ABSTRACT
The potential diagnostic value of chemiluminescence immunoassays (CLIAs) has been accepted in recent years, although their use for foot-and-mouth disease (FMD) diagnostics has not been reported. Full-length 3ABC and 2C proteins were expressed in bacteria and purified by affinity chromatography to develop a rapid and accurate approach to distinguish pigs infected with foot-and-mouth disease virus (FMDV) from vaccinated pigs. The recombinant proteins were then used as antigens to develop two CLIAs for the detection of antibodies against nonstructural viral proteins. The diagnostic performance of the two assays was compared by analyzing serum from pigs (naive pigs, n = 63; vaccinated, uninfected pigs, n = 532; naive, infected pigs, n = 117) with a known infection status. The 3ABC-2C CLIA had a higher accuracy rate, with a diagnostic sensitivity of 100% and a diagnostic specificity of 96.5%, than the 3ABC CLIA, which had a diagnostic sensitivity of 95.7% and a diagnostic specificity of 96.0%. The results of the 3ABC-2C CLIA also had a high rate of concordance with those of two commercial FMDV enzyme-linked immunosorbent assay (ELISA) kits used to assess serum collected from 962 pigs in the field (96.2% and 97.8%, respectively). The 3ABC-2C CLIA detected infection in serum samples from infected pigs earlier than the commercial ELISA kits. In addition, the 3ABC-2C CLIA produced results within 15 min. On the basis of these findings, the 3ABC-2C CLIA could serve as the foundation for the development of penside FMD diagnostics and offers an alternative method to detect FMDV infections.
KEYWORDS: 2C, 3ABC, chemiluminescence immunoassay, diagnosis, foot-and-mouth disease virus
INTRODUCTION
Foot-and-mouth disease (FMD) is a severe infectious illness that afflicts cloven-hoofed animals. The disease is caused by foot-and-mouth disease virus (FMDV), which belongs to the Aphthovirus genus of the Picornaviridae family (1). FMDV contains a positive-sense, single-stranded RNA genome that is translated into a polyprotein and is then cleaved by proteases into 4 structural proteins (SPs; VP1, VP2, VP3, and VP4) that form the viral capsid and 10 nonstructural proteins (NSPs; L, 2A, 2B, 2C, 3A, 3B, 3C, 3D, 3AB, and 3ABC) that play key roles in viral replication and host cell interactions (2–4).
Several diagnostic methods, such as virus isolation, reverse transcriptase PCR, a mucosal IgA enzyme-linked immunosorbent assay (ELISA), and NSP-based ELISAs, have been used to differentiate FMDV-infected animals from vaccinated animals (5). Among these methods, NSP-based ELISAs are the most widely used technique because they can detect seven different serotypes and discriminate infected from vaccinated animals (2, 6). Antibodies against NSPs 3ABC and 2C are the most important indicators of an FMDV infection. The 2C protein remains membrane bound and is not present in purified vaccines (7); furthermore, antibodies against NSP 2C are induced early after FMDV infection (8). Therefore, the ability to detect antibodies against 2C is helpful for diagnosing early infections (9). The presence of the 3ABC protein is the most reliable single indicator of infection because it produces abundant antibodies and persists in the body for a longer time than the other NSPs (10, 11). The presence of antibodies against the 2C protein and, to a lesser extent, the 3ABC protein has been used to distinguish potential carrier animals in the convalescent phase from vaccinated animals (7, 12). Diagnosis would be more accurate if researchers could simultaneously detect antibodies against both proteins.
The chemiluminescence immunoassay (CLIA) was developed in the late 1970s (13). In recent years, CLIA has been shown to perform extremely well in diagnosing human diseases, such as human immunodeficiency virus (HIV) infection (14, 15), cancer (based on the detection of tumor markers) (16), and hepatitis B (based on the detection of hepatitis B virus surface antigen [HBsAg]) (17, 18), due to its high sensitivity, high signal-to-noise ratio, and wide linear range. To date, no reports of studies that used CLIA to detect FMDV infections have been published.
We developed a CLIA platform for the simultaneous detection of antibodies against NSPs 2C and 3ABC to evaluate the performance of CLIAs in distinguishing FMDV-infected pigs from vaccinated pigs. The results showed that the CLIAs have a higher sensitivity and a shorter detection time than currently available ELISAs and may serve as a basis for the development of accurate and rapid penside FMD diagnostics.
RESULTS
Expression and characterization of the rmu3ABC and r2C proteins.
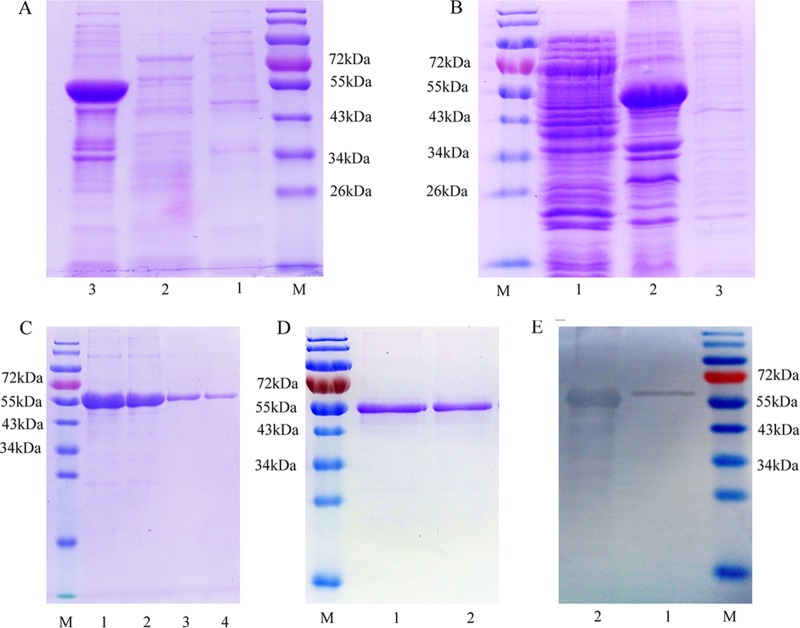
The His-tagged recombinant mutated 3ABC (rmu3ABC) protein was expressed in insoluble inclusion bodies and had an apparent molecular mass of 55 kDa on SDS-polyacrylamide gels (Fig. 1A). The His-tagged recombinant r2C (r2C) protein was also expressed in inclusion bodies and had an apparent molecular mass of 55 kDa (Fig. 1B). After washing and nickel affinity chromatography purification, we obtained highly pure rmu3ABC and r2C proteins (Fig. 1C and D). According to the results of the Western blot analysis, the FMDV-infected positive serum sample produced a strong signal for both purified proteins (Fig. 1E). In addition, the purified 3ABC and 2C proteins showed a good power to discriminate serum samples that were positive and weakly positive in an indirect ELISA, whereas the negative serum sample displayed almost no reactivity. On the basis of these results, the rmu3ABC and r2C proteins are suitable for use for detecting antibodies against NSPs from FMDV.
FIG 1.
(A) SDS-PAGE profile of the expressed rmu3ABC protein. Lane M, prestained protein ladder (Thermo Scientific, USA); lane 1, uninduced E. coli cell extract; lane 2, soluble protein fraction; lane 3, insoluble protein fraction. (B) SDS-PAGE profile of the expressed r2C protein. Lane M, prestained protein ladder; lane 1, soluble protein fraction; lane 2, insoluble protein fraction; lane 3, uninduced E. coli cell extract. (C) SDS-PAGE profile of the purified rmu3ABC protein determined using nickel affinity chromatography. Lane M, prestained protein ladder; lane 1, the proteins after washes with IB washing buffer; lane 2, unbound protein; lanes 3 and 4, the eluted purified protein. (D) SDS-PAGE profile showing the r2C protein affinity purified using Ni-NTA resin. Lane M, prestained protein ladder; lanes 1 and 2, the eluted purified 2C protein. (E) Western blots showing the reactivity of rmu3ABC and r2C proteins with sera from FMDV-infected animals. Lane M, prestained protein ladder; lane 1, affinity-purified rmu3ABC protein; lane 2, affinity-purified r2C protein.
Standardization of the CLIA.
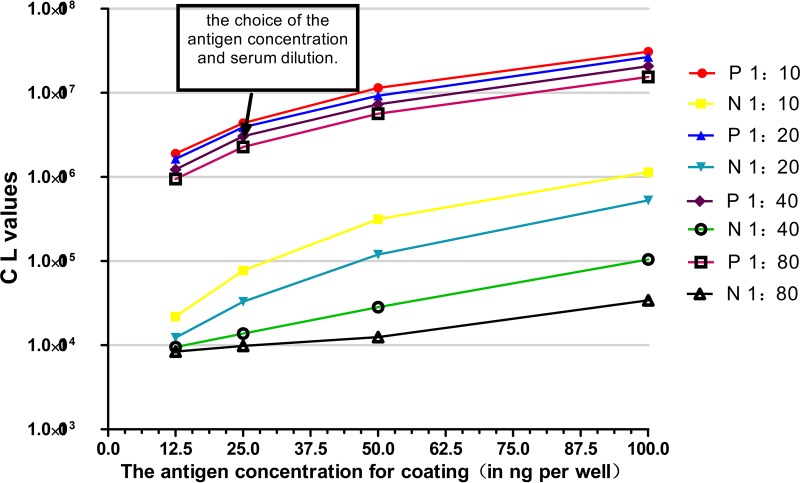
The optimum rmu3ABC protein coating concentration and the serum dilution were determined using a checkerboard titration method (Fig. 2). The concentration of the rmu3ABC antigen was fixed at 25 ng/well, and the test serum dilution was fixed at 1:40 to ensure that an acceptable signal-to-noise ratio was achieved and the chemiluminescence (CL) value of standard negative-control serum sample was less than 15,000. The 96-well CLIA plates were simultaneously coated with 25 ng/well r2C protein and 25 ng/well rmu3ABC protein to improve the diagnostic accuracy; the other detection conditions were the same as those used for the 3ABC CLIA.
FIG 2.
Results of checkerboard titration method used to optimize the rmu3ABC protein concentration and serum dilution. Standard positive and negative serum samples were serially diluted 2-fold, and the coating concentration was varied. An rmu3ABC protein coating concentration of 25 ng/well and a serum dilution of 1:40 were considered the best conditions. P, positive serum sample; N, negative serum sample.
Determining the cutoff value, Dsn, and Dsp of the 3ABC CLIA and 3ABC-2C CLIA.
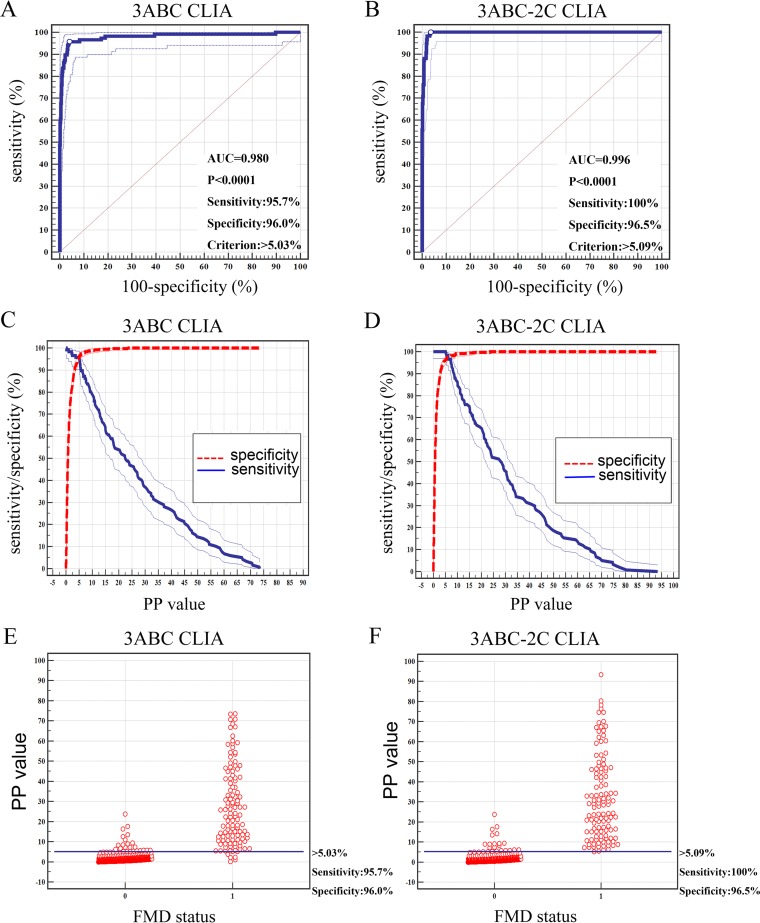
The percent positive (PP) values for the 712 serum samples collected from the pigs of known status (NSP-negative serum samples, n = 595; NSP-positive serum samples, n = 117) were used to estimate the cutoff value, diagnostic specificity (Dsp), and diagnostic sensitivity (Dsn) by analyzing the receiver operating characteristic (ROC) curve, the two-graph ROC (TG-ROC), and the interactive dot diagram (Fig. 3). The diagnostic performance of the 3ABC CLIA was compared with that of the 3ABC-2C CLIA. On the basis of the results of the ROC curve analysis, the cutoff value of the 3ABC CLIA was a PP value of 5.03%, which yielded a Dsn of 95.7% and a Dsp of 96.0%. The area under the curve (AUC) was 0.979 (standard error [SE] = 0.00878), with a 95% confidence interval of 0.966 to 0.988 and a Youden index value of 0.902. For the 3ABC-2C CLIA, the ROC curve analysis revealed a Dsn of 100% and a Dsp of 96.5% at the cutoff value of 5.09%, with an AUC of 0.996 (SE = 0.00144) and a 95% confidence interval of 0.988 to 0.999. The Youden index value was 0.965. Thus, the two CLIAs have good discriminatory powers. Moreover, based on the results of the ROC curve analysis, we concluded that the 3ABC-2C CLIA has a higher accuracy than the 3ABC CLIA.
FIG 3.
Determination of the cutoff values of the 3ABC CLIA and the 3ABC-2C CLIA. (A and B) Each point on the ROC plot represents a sensitivity-specificity pair corresponding to a particular decision threshold. (C and D) Plot-versus-criterion values. The 95% confidence intervals of Dsn and Dsp are plotted against the different criterion values. (E and F) Interactive dot diagram. 0, NSP-negative serum samples; 1, NSP-positive serum samples.
Comparison of the Ase of the 3ABC-2C CLIA with that of two commercial ELISA kits.
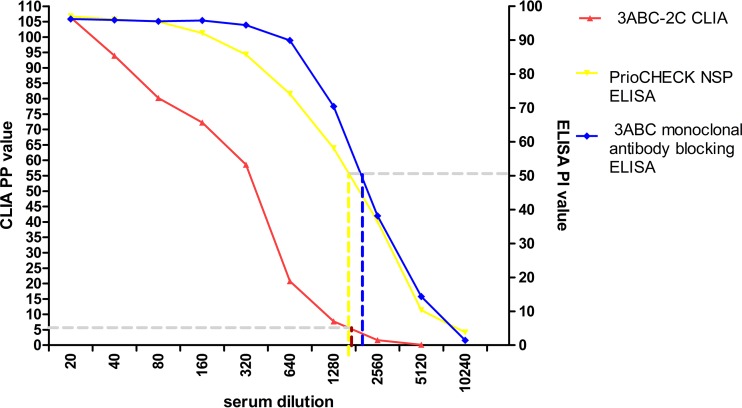
The detection limit for the standard positive serum using the 3ABC-2C CLIA was 1:2,560; the detection limit was also 1:2,560 for both the PrioCHECK FMDV NSP ELISA kit and the 3ABC monoclonal antibody blocking ELISA kit (Fig. 4). Therefore, with incubation times of 5 min each for the serum and the labeling antibody, the 3ABC-2C CLIA had the same analytical sensitivity (Ase) as the two commercial ELISA kits.
FIG 4.
The Ase of the 3ABC-2C CLIA was determined using 2-fold serially diluted standard positive pig serum samples and compared to that of the 3ABC monoclonal antibody blocking ELISA and the PrioCHECK FMDV NSP ELISA.
Estimation of repeatability and stability of the 3ABC-2C CLIA.
The intrarun coefficient of variation (CV) for the 3ABC-2C CLIA varied from 1.04% to 10.1%, and the interrun CV varied from 3.71% to 14.6%. As all CV values were <20%, the assay showed good reproducibility (Table 1).
TABLE 1.
Estimated precision of the 3ABC-2C CLIA using a panel of seven serum sample standards
| Serum sample | Intrarun reproducibility |
Interrun reproducibility |
||||
|---|---|---|---|---|---|---|
| Meana | SD | CVb | Meanc | SD | CV | |
| 1 | 129.00 | 3.21 | 2.49 | 137.85 | 9.01 | 6.53 |
| 2 | 85.70 | 5.70 | 6.65 | 86.95 | 3.23 | 3.71 |
| 3 | 26.40 | 0.27 | 1.04 | 27.21 | 1.47 | 5.43 |
| 4 | 12.00 | 0.15 | 1.32 | 13.05 | 0.94 | 7.21 |
| 5 | 9.85 | 0.37 | 3.83 | 10.55 | 1.06 | 10.04 |
| 6 | 5.05 | 0.50 | 10.10 | 5.61 | 0.54 | 9.77 |
| 7 | 0.68 | 0.01 | 2.58 | 0.63 | 0.09 | 14.60 |
Average percent positive for three replicates on the same plate.
CV, coefficient of variation.
Average percent positive for three replicates of detection on three different plates on different days.
Both the standard positive serum samples and the standard negative serum samples were tested on CLIA plates coated with rmu3ABC and r2C proteins that had been stored at 37°C for 15 days or at 4°C for 180 days. The CL values of standard positive serum samples exceeded 500,000, and the PP values for the negative serum samples did not exceed 1%. Therefore, the results met the validity index for the test. On the basis of these results, the CLIA plates coated with rmu3ABC and r2C can be stored for up to 15 days at 37°C and up to 180 days at 4°C without compromising performance.
Evaluation of diagnostic performance.
When the 595 serum samples from the animals with a known NSP-negative status were tested, the accuracy rates of the two CLIAs and the two commercial ELISA kits were all greater than 92%, with the exception of the 3ABC monoclonal antibody blocking ELISA kit, which had a lower accuracy rate of 88.9% for the naive serum samples. For the 117 NSP-positive serum samples, the two commercial ELISA kits had high accuracy rates of 90.6% (106/117) and 93.2% (109/117), respectively. However, the 3ABC-2C CLIA had the highest accuracy rate at 100% (117/117). For the serum samples collected from the 962 animals in the field, the overall concordance between the 3ABC-2C CLIA and the PrioCHECK FMDV NSP ELISA was estimated to be 97.8% (941/962 serum samples), and a concordance rate of 96.2% (925/962 serum samples) was obtained for the 3ABC-2C CLIA and the 3ABC monoclonal antibody blocking ELISA (Table 2).
TABLE 2.
Comparison of diagnostic performance of the 3ABC-2C CLIA with that of the 3ABC CLIA, 3ABC monoclonal antibody blocking ELISA, and PrioCHECK FMDV NSP ELISAa
| Sample source | Total no. of samples | 3ABC-2C CLIA |
3ABC CLIA |
3ABC ELISA |
PrioCHECK FMDV NSP ELISA |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of samples positive/total no. tested | No. of samples negative/total no. tested | Accuracy rate (%) | No. of samples positive/total no. tested | No. of samples negative/total no. tested | Accuracy rate (%) | No. of samples positive (concordant)/total no. tested | No. of samples negative (concordant)/total no. tested | Accuracy rate (%) | No. of samples positive (concordant)/total no. tested | No. of samples negative (concordant)/total no. tested | Accuracy rate (%) | ||
| Naive pigs | 63 | 5/63 | 58/63 | 92.1 | 5/63 | 58/63 | 92.1 | 7/63 | 56/63 | 88.9 | 3/63 | 60/63 | 95.2 |
| Vaccinated, uninfected pigs | 532 | 16/532 | 516/532 | 97.0 | 19/532 | 513/532 | 96.4 | 22/532 | 510/532 | 95.9 | 12/532 | 520/532 | 97.7 |
| Naive, infected pigs | 117 | 117/117 | 0/117 | 100 | 112/117 | 5/117 | 95.7 | 106/117 | 11/117 | 90.6 | 109/117 | 8/117 | 93.2 |
| Pigs in the field | 962 | 53/962 | 909/962 | 78 (47)/962 | 884 (878)/962 | 50 (41)/962 | 912 (900)/962 | ||||||
The rate of concordance of the results of the 3ABC-2C CLIA with those of the 3ABC ELISA was 96.2% (925/962 serum samples). The rate of concordance of the results of the 3ABC-2C CLIA with those of the PrioCHECK FMDV NSP ELISA was 97.8% (941/962 serum samples).
Comparison of early diagnostic performance.
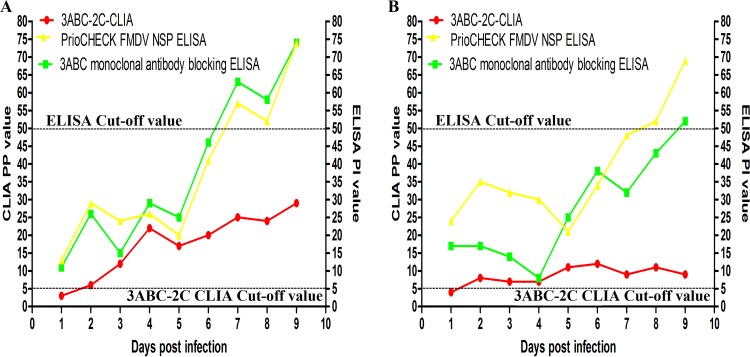
Two panels of serum samples collected between 1 day postinfection (dpi) and 9 dpi from 2 pigs infected with FMDV were used to determine the early diagnostic performance of the 3ABC-2C CLIA and the two commercial ELISA kits. The 3ABC-2C CLIA detected positive signals for the antibodies to NSPs of FMDV in sera from the 2 pigs at 2 dpi. However, antibodies to NSPs of FMDV were not detected by the two commercial ELISA kits until 7 dpi to 9 dpi (Fig. 5). Therefore, the 3ABC-2C CLIA generated an accurate diagnosis earlier than the two commercial ELISA kits.
FIG 5.
Comparison of the diagnostic performance in the early FMDV infection phase. (A) Results for a naive pig challenged with the A/GDMM/CHA/2013 FMDV strain; (B) results for a naive pig challenged with the O/China/99 FMDV strain.
DISCUSSION
Progress has been achieved in developing new and rapid penside FMD diagnostics in recent years. The development of reverse transcription–loop-mediated isothermal amplification (RT-LAMP) has been a big breakthrough for the molecular detection of FMD. RT-LAMP distinguishes FMDV serotypes (19, 20) and offers a rapid means of early field detection of FMD at the penside (21). However, this method also has a high false-positive rate, which is a serious limitation. Infrared thermography and analyses of air samples have also been used to detect early and subclinical infections, but these methods are not highly accurate (22, 23). Therefore, a rapid and sensitive method for the diagnosis of FMDV infection is needed.
There are three main challenges associated with the diagnosis of FMDV infection: (i) contamination of the inactivated vaccine with NSP is a substantial challenge for the differentiation of FMDV-infected animals from vaccinated animals, (ii) animals infected with FMDV are not detected by the current detection methods in the early infection phase, and (iii) the time-consuming and complex operations required by currently available methods seriously restrict their clinical applications.
NSP contamination in inactivated vaccines confounds the detection of the true disease status (24). For a method with an extremely high analytical sensitivity, even very low concentrations of antibodies against NSP that are elicited by the inactivated vaccine would be detected, leading to false-positive results. The serum reaction time was limited to 5 min to ensure that the Ase of the 3ABC-2C CLIA was similar to that of the commercial ELISA kits. However, if the detection time was prolonged, the Ase of the 3ABC-2C CLIA would be expected to improve. One solution to the problem of NSP contamination is the development of new vaccines for FMD. For example, a newly developed epitope-based vaccine does not appear to contain any NSP components (25). The 3ABC-2C CLIA would be a suitable assay to test serum from animals vaccinated with non-NSP-containing vaccines.
Notably, animals exposed to infection do not seroconvert against every NSP, and seroconversion occurs at a lower concentration in the early infection phase (26–28). An international study validating FMD antibody tests (26, 29) recommended the use of tests that detect antibodies to more than one NSP to increase diagnostic accuracy. Considering the high nonspecificity of the CLIA when using proteins expressed in bacteria, in preliminary experiments, we utilized two synthetic peptides based on the 2C and 3B epitopes to decrease the number of false-positive results generated by proteins produced in Escherichia coli. CLIA plates were simultaneously coated with peptides derived from 2C and 3B to detect sera with a known status; however, the rate of concordance of the results with those for the reference serum samples was low (data not shown). In contrast, the results of the 3ABC-2C CLIA and the 3ABC CLIA showed high rates of concordance with those of the commercial ELISA kits when the reference serum samples were tested. Notably, the 3ABC-2C CLIA detected NSP antibody-positive serum samples from the FMDV-infected pigs (2 dpi) earlier than the two ELISA methods (7 dpi to 9 dpi). However, the results obtained from the 3ABC CLIA and 2C CLIA (using the r2C protein as the coating antigen) were negative when serum samples were assessed at 2 dpi. These results may be attributed to the observation that antibodies against both the 2C and 3ABC proteins bind to the rmu3ABC and r2C proteins in the 3ABC-2C CLIA and, consequently, produced positive results. In addition, the positive signal obtained from the 2C-3AB ELISA (using the recombinant 2C-3AB protein as the coating antigen) was significantly higher than the signal obtained from the 3ABC ELISA for detecting infected animals in a previous study (9). Based on these results, we conclude that the simultaneous use of antibodies against 2C and to 3ABC improves the accuracy of detecting FMDV infection during the early phase of infection.
Detection methods that require lengthy incubation steps would limit the timely implementation of protective measures, particularly in regions with a high suspicion of FMDV infections, possibly resulting in outbreaks of FMD. The 3ABC-2C CLIA required only 15 min to obtain results, whereas the 3ABC monoclonal antibody blocking ELISA required 3.5 h and the PrioCHECK NSP FMDV ELISA required a full day. Although the detection time for the CLIA is extremely short, the Ase was not compromised and was similar to that of the two commercial ELISA kits. In addition, the overall cost of this newly developed CLIA was similar to that of the commercial ELISA methods, as the reagents and the luminometer used for the CLIA are not expensive. Therefore, the CLIA method may be able to replace ELISA for the diagnosis of FMD in the field.
CLIA-based methods are increasingly used in the clinical veterinary laboratory. In recent years, nanotechnology has been applied to enhance CLIA systems, further improving their sensitivity, and several CL probes have also been developed. The use of miniaturized and ultrasensitive charge-coupled devices in combination with CL probes may enable rapid, ultrasensitive, and high-throughput penside diagnosis (30). Therefore, the development of CLIA platforms for the diagnosis of FMD is expected to improve in the future, particularly if these modifications are adopted.
In conclusion, we established a rapid and accurate CLIA method for the detection of FMD that is capable of simultaneously identifying antibodies against 3ABC and 2C. Furthermore, the method required only 15 min to produce results, not including the coating time. On the basis of the results of preliminary assessments, the 3ABC-2C CLIA had high Dsp, Dsn, and Ase, as well as good repeatability and stability. Future studies will examine the design of CL probes to further the development of penside FMD diagnostics.
MATERIALS AND METHODS
NSP-negative serum samples.
Five hundred ninety-five FMDV NSP antibody-free serum samples were collected from clinically healthy pigs. Among these samples, 63 were collected from pigs with a known unvaccinated status; these did not contain antibodies to the O, A, or Asia1 FMDV SP (titer, <1:4), on the basis of the results of a liquid-phase blocking ELISA (LPBE) developed by Lanzhou Veterinary Research Institute (LVRI). Five hundred thirty-two samples (n = 532) were collected from 133 pigs that had been inoculated once with a trivalent inactivated vaccine (serotype A, O, and Asia1) at 7, 14, 21, and 28 days postvaccination. The antibody titers against SPs were known (1:16 to 1:256) in these samples. All of these samples (n = 595 [63 + 532]) were used to estimate the cutoff value and diagnostic specificity (Dsp).
NSP-positive serum samples.
Samples (n = 117, from 117 pigs) were collected from naive pigs that had been experimentally challenged with the O/China/99 FMDV strain at 9 days postinfection (dpi). All of these serum samples were collected and identified in 2010 and then stored at −20°C; they were used to estimate the cutoff value and diagnostic sensitivity (Dsn). All experiments were performed in the Animal Biological Safety Level 3 (ABSL-3) Laboratory at LVRI.
Serum samples from infected pigs.
Eighteen serum samples were collected from 2 pigs that had been experimentally challenged with the O/China/99 FMDV strain and the A/GDMM/CHA/2013 FMDV strain, respectively, every 24 h between 1 dpi and 9 dpi in the ABSL-3 Laboratory at LVRI. These samples were used to compare the performance of the 3ABC-2C CLIA, the 3ABC monoclonal antibody blocking ELISA from LVRI, China, and the PrioCHECK FMDV NSP ELISA for the early diagnosis of FMD. All procedures involving animals were approved by the Animal Ethics Committee of LVRI, Chinese Academy of Agricultural Sciences.
Collection of serum samples in the field.
Serum samples (n = 962) were collected from pigs in a region suspected to have experienced FMDV infections from 2010 to 2015. These samples were stored at the national FMD reference laboratory. The concordance between the results of the 3ABC-2C CLIA, the PrioCHECK NSP FMDV ELISA, and the 3ABC monoclonal antibody blocking ELISA kit was evaluated by assessing 962 randomly selected samples.
Plasmids and cloning.
Total RNA was extracted from the vesicle fluid of pigs infected with FMDV (A/GDMM/CHA/2013) using an RNeasy minikit (Qiagen, Hilden, Germany) and then reverse transcribed into cDNAs using Smart Moloney murine leukemia virus reverse transcriptase (TaKaRa, Japan) and oligo(dT) primers. The key protease active sites (amino acids 46 and 163) in the 3C protein must be modified to express the full-length 3ABC protein. Thus, a series of primers was designed according to the method described in a previous report (5). Briefly, the histidine at site 46 (CAC) was changed to tyrosine (TAC) using overlap extension PCR with two pairs of primers, 3ABC-F/3C-46-R and 3C-46-F/3ABC-R, for amplification (Table 3). Similarly, the cysteine at site 163 (TGT) was changed to glycine (GGT) using primers 3ABC-F/3C-163-R and 3C-163-F/3ABC-R (Table 3). After purification using an agarose gel DNA extraction kit (TaKaRa Japan), the full-length mutated 3ABC gene was cloned into a pProEXHTb (Novagen) plasmid using BamHI and XhoI restriction sites. For 2C cloning, we utilized a PrimeScript one-step reverse transcriptase PCR kit (TaKaRa Japan) to amplify the 2C gene, which was then cloned into a PET-SUMO (Novagen) plasmid using steps similar to those used in the method described above. We referred to the manufacturer's instructions for all cloning steps.
TABLE 3.
Primers used to amplify the 3ABC and 2C genes
| Primer name | Primer sequencea |
|---|---|
| 3ABC-F | 5′-CGGGATCCATCTCAATTCCTTCCCAAAAGTCC-3′ |
| 3ABC-R | 5′-CCGCTCGAGTCTCATGGTGTGGTTCGGGGT-3′ |
| 3C-46-F | 5′-CGTACCTCGTTACCTTTTCGC-3′ |
| 3C-46-R | 5′-GCGAAAAGGTAACGAGGTACG-3′ |
| 3C-163-F | 5′-AGGCTACGGTGGGGGAGC-3′ |
| 3C-163-R | 5′-GCTCCCCCACCGTAGCCT-3′ |
| 2C-R | 5′-CCGCTCGAGCTGCTTGAAAATCGGGTG-3′ |
| 2C-F | 5′-CGGGATCCCTCAAAGCACGTGACATCAA-3′ |
The restriction enzyme sites are underlined.
Expression, purification, and immunoreactivity of the recombinant proteins.
Chemically competent E. coli Rosetta(DE3) pLysS (Merck) cells were separately transformed with the recombinant 3ABC or 2C expression plasmid. Cultures of each clone were grown in 100 ml of LB medium and incubated at 37°C with shaking at 220 rpm in a shaker incubator until the optical density at 600 nm (OD600) was approximately 0.6. Then, protein expression was induced by treating the cultures with isopropyl-β-d-thiogalactoside (IPTG) at a final concentration of 1 mM, and the cells were further grown for 20 h at 16°C. Finally, the cells were harvested by centrifugation at 10,000 × g for 20 min. The recombinant 2C (r2C) protein and recombinant mutated 3ABC (rmu3ABC) protein were purified using the procedures described below. After ultrasonication, the cell pellet containing rmu3ABC was fully resuspended with inclusion body (IB) washing buffer (20 mM Tris, 500 mM NaCl, 1% Triton X-100, 10 mM EDTA, 6 M urea) and then centrifuged to remove the supernatant. This step was repeated 10 times to eliminate most vector proteins. The cell pellet containing r2C was washed with IB washing buffer (20 mM Tris, 500 mM NaCl, 1% Triton X-100, 10 mM EDTA, 2 M urea) 5 times. Inclusion bodies were solubilized with IB solubilization buffer (40 mM Na2PHO3, 500 mM NaCl, 8 M urea, and 20 mM β-mercaptoethanol) overnight at 4°C. Supernatants containing the protein of interest were further purified using nickel-nitrilotriacetic acid (Ni-NTA) metal affinity chromatography (GE Healthcare, Sweden) according to the manufacturer's instructions. The purity of the eluted proteins was measured using SDS-PAGE.
The immunoreactivity of the purified rmu3ABC and r2C proteins was tested by Western blotting and an indirect ELISA. For Western blotting, the purified rmu3ABC and r2C proteins were electrotransferred onto a nitrocellulose membrane at 200 mA for 2 h. After blocking in blocking buffer (phosphate-buffered saline [PBS] containing 5% skim milk and 0.01% Tween 20), high-titer FMDV-infected pig serum (collected at 60 dpi) was added at a 1:200 dilution, and the mixture was then incubated for 1 h at 37°C. After washing the membrane, a horseradish peroxidase-conjugated anti-pig IgG antibody (Sigma-Aldrich) was added at a dilution of 1: 2,000 and the mixture was incubated for 1 h at 37°C. Finally, diaminobenzidine (DAB; Sigma-Aldrich) was added to visualize the reaction. In addition, FMDV-infected pig serum and negative serum were tested in an indirect ELISA with the purified rmu3ABC and r2C proteins.
Development of the 3ABC CLIA.
High-titer FMDV-infected pig serum collected from experimentally infected pigs at 60 dpi served as a standard positive serum sample. The SP antibody titer in the serum was >1:1,024, based on an LPBE developed by LVRI, and the percent inhibition (PI) was 98%, according to the results obtained with the 3ABC monoclonal antibody blocking ELISA kit developed by LVRI. Serum from a clinically healthy naive pig was used as a standard negative serum sample. It had an SP antibody titer of <1:4 in the LPBE and a PI of 1%. These standard positive and negative serum samples served as internal controls. The optimum concentration for the rmu3ABC coating protein and the optimum serum dilution were determined by testing a series of rmu3ABC protein concentrations (100, 50, 25, and 12.5 ng/well) and 2-fold dilutions of the standard positive and negative serum samples (1:10, 1:20, 1:40, and 1:80) in a checkerboard titration format. Briefly, 96-well CLIA plates (C8 Lockwell Lumi white MaxiSorp; Thermo, Denmark) were coated with the rmu3ABC protein diluted in carbonate buffer (pH 9.6) and incubated at 4°C overnight. After 3 washes with PBS containing 0.01% Tween 20 (PBST), the test serum samples and the standard control positive and negative serum samples were diluted in serum dilution buffer (PBST, 1% casein, 10% horse serum, 1% E. coli lysate), and then 100 μl was transferred to each well of a coated plate, after which the plate was incubated at 37°C for 5 min. After washing, 100 μl of horseradish peroxidase-conjugated rabbit anti-pig immunoglobulin antibody (Sigma-Aldrich) was added at an appropriate concentration and the plate was incubated for 5 min at 37°C. Finally, after 5 washes, 50 μl each of solution A (luminol) and solution B (luminous enhancer) (Thermo Fisher Scientific, USA) was added. The reaction time was 5 min, and chemiluminescence (CL) signals were measured using a GloMax 96 microplate luminometer (Promega, USA).
Development of the 3ABC-2C CLIA.
After determination of the optimum concentration for the rmu3ABC protein coating (25 ng/well), 25 ng/well rmu3ABC proteins and 25 ng/well r2C proteins were simultaneously coated onto a 96-well CLIA plate. The remaining detection conditions were the same as those described above for the 3ABC CLIA. The mean CL values for the positive-control samples (mCLpos) and the test samples (mCLsample) were corrected by subtracting the mean CL value for the negative-control sample (mCLneg). The values for the test samples were expressed as percent positive (PP), which was determined using the following formula: PP = [(mCLsample − mCLneg) × 100]/(mCLpos − mCLneg).
The following indices had to be achieved to ensure the validity of the test: (i) the CL values of the standard positive serum samples had to exceed 500,000, and (ii) the PP values of the standard negative serum samples could not exceed 1%.
Cutoff value, Dsn, and Dsp.
Five hundred ninety-five NSP-negative serum samples collected from pigs were used to estimate the cutoff value and Dsp. Additionally, 117 NSP-positive serum samples were used to determine the cutoff value and Dsn using receiver operating characteristic (ROC) curve analysis with MedCalc software.
Ase, repeatability, and stability.
The analytical sensitivity (Ase) of the 3ABC-2C CLIA was determined using an endpoint dilution method. The standard positive pig control serum sample was serially diluted 2-fold over a range of 1:20 to 1:10,240. The detection limits of the 3ABC-2C CLIA and the commercial ELISA kits were compared.
Three replicates of seven serum samples of known status that represented various PI values ranging from 3% to 98% in the PrioCHECK FMDV NSP ELISA were run on the same plate and on three different plates on three different days to calculate the intrarun and within-laboratory interrun coefficients of variation (CVs).
After 200 μl of plate stabilizer (Sigma-Aldrich) was added to the CLIA plates coated with r2C and rmu3ABC and incubated for 30 min, the CLIA plates were placed at 37°C for 15 days and 4°C for 180 days. Then, the standard positive and negative serum samples were tested to identify the shelf life of the plates.
Comparison test.
The 3ABC monoclonal antibody blocking ELISA from LVRI, China, is the most widely used method for detecting FMDV infection in China. Additionally, the PrioCHECK FMDV NSP ELISA has seen widespread use worldwide. Both kits use a blocking format with fixed thresholds based on a PI rate of 50%. The diagnostic performance of the 3ABC-2C CLIA and the two commercial ELISA kits was compared by testing 712 serum samples from pigs of known status, as well as 962 serum samples from the field. Another 18 serum samples from 2 pigs challenged with FMDV collected at different time points (1 dpi to 9 dpi) were used to evaluate the diagnostic performance in the early infection phase. All serum samples were tested according to the manufacturer's instructions.
ACKNOWLEDGMENTS
This work was supported by grant no. 2015-4-20 from the Lanzhou Science and Technology Bureau (Science and Technology Bureau of Lanzhou) and grant no. 1604NKCA045 from the Key Science and Technology Foundation of Gansu Province.
We thank Guangqing Zhou and Xiumei Li for providing technical assistance with the CLIA methods, Shandian Gao for critically reading the manuscript, Jianliang Lv for providing the serum samples from animals with a known status, and the national FMD reference lab (China) for providing the serum samples from field animals.
Z.L. performed most of the experiments and wrote the manuscript. H.C., Y.Z., and J.S. designed the study, provided critical input on the experimental design, and assisted in preparing the manuscript. F.Z. and W.L. assisted with the experiments and data analysis. We all read and approved the final manuscript.
We have no competing interests to declare.
REFERENCES
- 1.Sharma GK, Mohapatra JK, Mahajan S, Matura R, Subramaniam S, Pattnaik B. 2014. Comparative evaluation of non-structural protein-antibody detecting ELISAs for foot-and-mouth disease sero-surveillance under intensive vaccination. J Virol Methods 207:22–28. doi: 10.1016/j.jviromet.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 2.Jaworski JP, Compaired D, Trotta M, Perez M, Trono K, Fondevila N. 2011. Validation of an r3AB1-FMDV-NSP ELISA to distinguish between cattle infected and vaccinated with foot-and-mouth disease virus. J Virol Methods 178:191–200. doi: 10.1016/j.jviromet.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Rueckert RR, Wimmer E. 1984. Systematic nomenclature of picornavirus proteins. J Virol 50:957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma LN, Zhang J, Chen HT, Zhou JH, Ding YZ, Liu YS. 2011. An overview on ELISA techniques for FMD. Virol J 8:419. doi: 10.1186/1743-422X-8-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma GK, Mohapatra JK, Pandey LK, Mahajan S, Mathapati BS, Sanyal A, Pattnaik B. 2012. Immunodiagnosis of foot-and-mouth disease using mutated recombinant 3ABC polyprotein in a competitive ELISA. J Virol Methods 185:52–60. doi: 10.1016/j.jviromet.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 6.Srisombundit V, Tungthumniyom N, Linchongsubongkoch W, Lekcharoensuk C, Sariya L, Ramasoota P, Lekcharoensuk P. 2013. Development of an inactivated 3C(pro)-3ABC (mu3ABC) ELISA to differentiate cattle infected with foot and mouth disease virus from vaccinated cattle. J Virol Methods 188:161–167. doi: 10.1016/j.jviromet.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Lubroth J, Brown F. 1995. Identification of native foot-and-mouth-disease virus nonstructural protein 2c as a serological indicator to differentiate infected from vaccinated livestock. Res Vet Sci 59:70–78. doi: 10.1016/0034-5288(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 8.Lubroth J, Grubman MJ, Burrage TG, Newman JFE, Brown F. 1996. Absence of protein 2C from clarified foot-and-mouth disease virus vaccines provides the basis for distinguishing convalescent from vaccinated animals. Vaccine 14:419–427. doi: 10.1016/0264-410X(95)00172-W. [DOI] [PubMed] [Google Scholar]
- 9.Lu Z, Zhang X, Fu Y, Cao Y, Tian M, Sun P, Li D, Liu Z, Xie Q. 2010. Expression of the major epitope regions of 2C integrated with the 3AB non-structural protein of foot-and-mouth disease virus and its potential for differentiating infected from vaccinated animals. J Virol Methods 170:128–133. doi: 10.1016/j.jviromet.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Robiolo B, Seki C, Fondevilla N, Grigera P, Scodeller E, Periolo O, La Torre J, Mattion N. 2006. Analysis of the immune response to FMDV structural and non-structural proteins in cattle in Argentina by the combined use of liquid phase and 3ABC-ELISA tests. Vaccine 24:997–1008. doi: 10.1016/j.vaccine.2005.08.071. [DOI] [PubMed] [Google Scholar]
- 11.Mackay DKJ, Forsyth MA, Davies PR, Berlinzani A, Belsham GJ, Flint M, Ryan MD. 1998. Differentiating infection from vaccination in foot-and-mouth disease using a panel of recombinant, non-structural proteins in ELISA. Vaccine 16:446–459. doi: 10.1016/S0264-410X(97)00227-2. [DOI] [PubMed] [Google Scholar]
- 12.Clavijo A, Wright P, Kitching P. 2004. Developments in diagnostic techniques for differentiating infection from vaccination in foot-and-mouth disease. Vet J 167:9–22. doi: 10.1016/S1090-0233(03)00087-X. [DOI] [PubMed] [Google Scholar]
- 13.Halmann M, Velan B, Sery T. 1977. Rapid identification and quantitation of small numbers of microorganisms by a chemiluminescent immunoreaction. Appl Environ Microbiol 34:473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso R, Roa PL, Suarez M, Bouza E. 2014. New automated chemiluminescence immunoassay for simultaneous but separate detection of human immunodeficiency virus antigens and antibodies. J Clin Microbiol 52:1467–1470. doi: 10.1128/JCM.03486-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui CJ, Liu P, Feng ZR, Xin RL, Yan CL, Li ZY. 2015. Evaluation of the clinical effectiveness of HIV antigen/antibody screening using a chemiluminescence microparticle immunoassay. J Virol Methods 214:33–36. doi: 10.1016/j.jviromet.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Kim J, Rho THD, Lee JH. 2014. Rapid chemiluminescent sandwich enzyme immunoassay capable of consecutively quantifying multiple tumor markers in a sample. Talanta 129:106–112. doi: 10.1016/j.talanta.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Song LW, Fang LL, Wu Y, Ge SX, Li H, Yuan Q, Zhang J, Xi NS. 2016. Evaluation of a novel chemiluminescent microplate enzyme immunoassay for hepatitis B surface antigen detection. J Virol Methods 228:55–59. doi: 10.1016/j.jviromet.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Ohne K, Kani S, Ohashi M, Shinkai N, Inoue T, Wakimoto Y, Tanaka Y. 2013. Clinical evaluation of a newly developed high-sensitive detection of hepatitis B virus surface antigen by a semi-automated immune complex transfer chemiluminescent enzyme immunoassay. Rinsho Byori 61:787–794. [PubMed] [Google Scholar]
- 19.Ding YZ, Zhou JH, Ma LN, Qi YN, Wei G, Zhang J, Zhang YG. 2014. A reverse transcription loop-mediated isothermal amplification assay to rapidly diagnose foot-and-mouth disease virus C. J Vet Sci 15:423–426. doi: 10.4142/jvs.2014.15.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki W, Mioulet V, Murray L, Madi M, Haga T, Misawa N, Horii Y, King DP. 2013. Development and evaluation of multiplex RT-LAMP assays for rapid and sensitive detection of foot-and-mouth disease virus. J Virol Methods 192:18–24. doi: 10.1016/j.jviromet.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Waters RA, Fowler VL, Armson B, Nelson N, Gloster J, Paton DJ, King DP. 2014. Preliminary validation of direct detection of foot-and-mouth disease virus within clinical samples using reverse transcription loop-mediated isothermal amplification coupled with a simple lateral flow device for detection. PLoS One 9:e105630. doi: 10.1371/journal.pone.0105630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gloster J, Ebert K, Gubbins S, Bashiruddin J, Paton DJ. 2011. Normal variation in thermal radiated temperature in cattle: implications for foot-and-mouth disease detection. BMC Vet Res 7:73. doi: 10.1186/1746-6148-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight-Jones TJD, Robinson L, Charleston B, Rodriguez LL, Gay CG, Sumption KJ, Vosloo W. 2016. Global foot-and-mouth disease research update and gap analysis: 4-diagnostics. Transbound Emerg Dis 63:42–48. doi: 10.1111/tbed.12523. [DOI] [PubMed] [Google Scholar]
- 24.Bergmann IE, Malirat V, Neitzert E, Beck E, Panizzutti N, Sanchez C, Falczuk A. 2000. Improvement of a serodiagnostic strategy for foot-and-mouth disease virus surveillance in cattle under systematic vaccination: a combined system of an indirect ELISA-3ABC with an enzyme-linked immunoelectrotransfer blot assay. Arch Virol 145:473–489. doi: 10.1007/s007050050040. [DOI] [PubMed] [Google Scholar]
- 25.Shao JJ, Wong CK, Lin T, Lee SK, Cong GZ, Sin FW, Du JZ, Gao SD, Liu XT, Cai XP, Xie Y, Chang HY, Liu JX. 2011. Promising multiple-epitope recombinant vaccine against foot-and-mouth disease virus type O in swine. Clin Vaccine Immunol 18:143–149. doi: 10.1128/CVI.00236-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brocchi E, Bergmann IE, Dekker A, Paton DJ, Sammin DJ, Greiner M, Grazioli S, De Simone F, Yadin H, Haas B, Bulut N, Malirat V, Neitzert E, Goris N, Parida S, Sorensen K, De Clercq K. 2006. Comparative evaluation of six ELISAs for the detection of antibodies to the non-structural proteins of foot-and-mouth disease virus. Vaccine 24:6966–6979. doi: 10.1016/j.vaccine.2006.04.050. [DOI] [PubMed] [Google Scholar]
- 27.Inoue T, Parida S, Paton DJ, Linchongsubongkoch W, Mackay D, Oh Y, Aunpomma D, Gubbins S, Saeki T. 2006. Development and evaluation of an indirect enzyme-linked immunosorbent assay for detection of foot-and-mouth disease virus nonstructural protein antibody using a chemically synthesized 2B peptide as antigen. J Vet Diagn Invest 18:545–552. doi: 10.1177/104063870601800604. [DOI] [PubMed] [Google Scholar]
- 28.Mohapatra AK, Mohapatra JK, Pandey LK, Sanyal A, Pattnaik B. 2014. Diagnostic potential of recombinant nonstructural protein 3B to detect antibodies induced by foot-and-mouth disease virus infection in bovines. Arch Virol 159:2359–2369. doi: 10.1007/s00705-014-2089-0. [DOI] [PubMed] [Google Scholar]
- 29.Paton DJ, de Clercq K, Greiner M, Dekker A, Brocchi E, Bergmann I, Sammin DJ, Gubbins S, Parida S. 2006. Application of non-structural protein antibody tests in substantiating freedom from foot-and-mouth disease virus infection after emergency vaccination of cattle. Vaccine 24:6503–6512. doi: 10.1016/j.vaccine.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 30.Roda A, Pasini P, Mirasoli M, Michelini E, Guardigli M. 2004. Biotechnological applications of bioluminescence and chemiluminescence. Trends Biotechnol 22:295–303. doi: 10.1016/j.tibtech.2004.03.011. [DOI] [PubMed] [Google Scholar]