Abstract
The ribonucleotide reductase gene tandem bnrdE/bnrdF in SPβ-related prophages of different Bacillus spp. isolates presents different configurations of intervening sequences, comprising one to three of six non-homologous splicing elements. Insertion sites of group I introns and intein DNA are clustered in three relatively short segments encoding functionally important domains of the ribonucleotide reductase. Comparison of the bnrdE homologs reveals mutual exclusion of a group I intron and an intein coding sequence flanking the codon that specifies a conserved cysteine. In vivo splicing was demonstrated for all introns. However, for two of them a part of the mRNA precursor molecules remains unspliced. Intergenic bnrdE–bnrdF regions are unexpectedly long, comprising between 238 and 541 nt. The longest encodes a putative polypeptide related to HNH homing endonucleases.
INTRODUCTION
Intervening sequences (IVSs) i.e. inteins or introns, are entities spliced from precursor polypeptide or mRNA, respectively. Inteins and several group I introns that encode site-specific endonucleases can be horizontally transferred into allelic IVS-free sites, a process known as homing (1).
Group I introns can be specifically detected by radiolabelling with GTP (2), since the first transesterification reaction in their splicing pathway involves covalent attachment of an exogenous guanosine cofactor to the 5′-end of the intron (3). Inspection of the ever-increasing number of sequenced bacterial and bacteriophage genomes confirms the very rare occurrence of IVSs and reveals their peculiar distribution within the phylogenetic tree, as well as their apparently specific occurrence in certain genes. All so far identified group I introns in protein coding sequences are confined to bacteriophages (4) and conform to the structural model of the A2 subgroup of group I introns (5). Most phage introns, as well as most intein coding sequences, are found within genes of DNA metabolism, an observation which has suggested either that IVSs play a regulatory role in DNA metabolism or, alternatively, a particular susceptibility of these loci for IVS insertion (6).
Although a strictly parasitic nature of IVSs is not incompatible with the presently available information, the paucity of experimental evidence for a possible role(s) of IVSs in the cell and, in particular, in the bacteriophage life cycle does not allow definite conclusions to be drawn. Analyses of the distribution of IVSs within related species offers a starting point for future functional studies, consisting of comparisons of physiological properties of IVS+ versus IVS– variants. So far, the identification of bacteriophage genes harboring introns and the screening of related isolates for homologous entities has revealed the existence of intronless variants. Of the 32 examined coliphage T4 close relatives, most were devoid of any of the three introns identified in T4 (4). DNA polymerase genes in four of five examined Bacillus subtilis HMU bacteriophages are interrupted by an intron. Although these introns have an extremely similar core, they encode HNH endonucleases that differ in size and amino acid sequence (7). Finally, half of the 62 Streptococcus thermophilus S3b-like bacteriophages contain a group I intron within a putative lysin gene, while in a few isolates, exhibiting a near identical core, the invading endonuclease ORF has apparently suffered a deletion (8).
The ribonucleotide reductase (RR) gene tandem bnrdE/bnrdF of the B.subtilis 168 prophage SPβ harbors two group I introns and an intein coding sequence (9). In the present contribution, comparison of SPβ-related prophages present in other Bacillus strains has revealed three new, non-homologous group I introns and four new IVS configurations with respect to that previously identified.
MATERIALS AND METHODS
Strains
Bacillus strains were kindly provided by A.A. Prozorov (M135, M137, M141, M1918, M1586 and M1321) and T.A. Trautner (BS30, BS41, BS121, BS129, BSG33-1 BSG40, BSG66 and BSG150).
PCR conditions
For the initial screening of different Bacillus strains, PCRs, performed with 1 ng genomic DNA and 20 pmol each primer, consisted of 30 cycles of incubation at 95°C for 30 s, 40°C for 1 min and 72°C for 2 min. Oligonucleotide pairs VL264/VL265, VL268/VL269 and VL262/VL263 were used. In strain 168 they amplify the bnrdE intron, the bnrdE intein coding sequence and the bnrdF intron, respectively (9). From the nucleotide sequence of these PCR products, new, specific oligonucleotides were synthesized. A search for possible IVSs integrated distant from the reported insertion sites in the RR genes was achieved by additional PCR reactions whose overlapping products entirely covered the bnrdE/bnrdF doublet. In these reactions the annealing temperature was raised to 45°C. The oligonucleotide pairs used were: strain M1918, VL308/VL311, VL314/VL317, VL319/VL321, VL292/VL293, VL322/VL323, VL330/VL329; strain M1321, VL308/VL311, VL314/VL318, VL320/VL321, VL292/VL293, VL347/VL329; strain M135, VL308/VL312, VL315/VL318, VL320/VL348, VL292/VL348, VL347/VL329; strain BSG40, VL308/VL313, VL316/VL318, VL320/348, VL292/VL348, VL347/VL32, VL329/VL33.
Oligonucleotides
VL262, AGCAACATCTTTCTAACATTGGCTC; VL263, CAGTAAGTTTAAAGCCCATGCGTAC; VL264, AGCATTAAATCTAAACAAACTAAGAGC; VL265, GCTCCAACAACTTTCGAGCATTTCCC; VL268, GGGAAATGCTCGAAAGTTGTTGGAGC; VL269, TGCTCTCGCAACTGCTGGAGCATTTAC; VL292, CGATGCCAGGTTTATCGGCTCAGAAT; VL293, GAAATTTAAAACTGCCTTTCTTTGA; VL308, ACGAAACAAGATCTACAGAAAATCA; VL311, ATTAAAGAGGTTTTCGATCACCATT; VL312, GTCTGGATGGAATACACTTAGATAA; VL313, TTCACAAAGTTTTCACTGCCATATT; VL314, CGAACACGTCTGAATGGTCTGCAAA; VL315, TAGATAATGCCTTCAGATATGCCGA; VL316, TAGAAATAAGGGGATAGTTGACCAA; VL317, GCTTTTTCATATCCATTCTCAGTTA; VL318, CGTTTAGACGGGTTTGCTACGATCA; VL319, ATGATACAACACAAGAAGACTATCA; VL320, CGCCTTTCGTTGAATATTAAAGTCA; VL321, AGGACAAGCATCCCTCTTGGGTCGT; VL329, CATTGAATACATCCTTTCTCGGTAA; VL347, AATAAAAAAATTCAGTATACAGCAGC; VL348, CTAGCCTTCTTTTGAAGAAACCTATTG.
Sequencing and sequence analysis
Sequencing was performed on an ABI Prism 377 DNA sequencer (Applied Biosystems). Unless otherwise stated, the nucleotide sequences of PCR products were determined on both strands by direct sequencing. Prior to sequencing, RT–PCR products were cloned in the pBAD-TOPO vector and propagated in TOP 10 Escherichia coli using a pBAD TOPO TA Cloning Kit (Invitrogen). Sequence analyses were performed with the University of Wisconsin Genetics Computer Group software (10).
RNA isolation and RT–PCR
Total RNA was isolated with a RNeasy Mini Kit (Qiagen) from cells grown to an OD600 of 0.4. Samples containing 10 µg total RNA were incubated with 10 U RNase-free DNase (Promega) for 45 min at 37°C in a 100 µl volume of reaction buffer (Promega). RT–PCR reactions were carried out with Ready-To-Go RT–PCR Beads (Amersham Pharmacia Biotech) using 3 µg repurified total RNA and 25 pmol each oligonucleotide. The cDNA synthesis was carried out by 20 min incubation at 42°C and stopped by 5 min heating at 95°C, a step ensuring template denaturation. The cDNA amplification procedure consisted of 31 cycles, each including 45 s melting at 95°C, followed by 1 min annealing at 50°C and 1 min extension at 72°C. The last cycle extension was prolonged for an additional 10 min. Oligonucleotide pairs used in RT–PCR reactions were: strain M1918, VL389/VL391 and VL394/VL395; strain M1321, VL389/VL391 and VL392/VL393; strain M135, VL392/VL393; strain BSG40, VL390/VL391, VL392/VL393 and VL394/VL395.
RESULTS
Identification of IVSs in RR genes of SPβ-related prophages
Sixteen Bacillus spp. strains (see Material and Methods) were screened for the presence of IVSs by low stringency PCR using oligonucleotides corresponding to sequences neighboring the insertion sites of the two introns and the intein coding sequence present in the SPβ RR locus (9). Seven strains were characterized by strong single bands for all of the three examined regions, while the remaining nine strains yielded either no products or weak multiple bands, suggesting that they were devoid of prophage-encoded RR genes or of the prophage itself (not presented).
Single-strand sequencing of relevant PCR products revealed four IVS configurations different from that of the reference 168 strain (Fig.1A). In strains with the same IVS pattern, sequences of given IVSs and their surrounding exons were 98–100% identical. Therefore, PCR products were sequenced on both strands for only one representative of each of the four newly identified IVS configurations. Control PCR experiments ruled out the presence of any IVS at analogous sites of host RR genes (not presented).
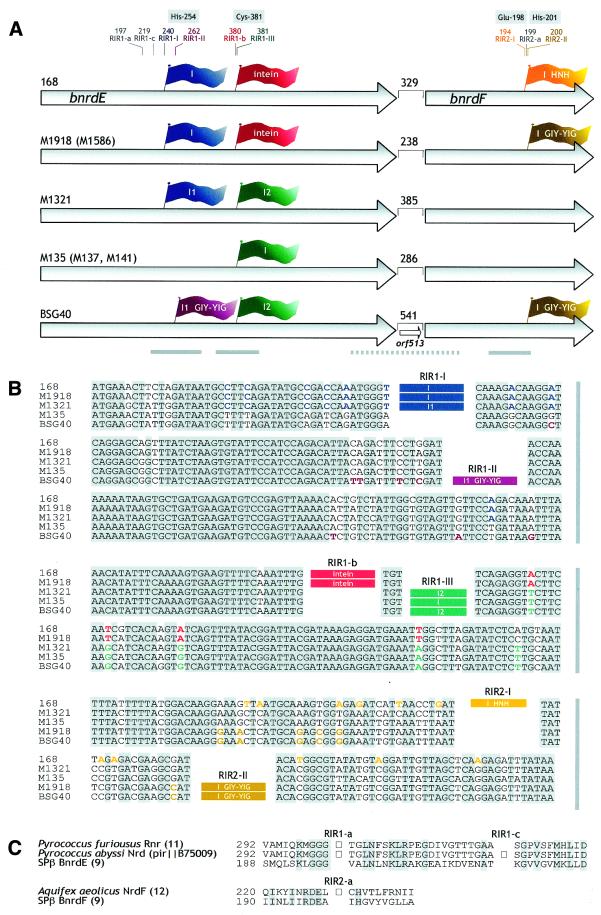
Figure 1.
(A) Schematic representation of IVS configurations in the bnrdE/bnrdF tandem of different Bacillus prophages. Arrows correspond to the bnrdE and bnrdF genes. The lengths of intergenic regions are given in nucleotides. A flag representing an IVS and its corresponding insertion site are in the same color. Occupied as well as unoccupied IVS insertion sites are represented by thin lines and annotated as follows: intron insertion sites, RIR1-I, RIR1-II, RIR1-III, RIR2-I and RIR2-II; insertion sites for intein DNA, RIR1-a, RIR1-b, RIR1-c and RIR2-a (13). Codons containing or preceding a given IVS insertion site refer to the sequence of hypothetical IVS– variants of the SPβ prophage bnrdE and bnrdF genes (9). Codons for some of the conserved and functionally important residues are boxed. I, intron. If two introns occupy the bnrdE gene, they are designated I1 and I2. GIY-YIG and HNH specify the intron homing endonuclease ORF present within the intron. Thick solid lines denote regions sequenced in all listed strains. Thick dotted lines represent segments sequenced in strains M1918, M1321, M135 and BSG40. Names of partly analyzed strains (see Results) are in parentheses. (B) Alignment of sequences surrounding IVS insertion sites in the bnrdE and bnrdF genes of Bacillus prophages. Conserved residues are shaded grey. IVSs more than 98% identical are specified by rectangles of the same color. Residues that are common to strains containing nearly identical IVSs, but absent from strains with an unrelated IVS, are colored accordingly. The intein DNA was deduced from the predicted intein sequence. (C) Alignment of sequences around archaeal intein insertion sites RIR1-a and RIR1-c, as well as the bacterial intein insertion site RIR2-a (13), with their unoccupied equivalents in the SPβ BnrdE and BnrdF proteins. Squares correspond to inteins.
All introns or intein coding sequences inserted at analogous positions in strains with different IVS configurations have the same length and exhibit >98% identity. Conversely, introns occupying different sites are more distantly related. The highest similarity (77% identity over 164 residues) was found between introns inserted at positions RIR2-I and RIR2-II (Fig. 1A and B) located in bnrdF. Introns occupying positions RIR1-I, RIR1-II and RIR1-III (Fig. 1A and B) exhibit 60–70% identity in 100–200 nt overlaps with introns identified in a variety of phages unrelated to SPβ.
The equivalents of the archaeal intein insertion sites RIR1-a and RIR1-c (13) could be identified in BnrdE, the large RR subunit of Bacillus prophages, thanks to a limited homology (Fig. 1A and C). Similarly, bacterial intein insertion site RIR2-a (13) was identified in BnrdF homologs (Fig. 1A and B). However, all these sites are not occupied by inteins in the strains here investigated.
Intron splicing
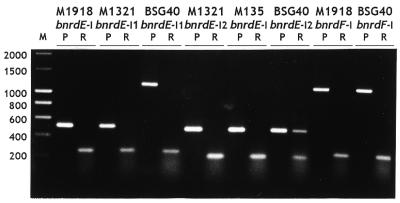
In vivo intron splicing was demonstrated by reverse transcription, using total RNA extracted from exponentially grown bacteria, followed by PCR amplification of synthesized cDNA. The size of the product obtained in each reaction confirmed the predicted intron excision from the relevant primary transcript (Fig. 2). Therefore, synthesis and splicing of the RR transcript occurs not only during SPβ infection of a non-lysogenic strain (9) but also in uninduced lysogenic strains, including 168 (not presented), possibly due to sporadic prophage induction (14).
Figure 2.
The in vivo splicing of intron RNA. Lanes P and R correspond to PCR products obtained on bacterial DNA and to RT–PCR products obtained with the total bacterial RNA extract, respectively. For each intron, the same oligonucleotide pair was used in PCR and RT–PCR. For any given strain, an aliquot of the same RNA preparation was used in different RT–PCR. Lane M, DNA sizing ladder (bp).
Cloning and sequencing of RT–PCR products, obtained from two independent experiments, determined each exon–intron junction. In all cases, 3′-terminal residues of introns and upstream exons, i.e. G and T, respectively, as well as the intron secondary structure predictions (Fig. 3 and Supplementary Material) were those characteristic of group IA introns (5). It appeared that unlike the other introns investigated, bnrdE-I2 present in strain BSG40 was not excised from roughly half of the mRNA precursors, since the relevant RT–PCR reaction revealed two products in comparable amounts (Fig. 2). RT–PCR amplification of strain M1321 mRNA with oligonucleotides specific for sequences neighboring the bnrdE-I1 insertion site also yielded an additional, but very faint, band of a size indistinguishable from that obtained by amplification of the relevant DNA segment. In each case, extracting RNA from independent cultures did not significantly alter the ratio of unspliced to spliced messenger (not presented). Sequences of the higher molecular weight RT–PCR products obtained from BSG40 and M1321 RNA corresponded to unspliced mRNA. The possibility of residual DNA contamination of RNA samples was eliminated by: (i) the absence of amplification products in control RT–PCR reactions in which RNA was added during the first denaturation step of PCR amplification (not presented); and (ii) the apparent absence of unspliced products in the internal control RT–PCR reactions spanning introns BSG40 bnrdE-I1, BSG40 bnrdF-I and M1321 bnrdE-I2 (Fig. 2).
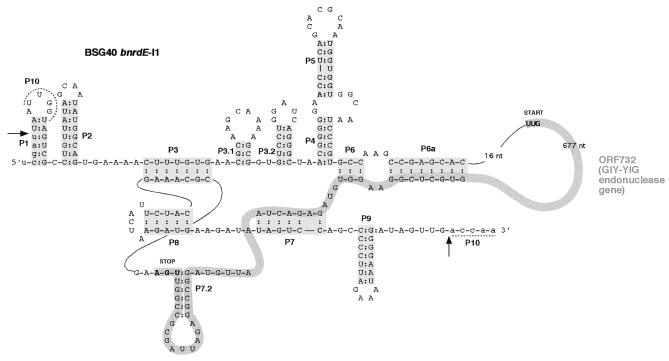
Figure 3.
Proposed secondary structure of the BSG40 bnrdE-I1 intron RNA. Arrows indicate splice boundaries between exons (lower case) and the intron (upper case). The base paired regions P1–P10 are represented by shaded boxes or dotted lines and numbered according to Burke et al. (15). The thick grey line represents the intron ORF. Its start and stop codons are specified.
Properties of IVSs
M1918 bnrdE-I, M1321 bnrdE-I1 and their homolog, the formerly identified 168 bnrdE-I (9), are 252 nt long and unoccupied by an ORF.
The bnrdE-I1 intron in strain BSG40 (Fig. 3) does not unambiguously fall into any of the group I intron subgroups (5). Indeed, BSG40 bnrdE-I1 is the only so far described phage intron with a single stem–loop between P7 and P3, a property characteristic of subgroup IA1 introns. Several peculiarities common to BSG40 bnrdE-I1 and to introns present in phages related to S3b (8) and SPO1 (7) distinguish them from the other phage introns. (i) The P9 region consists of a single stem–loop structure, a feature shared with a chloroplast intron psbA-2, considered as a subgroup IA3 intron (16). Incidentally, the absence of L9.2 is accompanied by absence of the AGY sequence in L7.2, a motif involved in the P12 interaction between and L9.2 and L7.2 (17). (ii) The additional stem–loops formed in the P3–P4 spacer probably compensate for lack of the 9.1 and 9.2 structures.
ORF732, encoded by the BSG40 bnrdE-I1 intron, begins in L6a (Fig. 3). Its stop codon overlaps with the P7.2 stem, while its putative start codon is preceded at an appropriate distance by a 12 nt stretch complementary to the 3′-end of 16S rRNA. Similarly, the Shine–Dalgarno sequence of the endonuclease ORF yosQ encoded by the SPβ bnrdF intron (9), comprises 11 residues. Such extended pairing with 16S rRNA, rare in both SPβ and B.subtilis, possibly affects translational initiation efficiency. The 243 residue polypeptide encoded by ORF732 contains a variant of the GIY-YIG motif (AVY-X10-YIG18). It exhibits nearly the same identity (33%) to the GIY-YIG homing endounclease I-TevI, encoded by the coliphage T4 thy intron (18), as to deduced products of Bacillus spp. BSG40 and M1918 bnrdF intron ORFs (see above). The spectrum of GIY-YIG homing endonuclease genes, comprising fungal mitochondrial introns as well as T4 phage intron and non-intron DNA (19), is now extended to introns of prophages of Gram-positive bacteria.
The putative 385 amino acid intein from strain M1918 and its highly similar (98.4% identity) counterpart from strain 168 have a rare glycyl residue at the penultimate position (13). They share 24–27% identity with inteins inserted at the analogous position, designated site RIR1-b, in RR of different prokaryotes and of an insect virus (13,20).
Each of the M135 bnrdE-I, M1321 bnrdE-I2 and BSG40 bnrdE-I2 introns spans 280 nt and does not contain an ORF.
M1918 bnrdF-I and BSG40 bnrdF-I of 988 nt have identical intron cores occupied by nearly identical (99.2%) 639 nt ORFs entirely located within L6a. The 212 residue product of these ORFs contains a variant of the GIY-YIG motif (YVY-X10-YIG18) that characterizes a family of homing endonucleases (reviewed in 19). Their closest homolog revealed by sequence similarity (33% identity) is the putative polypeptide encoded by the BSG40 bnrdE-I1 intron ORF732 (see above).
bnrdE–bnrdF intergenic region
In contrast to the B.subtilis 168 situation, where RR genes nrdE and nrdF are separated by 17 nt (21), the bnrdE–bnrdF intergenic spacers in SPβ and related prophages are unusually long, ranging from 238 to 541 nt (Fig. 1A). They share no homology between themselves. A similarity search for sequences deduced from the six frames of the bnrdE–bnrdF intergenic region of strains 168 (9), M135, M1918 and M1918 did not reveal any close homologs to known protein sequences.
The 513 nt ORF in the BSG40 bnrdE–bnrdF intergenic region is preceded by a questionable RBS and thus may represent a remnant of a functional gene. The C-terminal two-thirds of the deduced ORF513 product (residues 52–168) exhibit 40% identity with a putative HNH endonuclease encoded by the intron present in a putative lysin gene of S.thermophilus S3b-related bacteriophages (8). Interestingly, three of the four ORF513 closest relatives, ORF3.8 from phage T7 (22), ORF7903 of Yersinia enterocolitica phage φYe03-12 (accession no. AJ251805) and an unannotated ORF of the Rhizobium meliloti temperate phage 16-3 (23), are not harbored by an intron, but are adjacent to genes involved in DNA metabolism.
DISCUSSION
Sequencing of the RR locus of SPβ-like prophages in different isolates of bacilli revealed in each of them one to three IVSs. The variety of the observed IVS configurations implies that at least some of their elements had been independently acquired by different strains. This acquisition seems to be recent in view of the high degree of identity (98–100%) between splicing elements inserted at identical positions. Pairwise identities between corresponding exons, calculated as the mean of five sequenced bnrdEF exon segments representing together 1.3 kb, are lower, ranging from 89 to 93%. However, the short DNA segments flanking nearly identical IVSs are more similar to each other than to analogous sequences corresponding to unoccupied sites (Fig. 1B). This observation may be accounted for by assuming that independent homing events for nearly identical IVSs are accompanied by co-conversion of exon sequences (24).
Alignment of relevant nucleotide sequences reveals that the six identified IVS insertion sites in SPβ-like prophage RR genes are clustered in three domains comprising 3, 18 and 66 nt, respectively (Fig. 1B).
Curiously, each of the so far identified bnrdE homologs harbors an IVS immediately adjacent to the equivalent of the SPβ codon corresponding to Cys381 (Fig. 1A and B). This residue is putatively involved in the generation of a transient protein radical required for substrate activation (25,26). The narrow spacing between the intron insertion site RIR1-III and the back translated intein insertion site RIR1-b may be accounted for by homing events involving endonucleases recognizing similar target sequences, although the relevant introns are devoid of a homing endonuclease gene (see below). Therefore, not only can intein DNA and an intron coexist in the large RR subunit gene (9), they can compete for the same DNA region. The possibility that introns and inteins inserted in the same region of diverse phages have a similar impact on RR activity is not evident, as the splicing mechanisms of these elements are substantially different. The intron insertion sites RIR2-I and RIR2-II, corresponding to codons 194 and 200 of the SPβ bnrdF gene, respectively, as well as the unoccupied intein insertion site RIR2-a, are very close to the conserved iron ligands Glu198 and His201 (27). IVS insertion sites clustered in the 5′-part of bnrdE, i.e. RIR1-I, RIR1-II and analogs of RIR1-a and RIR1-c, may also correspond to a functionally important region of the gene product, as shown for insertion sites of many inteins, group I introns and archaeal introns (28–30). Indeed, His254 of BnrdE, located between RIR1-I and RIR1-II, corresponds to the B.subtilis NrdE His255 residue, whose substitution by Tyr is accompanied by a temperature-sensitive phenotype (21).
Examples of phage introns devoid of an endonuclease ORF (9,31) raised the question of their mode of spreading. The above described non-uniform distribution of such introns argues in favor of either their relatively recent acquisition or, in turn, the existence of an efficient intron loss mechanism. Transposition by reverse splicing has been considered a possible mechanism of ORF-free intron spread (31). Another possibility, based on analogy with an artificial mobile system (32), is that the endonuclease function required for homing initiation is provided in trans. Indeed, there are examples of endonucleases similar to homing enzymes that are not intron encoded or associated with an intein (19; see below).
Landthaler and Shub (31) found that a single primary transcript of Streptococcus bacteriophage Twort could give rise to different mRNA molecules via optional skipping of an exon located between two closely inserted and similar group I introns. The opposite situation, i.e. an optional intron, was found in the bnrdE gene of strains BSG40 and M1321. Interestingly, they remain partly unspliced under growth conditions that stimulate apparently complete splicing of identical or highly similar introns in different strain backgrounds. Therefore, splicing efficiency seems to be affected by interactions between the relevant unprocessed transcript and other molecules, like, for instance, RNA or proteins that may differ in the strains studied. Whether the formation of truncated and, likely, non-functional proteins by alternative mRNA processing based on optional exclusion of introns or exons (31) is exploited to regulate the phage life cycle remains an open question. It would be most interesting to find conditions that favor alternative RNA processing.
Comparison of the SPβ prophage intergenic bnrdE–bnrdF region to that of its 168 host revealed an additional domain in the prophage spacer, starting with AGGTACGT, a good 5′ splice site consensus for eukaryotic spliceosomal introns (33). A similar motif, AGGTATCT, is present at the analogous position in strain BSG40. This possibly fortuitous resemblance to eukaryotic intron 5′ splice sites may, on the other hand, hint at a remnant of a spliced element or a regulatory element that shares a common origin with spliceosomal introns (33). In addition, the bnrdE–bnrdF intergenic region of strain BSG40 contains ORF513, which apparently encodes an endonuclease. However, the postulated mobile nature of homing endonuclease genes (1) might have allowed the insertion of ORF513 or of its predecessor without a surrounding splicing element. Therefore, regardless of its origin, the location of ORF513 provides an additional argument for a particular susceptibility of the RR locus to different types of invasive DNA. Whether this susceptibility relies on a specific sequence context or on a functional impact cannot presently be resolved.
The variety of unrelated DNA sequences colonizing the RR locus in different prophages rather suggests a parasitic nature for these elements. Should spreading be the ultimate ‘goal’ of group I introns and inteins, then the conserved regions of widespread RR genes represent one of the most convenient targets. A possible regulatory role of IVSs would be expected to correlate with a wider and more uniform distribution in phage genomes. However, while in so far studied SPβ-related phages there are no intronless RR loci, such variants predominate in T4 coliphages (4). In addition, most known phage genomes are apparently devoid of introns.
Abundance of IVSs in the here described RR genes may, in part, be due to their belonging to a temperate phage. The possibility of transduction, transformation or conjugation between related isolates of bacilli may also contribute to the acquisition of new IVSs by a homing-independent pathway. Indeed, transformation of B.subtilis SPβ lysogens with DNA from close SPβ relatives may introduce new genetic markers in the SPβ prophage (34).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENT
I thank Prof. Dimitri Karamata for critical reading of the manuscript prior to submission.
DDBJ/EMBL/GenBank accession nos AJ309304–AJ309313
References
- 1.Lambowitz A.M. and Belfort,M. (1993) Introns as mobile genetic elements. Annu. Rev. Biochem., 62, 587–622. [DOI] [PubMed] [Google Scholar]
- 2.Garriga G. and Lambowitz,A.M. (1984) RNA splicing in neurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell, 39, 631–641. [DOI] [PubMed] [Google Scholar]
- 3.Cech T.R. (1990) Self-splicing of group I introns. Annu. Rev. Biochem., 59, 543–568. [DOI] [PubMed] [Google Scholar]
- 4.Edgell D.R., Belfort,M. and Shub,D.A. (2000) Barriers to intron promiscuity in bacteria. J. Bacteriol ., 182, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michel F. and Westhof,E. (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 6.Derbyshire V. and Belfort,M. (1998) Lightning strikes twice: intron-intein coincidence. Proc. Natl Acad. Sci. USA, 95, 1356–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodrich-Blair H. and Shub,D.A. (1994) The DNA polymerase genes of several HMU-bacteriophages have similar group I introns with highly divergent open reading frames. Nucleic Acids Res., 22, 3715–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foley S., Bruttin,A. and Brussow,H. (2000) Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J. Virol., 74, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazarevic V., Soldo,B., Düsterhöft,A., Hilbert,H., Mauël,C. and Karamata,D. (1998) Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPβ. Proc. Natl Acad. Sci. USA, 95, 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux J., Haeberli,P. and Smithies,O. (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res., 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riera J., Robb,F.T., Weiss,R. and Fontecave,M. (1997) Ribonucleotide reductase in the archaeon Pyrococcus furiosus: a critical enzyme in the evolution of DNA genomes? Proc. Natl Acad. Sci. USA, 94, 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deckert G., Warren,P.V., Gaasterland,T., Young,W.G., Lenox,A.L., Graham,D.E., Overbeek,R., Snead,M.A., Keller,M., Aujay,M., Huber,R., Feldman,R.A., Short,J.M., Olsen,G.J. and Swanson,R.V. (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature, 392, 353–358. [DOI] [PubMed] [Google Scholar]
- 13.Perler F.B. (2000) InBase, the Intein Database. Nucleic Acids Res., 28, 344–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warner F.D., Kitos,G.A., Romano,M.P. and Hemphill,H.E. (1977) Characterization of SPβ: a temperate bacteriophage from Bacillus subtilis 168M. Can. J. Microbiol., 23, 45–51. [Google Scholar]
- 15.Burke J.M., Belfort,M., Cech,T.R., Davies,R.W., Schweyen,R.J., Shub,D.A., Szostak,J.W. and Tabak,H.F. (1987) Structural conventions for group I introns Nucleic Acids Res., 15, 7217–7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holloway S.P., Deshpande,N.N. and Herrin,D.L. (1999) The catalytic group-I introns of the psbA gene of Chlamydomonas reinhardtii: core structures, ORFs and evolutionary implications. Curr. Genet., 36, 69–78. [DOI] [PubMed] [Google Scholar]
- 17.Michel F., Jaeger,L., Westhof,E., Kuras,R., Tihy,F., Xu,M.Q. and Shub,D.A. (1992) Activation of the catalytic core of a group I intron by a remote 3′ splice junction. Genes Dev., 6, 1373–1385. [DOI] [PubMed] [Google Scholar]
- 18.Chu F.K., Maley,G.F., Maley,F. and Belfort,M. (1984) Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc. Natl Acad. Sci. USA, 81, 3049–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belfort M. and Roberts,R.J. (1997) Homing endonucleases: keeping the house in order. Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietrokovski S. (1998) Identification of a virus intein and a possible variation in the protein-splicing reaction. Curr. Biol., 8, 634–635. [DOI] [PubMed] [Google Scholar]
- 21.Scotti C., Valbuzzi,A., Perego,M., Galizzi,A. and Albertini,A.M. (1996) The Bacillus subtilis genes for ribonucleotide reductase are similar to the genes for the second class I NrdE/NrdF enzymes of Enterobacteriaceae. Microbiology, 142, 2995–3004. [DOI] [PubMed] [Google Scholar]
- 22.Dunn J.J. and Studier,F.W. (1983) Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol., 166, 477–535. [DOI] [PubMed] [Google Scholar]
- 23.Semsey S., Papp,I., Buzas,Z., Patthy,A., Orosz,L. and Papp,P.P. (1999) Identification of site-specific recombination genes int and xis of the Rhizobium temperate phage 16-3. J. Bacteriol ., 181, 4185–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell-Pedersen D., Quirk,S.M., Aubrey,M. and Belfort,M. (1989) A site-specific endonuclease and co-conversion of flanking exons associated with the mobile td intron of phage T4. Gene, 82, 119–126. [DOI] [PubMed] [Google Scholar]
- 25.Mao S.S., Yu,G.X., Chalfoun,D. and Stubbe,J. (1992) Characterization of C439SR1, a mutant of Escherichia coli ribonucleotide diphosphate reductase: evidence that C439 is a residue essential for nucleotide reduction and C439SR1 is a protein possessing novel thioredoxin-like activity. Biochemistry, 31, 9752–9759. [DOI] [PubMed] [Google Scholar]
- 26.Licht S., Gerfen,G.J. and Stubbe,J. (1996) Thiyl radicals in ribonucleotide reductases. Science, 271, 477–481. [DOI] [PubMed] [Google Scholar]
- 27.Nordlund P., Sjöberg,B.M. and Eklund,H. (1990) Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature, 345, 593–598. [DOI] [PubMed] [Google Scholar]
- 28.Garrett R.A., Dalgaard,J., Larsen,N., Kjems,J. and Mankin,A.S. (1991) Archaeal rRNA operons. Trends Biochem. Sci., 16, 22–26. [DOI] [PubMed] [Google Scholar]
- 29.Dalgaard J.Z. (1994) Mobile introns and inteins: friend or foe? Trends Genet., 10, 306–307. [DOI] [PubMed] [Google Scholar]
- 30.Dalgaard J.Z., Klar,A.J., Moser,M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res., 25, 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landthaler M. and Shub,D.A. (1999) Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns and exon skipping by group I ribozymes. Proc. Natl Acad. Sci. USA, 96, 7005–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eddy S.R. and Gold,L. (1992) Artificial mobile DNA element constructed from the EcoRI endonuclease gene. Proc. Natl Acad. Sci. USA, 89, 1544–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazarevic V. (1998) Is there a relic of a spliceosomal intron in Bacillus subtilis temperate phage SPβ? Mol. Microbiol., 29, 1523–1526. [PubMed] [Google Scholar]
- 34.Stroynowski I.T. (1981) Integration of the bacteriophage φ3T-coded thymidylate synthetase gene into the Bacillus subtilis chromosome. J. Bacteriol ., 148, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.