Abstract
In non-small cell lung cancer (NSCLC), diagnosis of predictive biomarkers for targeted therapies is currently done in small tumor biopsies. However, tumor biopsies can be invasive, in some cases associated with risk, and tissue adequacy, both in terms of quantity and quality is often insufficient. The development of efficient and non-invasive methods to identify genetic alterations is a key challenge which circulating tumor cells (CTCs) have the potential to be exploited for. CTCs are extremely rare and phenotypically diverse, two characteristics that impose technical challenges and impact the success of robust molecular analysis. Here we introduce the clinical needs in this disease that mainly consist of the diagnosis of epidermal growth factor receptor (EGFR) activating alterations and anaplastic lymphoma kinase (ALK) rearrangement. We present the proof-of-concept studies that explore the detection of these genetic alterations in CTCs from NSCLC patients. Finally, we discuss steps that are still required before CTCs are routinely used for diagnosis of EGFR-mutations and ALK-rearrangements in this disease.
Keywords: Circulating tumor cells (CTC), non-small cell lung cancer (NSCLC), epidermal growth factor receptor (EGFR) mutations, anaplastic lymphoma kinase (ALK) rearrangement
Preface
On the basis that ISET filtration offers higher circulating tumor cells (CTCs) recovery and a broader coverage of the phenotypic heterogeneity of CTCs when compared to CellSearch, we utilized ISET for examining anaplastic lymphoma kinase (ALK)-rearrangement in CTCs. Our group published in 2013 a novel approach in detecting ALK-rearrangement in CTCs with high sensitivity and specificity in patients with non-small cell lung cancer (NSCLC) combining ISET filtration (isolation by size of epithelial tumor cells) and a fluorescent in situ hybridization (FISH) assay optimized for CTC characterization on filters called FA-FISH.
Clinical needs
In the last decade, the success of tyrosine kinase inhibitors (TKIs) in selected NSCLC patients has considerably transformed the management of this disease (1,2). NSCLC and in particular adenocarcinoma has been segmented into molecular subsets based on oncogenic ‘driver’ alterations (3-7). The two best characterized oncogene driver paradigms are the epidermal growth factor receptor (EGFR) activating alterations and ALK-rearrangement against which an increasing repertoire of TKI therapies is being developed (8). EGFR-activating alterations (the L858R point mutation in exon 21 and exon 19 deletions) present in about 15% of NSCLC patients confer sensitivity to first-line EGFR TKI therapy such as gefitinib, erlotinib or afatinib (9-15). Acquired resistance to first line EGFR TKIs usually developed within 12 months of treatment (15,16), the principal cause being the T790M mutation which is detectable in approximately 60% of patients (17-20). The ALK-gene rearrangement involves most often the echinoderm microtubule-associated protein-like 4 (EML4) loci and defines a unique molecular subset in 4% of NSCLC patients (21-24). In 2012, crizotinib was approved for the treatment of patients with previously advanced treated ALK-rearranged NSCLC and is now the standard of care in the first line setting (25-28). Acquired resistance develop after a median progression-free survival (PFS) of 8 to 9 months, the most frequently identified secondary mutations being the L1196M, which is analogous to T790M in EGFR, and G1296A (29,30). Additional ALK secondary mutations distributed throughout the kinase domain (1151Tins, L1152R, C1156Y, G1202R, S1206Y, I1171T, F1174C) have also been detected in crizotinib-resistant ALK-positive tumors (31-39). A third molecular entity also targetable by TKIs is a c-ros oncogene 1 (ROS1) fusion gene which has been more recently identified in approximately 1% of NSCLC and can benefit from crizotinib therapy (40,41).
In metastatic NSCLC, surgery is rarely a component of treatment. The availability of tumor tissue is a major hurdle to the identification of genetic alterations and screening of patients eligible for TKI therapies. Genetic profiling is most commonly performed on tumor biopsy which can be invasive, impractical and in some cases associated with risk. Furthermore tissue adequacy, both in terms of quantity and quality, is often insufficient. Importantly, a single biopsy sample may also not reflect the genetic diversity of a patient’s tumor (42,43). With the increasing number of EGFR and ALK inhibitors in current clinical development, the identification of alternative noninvasive options to tumor biopsy to diagnose predictive biomarkers of sensitivity to TKIs is becoming increasingly important. Moreover, at the time of disease progression, serial biopsies are needed to identify secondary resistance mutations and re-assess the changing molecular profile of tumors. Subjecting patients to serial biopsies to identify acquired resistance mutations is even more complicated and invasive. Therefore, monitoring the emergence of acquired resistance mutations and tumor evolution is also an important issue for the development of precision medicine in NSCLC (44).
Biological characteristics of CTCs in NSCLC
The development of efficient, non-invasive methods to identify molecular alterations is a key challenge which CTCs have the potential to meet (45). However CTCs are very rare (rate of one cell per 106 or 107 leucocytes) and their molecular characterization must rely on the combination of successive steps which include an initial enrichment process, the identification of CTCs, and the detection of the genetic alteration itself (46-48). The CellSearch platform is based on the detection of epithelial cells expressing EpCAM, and approved by the FDA as an aid to prognosis in patients with metastatic breast, prostate and colorectal cancers (49-51). CTC levels measured by CellSearch have been reported to be prognostic in NSCLC but the levels are very low in this cancer type and patients are frequently negative even in advanced states (52). We and others groups have reported that CTCs are identified in higher numbers using an enrichment technique based on blood filtration (ISET, isolation by size of epithelial tumor cells) compared to the CellSearch method in NSCLC most likely due to the fact that CTCs expressing markers of epithelial-mesenchymal transition (EMT) which have lost epithelial features can be missed by CellSearch (53-56). Using ISET, the prognostic value of CTCs was also reported in patients with resected NSCLC with CTC thresholds tenfold higher (50 CTC/10 mL) than by the CellSearch (55). Recently our group reported that differential total CTC counts and EMT characteristics can be observed according to different genetic subtypes of NSCLC (57). CTCs from EGFR-mutant and ALK-rearranged NSCLC patients express epithelial-mesenchymal transition characteristics, not seen in CTCs from patients with KRAS-mutant adenocarcinoma.
The biological characteristics of CTCs including rarity and phenotypical heterogeneity impose a number of limitations and technological challenges which currently impact on the success of robust molecular analysis. In NSCLC the studies cited above support the existence of a contingent of CTCs expressing mesenchymal characteristics and a low level of truly epithelial (both EpCAM and pan-keratin positive) CTCs. The absence of a universal detection assay capable to embrace the CTC phenotypical diversity imposes technological choices such that the results must take into account the technique used. Here we present studies reporting the detection of EGFR-mutations, ALK- and ROS1-rearrangements in CTCs from NSCLC, including the description of techniques used and results. Their level of validation in the context of routine clinical use is discussed.
Detection of gene-rearrangements
Detection of ALK-rearrangement
Crizotinib was approved with a companion diagnostic test, the Vysis ALK Break Apart FISH Probe Kit (Abott Molecular) (58). Therefore the Vysis FISH test performed on tumor biopsies has been the reference assay for years but it tends to be supplanted by immunohistochemistry (IHC) because of the greater technical facility of IHC. The detection of ALK-positive CTCs has been reported by four groups including our own (59-62). All groups used FISH testing, Paul Hofman’s group using both FISH and IHC (59). In 2012, Ilie et al. reported that ALK status could be determined in CTCs isolated from patients with NSCLC by IHC and FISH analysis. CTCs from 87 patients with NSCLC were isolated using ISET filtration and screened for their ALK status both in tumor biopsy samples and in CTCs. ALK-FISH was carried out using the Vysis Probe Kit on two spots of one ISET filter (each spot corresponding to the filtration of one milliliter of blood). IHC analyses were performed using the anti-ALK antibody (5A4 clone) on two other ISET spots. Five patients had ALK-rearrangement and strong ALK-protein expression in CTCs and in the corresponding tumor samples. FISH confirmed ALK-rearrangement with the break apart of the 5’ and 3’ probes and multiple signals per cell. Both ALK-FISH and ALK-immunoreactivity analyses showed negative results in CTCs and corresponding tumor samples for 82 patients negative in the tumor tissue. For the five patients with ALK-rearranged tumors, ALK-rearranged CTCs were detected by IHC and FISH in three different CTC samples collected per each patient. On the basis that ISET filtration offers higher CTC recovery and a broader coverage of the phenotypic heterogeneity of CTCs when compared to CellSearch, we utilized ISET for examining ALK-rearrangement in CTCs (60). Our group published in 2013 a novel approach in detecting ALK-rearrangement in CTCs with high sensitivity and specificity in patients with NSCLC combining ISET filtration and a FISH assay optimized for CTC characterization on filters called FA-FISH. Levels of four or more ALK-rearranged CTCs per 1 mL of blood were detected in all 18 ALK-positive patients tested, but no or only one ALK-rearranged CTC was detected in 14 ALK-negative patients.
Using the threshold of four ALK-rearranged CTC per 1mL blood, a sensitivity and specificity of 100% was met for predicting the ALK-rearrangement status present within the tumors. Diagnosis of ALK-rearrangement by FISH on paraffin-embedded tumor samples uses a threshold of 15% of ALK-rearranged cells, which represents two standard deviations above the mean cell count in negative tumor samples. Due to the rarity of CTCs in blood, we proposed using the number of ALK-rearranged CTCs per volume of blood rather than the overall percentage as a cutoff value for establishing the diagnosis of ALK-rearrangement. By combining four-color immunofluorescent-staining (IF) and FISH, ALK-rearranged CTCs were found to display a remarkably homogeneous mesenchymal phenotype. A unique ALK-FISH pattern (the break apart signal) was consistently identified in CTCs despite the inter-tumoral heterogeneity of ALK-rearrangements and the frequency of tumor cells harboring this rearrangement within tumors. In addition this unique ALK-rearrangement pattern was detected in CTCs of patients for whom it was not identified within the tumor biopsy. Although a single tumor biopsy sample might not be representative of the entire tumor, CTCs may originate from various metastatic sites. These findings suggested that CTCs that harbor this unique ALK-rearrangement and express a mesenchymal phenotype may result from the clonal selection of tumor cells that display migratory properties and higher invasive potential and may possibly contain highly metastatic cells, such as cancer stem cells. In 2016, using a sophisticated NanoVelcro Chip technology based on the selection of EpCAM positive CTCs, He et al. reported the detection of ALK-rearrangement in 21 ALK-positive patients in the tumor biopsy (61). All ALK-positive patients had at least three ALK-rearranged CTCs per one mL blood while 20 ALK-negative patients had no or two ALK-rearranged CTCs. The ALK-rearrangement status was consistent with that of the tumor. In addition the ALK-rearranged CTC ratio was found to correlate to the pTNM stage in ALK-positive patients. Although these results may appear similar to ours, it is important to note that ALK-rearranged CTCs were epithelial in this study. In our hands, ALK-rearranged CTCs collected on ISET filters expressed vimentin and N-cadherin at a level that was generally lower than that of leucocytes but were negative for both cytokeratins and E-cadherin. This result was consistent with the absence or very low level of CTCs by the CellSearch in our ALK-positive cohort. Another study published in 2016 by Tan et al. also reported the detection of ALK-rearranged CTCs and concordance between CTCs and tumor biopsies (62). After red blood cell lysis, the ClearCell FX system from Clearbridge Biomedics was used here for CTC enrichment. A fraction of the CTC-enriched fraction deposited on glass slides by cytospin was tested using the Vysis Probe kit after. Using this approach, the authors reported 3 to 15 ALK-rearranged CTCs per 1.88 mL in 14 ALK-positive patients and 0 to 2 ALK-rearranged CTCs per 1.88 mL blood in 12 ALK-negative patients and five healthy donors. There was no available information on the phenotype of the CTCs.
In ALK-rearranged NSCLC patients, treatment with crizotinib is marked by heterogeneity in the magnitude and duration of clinical response. Response durations vary from a few months to several years, and the long-term effectiveness of crizotinib is invariably limited by the development of acquired resistance (30). In our initial study several CTC subsets harboring distinct ALK-FISH patterns—including ALK-rearranged CTCs and CTCs with a gain of ALK-copy number (ALK-CNG)—were identified and were correlated with variable evolution on crizotinib treatment in the five examined patients (60) (Figure 1A). Based on this observation, we hypothesized that CTC subsets differing in ALK-FISH patterns might be associated with different clinical outcomes in ALK-rearranged patients treated by crizotinib. Therefore we recently evaluated whether these CTC subsets monitored on crizotinib in an extended cohort of 39 ALK-rearranged patients could inform on treatment benefit (64). CTCs were classified into distinct subsets according to the presence of ALK-rearrangement and/or ALK-CNG signals. As previously reported in tumors biopsies, no significant association between baseline numbers of ALK-rearranged or ALK-CNG CTCs, and PFS was observed. However, we observed a significant association between the decrease in the numbers of CTCs with ALK-CNG on crizotinib and a longer PFS. In multivariate analysis the dynamic change of CTCs with ALK-CNG was the strongest factor associated with PFS. Although not dominant, ALK-CNG has been reported to be one of the mechanisms of acquired resistance to crizotinib in tumor biopsies. This study shows that serial FISH analysis of CTCs could identify a predictive biomarker of therapeutic efficacy in ALK-rearranged NSCLC and could help to stratify patients at risk of early resistance.
Figure 1.
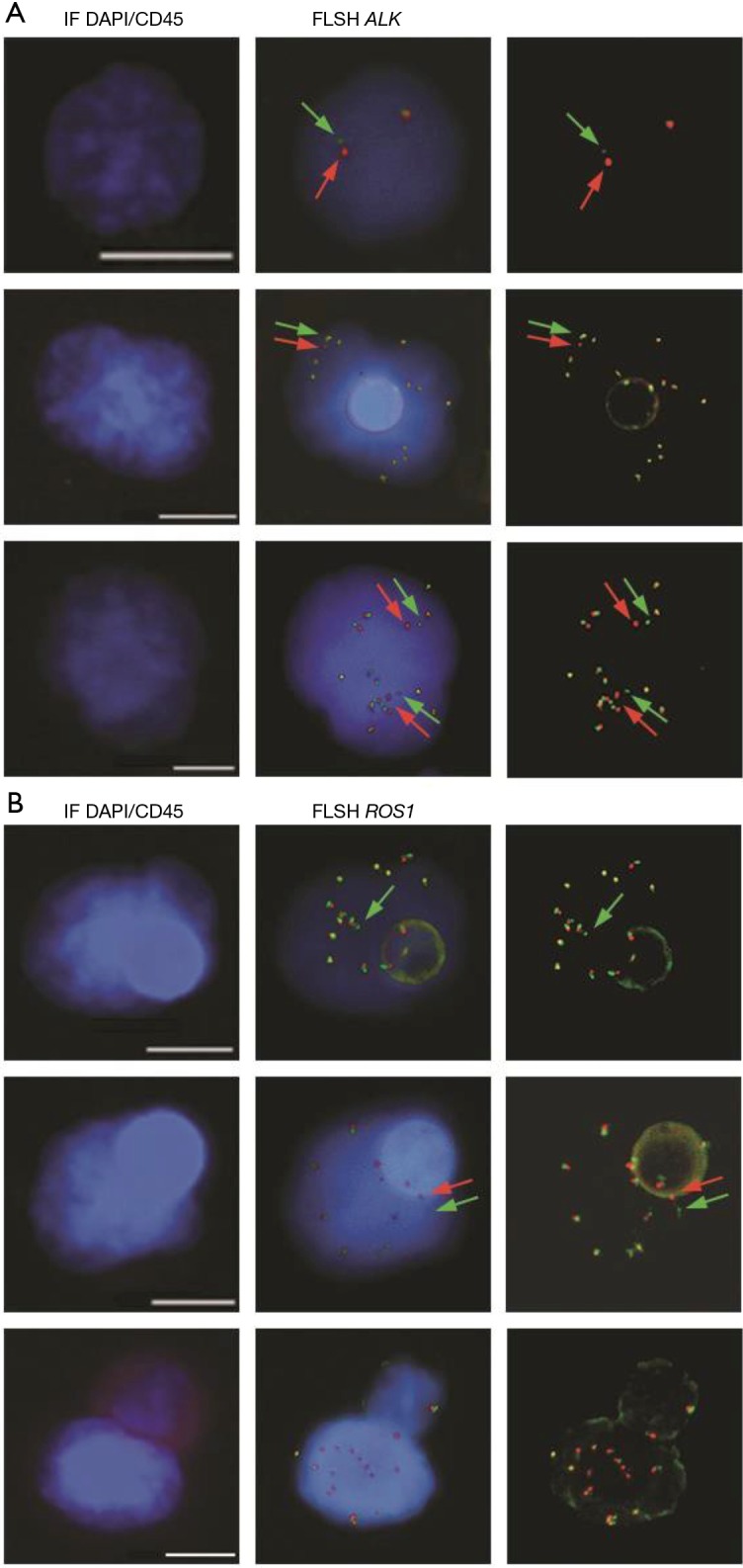
Examples of gene-rearrangement detection in filtration enriched-CTCs from NSCLC patients by combined fluorescent staining and FISH. (A) Example of ALK-rearrangement detection in NSCLC patients with an ALK-rearranged tumor; (B) example of ROS1-rearrangement detection in NSCLC patients with a ROS1-rearranged tumor. Adapted with permission from (63). Used under CC BY 4.0.
Detection of ROS1-rearrangement
Our group published the detection of ROS1-rearrangements in the CTCs of 4 patients with ROS1-rearrangement previously detected by FISH on tumor biopsy (65). Detection was performed using ISET filtration and FA-FISH (Figure 1B). In ROS1-rearranged patients, the median number of ROS1-rearranged CTCs at baseline was 34.5 (range, 24–55) per 3 mL of blood while in ROS1-negative NSCLC patients, median background hybridization of ROS1-rearranged CTCs was 7.5 (range, 7–11) per 3 mL blood. This study provides the proof-of-concept that CTCs can be used for non-invasive, sensitive and specific detection of ROS1-rearrangement in NSCLC patients. In this study we also evaluated the effect of crizotinib on ROS1-gene copy number; in the two patients who had tumor progression the number of ROS1-gene copies present in ROS1-rearranged cells increased significantly during treatment. ROS1-rearranged CTCs showed considerable heterogeneity of ROS1-gene abnormalities and elevated numerical chromosomal instability, which was hypothesized to promote the emergence of drug resistant CTC sub-clones with increased metastatic capacity, offering potential mechanisms of ROS1-inhibitor-therapy resistance in ROS1-rearranged NSCLC tumors.
Detection of mutations
In 2008, Maheswaran et al. reported the detection of EGFR-mutations in CTCs isolated from 27 NSCLC patients using the CTC-chip, a microfluidic device containing microposts coated with anti-EpCAM antibodies (66). EGFR mutational analysis was performed using allele-specific PCR amplification and was compared to the results from the isolated free plasma DNA and original tumor biopsy. They identified the expected EGFR-activating mutation in CTCs from 11 of 12 patients (92%) and in matched free plasma DNA from 4 of 12 patients (P=0.009). They also identified the T790M resistance mutation in CTCs from patients who had already received TKI therapy. Serial analysis also demonstrated that reduction in CTC number under treatment was associated with an improved radiological response and vice versa. Interestingly, in patients who developed progression different EGFR-activating mutations were identified in CTCs suggesting the emergence of different tumor subclones. Punnoose et al. reported on the clinical correlation between change in CTC number detected during therapy and treatment response (67). The study cohort consisted of 41 patients with relapsed or refractory NSCLC who were enrolled in a single arm phase II clinical trial of erlotinib and pertuzumab, but only eight patients had confirmed EGFR-activating mutations in the tumor tissue. Peripheral blood was analyzed for CTC enumeration using the CellSearch platform at baseline and changes in CTC levels were assessed for correlation with PDG-PET and CT imaging and survival endpoints. The analysis reported a statistically significant correlation between high baseline CTC counts and patient response to treatment according to RECIST (P=0.009). EGFR mutational analysis was performed on CTCs captured by the CellSearch profile kit. After DNA extraction and a pre-amplification step, mutations were detected by Taqman PCR using multiplex gene-specific primers. Only one EGFR genetic alteration (exon 19 deletion) was detected in CTCs while eight patients had confirmed mutations in archival tumor tissue. In 2014, Marchetti et al. reported the results of a multi-center trial of erlotinib treatment in 37 advanced NSCLC patients with activating EGFR-mutations in tumor tissue. CTCs were obtained from the CellSearch and subjected to ultra-deep next generation sequencing (NGS) (68). EGFR-mutations were in 31 (84%) of the CTC samples examined, 25 (81%) had in frame exon 19 deletions and 6 (19%) had point mutations at exon 21. In 29 (91%) of the 31 cases the mutation type detected by NGS in CTCs corresponded to that found in matching tumor tissue by Sanger sequencing. Interestingly in 4 cases, double or multiple mutations were observed by NGS in CTCs suggesting CTC heterogeneity. No mutations were detected in control samples (CTC from 10 breast cancer patients, 12 healthy subjects). Gorges et al. reported the detection of mutations in the EGFR and KRAS genes in CTCs isolated by the Gilupi CellCollector in vivo system (69). In this study the authors provided a proof-of-principle that CTCs captured by this device were suitable for molecular analysis. Captured CTCs from two patients with known mutations in the primary tumor were analyzed using Digital PCR after whole genome amplification. The same KRAS- and EGFR-mutations found in the primary tumors were detected when one to five CTCs were detected by the system. Recently, Zhang et al. reported the L1196M resistance mutation in short-term cultured CTCs from ALK-rearranged NSCLC (70).
Clinical use of CTCs: ready for prime-time?
Thus there is still very little data on the detection of these gene aberrations in CTCs from NSCLC patients. All the exploratory studies presented above have only involved a small number of patients. In most cases, different CTC enrichment techniques were used which implies that CTCs identified positive for a genetic biomarker possibly differed from one study to another and the results cannot be compared. Beyond this proof-of-concept studies, there are still many steps that should be taken before CTC assays can be used in routine clinical diagnosis. First of all, because the studies presented above used different CTC enrichment techniques, it is important to identify for each genetic biomarker the most appropriate CTC enrichment technique prior to CTC identification and downstream molecular assays. Such a comparison of test performance, validation of selected assays in cohorts of positive and negative patients and their further clinical qualification will require important collaborative efforts. Such projects can only be carried out within the framework of collaborative groups, for example, the Cancer-ID European consortium whose purpose is to validate and standardize liquid biopsy assays.
Regarding FISH assays, the automation of microscopy analysis is a necessary step to improve reproducibility of analyses, reduce risks of errors, inter-operator differences, and progress towards a standardization and validation of biomarker detection. Our group recently reported a semi-automated method established to analyze filtration-enriched CTCs according to combined fluorescent staining and FISH (63). This method relies on the detection of molecular biomarkers by establishing FISH scanning parameters (z-stacking, step, i.e., distance between two z-stacks, exposure time) for optimal FISH signal identification in filtration-enriched CTCs. For using CTC FISH assays in routine clinical diagnosis, another important point that increases the cumbersome nature of the analytical process is the necessity of the validation of FISH signals by an experienced cytogeneticist.
Mutational analyses have been mainly performed on DNA extracted from enriched CTC populations and in most studies the sensitivity of detection was limited by the presence of leukocyte DNA. The analysis of CTCs at the single cell level will overcome this limitation but imposes important technical challenges for individual CTC isolation, whole genome amplification (WGA), PCR for targeted mutations and NGS. WGA is a process which is prone to amplification bias, polymerase errors and results in nucleotide variations errors and allelic distortion. The distinction of true somatic variants present in CTCs from WGA errors is essential and requires a very rigorous and careful assessment.
Assays using circulating tumor DNA (ctDNA) assays for detecting gene aberrations (i.e., mutations, gene rearrangements and gain of copy numbers) are being rapidly implemented in the clinic and several studies have shown the specificity and sensitivity of EGFR-mutations detection in ctDNA from NSCLC patients (71-73). Although ctDNA assays indeed offer advantages in terms of simplicity, the molecular analysis of CTCs can provide unique additional information such as the morphology and phenotype. The presence of multiple and different genetic alterations can be identified within the same CTC offering the possibility to infer tumor heterogeneity and evolution. Although the utilization of CTCs is less eminent, CTCs and ctDNA are anticipated to be complementary in their clinical utility. Moreover while tumor tissue biopsies presently remain the gold standard for CTCs and ctDNA analyzes, it is important to question their use as a reference (74). CTCs and ctDNA can be derived from lesions that were not biopsied and may contain a different genetic composition than the tumor biopsies used as a reference.
Although many steps have still to be evaluated and validated, it is anticipated that the progression from “bench-to-bedside” of the molecular analysis of CTCs would be critical to aid the personalized treatment of patients with NSCLC. When considering the path that research in this field will follow over the next 5 years there are some key elements which come to mind; firstly, the validation of CTC detection and characterization techniques on large patient cohorts is necessary and secondly the development of automated platforms to simplify and standardize techniques to aid their passage to widespread clinical utility. Currently several research teams are focusing on the use of highly sophisticated automated platforms and we would hope that over the next 5 years clinical validation of their diagnostic role in NSCLC will be achieved. Thirdly the development of single cell sequencing technologies offers the opportunity to distinguish complex tumor genomes from single CTCs and along intra-tumor heterogeneity and tumor evolution. Ultimately the overall objective is to optimize the tailoring of personalized therapies to NSCLC patients with CTCs potentially utilized in a diagnostic role and predictive assessments.
Acknowledgements
The authors are grateful to the patients and their families.
Funding: EP is supported by the LabEx LERMIT (grant No. ANR-10-LABX-0033-LERMIT) and the Fondation pour la Recherche Médicale (grant No. FDT20150532072). VF is supported by the Fondation pour la Recherche Médicale (grants No. FDT20160435543). CC is supported by the Agence Nationale pour la Recherche (ANR-CE17-0006-01). The authors are grateful for the research support of the Fondation de France (grant No. 201300038317), the Fondation ARC pour la Recherche sur le Cancer (grant No. 20131200417), Innovative Medicines Initiative 11th Call CANCER ID (IMI-JU-11-2013, 115749), Institut National du Cancer (PRT-K14-032) and Agence Nationale de la Recherche (ANR-CE17-0006-01).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Jänne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov 2009;8:709-23. 10.1038/nrd2871 [DOI] [PubMed] [Google Scholar]
- 2.Hiley CT, Le Quesne J, Santis G, et al. Challenges in molecular testing in non-small-cell lung cancer patients with advanced disease. Lancet 2016;388:1002-11. 10.1016/S0140-6736(16)31340-X [DOI] [PubMed] [Google Scholar]
- 3.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science 2002;297:63-4. 10.1126/science.1073096 [DOI] [PubMed] [Google Scholar]
- 4.Kerr KM, Bubendorf L, Edelman MJ, et al. Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non-small-cell lung cancer. Ann Oncol 2014;25:1681-90. 10.1093/annonc/mdu145 [DOI] [PubMed] [Google Scholar]
- 5.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shames DS, Wistuba II. The evolving genomic classification of lung cancer. J Pathol 2014;232:121-33. 10.1002/path.4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massard C, Michiels S, Ferte C, et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov 2017;7:586-95. 10.1158/2159-8290.CD-16-1396 [DOI] [PubMed] [Google Scholar]
- 8.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn 2013;15:415-53. 10.1016/j.jmoldx.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 9.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 10.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. 10.1073/pnas.0405220101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 13.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. 10.1016/S1470-2045(09)70364-X [DOI] [PubMed] [Google Scholar]
- 14.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 15.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 16.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. 10.1200/JCO.2010.33.4235 [DOI] [PubMed] [Google Scholar]
- 17.Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol 2010;28:357-60. 10.1200/JCO.2009.24.7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortot AB, Janne PA. Molecular mechanisms of resistance in epidermal growth factor receptor- mutant lung adenocarcinomas. Eur Respir Rev 2014;23:356-66. 10.1183/09059180.00004614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacher AG, Janne PA, Oxnard GR. Management of acquired resistance to epidermal growth factor receptor kinase inhibitors in patients with advanced non-small cell lung cancer. Cancer 2014;120:2289-98. 10.1002/cncr.28723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 22.Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol 2013;31:1105-11. 10.1200/JCO.2012.44.5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solomon B, Soria JC. The continuum of care for ALK-positive NSCLC: from diagnosis to new treatment options - an overview. Ann Oncol 2016;27 Suppl 3:iii1-iii3. [DOI] [PubMed] [Google Scholar]
- 24.Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016;387:1415-26. 10.1016/S0140-6736(16)00004-0 [DOI] [PubMed] [Google Scholar]
- 25.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK- positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 2012;13:1011-9. 10.1016/S1470-2045(12)70344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK- positive lung cancer. N Engl J Med 2013;368:2385-94. 10.1056/NEJMoa1214886 [DOI] [PubMed] [Google Scholar]
- 27.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. 10.1056/NEJMoa1408440 [DOI] [PubMed] [Google Scholar]
- 28.Blackhall F, Cappuzzo F. Crizotinib: from discovery to accelerated development to front-line treatment. Ann Oncol 2016;27 Suppl 3:iii35-iii41. 10.1093/annonc/mdw304 [DOI] [PubMed] [Google Scholar]
- 29.Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. 10.1056/NEJMoa1007478 [DOI] [PubMed] [Google Scholar]
- 30.Dagogo-Jack I, Shaw AT. Crizotinib resistance: implications for therapeutic strategies. Ann Oncol 2016;27 Suppl 3:iii42-iii50. 10.1093/annonc/mdw305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katayama R, Khan TM, Benes C, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci U S A 2011;108:7535-40. 10.1073/pnas.1019559108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. 10.1158/1078-0432.CCR-11-2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. 10.1126/scitranslmed.3003316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gainor JF, Shaw AT. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J Clin Oncol 2013;31:3987-96. 10.1200/JCO.2012.45.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. 10.1158/1078-0432.CCR-13-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. 10.1158/2159-8290.CD-13-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyokawa G, Hirai F, Inamasu E, et al. Secondary mutations at I1171 in the ALK gene confer resistance to both Crizotinib and Alectinib. J Thorac Oncol 2014;9:e86-7. 10.1097/JTO.0000000000000358 [DOI] [PubMed] [Google Scholar]
- 38.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014;11:473-81. 10.1038/nrclinonc.2014.104 [DOI] [PubMed] [Google Scholar]
- 39.Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second- Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-1133. 10.1158/2159-8290.CD-16-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. 10.1200/JCO.2011.35.6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865-75. 10.1634/theoncologist.2013-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 44.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev 2008;18:73-9. 10.1016/j.gde.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 45.Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol 2014;11:129-44. 10.1038/nrclinonc.2013.253 [DOI] [PubMed] [Google Scholar]
- 46.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 47.Young R, Pailler E, Billiot F, et al. Circulating tumor cells in lung cancer. Acta Cytol 2012;56:655-60. 10.1159/000345182 [DOI] [PubMed] [Google Scholar]
- 48.Ross K, Pailler E, Faugeroux V, et al. The potential diagnostic power of circulating tumor cell analysis for non-small-cell lung cancer. Expert Rev Mol Diagn 2015;15:1605-29. 10.1586/14737159.2015.1111139 [DOI] [PubMed] [Google Scholar]
- 49.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 50.de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- 51.Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 52.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. 10.1200/JCO.2010.28.7045 [DOI] [PubMed] [Google Scholar]
- 53.Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer 2011;105:847-53. 10.1038/bjc.2011.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou JM, Krebs M, Ward T, et al. Circulating tumor cells as a window on metastasis biology in lung cancer. Am J Pathol 2011;178:989-96. 10.1016/j.ajpath.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. 10.1002/ijc.25819 [DOI] [PubMed] [Google Scholar]
- 56.Lecharpentier A, Vielh P, Perez-Moreno P, et al. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer 2011;105:1338-41. 10.1038/bjc.2011.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindsay CR, Faugeroux V, Michiels S, et al. A Prospective Examination of Circulating Tumor Cell Profiles in Non-Small-Cll Lung Cancer Molecular Subgroups. Ann Oncol 2017. [Epub ahead of print]. 10.1093/annonc/mdx156 [DOI] [PubMed] [Google Scholar]
- 58.Kerr KM, Lopez-Rios F. Precision medicine in NSCLC and pathology: how does ALK fit in the pathway? Ann Oncol 2016;27 Suppl 3:iii16-iii24. 10.1093/annonc/mdw302 [DOI] [PubMed] [Google Scholar]
- 59.Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. 10.1093/annonc/mds137 [DOI] [PubMed] [Google Scholar]
- 60.Pailler E, Adam J, Barthelemy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. 10.1200/JCO.2012.44.5932 [DOI] [PubMed] [Google Scholar]
- 61.He W, Xu D, Wang Z, et al. Detecting ALK-rearrangement of CTC enriched by nanovelcro chip in advanced NSCLC patients. Oncotarget 2016. [Epub ahead of print]. [Google Scholar]
- 62.Tan CL, Lim TH, Lim T, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 2016;7:23251-62. 10.18632/oncotarget.8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pailler E, Oulhen M, Billiot F, et al. Method for semi-automated microscopy of filtration-enriched circulating tumor cells. BMC Cancer 2016;16:477. 10.1186/s12885-016-2461-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pailler E, Oulhen M, Borget I, et al. Circulating Tumor Cells with Aberrant ALK Copy Number Predict Progression-Free Survival during Crizotinib Treatment in ALK-Rearranged Non–Small Cell Lung Cancer Patients. Cancer Res 2017;77:2222-30. 10.1158/0008-5472.CAN-16-3072 [DOI] [PubMed] [Google Scholar]
- 65.Pailler E, Auger N, Lindsay CR, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol 2015;26:1408-15. 10.1093/annonc/mdv165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. 10.1056/NEJMoa0800668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. 10.1158/1078-0432.CCR-11-3148 [DOI] [PubMed] [Google Scholar]
- 68.Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One 2014;9:e103883. 10.1371/journal.pone.0103883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gorges TM, Penkalla N, Schalk T, et al. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In Vivo Device for Capturing Circulating Tumor Cells. Clin Cancer Res 2016;22:2197-206. 10.1158/1078-0432.CCR-15-1416 [DOI] [PubMed] [Google Scholar]
- 70.Zhang Z, Shiratsuchi H, Palanisamy N, et al. Expanded CTCs from a Patient with ALK Positive Lung Cancer Present EML4-ALK Rearrangement along with Resistance Mutation and Enable Drug Sensitivity Testing: A Case Study. J Thorac Oncol 2017;12:397-402. 10.1016/j.jtho.2016.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 72.Jovelet C, Ileana E, Le Deley MC, et al. Circulating Cell-Free Tumor DNA Analysis of 50 Genes by Next-Generation Sequencing in the Prospective MOSCATO Trial. Clin Cancer Res 2016;22:2960-8. 10.1158/1078-0432.CCR-15-2470 [DOI] [PubMed] [Google Scholar]
- 73.Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 2017;28:784-790. [DOI] [PubMed] [Google Scholar]
- 74.Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. 10.1200/JCO.2016.66.7162 [DOI] [PMC free article] [PubMed] [Google Scholar]


