Abstract
Multi-drug resistant bacteria (particularly those producing extended-spectrum β-lactamases) have become a major health concern. The continued exposure to antibiotics, biocides, chemical preservatives, and metals in different settings such as the food chain or in the environment may result in development of multiple resistance or co-resistance. The aim of the present study was to determine multiple resistances (biocides, antibiotics, chemical preservatives, phenolic compounds, and metals) in bacterial isolates from seafoods. A 75.86% of the 87 isolates studied were resistant to at least one antibiotic or one biocide, and 6.90% were multiply resistant to at least three biocides and at least three antibiotics. Significant (P < 0.05) moderate or strong positive correlations were detected between tolerances to biocides, between antibiotics, and between antibiotics with biocides and other antimicrobials. A sub-set of 30 isolates selected according to antimicrobial resistance profile and food type were identified by 16S rDNA sequencing and tested for copper and zinc tolerance. Then, the genetic determinants for biocide and metal tolerance and antibiotic resistance were investigated. The selected isolates were identified as Pseudomonas (63.33%), Acinetobacter (13.33%), Aeromonas (13.33%), Shewanella, Proteus and Listeria (one isolate each). Antibiotic resistance determinants detected included sul1 (43.33% of tested isolates), sul2 (6.66%), blaTEM (16.66%), blaCTX−M (16.66%), blaPSE (10.00%), blaIMP (3.33%), blaNDM−1 (3.33%), floR (16.66%), aadA1 (20.0%), and aac(6′)-Ib (16.66%). The only biocide resistance determinant detected among the selected isolates was qacEΔ1 (10.00%). A 23.30 of the selected isolates were able to grow on media containing 32 mM copper sulfate, and 46.60% on 8 mM zinc chloride. The metal resistance genes pcoA/copA, pcoR, and chrB were detected in 36.66, 6.66, and 13.33% of selected isolates, respectively. Twelve isolates tested positive for both metal and antibiotic resistance genes, including one isolate positive for the carbapenemase gene blaNDM−1 and for pcoA/copA. These results suggest that exposure to metals could co-select for antibiotic resistance and also highlight the potential of bacteria on seafoods to be involved in the transmission of antimicrobial resistance genes.
Keywords: antimicrobial resistance, biocides, antibiotics, metals, seafood
Introduction
The global world fish market reached 175 million tons in 2016 (Food and Agriculture Organization (FAO), 2016). There is a growing concern in the fisheries sector about the increasing prevalence of multidrug-resistant bacterial strains in the food chain (EFSA, 2010; ECDC, 2012, 2013; EFSA-ECDC, 2013; Watts et al., 2017). Multidrug-resistant bacteria carrying extended-spectrum (ESBL) β-lactamase genes can now be found in healthy humans as well as in various animal species, food and even in environmental samples, and have become a threat in hospital settings. Carbapenemase-producing Enterobacteriaceae are particularly of concern because they tend to spread, making infection treatment difficult (Iovleva and Doi, 2017). Resistance genes for antimicrobials such as β-lactams, quinolones, and fluoroquinolones may be associated with plasmids. Plasmids of fluoroquinolone resistance in Enterobacteriaceae are widespread, and are often associated with resistance to β-lactams in strains with multidrug resistant phenotypes (Crémet et al., 2011). Some genes, such as the variant aac(6′)-Ib-cr simultaneously confer resistance to aminoglycosides, and fluoroquinolones (Kim et al., 2011). Others like the efflux pump oqxAB described in Escherichia coli, confer resistance to various antibiotics and biocides (Hansen et al., 2007; Wong and Chen, 2013).
Different types of selective pressure (such as antibiotics, biocides, or heavy metals) could play a role in the prevalence of antimicrobial resistance in the food chain. Biocides may co-select strains resistant to antibiotics of clinical use, as verified in the case of triclosan, and others (Chuanchuen et al., 2001; Braoudaki and Hilton, 2004; Ortega-Morente et al., 2013). Biocides are widely used in disinfecting equipment and facilities, including fish farming, and fish processing facilities (Directive 98/8/CE). In Gram-negative bacteria, several genes for resistance to biocides belonging to the group of quaternary ammonium compounds (QACs) as qacE, qacEΔ1, qacF, qacG, and qacH have been described. Among them, qacEΔ1 is the most widespread, as it is found in various groups of Gram-negative bacteria (Kücken et al., 2000). Association of qacEΔ1 (and less frequently also qacH and qacF) with class I integrons together with antibiotic resistance genes has been reported (Mulvey et al., 2006; De Toro et al., 2011). A study of microbial populations in environments contaminated with QACs related the increased incidence of resistance to QACs with a higher incidence of class I integrons (Gaze et al., 2005), suggesting that exposure to QACs could also selected for antibiotic resistance associated with such integrons. Other antimicrobials such as lysozyme-EDTA combinations and chemical preservatives such as sodium lactate and trisodium phosphate also deserve attention because of their wide use and potential applications in the food industry for decontamination and food preservation (Lucera et al., 2012; Bjornsdottir-Butler et al., 2015; Wang et al., 2015). There is also a growing interest in extending the use of plant essential oils or their antimicrobial compounds (such as carvacrol or thymol) for disinfection and food preservation (Lucera et al., 2012; Patel, 2015; Wang et al., 2015). One study showed that exposure to pine oil induced a decreased susceptibility to a range of antimicrobial compounds (including antibiotics and biocides; Moken et al., 1997). Yet, little is known about development of resistance/tolerance to these compounds, and co-resistance to antibiotics.
Metal salts are used for decontamination in fish farming. Metals such as copper (Cu) and zinc (Zn) are essential micronutrients in living things, but they can become toxic if they are above a certain concentration. Copper and zinc are frequently used in aquaculture and also as antifouling paints on boats (Yebra et al., 2004; Watermann et al., 2005; Guardiola et al., 2012). Copper can be released into the environment by both human activities and natural processes. Zinc is rarely found in nature in its metallic state, but many minerals contain zinc as a main component. The main anthropogenic sources of zinc are mining, zinc production facilities, iron, and steel production, corrosion of galvanized structures, coal, and fuel combustion, waste disposal and incineration, and the use of fertilizers, and pesticides containing zinc (WHO, 2001). Fish can accumulate and transmit heavy metals along the food chain. The genes that control resistance to metals may be associated with plasmids, which provide bacteria a competitive advantage over other organisms when specific metals are present (Trevors et al., 1985; Hobman and Crossman, 2014). The appearance of multidrug resistance plasmids carrying resistance to heavy metals is alarming and requires additional monitoring (Gómez-Sanz et al., 2013). The co-location of metal-resistance and antimicrobial-resistance genes can facilitate their persistence, co-selection, and dissemination (Gómez-Sanz et al., 2013; Wales and Davies, 2015).
The aim of this study was to provide insights on resistance to clinically relevant antibiotics in bacterial strains isolated from seafood sold at supermarkets and fishmarket. Since exposure to other types of antimicrobials such as biocides and metals could co-select for antibiotic resistance, we hypothesized that antibiotic-resistant strains from seafoods could also be phenotypically resistant, and/or carry genetic determinants of resistance to other antimicrobials, thus increasing the risk of transmission of antibiotic resistance through the seafood production chain. Therefore, susceptibility of bacterial strains isolated from different seafood samples against various biocides, antibiotics, and metals was evaluated, and the presence of resistance genes in multiresistant strains was determined. A special emphasis was made on extended-spectrum β-lactamase genes because of the risks that ESBL-producing bacteria may pose to human health.
Materials and methods
Bacterial isolation
A total of 22 seafood samples from 16 different fish and seafood species purchased at supermarkets and fishmarket in the province of Jaen (Spain) during the years 2013 and 2014 were investigated (Table 1). Unless indicated, samples consisted of unprocessed whole specimens from sea fishing, and were sold over the counter on ice. Samples (25 g each) were suspended in 225 ml of buffered peptone water supplemented with 10 g of NaCl/liter and homogenized by stomaching with as Stomacher 400 (Sewald, UK). The samples were analyzed following standard procedures for microbiological analysis of food by plating serial dilutions on Trypticase Soya Agar (TSA, Scharlab, Barcelona, Spain) supplemented with 10 g of NaCl/liter (saline TSA, STSA). After 24–48 h incubation at 30°C, colonies were purified by streaking on TSA (without added NaCl), and the pure cultures were examined by Gram-staining, catalase and oxidase tests, and stored at −80°C in Trypticase Soya Broth (TSB, Scharlab) supplemented with 20% glycerol.
Table 1.
Sensitivity to antibiotics, biocides and other antimicrobials in bacterial isolates recovered from seafoods.
| Food source and isolate | Viable counts (Log10 CFU/g) | RESISTANCE PHENOTYPE | BC | CT | HDP | TC | CF | CHX | PHMG | OX | CVC | THY | SL | TSP | L | LE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anchovy (Engraulis encrasicholus) | 4.27; 3.80; 4.32 | |||||||||||||||
| B2 | AMP, CTX, CM, NA, TM/STX, TC | 25 | 7.5 | 25 | >250 | 25 | 5 | 25 | 25 | 0.050 | 0.250 | 5.00 | 1.00 | >0.01 | ABC | |
| B6 | 25 | 2.5 | 25 | 25 | 5 | 20 | 2.5 | 7.5 | 0.010 | 0.010 | 3.00 | 0.75 | >0.01 | ABC | ||
| B7 | AMP, CM, NA, TM/STX | 25 | 7.5 | 50 | 5 | 25 | 5 | 5 | 2.5 | 0.050 | 0.250 | 5.00 | 0.75 | >0.01 | ABC | |
| B22 | 25 | 2.5 | 25 | 25 | 25 | 15 | 5 | 25 | 0.010 | 0.010 | 3.00 | 0.75 | >0.01 | ABC | ||
| B24 | 25 | 2.5 | 25 | 25 | 25 | 20 | 5 | 10 | 0.010 | 0.010 | 3.00 | 0.75 | >0.01 | ABC | ||
| 4B1 | AMP, NA, TM/STX | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 25 | 0.100 | 0.010 | 5.00 | 0.75 | >0.01 | ABC | |
| 4B21 | AMP, TM/STX | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 25 | 0.100 | 0.050 | 5.00 | 0.75 | >0.01 | ABC | |
| 4B22 | NA, CHX | 25 | 0.5 | 25 | 25 | 5 | 50 | 7.5 | 10 | 0.010 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | |
| 4B23 | AMP, NA, CHX | 25 | 2.5 | 25 | 25 | 25 | 50 | 7.5 | 10 | 0.100 | 0.125 | 5.00 | 0.75 | >0.01 | ABC | |
| 4B31 | AMP, CTX, IMP, CM, TM/STX, BC, CT, HDP, PHMG, TC | 250 | 25 | >250 | >250 | 25 | 20 | 100 | 25 | 1.00 | 0.125 | 5.00 | 1.00 | >0.01 | ABC | |
| 4B41 | NA | 25 | 0.5 | 25 | 5 | 5 | 1 | 5 | 25 | 0.010 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | |
| 4B43 | AMP, CTX, CM, NA, TM/STX | 25 | 2.5 | 50 | 5 | 25 | 10 | 25 | 25 | 1.000 | 0.125 | 5.00 | 0.75 | >0.01 | ABC | |
| Sardine (Sardina pilchardus) | 4.63; 4.50 | |||||||||||||||
| 4Sd1 | AMP, NA, TM/STX, CHX, OX | 25 | 5 | 25 | 50 | 25 | 50 | 25 | >250 | 0.100 | 0.050 | 5.00 | 1.00 | >0.01 | ABC | |
| 4Sd3 | BC | 250 | 2.5 | 25 | 5 | 5 | 5 | 2.5 | 7.5 | 0.100 | 0.010 | 3.00 | 0.50 | >0.01 | ABC | |
| 4Sd21 | CHX, PHMG, OX | 25 | 5 | 25 | 50 | 25 | 50 | 100 | >250 | 0.050 | 0.010 | 5.00 | 1.50 | >0.01 | C | |
| 4Sd22 | TM/STX, TC | 50 | 7.5 | 25 | >250 | 25 | 15 | 25 | 75 | 0.100 | 0.050 | 5.00 | 0.75 | >0.01 | ABC | |
| 4Sd41 | AMP, CTX, CM, S, CHX, PHMG, TC | 50 | 7.5 | 25 | >250 | 25 | 50 | 100 | 250 | 0.100 | 0.250 | 5.00 | 0.75 | >0.01 | ABC | |
| 4Sd42 | AMP, NA, TM/STX | 25 | 2.5 | 25 | 5 | 25 | 5 | 25 | 100 | 0.050 | 0.125 | 5.00 | 0.50 | >0.01 | ABC | |
| 4Sd51 | AMP, S | 50 | 0.5 | 25 | 50 | 25 | 10 | 5 | 250 | 0.050 | 0.010 | 5.00 | 1.50 | >0.01 | ABC | |
| 4Sd52 | AMP | 50 | 2.5 | 25 | 50 | 25 | 20 | 7.5 | 250 | 0.025 | 0.010 | 5.00 | 0.75 | >0.01 | ABC | |
| 4Sd53 | AMP, CHX | 25 | 2.5 | 25 | 25 | 25 | 50 | 25 | 250 | 0.100 | 0.250 | 3.00 | 1.00 | >0.01 | ABC | |
| 4Sd54 | 50 | 0.5 | 25 | 5 | 5 | 1 | 5 | 100 | 0.025 | 0.010 | 1.00 | 0.25 | >0.01 | ABC | ||
| Blue whiting (Micromesistius poutassou) | 3.78 | |||||||||||||||
| 4Ba1 | AMP, TM/STX | 25 | 2.5 | 25 | 5 | 25 | 10 | 7.5 | 250 | 0.050 | 0.250 | 3.00 | 0.50 | >0.01 | ABC | |
| 4Ba2 | 5 | 0.5 | 5 | 5 | 5 | 1 | 0.5 | 5 | 0.025 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | ||
| 4Ba31 | AMP, TM/STX | 5 | 0.5 | 5 | 5 | 5 | 1 | 5 | 50 | 0.010 | 0.010 | 1.00 | 0.25 | >0.01 | ABC | |
| 4Ba32 | 5 | 0.5 | 5 | 5 | 5 | 1 | 0.5 | 5 | 0.010 | 0.010 | 1.00 | 0.25 | >0.01 | ABC | ||
| 4Ba33 | IMP | 25 | 2.5 | 5 | 25 | 25 | 15 | 7.5 | 250 | 0.010 | 0.250 | 3.00 | 0.50 | >0.01 | ABC | |
| 4Ba41 | 25 | 2.5 | 25 | 25 | 25 | 15 | 5 | 50 | 0.025 | 0.250 | 5.00 | 0.75 | >0.01 | ABC | ||
| 4Ba42 | AMP, TM/STX | 25 | 2.5 | 25 | 5 | 25 | 10 | 7.5 | 50 | 0.025 | 0.010 | 3.00 | 0.50 | >0.01 | ABC | |
| Athlantic horse mackerel (Trachurus trachurus) | 6.25 | |||||||||||||||
| J2 | AMP, CTX, CM, S, NA, TM/STX, CT, HDP, TC | 5 | 25 | >250 | 250 | 25 | 10 | 0.5 | 5 | 0.100 | 0.250 | 5.00 | 1.00 | >0.01 | ABC | |
| J4 | AMP, CTX, CM, TM/STX, BC, CT, HDP, TC | >250 | 25 | >250 | 250 | 25 | 10 | 25 | 100 | 0.100 | 0.250 | 3.00 | 1.00 | >0.01 | ABC | |
| J5 | NA | 5 | 0.5 | 5 | 5 | 5 | 1 | 0.5 | 0.5 | 0.025 | 0.250 | 5.00 | 1.50 | >0.01 | ABC | |
| J6 | CM | 25 | 2.5 | 50 | 25 | 25 | 5 | 5 | 25 | 0.010 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | |
| J7 | AMP, S, BC | 250 | 7.5 | 50 | 25 | 25 | 5 | 2.5 | 75 | 0.100 | 0.125 | 3.00 | 0.75 | >0.01 | ABC | |
| Mackerel (Scomber scombrus) | 4.93 | |||||||||||||||
| Cb22 | AMP, IMP, CHX | 25 | 0.5 | 25 | 50 | 25 | 50 | 10 | 250 | 0.025 | 0.250 | 5.00 | 3.00 | >0.01 | ABC | |
| Cb27 | AMP, CTX, IMP, CM, NA, TM/STX, TC | 50 | 7.5 | 25 | 250 | 25 | 10 | 25 | 250 | 0.050 | 0.250 | 5.00 | 1.50 | >0.01 | ABC | |
| Cb212 | AMP, CTX, CM, NA, TM/STX, TC | 50 | 7.5 | 25 | >250 | 25 | 10 | 25 | 250 | 0.100 | 0.250 | 5.00 | 1.50 | >0.01 | ABC | |
| Sole (Solea spp.) | 4.99; 5.62 | |||||||||||||||
| Le1 | AMP | 5 | 2.5 | 25 | 25 | 25 | 20 | 0.5 | 5 | 0.010 | 0.010 | 3.00 | 0.75 | >0.01 | ABC | |
| Le2 | 25 | 2.5 | 50 | 50 | 25 | 15 | 5 | 75 | 0.010 | 0.010 | 3.00 | 0.75 | >0.01 | ABC | ||
| Le4 | AMP | 25 | 2.5 | 50 | 50 | 25 | 15 | 5 | 75 | 0.010 | 0.010 | 3.00 | 0.75 | >0.01 | ABC | |
| Le5 | CHX | 25 | 2.5 | 50 | 50 | 25 | 50 | 5 | 50 | 0.010 | 0.010 | 3.00 | 0.75 | >0.01 | ABC | |
| Hake (Merluccius capensis; sliced) | 5.84 | |||||||||||||||
| M135 | CHX | 5 | 0.5 | 25 | 5 | 5 | 50 | 0.5 | 5 | 0.010 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | |
| Blue shark (Prionace glauca; sliced) | 3.62 | |||||||||||||||
| T213 | 25 | 2.5 | 25 | 5 | 25 | 5 | 2.5 | 50 | 0.025 | 0.010 | 3.00 | 1.00 | 0.0025 | ABC | ||
| T215 | S, CHX | 50 | 7.5 | 50 | 50 | 25 | 50 | 25 | 250 | 0.025 | 0.010 | 5.00 | 1.50 | >0.01 | ABC | |
| Gilthead seabream (Sparus aurata) | 6.44 | |||||||||||||||
| Do11 | AMP, IMP, CHX | 25 | 2.5 | 25 | 25 | 25 | 50 | 7.5 | 250 | 0.025 | 0.250 | 3.00 | 3.00 | >0.01 | ABC | |
| Do15 | 25 | 2.5 | 50 | 50 | 25 | 20 | 10 | 250 | 0.010 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | ||
| Do24 | AMP, IMP, CHX | 25 | 2.5 | 25 | 50 | 25 | 50 | 5 | 100 | 0.025 | 0.010 | 5.00 | 3.00 | >0.01 | ABC | |
| Do26 | S, CHX | 25 | 2.5 | 50 | 50 | 25 | 50 | 25 | 75 | 0.010 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | |
| Sea bass (Dicentrarchus labrax; aquaculture) | 5.88; 5.70 | |||||||||||||||
| Lbi14 | CHX | 25 | 2.5 | 50 | 50 | 25 | 50 | 25 | 50 | 0.010 | 0.010 | 3.00 | 0.75 | >0.01 | ABC | |
| Lbi15 | AMP, CTX, IMP, CM, NA, TM/STX, TC | 50 | 7.5 | 50 | >250 | 25 | 20 | 25 | 100 | 0.100 | 0.250 | 5.00 | 1.50 | >0.01 | ABC | |
| Lbi16 | AMP, CTX, CAZ, IMP, CM, S, NET, CIP, NA, TM/STX, PHMG, TC, OX | 75 | 5 | 50 | 250 | 25 | 10 | 100 | >250 | 0.100 | 0.250 | 3.00 | 1.00 | >0.01 | ABC | |
| Lbi25 | AMP, CTX, IMP, CM, S, NA, TM/STX, BC, CT, HDP, PHMG, TC, OX | >250 | 25 | 250 | 250 | 25 | 10 | 100 | >250 | 0.100 | 0.250 | 3.00 | 0.25 | >0.01 | ABC | |
| Salmon (Salmo salar; slices packed in trays; aquaculture) | 7.85; 7.47 | |||||||||||||||
| S11 | AMP, CTX, CAZ, CM, NA | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 250 | 0.010 | 0.025 | 5.00 | 1.00 | 0.0025 | ABC | |
| S12 | AMP, CTX, CM, S | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 250 | 0.010 | 0.010 | 3.00 | 1.00 | 0.0025 | ABC | |
| S13 | AMP, CAZ, CM | 25 | 2.5 | 25 | 5 | 25 | 20 | 25 | 250 | 0.010 | 0.010 | 3.00 | 0.75 | 0.0025 | ABC | |
| S14 | AMP, CAZ, CM, S, NA | 25 | 2.5 | 25 | 5 | 25 | 20 | 25 | 250 | 0.010 | 0.025 | 3.00 | 1.00 | 0.0025 | ABC | |
| S15 | AMP, CTX, CM, NA | 25 | 2.5 | 25 | 5 | 25 | 20 | 25 | 250 | 0.010 | 0.025 | 3.00 | 1.00 | 0.0025 | ABC | |
| S16 | AMP, CTX, CM, TE | 25 | 2.5 | 25 | 5 | 25 | 15 | 25 | 250 | 0.010 | 0.025 | 3.00 | 1.00 | 0.0025 | ABC | |
| S21 | AMP, CM, NA | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 250 | 0.010 | 0.025 | 3.00 | 1.00 | 0.0025 | ABC | |
| S22 | AMP, CTX, CM, S, NA | 25 | 2.5 | 25 | 5 | 25 | 20 | 25 | 250 | 0.010 | 0.025 | 3.00 | 1.00 | 0.0025 | ABC | |
| S23 | AMP, CTX, CM, NA | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 250 | 0.010 | 0.025 | 5.00 | 1.50 | 0.0025 | ABC | |
| S24 | AMP, CTX, CM, NA | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 250 | 0.010 | 0.025 | 5.00 | 1.50 | 0.0025 | ABC | |
| S25 | AMP, CTX, CM, S, NA | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 250 | 0.010 | 0.025 | 3.00 | 1.50 | 0.0025 | ABC | |
| S26 | AMP, CM, NA | 25 | 2.5 | 25 | 5 | 25 | 10 | 25 | 250 | 0.010 | 0.025 | 3.00 | 1.50 | 0.0025 | ABC | |
| Squid (Loligo spp.) | 4.44 | |||||||||||||||
| C121 | 25 | 2.5 | 25 | 25 | 25 | 5 | 5 | 25 | 0.025 | 0.010 | 3.00 | 1.50 | >0.01 | ABC | ||
| C123 | 25 | 2.5 | 25 | 25 | 25 | 5 | 2.5 | 25 | 0.025 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | ||
| C125 | 25 | 2.5 | 25 | 25 | 25 | 5 | 2.5 | 25 | 0.025 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | ||
| C126 | 25 | 2.5 | 25 | 25 | 25 | 5 | 5 | 25 | 0.025 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | ||
| Juvenile squid (Loligo vulgaris) | 4.49 | |||||||||||||||
| 4C1 | CAZ, S, NA | 25 | 0.5 | 25 | 25 | 5 | 5 | 5 | 250 | 0.025 | 0.010 | 1.00 | 0.75 | >0.01 | ABC | |
| 4C4 | AMP, TM/STX | 25 | 2.5 | 5 | 5 | 5 | 1 | 0.5 | 5 | 0.025 | 0.010 | 1.00 | 0.25 | >0.01 | ABC | |
| 4C21 | CTX, CAZ, S, NA | 25 | 2.5 | 25 | 25 | 5 | 5 | 5 | 250 | 0.025 | 0.010 | 1.00 | 0.75 | >0.01 | ABC | |
| 4C22 | AMP, TM/STX | 25 | 5 | 25 | 5 | 25 | 10 | 7.5 | 75 | 0.100 | 0.250 | 3.00 | 0.50 | >0.01 | ABC | |
| 4C23 | CTX, CAZ, S, NA | 25 | 2.5 | 25 | 5 | 5 | 5 | 5 | 100 | 0.025 | 0.010 | 1.00 | 0.75 | >0.01 | ABC | |
| 4C31 | 25 | 2.5 | 25 | 25 | 25 | 15 | 7.5 | 250 | 0.025 | 0.250 | 3.00 | 0.50 | >0.01 | ABC | ||
| 4C32 | AMP, CTX, IMP, CM, NA, BC, CT, HDP, CHX, PHMG, TC | >250 | 25 | >250 | >250 | 25 | 50 | 100 | 250 | 0.100 | 0.125 | 5.00 | 1.00 | >0.01 | ABC | |
| 4C51 | CAZ, S | 5 | 0.5 | 5 | 5 | 5 | 1 | 5 | 5 | 0.010 | 0.010 | 1.00 | 0.25 | >0.01 | ABC | |
| 4C52 | AMP, CTX, C, NA, TM/STX, HDP, TC | 75 | 10 | 250 | >250 | 25 | 15 | 25 | 250 | 0.100 | 0.010 | 5.00 | 0.75 | >0.01 | ABC | |
| Mussels (Mytilus edulis) | 3.65 | |||||||||||||||
| Mj2 | AMP, IMP, S, CHX | 25 | 5 | 25 | 50 | 25 | 50 | 7.5 | 250 | 0.025 | 0.250 | 5.00 | 3.00 | >0.01 | ABC | |
| Prawns (Penaeus spp.; boiled and frozen; aquaculture) | 4.60 | |||||||||||||||
| L122 | AMP, CAZ | 25 | 2.5 | 25 | 25 | 25 | 5 | 5 | 50 | 0.025 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | |
| L123 | AMP, CTX, CAZ, IMP, CM, S, NET, TE, CIP, TM/STX | 25 | 2.5 | 25 | 5 | 25 | 5 | 0.5 | 50 | 0.010 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | |
| L124 | AMP, IMP, CHX | 25 | 5 | 25 | 50 | 25 | 50 | 10 | 250 | 0.025 | 0.250 | 5.00 | 3.00 | >0.01 | ABC | |
| L125 | 25 | 2.5 | 25 | 5 | 5 | 5 | 2.5 | 250 | 0.010 | 0.250 | 3.00 | 1.00 | >0.01 | ABC | ||
| Norway lobster (Nephrops norvergicus; boiled and frozen) | 4.47 | |||||||||||||||
| Cg11 | 25 | 2.5 | 25 | 5 | 5 | 5 | 2.5 | 250 | 0.010 | 0.010 | 3.00 | 3.00 | >0.01 | ABC | ||
| Cg12 | 5 | 2.5 | 25 | 5 | 5 | 5 | 0.5 | 5 | 0.010 | 0.010 | 3.00 | 1.50 | >0.01 | ABC | ||
| Cg14 | 25 | 2.5 | 25 | 5 | 25 | 5 | 2.5 | 25 | 0.010 | 0.010 | 3.00 | 1.00 | >0.01 | ABC | ||
| Cg21 | 25 | 2.5 | 25 | 5 | 5 | 5 | 2.5 | 250 | 0.025 | 0.010 | 3.00 | 3.00 | >0.01 | ABC | ||
| Cg22 | AMP, CM, NA, TM/STX, TC | 25 | 5 | 25 | 250 | 25 | 5 | 25 | 250 | 0.050 | 0.250 | 5.00 | 1.50 | >0.01 | ABC |
In bold, isolates selected for further study.
AMP, ampicillin; CTX, cefotaxime, CAZ, ceftazidime; IMP, imipenen; CIP, ciprofloxacin; NA, nalidixic acid; CM, chloramphenicol; S, streptomycin; NET, netilmicin; TE, tetracycline; TM/STX, trimethoprim/solfamethoxazole. The minimum inhibitory concentrations (MICs) for biocides (in mg/l, or in microliters of commercial solution per liter in the case of OX), phenolic compounds and preservatives (in %) are indicated. BC, benzalkonium chloride; CT, cetrimide; CHX, chlorhexidine; HDP, hexadecylpyridinium chloride; TC, triclosan; PHMG, polyhexamethylen guanidium chloride; OX, P3 Oxonia (in microliters of commercial solution per; CVC, carvacrol; THY, thymol (THY); SL, sodium lactate; TSP, trisodium phosphate; L, lysozyme (L). LE, lysozyme-EDTA combinations (A, B, C indicate the different LE inhibitory combinations).
Determination of resistance to antibiotics, biocides, and other antimicrobial compounds
A collection of 87 bacteria randomly isolated from the different seafood samples were screened for sensitivity to biocides, antibiotics, and other antimicrobial compounds as described below. Benzalkonium chloride (BC), cetrimide (CT), hexadecylpyridinium chloride (HDP), chlorhexidine digluconate (CHX), triclosan (TC), and hexachlorophene (CF) were from Sigma-Aldrich (Madrid, Spain). The commercial solution of benzalkonium chloride contained 50% (wt/v) of the active compound. Triclosan and hexachlorophene were dissolved (10% wt/v) in 96% ethanol. HDP (5% wt/v), CT (10% wt/v), and CHX (20% wt/v) were dissolved aseptically in sterile distilled water. Poly-(hexamethylen guanidinium) hydrochloride (PHMG) solution (containing 7.8% of PHMG, by weight) was a kind gift of Oy Soft Protector Ltd (Espoo, Finland). P3 oxonia (OX, containing 25–35% hydrogen peroxide, 0.83–2.5 N acetic acid, and 0.26–0.66 N peracetic acid) was supplied by ECOLAB (Barcelona, Spain). Biocide solutions were stored at 4°C for ≤7 days. Carvacrol (CVC), thymol (THY), sodium lactate (SL), trisodium phosphate (TSP), lysozyme (L), and ethylenediaminetetraacetic acid (EDTA) were from Sigma-Aldrich. Solutions containing 100 mg/l lysozyme and 5 mM EDTA were combined in different proportions to yield the following final concentrations: A, 30 mg/l lysozyme plus 3.5 mM EDTA; B, 50 mg/l lysozyme plus 2.5 mM EDTA; C, 70 mg/l lysozyme plus 1.5 mM EDTA. Minimum inhibitory concentrations (MIC's) were determined by the broth microdilution method on 96-well, flat-bottom microtiter plates (Becton Dickinson Labware, Franklin Lakes, NJ). Briefly, serial dilutions of each antimicrobial were inoculated (1%, vol/vol) with overnight cultures of bacterial strains grown in Trypticase Soya Broth (TSB; Scharlab). Growth and sterility controls were included for each isolate. Microtiter plates were incubated at 30°C. Optical density (OD 595 nm) readings were performed with an iMark Microplate Reader (BioRad, Madrid) after 24–48 h incubation. All assays were done in triplicate.
Antibiotic resistance was determined by the disk diffusion method as described by the Clinical and Laboratory Standards Institute CLSI (2015) on cation-adjusted Mueller-Hinton agar (Fluka, Sigma-Aldrich, Madrid, Spain). Disks containing ampicillin (AMP, 10 μg), ceftazidine (CAZ, 30 μg), cefotaxime (CTX, 30 μg), imipenem (IMP, 10 μg), streptomycin (S, 10 μg), tetracycline (TE, 30 μg), ciprofloxacin (CIP, 5 μg), nalidixic acid (NA, 30 μg), and trimethoprim/sulfamethoxazole (TM/STX, 1.25/23.75 μg) were supplied by Biomérieux (Madrid, Spain). Chloramphenicol (CM, 30 μg) was from BBL (Madrid, Spain).
Identification of antimicrobial-tolerant isolates
From the preliminary screening on antimicrobial resistance, 30 isolates were selected for further study based on food source, antibiotic resistance and biocide tolerance. Selected isolates were resistant to at least three antibiotics or at least to one antibiotic and one biocide. The 30 isolates were identified by 16S rDNA sequencing. DNA was extracted with a bacterial genomic DNA extraction kit (GenEluteTM, Sigma-Aldrich) and 16 S rDNA was amplified as described by Abriouel et al. (2005). PCR amplification products were purified using a GFX PCR DNA and Gel Band Purification Kit (GE-Healthcare, Spain), and then sequenced according to Weisburg et al. (1991) in a CEQ 2000 XL DNA Analysis System (Beckman Coulter, CA, USA). The DNA sequences obtained were searched for homology by using the BLAST algorithm available at the National Center for Biotechnology Information (NCBI, USA).
Determination of tolerance to metals
The selected 30 isolates were tested for tolerance to copper and zinc metals as follows. Mueller–Hinton II agar plates (Sigma-Aldrich, Madrid, Spain) were supplemented with CuSO4•5H2O (PanReac, Barcelona, Spain) (4, 8, 12, 16, 20, 24, 32, and 36 mM, adjusted to pH 7.2) or ZnCl2 (PanReac) (2, 4, 8, and 16 mM, adjusted to pH 6.5) according to Cavaco et al. (2011). Then, plates were inoculated with 2 μl from overnight cultures of bacterial strains diluted 10-folds in sterile saline solution. The plates were incubated at 37°C under aerobic conditions and inspected for bacterial growth after 24 h. The lowest metal concentration that inhibited growth of the inoculated bacterial strains was taken as the MIC.
Investigation of genetic determinants of resistance
The selected 30 isolates were investigated for the presence of genetic determinants of resistance. The presence of the biocide resistance genes qacE and qacEΔ1 and their possible association with class I integrons was investigated by PCR according to Chuanchuen et al. (2007). Specifically, the forward primer qacEF was used in combination with reverse primers qacER and sulR for amplification of qacE, qacEΔ1 and the 3′ coding sequence (3′ CS).
The following extended-spectrum β-lactamase genetic determinants were investigated by PCR: blaTEM (Sáenz et al., 2004), blaPSE (Chiu et al., 2006), blaCTX−M, and blaCTX−M−2 (Bertrand et al., 2006), as well as carbapenemases blaIMP, blaNDM−1, blaOXA−23, and blaVIM−2 (Ramakrishnan et al., 2014). Other antimicrobial resistance genes investigated by PCR were the aminoglycoside resistance genes aadA1 (Guerra et al., 2001), and aac(6′)-Ib (Park et al., 2006), the phenicol resistance genes floR (Chiu et al., 2006) and cmlA (Sáenz et al., 2004), and the sulfonamide and trimethoprim resistance genes sul1 (Guerra et al., 2001), sul2 (Sáenz et al., 2004), and sul3 (Sáenz et al., 2010).
The gene for the multicopper oxidase found in the plasmid-borne operons pcoABCDRSE (described in Pseudomonas syringae) and copABCDRS (E. coli) was investigated using the primers and PCR conditions described for pcoA/copA by Badar et al. (2014). The additional genes copB, copC, copD (Kamika and Momba, 2013; Badar et al., 2014) and pcoR (Brown et al., 1992) were also investigated. The chromosomal cueAR operon, encoding a putative P1-type ATPase and a MerR-type regulatory protein involved in copper homeostasis in Pseudomonas putida was investigated according to Adaikkalam and Swarup (2002). The czcD gene involved in the regulation of the CZC zinc, cobalt, and cadmium efflux system was investigated by PCR according to Medardus et al. (2014). The chromate resistance gene chrB was investigated as described by Chihomvu et al. (2015) and Nies et al. (1990).
Statistical analysis
The relationships between resistances to the different antimicrobials tested were studied by Principal component analysis with Pearson correlation coefficient (r) by using IBM SPSS Statistics 22 (IBM Corporation, Armonk, New York, USA) and Mystat statistics and graphics package (Systat Software, Hounslow, London, UK; evaluation version 2015.1). Positive correlations were defined as very weak (0.00–0.19), weak (0.20–0.39), moderate (0.4–0.59), strong (0.60–0.79) or very strong (0.80–0.99), with a P significance of <0.05.
Results
Microbial load and bacterial isolation
The microbial load (aerobic mesophiles) of the different seafood samples is shown in Table 1. Most samples had viable cell counts comprised between 4 and 7 log CFU/g. Lowest viable counts were reported for blue shark, and highest counts were found in refrigerated raw salmon slices packed in trays.
After viable cell counting, bacterial colonies grown on saline TSA from highest dilutions were repurified by streaking on TSA without added salt. This was done so in order to avoid possible interference of added salt in growth media with antimicrobial resistance tests. A total of 87 bacterial colonies isolated at random were selected, representing the different seafood products sampled (Table 1). Eighty two of them were Gram-negatives (including mainly bacilli), while the remaining five were Gram-positive (including four cocci and one rod).
Antimicrobial resistance
The 87 isolates were tested for sensitivity to antibiotics, biocides and other antimicrobials (cavacrol, thymol, sodium lactate, trisodium phosphate, lysozyme, and different lysozyme-EDTA combinations; Table 1). The incidence of resistance to β-lactam antibiotics was highest for AMP (57.47% of isolates), followed by CTX (27.59%), IMP (14.94%), and CAZ (11.49%). Resistance to protein synthesis inhibitors was detected mostly for CM (33.33%) and S (20.69%), while resistance to TE (2.30%) and NET (2.30%) was low. Remarkably, a 35.63% of isolates were resistant to NA, but only 2.30% were resistant to CIP. A 27.58% of isolates were resistant to TM/STX. A 39.08% of isolates were resistant to three or more antibiotics, and 18.40% were resistant to five or more. Two isolates were resistant to 10 out of the 11 antibiotics tested.
Bacterial isolates were tested for biocide tolerance in two groups (Gram-positives, and Gram-negatives) since Gram-negative bacteria in general have greater tolerance to biocides because of the outer membrane permeability barrier. Only low percentages of the Gram negative isolates showed high tolerance levels to the biocides BC (5.75%; MIC ≥ 250 mg/l), CT (5.75%; MIC ≥ 25 mg/l), HDP (6.90%; MIC ≥ 250 mg/l), PHMG (6.90%; MIC ≥ 100 mg/l), and OX (4.60%; MIC > 250 μl/l) (Table 1). Higher percentages of biocide-tolerant isolates were obtained for CHX (19.54%; MIC ≥ 50 mg/l) and TC (16.09%; MIC ≥ 250 mg/l). Among the Gram positive isolates, only one had a high MIC of 250 mg/l for BC. A total of seven isolates showed high tolerance to three or more biocides (Table 1).
Isolates showed large differences in sensitivities to carvacrol and thymol (Table 1). Only two isolates had MICs higher than 0.25% for carvacrol, and 16.39% of isolates required 0.1% for inhibition. By contrast, 43.68% of isolates were inhibited by very low carvacrol concentration (0.01%). Regarding thymol, 26.44% of isolates had a MIC of 0.25%, but none required concentrations higher than this value for inhibition. There was also a high percentage of isolates (55.17%) that were inhibited by a low concentration of thymol (0.025%).
Most isolates were inhibited by sodium lactate at 3% (55.17%) or 5% (35.63%), while the rest (9.20%) were inhibited at 1% (Table 1). However, isolates were more heterogeneous in sensitivity to trisodium phosphate (TSP). About 80% of isolates were inhibited at TSP concentrations in the range of 0.75–1.5%, and only a small percentage (8.04%) required a TSP concentration of 3% for inhibition.
Most isolates (85.06%) were resistant to the highest concentration of lysozyme tested (100 mg/l), and only a low percentage (14.94%) were inhibited by a low lysozyme concentration (2.5 mg/l; Table 1). When lysozyme was tested in combination with EDTA at different proportions, all combinations were effective against most isolates, except for one isolate that only was inhibited by lysozyme-EDTA combination C containing a higher proportion of lysozyme to EDTA.
Correlations between antimicrobial resistances
Of the 87 isolates, 66 (75.86%) were resistant to antibiotics and/or tolerant to biocides. Comparing biocides and antibiotics, of the 29 isolates tolerant to at least one biocide (33.33%), 27 (31.03%) were also resistant to at least one antibiotic (Table 1). Six isolates (6.90%) were tolerant to at least three biocides and at least three antibiotics. The correlations between the different antimicrobials tested for the 87 bacterial isolates are shown in Figure 1 and Table 2. Only the statistically significant (P < 0.05) positive correlations that were moderate, strong or very strong are described below. The statistically significant (P < 0.05) weak correlations are also listed in Table 2, but are not described in the text.
Figure 1.
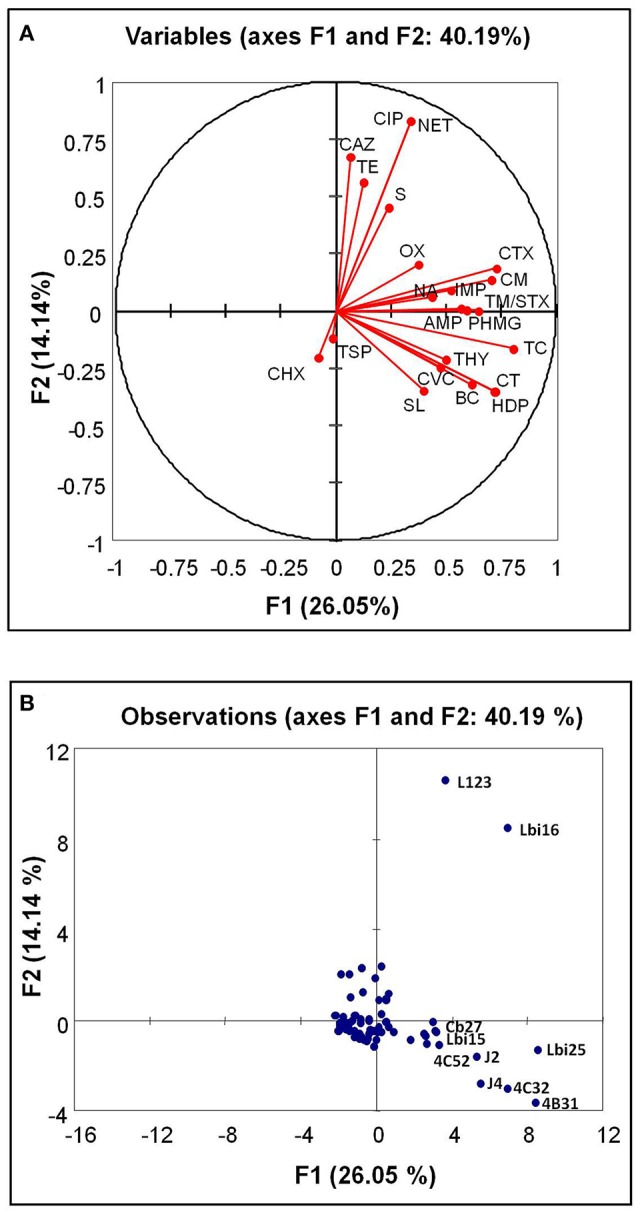
Biplot for biocide tolerance and antimicrobial resistance (scores) in the 87 bacterial isolates (variables) from seafoods. Antimicrobials (A), red dots, and isolates (B), blue dots are indicated. In (B), the letters indicate the bacterial isolates with an outstanding high number of antimicrobial resistance traits. BC, benzalkonium chloride; CT, cetrimide; HDP, hexadecylpyridinium chloride; TC, triclosan; CF, hexachlorophene; PHMG, poly-(hexamethylen guanidinium) hydrochloride; OX, P3 oxonia; AMP, ampicillin; CTX, cefotaxime; CAZ, ceftazidime; IMP, imipenem; CM, chloramphenicol; S, streptomycin; TE, tetracycline; NA, nalidixic acid; TM/STX, trimethoprim-sulfamethoxazole; CVC, carvacrol; THY, thymol; SL, sodium lactate; TSP, trisodium phosphate.
Table 2.
Correlations between antibiotic resistance and tolerance to biocides and other antimicrobials in the 87 bacterial isolates recovered from seafood.
| Variables | BC | CT | CHX | HDP | TC | PHMG | OX | AMP | CTX | CAZ | IMP | CIP | NA | CM | S | NET | TE | TM/ STX | CVC | THY | SL | TSP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BC | 1.000 | |||||||||||||||||||||
| CT | 0.788 | 1.000 | ||||||||||||||||||||
| CHX | 0.003 | 0.003 | 1.000 | |||||||||||||||||||
| HDP | 0.712 | 0.907 | −0.020 | 1.000 | ||||||||||||||||||
| TC | 0.429 | 0.564 | −0.137 | 0.621 | 1.000 | |||||||||||||||||
| PHMG | 0.517 | 0.517 | 0.095 | 0.463 | 0.498 | 1.000 | ||||||||||||||||
| OX | 0.182 | 0.182 | 0.169 | 0.157 | 0.203 | 0.590 | 1.000 | |||||||||||||||
| AMP | 0.212 | 0.212 | −0.045 | 0.234 | 0.313 | 0.142 | 0.078 | 1.000 | ||||||||||||||
| CTX | 0.290 | 0.400 | −0.239 | 0.441 | 0.570 | 0.339 | 0.110 | 0.427 | 1.000 | |||||||||||||
| CAZ | −0.089 | −0.089 | −0.178 | −0.098 | −0.060 | 0.044 | 0.093 | 0.018 | 0.181 | 1.000 | ||||||||||||
| IMP | 0.312 | 0.312 | 0.363 | 0.268 | 0.343 | 0.395 | 0.216 | 0.295 | 0.246 | 0.051 | 1.000 | |||||||||||
| CIP | −0.038 | −0.038 | −0.076 | −0.042 | 0.142 | 0.261 | 0.333 | 0.132 | 0.249 | 0.426 | 0.366 | 1.000 | ||||||||||
| NA | 0.023 | 0.126 | −0.125 | 0.176 | 0.327 | 0.082 | 0.180 | 0.349 | 0.507 | 0.183 | 0.025 | 0.046 | 1.000 | |||||||||
| CM | 0.244 | 0.349 | −0.287 | 0.385 | 0.553 | 0.289 | 0.078 | 0.559 | 0.764 | 0.127 | 0.182 | 0.217 | 0.543 | 1.000 | ||||||||
| S | 0.118 | 0.118 | −0.037 | 0.085 | 0.085 | 0.197 | 0.159 | 0.095 | 0.320 | 0.439 | 0.104 | 0.300 | 0.153 | 0.181 | 1.000 | |||||||
| NET | −0.038 | −0.038 | −0.076 | −0.042 | 0.142 | 0.261 | 0.333 | 0.132 | 0.249 | 0.426 | 0.366 | 1.000 | 0.046 | 0.217 | 0.300 | 1.000 | ||||||
| TE | −0.038 | −0.038 | −0.076 | −0.042 | −0.067 | −0.042 | −0.034 | 0.132 | 0.249 | 0.185 | 0.151 | 0.488 | −0.114 | 0.217 | 0.111 | 0.488 | 1.000 | |||||
| TM/STX | 0.179 | 0.290 | −0.239 | 0.339 | 0.570 | 0.136 | 0.233 | 0.479 | 0.310 | −0.061 | 0.174 | 0.249 | 0.293 | 0.327 | −0.061 | 0.249 | 0.077 | 1.000 | ||||
| CVC | 0.368 | 0.368 | −0.068 | 0.350 | 0.279 | 0.335 | 0.042 | 0.216 | 0.298 | −0.083 | 0.179 | −0.004 | 0.101 | 0.250 | −0.061 | −0.004 | −0.051 | 0.373 | 1.000 | |||
| THY | 0.216 | 0.276 | 0.125 | 0.221 | 0.493 | 0.221 | 0.118 | 0.341 | 0.205 | −0.166 | 0.444 | 0.068 | 0.169 | 0.230 | 0.005 | 0.068 | −0.099 | 0.388 | 0.167 | 1.000 | ||
| SL | 0.054 | 0.135 | 0.212 | 0.178 | 0.371 | 0.178 | 0.084 | 0.408 | 0.195 | −0.331 | 0.239 | −0.066 | 0.265 | 0.291 | −0.173 | −0.066 | −0.066 | 0.278 | 0.273 | 0.388 | 1.000 | |
| TSP | −0.107 | −0.088 | 0.368 | −0.104 | −0.032 | −0.069 | −0.049 | 0.109 | −0.039 | −0.142 | 0.426 | −0.019 | −0.049 | −0.015 | −0.032 | −0.019 | −0.019 | −0.225 | −0.077 | 0.171 | 0.358 | 1.000 |
BC, benzalkonium chloride; CT, cetrimide; CHX, chlorhexidine; HDP, hexadecylpyridinium chloride; TC, triclosan; PHMG, polyhexamethylen guanidium chloride; OX, P3 Oxonia; AMP, ampicillin; CTX, cefotaxime, CAZ, ceftazidime; IMP, imipenen; CIP, ciprofloxacin; NA, nalidixic acid; CM, chloramphenicol; S, streptomycin; NET, netilmicin; TE, tetracyclin; TM/STX, trimethoprim/solfamethoxazole; CVC, carvacrol; THY, thymol; SL, sodium lactate; TSP, trisodium phosphate.
Data in bold indicate significant correlations at P < 0.05 level.
The following pairs of biocides showed positive correlations that were very strong (CT-HDP), strong (BC-CT, BC-HDP, and HDP-TC) or moderate (BC-TC, BC-PHMG, CT-TC, CT-PHMG, HDP-PHMG, TC-PHMG, and PHMG-OX).
For antibiotics, the following positive correlations were very strong (CIP-NET), strong (CTX-CM) or moderate (AMP-CTX, AMP-CM, AMP-TM/STX, CTX-NA, CAZ-CIP, CAZ-SM, CAZ-NET, CIP-TE, NA-CM, and TE-NET).
For the rest of antimicrobials tested (CVC, THY, SL, TSP), the only significant (P < 0.05) correlations detected were weak (Table 2).
Interestingly, significant (P < 0.05) positive correlations were also detected between antimicrobials belonging to the different groups tested. For example, moderate positive correlations were detected for the biocides CT, HDP, and TC with the antibiotic CTX, and also for TC with CM and TM/STX and with the phenolic compound THY (Table 2). A few antibiotics showed moderate positive correlations with the rest of antimicrobials tested, as in the case of AMP with SL and IMP with THY and TSP.
From the preliminary general study, 30 isolates were selected for further analysis regarding metal resistance, identification, and study of the genetic determinants of resistance.
Identification of selected isolates
The 30 isolates selected for further study were identified by 16s rDNA sequencing (Table 3). Most of them (96.66%) were identified as Gram negative bacteria: Pseudomonas brassicacearum (10.00%), Pseudomonas poae (16.67%), P. putida (3.33%), Pseudomonas synxantha (26.67%), Pseudomonas spp. (6.67%), Acinetobacter calcoaceticus (10.00%), Acinetobacter oleivorans (3.33%), Aeromonas salmonicida (10.00%), Aeromonas spp. (3.33%), Shewanella baltica (3.33%), and Proteus mirabilis (3.33%). The only Gram positive isolate identified belonged to Listeria innocua (3.33%).
Table 3.
Characterization of selected isolates.
| Isolate | Food source | Genetics determinants of resistance | CuSO4 (MIC) | ZnCl2 (MIC) |
|---|---|---|---|---|
| Pseudomonas brassicacearum 4Sd1 | Sardine | sul1, chrB | 16 | 4 |
| Pseudomonas brassicacearum 4B1 | Anchovy | chrB | 16 | 4 |
| Pseudomonas brassicacearum 4B43 | Anchovy | 16 | 4 | |
| Pseudomonas poae B2 | Anchovy | sul1, floR | 20 | 16 |
| Pseudomonas poae J7 | Athlantic horse mackerel | floR, pcoR, chrB | 32 | 8 |
| Pseudomonas poae 4Sd41 | Sardine | 36 | 16 | |
| Pseudomonas poae 4Sd42 | Sardine | blaPSE, aadA1, pcoR, chrB | 12 | 4 |
| Pseudomonas poae 4C52 | Squid | blaCTX−M, aadA1, pcoA/copA | 32 | 4 |
| Pseudomonas putida Cg22 | Norway lobster | blaTEM, pcoA/copA | 32 | 16 |
| Pseudomonas synxantha J2 | Athlantic horse mackerel | sul1, blaCTX−M | 36 | 16 |
| Pseudomonas synxantha J4 | Athlantic horse mackerel | sul1, floR, pcoA/copA | 20 | 16 |
| Pseudomonas synxantha Cb27 | Mackerel | aac(6′)-Ib, pcoA/copA | 36 | 16 |
| Pseudomonas synxantha Lbi15 | Sea bass | aadA1, pcoA/copA | 20 | 8 |
| Pseudomonas synxantha Lbi16 | Sea bass | pcoA/copA | 24 | 8 |
| Pseudomonas synxantha Lbi25 | Sea bass | sul1, sul2, floR, aadA1, pcoA/copA | 24 | 16 |
| Pseudomonas synxantha 4C32 | Squid | sul1, blaPSE, blaCTX−M, pcoA/copA | 36 | 16 |
| Pseudomonas synxantha 4B31 | Anchovy | blaNDM−1, pcoA/copA | 36 | 16 |
| Pseudomonas spp. B7 | Anchovy | sul1, sul2, floR, blaIMP, aadA1, pcoA/copA | 36 | 8 |
| Pseudomonas spp. Cb212 | Mackerel | pcoA/copA | 36 | 16 |
| Shewanella baltica 4Sd53 | Sardine | sul1, aac(6′)-Ib | 16 | 4 |
| Acinetobacter calcoaceticus S14 | Salmon | qacEΔ1, sul1, blaTEM, aac(6′)-Ib | 32 | 16 |
| Acinetobacter calcoaceticus S22 | Salmon | qacEΔ1, sul1, blaTEM, aac(6')-Ib | 32 | 16 |
| Acinetobacter calcoaceticus S25 | Salmon | qacEΔ1, sul1, aac(6′)-Ib | 32 | 16 |
| Acinetobacter oleivorans L123 | Prawn | blaTEM, blaCTX−M | 24 | 4 |
| Aeromonas salmonicida Cb22 | Mackerel | sul1, blaTEM | 20 | 8 |
| Aeromonas salmonicida Mj2 | Mussel | 12 | 8 | |
| Aeromonas salmonicida L124 | Shrimp | 32 | 8 | |
| Aeromonas spp. Do11 | Gilthead seabream | sul1, blaTEM, blaCTX−M | 24 | 4 |
| Proteus mirabilis T215 | Blue shark | qacEΔ1, blaCTX−M, aadA1 | 32 | 16 |
| Listeria innocua 4C21 | Squid | blaPSE | 12 | 4 |
MIC, minimum inhibitory concentration (in mM).
Genetic determinants of biocide tolerance and antibiotic resistance
Results obtained on the genetic determinants of resistance for the selected isolates are shown in Table 3. Twenty seven out of the 30 isolates tested positive for at least one of the genetic determinants studied. The only QAC resistance determinant detected was qacEΔ1. It was found in three isolates of A. calcoaceticus from salmon and in one Proteus mirabilis isolate from blue shark. The genetic determinant for sulfonamide resistance sul1 was detected in 13 isolates, two of which also tested positive for sul2. In addition, three A. calcoaceticus isolates positive for sul1 also were positive for qacEΔ1. However, PCR experiments using a qacEF primer and a sulR primer did not yield any amplification, suggesting that both genetic determinants were not physically close as in class I integrons.
Among the β-lactamase genes tested, blaTEM and blaCTX−M were the most frequent. Of the five isolates positive for blaTEM, three belonged to genus Acinetobacter isolated from salmon, one to Aeromonas and one to Pseudmonas. Five isolates tested positive for blaCTX−M: Pseudomonas (3), Acinetobacter and Proteus. The genetic determinant blaPSE was detected in three isolates, including two Pseudomonas and the L. innocua isolate obtained from squid. Furthermore, blaIMP and blaNDM−1 were detected in one Pseudomonas isolate each, both from anchovies. The remaining β-lactamase genes investigated were not detected.
The phenicol resistance determinant floR was detected in five isolates (all of them belonging to genus Pseudomonas) from different sources: anchovies, black mackerel and sea bass. By contrast, cmlA was not detected in any isolate. The aminoglycoside resistance determinant aadA1 was detected in six isolates, five of which belonged to genus Pseudomonas and one to Proteus, while aac(6′)-Ib was detected in five isolates (including three from genus Acinetobacter isolated from salmon, one Shewanella and one Pseudomonas).
Resistance to copper and zinc
A 26.60% of the 30 isolates tested were able to grow in the presence of 24 mM CuSO4, and still 23.30% were able to grow at 32 mM of the copper salt (Table 3). Regarding zinc tolerance, a 46.60% of the isolates were able to grow in the presence of 8 mM ZnCl2. However, all isolates tested were inhibited by 16 mM ZnCl2.
Study of the genetic determinants for metal resistance (Table 3) yielded positive results for the plasmid-borne multicopper oxidase gene pcoA/copA (36.66% of selected isolates) and the DNA binding repressor protein gene pcoR (6.66%), and for the zinc-chromate resistance gene chrB (13.33%). The remaining metal resistance genes investigated were not detected.
Discussion
In the present study, biocide tolerance and antibiotic resistance were detected among bacteria isolated from seafoods. Biocides are used for many different purposes, including health care products and in disinfection processes in the food industry. For example, benzalkonium chloride is used for water treatment, general site disinfection, fish parasite removal, and prevention of infectious disease in fish and shellfish. As a result, large amounts of biocides arrive to waters. The impact of triclosan on aquatic bacterial communities has been described (Dann and Hontela, 2011; McNamara et al., 2014). Previous contact with biocides as well as natural background resistance could explain the biocide tolerances observed in the present study. It is also worth noting that there were positive correlations not only for tolerance to biocides of the same chemical group but also between biocides from different groups. However, there were differences between polyguanides, since poly-(hexamethylen guanidinium) hydrochloride showed positive correlation with several other biocides while chlorhexidine did not. These results could be explained by differences in chemical formula, mechanisms of adaptation (including intrinsic resistance), and also by the development of specific mechanisms of tolerance upon exposure to multiple biocides.
A relatively high percentage of isolates were resistant to at least one biocide and at least one antibiotic, and there were significant (P < 0.05) positive correlations between biocide tolerance and antibiotic resistance. These results reinforce the general concern that the use of biocides may co-select for antibiotic resistance (SCENIHR, 2009; Ortega-Morente et al., 2013; Wales and Davies, 2015). Cross-resistance between antibiotics and biocides and between different biocides has been reported for different bacteria, like for example Pseudomonas aeruginosa (Lambert et al., 2001; Lavilla Lerma et al., 2015). Furthermore, previous studies have shown that adaptation to biocides by repeated exposure results in an increased resistance to antibiotics (Gadea et al., 2016, 2017a). It is worth noting that there were also some positive correlations between tolerance/resistance to biocides, antibiotics, and other antimicrobials. For example, the phenolic biocide triclosan showed positive correlation with a lower sensitivity to the phenolic compound thymol (but not with carvacrol), and the antibiotic ampicillin also showed positive correlation with thymol. Previous studies have shown that exposure to plant essential oils (which are rich in phenolic compounds) such as pine oil resulted in the selection of mutants with deregulated mar operon that had decreased susceptibility to a range of antimicrobial compounds (including antibiotics and biocides) as a consequence of reduced cell permeability and increased efflux pump activity (Moken et al., 1997; Ortega-Morente et al., 2013). It is worth mentioning that the chemical preservatives sodium lactate and trisodium phosphate only showed moderate positive correlations with the antibiotics ampicillin and imipenem, but not with biocides. A recent study indicated that bacteria adapted to quaternary ammonium compounds under laboratory conditions showed a generalized increased tolerance to preservatives (such as 4-hydroxybenzoic acid, thyme, and clove oil, sodium, and potassium nitrates, potassium sorbate), while the opposite was observed in the case of triclosan (Gadea et al., 2017b).
Sulfonamides potentiated with trimethoprim or ormethoprim and florfenicol are some of the antibiotics commonly used in aquaculture (Hernández Serrano, 2005). In the present study, the sulfonamide resistance sul1 gene was the genetic determinant detected most frequently. Sulfonamide resistance is most often linked to Class I integrons. These mobile genetic elements tend to accumulate different antibiotic resistance genes and also biocide tolerance genes like qacEΔ1. However, qacEΔ1 was detected in combination with sul1 only in the A. calcoaceticus isolates, and PCR amplification with a forward primer for qacEΔ1 and a reverse primer for sul1 did not suggest a close physical location of the two genes unless they were in opposite direction. Class 1 resistance integrons are located on mobile elements like transposons and plasmids and are widely distributed among clinical strains and also in environmental isolates, and play an important role as reservoirs of antimicrobial resistance genes (L'Abée-Lund and Sørum, 2001; Stokes and Gillings, 2011; Koczura et al., 2013, 2014). Remarkably, one study carried out at aquaculture facilities in the northern Baltic Sea (Finland) reported that antibiotic resistance genes for sulfonamides (sul1 and sul2) and trimethoprim (dfrA1) and an integrase gene for a class 1 integron (intI1) persisted in sediments below fish farms at very low antibiotic concentrations during a 6-year observation period (Muziasari et al., 2014). Presumably, antimicrobial resistance genes could spread in marine sediments to other bacteria that colonize non-aquaculture fish and from these to seafood processing environments. This could explain the finding of sul1 in bacteria isolated in the present study from fish like sardines, anchovies, Athlantic horse mackerel, sea bass, gilthead seabream, and salmon, or from squid. Furthermore, in a number of cases, bacterial isolates carrying sul1 also tested positive for the florfenicol resistance gene floR, which can also be associated with Class 1 integrons (Toleman et al., 2007; Lin et al., 2016).
Aquaculture heavily depends on the use of antibiotics (Hernández Serrano, 2005), and several studies have reported on antimicrobial resistance in bacteria from aquaculture ecosystems (e.g., Shah et al., 2014; Huang et al., 2015; Xiong et al., 2015; Watts et al., 2017). Remarkably, one of the two isolates from the present study with broadest spectra of antimicrobial resistance (identified as A. oleivorans) was isolated from prawns grown in aquaculture. The A. oleivorans strain from the present study was resistant to 10 antimicrobials and tested positive for the genetic determinants blaTEM and blaCTX−M. Since the prawns were boiled and frozen and then sold unfrozen over the counter on ice, there is a possibility that this strain arrived to the food by cross contamination during handling. Boiled prawns are ready to eat, therefore the bacteria with multiple antibiotic resistance together with their antibiotic resistance genes could be passed from prawns to humans. Acinetobacters are inhabitants of soil and water in addition to being opportunistic pathogens for humans, and the human-pathogenic strains are known to exhibit both intrinsic and acquired resistance to a wide variety of antimicrobials (Doughari et al., 2011). It is also worth noting that the remaining acinetobacters detected in the present study (all of them identified as A. oleivorans) were isolated from raw salmon slices, also grown in aquaculture. They were resistant to a lower number of antibiotics (4 and 6), but all of them tested positive for the genetic determinants qacEΔ1, sul1 and aac(6′)-Ib, and two also were positive for the extended-spectrum β-lactamase gene blaTEM. A. oleivorans was described as a diesel-oil and n-hexadecane-degrading bacterium isolated from a rice paddy (Kang et al., 2011). So far, antibiotic resistance of non-pathogenic Acinetobacter species has been weakly explored, but results from the present study suggest that they could be an important reservoir of antimicrobial resistance traits.
A 14.94% of the 87 isolates from the present study were resistant to imipenem. Carbapenems were the last β-lactams retaining nearuniversal anti-Gram-negative activity, but carbapenemase genes are spreading, conferring resistance (Nordmann et al., 2011). Furthermore, among the metallo-β-lactamases investigated in the present study, the imipenemase (IMP), and the New Delhi metallo-β-lactamase NDM-1 gene were detected in two isolates, both belonging to Pseudomonas. Several metallo-β-lactamases encoded by mobile DNA have emerged in important Gram-negative pathogens (Cornaglia et al., 2011). NDM has been reported mainly in Klebsiella pneumoniae and E. coli, but it has also been found in a variety of other members of the Enterobacteriaceae, including Acinetobacter spp., in Pseudomonas spp., and in Vibrio cholerae (Poirel et al., 2011; Walsh et al., 2011; Mataseje et al., 2016). In Pseudomonas aeruginosa, the chromosomal blaNDM−1 gene has been reported to be located within a Class 1 integron bearing insertion sequence (IS) common region 1 (ISCR1) in a Tn402-like structure (Janvier et al., 2013; Jovcic et al., 2013). Remarkably, the integron also contained the QAC resistance determinant qacEΔ1 and the sulfonamide resistance gene sul1. A similar gene array has been reported for the blaNDM−1 regions from E. coli transferable plasmid pNDM15-1078 (Mataseje et al., 2016). Furthermore, blaNDM−1 frequently appears in association with bleMBL gene that confers resistance to bleomycin, a glycopeptide antibiotic that is naturally produced by Streptomyces verticillus. It is possible that bleomycin-like molecules contribute to selective pressure, leading to the further spread of NDM producers in the environment (Mataseje et al., 2016).
All Aeromonas isolates from the present study were resistant to chlorhexidine, ampicillin and imipenem. In addition two of them tested positive for sul1 and blaTEM and one also was positive for blaCTX−M. Aeromonas are widely distributed in aquatic environments (Holmes et al., 1996). The genus includes species pathogenic for fish (like A. salmonicida and others) and humans. In one study on ampicillin-resistant isolates from estuarine waters, Henriques et al. (2006) detected the presence of the integrase gene (along with other genes associated with Class I integrons) in Aeromonas strains. The authors also detected the presence of β-lactamase genes blaTEM, blaSHV, blaCphA, and blaOXA−B in Aeromonas strains. Remarkably, the blaOXA−B detected in Aeromonas sp. and A. hydrophila was associated with Class I integrons. Another study on Aeromonas isolated from nine freshwater trout farms in Australia reported the presence of sul1 together with other antibiotic resistance genes typically associated with Class I integrons (Ndi and Barton, 2011). However, the β-lactamase genes investigated (blaTEM and blaSHV) were not detected. Further studies should be carried out in order to determine the possible association of the β-lactamase genes detected in the Aeromonas isolates from present study with Class I integrons.
In the present study, one isolate resistant to cefotaxime, ceftazidime, streptomycin and nalidixic acid was identified as Listeria innocua. This isolate tested positive for the β-lactamase resistance gene blaPSE. Previous studies have reported on antimicrobial resistance in Listeria spp. isolated from raw fish and open-air fish market environment (Jamali et al., 2015). The authors reported a high resistance of L. monocytogenes to tetracycline and penicillin G in agreement with other studies (Rodas-Suárez et al., 2006; Fallah et al., 2013). Another work reported a high resistance level to ampicillin, cefotaxime (100%), and penicillin (57%) in seafood isolates of L. monocytogenes (Abdollahzadeh et al., 2016). We could speculate that the L. innocua isolated in the present study from squid originated from the seafood handling and processing environment. Although this is a non-pathogenic species, the results would suggest that different species of Listeria in seafood processing environments can act as reservoirs for antimicrobial resistance traits.
Exposure to low concentrations of antibiotics, disinfectants, chemical pollutants, and metals can act as a selective force leading to resistance processes among indigenous bacterial populations (Martinez, 2009). There is a concern that exposure to metals may co-select for antibiotic resistance. In the present study, isolates capable of growing at high concentrations of copper sulfate, and zinc chloride included representatives of Pseudomonas, Aeromonas, Acinetobacter, and Proteus. The observed metal tolerance could be due to direct exposure to these metals in the environment. Furthermore, the multicopper oxidase gene pcoA/copA was detected in several Pseudomonas strains that were also positive for different antibiotic resistance genes (blaCTX−M, blaTEM, blaPSE, blaNDM−1, blaIMP, aadA1, aac(6′)-Ib, sul1, sul2, floR). The copper-inducible system copABCDRS was first described within the plasmid pPT23D in P. syringae (Cha and Cooksey, 1991) and is homologous to the pco system found on the conjugative plasmid pRJ1004 from E. coli. In addition to the plasmid-borne system, chromosomal P-type ATPases are responsible for conferring tolerance to metals like copper, zinc, cobalt, chromium, and cadmium (Teitzel and Parsek, 2003). As indicated by Berendonk et al. (2015), selective pressures present in natural environments such as rivers and lakes as a result of human practices are leading to the occurrence of an increasing number of multi-drug resistant environmental Pseudomonas isolates (Berendonk et al., 2015). Nevertheless, in the study of the resistome of P. aeruginosa E67, an epiphytic isolate from a metal-contaminated estuary, physical links between metal and antibiotic resistance genes were not identified, suggesting a predominance of cross-resistance associated with multidrug efflux pumps (Teixeira et al., 2016). Bacterial multidrug efflux pumps can accommodate a variety of antimicrobials, including dyes, antibiotics, and biocides (Poole, 2007). Nevertheless, a possible association of plasmidic pcoA/copA with antibiotic resistance genes in Pseudomonas deserves to be further investigated.
In conclusion results from the present study clearly indicate that bacteria from seafoods carry resistance traits against diverse antimicrobials. The possible role of these bacteria in spread of antimicrobial resistance through the food chain deserves further investigation. It is also important to find alternative ways to manage antimicrobial resistance. Biological methods such as the use of probiotics (Tan et al., 2016; Banerjee and Ray, 2017) or the application of bacteriophages (Letchumanan et al., 2016; Torres-Barceló et al., 2016; Parmar et al., 2017) could be promising alternatives to classical antimicrobials.
Author contributions
JR carried out seafood sampling, microbiological analysis and PCR investigation of genetic determinants of resistance. MG participated in planning and supervision of the experimental work and contributed with strain identification and preparation of PCR experiments. RP contributed with data analysis and preparation of graphical material for the manuscript. RL and AG carried out global analysis of the results and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by research project P08-AGR-4295 (CICE, FEDER) and University of Jaén (Research Structure AGR230). We also acknowledge the Campus de Excelencia Internacional Agroalimentario CeiA3.
References
- Abdollahzadeh E., Ojagh S. M., Hosseini H., Ghaemi E. A., Irajian G., Naghizadeh H. M. (2016). Antimicrobial resistance of Listeria monocytogenes isolated from seafood and humans in Iran. Microb. Pathog. 100, 70–74. 10.1016/j.micpath.2016.09.012 [DOI] [PubMed] [Google Scholar]
- Abriouel H., Lucas R., Ben Omar N., Valdivia E., Maqueda M., Martinez-Canamero M., et al. (2005). Enterocin AS-48RJ: a variant of enterocin AS-48 chromosomally encoded by Enterococcus faecium RJ16 isolated from food. Syst. Appl. Microbiol. 28, 383–397. 10.1016/j.syapm.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Adaikkalam V., Swarup S. (2002). Molecular characterization of an operon, cueAR, encoding a putative P1-type ATPase and a MerR-type regulatory protein involved in copper homeostasis in Pseudomonas putida. Microbiology 148, 2857–2867. 10.1099/00221287-148-9-2857 [DOI] [PubMed] [Google Scholar]
- Badar U., Ahmed N., Shoeb E., Gadd G. M. (2014). Identification of the pco operon in Enterobacter species isolated from contaminated soil. Int. J. Adv. Res. 2, 227–233. 10.21474/IJAR01 [DOI] [Google Scholar]
- Banerjee G., Ray A. K. (2017). The advancement of probiotics research and its application in fish farming industries. Res. Vet. Sci. 115, 66–77. 10.1016/j.rvsc.2017.01.016 [DOI] [PubMed] [Google Scholar]
- Berendonk T. U., Manaia C. M., Merlin C., Fatta-Kassinos D., Cytryn E., Walsh F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317. 10.1038/nrmicro3439 [DOI] [PubMed] [Google Scholar]
- Bertrand S., Weill F. X., Cloeckaert A., Vrints M., Mairiaux E., Praud K., et al. (2006). Clonal emergence of extended-spectrum β-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J. Clin. Microbiol. 44, 2897–2903. 10.1128/JCM.02549-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsdottir-Butler K., Green D. P., Bolton G. E., McClellan-Green P. D. (2015). Control of histamine-producing bacteria and histamine formation in fish muscle by trisodium phosphate. J. Food Sci. 80, M1253–M1258. 10.1111/1750-3841.12875 [DOI] [PubMed] [Google Scholar]
- Braoudaki M., Hilton A. C. (2004). Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J. Clin. Microbiol. 42, 73–78. 10.1128/JCM.42.1.73-78.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Rouch D. A., Lee B. T. O. (1992). Copper resistance systems in bacteria. Plasmid 27, 41–51. 10.1016/0147-619X(92)90005-U [DOI] [PubMed] [Google Scholar]
- Cavaco L. M., Hasman L., Aarestrup F. M. (2011). Zinc resistance of Staphylococcus aureus of animal origin is strongly associated with methicillin resistance. Vet. Microbiol. 150, 344–348. 10.1016/j.vetmic.2011.02.014 [DOI] [PubMed] [Google Scholar]
- Cha J. S., Cooksey D. A. (1991). Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Microbiology 88, 8915–8919. 10.1073/pnas.88.20.8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihomvu P., Stegmann P., Pillay M. (2015). Characterization and structure prediction of partial length protein sequences of pcoA, pcoR and chrB genes from heavy metal resistant bacteria from the Klip River, South Africa. Int. J. Mol. Sci. 16, 7352–7374. 10.3390/ijms16047352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. H., Su L. H., Chu C. H., Wang M. H., Yeh C. M., Weill F. X., et al. (2006). Detection of multidrug-resistant Salmonella enterica serovar typhimurium phage types DT102, DT104, and U302 by multiplex PCR. J. Clin. Microbiol. 44, 2354–2358. 10.1128/JCM.00171-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuanchuen R., Beinlich K., Hoang T. T., Becher A., Karkhoff-Schweizer R. R., Schweizer H. P. (2001). Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 45, 428–432. 10.1128/AAC.45.2.428-432.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuanchuen R., Khemtong S., Padungtod P. (2007). Occurrence of qacE/qacEΔ1 genes and their correlation with class 1 integrons in Salmonella enterica isolates from poultry and swine. S. East Asian. J. Trop. Med. Pub. Health 38, 855–862. [PubMed] [Google Scholar]
- Cornaglia G., Giamarellou H., Rossolini G. M. (2011). Metallo- β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11, 381–393. 10.1016/S1473-3099(11)70056-1 [DOI] [PubMed] [Google Scholar]
- Crémet L., Caroff N., Dauvergne S., Reynaud A., Lepelletier D., Corvec S. (2011). Prevalence of plasmid-mediated quinolone resistance determinants in ESBL Enterobacteriaceae clinical isolates over a 1-year period in a French hospital. Pathol. Biol. 59, 151–156. 10.1016/j.patbio.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Dann A. B., Hontela A. (2011). Triclosan: environmental exposure, toxicity and mechanisms of action. J. Appl. Toxicol. 31, 285–311. 10.1002/jat.1660 [DOI] [PubMed] [Google Scholar]
- De Toro M., Sáenz Y., Cercenado E., Rojo-Bezares B., García-Campello M., Undabeitia, et al. (2011). Genetic characterization of the mechanisms of resistance to amoxicillin/clavulanate and third-generation cephalosporins in Salmonella enterica from three Spanish hospitals. Int. Microbiol. 14, 173–181. 10.2436/20.1501.01.146 [DOI] [PubMed] [Google Scholar]
- CLSI (2015). Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement, Vol. 35 Wayne, PA: Clinical and Laboratory Standards Institute Performance Standards for Document M100–S25. [Google Scholar]
- Doughari H. J., Ndakidemi P. A., Human I. S., Benade S. (2011). The ecology, biology and pathogenesis of Acinetobacter spp: an overview. Microb. Environ. 26, 101–112. 10.1264/jsme2.ME10179 [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (ECDC) (2012). Antimicrobial resistance surveillance in Europe 2011. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). ECDC, Stockholm. Available online at: http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-surveillance-europe-2011.pdf
- European Centre for Disease Prevention and Control (ECDC) (2013). Epidemiological Update: Multistate Outbreak of Salmonella Stanley Infection. Solna: European Centre for Disease Prevention and Control (ECDC). [Google Scholar]
- European Food Safety Authority (EFSA) (2010). The community summary report on antimicrobial resistance in zoonotic and indicator bacteria from animals and food in the European Union in 2004–2007. EFSA J. 8:1309 10.2903/j.efsa.2010.1309 [DOI] [Google Scholar]
- European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC) (2013). The european union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2011. EFSA J. 11:3196 10.2903/j.efsa.2013.3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallah A. A., Saei-Dehkordi S. S., Mahzounieh M. (2013). Occurrence and antibiotic resistance profiles of Listeria monocytogenes isolated from seafood products and market and processing environments in Iran. Food Cont. 34, 630–636. 10.1016/j.foodcont.2013.06.015 [DOI] [Google Scholar]
- Food and Agriculture Organization (FAO) (2016). Globefish Highlights-Issue 3/2016. Rome: FAO. [Google Scholar]
- Gadea R., Fernández Fuentes M. A., Pérez Pulido R., Gálvez A., Ortega E. (2016). Adaptive tolerance to phenolic biocides in bacteria from organic foods: effects on antimicrobial susceptibility and tolerance to physical stresses. Food Res. Int. 85, 131–143. 10.1016/j.foodres.2016.04.033 [DOI] [PubMed] [Google Scholar]
- Gadea R., Fernández Fuentes M. Á., Pérez Pulido R., Gálvez A., Ortega E. (2017a). Effects of exposure to quaternary-ammonium-based biocides on antimicrobial susceptibility and tolerance to physical stresses in bacteria from organic foods. Food Microbiol. 63, 58–71. 10.1016/j.fm.2016.10.037 [DOI] [PubMed] [Google Scholar]
- Gadea R., Glibota N., Pérez Pulido R., Gálvez A., Ortega E. (2017b). Effects of exposure to biocides on susceptibility to essential oils and chemical preservatives in bacteria from organic foods. Food Cont. 80, 176–182. 10.1016/j.foodcont.2017.05.002 [DOI] [Google Scholar]
- Gaze W. H., Abdouslam N., Hawkey P. M., Wellington E. M. H. (2005). Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob. Agents Chemother. 49, 1802–1807. 10.1128/AAC.49.5.1802-1807.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sanz E., Kadlec K., Feßler A. T., Zarazaga M., Torres C., Schwarz S. (2013). Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob. Agents Chemother. 57, 3275–3282. 10.1128/AAC.00171-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardiola F. A., Cuesta A., Meseguer J., Esteban M. A. (2012). Risks of using antifouling biocides in aquaculture. Int. J. Mol. Sci. 13, 1541–1560. 10.3390/ijms13021541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra B., Soto S. M., Argüelles J. M., Mendoza C. (2001). Multidrug resistance is mediated by large plasmids carrying a Class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:−]. Antimicrob. Agents Chemother. 45, 1305–1308. 10.1128/AAC.45.4.1305-1308.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. S., Jensen L. B., Sørensen H. I. (2007). Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother. 60, 145–147. 10.1093/jac/dkm167 [DOI] [PubMed] [Google Scholar]
- Henriques I. S., Fonseca F., Alves A., Saavedra M. J., Correia A. (2006). Occurrence and diversity of integrons and β-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res. Microbiol. 157, 938–947. 10.1016/j.resmic.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Hernández Serrano P. (2005). Responsible Use of Antibiotics in Aquaculture. FAO Fisheries Technical Paper 469, FAO, Rome. [Google Scholar]
- Hobman J. L., Crossman L. C. (2014). Bacterial antimicrobial metal ion resistance. J. Med. Microbiol. 64, 471–497. 10.1099/jmm.0.023036-0 [DOI] [PubMed] [Google Scholar]
- Holmes P., Nicholls L. M., Sartory D. P. (1996). The ecology of mesophilic aeromonas in the aquatic environment, in The Genus Aeromonas, eds Austin B., Altwegg M., Gosling P. J., Joseph S. (New York, NY: John Wiley and Sons; ), 127–150. [Google Scholar]
- Huang Y., Zhang L., Tiu L., Wang H. H. (2015). Characterization of antibiotic resistance in commensal bacteria from an aquaculture ecosystem. Front. Microbiol. 6:914 10.3389/fmicb.2015.00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovleva A., Doi Y. (2017). Carbapenem-resistant Enterobacteriaceae. Clin. Lab. Med. 37, 303–315. 10.1016/j.cll.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamali H., Paydar M., Ismail S., Looi C. Y., Wong W. F., Radmehr B., et al. (2015). Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol. 15:144. 10.1186/s12866-015-0476-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier F., Jeannot K., Tesse S., Robert-Nicoud M., Delacour H., Rapp C., et al. (2013). Molecular characterization of blaNDM-1 in a sequence type 235 Pseudomonas aeruginosa isolate from France. Antimicrob. Agents Chemother. 57, 3408–3411. 10.1128/AAC.02334-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovcic B., Lepsanovic Z., Begovic J., Rakonjac B., Perovanovic J., Topi Sirovic L., et al. (2013). The clinical isolate Pseudomonas aeruginosa MMA83 carries two copies of the blaNDM-1 gene in a novel genetic context. Antimicrob. Agents Chemother. 57, 3405–3407. 10.1128/AAC.02312-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamika I., Momba M. N. B. (2013). Assessing the resistance and bioremediation ability of selected bacterial and protozoan species to heavy metals in metal-rich industrial wastewater. BMC Microbiol. 13:28. 10.1186/1471-2180-13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. S., Jung J., Jeon C. O., Park W. (2011). Acinetobacter oleivorans sp. nov. is capable of adhering to and growing on diesel-oil. J. Microbiol. 49, 29–34. 10.1007/s12275-011-0315-y [DOI] [PubMed] [Google Scholar]
- Kim Y. T., Jang J. H., Kim H. C., Kim H., Lee K. R., Park K. S., et al. (2011). Identification of strain harboring both aac(6′)-Ib and aac(6′)-Ib-cr variant simultaneously in Escherichia coli and Klebsiella pneumonia. BMB Rep. 44, 262–266. 10.5483/BMBRep.2011.44.4.262 [DOI] [PubMed] [Google Scholar]
- Koczura R., Mokracka J., Barczak A., Krysiak N., Kubek M., Kaznowski A. (2013). Association between the presence of class 1 integrons virulence genes and phylogenetic groups of Escherichia coli isolates from river water. Microb. Ecol. 65, 84–90. 10.1007/s00248-012-0101-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczura R., Semkowska A., Mokracka J. (2014). Integron-bearing Gram-negative bacteria in lake waters. Lett. Appl. Microbiol. 59, 514–519. 10.1111/lam.12307 [DOI] [PubMed] [Google Scholar]
- Kücken D., Feucht H. -H., Kaulfers P. -M. (2000). Association of qacE and qacEΔ1 with multiple resistance to antibiotics and antiseptics in clinical isolates of Gram-negative bacteria. FEMS Microbiol. Lett. 183, 95–98. 10.1016/S0378-1097(99)00636-9 [DOI] [PubMed] [Google Scholar]
- L'Abée-Lund T. M., Sørum H. (2001). Class 1 integrons mediate antibiotic resistance in the fish pathogen Aeromonas salmonicida worldwide. Microbial. Drug Res. 7, 263–272. 10.1089/10766290152652819 [DOI] [PubMed] [Google Scholar]
- Lambert R. J. W., Joynson J., Forbes B. (2001). The relationships and susceptibilities of some industrial, laboratory and clinical isolates of Pseudomonas aeruginosa to some antibiotics and biocides. J. Appl. Microbiol. 91, 972–984. 10.1046/j.1365-2672.2001.01460.x [DOI] [PubMed] [Google Scholar]
- Lavilla Lerma L., Benomar N., Casado Mu-oz M. C., Gálvez A., Abriouel H. (2015). Correlation between antibiotic and biocide resistance in mesophilic and psychrotrophic Pseudomonas spp. isolated from slaughterhouse surfaces throughout meat chain production. Food Microbiol. 51, 33–44. 10.1016/j.fm.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Letchumanan V., Chan K.-G., Pusparajah P., Saokaew S., Duangjai A., Goh B.-H., et al. (2016). Insights into bacteriophage application in controlling Vibrio species. Front. Microbiol. 7:1114. 10.3389/fmicb.2016.01114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Wu X., Yan Q., Ma Y., Huang L., Qin Y., et al. (2016). Incidence of antimicrobial-resistance genes and integrons in antibiotic-resistant bacteria isolated from eels and aquaculture ponds. Dis. Aquat. Organ. 120, 115–123. 10.3354/dao03013 [DOI] [PubMed] [Google Scholar]
- Lucera A., Costa C., Conte A., Del Nobile M. A. (2012). Food applications of natural antimicrobial compounds. Front. Microbiol. 3:287. 10.3389/fmicb.2012.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902. 10.1016/j.envpol.2009.05.051 [DOI] [PubMed] [Google Scholar]
- Mataseje L. F., Peirano G., Church D. L., Conly J., Mulvey M., Pitout J. D. (2016). Colistin-nonsusceptible Pseudomonas aeruginosa sequence type 654 with blaNDM-1 arrives in North America. Antimicrob. Agents Chemother. 60, 1794–1800. 10.1128/AAC.02591-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P. J., LaPara T. M., Novak P. J. (2014). The impacts of triclosan on anaerobic community structures, function, and antimicrobial resistance. Environ. Sci. Technol. 48, 7393–7400. 10.1021/es501388v [DOI] [PubMed] [Google Scholar]
- Medardus J. J., Molla B. Z., Nicol M., Morrow W. M., Rajala-Schultz P. J., Kazwala R., et al. (2014). In-feed use of heavy metal micronutrients in U.S. swine production systems and its role in persistence of multidrug-resistant Salmonellae. Appl. Environ. Microbiol. 80, 2317–2325. 10.1128/AEM.04283-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moken M. C., McMurry L. M., Levy S. B. (1997). Selection of multiple antibiotic resistant (Mar) mutants of Escherichia coli by using the disinfectant pine oil: roles of the mar and acrAB loci. Antimicrob. Agents Chemother. 41, 2770–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. R., Boyd D., Olson A. B. (2006). The genetics of Salmonella genomic island 1. Microb. Infect. 8, 1915–1922. 10.1016/j.micinf.2005.12.028 [DOI] [PubMed] [Google Scholar]
- Muziasari W. I., Managaki S., Pärnänen K., Karkman A., Lyra C., Tamminen M., et al. (2014). Sulphonamide and trimethoprim resistance genes persist in sediments at Baltic Sea aquaculture farms but are not detected in the surrounding environment. PLoS ONE 9:e92702 10.1371/journal.pone.0092702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndi O. L., Barton M. D. (2011). Incidence of class 1 integron and other antibiotic resistance determinants in Aeromonas spp. from rainbow trout farms in Australia. J. Fish Dis. 34, 589–599. 10.1111/j.1365-2761.2011.01272.x [DOI] [PubMed] [Google Scholar]
- Nies A., Nies D. H., Silver S. (1990). Nucleotide sequence and expression of a plasmid encoded chromate resistance determinant from Alcaligenes eutrophus. J. Biol. Chem. 265, 5648–5653. [PubMed] [Google Scholar]
- Nordmann P., Poirel L., Walsh T. R., Livermore D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19, 588–595. 10.1016/j.tim.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Ortega-Morente E., Fernández-Fuentes M. A., Grande-Burgos M. J., Abriouel H., Pérez-Pulido R., Gálvez A. (2013). Biocide tolerance in bacteria. Int. J. Food Microbiol. 162, 13–25. 10.1016/j.ijfoodmicro.2012.12.028 [DOI] [PubMed] [Google Scholar]
- Park C. H., Robicsek A., Jacoby G. A., Sahm D., Hooper D. C. (2006). Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50, 3953–3955. 10.1128/AAC.00915-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K. M., Hathi Z. J., Dafale N. A. (2017). Control of multidrug-resistant gene flow in the environment through bacteriophage intervention. Appl. Biochem. Biotechnol. 181, 1007–1029. 10.1007/s12010-016-2265-7 [DOI] [PubMed] [Google Scholar]
- Patel S. (2015). Plant essential oils and allied volatile fractions as multifunctional additives in meat and fish-based food products: a review. Food Addit. Contam. A Chem. Anal. Control Expo. Risk Assess. 32, 1049–1164. 10.1080/19440049.2015.1040081 [DOI] [PubMed] [Google Scholar]
- Poirel L., Dortet L., Bernabeu S., Nordmann P. (2011). Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55, 5403–5407. 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2007). Efflux pumps as antimicrobial resistance mechanisms. Annals Med. 39, 162–176. 10.1080/07853890701195262 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan K., Rajagopalan S., Nair S., Kenchappa P., Chandrakesan S. D. (2014). Molecular characterization of metallo β-lactamase producing multidrug resistant Pseudomonas aeruginosa from various clinical samples. Indian. J. Pathol. Microbiol. 57, 579–582. 10.4103/0377-4929.142670 [DOI] [PubMed] [Google Scholar]
- Rodas-Suárez O., Flores-Pedroche J., Betancourt-Rule J., Qui-ones-Ramírez E. I., Vázquez-Salinas C. (2006). Occurrence and antibiotic sensitivity of Listeria monocytogenes strains isolated from oysters, fish, and estuarine water. Appl. Environ. Microbiol. 72, 7410–7412. 10.1128/AEM.00956-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scientific Committee on Emerging and Newly Identified Health Risks (2009). Assessment of the Antibiotic Resistance Effects of Biocides. Avalaible online at: http://ec.europa.eu/health/archive/ph_risk/committees/04_scenihr/docs/scenihr_o_021.pdf (Accessed May 2, 2017) [Google Scholar]
- Sáenz Y., Briñas L., Domínguez E., Ruiz J., Zarazaga M., Vila J., et al. (2004). Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 48, 3996–4001. 10.1128/AAC.48.10.3996-4001.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz Y., Vinué L., Ruiz E., Somalo S., Martínez S., Rojo-Bezares B., et al. (2010). Class 1 integrons lacking qacEΔ1 and sul1 genes in Escherichia coli isolates of food, animal and human origin. Vet. Microbiol. 144, 493–497. 10.1016/j.vetmic.2010.01.026 [DOI] [PubMed] [Google Scholar]
- Shah S. Q., Cabello F. C., L'abée-Lund T. M., Tomova A., Godfrey H. P., Buschmann, et al. (2014). Antimicrobial resistance and antimicrobial resistance genes in marine bacteria from salmon aquaculture and non-aquaculture sites. Environ. Microbiol. 16, 1310–1320. 10.1111/1462-2920.12421 [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Gillings M. (2011). Gene flow mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 35, 790–819. 10.1111/j.1574-6976.2011.00273.x [DOI] [PubMed] [Google Scholar]
- Tan L. T.-H., Chan K.-G., Lee L.-H., Goh B.-H. (2016). Streptomyces bacteria as potential probiotics in aquaculture. Front. Microbiol. 7:79. 10.3389/fmicb.2016.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitzel G. M., Parsek M. R. (2003). Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69, 2313–2320. 10.1128/AEM.69.4.2313-2320.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira P., Tacão M., Alves A., Henriques I. (2016). Antibiotic and metal resistance in a ST395 Pseudomonas aeruginosa environmental isolate: a genomics approach. Mar. Pollut. Bull. 110, 75–81. 10.1016/j.marpolbul.2016.06.086 [DOI] [PubMed] [Google Scholar]
- Toleman M. A., Bennett P. M., Bennett D. M., Jones R. N., Walsh T. R. (2007). Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg. Infect. Dis. 13, 559–565. 10.3201/eid1304.061378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Barceló C., Franzon B., Vasse M., Hochberg M. E. (2016). Long-term effects of single and combined introductions of antibiotics and bacteriophages on populations of Pseudomonas aeruginosa. Evol. Appl. 9, 583–595. 10.1111/eva.12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevors J. T., Oddie K. M., Belliveau B. H. (1985), Metal resistance in bacteria. FEMS Microbiol. Rev. 32, 39–54. 10.1111/j.1574-6968.1985.tb01181.x [DOI] [Google Scholar]
- Wales A. D., Davies R. H. (2015). Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4, 567–604. 10.3390/antibiotics4040567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. R., Weeks J., Livermore D. M., Toleman M. A. (2011). Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11, 355–362. 10.1016/S1473-3099(11)70059-7 [DOI] [PubMed] [Google Scholar]
- Wang W., Li M., Li Y. (2015). Intervention strategies for reducing Vibrio parahaemolyticus in seafood: a review. J. Food Sci. 80, R10–R19. 10.1111/1750-3841.12727 [DOI] [PubMed] [Google Scholar]
- Watermann B. T., Daehne B., Sievers S., Dannenberg R., Overbeke J. C., Klijnstra J. W., et al. (2005). Bioassays and selected chemical analysis of biocide-free antifouling coatings. Chemosphere 60, 1530–1541. 10.1016/j.chemosphere.2005.02.066 [DOI] [PubMed] [Google Scholar]
- Watts J. E. M., Schreier H. J., Lanska L., Hale M. S. (2017). The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions. Mar. Drugs 15:e158. 10.3390/md15060158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. 10.1128/jb.173.2.697-703.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2001). Environmental Health Criteria 221. Geneva: Zinc World Health Organization. [Google Scholar]
- Wong M. H., Chen S. (2013). First detection of oqxAB in Salmonella spp. isolated from food. Antimicrob. Agents Chemother. 57, 658–660. 10.1128/AAC.01144-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Sun Y., Zhang T., Ding X., Li Y., Wang M., et al. (2015). Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China. Microb. Ecol. 70, 425–432. 10.1007/s00248-015-0583-x [DOI] [PubMed] [Google Scholar]
- Yebra D. M., Kiil S., Dam-Johansen K. (2004). Antifouling technology-past, present and future step toward efficient and environmentally friendly antifouling coatings. Prog. Org. Coat. 50, 75–104. 10.1016/j.porgcoat.2003.06.001 [DOI] [Google Scholar]


