.1038/nbt0402-387 [DOI] [PubMed] [Google Scholar]
- 43.Heitzer E, Auer M, Gasch C, et al. Complex Tumor Genomes Inferred from Single Circulating Tumor Cells by Array-CGH and Next-Generation Sequencing. Cancer Res 2013;73:2965-75. 10.1158/0008-5472.CAN-12-4140 [DOI] [PubMed] [Google Scholar]
- 44.Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 2017;23:114-9. 10.1038/nm.4239 [DOI] [PubMed] [Google Scholar]
- 45.Klein CA, Schmidt-Kittler O, Schardt JA, et al. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci U S A 1999;96:4494-9. 10.1073/pnas.96.8.4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martelotto LG, Baslan T, Kendall J, et al. Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat Med 2017;23:376-85. 10.1038/nm.4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zong C, Lu S, Chapman AR, et al. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 2012;338:1622-6. 10.1126/science.1229164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dean FB, Hosono S, Fang L, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A 2002;99:5261-6. 10.1073/pnas.082089499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosono S, Faruqi AF, Dean FB, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res 2003;13:954-64. 10.1101/gr.816903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spits C, Le Caignec C, De Rycke M, et al. Whole-genome multiple displacement amplification from single cells. Nat Protocols 2006;1:1965-70. 10.1038/nprot.2006.326 [DOI] [PubMed] [Google Scholar]
- 51.Hou Y, Wu K, Shi X, et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. GigaScience 2015;4:37. 10.1186/s13742-015-0068-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulmer A, Schmidt-Kittler O, Fischer J, et al. Immunomagnetic enrichment, genomic characterization, and prognostic impact of circulating melanoma cells. Clin Cancer Res 2004;10:531-7. 10.1158/1078-0432.CCR-0424-03 [DOI] [PubMed] [Google Scholar]
- 53.Dong X, Zhang L, Milholland B, et al. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat Methods 2017;14:491-3. 10.1038/nmeth.4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng Q, Vijaya Satya R, Lewis M, et al. Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics 2015;16:589. 10.1186/s12864-015-1806-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou Y, Song L, Zhu P, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 2012;148:873-85. 10.1016/j.cell.2012.02.028 [DOI] [PubMed] [Google Scholar]
- 56.Gasch C, Bauernhofer T, Pichler M, et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin Chem 2013;59:252-60. 10.1373/clinchem.2012.188557 [DOI] [PubMed] [Google Scholar]
- 57.Kolodziejczyk AA, Kim JK, Svensson V, et al. The technology and biology of single-cell RNA sequencing. Mol Cell 2015;58:610-20. 10.1016/j.molcel.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 58.Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015;349:1351-6. 10.1126/science.aab0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramsköld D, Luo S, Wang YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30:777-82. 10.1038/nbt.2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denis JA, Patroni A, Guillerm E, et al. Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery. Mol Oncol 2016;10:1221-31. 10.1016/j.molonc.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coumans FA, Ligthart ST, Uhr JW, et al. Challenges in the Enumeration and Phenotyping of CTC. Clin Cancer Res 2012;18:5711-8. 10.1158/1078-0432.CCR-12-1585 [DOI] [PubMed] [Google Scholar]
- 62.Fischer JC, Niederacher D, Topp SA, et al. Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proc Natl Acad Sci U S A 2013;110:16580-5. 10.1073/pnas.1313594110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 64.Demeulemeester J, Kumar P, Møller EK, et al. Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing. Genome Biol 2016;17:250. 10.1186/s13059-016-1109-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pantel K, Angstwurm M, Riethmüller G, et al. Frequency and prognostic significance of isolated tumour cells in bone marrow of patients with non-small-cell lung cancer without overt metastases. Lancet 1996;347:649-53. 10.1016/S0140-6736(96)91203-9 [DOI] [PubMed] [Google Scholar]
- 66.Crosbie PA, Shah R, Krysiak P, et al. Circulating Tumor Cells Detected in the Tumor-Draining Pulmonary Vein Are Associated with Disease Recurrence after Surgical Resection of NSCLC. J Thorac Oncol 2016;11:1793-7. 10.1016/j.jtho.2016.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature 2016;529:298-306. 10.1038/nature17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luzzi KJ, MacDonald IC, Schmidt EE, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol 1998;153:865-73. 10.1016/S0002-9440(10)65628-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cameron MD, Schmidt EE, Kerkvliet N, et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res 2000;60:2541-6. [PubMed] [Google Scholar]
- 70.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002;2:563-72. 10.1038/nrc865 [DOI] [PubMed] [Google Scholar]
- 71.Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell 2013;23:573-81. 10.1016/j.ccr.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23-8. 10.1126/science.959840 [DOI] [PubMed] [Google Scholar]
- 73.Fidler IJ, Hart IR. Biological diversity in metastatic neoplasms: origins and implications. Science 1982;217:998-1003. 10.1126/science.7112116 [DOI] [PubMed] [Google Scholar]
- 74.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 76.Beca F, Beck AH. Precision Cancer Diagnostics: Tracking Genomic Evolution in Clinical Trials. PLoS Med 2016;13:e1002177. 10.1371/journal.pmed.1002177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational implications of tumor heterogeneity. Clin Cancer Res 2015;21:1258-66. 10.1158/1078-0432.CCR-14-1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alizadeh AA, Aranda V, Bardelli A, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med 2015;21:846-53. 10.1038/nm.3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zellmer VR, Zhang S. Evolving concepts of tumor heterogeneity. Cell Biosci 2014;4:69. 10.1186/2045-3701-4-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwartz R, Schäffer AA. The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet 2017;18:213-29. 10.1038/nrg.2016.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis A, Navin NE. Computing tumor trees from single cells. Genome Biol 2016;17:113. 10.1186/s13059-016-0987-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jamal-Hanjani M, Hackshaw A, Ngai Y, et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol 2014;12:e1001906. 10.1371/journal.pbio.1001906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature 2011;472:90-4. 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwarz RF, Trinh A, Sipos B, et al. Phylogenetic Quantification of Intra-tumour Heterogeneity. PLoS Comput Biol 2014;10:e1003535. 10.1371/journal.pcbi.1003535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klein CA, Blankenstein TJ, Schmidt-Kittler O, et al. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet 2002;360:683-9. 10.1016/S0140-6736(02)09838-0 [DOI] [PubMed] [Google Scholar]
- 86.de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. 10.1126/science.1253462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O'Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007;445:106-10. 10.1038/nature05372 [DOI] [PubMed] [Google Scholar]
- 88.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007;445:111-5. 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- 89.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res 2007;67:1030-7. 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- 90.Schatton T, Murphy GF, Frank NY, et al. Identification of cells initiating human melanomas. Nature 2008;451:345-9. 10.1038/nature06489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang ZF, Ho DW, Ng MN, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008;13:153-66. 10.1016/j.ccr.2008.01.013 [DOI] [PubMed] [Google Scholar]
- 92.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature 2004;432:396-401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- 93.Meng S, Tripathy D, Frenkel EP, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res 2004;10:8152-62. 10.1158/1078-0432.CCR-04-1110 [DOI] [PubMed] [Google Scholar]
- 94.Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target? Nat Rev Cancer 2010;10:871-7. 10.1038/nrc2933 [DOI] [PubMed] [Google Scholar]
- 95.Wikman H, Vessella R, Pantel K. Cancer micrometastasis and tumour dormancy. Apmis 2008;116:754-70. 10.1111/j.1600-0463.2008.01033.x [DOI] [PubMed] [Google Scholar]
- 96.Lindemann F, Schlimok G, Dirschedl P, et al. Prognostic significance of micrometastatic tumour cells in bone marrow of colorectal cancer patients. Lancet 1992;340:685-9. 10.1016/0140-6736(92)92230-D [DOI] [PubMed] [Google Scholar]
- 97.Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer 2004;4:448-56. 10.1038/nrc1370 [DOI] [PubMed] [Google Scholar]
- 98.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer 2014;14:611-22. 10.1038/nrc3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Joosse SA, Pantel K. Genetic traits for hematogeneous tumor cell dissemination in cancer patients. Cancer Metastasis Rev 2016;35:41-8. 10.1007/s10555-016-9611-7 [DOI] [PubMed] [Google Scholar]
- 100.Sienel W, Mecklenburg I, Dango S, et al. Detection of MAGE-A transcripts in bone marrow is an independent prognostic factor in operable non-small-cell lung cancer. Clin Cancer Res 2007;13:3840-7. 10.1158/1078-0432.CCR-06-2507 [DOI] [PubMed] [Google Scholar]
- 101.Sugio K, Kase S, Sakada T, et al. Micrometastasis in the bone marrow of patients with lung cancer associated with a reduced expression of E-cadherin and beta-catenin: risk assessment by immunohistochemistry. Surgery 2002;131:S226-31. 10.1067/msy.2002.119793 [DOI] [PubMed] [Google Scholar]
- 102.Saito T, Kobayashi M, Harada R, et al. Sensitive detection of small cell lung carcinoma cells by reverse transcriptase-polymerase chain reaction for prepro-gastrin-releasing peptide mRNA. Cancer 2003;97:2504-11. 10.1002/cncr.11378 [DOI] [PubMed] [Google Scholar]
- 103.Ni X, Zhuo M, Su Z, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A 2013;110:21083-8. 10.1073/pnas.1320659110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baccelli I, Schneeweiss A, Riethdorf S, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 2013;31:539-44. 10.1038/nbt.2576 [DOI] [PubMed] [Google Scholar]
- 105.Thiery JP, Acloque H, Huang RYJ, et al. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009;139:871-90. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 106.Edelman GM, Gallin WJ, Delouvée A, et al. Early epochal maps of two different cell adhesion molecules. Proc Natl Acad Sci U S A 1983;80:4384-8. 10.1073/pnas.80.14.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science 2013;342:1234850. 10.1126/science.1234850 [DOI] [PubMed] [Google Scholar]
- 108.Kim KK, Kugler MC, Wolters PJ, et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A 2006;103:13180-5. 10.1073/pnas.0605669103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell 2013;23:272-3. 10.1016/j.ccr.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 110.Brabletz T, Jung A, Reu S, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A 2001;98:10356-61. 10.1073/pnas.171610498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2:442-54. 10.1038/nrc822 [DOI] [PubMed] [Google Scholar]
- 112.Hou JM, Krebs M, Ward T, et al. Circulating Tumor Cells as a Window on Metastasis Biology in Lung Cancer. Am J Pathol 2011;178:989-96. 10.1016/j.ajpath.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joosse SA, Hannemann J, Spotter J, et al. Changes in keratin expression during metastatic progression of breast cancer: impact on the detection of circulating tumor cells. Clin Cancer Res 2012;18:993-1003. 10.1158/1078-0432.CCR-11-2100 [DOI] [PubMed] [Google Scholar]
- 114.Kishi S, Bayliss PE, Hanai J. A prospective epigenetic paradigm between cellular senescence and epithelial-mesenchymal transition in organismal development and aging. Transl Res 2015;165:241-9. 10.1016/j.trsl.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 115.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013;19:1438-49. 10.1038/nm.3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580-4. 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kallergi G, Papadaki MA, Politaki E, et al. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res 2011;13:R59. 10.1186/bcr2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lecharpentier A, Vielh P, Perez-Moreno P, et al. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer 2011;105:1338-41. 10.1038/bjc.2011.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Balasubramanian P, Lang JC, Jatana KR, et al. Multiparameter analysis, including EMT markers, on negatively enriched blood samples from patients with squamous cell carcinoma of the head and neck. PLoS One 2012;7:e42048. 10.1371/journal.pone.0042048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Armstrong AJ, Marengo MS, Oltean S, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 2011;9:997-1007. 10.1158/1541-7786.MCR-10-0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krebs MG, Hou JM, Sloane R, et al. Analysis of Circulating Tumor Cells in Patients with Non-small Cell Lung Cancer Using Epithelial Marker-Dependent and -Independent Approaches. J Thorac Oncol 2012;7:306-15. 10.1097/JTO.0b013e31823c5c16 [DOI] [PubMed] [Google Scholar]
- 122.de Wit S, van Dalum G, Lenferink AT, et al. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep 2015;5:12270. 10.1038/srep12270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jahchan NS, Lim JS, Bola B, et al. Identification and targeting of long-term tumor-propagating cells in small cell lung cancer. Cell reports 2016;16:644-56. 10.1016/j.celrep.2016.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Williamson SC, Metcalf RL, Trapani F, et al. Vasculogenic mimicry in small cell lung cancer. Nat Commun 2016;7:13322. 10.1038/ncomms13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hendrix MJ, Seftor EA, Hess AR, et al. Vasculogenic mimicry and tumour-cell plasticity: lessons from melanoma. Nat Rev Cancer 2003;3:411-21. 10.1038/nrc1092 [DOI] [PubMed] [Google Scholar]
- 126.Hendrix MJ, Seftor EA, Seftor RE, et al. Tumor cell vascular mimicry: Novel targeting opportunity in melanoma. Pharmacol Ther 2016;159:83-92. 10.1016/j.pharmthera.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wagenblast E, Soto M, Gutierrez-Angel S, et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature 2015;520:358-62. 10.1038/nature14403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brandt B, Junker R, Griwatz C, et al. Isolation of prostate-derived single cells and cell clusters from human peripheral blood. Cancer Res 1996;56:4556-61. [PubMed] [Google Scholar]
- 129.Kats-Ugurlu G, Roodink I, de Weijert M, et al. Circulating tumour tissue fragments in patients with pulmonary metastasis of clear cell renal cell carcinoma. J Pathol 2009;219:287-93. 10.1002/path.2613 [DOI] [PubMed] [Google Scholar]
- 130.Molnar B, Ladanyi A, Tanko L, et al. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clin Cancer Res 2001;7:4080-5. [PubMed] [Google Scholar]
- 131.Zhang D, Zhao L, Zhou P, et al. Circulating tumor microemboli (CTM) and vimentin+ circulating tumor cells (CTCs) detected by a size-based platform predict worse prognosis in advanced colorectal cancer patients during chemotherapy. Cancer Cell Int 2017;17:6. 10.1186/s12935-016-0373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Duda DG, Duyverman AM, Kohno M, et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proc Natl Acad Sci U S A 2010;107:21677-82. 10.1073/pnas.1016234107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liotta LA, Kleinerman J, Saidel GM. Quantitative relationships of intravascular tumor cells, tumor vessels, and pulmonary metastases following tumor implantation. Cancer Res 1974;34:997-1004. [PubMed] [Google Scholar]
- 134.Fidler IJ. Immune stimulation-inhibition of experimental cancer metastasis. Cancer Res 1974;34:491-8. [PubMed] [Google Scholar]
- 135.Ludatscher RM, Luse SA, Suntzeff V. An electron microscopic study of pulmonary tumor emboli from transplantable Morris hepatoma 5123. Cancer Res 1967;27:1939-52. [PubMed] [Google Scholar]
- 136.Sugino T, Kusakabe T, Hoshi N, et al. An Invasion-Independent Pathway of Blood-Borne Metastasis: A New Murine Mammary Tumor Model. Am J Pathol 2002;160:1973-80. 10.1016/S0002-9440(10)61147-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Küsters B, Kats G, Roodink I, et al. Micronodular transformation as a novel mechanism of VEGF-A-induced metastasis. Oncogene 2007;26:5808-15. 10.1038/sj.onc.1210360 [DOI] [PubMed] [Google Scholar]
- 138.Läubli H, Stevenson JL, Varki A, et al. L-selectin facilitation of metastasis involves temporal induction of Fut7-dependent ligands at sites of tumor cell arrest. Cancer Res 2006;66:1536-42. 10.1158/0008-5472.CAN-05-3121 [DOI] [PubMed] [Google Scholar]
- 139.Gasic GJ, Gasic TB, Galanti N, et al. Platelet-tumor-cell interactions in mice. The role of platelets in the spread of malignant disease. Int J Cancer 1973;11:704-18. 10.1002/ijc.2910110322 [DOI] [PubMed] [Google Scholar]
- 140.Borsig L, Wong R, Hynes RO, et al. Synergistic effects of L- and P-selectin in facilitating tumor metastasis can involve non-mucin ligands and implicate leukocytes as enhancers of metastasis. Proc Natl Acad Sci U S A 2002;99:2193-8. 10.1073/pnas.261704098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Marusyk A, Tabassum DP, Altrock PM, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 2014;514:54-8. 10.1038/nature13556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tabassum DP, Polyak K. Tumorigenesis: it takes a village. Nat Rev Cancer 2015;15:473-83. 10.1038/nrc3971 [DOI] [PubMed] [Google Scholar]
- 143.Cleary AS, Leonard TL, Gestl SA, et al. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 2014;508:113-7. 10.1038/nature13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Calbo J, van Montfort E, Proost N, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011;19:244-56. 10.1016/j.ccr.2010.12.021 [DOI] [PubMed] [Google Scholar]
- 145.Amo L, Tamayo-Orbegozo E, Maruri N, et al. Involvement of platelet-tumor cell interaction in immune evasion. Potential role of podocalyxin-like protein 1. Front Oncol 2014;4:245. 10.3389/fonc.2014.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lou XL, Sun J, Gong SQ, et al. Interaction between circulating cancer cells and platelets: clinical implication. Chin J Cancer Res 2015;27:450-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gay LJ, Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat Rev Cancer 2011;11:123-34. 10.1038/nrc3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tsuji T, Ibaragi S, Shima K, et al. Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer Res 2008;68:10377-86. 10.1158/0008-5472.CAN-08-1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liotta LA, Saidel MG, Kleinerman J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res 1976;36:889-94. [PubMed] [Google Scholar]
- 150.Au SH, Storey BD, Moore JC, et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc Natl Acad Sci U S A 2016;113:4947-52. 10.1073/pnas.1524448113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rink M, Chun FK, Dahlem R, et al. Prognostic role and HER2 expression of circulating tumor cells in peripheral blood of patients prior to radical cystectomy: a prospective study. Eur Urol 2012;61:810-7. 10.1016/j.eururo.2012.01.017 [DOI] [PubMed] [Google Scholar]
- 152.Rack B, Schindlbeck C, Jückstock J, et al. Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients. J Natl Cancer Inst 2014;106(5). 10.1093/jnci/dju066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ilie M, Hofman V, Long-Mira E, et al. “Sentinel” Circulating Tumor Cells Allow Early Diagnosis of Lung Cancer in Patients with Chronic Obstructive Pulmonary Disease. PLOS ONE 2014;9:e111597. 10.1371/journal.pone.0111597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nalejska E, Maczynska E, Lewandowska MA. Prognostic and predictive biomarkers: tools in personalized oncology. Mol Diagn Ther 2014;18:273-84. 10.1007/s40291-013-0077-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 2011;17:827-35. 10.1158/1078-0432.CCR-10-0445 [DOI] [PubMed] [Google Scholar]
- 156.Carlsson A, Nair VS, Luttgen MS, et al. Circulating tumor microemboli diagnostics for patients with non-small-cell lung cancer. J Thorac Oncol 2014;9:1111-9. 10.1097/JTO.0000000000000235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. Jama 2014;311:1998-2006. 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. N Engl J Med 2008;359:366-77. 10.1056/NEJMoa0800668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR Mutations in Circulating Tumor Cell Preparations from NSCLC Patients by Next Generation Sequencing: Toward a Real-Time Liquid Biopsy for Treatment. PLOS ONE 2014;9:e103883. 10.1371/journal.pone.0103883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pailler E, Adam J, Barthelemy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. 10.1200/JCO.2012.44.5932 [DOI] [PubMed] [Google Scholar]
- 162.Faugeroux V, Pailler E, Auger N, et al. Clinical Utility of Circulating Tumor Cells in ALK-Positive Non-Small-Cell Lung Cancer. Front Oncol 2014;4:281. 10.3389/fonc.2014.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pailler E, Auger N, Lindsay CR, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol 2015;26:1408-15. 10.1093/annonc/mdv165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.George J, Lim JS, Peifer M, et al. Comprehensive genomic characterization of small cell lung cancer. Cancer Research. 2015;75:LB-168 10.1158/1538-7445.AM2015-LB-168 [DOI] [Google Scholar]
- 165.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. 10.1038/ng.2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Naito T, Tanaka F, Ono A, et al. Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J Thorac Oncol 2012;7:512-9. 10.1097/JTO.0b013e31823f125d [DOI] [PubMed] [Google Scholar]
- 167.Hiltermann TJ, Pore MM, van den Berg A, et al. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann Oncol 2012;23:2937-42. 10.1093/annonc/mds138 [DOI] [PubMed] [Google Scholar]
- 168.Punnoose EA, Atwal S, Liu W, et al. Evaluation of Circulating Tumor Cells and Circulating Tumor DNA in Non–Small Cell Lung Cancer: Association with Clinical Endpoints in a Phase II Clinical Trial of Pertuzumab and Erlotinib. Clin Cancer Res 2012;18:2391. 10.1158/1078-0432.CCR-11-3148 [DOI] [PubMed] [Google Scholar]
- 169.Juan O, Vidal J, Gisbert R, et al. Prognostic significance of circulating tumor cells in advanced non-small cell lung cancer patients treated with docetaxel and gemcitabine. Clin Transl Oncol 2014;16:637-43. 10.1007/s12094-013-1128-8 [DOI] [PubMed] [Google Scholar]
- 170.Muinelo-Romay L, Vieito M, Abalo A, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers 2014;6:153-65. 10.3390/cancers6010153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dorsey JF, Kao GD, MacArthur KM, et al. Tracking viable circulating tumor cells (CTCs) in the peripheral blood of non–small cell lung cancer (NSCLC) patients undergoing definitive radiation therapy: Pilot study results. Cancer 2015;121:139-49. 10.1002/cncr.28975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.de Bono JS, Attard G, Adjei A, et al. Potential applications for circulating tumor cells expressing the insulin-like growth factor-I receptor. Clin Cancer Res 2007;13:3611-6. 10.1158/1078-0432.CCR-07-0268 [DOI] [PubMed] [Google Scholar]
- 173.Riethdorf S, Muller V, Zhang L, et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res 2010;16:2634-45. 10.1158/1078-0432.CCR-09-2042 [DOI] [PubMed] [Google Scholar]
- 174.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 2015;29:1447-62. 10.1101/gad.263145.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin JJ, Shaw AT. Resisting Resistance: Targeted Therapies in Lung Cancer. Trends in Cancer 2016;2:350-64. 10.1016/j.trecan.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Maione P, Sacco PC, Sgambato A, et al. Overcoming resistance to targeted therapies in NSCLC: current approaches and clinical application. Ther Adv Med Oncol 2015;7:263-73. 10.1177/1758834015595048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nicolazzo C, Raimondi C, Mancini M, et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Scientific Reports 2016;6:31726. 10.1038/srep31726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lianidou ES. Gene expression profiling and DNA methylation analyses of CTCs. Mol Oncol 2016;10:431-42. 10.1016/j.molonc.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 180.Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017;31:172-9. 10.1016/j.ccell.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 181.Yu M, Bardia A, Aceto N, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014;345:216. 10.1126/science.1253533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rothwell DG, Smith N, Morris D, et al. Genetic profiling of tumours using both circulating free DNA and circulating tumour cells isolated from the same preserved whole blood sample. Mol Oncol 2016;10:566-74. 10.1016/j.molonc.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]