Abstract
The product of the retinoblastoma susceptibility gene, the Rb protein, functions partly through transcriptional repression of E2F-regulated genes. Repression by Rb is mediated, at least in part, by a histone deacetylase complex, whose enzymatic activity relies on HDAC1, HDAC2 or HDAC3. Recently, we have shown that the Rb-associated histone deacetylase complex contains RbAp48 protein, which interacts with HDAC1 and HDAC2. RbAp48 could favour the deacetylation of histones since it binds directly to histone H4. In agreement with that, we show that transcriptional repression of E2F activity requires the presence of RbAp48. HDAC3 was thought not to interact with RbAp48. However, we found that it shared with HDAC1 the ability to favour the recruitment of RbAp48 to Rb. This latter effect was unlikely to be due to activation of Rb function, since HDAC3 did not increase Rb–E2F1 interaction. Rather, we found, surprisingly, that HDAC3 could physically interact with RbAp48 both in vitro and in living cells. Taken together, our data suggest a model in which Rb mediates the recruitment to E2F-regulating promoters of a repressive complex containing either HDAC1, HDAC2 or HDAC3 and the histone-binding protein RbAp48.
INTRODUCTION
The E2F transcription factor regulates progression into S phase of the cell cycle by activating many S phase-specific genes, such as DNA polymerase α, cdc6, cyclin E and DHFR, at the end of G1. E2F is composed of heterodimers between the so-called E2F proteins (E2F1–E2F6) and their partner DP proteins (DP1 and DP2) (reviewed in 1,2). E2F1–E2F5 all share a dimerization/DNA-binding domain and a transcriptional activation domain and specifically bind a member of the pocket protein family, which is composed of retinoblastoma protein (Rb) and its two cousins, p107 and p130.
The founding member of the family, Rb, is recruited to E2F-responsive genes through direct binding to E2F1, E2F2 or E2F3. Phosphorylation of Rb at the end of G1 by the concerted action of cyclin/cdks results in functional inactivation of the protein and the appearance of free E2F–DP heterodimers able to activate transcription (3).
At the beginning of G1, Rb is recruited to E2F-regulated genes and represses their transcription. A large body of evidence indicates that transcriptional repression by Rb is crucial for its anti-proliferative effects: in some instances, a basal non-repressed transcription of E2F-regulated genes is sufficient to promote cell growth and cell transformation (4–7).
Various mechanisms for transcriptional repression by Rb have been suggested (8,9). However, many recent studies have shown that Rb represses transcription, at least in part, through the recruitment of histone deacetylases (10–12). Histone deacetylases are thought to create a ‘closed’ chromatin structure through deacetylation of nucleosomal histone N-terminal tails (for a recent review, see 13). Also, it is now known that many other proteins than histones are acetylated in live cells, such as p53, the acetyltransferase SRC1 and transcription factor E2F1 itself (reviewed in 14). These acetylated proteins, in particular E2F1, could be important substrates for Rb-associated histone deacetylases.
Recently, chromatin immunoprecipitation experiments have shown that histones on E2F-regulated promoters evolve from a hypoacetylated to a hyperacetylated state as cells progress towards S phase (15). This result indicates that histones are likely to be real substrates of the histone deacetylase complex recruited by Rb. This complex could be targeted to histones through the protein RbAp48 (16), which interacts physically with histone H4 (17,18). RbAp48 was proposed to be assembled in the complex through its interaction with the histone deacetylase HDAC1 (16).
Recently, histone deacetylase HDAC3 was also shown to interact with Rb (19). Interestingly, HDAC3 is believed not to be associated with RbAp48 in live cells (20). This suggests that there could be two types of histone deacetylase complexes associated with Rb: one depending on HDAC1 or HDAC2 and targeted to histones by the presence of RbAp48 and the other depending on HDAC3, devoid of histone targeting and perhaps specific for non-histones proteins, such as E2F1.
We here show that RbAp48 is required for transcriptional repression of E2F activity. Surprisingly, we found that HDAC3, as HDAC1, favours its recruitment to Rb. HDAC3 is likely to function as a bridge between RbAp48 and Rb since it interacts in vitro as well as in living cells with RbAp48. Taken together, these results suggest that the Rb-associated repressive complex contains HDAC1, HDAC2 or HDAC3 and RbAp48.
MATERIALS AND METHODS
Cell culture and transfection
SAOS-2 and NIH 3T3 cells were maintained in DMEM supplemented with 10% FCS and antibiotics. For transient transfection experiments, 106 SAOS-2 cells were plated in 10 cm Petri dishes. Transfection was performed the following day by calcium/phosphate co-precipitation using standard procedures. Cells were harvested 24 h later.
Vectors
Details of the construction of pGEX2T-RbAp48 are available upon request. The PCMV Neo Bam Rb 379–928, pCMV Neo Bam E2F1 and pCMV HA-HDAC1 expression vectors have been described previously (12). pCMV Flag-HDAC3 was a kind gift from Dr E. Seto (H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL) (21). The PCMV HA-RbAp48 expression vector has also been described (16). Vectors allowing in vitro translation of HDAC1 and HDAC3 were kind gifts from Drs S.L. Schreiber (Harvard University, Cambridge, MA) and E. Seto, respectively. PCMV F-HDAC1 was a kind gift from Dr S.L. Schreiber (22). The E2F–GFP reporter vector was constructed as described (23). Plasmids expressing GST–E2F1 359–437 (GST-E2F1-AD) and GST–CREB 88–160 (GST-CREB-AD) were kind gifts from Dr T. Kouzarides (Wellcome/CRC Institute, Cambridge, UK) and M. Montminy (Harvard Medical School, Boston, MA), respectively.
Immunoprecipitations
Transiently transfected cells (one dish) were lysed in 500 µl of lysis buffer (50 mM Tris, pH 8.0, 300 mM NaCl, 0.4% NP40, 10 mM MgCl2) supplemented with a cocktail of protease inhibitors (Complete; Roche Diagnostics) and phosphatase inhibitors (10 mM NaF, 1 mM NaOV; both from Sigma). Extracts were cleared by centrifugation (20 000 g for 15 min), then diluted with 500 µl of dilution buffer (50 mM Tris, pH 8.0, 0.4% NP40, 2.5 mM CaCl2) supplemented with a cocktail of protease and phosphatase inhibitors and with DNase I (Worthington). Extracts were precleared by a 30 min incubation with 20 µl of protein A/protein G beads at 4°C on a rotating wheel. Antibodies (as indicated in the figure legends) were then added to the precleared extracts. After 1 h at 4°C, 10 µl of protein A/protein G beads were added and extracts were incubated for 1 h at 4°C on a rotating wheel. After extensive washing, bound proteins were analysed by western blotting. For immunoprecipitation of endogenous proteins, HeLa nuclear extracts (50 or 200 µl; Computer Cell Culture Center, Belgium) were diluted in 500 µl of washing buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.4% NP40, 5 mM MgCl2) and subjected to immunoprecipitation as described above.
GST pull downs
GST fusion proteins were expressed in Escherichia coli and purified essentially as described (16). For GST pull downs, 10 µl of beads [corresponding to ∼2 µg fusion proteins and prewashed with ELB buffer (50 mM HEPES, pH 7.9, 250 mM NaCl, 0.1% NP40)] were incubated for 10 min at room temperature with 10 µl of unprogrammed reticulocyte lysate (Promega). Beads were then diluted in 200 µl of ELB buffer, supplemented with 1 mM DTT and BSA (150 µg/ml), and incubated for 10 min at room temperature. 35S-labelled in vitro translated proteins were then added and incubated for 1 h at room temperature. After extensive washing, bound proteins were detected by SDS–PAGE, followed by autoradiography.
Microinjections
NIH 3T3 cells were grown on coverslips. Antibodies and plasmids were microinjected in a buffer containing 10 mM Tris, pH 7.4, 100 mM KCl. NIH 3T3 cells were microinjected with a mixture of Dextran–Rhodamine (10%; Molecular Probes) as a microinjection marker, an E2F-driven GFP reporter construct (2 µg/µl final concentration), a monoclonal antibody (250 ng/µl final) against either RbAp48 (11G10; Genetex) or HA (12CA5; Roche Diagnostics) as a negative control and with or without pcDNA3-Flag-p48 or the empty vehicle vector, at 2 µg/µl final concentration. Cells were fixed 24–36 h later using 4% paraformaldehyde. Red cells (microinjected cells) and green cells (GFP-positive cells) were then counted.
RESULTS
We have previously shown that RbAp48 is physically present within the Rb-associated histone deacetylase complex. We next wanted to investigate the functional role of RbAp48 in E2F regulation.
RbAp48 is an abundant protein and all our experiments aimed at studying the role of RbAp48 by overexpressing exogenous protein from a transiently transfected template failed (data not shown). Thus, in order to test whether RbAp48 is important for transcriptional repression of E2F activity, we inactivated it by microinjection of an anti-RbAp48 antibody. In microinjection experiments we used antibody 11G10, which is specific for RbAp48, since it does not recognise the highly related protein RbAp46 (16). Microinjection of an E2F–GFP reporter construct into Rb-positive NIH 3T3 cells [performed as described (23), except that microinjected cells were growing exponentially] led to between 15 and 20% GFP-positive cells, likely reflecting cells around the G1/S transition, in which E2F is active (Table 1). Strikingly, co-injection of a RbAp48-specific antibody led to a major increase in the percentage of green cells (up to 50–70%). This increase was specific for E2F-regulated promoters and did not reflect a general increase in transcription, since two control promoters, CMV and p21, were not affected by addition of anti-RbAp48 antibody. Thus, inactivation of RbAp48 resulted in activation of E2F activity. Importantly, this increase was reversed, at least in part, by co-injection of an expression vector for RbAp48 (Table 2). These data indicate that RbAp48 is required for repression of E2F activity. To our knowledge, they are the first direct demonstration of the involvement of RbAp48 in transcriptional repression.
Table 1. Microinjection of an anti-RbAp48 antibody stimulates transcription from a E2F–GFP reporter construct.
| Reporter vector |
Microinjected antibodya |
No. of microinjected cells (red cells)b |
GFP-positive cellsb (no.) |
GFP-positive cellsb (%) |
| E2F-GFP | Anti-HA | 73 | 13 | 17.7 |
| Anti-RbAp48 | 61 | 37 | 60.6 | |
| CMV-GFP | Anti-HA | 65 | 55 | 84.6 |
| Anti-RbAp48 | 84 | 73 | 87.0 | |
| p21-GFP | Anti-HA | 52 | 26 | 50 |
| Anti-RbAp48 | 64 | 35 | 54.7 |
aAnti-RbAp48, 11G10 (Genetex); anti-HA, 12CA5 (Roche Diagnostics).
bMean of independent experiments.
Table 2. Co-injection of a RbAp48 expression vector partially restores repression of E2F activity.
| Microinjected antibodya |
Microinjected expression vector |
No. of microinjected cells |
|
GFP-positive cells (no.) |
|
GFP-positive cells (%) |
|
| Exp. 1 | Exp. 2 | Exp. 1 | Exp. 2 | Exp. 1 | Exp. 2 | ||
| Anti-HA | pCMV | 63 | 67 | 8 | 14 | 12.7 | 21 |
| Anti-RbAp48 | pCMV | 71 | 85 | 23 | 55 | 32 | 65 |
| Anti-RbAp48 | pCMV-RbAp48 | 54 | 60 | 14 | 28 | 26 | 46 |
aAnti-RbAp48, 11G10 (Genetex); anti-HA, 12CA5 (Roche Diagnostics).
We previously proposed that RbAp48 could be recruited to E2F-regulated promoters through a direct interaction with the histone deacetylase HDAC1, which binds Rb (16). Interestingly, Rb also binds the histone deacetylase HDAC3 (19), which is not believed to interact with RbAp48 (20). This could provide a molecular basis for potential functional differences between these two Rb-associated histone deacetylases. We thus tested the effect of HDAC3 on the Rb–RbAp48 interaction. We transfected SAOS-2 cells with expression vectors for the Rb pocket domain (Rb 379–928, Rb), HA-tagged RbAp48 (HA-RbAp48) and Flag-tagged HDAC3 (F-HDAC3). As previously shown (16), immunoprecipitation of Rb in the absence of exogenous histone deacetylases led to specific co-immunoprecipitation of a small amount of fast migrating transfected RbAp48 (Fig. 1, lane 3). In the presence of exogenous HDAC3 much more RbAp48 was found associated with Rb (lane 7). This increase was not due to better production of transfected Rb (Fig. 1, bottom, compare lanes 3 and 7) or RbAp48 (Fig. 1, top, compare lanes 4 and 8). These results indicate that, as for HDAC1, the presence of exogenous HDAC3 led to a substantial increase in Rb–RbAp48 interaction in transfected cells.
Figure 1.
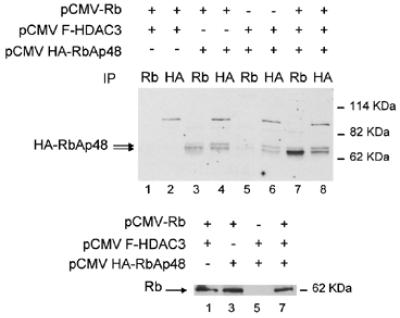
Exogenous HDAC3 increases the Rb–RbAp48 interaction. SAOS-2 cells were transfected with 5 µg of the indicated expression vectors [the Rb expression vector encodes the Rb pocket domain (379–928)] by the calcium phosphate co-precipitation method. The amount of promoter in the transfection was kept constant using empty vectors. Twenty-four hours after transfection, total cell lysates were prepared and immunoprecipitated as described (16), using 1 µg of the indicated antibody [anti-Rb, antibody C15G (Santa Cruz Biotechnologies); anti-HA, antibody 12CA5 (Roche Diagnostics)]. Immunoprecipitates were subjected to western blot analysis using the anti-HA antibody (top) or an anti-Rb antibody (antibody XZ55; Pharmingen) (bottom) using standard procedures. The arrows indicate the position of the exogenous proteins. Note that exogenous Rb migrates at ∼60 kDa, because the expression vector used in these experiments expressed a version of Rb deleted for the first 378 amino acids of the molecule.
Since HDAC3, in contrast to HDAC1, is thought not to interact with RbAp48 in live cells (20), we reasoned that the increase in Rb–RbAp48 interaction could result from an indirect effect of HDAC3, leading for example to hyperactive Rb. To test this idea, we investigated the effect of exogenous HDAC3 on the ability of Rb to interact with E2F1. This E2F1–Rb interaction is direct and is dependent upon active hypophosphorylated Rb. As expected, immunoprecipitation of Rb from transfected cells led to specific co-immunoprecipitation of transfected E2F1 (Fig. 2, top, lane 4). In the presence of exogenous HDAC3, the amount of E2F1 immunoprecipitated with Rb did not increase (lane 1), but rather, if anything, decreased slightly. This cannot be explained by a lack of expression of transfected E2F1 (bottom, lane 1) or Rb (middle, lane 1). These data indicate that the presence of exogenous HDAC3 does not change the ability of Rb to contact E2F1 and suggest that it does not induce any major changes in the biochemical activity of exogenous Rb. Thus, the results from Figure 1 are unlikely to be due to an effect of HDAC3 on the activity of Rb and rather suggest that HDAC3, like HDAC1, is able to physically bridge Rb and RbAp48. Consistent with this hypothesis, RbAp48 and HDAC3 have been shown to bind the same domain of Rb, which contacts the so-called ‘LXCXE’ motif (16,19). This motif is shared by many viral and cellular proteins and is responsible for direct interactions with the pocket domain of Rb (24).
Figure 2.
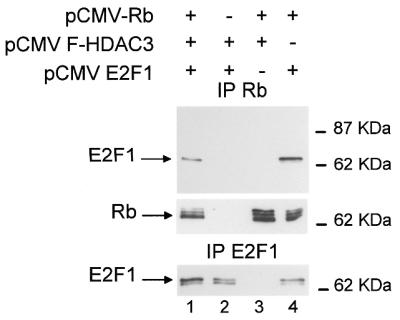
Exogenous HDAC3 does not increase the E2F1–Rb interaction. SAOS-2 cells were transiently transfected by calcium phosphate co-precipitation with the indicated expression vectors. Total cell lysates were immunoprecipitated using the anti-Rb antibody (top and middle) or an anti-E2F1 antibody (antibody KH95; Santa Cruz Biotechnologies) (bottom). Immunoprecipitates were subjected to western blot analysis using the anti-E2F1 antibody (top and bottom) or the anti-Rb antibody (middle).
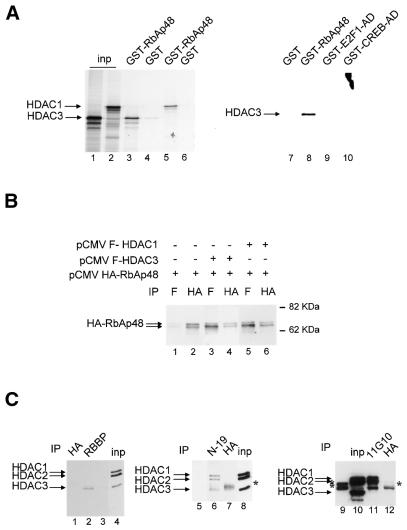
This latter explanation would require a physical interaction between HDAC3 and RbAp48. We tested this possibility in vitro by GST pull down experiments (Fig. 3A). We found that in vitro translated HDAC1 was retained on GST–RbAp48 beads (lane 5), but not on control GST beads (lane 6), indicating that HDAC1 can bind RbAp48 in vitro. This result was expected, since the physical interaction between HDAC1 and RbAp48 is well documented (25). In vitro translated HDAC3 was also specifically retained on RbAp48 beads (compare lanes 3 and 4). In similar experiments, HDAC3 did not interact with GST–E2F1-AD or GST–CREB-AD fusion proteins (compare lanes 9 and 10 with lane 8). Thus, HDAC3 shares with HDAC1 the ability to interact with RbAp48, at least in vitro. Furthermore, the amounts of HDAC3 or HDAC1 specifically retained on GST–RbAp48 beads are similar, suggesting that the affinity of RbAp48 for HDAC3 is comparable to its affinity for HDAC1.
Figure 3.
Physical association between HDAC3 and RbAp48. (A) 35S-labelled in vitro translated HDAC1 (lanes 5 and 6) or HDAC3 (lanes 3–4 and 7–10) was subjected to GST pull down analysis using beads harbouring 1 µg GST–RbAp48 fusion protein (lanes 3, 5 and 8), control GST (lanes 4, 6 and 7), GST–E2F1 AD (lane 9) or GST–CREB AD (lane 10), as indicated. After extensive washing, bound proteins were analysed by SDS–PAGE followed by autoradiography. In lanes 1 and 2, 10% of the amount of in vitro translated HDAC3 or HDAC1 used in the pull down reaction was directly loaded. (B) SAOS-2 cells were transiently transfected by calcium phosphate co-precipitation with the indicated expression vector and total cell extracts were immunoprecipitated with the indicated antibody (anti-Flag M2 antibody, antibody F; Sigma). Immunoprecipitates were subjected to western blot analysis using the anti-HA antibody. The arrows indicate the position of the two RbAp48 bands. (C) Hela nuclear extracts [50 (lanes 1, 2, 6 and 7) or 200 µl (lanes 11–12)] were immunoprecipitated with 1 µg of either an anti-RbAp48 antibody [lane 2, antibody RBBP (Transduction Laboratories); lane 6, antibody N19 (Santa-Cruz); lane 11, antibody 11G10 (Genetex)] or control anti-HA antibody [lanes 1, 7 and 12, antibody 12CA5 (Roche Diagnostics)]. In lanes 4, 8 and 10, 4 µl of HeLa nuclear extracts were directly loaded. In lanes 3, 5 and 9, 1 µg RBBP, N19 and 11G10 antibodies, respectively, were loaded, to monitor the migration of immunoglobulins. Immunoprecipitates were tested for the presence of HDAC1, HDAC2 and HDAC3 by western blotting using an anti-HDAC antibody (Transduction Laboratories). The stars indicate bands due to the immunoglobulin heavy chains from the anti-RbAp48 antibody (lanes 9 and 11) or the anti-HA antibody (lanes 7 and 12). Note that at longer exposures HDAC1 and HDAC2 could be detected in RBBP immunoprecipitates (lane 2). Also, the amount of HDAC3 in N-19 immunoprecipitates (lane 6) is likely to be overestimated due to co-migration with the immunoglobulin heavy chains, which were weakly detected (lane 5).
To test whether these proteins can interact in living cells, we performed co-immunoprecipitation experiments on transfected cells (Fig. 3B). We found that HA–RbAp48 was present in the flag immunoprecipitates only when a Flag–HDAC1 or Flag–HDAC3 expression vector was included in the transfection (lanes 3 and 5), but not in their absence (lane 1). This difference was not due to a difference in the expression levels of exogenous RbAp48 (compare lanes 2, 4 and 6). Thus, this result indicates that RbAp48 interacts with HDAC3 in transfected cells.
We also tested co-immunoprecipitation of endogenous proteins. Immunoprecipitation of RbAp48 with an anti-RbAp48 antibody (RBBP; Transduction Laboratories) led to specific co-immunoprecipitation of HDAC3, indicating that both proteins are physically associated (Fig. 3C, left). HDAC1 and HDAC2 were also detected in RBBP immunoprecipitates at longer exposures (data not shown). Co-immunoprecipitation of HDAC3 and RbAp48 was confirmed by immunoprecipitation using other anti-RbAp48 antibodies (middle and right), although in these cases less HDAC3 was co-immunoprecipitated compared to HDAC1 and HDAC2 (Fig. 3C). This discrepancy could reflect a difference in the accessibility of the various epitopes depending on the multimolecular complex. Alternatively, strong co-immunoprecipitation of HDAC3 using antibody RBBP could be due to RbAp46, a protein which is highly related to RbAp48 and which is recognised by this antibody. Whatever the explanation, these results indicate that endogenous HDAC3 and RbAp48 are present within the same multimolecular complex.
DISCUSSION
In this paper we show that: (i) RbAp48 is required for transcriptional repression of E2F-regulated promoters; (ii) HDAC3 interacts physically with RbAp48; (iii) HDAC3 shares with HDAC1 the ability to mediate Rb–RbAp48 interaction.
These data stand in contrast to what was found in a previous study (20), which could not detect any physical interaction between HDAC3 and RbAp48. This discrepancy could reflect the different cell type used in their experiments compared to ours. Our results (Fig. 3C) indeed indicate that most of the endogenous HDAC3 is likely to be free of RbAp48. However, our results show that HDAC3 very efficiently mediates formation of a ternary complex with Rb and RbAp48 (Fig. 1). The direct Rb–RbAp48 interaction, previously shown in vitro (26), could help to stabilise this ternary complex.
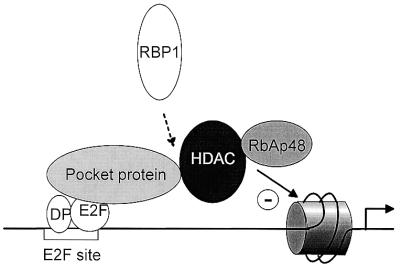
Taken together, our data indicate that HDAC3 has the ability to bind RbAp48 and to mediate recruitment of RbAp48 to Rb. We did not detect any interaction in vitro between Rb and HDAC3 (data not shown), suggesting that HDAC3 interacts with Rb through another protein, which has been proposed to be RBP1 (19). Also, contrary to what was previously found using purified proteins (26), we found no evidence for direct contacts between RbAp48 and Rb (data not shown). This led us to propose a model (Fig. 4) in which HDAC1, HDAC2 or HDAC3 is targeted to E2F-regulated promoters through either a direct (HDAC1 or HDAC2) or indirect (HDAC3) interaction with Rb. In turn, these three proteins recruit RbAp48 protein.
Figure 4.
Model of transcriptional repression by Rb through the recruitment of histone deacetylases. In G0 or during the beginning of the G1 phase of the cell cycle Rb protein (or one of its cousins, ‘Pocket protein’ in the figure) binds to E2F sites (15). It recruits a histone deacetylase (‘HDAC’) through a direct (HDAC1 or HDAC2) or indirect (HDAC3, through RBP1) interaction. These three deacetylases share the ability to recruit the histone-binding protein RbAp48, leading to deacetylation of histones present on the promoter.
If this model is correct, all three histone deacetylases could be involved in transcriptional repression by Rb protein. In transient transfection experiments, we did not detect any effect of expression of exogenous HDAC3 on transcriptional repression by Rb (data not shown). In similar experiments, HDAC1 has been shown to cooperate with Rb (12). This could suggest that HDAC1 is more efficient as a co-repressor for Rb than HDAC3. An alternative explanation could be that HDAC3 is not limiting in this type of experiment. The relative contributions of these histone deacetylases to transcriptional control of E2F-regulated promoters is presently unknown and might await the availability of knock-out mice. However, recent results suggest that HDAC3 plays a prominent role in transcriptional repression of specific promoters, since it is a major component of the N-CoR and SMRT complexes (27–30). Furthermore, HDAC3 was recently shown to be present in the same complex as members of the SWI–SNF complex (31). Strikingly, the catalytic subunits of the SWI–SNF complex, BRG1 and hbrm, are involved in transcriptional repression by Rb (32,33).
Our results (Table 1) indicate that RbAp48 is required for transcriptional repression of E2F-regulated promoters. To our knowledge, our data are the first direct evidence showing that RbAp48 is involved in transcriptional repression. What could be the role of RbAp48? RbAp48 interacts directly with histone H4, suggesting that it could direct histone deacetylases to histones (17,18). It could be envisioned that the presence of RbAp48 increases the efficiency of HDAC3-mediated deacetylation. Unfortunately, purified recombinant HDAC3 does not harbour any histone deacetylase activity, making this interesting hypothesis difficult to test practically.
Nevertheless, RbAp48 is found in many chromatin-related complexes (34–37). Thus, although a role of RbAp48 in the deacetylation of nucleosomal histones has never been proven, its presence suggests that histones are bona fide substrates of histone deacetylase complexes. The fact that RbAp48 can be recruited through a physical interaction with HDAC3 (Fig. 3) argues against the existence of a Rb-associated deacetylase complex that would not be targeted to histones and that would deacetylate only non-histone proteins. Consistent with that, targeting Rb to a transiently transfected DNA is sufficient to induce histone deacetylation (10). Furthermore, histones present within E2F-regulated promoters are hypoacetylated when the Rb-related protein p130 is bound to the promoter (15).
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Drs A. Harel-Bellan, E. Seto, S. L. Schreiber and H. Richard-Foy for the kind gift of materials. This work was supported by a grant from the Ligue Nationale Contre le Cancer to D.T., as an Equipe Labellisée. E.N. is the recipient of a studentship from the MENRT.
References
- 1.Muller H. and Helin,K. (2000) The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta, 1470, M1–M12. [DOI] [PubMed] [Google Scholar]
- 2.Black A.R. and Azizkhan-Clifford,J. (1999) Regulation of E2F: a family of transcription factors involved in proliferation control. Gene, 237, 281–302. [DOI] [PubMed] [Google Scholar]
- 3.Kaelin W.G.Jr (1999) Functions of the retinoblastoma protein. Bioessays, 21, 950–958. [DOI] [PubMed] [Google Scholar]
- 4.Krek W., Xu,G. and Livingston,D.M. (1995) Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell, 83, 1149–1158. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H.S., Postigo,A.A. and Dean,D.C. (1999) Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta and contact inhibition. Cell, 97, 53–61. [DOI] [PubMed] [Google Scholar]
- 6.Field S.J., Tsai,F.Y., Kuo,F., Zubiaga,A.M., Kaelin,W.G.Jr, Livingston,D.M., Orkin,S.H. and Greenberg,M.E. (1996) E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell, 85, 549–561. [DOI] [PubMed] [Google Scholar]
- 7.Yamasaki L., Jacks,T., Bronson,R., Goillot,E., Harlow,E. and Dyson,N.J. (1996) Tumor induction and tissue atrophy in mice lacking E2F-1. Cell, 85, 537–548. [DOI] [PubMed] [Google Scholar]
- 8.Hagemeier C., Cook,A. and Kouzarides,T. (1993) The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res., 21, 4998–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weintraub S.J., Chow,K.N., Luo,R.X., Zhang,S.H., He,S. and Dean,D.C. (1995) Mechanism of active transcriptional repression by the retinoblastoma protein. Nature, 375, 812–815. [DOI] [PubMed] [Google Scholar]
- 10.Luo R.X., Postigo,A.A. and Dean,D.C. (1998) Rb interacts with histone deacetylase to repress transcription. Cell, 92, 463–473. [DOI] [PubMed] [Google Scholar]
- 11.Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- 12.Magnaghi-Jaulin L., Groisman,R., Naguibneva,I., Robin,P., Lorain,S., Le Villain,J.P., Troalen,F., Trouche,D. and Harel-Bellan,A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- 13.Davie J.R. and Spencer,V.A. (1999) Control of histone modifications. J. Cell. Biochem., 32/33 (suppl.), 141–148. [DOI] [PubMed] [Google Scholar]
- 14.Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi Y., Rayman,J.B. and Dynlacht,B.D. (2000) Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev., 14, 804–816. [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolas E., Morales,V., Magnaghi-Jaulin,L., Harel-Bellan,A., Richard-Foy,H. and Trouche,D. (2000) RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J. Biol. Chem., 275, 9797–9804. [DOI] [PubMed] [Google Scholar]
- 17.Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell, 87, 95–104. [DOI] [PubMed] [Google Scholar]
- 18.Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1998) Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol., 8, 96–108. [DOI] [PubMed] [Google Scholar]
- 19.Lai A., Lee,J.M., Yang,W.M., DeCaprio,J.A., Kaelin,W.G.Jr, Seto,E. and Branton,P.E. (1999) RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol. Cell. Biol., 19, 6632–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emiliani S., Fischle,W., Van Lint,C., Al-Abed,Y. and Verdin,E. (1998) Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl Acad. Sci. USA, 95, 2795–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W.M., Yao,Y.L., Sun,J.M., Davie,J.R. and Seto,E. (1997) Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem., 272, 28001–28007. [DOI] [PubMed] [Google Scholar]
- 22.Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- 23.Ait-Si-Ali S., Polesskaya,A., Filleur,S., Ferreira,R., Duquet,A., Robin,P., Vervish,A., Trouche,D., Cabon,F. and Harel-Bellan,A. (2000) CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene, 19, 2430–2437. [DOI] [PubMed] [Google Scholar]
- 24.Harbour J.W. and Dean,D.C. (2000) Chromatin remodeling and Rb activity. Curr. Opin. Cell Biol., 12, 685–689. [DOI] [PubMed] [Google Scholar]
- 25.Hassig C.A., Tong,J.K., Fleischer,T.C., Owa,T., Grable,P.G., Ayer,D.E. and Schreiber,S.L. (1998) A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl Acad. Sci. USA, 95, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian Y.W., Wang,Y.C., Hollingsworth,R.E.Jr, Jones,D., Ling,N. and Lee,E.Y. (1993) A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature, 364, 648–652. [DOI] [PubMed] [Google Scholar]
- 27.Wen Y.D., Perissi,V., Staszewski,L.M., Yang,W.M., Krones,A., Glass,C.K., Rosenfeld,M.G. and Seto,E. (2000) The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl Acad. Sci. USA, 97, 7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urnov F.D., Yee,J., Sachs,L., Collingwood,T.N., Bauer,A., Beug,H., Shi,Y.B. and Wolffe,A.P. (2000) Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-erbA yields a chromatin infrastructure-dependent transcriptional repression pathway. EMBO J., 19, 4074–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenther M.G., Lane,W.S., Fischle,W., Verdin,E., Lazar,M.A. and Shiekhattar,R. (2000) A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev., 14, 1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Wang,J., Nawaz,Z., Liu,J.M., Qin,J. and Wong,J. (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J., 19, 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Underhill C., Qutob,M.S., Yee,S.P. and Torchia,J. (2000) A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem., 275, 40463–40470. [DOI] [PubMed] [Google Scholar]
- 32.Trouche D., Le Chalony,C., Muchardt,C., Yaniv,M. and Kouzarides,T. (1997) RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl Acad. Sci. USA, 94, 11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H.S., Gavin,M., Dahiya,A., Postigo,A.A., Ma,D., Luo,R.X., Harbour,J.W. and Dean,D.C. (2000) Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb- hSWI/SNF and Rb-hSWI/SNF. Cell, 101, 79–89. [DOI] [PubMed] [Google Scholar]
- 34.Parthun M.R., Widom,J. and Gottschling,D.E. (1996) The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell, 87, 85–94. [DOI] [PubMed] [Google Scholar]
- 35.Tyler J.K., Bulger,M., Kamakaka,R.T., Kobayashi,R. and Kadonaga,J.T. (1996) The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol., 16, 6149–6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Sun,Z.W., Iratni,R., Erdjument-Bromage,H., Tempst,P., Hampsey,M. and Reinberg,D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell, 1, 1021–1031. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Balbas M.A., Tsukiyama,T., Gdula,D. and Wu,C. (1998) Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl Acad. Sci. USA, 95, 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]