Abstract
In this study, the re-sequencing data from 3,000 rice genomes project (3 K RGP) was used to analyze the allelic variation at the rice blast resistance (R) Pid3 locus. A total of 40 haplotypes were identified based on 71 nucleotide polymorphic sites among 2621 Pid3 homozygous alleles in the 3k genomes. Pid3 alleles in most japonica rice accessions were pseudogenes due to premature stop mutations, while those in most indica rice accessions were identical to the functional haplotype Hap_6, which had a similar resistance spectrum as the previously reported Pid3 gene. By sequencing and CAPS marker analyzing the Pid3 alleles in widespread cultivars in China, we verified that Hap_6 had been widely deployed in indica rice breeding of China. Thus, we suggest that the priority for utilization of the Pid3 locus in rice breeding should be on introducing the functional Pid3 alleles into japonica rice cultivars and the functional alleles of non-Hap_6 haplotypes into indica rice cultivars for increasing genetic diversity.
Introduction
Rice (Oryza sativa L.) is a staple food for nearly half of the world’s population. It also represents a model for functional genome research among the crop plants. Rice was the first crop plant to be fully sequenced1 and so far, has at least four different reference genomes in two subspecies (Oryza sativa subsp. indica and Oryza. sativa subsp. japonica)2–4. Moreover, a lot of rice cultivars within different subpopulations and wild relatives in different Oryza species have their own genome assemblies5–7. Recently, with the cost reduction of sequencing, more studies on rice have been undertaken for exploring allelic variants through next generation sequencing (NGS)8–15. Particularly, the 3,000 rice genomes project (3 K RGP) has completed re-sequencing a core collection of 3,000 rice cultivars from 89 countries with an average sequencing depth of 14×, from which a total of 18.9 million single nucleotide polymorphisms (SNPs) were discovered when compared to the reference genome of Nipponbare, providing a complete picture of the total genetic diversity in the O. sativa gene pool16–18. With these whole genome sequences and large amount of re-sequencing data in hand, works for comparative rice genome researches5, 10, 19 and genome-wide association studies (GWAS) on important traits, such as grain size, grain weight, flowering time, metabolites, disease resistance and abiotic stress tolerance have been widely conducted8, 12, 15, 20–25. However intensive studies on allelic functional and nonfunctional variations of a certain type of genes or a specific gene locus were rarely reported.
Rice blast, caused by the filamentous ascomycete Magnaporthe oryzae (M. oryzae), is the most devastating rice fungus disease worldwide. It has been proven that deployment of cultivars with resistance (R) genes is the most effective and eco-friendly approach for the control of rice blast26. To date, at least 69 rice blast R loci have been identified, of which 16 loci harboring more than 30 R genes/alleles have been cloned and functionally analyzed in detail27–30. It is important to note that almost all cloned rice blast R genes encode nucleotide-binding site leucine-rich repeats (NBS-LRR) proteins except for Pid2 31 and pi21 32; the former encodes a receptor-like kinase and the later a proline-rich protein. Likewise, a number of NBS-LRR genes cloned from maize, sorghum, and brachypodium were also proved being blast resistant in rice30, 33, 34. In recent years, a trend has become clear: a significant number of newly cloned rice blast R genes have finally been verified as being allelic to one of the previously cloned rice blast R genes, and fewer represent a new rice blast R locus29, 35. Considering that there are more than 400 NBS–LRR gene sequences identified in a rice genome, and that allelic rice blast R genes may confer distinct resistance spectra to M. oryzae isolates27, 29, we believe that allele mining of cloned rice blast R genes in rice germplasms would reveal more favorable R alleles for rice blast resistance breeding13.
However, the majority of the cloned rice blast R genes are clustered28, 36, 37 as most of NBS-LRR genes present in diverse multigene families30. Moreover, the clustered NBS-LRR genes usually fall into heterogeneous groups based on their structural similarity. For example, in the 76-kb chromosomal region containing the rice blast R gene Pi9 locus, six tandemly arranged NBS-LRR type putative genes were identified. The identities among the six paralogs ranged from 63.8 to 98.6% and only the Nbs2-Pi9 was proved to be the Pi9 gene37. The other example is Pi5, whose blast resistance function is actually conferred by two NBS-LRR genes, Pi5-1 and Pi5-2 38. The ~90-kb sequences of the Pi5 locus are significantly diverged between resistant and susceptible rice cultivars; the susceptible cultivar Nipponbare completely lacks the corresponding allele of Pi5-2 38. Similar statuses were also found at other rice blast R loci, like Pik 39, Pia 40, Pi37 41, Pb1 42, and Pit 43. These duplicated sequences have diverged through accumulated mutation, which increase the complexity of NBS-LRR gene sequences. Therefore, it is difficult to identify alleles of cloned NBS-LRR type rice blast R genes through allele mining approach based on either traditional PCR or NGS data analyzing. However, at a few rice blast R loci, the structure of NBS-LRR genes are rather simple, with only single NBS-LRR gene. Allele mining at these loci is feasible.
The rice blast R gene Pid3 was initially identified in the indica variety Digu by performing a genome-wide comparison of paired NBS-LRR genes and their pseudogene alleles between 93-11 (indica) and Nipponbare (japonica) on the premise of the verification of obvious different resistance of indica and japonica varieties to M. oryzae strains collected from south and north China44. Pid3 is a typical CC-NBS-LRR protein of 924 amino acids with no intron. Alleles in most japonica varieties were identified as pseudogenes due to the presence of a nonsense mutation at the nucleotide position 2209 starting from the translation initiation site; however, this pseudogene mutation did not occur in tested indica varieties, including African cultivated rice varieties and AA genome-containing wild rice species44. Then, a number of Pid3 alleles or orthologs were cloned by map-based cloning45 and sequencing-based allele mining from indica and wild rice accessions, of which five had been verified to confer differential resistance spectra to a set of M. oryzae isolates29, 46. In this study, mainly based on the 3 K RGP sequencing data, a total of 40 haplotypes were identified according to 71 nucleotide polymorphic sites in 2621 Pid3 homozygous alleles. Finally, by PCR-based allele mining and gene transformation, we disclosed a functional Pid3 allele, which has been widely deployed in indica rice cultivars and especially in hybrid rice in China. With the above overview, we may propose different strategies in application of the functional Pid3 alleles to indica and japonica rice breeding.
Results
The nonsense mutation of Pid3 alleles at the position 2209
We previously revealed that Pid3 alleles in 29 out of 32 japonica varieties were identified as pseudogenes due to the presence of a nonsense mutation (CAG to TAG) at the nucleotide position 2209, whereas none of the varieties in 32 indica collection contained this mutation44. To figure out the distribution of nonsense mutation of Pid3 alleles in the 3 K RGP sequencing data18, we checked the corresponding position 13055819 on chromosome 6, where “G” represents “C” and “A” represents “T”, since Pid3 coding sequence is on the “-” chain of the sequencing data. A total of 2953 Pid3 alleles at this position were identified, of which 22 out of 1732 indica, 715 out of 859 japonica and 40 out of 362 other rice accessions were “A”, indicating Pid3 alleles in most japonica and scarcely in indica rice accessions were nonfunctional due to the nonsense mutation at the position 2209. The detailed information was shown in Fig. 1. To verify the general survey based on the sequencing data, we used the CAPS marker44 to test nearly 300 varieties, including 149 widely cultivated japonica varieties in north China and 140 indica varieties, most of which were backbone parents of hybrid rice cultivars widely used in China. We found the nonsense mutation in 91.9% of the 149 japonica cultivars but neither of the 140 indica cultivars. The details for these cultivars were listed in Supplementary Table S1. Both of the results further confirmed our previous report that the pseudogenization of Pid3 has prevailed in japonica.
Figure 1.
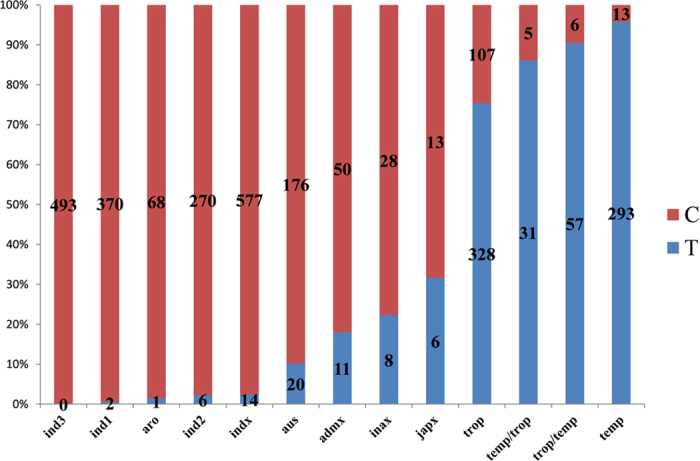
The nonsense mutation of Pid3 alleles at the nucleotide position 2209. Blue “T” represents nonsense mutation. Ind1, ind2 and ind3 are three subgroups of indica rice, indx corresponds to other indica varieties, temp is temperate japonica, trop is tropical japonica, temp/trop and trop/temp are admixed temperate and tropical japonica varieties, japx is other japonica varieties, aus is aus, inax is admixed aus and indica, aro is aromatic, and admix is all other unassigned varieties18.
Nucleotide polymorphisms of Pid3 alleles
The coding sequence of Pid3 is located in a region (13055253-13058027) on chromosome 6 according to the Nipponbare genome in the 3 K RGP sequencing data. In this region, only 16 Pid3 alleles showed obvious InDel polymorphisms at one singleton nucleotide site, which is an 8-bp (ATATATTC) insertion at the position 13055566, corresponding to the nucleotide position 2461 starting from the translation initiation site of Pid3 gene. Of the 16 Pid3 allelic loci, three showed heterozygous insertions, and all alleles were from indica subpopulation excepting one allele from japonica (Supplementary Table S2). After eliminating alleles with heterozygous sequences and ambiguous single site deletion probably caused by insufficient sequencing coverage, we have obtained a total of 2621 Pid3 alleles for subsequent analyses.
In total, 71 nucleotide polymorphic sites were detected in the 2775-bp coding region of these 2621 Pid3 alleles (Fig. 2). It was consistent to our previous finding29 that the Pid3 gene processed lower nucleotide polymorphism (π = 0.00255 in this study), belonging to the conserved type of plant NBS-LRR class R genes47. Among the surveyed six subpopulations, indica has the lowest nucleotide polymorphism (π = 0.00102), while aromatic basmati/sadri (aro) group has the highest (π = 0.00265). The values of Tajima’s D were mostly negative but did not significantly deviate from the neutral model (Table 1). To figure out detailed information, three domains including CC, NBS and LRR were defined as those in our former study29, 44. Meanwhile, we assigned Pid3-W5, a functional Pid3 ortholog cloned from Oryza. rufipogon 29, 46 as the reference, and calculated the Ka/Ks ratio between each Pid3 haplotype and Pid3-W5 (Supplementary Table S3). On average, the nucleotide polymorphisms in NBS domain were comparable with those in LRR domain, but much higher than those in CC domain. However, most of the ratios of πnon/πsyn and Ka/Ks in NBS domain were far less than 1, while they were much greater than 1 in LRR domain, indicating that the nucleotide polymorphisms in NBS domain were affected by purifying selection, while polymorphisms in LRR domain mainly affected by positive selection (Table 1 and Supplementary Table S3).
Figure 2.
Summary of DNA variations in the 2775-bp coding region of 2621 Pid3 alleles. Numbers in brackets represent rice accessions belonging to specific haplotypes. Site 1 corresponds to the first position of the start codon. Dots represent nucleotide variants identical to the Pid3 sequence in Digu. The boxed nucleotide is the premature mutation.
Table 1.
Polymorphism, neutral test and haplotype analysis of Pid3 alleles.
| Population | Number | Region | S | π | Tajima’s D | πnon | πsyn | πnon/πsyn | Nhap | Hd |
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 2621 | Coding | 71 | 0.00255 | −0.47095 | 0.00222 | 0.00372 | 0.5968 | 40 | 0.680 |
| CC | 6 | 0.00143 | −0.61872 | 0.00117 | 0.00224 | 0.5223 | 8 | 0.275 | ||
| NBS | 25 | 0.00254 | −0.16943 | 0.00133 | 0.00699 | 0.1903 | 22 | 0.615 | ||
| LRR | 28 | 0.00251 | −0.60729 | 0.00317 | 0.00051 | 6.2157 | 20 | 0.579 | ||
| Indica | 1544 | Coding | 44 | 0.00102 | −1.25381 | 0.00090 | 0.00143 | 0.6294 | 18 | 0.384 |
| CC | 4 | 0.00148 | −0.16974 | 0.00127 | 0.00213 | 0.5962 | 5 | 0.299 | ||
| NBS | 14 | 0.00078 | −1.09871 | 0.00034 | 0.00238 | 0.1429 | 11 | 0.247 | ||
| LRR | 18 | 0.00093 | −1.25942 | 0.00121 | 0.00005 | 24.2000 | 10 | 0.213 | ||
| Japonica | 784 | Coding | 54 | 0.00198 | −0.74996 | 0.00200 | 0.00191 | 1.0471 | 19 | 0.614 |
| CC | 6 | 0.00164 | −0.70577 | 0.00123 | 0.00290 | 0.4241 | 7 | 0.275 | ||
| NBS | 18 | 0.00163 | −0.67529 | 0.00128 | 0.00294 | 0.4354 | 12 | 0.549 | ||
| LRR | 21 | 0.00245 | −0.36720 | 0.00292 | 0.00096 | 3.0417 | 9 | 0.343 | ||
| Aus | 163 | Coding | 39 | 0.00236 | −0.14566 | 0.00198 | 0.00364 | 0.5440 | 10 | 0.706 |
| CC | 2 | 0.00047 | −0.84252 | 0.00042 | 0.00065 | 0.6462 | 3 | 0.140 | ||
| NBS | 12 | 0.00202 | 0.07995 | 0.00056 | 0.00738 | 0.0759 | 8 | 0.656 | ||
| LRR | 18 | 0.00300 | −0.02360 | 0.00365 | 0.00093 | 3.9247 | 7 | 0.642 | ||
| Inax | 29 | Coding | 23 | 0.00178 | −0.55900 | 0.00150 | 0.00270 | 0.5556 | 4 | 0.461 |
| CC | 1 | 0.00023 | −1.14923 | 0.00031 | 0 | — | 2 | 0.069 | ||
| NBS | 8 | 0.00214 | 0.41815 | 0.00118 | 0.00565 | 0.2088 | 4 | 0.461 | ||
| LRR | 10 | 0.00154 | −1.16007 | 0.00203 | 0 | — | 3 | 0.431 | ||
| Aro | 56 | Coding | 30 | 0.00265 | 0.41230 | 0.00233 | 0.00372 | 0.6263 | 5 | 0.561 |
| CC | 1 | 0.00012 | −1.09119 | 0 | 0.00048 | 0.0000 | 2 | 0.036 | ||
| NBS | 10 | 0.00127 | −1.04112 | 0.00017 | 0.00528 | 0.0322 | 5 | 0.561 | ||
| LRR | 14 | 0.00408 | 1.20787 | 0.00473 | 0.00202 | 2.3416 | 4 | 0.544 | ||
| Admx | 45 | Coding | 37 | 0.00240 | −0.80080 | 0.00186 | 0.00422 | 0.4408 | 9 | 0.549 |
| CC | 2 | 0.00071 | −0.97392 | 0.00038 | 0.00171 | 0.2222 | 3 | 0.129 | ||
| NBS | 14 | 0.00238 | −0.61379 | 0.00108 | 0.00710 | 0.1521 | 7 | 0.536 | ||
| LRR | 17 | 0.00267 | −1.02375 | 0.00301 | 0.00156 | 1.9295 | 8 | 0.541 |
Indica, including ind1, ind2, ind3 and indx varieties; Japonica, including temp, trop, temp/trop, trop/temp and japx varieties; Aus, aus varieties; Inax, admixed aus and indica varieties; Aro, aromatic varieties; Admx, all other unassigned varieties. S, number of segregating sites; π, nucleotide diversity; πnon, average nonsynonymous site diversity; πsyn, average synonymous site diversity; πnon/πsyn, ratio of nonsynonymous site diversity over synonymous site diversity; Nhap, number of haplotype; Hd, haplotype diversity; *Statistical significance P < 0.05.
Haplotype analysis of Pid3 alleles
Based on the 71 nucleotide polymorphic sites, a total of 40 haplotypes of Pid3 alleles were identified (Fig. 2 and Table 1). The average haplotype diversity (hd) of the Pid3 coding region was 0.680. The aus/boro subpopulation had the highest hd (0.706), while the indica subpopulation has the lowest hd (0.384). The number of rice accessions in each haplotype varied significantly; Hap_6 was the largest haplotype shared by as many as 1376 rice accessions, whereas, in contrast to Hap_6, there were nine haplotypes each was carried by only one accession (Fig. 2). Besides the 14 haplotypes that had the premature stop codon at the above mentioned nucleotide position 2209, Hap_11, Hap_36 and Hap_2 were newly identified pseudogenization types with a premature stop codon at nucleotide position 885, 1088 and 1766, respectively. It is noteworthy that Hap_36 owned both of the premature stop codons at 1088 and 2209 simultaneously.
A haplotype flowchart was constructed to describe the evolutionary relationships and mutational steps of these 31 haplotypes, which were identified in at least two rice accessions (Fig. 3). Meanwhile, the components of each haplotype were also taken into account. The flowchart analysis illustrated that the haplotypes of Pid3 could be roughly divided into three groups. Group I contains Hap_9 and twelve other haplotypes, in which most carriers are japonica accessions. All of these haplotypes have premature stop codon at 2209. Accordingly, in Group II, the predominant haplotype is Hap_6 but most of its carriers are indica accessions. The nucleotide diversities within group I and group II are much lower, and there only exist a few SNPs, as compared with Hap_9 and Hap_6, respectively. The remaining haplotypes belong to group III, in which no predominant haplotype exists, and the carriers of this group are diverse. It is notable that although Hap_2, shared mostly by japonica accessions, is not in group I, it is still a pseudogene due to the premature stop codon at 1766 as mentioned above. Indeed, out of the 95 Pid3 alleles belonging to Hap_2, 86 were identified from tropical japonica. Moreover, the Pid3 haplotypes from tropical and temperate japonica are significantly different (Supplementary Figure S1). Ten haplotypes and eleven haplotypes were identified from 287 temperate japonica and 392 tropical japonica rice accessions, respectively, and only five haplotypes were found in both subgroups. Pid3 alleles in most temperate japonica belong to Hap_9, whereas Hap_9, Hap_7 and Hap_2 were mainly shared by tropical japonica rice accessions (Supplementary Figure S1).
Figure 3.
The flowchart of 31 haplotypes of Pid3. Each colored circle represents a unique haplotype. The size of the circle corresponds to the frequency of each haplotype. Each dot on a solid line represents one SNP between two haplotypes.
In a previous study, we completely cloned 30 Pid3 orthologs from 17 wild rice and 10 cultivated rice accessions by allele mining (wild rice accessions W1, W11, W13 were heterozygous)29. By comparing the sequences of these 30 orthologs with the above identified 40 haplotypes, we found that, out of the 10 Pid3 orthologs cloned from cultivated rice, eight were identical to the newly discovered haplotypes (Pid3-I1 = Hap_14, Pid3-I2 = Hap_6, Pid3-I3 = Hap_20, Pid3-I4 = Hap_21, Pid3-J1/J2 = Hap_9, Pid3-J3/J5 = Hap_8). Meanwhile, out of the 20 Pid3 orthologs from wild rice accessions, only four were identical to those haplotypes (Pid3-W5 = Hap_13, Pid3-W11-1 = Hap_9, Pid3-W12 = Hap_6, Pid3-W15 = Hap_18) (Supplementary Table S4). For the 10 cultivated rice orthologs, a total of 26 SNPs were identified, of which only the SNPs at the position 46 and 994 were not included by the above identified 71 SNPs. In the 20 Pid3 orthologs from wild rice, a total of 101 SNPs were characterized, of which only 35 could be included by the above described 40 haplotypes (Supplementary Table S4).
The closest wild relatives of O. sativa are O. nivara and O. rufipogon, although which of them is the immediate progenitor of the cultivated rice remains controversial9. To investigate the domesticated history of Pid3, the 40 cultivated and 20 wild rice haplotypes were aligned (Fig. 4). It could be inferred that the haplotypes Hap_6 and Hap_9 were the ancestral types in cultivated rice, as they existed in all six cultivated rice subpopulations (Fig. 3) and two wild rice accessions, W12 and W11 (Fig. 4). In addition, they could be domesticated independently from different wild rice accessions, and the other haplotypes in group I and group II mentioned above might originate from Hap_9 and Hap_6, respectively. The haplotypes in group III might originate from a third type of wild rice accessions, because these haplotypes were much different from Hap_6 and Hap_9, and most of them were similar to those from wild rice accessions.
Figure 4.
Phylogenetic analysis of Pid3 haplotypes. The phylogenetic tree was generated by MEGA5 software using neighbor joining method, with the numbers associated with the interior branches indicating bootstrap values (1000 replications). The scale shows nucleotide substitutions per site.
Analysis of predicted Pid3 proteins
A total of 44 amino acid variations caused by the 71 nucleotide polymorphic sites, leads to 32 different predicted proteins (the original Pid3 haplotype Hap_9 = Hap_24 = Hap_31 = Hap_33 = Hap_38 and Hap_6 = Hap_12 = Hap_16 = Hap_17 = Hap_34). Of them, 20 encode complete CC-NBS-LRR proteins with 924 amino acids, and 12 show premature transcription termination at the position 295, 363, 589 and 737, respectively (Fig. 5). Most predicted proteins encoded by Pid3 alleles are different from Pid3 itself at nine positions, including 44, 259, 571, 577, 625, 815, 856, 894 and 896. It is noteworthy that besides the premature site between full length and truncated proteins at the position 737, there are five other completely different sites (153, 204, 515, 669 and 670) among them (Fig. 5), but this phenomenon was not found in other truncated proteins, which were premature at the position 295 and 589.
Figure 5.
Amino acid variation of Pid3 in 40 haplotypes. “.” represents the same base as Pid3 in Digu; “*” represents terminators.
We also compared Pid3 protein sequences in different cultivated rice growing areas, including indica/japonica growing areas in East Asia and Southeast Asia, and indica/japonica/aus/aro growing areas in South Asia. The sequence comparison revealed that most amino acid variations were found in the LRR region, and indica subgroups had lower diversity in all three areas (Supplementary Figure S2). For indica and japonica subgroups in East Asia and Southeast Asia, Pid3 haplotypes had no obvious difference. However, two stop codons at the position 589 and 737, five common variant amino acids, T153M, G204S, R515H in NBS domain, V669F, G670D in LRR region were found between indica and japonica subgroups in these two areas. We can infer that these variants are related to indica -japonica differentiation and play an important role in Pid3 function. Most aus and aro rice cultivars were found in South Asia. In this area, except for the 15 aus cultivars belonging to the japonica predominant Hap_9, most of the remaining haplotypes were extremely similar to indica haplotypes.
The Pid3 ortholog (Hap_6) present a similar resistance spectrum as Pid3 gene
In a previous study, we evaluated the resistance of 11 Pid3 orthologs by rice genetic transformation and blast inoculation, and found that five Pid3 orthologs were functional rice blast R genes, including Pid3-I1(Hap_14), Pid3-I3 (Hap_20/Pid3/Pi25) from indica varieties and Pid3-W3, Pid3-W4, Pid3-W5 (Hap_13/Pid3-A4) from wild rice accessions29. However, although it is the most popular haplotype in cultivated rice accessions, the rice blast resistance of the Pid3 ortholog (Hap_6) had not been verified yet. In this study, Pid3-I2 from the indica variety 93-11, a widely used inbred cultivar and backbone parent of hybrid rice in China, was chosen as the representative of Hap_6 for blast resistance testing. First, the entire 2775-bp coding region of Pid3-I2 was inserted into the binary vector pZH01 under the (CAMV) 35 S promoter control and transformed into the susceptible rice variety TP309, which was the same recipient used for 11 Pid3 orthologs in our previous study29. Next, we performed a genetic complementation test of Pid3-I2 as previously described44. A 6236-bp 93-11’s DNA fragment, including the Pid3-I2 coding region, 3010-bp upstream region, and 451-bp downstream region, was sub-cloned into the binary vector pMNDRBBin6, which was then introduced into TP309 as well. Finally, we obtained nine and eleven independent primary transgenic plants (T0) for these two constructs, respectively. All 20 transgene-positive plants were confirmed to be resistant to the M. oryzae isolate Zhong-10-8-14, which was the same isolate employed in our previous study (Fig. 6). Co-segregation of the transgene and the blast resistance was confirmed in selfed progenies (T1) of the two types of T0 lines, respectively. The results suggested that Pid3-I2/Hap_6 was indeed functional rice blast R gene. We then inoculated Pid3 and Pid3-I2/Hap_6 homozygous T2 transgenic plants, respectively, with 125 M. oryzae isolates collected from China. The testing revealed that compared to the susceptible recipient TP309, Pid3-I2/Hap_6 transgenic lines conferred resistance to 28 isolates, with a resistance frequency 22.4%, which is the same as that of the Pid3 transgenic plants (Supplementary Tables S5 and S6).
Figure 6.

Complementation test and overexpression of haplotype Hap_6. The primary transgenic lines are inoculated by M. oryzae isolate Zhong-10-8-14. The susceptible variety TP309 is showed as control. Hap_6 represents Pid3-I2 transgenic plant driven by itself promoter, and Hap_6-OE represents Pid3-I2 transgenic plant driven by CAMV 35 S promoter.
Geographic distribution of the known functional Pid3 alleles in cultivated rice accessions
At present, of the total of 42 haplotypes of Pid3 identified in cultivated rice accessions, only four haplotypes, including Hap_6, Hap_13, Hap_14, and Hap_20 were confirmed to be functional in rice blast resistance. Of the remaining 38 haplotypes, 16 were identified as pseudogenes due to the presence of premature stop codons at different positions (Fig. 2), while the other 22 remained to be further elucidated. Based on the information of the 3 K RGP, the worldwide geographic distributions of the three types of Pid3 haplotypes were presented in Table 2. Obviously, Hap_6 is the most common haplotype with the widest geographic distribution in the indica-cultivated area, whereas in most japonica-cultivated area, such as Japan, South Korea and Europe, functional haplotypes of Pid3 are almost nonexistent. In Southeast Asia countries, such as Thailand, Vietnam, Cambodia and Myanmar, Hap_20 distributes widely, while Hap_14 is only found in four South Asia countries: India, Pakistan, Nepal and Bangladesh. Finally, Hap_13, the rarest functional haplotype in cultivated rice accessions, is only found in China, India and Bangladesh. Although the number of haplotypes (whose functions have not been determined yet) is up to 22, these haplotypes are distributed scarcely in most countries and areas.
Table 2.
Geographic distributions of three types of haplotypes of Pid3.
| Continents | Countries/Regions | Total rice accessions | Functional haplotypes | Non-functional haplotypes | Function not determined haplotypes | |||
|---|---|---|---|---|---|---|---|---|
| Hap_6 | Hap_20 | Hap_14 | Hap_13 | |||||
| Asia | China | 439 | 265 | 13 | 0 | 2 | 112 | 47 |
| India | 351 | 241 | 6 | 3 | 6 | 43 | 52 | |
| Indonesia | 210 | 112 | 9 | 0 | 0 | 84 | 5 | |
| Philippines | 195 | 110 | 3 | 0 | 0 | 72 | 10 | |
| Bangladesh | 142 | 80 | 0 | 12 | 1 | 2 | 47 | |
| Thailand | 131 | 77 | 28 | 0 | 0 | 19 | 7 | |
| Laos | 114 | 51 | 5 | 0 | 0 | 54 | 4 | |
| Malaysia | 63 | 25 | 4 | 0 | 0 | 19 | 15 | |
| Myanmar | 57 | 38 | 12 | 0 | 0 | 4 | 3 | |
| Cambodia | 51 | 28 | 17 | 0 | 0 | 3 | 3 | |
| Japan | 51 | 3 | 0 | 0 | 0 | 44 | 4 | |
| Vietnam | 47 | 30 | 7 | 0 | 0 | 7 | 3 | |
| Sri Lanka | 45 | 34 | 0 | 0 | 0 | 3 | 8 | |
| Nepal | 43 | 31 | 0 | 2 | 0 | 6 | 4 | |
| Taiwan | 34 | 22 | 0 | 0 | 0 | 12 | 0 | |
| South Korea | 32 | 5 | 0 | 0 | 0 | 27 | 0 | |
| Pakistan | 29 | 11 | 0 | 4 | 0 | 4 | 10 | |
| Bhutan | 16 | 6 | 0 | 0 | 0 | 9 | 1 | |
| Africa | Madagascar | 66 | 38 | 0 | 0 | 0 | 27 | 1 |
| Senegal | 22 | 14 | 1 | 0 | 0 | 1 | 6 | |
| Ivory Coast | 21 | 5 | 0 | 0 | 0 | 15 | 1 | |
| Sierra Leone | 18 | 6 | 0 | 0 | 0 | 3 | 9 | |
| Liberia | 12 | 4 | 1 | 0 | 0 | 7 | 0 | |
| Nigeria | 12 | 8 | 0 | 0 | 0 | 3 | 1 | |
| Europe | Italy | 37 | 2 | 0 | 0 | 0 | 35 | 0 |
| Portugal | 22 | 0 | 1 | 0 | 0 | 21 | 0 | |
| France | 10 | 1 | 0 | 0 | 0 | 9 | 0 | |
| Spain | 10 | 0 | 0 | 0 | 0 | 10 | 0 | |
| South America | Colombia | 24 | 12 | 2 | 0 | 0 | 8 | 2 |
| Brazil | 22 | 8 | 0 | 0 | 0 | 10 | 4 | |
| Argentina | 11 | 1 | 0 | 0 | 0 | 10 | 0 | |
| North America | United States | 47 | 3 | 0 | 0 | 0 | 43 | 1 |
| Oceania | Australia | 13 | 4 | 0 | 0 | 0 | 9 | 0 |
Only county/region containing more than 10 rice accessions were included; Non-functional haplotypes, including Hap_2, Hap_7, Hap_8, Hap_9, Hap_11, Hap_24, Hap_30, Hap_31, Hap_32, Hap_33, Hap_35, Hap_36, Hap_37, Hap_39, Hap_40; Function not determined haplotypes, including Hap_1, Hap_5, Hap_10, Hap_15, Hap_17, Hap_18, Hap_19, Hap_21, Hap_22, Hap_25, Hap_26, Hap_29.
Hap_6 has been widely employed in hybrid rice breeding in China
To investigate the distribution of the known functional Pid3 haplotypes in cultivated rice varieties in China, we first sequenced the respective allelic Pid3 coding regions of the 12 widely cultivated japonica varieties in China, which would not contain the premature mutation at the position 2209 as testified by the CAPS marker (Supplementary Table S1). Sequence comparison confirmed that in these japonica varieties Pid3 alleles were identical to Hap_6, suggesting that Hap_6 of Pid3 might be introduced into minor japonica varieties by rice breeders. Next, we investigated whether Pid3 alleles in widely cultivated indica varieties in China were identical to Hap_6 or not. We focused on backbone parental lines of hybrid rice varieties in China. We chose nine restorer lines (Minghui 63, Shuhui 527, Gui 99, Fuhui 838, Xianhui 207, Miyang 46, CDR22, IR24, Mianhui 725) and nine male sterility lines (II-32A, Zhenshan 97A, Jin23A, Tianfeng A, V20A, Gang 46 A, Peiai64S, Y58S, Guangzhan 63-4S) to fully sequence their Pid3 alleles because they are most frequently used parents of hybrid rice in China (http://www.ricedata.cn/variety/). For example, the most popular male sterility line in hybrid rice breeding in China is II-32A, from which more than 200 hybrids have been released in recent 20 years (http://www.ricedata.cn/variety/). The results showed that out of the 18 lines, 16 have Pid3 alleles identical to Hap_6, while the alleles of remaining two lines, Tianfeng A and V20A, were identical to Hap_21. In addition, these backbone parental lines all conferred resistance to the M. oryzae strain Zhong-10-8-14, and the transcripts of the Pid3 alleles could be obviously detected in these lines (Supplementary Figure S3). These results demonstrated that Hap_6 of Pid3 has been widely utilized for hybrid rice breeding in China.
Moreover, by using re-sequencing data of hybrid rice12, we investigated nucleotide polymorphisms of Pid3 in 1495 hybrid rice varieties, which included 1,439 hybrid varieties from indica-indica crosses, 18 from indica-japonica crosses, and 38 from japonica-japonica crosses12. A total of 11 nucleotide polymorphism sites were identified in these hybrid rice varieties, all of which were included in the 71 sites (Supplementary Table S7). Only 88 hybrid rice varieties were found containing heterozygous sequences of Pid3; the remaining 1407 Pid3 alleles belonged to four haplotypes, of which Hap_H1 was the most common haplotype shared by 1392 hybrid rice varieties. Because of low sequencing coverage (approximate 2×)12, it was impossible to get full sequences of Pid3 in these 1407 hybrid rice varieties, though all variations of the Hap_H1 at the 11 nucleotide polymorphism sites were identical to Hap_6. As a result, we have reason to believe that the Pid3 alleles of Hap_6 have prevailed in hybrid rice varieties in China.
Discussion
So far, a great quantity of rice blast R genes have been identified and cloned, and almost all the cloned blast R genes have been applied to rice blast resistance breeding via R gene- self based or -tightly linked markers48–51. However, considering the possibility of a variety of functional alleles of the known blast R genes in rice populations, before a specific R gene is used for introgression, an accurate evaluation of its alleles in recurrent parental lines is in need. Moreover, some superior alleles, if any, could be identified by precise evaluation of the cloned R loci in rice germplasms29, 46, 52. Usually, there are three ways can be taken to evaluate a cloned R locus: first, certain markers which are always used in the MAS procedure for R loci can be applied. Then, the coding sequence fragment(s) amplified based on the cloned R gene should be examined. Finally, the complete R gene coding sequence(s) of every donor should be evaluated. However, due to the complicated and variable structure of these NBS-LRR type R genes, it is impossible to obtain accurate distribution of these cloned R genes just by markers and CDS fragments since single SNP could lead to the loss of function44, 53. Therefore, it is necessary to get the entire R gene sequences for their function assessment.
For the majority of rice blast R genes, it is rather difficult to obtain full sequences of alleles/orthologs by allele mining approach, due to the complexity of gene structure and vulnerable variations. However, the Pid3 locus, as mentioned above, is a typical NBS-LRR type gene, and it is relatively uncomplicated, because it is single-copy and intronless. Moreover, our former study29 has revealed that alleles/orthologs of Pid3 in other rice germplasms, even from wild rice lines, contained no InDel or structure variations (SVs), so it is practical for evaluation of Pid3 in rice resources by exploring the existing NGS data. In this study, we analyzed nearly 3,000 Pid3 alleles in cultivated rice accessions mainly based on the 3 K RGP sequencing data, in which each genome had an average sequencing depth of 14× with averaged genome coverage and mapping rates of 94.0% and 92.5%, respectively. Except for 16 alleles with an 8-bp insertion at the position 2461, the remaining alleles revealed no obvious InDel polymorphisms. In the coding region of the 2621 homozygous Pid3 alleles, a total of 71 polymorphic sites were identified. By comparing sequences of Pid3 alleles obtained from the PCR-based allele mining approach29, we found that most polymorphisms of Pid3 in cultivated rice accessions were included in these 71 sites. Recently, in another study, the sequence variations of Pid3 in 80 Yunnan rice landraces were analyzed by PCR-based allele mining approach54, in which a total of 39 nucleotide variations were found in the coding region of Pid3 alleles and no InDel or SV variations were identified. By comparing the positions of nucleotide variation, we found in that study, except for 8 sites, the remaining 31 were all involved in the 71 sites. Moreover, the haplotype 8 with the highest frequency (28.8%) in that study is identical to the most common Hap_6 in our work. These results demonstrated that it was feasible to analyze the sequence variations of Pid3 locus by utilization of the 3 K RGP sequencing data.
Some studies have shown that rice blast resistance is also correlated with the changes of some R gene expressions42, 43. In this study, our analyzing was only focused on the Pid3 coding sequences. Because most sequence variations existed in the promoter regions, they are difficult to be further judged for their relationship with the expression changes of the corresponding Pid3 alleles. Nevertheless, in some cultivars we checked the expression levels of Pid3 alleles (Supplementary Figure S3) since it is possible that the loss of blast resistance in certain haplotypes might be caused by sequence variations in their promoter regions.
Of note, in this work, a total of 2953 Pid3 alleles were tested for the premature mutation at the nucleotide position 2209. It was found that 22 (1.3%) of 1732 indica, 715 (83.2%) of 859 japonica and 40 (11%) of 362 other types carried the premature mutation. Moreover, haplotype analysis demonstrated that although Hap_2 (which was carried by 95 rice accessions and made up mostly of japonica lines) did not contain the premature mutation at the position 2209, it was still a pseudogene due to the premature stop codon at the position 1766. In addition, we checked the premature mutation at the position 2209 in 149 widely cultivated japonica varieties in north China with CAPS marker. The results showed that except for 12 accessions, all the other japonica cultivars carried the premature mutation. These results clearly demonstrated that Pid3 alleles in most of these japonica rice cultivars were non-functional, leaving a great opportunity for utilization of functional Pid3 alleles to improve their blast resistance. For example, the japonica variety Kongyu 131, the most important cultivar in north China (http://www.ricedata.cn/variety/), has not contained a functional Pid3 allele yet (Supplementary Table S1).
Similarly, in 2621 cultivated rice lines, merely 40 haplotypes of Pid3 were identified, and most haplotypes in group I and group II were similar to Hap_9 and Hap_6, respectively, and distinguished only by one to two SNPs. Until now, we have verified four functional Pid3 alleles (Hap_6, Hap_13, Hap_14 and Hap_20) from cultivated rice. Of them, Hap_14 has the broadest resistance spectrum29. For the remaining 16 haplotypes which encode full length CC-NBS-LRR proteins, it is important to assay their resistance functionality and spectrum. We may suggest that due to merely amino acids variations, most of them probably have resistance function but the spectrum are not distinct from those verified alleles. It is noteworthy that, recently, we made a reverse mutation (T-G) at the position 2209 in Hap_9, and found that this point mutant construct had no resistance function yet. So the remaining five amino acid variations at the position 153, 204, 515, 669 and 670 are probably important to its function (Fig. 5). Moreover, in contrast to wild rice accessions where a total of 101 polymorphic sites were identified in just 17 wild lines, there were only 71 polymorphic sites detected in a total of 2621 cultivated rice lines. This contrasting picture unambiguously showed that the genetic diversity at the Pid3 locus in cultivated rice lines has been restricted greatly. In order to explore more superior alleles at the Pid3 locus, we must focus on wild rice accessions in future. In our previous study, three orthologs of Pid3 with broad resistance spectrum were cloned from wild rice accessions29. In fact, many rice R genes have been identified from wild rice, including the two well-known rice R genes, Xa21 for resistance to Xanthomonas oryzae and Pi9 for resistance to M. oryzae originated from the wild rice O. longistaminat and O. minuta respectively37, 55. They both showed broader-spectrum resistances to pathogens and have been widely applied to rice breeding. Finally, considering that Hap_6, the most common haplotype of Pid3, has been widely deployed in Chinese indica rice breeding especially in the major hybrid rice cultivars56, here we would suggest that it is the time to introduce some novel blast resistance genes (alleles) into the rice varieties with indica background57.
Materials and Methods
Re-sequencing data of Pid3 alleles
Data of SNPs and InDels at the Pid3 locus in 3,000 rice accessions were downloaded from the Rice SNP-Seek Database18 (http://oryzasnp.org/iric-portal/index.zul) and the RMB database58 (http://www.rmbreeding.cn/snp3k). The 2775-bp coding sequence of Pid3 corresponds to the region (13055256-13058027) on chromosome 6 of the Nipponbare genome in the 3,000 rice genome project sequencing data. Sequence variations at the Pid3 locus in 1495 hybrid rice varieties were obtained from the RiceHap4 database12 (http://202.127.18.228/RiceHap4/index.php).
Plant materials
A set of 289 cultivated varieties including 140 indica varieties and 149 japonica varieties (Supplementary Table S1) were selected from China for detection of the nonsense mutation of Pid3 at the nucleotide position 2209 by the CAPS marker44. All of the rice varieties were kept in our lab. The susceptible recipient TP309 was used for transformation of the Hap_6 of Pid3. The varieties were cultivated in the experimental field of the Hunan Hybrid Rice Research Center in Changsha under normal growing conditions.
M. oryzae isolates
125 M. oryzae isolates used in this study were collected from rice fields around China, and were kindly provided by Dr.Yunliang Peng of Sichuan Academy of Agricultural Sciences59 and by Dr. Cailin Lei of Institute of Crop Sciences, Chinese Academy of Agricultural Sciences44. The diagnostic isolate of M. oryzae Zhong-10-8-14 was used for the phenotypic evaluation of the backbone parental lines of hybrid rice varieties in China, and the remaining isolates were used to assay the resistance spectra of Pid3 and Hap_6.
Detection of the nonsense mutation by the CAPS marker
Genomic DNA were extracted from fresh leaves of the 289 rice varieties using modified CTAB method of DNA isolation. A 658-bp fragment was amplified using the primer pair (Pid3CF: 5′-TACTACTCATGGAAGCTAGTTCTC-3′ and Pid3CR: 5′-ACGTCACAAATCATTCGCTC-3′). PCR amplification was carried out using the following profile: initial DNA denaturation at 95 °C for 4 min; followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 30 s; and final extension at 72 °C for 5 min. PCR products were digested with the restriction endoenzyme BamHI. The absence of a 506-bp restriction fragment was considered to represent the nonsense mutation at the position 2209.
Sequence analysis
Sequences were aligned using CLUSTAL X version 2.060 and adjusted manually with Microsoft office excel 2010. Nucleotide diversity π (average number of nucleotide differences per site), πnon/πsyn (average ratio of non-synonymous site diversity over synonymous site diversity) and haplotype diversity analysis were calculated using DNASP v5.061. Haplotype flowchart was constructed with the computer program Network 5.0 (http://www.fluxus-engineering.com/sharenet.htm). DNASP v5.0 was also used to perform Tajima’s D test and sliding-window analysis of Pid3 alleles.
DNA sequencing
DNA was extracted from fresh leaves of the 18 indica and 12 japonica rice varieties. Primers (Pid3SF: 5′-AGTAACACCCAAGGATAGGATAG-3′ and Pid3SR: 5′-GAACGACAAGTGCGACATGATTG-3′) that amplified the full coding sequence of Pid3 were designed according to Pid3 sequence in rice variety Digu. PCR amplification was carried out using the following profile: initial DNA denaturation, 95 °C for 4 min; followed by 30 cycles of denaturation, 95 °C for 30 s; annealing, 58 °C for 30 s; extension, 72 °C for 3 min; and final extension at 72 °C for 5 min. The PCR products were sequenced by TsingKe Biology Technology.
Vector construction and Rice transformation
For the Hap_6 overexpression test, primer pair (Pid3OF: 5′-TTTCTAGAAGTAACACCCAAGGATAGGATAG-3′ and Pid3OR: 5′-CTGTCGACGAACGACAAGTGCGACATGATTG-3′) were designed to amplify the coding sequence of Pid3 allele from genomic DNA of cultivar 93-11. An XbaI and an SalI recognition site (underlined) with two protecting bases (TT and CT) were added to their 5′ ends, respectively, then the PCR product was cloned into the binary vector pZH01 through the XbaI and SalI cloning sites. For the Hap_6 complementation test, the 6236-bp genomic sequence of Pid3 allele containing the promoter region and the full coding region was amplified from genomic DNA of 93-11 using the primer pair (Pid3FF: 5′-GGGTACCCCACACATTGTACACCTACGACCAC-3′ and Pid3FR: 5′-CCCCGGGGGAACGACAAGTGCGACATGATTG-3′), and then cloned into the binary vector pMNDRBBin662 through the KpnI and XmaI cloning sites (underlined). After sequence verification the final constructs were introduced into Agrobacterium tumefaciens LBA4404. The callus of susceptible japonica variety TP309 was transformed according to published methods63. The resistance of the primary transgenic lines (T0) was challenged by inoculation with the M. oryzae strain Zhong-10-8-14.
Expression analysis of Pid3 alleles
RNA was isolated from leaf tissue with the TRIzol reagent (Invitrogen, Carlsbad, CA), and cDNA was synthesized from poly(A) + RNA using a cDNA synthesis kit (Transgen, Beijing). RT-PCR was performed with the specific primer pair Pid3C for 30 cycles of amplification. PCR amplification was as follows: 95 °C for 4 min; followed by 30 cycles of denaturation, 95 °C for 30 s; annealing, 58 °C for 30 s; extension, 72 °C for 30 s; and final extension at 72 °C for 5 min. Transcription of the actin gene was used to normalize the cDNA levels with the primer pair 5′-AGCAACTGGGATGATATGGA-3′ and 5′-CAGGGCGATGTAGGAAAGC-3′. Amplification of the actin gene was conducted for 27 cycles and the annealing temperature was 57 °C.
Fungal inoculation
Six to eight plants were tested for each cultivar. Disease reaction to blast followed the modified standard pathogenicity assay as previously described31. Specifically, Rice seedlings at the four-leaf stage were inoculated by spraying a spore suspension (5 × 104 spores/ml) of the M.oryzae isolates onto the leaves in a plastic bag. After inoculation plastic bags were sealed to maintain at 25 °C and 100% humidity in the dark for 24 h. Subsequently, plants were moved to the greenhouse (the humidity was maintained 70–85%, the temperature was 23/28 °C, and the lighting was 14/10 h for light/dark). and were allowed to grow to permit the development of expected disease symptoms. The disease reaction was examined one week after inoculation with the susceptible variety, TP309, as a control. The disease reaction was rated as 0–5, 0–3 as resistance and 4–5 as susceptible based on visual number and amount of lesions at the second youngest leaf 52.
Electronic supplementary material
Acknowledgements
We thank Dr. Yunliang Peng (Sichuan Academy of Agricultural Sciences) and Dr. Cailin Lei (Chinese Academy of Agricultural Sciences) for kindly providing M. oryzae isolates. The research was supported by funds from NSFC (31401723), Transgenic Projects from the Chinese Ministry of Agriculture (2016ZX08009-003-001) and Hunan Provincial Key Research and Development Program (2016JC2025).
Author Contributions
Lihuang Zhu and Qiming Lv conceived and designed the experiment, Qiming Lv, Xiao Xu, Zhiyuan Huang, Hai Liu, Li Tang, Junjie Xing, Zhirong Peng carried out the experiments, Yeyun Xin, Xiaobing Li, Tianqing Zheng collected the data, Qiming Lv, Lihuang Zhu, Tianqing Zheng, Zhuangzhi Zhou and Chunchao Wang analyzed the data, Lihuang Zhu and Qiming Lv wrote the manuscript. All authors have read the manuscript and agree with its content.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10617-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tianqing Zheng, Email: zhengtianqing@caas.cn.
Lihuang Zhu, Email: lhzhu@genetics.ac.cn.
References
- 1.Goff SA, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 3.International Rice Genome Sequencing P. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, et al. Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc Natl Acad Sci USA. 2016;113:E5163–71. doi: 10.1073/pnas.1611012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacquemin J, Bhatia D, Singh K, Wing RA. The International Oryza Map Alignment Project: development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr Opin Plant Biol. 2013;16:147–56. doi: 10.1016/j.pbi.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Sakai H, et al. Construction of pseudomolecule sequences of the aus rice cultivar Kasalath for comparative genomics of Asian cultivated rice. DNA Res. 2014;21:397–405. doi: 10.1093/dnares/dsu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schatz MC, et al. Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol. 2011;30:105–11. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 9.Huang X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subbaiyan GK, et al. Genome-wide DNA polymorphisms in elite indica rice inbreds discovered by whole-genome sequencing. Plant Biotechnol J. 2012;10:623–34. doi: 10.1111/j.1467-7652.2011.00676.x. [DOI] [PubMed] [Google Scholar]
- 11.Yang W, et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun. 2014;5 doi: 10.1038/ncomms6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun. 2015;6 doi: 10.1038/ncomms7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung H, et al. Allele mining and enhanced genetic recombination for rice breeding. Rice (N Y) 2015;8 doi: 10.1186/s12284-015-0069-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao H, et al. RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res. 2015;43:D1018–22. doi: 10.1093/nar/gku894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCouch SR, et al. Open access resources for genome-wide association mapping in rice. Nat Commun. 2016;7 doi: 10.1038/ncomms10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JY, Wang J, Zeigler RS. The 3,000 rice genomes project: new opportunities and challenges for future rice research. Gigascience. 2014;3 doi: 10.1186/2047-217X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Project RG. The 3,000 rice genomes project. Gigascience. 2014;3 doi: 10.1186/2047-217X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov N, et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–7. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo L, Gao Z, Qian Q. Application of resequencing to rice genomics, functional genomics and evolutionary analysis. Rice (N Y) 2014;7 doi: 10.1186/s12284-014-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42:961–7. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2011;44:32–9. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet. 2014;46:714–21. doi: 10.1038/ng.3007. [DOI] [PubMed] [Google Scholar]
- 23.Begum H, et al. Genome-wide association mapping for yield and other agronomic traits in an elite breeding population of tropical rice (Oryza sativa) PLoS One. 2015;10 doi: 10.1371/journal.pone.0119873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yano K, et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet. 2016;48:927–34. doi: 10.1038/ng.3596. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Jia MH, Ghai P, Lee FN, Jia Y. Genome-Wide Association of Rice Blast Disease Resistance and Yield-Related Components of Rice. Mol Plant Microbe Interact. 2015;28:1383–92. doi: 10.1094/MPMI-06-15-0131-R. [DOI] [PubMed] [Google Scholar]
- 26.Skamnioti P, Gurr SJ. Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 2009;27:141–50. doi: 10.1016/j.tibtech.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Devanna NB, Vijayan J, Sharma TR. The blast resistance gene Pi54 of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J, et al. Pi64, Encoding a Novel CC-NBS-LRR Protein, Confers Resistance to Leaf and Neck Blast in Rice. Mol Plant Microbe Interact. 2015;28:558–68. doi: 10.1094/MPMI-11-14-0367-R. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, et al. Excavation of Pid3 orthologs with differential resistance spectra to Magnaporthe oryzae in rice resource. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, et al. A genome-wide survey reveals abundant rice blast R genes in resistant cultivars. Plant J. 2015;84:20–8. doi: 10.1111/tpj.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006;46:794–804. doi: 10.1111/j.1365-313X.2006.02739.x. [DOI] [PubMed] [Google Scholar]
- 32.Fukuoka S, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, et al. Allele-mining of rice blast resistance genes at AC134922 locus. Biochem Biophys Res Commun. 2014;446:1085–90. doi: 10.1016/j.bbrc.2014.03.056. [DOI] [PubMed] [Google Scholar]
- 34.Yang S, et al. Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc Natl Acad Sci USA. 2013;110:18572–7. doi: 10.1073/pnas.1318211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasudevan K, Gruissem W, Bhullar NK. Identification of novel alleles of the rice blast resistance gene Pi54. Sci Rep. 2015;5 doi: 10.1038/srep15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Lin F, Wang L, Pan Q. The in silico map-based cloning of Pi36, a rice coiled-coil nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics. 2007;176:2541–9. doi: 10.1534/genetics.107.075465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu S, et al. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics. 2006;172:1901–14. doi: 10.1534/genetics.105.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SK, et al. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics. 2009;181:1627–38. doi: 10.1534/genetics.108.099226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai C, et al. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011;189:321–34. doi: 10.1111/j.1469-8137.2010.03462.x. [DOI] [PubMed] [Google Scholar]
- 40.Okuyama Y, et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011;66:467–79. doi: 10.1111/j.1365-313X.2011.04502.x. [DOI] [PubMed] [Google Scholar]
- 41.Lin F, et al. The blast resistance gene Pi37 encodes a nucleotide binding site leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics. 2007;177:1871–80. doi: 10.1534/genetics.107.080648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi N, et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010;64:498–510. doi: 10.1111/j.1365-313X.2010.04348.x. [DOI] [PubMed] [Google Scholar]
- 43.Hayashi K, Yoshida H. Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 2009;57:413–25. doi: 10.1111/j.1365-313X.2008.03694.x. [DOI] [PubMed] [Google Scholar]
- 44.Shang J, et al. Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site–leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics. 2009;182:1303–11. doi: 10.1534/genetics.109.102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, et al. A Pid3 allele from rice cultivar Gumei2 confers resistance to Magnaporthe oryzae. J Genet Genomics. 2011;38:209–16. doi: 10.1016/j.jgg.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Lv Q, et al. Functional analysis of Pid3-A4, an ortholog of rice blast resistance gene Pid3 revealed by allele mining in common wild rice. Phytopathology. 2013;103:594–9. doi: 10.1094/PHYTO-10-12-0260-R. [DOI] [PubMed] [Google Scholar]
- 47.Yang S, et al. Genetic variation of NBS-LRR class resistance genes in rice lines. Theor Appl Genet. 2008;116:165–77. doi: 10.1007/s00122-007-0656-4. [DOI] [PubMed] [Google Scholar]
- 48.Ellur RK, et al. Improvement of Basmati rice varieties for resistance to blast and bacterial blight diseases using marker assisted backcross breeding. Plant Sci. 2016;242:330–41. doi: 10.1016/j.plantsci.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 49.Tanweer FA, et al. Current advance methods for the identification of blast resistance genes in rice. C R Biol. 2015;338:321–34. doi: 10.1016/j.crvi.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Ni D, et al. Marker-assisted selection of two-line hybrid rice for disease resistance to rice blast and bacterial blight. Field Crops Research. 2015;184:1–8. doi: 10.1016/j.fcr.2015.07.018. [DOI] [Google Scholar]
- 51.Ashkani S, et al. Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das A, et al. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct Integr Genomics. 2012;12:215–28. doi: 10.1007/s10142-012-0284-1. [DOI] [PubMed] [Google Scholar]
- 53.Bryan GT, et al. tA single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell. 2000;12:2033–46. doi: 10.1105/tpc.12.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, et al. Sequence variation of Pid3 for rice blast resistance in Yunnan rice landrace. Chin. J Rice Sci. 2016;30:17–26. [Google Scholar]
- 55.Ronald PC, et al. Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol Gen Genet. 1992;236:113–20. doi: 10.1007/BF00279649. [DOI] [PubMed] [Google Scholar]
- 56.Xie HG, et al. Development of hybrid rice variety FY7206 with blast resistance gene Pid3 and cold tolerance gene Ctb1. Rice. Science. 2016;23:266–73. [Google Scholar]
- 57.Wang HM, et al. Development and validation of CAPS markers for Marker-Assisted selection of rice blast resistance gene. Pi25. Acta Agronomica Sinica. 2012;38:1960–68. doi: 10.3724/SP.J.1006.2012.01960. [DOI] [Google Scholar]
- 58.Zheng TQ, et al. Rice functional genomics and breeding database (RFGB): 3K-rice SNP and InDel sub-database (in Chinese) Chin Sci Bull. 2015;60:367–71. doi: 10.1360/N972014-01231. [DOI] [Google Scholar]
- 59.Bai, Y. L. Virulence to hybrid rice in Magnaporthe oryzae from Sichuan Province. Master dissertation, Sichuan Agricultural University, Chengdu City, Sichuan Province, China (2011).
- 60.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 61.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–7. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 62.Lu HJ, Zhou XR, Gong ZX, Upadhyaya NM. Generation of selectable marker-free transgenic rice using double right-border (DRB) binary vectors. Functional Plant Biology. 2001;28:241–48. doi: 10.1071/PP00129. [DOI] [Google Scholar]
- 63.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–82. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






