Abstract
Telomeres are important structures for DNA replication and chromosome stability during cell growth. Telomere length has been correlated with the division potential of human cells and has been found to decrease with age in healthy individuals. Nevertheless, telomere lengths within the same cell are heterogeneous and certain chromosome arms typically have either short or long telomeres. Both the origin and the physiological consequences of this heterogeneity in telomere length remain unknown. In this study we used quantitative telomeric FISH combined with a method to identify the parental origin of chromosomes to show that significant differences in relative telomere intensities are frequently observed between chromosomal homologs in short-term stimulated cultures of peripheral blood lymphocytes. These differences appear to be stable for at least 4 months in vivo, but disappear after prolonged proliferation in vitro. The telomere length differences are also stable during in vitro growth of telomerase-negative fibroblast cells but can be abolished by exogenous telomerase expression in these cells. These findings suggest the existence of a mechanism maintaining differences in telomere length between chromosome homologs that is independent of telomere length itself.
INTRODUCTION
Telomeres are specialized structures found at the ends of chromosomes whose primary structure is the same for all vertebrates (T2AG3)n. They typically shorten at each cell division, due to the inability of conventional DNA polymerases to replicate the end of linear molecules (1). Under normal conditions, cells unable to maintain telomere length stop dividing as soon as their telomeres fall below a critical minimum size (2,3). In vitro experiments showed that induction of telomerase expression was sufficient to restore the division potential of some types of cells with shortened telomeres (4). In vivo cells undergoing multiple cell divisions, such as tumor cells, must find an efficient mechanism to maintain telomere length. Usually this involves activation of expression of TERT (5,6), the catalytic subunit of the telomerase complex, which elongates the 3′-ends of chromosomes (7).
In both mice and humans telomere length seems to be, at least partially, genetically determined (8,9). Estimation of telomere length in peripheral blood lymphocytes (PBLs) from healthy human donors has shown an inverse correlation between telomere length and age (10), leading to speculations on a relationship between telomere length and lifespan. In mice, however, telomere length seems not to be correlated with aging (11) and the phenotypic consequences of disrupting the telomerase gene are only observed after several generations (12).
The distribution of telomere sizes has classically been determined through telomere restriction fragment (TRF) analysis, after enzymatic digestion of genomic DNA and Southern blotting. Recently, fluorescence in situ hybridization (FISH) techniques have been introduced to estimate individual telomere sizes (13,14). In particular, the use of telomere-specific PNA (peptide nucleic acids) probes permitted the demonstration of heterogeneity between telomere lengths within the same cell (14). It was subsequently suggested that human chromosome 17 short arms are associated with short telomeres (15). Since individual homologs were not distinguished, it remained possible that telomere length differences existed between homologs.
Human chromosomes are highly polymorphic with regard to their telomeres. Most sub-telomeric sequences derived from chromosome-specific human half-YACs are found, either through FISH or by PCR, to be common to several other chromosome ends (16–18). Differences in size (sometimes implicating hundreds of kilobases) between sub-telomeric alleles (16pter and 2qter) have also been shown to exist in normal populations (19,20). Polymorphism, involving duplication of a large block of sub-telomeric sequence, has recently been observed in several human chromosomes (21,22). These large insertion/deletion polymorphisms can be consistently detected by FISH using cosmid probes carrying the corresponding sequences (21,22).
In this report, we have combined quantitative telomere PNA-FISH and conventional FISH techniques to obtain homolog-specific telomere length estimations. We monitored the presence of telomere length differences between homologous chromosomes in stimulated PBLs from human donors. The observed differences in telomere length between homologs were also followed in vitro in cultured telomerase-negative fibroblast cells. Finally, the effect of exogenous telomerase expression on these differences was analyzed.
MATERIALS AND METHODS
Cell lines and culture conditions
WI-38 (a human diploid fibroblast cell line) cells were obtained from ATCC, at population doubling (PD) 30 and were kept in culture following ATCC recommendations, with 1/4 dilutions (+2PD), until cells reached senescence. WI-38+hTERT cells (PD48) were a gift of Judith Campisi (Lawrence Berkeley National Laboratory, Berkeley, CA) and were kept in culture under the same conditions as the wild-type WI-38 strain. At the indicated PDs, cultures were harvested for DNA extraction or TRAP assay. For metaphase chromosome preparations, 10 µg/ml colcemid (Eurobio) was added to culture flasks 1 h before harvesting, followed by KCl hypotonic shock and methanol/acetic acid (3:1) fixation. Fixed pellets were kept at –20°C until use.
Human lymphoblastoid cell lines from the CEPH collection were cultured in RPMI with 10% FCS. Flasks were split for simultaneous metaphase chromosome preparation and DNA extraction. For correlation studies between telomere fluorescence intensity and age, aliquots were anonymously obtained from fresh blood samples submitted to CEPH; only the date of birth was registered. For reproducibility studies of single telomere intensity estimations, two or three blood samples were obtained from five healthy volunteers over a period of 4 months. Whole blood (0.5 ml) was diluted in 5 ml of RPMI + 10% FCS and incubated in 14 ml tubes with 30 µg/ml PHA-C (Biosepra) at 37°C for 72 h. Metaphase chromosomes were prepared as described above. For extended proliferation studies, two purified PBL frozen samples from the CEPH collection were thawed out in RPMI + 10% FCS and stimulated 24 h later with 30 µg/ml PHA-P (Difco). After 24 h, the medium was replaced by IL-2 conditioned medium (23) and cultures were maintained for 10 more days. Metaphase chromosome preparations were obtained on days 2 and 11 after the beginning of stimulation.
TRF analysis
DNA was extracted using a Wizard kit (Promega) and aliquots (5 µg) digested overnight with the restriction enzymes RsaI and HinfI. Digested DNA fragments were separated in 0.65% TAE agarose gels and blotted onto positively charged nylon membranes. Telomere restriction fragments were revealed through hybridization with a 32P-labeled (CCCTAA)5 oligonucleotide probe. Signals were visualized and quantified in a phosphorimager (Bio-Rad). Intensity profiles of the smears were obtained and telomere mean sizes were calculated at the peak of signal intensity.
Telomerase assay
Telomerase activity was detected using a TRAPeze kit (Appligene). An equivalent number of cells were processed at indicated PDs for wild-type WI-38 and WI-38+hTERT fibroblasts. Products were separated by PAGE (10%) and quantified in a phosphorimager.
DNA probes
Cosmids ICRF10 (21) and f7501 (22) were obtained from Drs Gilles Vergnaud (IGM, Orsay, France) and Barbara Trask (Human Genome Center, Lawrence Livermore National Laboratory), respectively. BAC b253C3, from the CEPH collection, was occasionally used in FISH experiments to assign chromosome 8 homologs.
FISH
Metaphase spreads were prepared the day before telomere PNA-FISH experiments, which were carried out as described (24) using a (CCCTAA)3-Cy3 PNA probe (PerSeptive Biosystems). For identification of specific telomeres, a second FISH step was performed on the same slides using standard methods (25) with slight modifications. Briefly, after image acquisition and storage of telomeric signals, slides were washed in BN buffer [100 mM NaHCO3, 0.05% v/v Igepal (Sigma)] and denatured in 70% formamide, 2× SSC at 70°C for 2 min. After dehydration, slides were covered with 40 µl of hybridization buffer (50% formamide, 10% dextran sulfate, 2× SSC) containing 80 ng biotin- or digoxigenin-labeled cosmids, 26 µg Cot1 DNA and 100 µg sonicated salmon sperm DNA. Slides were incubated at 37°C overnight and then washed. Biotin was revealed with Texas red–avidin (Vector) and digoxigenin with FITC-conjugated anti-digoxin (Sigma). Chromosomes were counterstained with DAPI.
Image capture
After PNA hybridization, fluorescence signals were visualized under an epifluorescence microscope (Axioplan2; Zeiss) equipped with a computer piloted filter wheel. After localization of metaphases, blue (DAPI) and red (Cy3) fluorescence signals were captured by a CCD camera (Photometrics-Sensys) using Smart-Capture software (Vysis) (settings: red, gain = 3; blue, gain = 1; binning = 4). A flat field template was used to correct for unevenness in field illumination and fluorescent beads were regularly used to check for UV light intensity decay (Fluoresbrite YG, 2 µm; Polyscience). Fixed exposure times of 1 or 2 s were sufficient to reveal all telomere signals in a metaphase spread, thus avoiding signal saturation and eliminating the need for time corrections as well as biases due to the presence of other stained objects in the same field (i.e. interphase nuclei). Original (not normalized) black and white (B&W) images were saved and used for quantitative analysis. Merged pseudo-color images were used to identify chromosomes based on DAPI simulated G-banding. For conventional FISH images, automatic exposure was allowed and normalization and enhancement procedures were used to improve G-banding and specific signal detection. To estimate total telomere fluorescence intensity, 20–40 metaphases per preparation were captured/stored. For single telomere analysis in WI-38 cells, 40–100 metaphases per preparation were captured/stored. With PBLs, 30–50 metaphases per individual and per preparation were captured/stored. Depending on the slides, 40–90% of the pre-localized metaphases were retrieved after the second FISH hybridization and analyzed.
Quantitative analysis of digital images
Iplab Spectrum P software (Skanalytics) was used for the quantitative analysis of images. Unmodified B&W images were used to estimate the mean pixel value for telomeric signals after manual (for single homolog-specific telomere measurement) or automatic (when the mean intensity for all telomeres in a metaphase was measured) segmentation protocols. Mean metaphase intensities were obtained by subtracting the mean pixel value associated with the interstitial regions of chromosomes (background) from the mean pixel value for all telomeres in the metaphase. The relative intensities of individual telomeres were obtained by dividing the mean pixel value associated with that telomere by the mean pixel value of all telomeres in the metaphase.
Statistical analysis
Regression analysis was used to test relationships between age and telomere fluorescence or mean TRF size. Differences between individual telomeres were tested either by paired t-test comparisons or analysis of variance after logarithmic transformation of the relative intensities. The Kolmogorov–Smirnov two-sample test was used to test for differences in the distributions of relative telomere intensities.
RESULTS
Telomere fluorescence intensities reflect telomere length
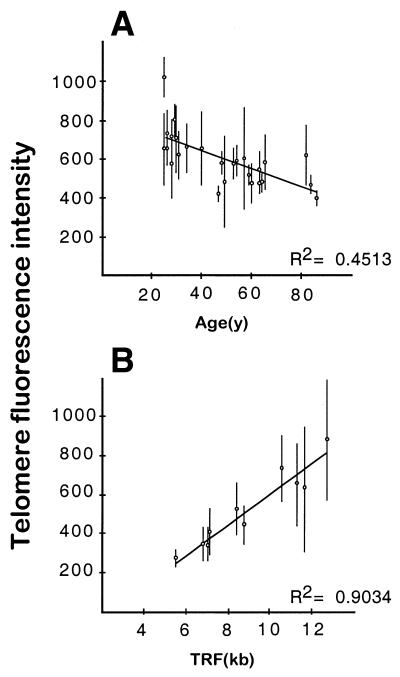
The fluorescence intensities of different telomeres, revealed through PNA-FISH, were heterogeneous within the same metaphase spread, as observed by Lansdorp et al. (14). The intensities of signals corresponding to sister chromatids were highly correlated (r = 0.87; not shown), indicating similar local hybridization efficiencies for predicted similar telomere lengths. In all experiments, metaphase intensities also varied from metaphase to metaphase on the same slide (coefficient of variation 15–25%). This variation may result from factors such as different replicative histories of cells, differences in hybridization efficiencies across the slide and/or UV source fluctuations (15). When mean fluorescence intensities for all chromosomes were calculated from around 20 metaphases, differences between individuals were also observed. In the case of PBLs, mean metaphase intensities were inversely correlated with donor age (r = –0.67; Fig. 1A), whereas in the case of lymphoblastoid cell lines they were directly correlated with telomere length, as determined by TRF analysis (r = 0.95; Fig. 1B). Taken together, these results indicate that, in our hands, telomere fluorescence intensities obtained by PNA-FISH do reflect telomere length.
Figure 1.
Telomere fluorescence intensities obtained through PNA-FISH reflect telomere length. (A) Regression of mean fluorescence intensities, measured by PNA-FISH on PBL metaphase spreads, on age (in years) (n = 25). Vertical bars represent standard deviations from the mean metaphase intensity (n = 15–25, exposure time 2 s). (B) Regression of mean fluorescence intensities, measured on metaphase spreads from 10 lymphoblastoid cell lines, on the sizes (in kb) of telomere restriction fragments as detected by Southern blot. Vertical bars represent standard deviations from the mean (n = 15–25, exposure time 2 s). Fluorescence intensity is expressed as mean pixel values.
Differences in telomere intensity between homologs are frequently detected in human PBLs
A second two-color FISH procedure, using probes f7501 and ICRF10 and applied to PNA-hybridized slides, allowed us to distinguish between some homologous chromosomes carrying insertion/deletion polymorphisms in sub-telomeric regions. This procedure was not likely to interfere with telomere intensity measurements since PNA-FISH images were captured and stored for later analysis before the second hybridization step.
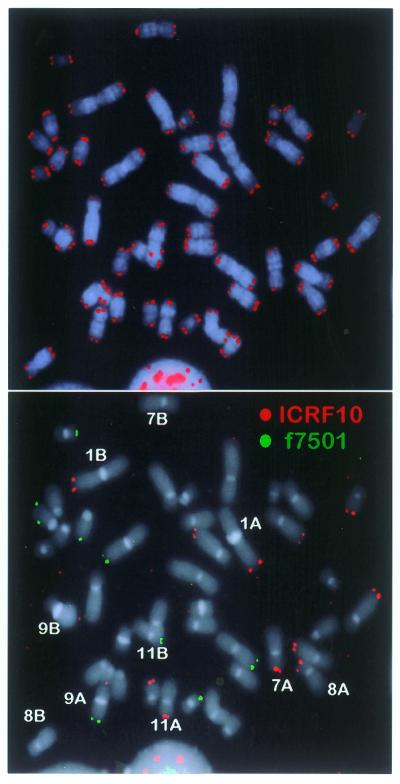
The number of chromosome pairs accessible to our analysis in stimulated PBLs from five healthy human donors varied from two to five depending on the donor. Homologs of chromosome 11 pairs could be distinguished in all donors since all of them were hemizygous for f7501 (4/5) and/or ICRF10 (4/5). Other chromosomes found to carry hemizygosities were 8 (3/5), 9 (2/5) and 1, 5, 7, 9, 15 and 16 (1/5) (Fig. 2). When heteromorphisms were present, such as obvious differences in heterochromatin size on chromosomes 1 (2/5) and 9 (1/5), they were also exploited to distinguish between homologs.
Figure 2.
Identification of homolog-specific telomere signals. Metaphase spread after hybridization with telomeric PNA (top) followed by a second hybridization step with polymorphic subtelomeric probes, f7501 and ICRF10 (bottom). Chromosomes are counterstained with DAPI. In this particular donor, the second step allows distinction between homologs for chromosome pairs 7, 8, 9 and 11. Homologs for chromosome pair 1 can be distinguished by the size of their heterochromatin.
Homolog-specific telomere signal intensities showed high variability (coefficient of variation 40–50%) between metaphases on the same preparation from the same donor (not shown). This variability was greatly reduced by normalization of the intensity of single telomeres within a metaphase with respect to the mean fluorescence intensity of all telomeres, which commonly resulted in a coefficient of variation for homolog-specific relative intensities of <10% (not shown). Furthermore, relative telomere intensities thus obtained for every telomere and from more than 20 metaphases showed a near to normal distribution, unlike the distribution obtained when absolute values are used (not shown).
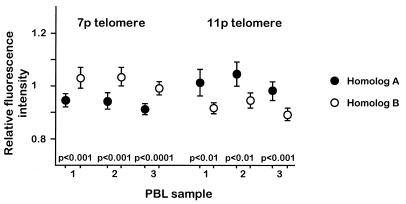
Individual homolog-specific relative telomere intensities obtained in different experiments with the same stimulated PBL sample were very similar (not shown). In most donors, paired comparisons between homologs frequently revealed significant differences (Table 1). The extreme medians for telomere-specific relative intensities were 0.88 and 1.10 and the maximum observed difference between two homologs was 0.19 (15p, Table 1). While the mean metaphase intensities were correlated with age (P < 0.05), there was no correlation between age and the degree of dispersion in the distribution of relative intensities when all telomeres were considered (not shown). Differences were reproducibly found in metaphase spreads of stimulated PBLs from the same donors isolated at 1–2 month intervals over a period of 4 months (Fig. 3).
Table 1. Differences in telomere relative intensities between homologous chromosomes are frequently detected in healthy donors.
 |
Fresh PBLs from five healthy donors were stimulated with PHA for 72 h and metaphase spreads were analyzed by telomere-specific PNA-FISH. A second hybridization allowed certain pairs of homologous chromosomes to be distinguished. The medians of the relative intensities measured on identifiable homologous telomeres are shown. The 95% confidence limits for the mean are also shown. Paired comparisons yielding statistically significant differences (P < 0.01) between homologs are boxed. n, average number of metaphases analyzed. Exposure time 2 s.
Figure 3.
Relative fluorescence intensity differences between homologous telomeres detected in short-term stimulated human PBLs are consistent. The circles represent mean relative fluorescence intensities measured on telomeres for both homologs of chromosomes 7p and 11p from the same donor. PBL samples were obtained over a period of 4 months. P values from paired t-tests between homologous positions are also shown. The number of metaphases analyzed in each sample varied from 25 to 40. Vertical bars indicate the 95% confidence intervals.
When the same locus was accessible to analysis in several donors, both high and low relative values were observed. Given the small number of observations and the lack of markers linked to many telomere locations, it was not possible to establish associations with high or low relative telomere lengths. However, in the case of chromosome 11, where eight pter alleles were distinguished by adjacent f7501 or ICRF10 sequences, all f7501-positive alleles yielded low relative telomere intensities whereas ICRF10-positive alleles yielded higher values (mean 0.93 versus 1.00).
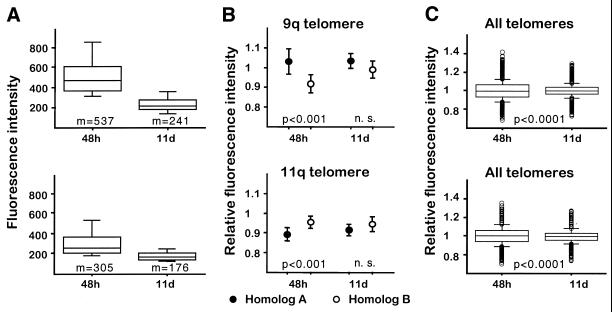
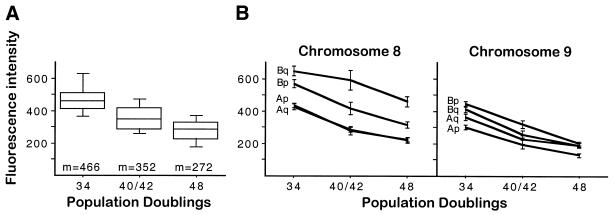
We finally looked at telomere length differences between homologous chromosomes after prolonged in vitro proliferation in two other PBL samples. In both cases, as expected, the mean metaphase intensities decreased with time (Fig. 4A). Significant differences in relative telomere intensities between homologs detected at 48 h disappeared after 11 days of in vitro proliferation (Fig. 4B), a time at which the overall distribution of telomere relative intensities was significantly more homogeneous (P < 0.0001) (Fig. 4C).
Figure 4.
Extended lymphocyte proliferation following in vitro stimulation leads to homogenization of telomere relative intensities. (A) The box plots represent the distribution of mean fluorescence intensities of metaphase spreads (n = 25–30) from two different donors (top and bottom), 48 h and 11 days after stimulation of PBLs in vitro. The five horizontal lines in each box indicate the 10th, 25th, 50th, 75th and 90th percentiles. Fluorescence intensity is expressed as mean pixel values. Exposure time 1 s. (B) The circles represent the mean relative intensities obtained for the telomeres on chromosomes 9q and 11q in the same metaphases measured in (A). The results of paired t-tests carried out between homologous positions are also shown. n. s., not statistically significant. (C) Box plot representing the distribution of relative intensity measurements obtained for all telomeres in the same metaphases. Small circles represent the observations below the 10th and above the 90th percentiles. In each case, the distributions are significantly different as revealed by the Kolmogorov–Smirnov two-sample test.
Telomere length differences between homologous chromosomes are stable in telomerase-negative, mortal fibroblasts replicating in vitro
To evaluate the impact of telomere shortening on telomere length differences between homologous chromosomes, we conducted experiments with telomerase-negative human fibroblasts. WI-38 fibroblast cells were maintained in culture for 6 weeks, during which they underwent 18 PDs before growth stopped. Since cells were estimated to have undergone 34 PDs before the beginning of the experiments, senescence was reached after about PD52, in agreement with previous observations (26). Telomere length, as estimated by TRF analysis, decreased during growth (from 8.0 to 5.7 kb, i.e. 140 bp/cell division; not shown), as did the mean metaphase fluorescence intensity (Fig. 5A).
Figure 5.
Overall and individual telomere fluorescence intensities in WI-38 cells during growth. (A) The box plot represents the distribution of mean fluorescence intensities measured on metaphase spreads of WI-38 cells at three different points (population doublings) during in vitro growth. (B) Mean fluorescence intensities obtained for individual telomeres on chromosomes 8 and 9 in the same metaphases. Vertical bars represent standard errors for the data. Fluorescence intensities are expressed as mean pixel values. The number of measured metaphases analyzed varied from 40 (for PD40/42) to 100 (for PD34). Exposure time 2 s.
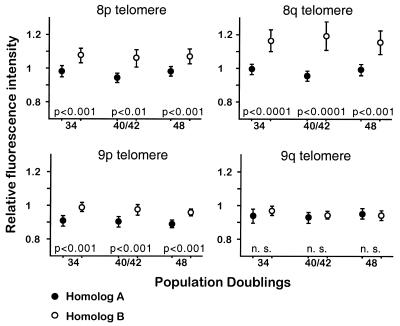
In these cells, homologs for chromosomes 8 and 9 were distinguishable because of their hemizygosities for ICRF10 on 8pter and 9qter. As expected, the mean fluorescence intensity for individual telomeres decreased with increasing PDs (Fig. 5B). On the other hand, relative telomere intensities for individual homologs remained stable throughout the culture period (Fig. 6). Paired comparisons between homologs revealed significant differences in three of the four loci at all studied time points (Fig. 6). Relative intensities for p and q telomeres on homolog 8B (negative for ICRF10) were consistently higher than those estimated on homolog 8A. In addition, the p telomere on homolog 9A (positive for ICRF10) always yielded lower relative intensities than its counterpart. In fact, the p telomere on chromosome 9A was consistently as short (mean 0.901) as both 17p telomeres (mean 0.908) in this cell line (P > 0.1).
Figure 6.
Differences between the relative fluorescence intensities of homologous telomeres are stable in WI-38 fibroblasts during in vitro growth. The circles indicate the mean relative intensities obtained for each telomere on both homologs of chromosomes 8 and 9, at different population doublings. The bars represent the 95% confidence limits. The results of paired t-tests carried out between homologous positions are also shown. n. s., not statistically significant.
Telomere length differences between homologs are abolished in WI-38 fibroblasts expressing exogenous telomerase
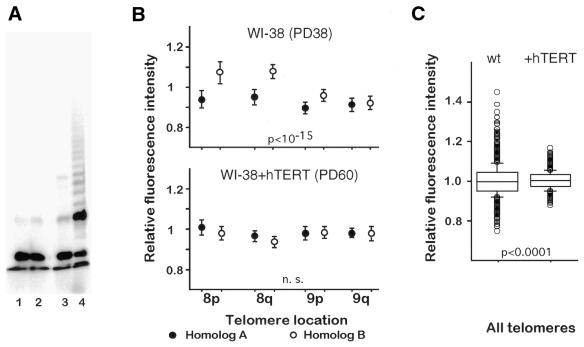
We next examined the impact of telomerase activity on telomere length differences in WI-38 fibroblasts transfected with hTERT. As expected, telomerase activity was present in WI-38+hTERT but not in wild-type WI-38 cells (Fig. 7A). WI-38+hTERT fibroblasts, which had been transfected at about PD30, were analyzed at PD50, 60 and 70, i.e. near and beyond the senescence point for wild-type WI-38 cells, which were again analyzed in parallel at PD38. WI-38+hTERT cells (PD60) had slightly longer telomeres than WI-38 cells (PD38), as revealed by TRF analysis (8.0 versus 7.3 kb) and PNA-FISH (mean metaphase intensity 429 ± 55 versus 376 ± 26; not shown). Paired comparisons between homolog-specific relative telomere intensities revealed that differences initially detected in wild-type cells were no longer present in WI-38+hTERT cells (not shown). Moreover, an analysis of variance including all homolog-specific estimations showed that the telomere lengths at these eight locations had been homogenized in the telomerase-expressing cells (Fig. 7B). Comparisons between single relative length estimations before and after transduction showed that, for instance, the telomere on homolog 9A, which was associated with a relatively low telomere intensity in wild-type WI-38 cells, exhibited a higher relative value in WI-38+hTERT cells (0.921 versus 0.976, respectively, P < 0.001). This is in contrast to the situation observed with the q telomere on homolog 8B, for which relative intensities were much lower in the transfected cell line than in wild-type cells (1.068 versus 0.941, P < 10–7).
Figure 7.
Expression of telomerase in WI-38 cells leads to homogenization of relative telomere intensities. (A) Detection of telomerase activity by a TRAP assay in WI-38+hTERT cells (lane 4) but not in wild-type WI-38 cells (lane 2). In lanes 3 and 1 samples were heat inactivated (negative control). (B) Circles indicate the means of relative fluorescence intensities obtained for each telomere on both homologs of chromosomes 8 and 9, in wild-type WI-38 (PD38) and WI-38+hTERT (PD60) cells. The differences in relative telomere length between all identified telomeres are highly significant in the wild-type cells (analysis of variance, P < 10–15) whereas they are not significant (n. s.) in cells expressing telomerase. The same result was observed with WI-38+hTERT cells at PD50 and 70. (C) Box plot representing the distribution of relative intensity measurements obtained for all telomeres in ∼25 metaphases. Circles represent observations below the 10th and above the 90th percentiles. Both distributions differ significantly, as revealed by the Kolmogorov–Smirnov two-sample test.
To determine whether telomere length homogenization affected all telomeres in the cell, we looked at the distribution of relative telomere intensities for all telomeres in WI-38+hTERT cells, obtained from more than 20 metaphases. The distribution was normal and exhibited markedly less dispersion than the distribution obtained for wild-type cells, from which it differed significantly (Fig. 7C). Together, these results suggest that in WI-38+hTERT cells short telomeres had been more efficiently elongated than long ones.
DISCUSSION
The observation made by others (14,15) that telomere length heterogeneity is not randomly distributed among chromosomes is confirmed in our study. The present work shows, in addition, that significant length differences among homologous chromosomes may be observed when these are distinguishable. Such differences are consistently observed both during in vitro growth of telomerase-negative fibroblasts and in several short-term stimulated PBL samples from the same donor. Since variations were also detected between individuals, length differences may correspond to homolog-specific telomere length polymorphisms. This possibility had already been suggested to Martens et al. who observed a marked variability in mean telomere lengths for chromosome 11p in individuals from their population sample (15). Our results provide definitive proof that certain 11p arms are associated with long telomeres as compared to their homologs. Moreover, telomere length polymorphism may not be restricted to the short arm of chromosome 11, since length differences were frequently detected between other homologous chromosome ends. Telomere length polymorphism might thus fully explain the telomere length heterogeneity observed within human cells.
The consequences of telomere length heterogeneity are not fully understood. Although telomere length is related to cellular senescence, no such function has been attributed to a specific telomere. The accumulation of several short telomeres (rather than the length of the shortest one) seems to be the signal that triggers senescence in telomerase-negative cells (27). Should the initial length of a given telomere be predetermined, the occurrence of particularly short telomeres in a given individual may affect the replication capacity of his/her telomerase-negative cells. Whether or not this would in turn affect lifespan remains highly speculative. On the other hand, inasmuch as telomeres are required for chromosome stability, chromosomes with short telomeres might be more likely to be involved in rearrangement or loss events related to oncogenesis (15). Considering the potential for every chromosome arm to carry short or long telomeres, the probability for a given chromosome to undergo such events would vary between individuals in the general population.
It is not known whether telomere length differences between homologs were present when WI-38 cells were initially isolated from fetal tissue (28). Nonetheless, our experiments show that, in the absence of telomerase, these differences are stable throughout at least one-third of the in vitro lifespan of the WI-38 cell line. This observation is in keeping with the proposed mechanisms for cell division-dependent erosion of telomeres, by which most telomeres undergo a comparable attrition during in vitro growth (29–31). It is not excluded, however, that in cell cultures closer to senescence telomere length heterogeneity will be reduced due to accumulation of short telomeres (27).
Interestingly, forced expression of exogenous telomerase in WI-38 fibroblast cells abolished telomere length differences, apparently through a more efficient elongation of short telomeres. This result is consistent with reported observations using fibroblast cells in which limiting amounts of exogenous telomerase led to a preferential elongation of short telomeres (32,33). These results may correspond to our observations in long-term culture of stimulated PBLs, in which the endogenous expression of hTERT, which normally follows in vitro lymphocyte activation, does not prevent overall telomere shortening due to proliferation (34,35). In this case a preferential activity of telomerase towards short telomeres could explain the observed telomere length homogenization in those cells. Although the accumulation of short telomeres in pre-senescent T cells might eventually lead to telomere homogenization, it is unlikely that the cells we examined here were close to senescence, the lifespan of in vitro stimulated PBLs usually being much longer than 11 days (23).
The telomere length homogenization observed in our experiments may be accounted for by current models of telomerase control (36) and, in particular, by telomere length-dependent cis-inhibition mechanisms similar to those in yeast (37). There is no direct evidence, however, that in humans negative control mechanisms dependent on telomere length occur in vivo. In fact, the prevalent telomere length heterogeneity detected in short-term stimulated PBLs (14,15; present results) clearly contrasts with the documented telomerase activity at several stages of B and T cell development (38–40). Perhaps, telomere length differences between chromosomes are already present in the zygote and are simply perpetuated through uniform telomere shortening and very tight regulation of telomerase activity. Alternatively, the telomeric length differences could be maintained by still unidentified length-independent modulating factors, which promote cis-activation/inhibition of telomerase during development. Further studies are needed to ascertain whether telomere length differences between homologous chromosomes are inherited polymorphisms or are the result of epigenetic phenomena, as is the case for the inactivated chromosome X (41).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the CEPH Cell Culture Laboratory group for their technical support and Drs J. Campisi, B. Trask and G. Vergnaud for providing some of the material used here. Thanks also to Dr L. Pascoe (CEPH) for discussions concerning data statistical analysis and for reviewing the manuscript.
References
- 1.Zakian V.A. (1995) Telomeres: beginning to understand the end. Science, 270, 1601–1607. [DOI] [PubMed] [Google Scholar]
- 2.Allsopp R.C. and Harley,C.B. (1995) Evidence for a critical telomere length in senescent human fibroblasts. Exp. Cell Res., 219, 130–136. [DOI] [PubMed] [Google Scholar]
- 3.Allsopp R.C., Vaziri,H., Patterson,C., Goldstein,S., Younglai,E.V., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA, 89, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Lichtsteiner,S. and Wright,W.E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 5.Hahn W.C., Counter,C.M., Lundberg,A.S., Beijersbergen,R.L., Brooks,M.W. and Weinberg,R.A. (1999) Creation of human tumour cells with defined genetic elements. Nature, 400, 464–468. [DOI] [PubMed] [Google Scholar]
- 6.Meyerson M., Counter,C.M., Eaton,E.N., Ellisen,L.W., Steiner,P., Caddle,S.D., Ziaugra,L., Beijersbergen,R.L., Davidoff,M.J., Liu,Q., Bacchetti,S., Haber,D.A. and Weinberg,R.A. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- 7.Cech T.R., Nakamura,T.M. and Lingner,J. (1997) Telomerase is a true reverse transcriptase. A review. Biokhimiia, 62, 1202–1205. [PubMed] [Google Scholar]
- 8.Zhu L., Hathcock,K.S., Hande,P., Lansdorp,P.M., Seldin,M.F. and Hodes,R.J. (1998) Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl Acad. Sci. USA, 95, 8648–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slagboom P.E., Droog,S. and Boomsma,D.I. (1994) Genetic determination of telomere size in humans: a twin study of three age groups. Am. J. Hum. Genet., 55, 876–882. [PMC free article] [PubMed] [Google Scholar]
- 10.Hastie N.D., Dempster,M., Dunlop,M.G., Thompson,A.M., Green,D.K. and Allshire,R.C. (1990) Telomere reduction in human colorectal carcinoma and with ageing. Nature, 346, 866–868. [DOI] [PubMed] [Google Scholar]
- 11.Hemann M.T. and Greider,C.W. (2000) Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res., 28, 4474–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blasco M.A., Lee,H.W., Hande,M.P., Samper,E., Lansdorp,P.M., De Pinho,R.A. and Greider,C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- 13.Henderson S., Allsopp,R., Spector,D., Wang,S.S. and Harley,C. (1996) In situ analysis of changes in telomere size during replicative aging and cell transformation. J. Cell Biol., 134, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lansdorp P.M., Verwoerd,N.P., van de Rijke,F.M., Dragowska,V., Little,M.T., Dirks,R.W., Raap,A.K. and Tanke,H.J. (1996) Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet., 5, 685–691. [DOI] [PubMed] [Google Scholar]
- 15.Martens U.M., Zijlmans,J.M., Poon,S.S., Dragowska,W., Yui,J., Chavez,E.A., Ward,R.K. and Lansdorp,P.M. (1998) Short telomeres on human chromosome 17p. Nature Genet., 18, 76–80. [DOI] [PubMed] [Google Scholar]
- 16.Flint J., Bates,G.P., Clark,K., Dorman,A., Willingham,D., Roe,B.A., Micklem,G., Higgs,D.R. and Louis,E.J. (1997) Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum. Mol. Genet., 6, 1305–1313. [DOI] [PubMed] [Google Scholar]
- 17.Brown W.R., MacKinnon,P.J., Villasante,A., Spurr,N., Buckle,V.J. and Dobson,M.J. (1990) Structure and polymorphism of human telomere-associated DNA. Cell, 63, 119–132. [DOI] [PubMed] [Google Scholar]
- 18.Weber B., Allen,L., Magenis,R.E. and Hayden,M.R. (1991) A low-copy repeat located in subtelomeric regions of 14 different human chromosomal termini. Cytogenet. Cell Genet., 57, 179–183. [DOI] [PubMed] [Google Scholar]
- 19.Wilkie A.O., Higgs,D.R., Rack,K.A., Buckle,V.J., Spurr,N.K., Fischel-Ghodsian,N., Ceccherini,I., Brown,W.R. and Harris,P.C. (1991) Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell, 64, 595–606. [DOI] [PubMed] [Google Scholar]
- 20.Macina R.A., Negorev,D.G., Spais,C., Ruthig,L.A., Hu,X.L. and Riethman,H.C. (1994) Sequence organization of the human chromosome 2q telomere. Hum. Mol. Genet., 3, 1847–1853. [DOI] [PubMed] [Google Scholar]
- 21.Monfouilloux S., Avet-Loiseau,H., Amarger,V., Balazs,I., Pourcel,C. and Vergnaud,G. (1998) Recent human-specific spreading of a subtelomeric domain. Genomics, 51, 165–176. [DOI] [PubMed] [Google Scholar]
- 22.Trask B.J., Friedman,C., Martin-Gallardo,A., Rowen,L., Akinbami,C., Blankenship,J., Collins,C., Giorgi,D., Iadonato,S., Johnson,F., Kuo,W.L., Massa,H., Morrish,T., Naylor,S., Nguyen,O.T., Rouquier,S., Smith,T., Wong,D.J., Youngblom,J. and van den Engh,G. (1998) Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum. Mol. Genet., 7, 13–26. [DOI] [PubMed] [Google Scholar]
- 23.Boucher N., Dufeu-Duchesne,T., Vicaut,E., Farge,D., Effros,R.B. and Schachter,F. (1998) CD28 expression in T cell aging and human longevity. Exp. Gerontol., 33, 267–282. [DOI] [PubMed] [Google Scholar]
- 24.Zijlmans J.M., Martens,U.M., Poon,S.S., Raap,A.K., Tanke,H.J., Ward,R.K. and Lansdorp,P.M. (1997) Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl Acad. Sci. USA, 94, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinkel D., Straume,T. and Gray,J.W. (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl Acad. Sci. USA, 83, 2934–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S.H., Kaminker,P. and Campisi,J. (1999) TIN2, a new regulator of telomere length in human cells. Nature Genet., 23, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martens U.M., Chavez,E.A., Poon,S.S., Schmoor,C. and Lansdorp,P.M. (2000) Accumulation of short telomeres in human fibroblasts prior to replicative senescence. Exp. Cell Res., 256, 291–299. [DOI] [PubMed] [Google Scholar]
- 28.Hayflick L. and Moorhead,P.S. (1961) The serial cultivation of human diploid cell strains. Exp. Cell Res., 25, 585–621. [DOI] [PubMed] [Google Scholar]
- 29.Reveal P.M., Henkels,K.M. and Turchi,J.J. (1997) Synthesis of the mammalian telomere lagging strand in vitro. J. Biol. Chem., 272, 11678–116781. [DOI] [PubMed] [Google Scholar]
- 30.von Zglinicki T., Pilger,R. and Sitte,N. (2000) Accumulation of single-strand breaks is the major cause of telomere shortening in human fibroblasts. Free Radic. Biol. Med., 28, 64–74. [DOI] [PubMed] [Google Scholar]
- 31.Huffman K.E., Levene,S.D., Tesmer,V.M., Shay,J.W. and Wright,W.E. (2000) Telomere shortening is proportional to the size of the 3′ G-rich telomeric overhang. J. Biol. Chem., 275, 1095–1098. [DOI] [PubMed] [Google Scholar]
- 32.Steinert S., Shay,J.W. and Wright,W.E. (2000) Transient expression of human telomerase extends the life span of normal human fibroblasts. Biochem. Biophys. Res. Commun., 273, 1095–1098. [DOI] [PubMed] [Google Scholar]
- 33.Ouellette M.M., Liao,M., Herbert,B.S., Johnson,M., Holt,S.E., Liss,H.S., Shay,J.W. and Wright,W.E. (2000) Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem., 275, 10072–10076. [DOI] [PubMed] [Google Scholar]
- 34.Yamada O., Motoji,T. and Mizoguchi,H. (1996) Up-regulation of telomerase activity in human lymphocytes. Biochim. Biophys. Acta, 1314, 260–266. [DOI] [PubMed] [Google Scholar]
- 35.Bodnar A.G., Kim,N.W., Effros,R.B. and Chiu,C.P. (1996) Mechanism of telomerase induction during T cell activation. Exp. Cell Res., 228, 58–64. [DOI] [PubMed] [Google Scholar]
- 36.Blackburn E.H. (2000) Telomere states and cell fates. Nature, 408, 53–56. [DOI] [PubMed] [Google Scholar]
- 37.Marcand S., Brevet,V. and Gilson,E. (1999) Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J., 18, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu B.T., Lee,S.C., Marin,E., Ryan,D.H. and Insel,R.A. (1997) Telomerase is up-regulated in human germinal center B cells in vivo and can be re-expressed in memory B cells activated in vitro. J. Immunol ., 159, 1068–1071. [PubMed] [Google Scholar]
- 39.Weng N.P., Granger,L. and Hodes,R.J. (1997) Telomere lengthening and telomerase activation during human B cell differentiation. Proc. Natl Acad. Sci. USA, 94, 10827–10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng N., Levine,B.L., June,C.H. and Hodes,R.J. (1997) Regulation of telomerase RNA template expression in human T lymphocyte development and activation. J. Immunol ., 158, 3215–3220. [PubMed] [Google Scholar]
- 41.Surralles J., Hande,M.P., Marcos,R. and Lansdorp,P.M. (1999) Accelerated telomere shortening in the human inactive X chromosome. Am. J. Hum. Genet., 65, 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.