ABSTRACT
The present study aimed to increase the processivity of Sulfolobus solfataricus DNA polymerase Dpo4. Protein engineering and bioinformatics were used to compile a library of potential Dpo4 mutation sites. Ten potential mutants were identified and constructed. A primer extension assay was used to evaluate the processivity of Dpo4 mutants. Thumb (A181D) and finger (E63K) domain mutants showed a processivity of 20 and 19 nucleotides (nt), respectively. A little finger domain mutant (I248Y) exhibited a processivity of 17 nt, only 1 nt more than wild-type Dpo4. Furthermore, the A181D mutant showed lower fidelity and higher nucleotide incorporation efficiency (4.74 × 10−4 s−1 μM−1) than E63K and I248Y mutants. When tasked with bypassing damage, the A181D mutant exhibited a 3.81-fold and 2.62-fold higher catalytic efficiency (kcat/Km) at incorporating dCTP and dATP, respectively, than wild-type Dpo4. It also showed a 55% and 91.5% higher catalytic efficiency when moving beyond the damaged 8-oxoG:C and 8-oxoG:A base pairs, respectively, compared to wild-type Dpo4. Protein engineering and bioinformatics methods can effectively increase the processivity and translesion synthesis ability of Dpo4.
IMPORTANCE DNA polymerases with poor fidelity can be exploited to store data and record changes in response to the intracellular environment. Sulfolobus solfataricus Dpo4 is such an enzyme, although its use is hindered by its low processivity. In this work, we used a bioinformatics and protein engineering approach to generate Dpo4 mutants with improved processivity. We identified the Dpo4 thumb domain as the most relevant in controlling processivity.
KEYWORDS: Dpo4, computational simulation, processivity, binding energy, protein engineering
INTRODUCTION
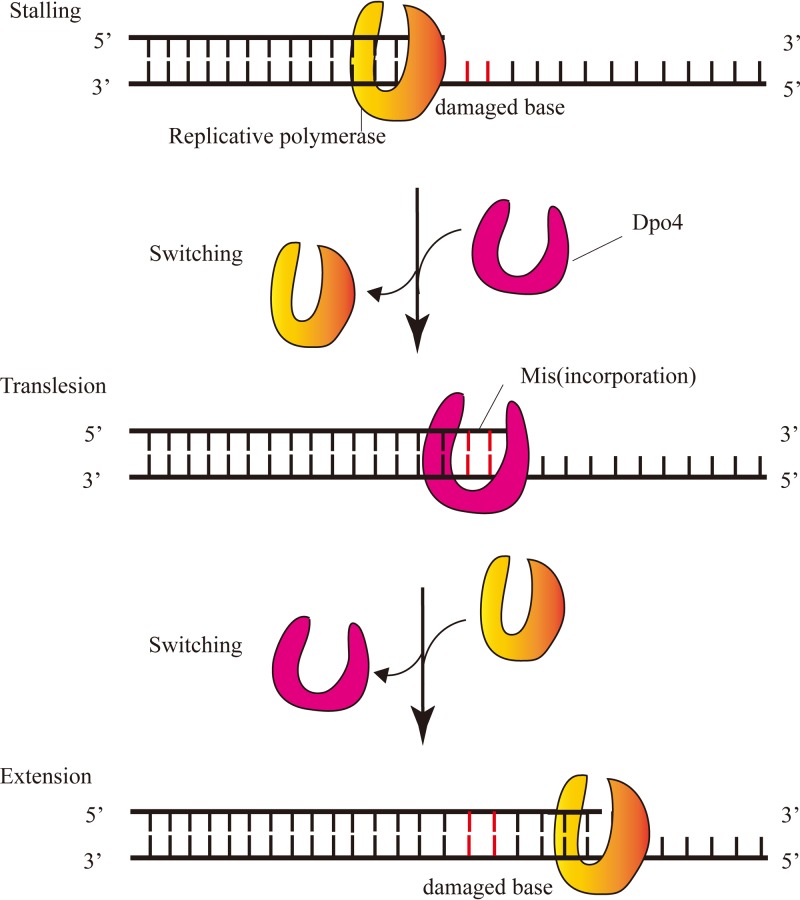
Y-family DNA polymerases are known for translesion synthesis (1, 2), which is the ability to tolerate a wide range of lesions during DNA synthesis (Fig. 1). This property protects cells from death caused by incomplete replication (3). There are various types of Y-family DNA polymerases in prokaryotes (4), archaea (5, 6), and eukaryotes (7, 8), such as DinB in Escherichia coli, Dpo4 in Sulfolobus solfataricus P2, and Pol η in Saccharomyces cerevisiae (9, 10). Y-family DNA polymerases are characterized by low fidelity and low processivity. For Dpo4, a model Y-family DNA polymerase, fidelity was reported in the range of 3 × 10−4 to 8 × 10−3, and processivity was only 1 to 2 nucleotides (nt) per binding cycle (11). Another study on Dpo4 processivity with an excess of DNA template compared to polymerase reported a value of 16 nt at 37°C (12). Recently, engineered DNA polymerases have been proposed as new tools for signal recording in neuroscience. These polymerases are ion sensitive, thus causing more or less nucleotide misincorporation during DNA replication in relation to timely changes in ion concentration (state of intracellular environment). Accordingly, the level of misincorporation in DNA can reveal the amplitude of intracellular environmental signals present at the time of synthesis and allow the DNA to record such signals (13). Y-family DNA polymerases are the most suitable such enzymes given their low fidelity, which can result in abundant misincorporation. However, their low processivity hinders their use in this context because it limits the number of nucleotides polymerized during a single-turnover reaction. Therefore, if they are to be used for signal recording, it is paramount that processivity of Y-family DNA polymerases is markedly improved.
FIG 1.
Translesion synthesis of Dpo4 in vivo. The replicative polymerase was stalled at a damaged base (red line), and then Y-family polymerase Dpo4 replaced the replicative polymerase to catalyze correct or incorrect base incorporation opposite the damaged base. The replicative polymerase replaced Dpo4 to complete DNA replication after translesion synthesis.
According to a previous study (14), we hypothesized that increasing the affinity of Dpo4 for DNA substrates could effectively improve processivity. This was achieved by linking the polymerase to a sequence for a nonspecific double-stranded DNA (dsDNA)-binding protein, such as β-sliding clamp, thioredoxin, proliferating cell nuclear antigen (PCNA), and S. solfataricus nonspecific DNA-binding protein 7d (Sso7d) (15). It was reported that the β-sliding clamp allowed Pol IV polymerase (Dpo4) to form a stable initiation complex, leading to a dramatic increase in processivity to an average of 300 to 400 nt (16). Thioredoxin fused to T7 DNA polymerase increased the affinity 80-fold, achieving a processivity of over several thousand nucleotides (17, 18). Polymerase activity of Dbh and processivity of Dpo4 can be improved by forming a heterodimer complex with PCNA subunits 1 and 2, which are homologous to each other (19, 20). Sso7d can significantly increase the processivity of family A and B polymerases (15). In addition to protein binding, a site-directed mutagenesis method has been reported to enhance processivity in telomerases, as evidenced by the L813Y telomerase mutant (21). The double-substitution mutation FCA720WCG at positions 720 and 722 of the yeast telomerase reverse transcriptase (TERT) produced an increase in telomerase processivity (22). However, residue mutagenesis has not been applied toward improving processivity of Y-family DNA polymerases.
Based on previous mutational studies demonstrating an increase in processivity in polymerases, telomerase, and TERT, we hypothesized that the processivity of Dpo4 would be similarly affected. Dpo4 shares a similar conformation with the replicative polymerase, that is, a conserved right-hand structure composed of palm, finger, and thumb domains, plus a unique “little finger” (LF) domain at the C terminus (6, 23). In the interaction between Dpo4 and DNA, the thumb domain contacts the backbone of the primer and the template strands through the minor groove. The LF domain seems to improve attachment of the polymerase to DNA. The palm domain mainly catalyzes DNA synthesis. The finger domain of Dpo4 contacts the template and the replicating base pair directly (6). Thus, key residues located in the four domains are likely to determine processivity, and mutating them can contribute to the activity of Y-family DNA polymerases (24, 25). During catalysis, Dpo4 undergoes a conformational change that loosens binding to the DNA template and results in low processivity (26). This suggests that the increasing binding between Dpo4 and DNA can increase processivity. All glycine mutants of the linker region between the thumb and the LF domains of Dpo4 showed a 250-fold weaker DNA-binding affinity than wild-type Dpo4 (wtDpo4) (27). This indicates that mutations of key residues substantially affects the protein's binding affinity for DNA. Binding affinity also can influence steady-state kinetic parameters and processivity, such as those of the Klenow fragment. Mutations at positions 849 and 668 of the latter affected its catalytic function by altering binding to the DNA substrate (28, 29). In this study, Dpo4 was selected as a model for signal storage owing to its low fidelity, which can result in substantial misincorporation of nucleotides. Meanwhile, the low processivity was the main obstacle, and the directed revolution was an alternative method to improve processivity. A library of nonconserved residues was picked using bioinformatics and protein engineering tools. The binding energy of potential mutants to DNA then was correctly calculated by computational simulation. Finally, the processivity of mutants was compared to determine the relationship between processivity and binding energy.
RESULTS
Determination of potential residue sites for targeted mutants.
To determine the nonconserved residues of Dpo4 critical for DNA binding, residues within 6 Å of the active site of Dpo4 (PDB entry 3QZ7) first were analyzed with PyMOL, and a total of 74 residues were obtained (see Fig. S1 in the supplemental material). We then determined which of them were conserved by aligning 38 homologous sequences with identity of more than 40%; 16 such residues are highlighted in blue in Fig. S2. Additionally, Table S1 shows 58 nonconserved residues with the corresponding mutation frequencies. Some residues, such as S40, A42, A44, and S103, were relatively conserved and therefore not suitable for mutagenesis. Some residues mutated at a significantly higher rate than the original (F37T, F/8 to T/21). We hypothesized that these improved Dpo4 properties and thus selected F33Y, F37T, I59M, E63K, A181D, V183I, N188S, I189V, E219K, I248Y, H285T, and V289I as possible mutants. We also chose A220S and M76I as controls with lower mutation frequencies than original residues and in direct contact with DNA. In total, 14 mutants were selected. From the structure of DNA-protein interactions, we inferred that residues F33, F37, I59, E63, and M76 were located in the finger domain, making them likely to interact with the template base and the incoming nucleotides. Residues A181, V183, N188, I189, E219, and A220 were located in the thumb and I248, H285, and V289 in the LF domain, indicating that they were in direct contact with dsDNA. Of the 14 candidate sites, some were very close to each other and had the same effect on DNA; for example, residues N188 and I189 in the thumb domain interacted with the phosphate group by van der Waals forces. Hence, we only picked 10 sites for further studies: F33Y, F37T, I59M, E63K, M76I, A181D, N188S, A220S, I248Y, and V289I.
Processivity of Dpo4 mutants in the presence of a trap.
To predict the processivity of the mutants, 10 of them and wtDpo4 were used to calculate the binding energy of the polymerase to DNA (Table S2). The (ΔGbind)mutants/(ΔGbind)wt value was used to compare the binding energies of the mutants to that of wtDpo4. If the value was greater than 1, the binding energy was higher for the mutated Dpo4 than for wtDpo4. Conversely, the binding energy of the mutated Dpo4 decreased when the value was less than 1. Mutants F37T and A220S displayed a 0.99-fold decrease in their binding energy to DNA relative to wtDpo4. In contrast, the binding energy of eight mutants was 1.02- to 1.05-fold higher than that for wtDpo4, particularly in mutants I59M and I248Y. Subsequently, 10 mutants were constructed by site-directed mutagenesis, expressed, and purified as described earlier for wtDpo4 (Fig. S3). Furthermore, changes in binding affinity were consistent with the binding energy calculated by computational simulation for all tested mutants (Table S2 and Fig. S4). These mutants presented stronger binding to DNA and hence were likely to increase processivity. Finally, their processivity was detected and compared.
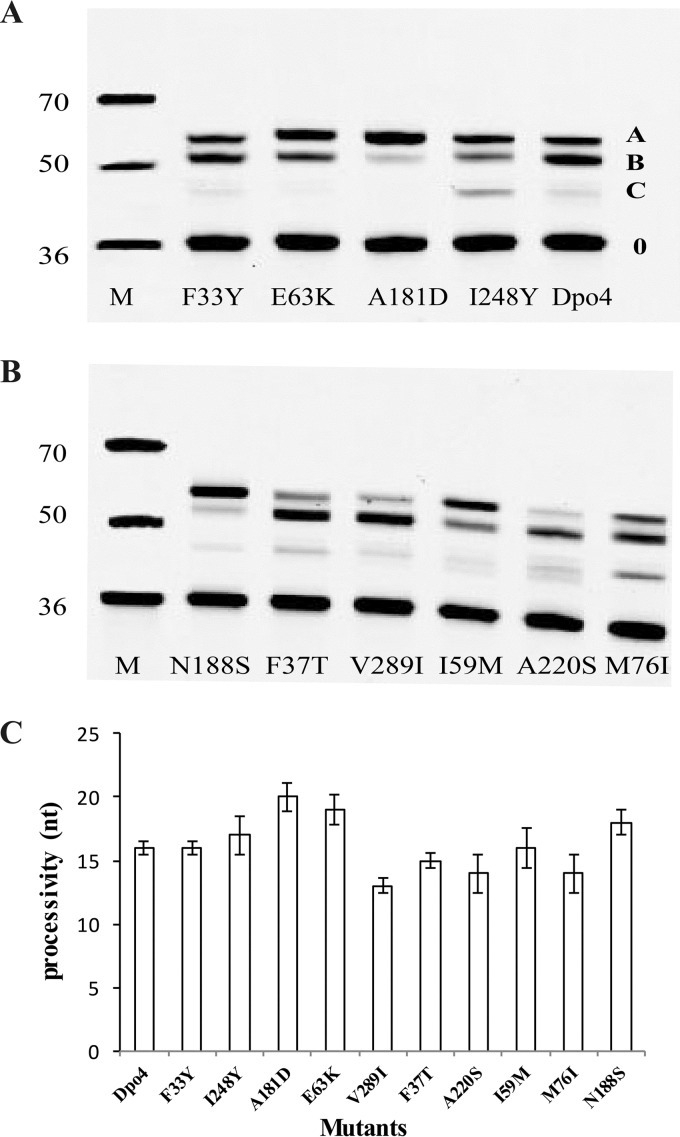
Processivity was evaluated as the average extension length of products in the presence of single-stranded M13mp18 DNA (ssDNA) as a trap, which limited DNA polymerase rebinding to the DNA. The elongation properties of wtDpo4 and mutants can be estimated by the size of product bands on gels. There were only three main extension bands, namely, A, B, and C, but the extension length and the concentration of each product was different for each DNA polymerase (Fig. 2A and B). Thus, to calculate the average length, the extension product lengths were multiplied by their relative concentration. For example, the A, B, and C extension products of wtDpo4 were 7, 15, and 20 nt in length, respectively, and the corresponding concentrations were 3.5, 55.8, and 40.7%. Accordingly, the average processivity of wtDpo4 was 16 nt. Similarly, mutants A181D, E63K, I248Y, and N188S yielded average lengths of 20, 19, 17, and 18 nt, respectively. Mutants F33Y and I59M showed a processivity of 16 nt, the same as that of wtDpo4. Mutants A220S and M76I displayed a processivity of 14 nt and mutants F37T and V289I of 15 and 13 nt, respectively (Fig. 2C; Table S3). In summary, mutants A181D, E63K, I248Y, and N188S increased processivity, whereas mutants A220S, M76I, F37T, and V289I decreased it. Mutants F33Y and I59M exhibited no change in processivity. We found that mutations E63K, A181D, N188S, and I248Y were located on the finger, thumb, and LF domains, respectively. We selected mutants E63K, A181D, and I248Y to determine the influence of different domain mutations on the properties of Dpo4.
FIG 2.
Processivity of Dpo4 and mutants. A constant concentration (10 nM) of Cy3-labeled 36-mer/M13mp18 was added to a reaction buffer. A constant concentration of DNA polymerase (100 nM) was added to initiate DNA synthesis and a 200-fold excess of 50-mer ssDNA as a polymerase trap at 37°C for 5 min, and products formed were separated by PAGE. All reactions were done in triplicate, and the indicated data points are shown as means ± standard deviations. (A and B) The bands of products were separated by PAGE, and the A, B, and C bands are from primer extension by wtDpo4 and mutants. M, molecular mass markers. (C) The processivity value calculated by the method in combination with the length and intensity of the three main bands.
Fidelity and nucleotide incorporation efficiency of Dpo4 mutants.
The percent incorporation of correct and incorrect nucleotides in the E63K, A181D, and I248Y mutants and in wtDpo4 is shown in Table 1. In the presence of the correct nucleotides, both wtDpo4 and the mutants showed maximal primer extension with TA, TC, and TT templates but not with the TG template (Fig. S5). The relative incorporation of dTTP, dATP, and dGTP with respect to TA, TT, and TC was 41.06%, 80.11%, and 68.25% (wtDpo4); 25.14%, 25.57%, and 35.27% (E63K); 59.23%, 80.76%, and 57.06% (A181D); 25.93%, 19.50%, and 42.91% (I248Y). We found that of the three mutants, A181D showed the highest incorporation of correct nucleotides. There were also a number of significant misincorporations by wtDpo4 and its mutated forms with different templates. We evaluated the fidelity of wild-type and mutated Dpo4 by measuring the percent incorporation of incorrect nucleotides, which were listed in Table 1. In all three mutants and for all four deoxynucleoside triphosphates (dNTPs), Dpo4 incorporated incorrect nucleotides mostly in the TA and TC templates. The A181D mutant exhibited more types of misincorporation than E63K and I248Y, including an additional C:C base pair in TC. The E63K mutant displayed a higher percent incorporation than the other two mutants in the TA template. The overall incorporation by the I248Y mutant was lower than that of mutants A181D and E63K, except for the G:T base pair in the TT template. In conclusion, fidelity was highest in the I248Y mutant and lowest in the A181D mutant.
TABLE 1.
Percent incorporation of correct and incorrect nucleotides
| Dpo4 variant and incorrect nucleotide | Incorporation (%) with templatea: |
||
|---|---|---|---|
| dA | dT | dC | |
| Wild type | |||
| A | 16.38 ± 4.83 | 80.11 ± 5.43* | 22.07 ± 7.34 |
| T | 41.06 ± 6.03* | 10.27 ± 1.46 | |
| G | 12.34 ± 1.85 | 68.25 ± 7.63* | |
| C | 12.27 ± 2.11 | ||
| A181D | |||
| A | 12.85 ± 3.62 | 25.57 ± 3.45* | 11.73 ± 2.12 |
| T | 25.14 ± 2.32* | 17.07 ± 3.65 | |
| G | 5.49 ± 2.82 | 12.31 ± 2.51 | 35.27 ± 5.37* |
| C | 6.15 ± 1.53 | 14.56 ± 3.21 | |
| E63K | |||
| A | 16.27 ± 5.49 | 80.76 ± 9.41* | 9.88 ± 2.23 |
| T | 59.23 ± 9.87* | 7.23 ± 0.56 | |
| G | 17.33 ± 4.52 | 57.06 ± 6.85* | |
| C | 21.6 ± 3.47 | 24.02 ± 1.73 | |
| I248Y | |||
| A | 7.41 ± 2.34 | 19.50 ± 1.23* | 4.93 ± 0.31 |
| T | 25.93 ± 1.45* | 6.2 ± 0.89 | |
| G | 9.9 ± 2.38 | 24.43 ± 5.78 | 42.91 ± 8.77* |
| C | 13.93 ± 3.55 | ||
Entries marked with an asterisk indicate the percent incorporation of correct nucleotides by wtDpo4 and mutants.
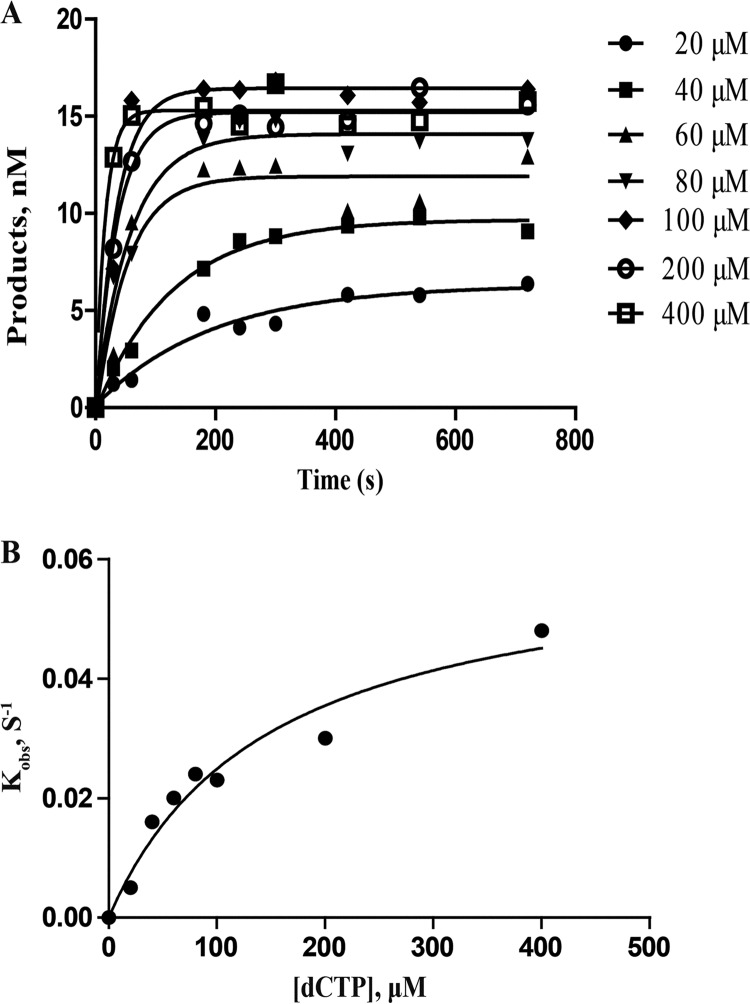
The template TG*, which has a damaged 8-oxoG base that is recognized by Dpo4, has been studied extensively (3, 30, 31). To further assess the nucleotide incorporation efficiency of the template TG* by wild-type and mutated Dpo4, maximum incorporation rate (kp), equilibrium dissociation constant (Kd,dCTP), and nucleotide incorporation efficiency (kp/Kd,dCTP) were measured at various dCTP concentrations, as shown in Fig. 3 and Table 2. In a study of wtDpo4 and A181D, E63K, and I248Y mutants, E63K showed the highest incorporation speed, with a kp of 0.171 s−1, whereas the I248Y mutant displayed the strongest affinity for dCTP, with a Kd,dNTP of 100.30 μM (Table 2). Finally, the A181D mutant presented the highest nucleotide incorporation efficiency (kp/Kd,dCTP), with a value of 4.74 × 10−4 s−1 μM−1, effectively balancing incorporation speed with affinity for dCTP opposite the TG* template. Based on the incorporation efficiency of A181D, it was used for additional kinetic assays.
FIG 3.
Nucleotide incorporation efficiency of dCTP incorporation onto 15-mer/TG*. A preincubated reaction mixture of wtDpo4 or mutants (120 nM) and 15-mer/TG* (30 nM) was rapidly mixed with dCTP (0 to 400 μM) for various times before being quenched. (A) Dependence of product concentration on time was plotted and fit to equation 1 (see Materials and Methods) to determine kobs. (B) kobs was plotted against dCTP concentration, and data were fit to equation 2 (see Materials and Methods), which determined a kp of 0.062 ± 0.009 s−1 and a Kd,dCTP of 148.40 ± 18.36 μM.
TABLE 2.
Nucleotide incorporation efficiency of wtDpo4 and mutants
| Dpo4 variant | kp (s−1) | Kd (μM) | kp/Kd (s−1 μM−1) |
|---|---|---|---|
| Wild type | 0.062 ± 0.009 | 148.40 ± 18.36 | 4.18 × 10−4 |
| A181D | 0.116 ± 0.004 | 244.50 ± 41.19 | 4.74 × 10−4 |
| E63K | 0.171 ± 0.003 | 390.80 ± 10.76 | 4.37 × 10−4 |
| I248Y | 0.021 ± 0.004 | 100.30 ± 5.81 | 2.09 × 10−4 |
Steady-state kinetic parameters of the A181D mutant.
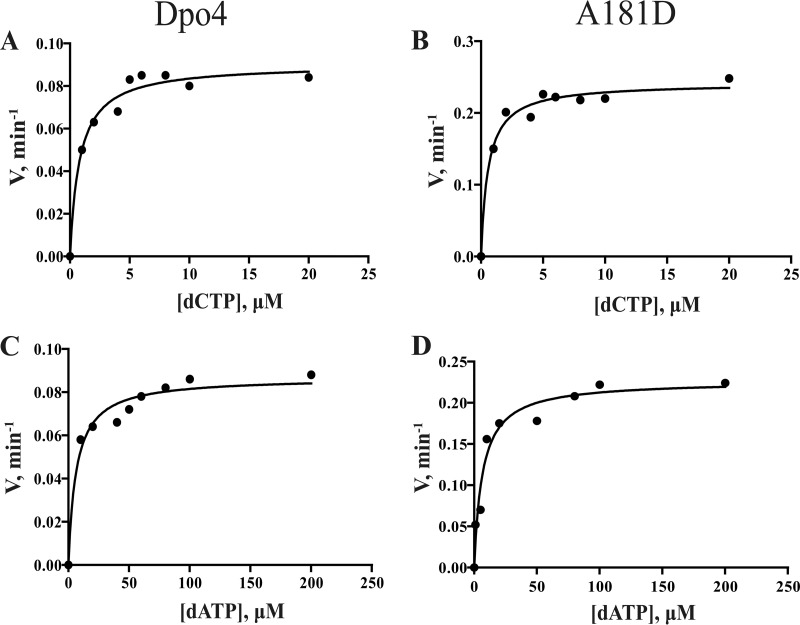
The steady-state kinetics of dCTP and dATP opposite the damaged base 8-oxoG are shown in Fig. 4 and Table 3. The kcat of the A181D mutant was 2.69-fold higher than that of wtDpo4 for dCTP and 2.61-fold for dATP. The Km of the A181D mutant was 0.71-fold that of wtDpo4 for dCTP incorporation and 1.02-fold for dATP. The catalytic efficiency (kcat/Km) of the A181D mutant was 3.81-fold and 2.62-fold higher than that of wtDpo4 for dCTP and dATP, respectively.
FIG 4.
Steady-state kinetic analysis of dNTP incorporation by Dpo4 and mutant A181D. Dpo4 or mutant A181D (2 nM) and 20 nM DNA substrate (15-mer primer/TG*) were used in the reaction mixture with various concentrations of dATP or dCTP. Incubation proceeded for 5 min at 37°C, and products formed were separated by PAGE. (A) dCTP incorporation opposite 8-oxoG catalyzed by wtDpo4. (B) dCTP incorporation opposite 8-oxoG by the mutant A181D. (C) dATP incorporation opposite 8-oxoG by wtDpo4. (D) dATP incorporation opposite 8-oxoG by the mutant A181D. kcat and Km values (estimated using the fit-to-hyperbolic-equations tool in Prism [GraphPad, San Diego, CA]) are listed in Table 3.
TABLE 3.
Steady-state kinetics of incorporation of dCTP and dATP opposite 8-oxoG by wtDpo4 and mutant A181D
| Dpo4 version and dNTP | kcat (min−1) | Km (μM) | kcat/Km (μM−1 min−1) |
|---|---|---|---|
| Wild type | |||
| dCTP | 0.0090 ± 0.0004 | 0.82 ± 0.03 | 0.0110 ± 0.0008 |
| dATP | 0.0087 ± 0.0010 | 6.54 ± 0.09 | 0.0013 ± 0.0002 |
| A181D | |||
| dCTP | 0.0242 ± 0.0049 | 0.58 ± 0.04 | 0.0419 ± 0.0093 |
| dATP | 0.0227 ± 0.0024 | 6.67 ± 0.24 | 0.0034 ± 0.0004 |
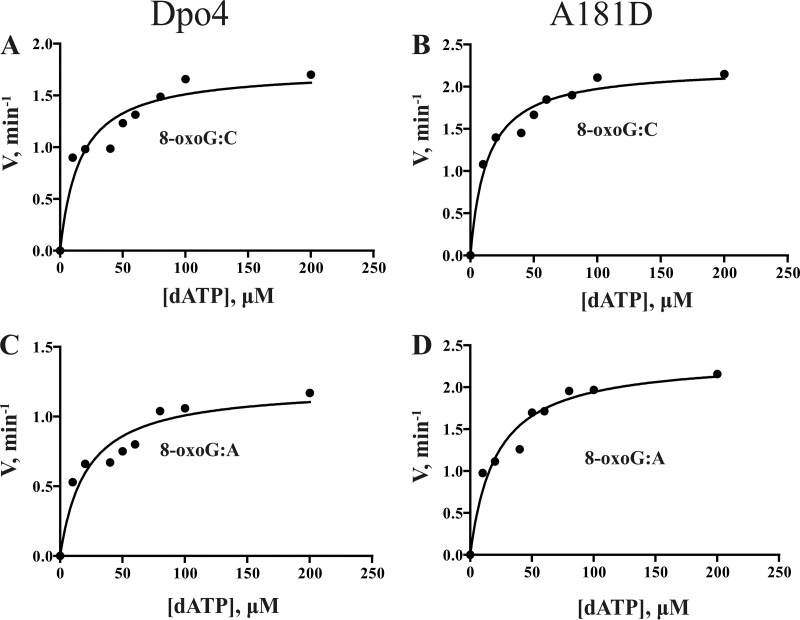
To evaluate primer extension beyond 8-oxoG, a steady-state kinetic analysis was performed on insertion of the next base (dATP) following an 8-oxoG:C pair as well as an 8-oxoG:A pair (Fig. 5 and Table 4). For the 8-oxoG:C base pair, the kcat of the A181D mutant was 1.27-fold, Km was 0.75-fold, and catalytic efficiency was 1.55-fold higher than those of wtDpo4. For the 8-oxoG:A base pair, the kcat of the A181D mutant was 1.93-fold, Km was 0.99-fold, and catalytic efficiency was 1.92-fold higher than those of wtDpo4.
FIG 5.
Steady-state kinetic analysis of the next base (dATP) insertion after 8-oxoG:C and 8-oxoG:A by wtDpo4 and mutant A181D. Dpo4 or mutant A181D (2 nM) and 20 nM DNA substrates (8-oxoG:C/8-oxoG:A) were used in the reaction mixture with various concentrations of dATP (0 to 200 μM). Incubation proceeded for 5 min at 37°C, and products formed were separated by PAGE. (A) dATP incorporation after 8-oxoG:C catalyzed by wtDpo4. (B) dATP incorporation after 8-oxoG:C by the mutant A181D. (C) dATP incorporation after 8-oxoG:A by wtDpo4. (D) dATP incorporation after 8-oxoG:A by the mutant A181D. kcat and Km values (estimated using the fit-to-hyperbolic-equations tool in Prism [GraphPad, San Diego, CA]) are listed in Table 4.
TABLE 4.
Steady-state kinetics of next base (dATP) insertion after 8-oxoG:C and 8-oxoG:A by wtDpo4 and mutant A181D
| Dpo4 variant and template | kcat (min−1) | Km (μM) | kcat/Km (μM−1 min−1) |
|---|---|---|---|
| WT | |||
| 8-oxoG:C | 0.1754 ± 0.0056 | 16.11 ± 1.01 | 0.0109 ± 0.0008 |
| 8-oxoG:A | 0.1220 ± 0.0088 | 21.05 ± 2.76 | 0.0059 ± 0.0006 |
| A181D | |||
| 8-oxoG:C | 0.2234 ± 0.0187 | 13.26 ± 0.93 | 0.0169 ± 0.0020 |
| 8-oxoG:A | 0.2349 ± 0.0185 | 20.91 ± 1.42 | 0.0113 ± 0.0013 |
Based on the comparison of steady-state kinetic parameters between wtDpo4 and the A181D mutant, we conclude that the latter could bypass the 8-oxoG lesion more efficiently than wtDpo4 when both dATP and dCTP were incorporated opposite the damaged base. The A181D mutant was also better at catalyzing dATP incorporation beyond the 8-oxoG:C and 8-oxoG:A base pairs. Taken together, these results indicate that the A181D mutant had a greater ability than wtDpo4 to bypass translesions, possibly as a consequence of a faster incorporation speed.
DISCUSSION
The S. solfataricus Dpo4 thumb, finger, and LF domains can establish direct contact with substrate DNA. Accordingly, altering the domains' structure is likely to have an effect on the properties of Dpo4. In this study, three mutants with residue alterations in different domains and increased binding energy and processivity were generated. They included the A181D, E63K, and I248Y mutations on the thumb, finger, and LF domains, respectively. Of these, the A181D mutant presented the highest processivity (Fig. 2) and more types of misincorporation (Table 1; see also Fig. S5 in the supplemental material), indicating the lowest fidelity. Incorporation efficiency by the A181D mutant was 8.5% and 126.8% higher than that for E63K and I248Y mutants, respectively (Table 2). In particular, the A181D mutant exhibited a 3.81-fold (dCTP) and a 2.62-fold (dATP) higher incorporation rate opposite the damaged base 8-oxoG (Table 3). It also increased catalytic efficiency by 55% and 91.5% when bypassing 8-oxoG:C and 8-oxoG:A base pairs, respectively (Table 4). These results demonstrate that the thumb domain mutation can effectively improve the catalytic properties of Dpo4.
The processivity of thumb domain mutants can be accurately predicted by calculating the binding energy of Dpo4 to DNA substrates using in silico methods (Table S2). Computer-based free energy calculations enable a detailed investigation of the energetic factors that determine binding affinity (32). The essence of processivity is that the polymerase can retain substrate affinity during multiple rounds of catalysis. Accordingly, increasing the affinity can effectively improve processivity (14, 33). For example, processivity of a TERT mutant was increased by augmenting its affinity for a telomeric DNA primer (21). Conversely, polymerases with decreased binding affinity resulted in decreased processivity, as exemplified by a Klenow fragment mutant (34). Therefore, we sought to confirm if it was possible to predict processivity knowing the binding energy. We selected various single-residue mutations on the thumb, finger, and LF domains, which affected the binding affinity of the Pol IV polymerase for DNA and, consequently, its processivity (21, 35, 36). Mutants A181D and N188S exhibited improved processivity when binding energy increased, whereas the A220S mutant showed diminished processivity when binding energy decreased. Instead, the finger domain mutant M76I and the LF mutant V289I showed increased binding energy (Table S2) but also lower processivity (Fig. 2).
Mutations in Dpo4 domains are likely to affect its properties. For Y-family DNA polymerases, mutations within the active site, palm, or finger domains affect translesion synthesis (37, 38) and catalytic efficiency (24), whereas changes in the LF domain mainly influence processivity, fidelity, and lesion bypassing (39). In addition, thumb domain mutants of yeast mitochondrial RNA polymerase (40) and T7 DNA polymerase (41) showed altered processive synthesis. Considering a similar general mechanism for DNA elongation (42), it is not surprising that Dpo4 mutants E63K (finger), A181D (thumb), and I248Y (LF) all exhibited increased processivity and misincorporation. Additionally, the A181D mutant displayed significantly increased translesion ability compared to wtDpo4. Furthermore, the increase in processivity was higher in thumb mutants than in other domain mutants. As we know, finger and thumb domains both may control processivity (43). However, the relaxed interaction between thumb domain and DNA could account for the notably low processivity of polymerase η (41). Similarly, reduced processivity during transcription was observed in a T7 RNA polymerase mutation with a deletion of the thumb subdomain (44). In contrast, there are fewer reports about the finger domain having a similar impact, indicating that thumb was the main domain to control processivity. Thus, thumb domain mutants A181D and N188S significantly increased processivity. Among five finger domain mutants, only E63K resulted in increased processivity, whereas the LF domain I248Y mutant showed only a minor improvement.
The present findings indicate that the A181D mutant is more suitable than others for molecular signal recording. Dpo4 was shown to be appropriate for signals recording as an error-prone polymerase and a far better sensor for Mn2+ concentration, especially in the presence of the T template (45). The statistical feasibility of molecular signal recording depends on several biochemical parameters, including the single-base elongation time (τc), average pause time (τp), and pause probability (P) (13). The τc parameter depends on replication velocity, which was modulated by the polymerase's affinity for DNA and its processivity (46, 47). A key problem for a signal recording machine is that polymerases do not incorporate nucleotides at a constant rate, which is attributed to pausing at specific sites on the DNA (48). An ideal situation for molecular signal recording is represented by lack of pausing, thus preventing dissociation of the polymerase (49). Pausing can be avoided by the TLS polymerase through translesion and low fidelity (50). In our study, three mutants, E63K, A181D, and I248Y, prefer to incorporate wrong nucleotides into the T template compared to wtDpo4 in Mg2+ (Table 1), supporting a more sensitive tool to record signal, as the misincorporation could be further affected with Mn2+ (51). These mutants showed increased processivity and affinity of the polymerase for DNA (Fig. 2; Table S2), resulting in shorter single-base elongation times, particularly by the A181D mutant. In addition, the A181D mutant presented higher translesion ability than wtDpo4 (Tables 3 and 4), suggesting that A181D decreases pause time and pause probability. It is postulated that the shorter the elongation and pausing times, the higher the resolution and accuracy of molecular signal recording.
MATERIALS AND METHODS
Protein expression and purification.
The gene encoding Dpo4 was synthesized by Synbio-tech (Suzhou, China) and then cloned in the expression plasmid pET28a. Dpo4 was expressed in E. coli BL-21 and purified to electrophoretic homogeneity as described previously (52).
Mutation site identification and homology modeling.
A Dpo4 crystal structure (PDB code 3QZ7) was selected as a model of the binary complex of polymerase and substrate and analyzed with PyMOL (www.pymol.org). Amino acids located within 6 Å of the DNA substrates were identified as potential mutation sites. Homologous proteins with sequence identity over 40% were identified by BLAST searches against the Dpo4 sequence template (NCBI). Multiple-sequence alignment was conducted to determine the nonconserved residues in the candidates mentioned above. Moreover, mutated Dpo4 structures were constructed with SWISS-MODEL (http://swissmodel.expasy.org), and the binary complex of mutated Dpo4 and substrate was generated with PyMOL.
Molecular dynamics and binding energy calculation.
Molecular dynamics (MD) simulations were performed with the GROMACS 4.5.5 toolkit by following three previously described steps: energy minimization, system equilibration, and production protocols (53, 54). The binary complex of polymerase and DNA was prepared by adding the GROMOS 9643a1 force field, placing it in a cubic box, and filling the box with atomistic TIP3P water. A two-step energy minimization process then was performed; this included the steepest descent method with an optimization of 800 steps and the conjugate gradient method with an optimization of 1,200 steps with solute molecule constraint. A further energy minimization step was performed during the steepest descent method, switching to a conjugate gradient every 800 steps for a total of 4,000 steps without any constraint. Simulation equilibration was conducted at a target pressure of 1 × 105 pascals and 300 K. After the two equilibration phases, a 500-ps simulation with a time step of 2 fs with solute molecule constraint and a 5-ns simulation without solute molecule constraint were performed. Samples were collected at 2,500-step intervals. Particle mesh Ewald summation was applied to compute long-range coulombic interactions, and the linear constraint solver method was used to constrain all covalent bonds in the MD simulation. The molecular mechanics Poisson-Boltzmann surface area method was used to predict the binding affinities of polymerase and DNA with the trajectory of MD (55).
Template-primer DNA preparation.
All oligonucleotides used in this work were synthesized by Synbio-tech and purified by high-performance liquid chromatography (Table 5). The primers used for this study were synthesized with a cyanine (Cy3) label for fluorescence visualization. Circular single-stranded M13mp18 DNA (ssDNA), used in the processivity assay, was purchased from Bayou Biolabs (Harahan, LA). Primers were annealed to the template in a 1:1.5 ratio in buffer containing 10 mM HEPES-NaOH (pH 7.4), 1 mM dithiothreitol (DTT), 50 mM NaCl, 100 μg/ml bovine serum albumin, and 0.10% Triton X-100. The annealed complex was heated at 95°C for 5 min, cooled to 60°C at a rate of 0.1°C/s, incubated at 60°C for 10 min, cooled again slowly to 5°C, and then stored at −20°C until use, when it was thawed on ice.
TABLE 5.
DNA sequences used in this study
| Oligonucleotide | Sequence (5′–3′)a |
|---|---|
| 15-mer primer | 5′-Cy3-CGTACTCGTAGGCAT-3′ |
| 16-mer primer | 5′-Cy3-CGTACTCGTAGGCATC-3′ |
| 16-mer primer | 5′-Cy3-CGTACTCGTAGGCATA-3′ |
| 36-mer primer | 5′-Cy3-ACGCCTGTAGCATTCCACAGACAACCCTCATAGTTA-3′ |
| 2AP-DNA | 5′-CCGTGCCTACCTGAACAGCTaTaCGCaCTAATGCCTACGA-3′ |
| 3′-GGCACGGATGGACTTGTCGaTaTGCGTGATTACGGATGCT-5′ | |
| Unlabeled DNA | 5′-CCGTGCCTACCTGAACAGCTATACGCACTAATGCCTACGA-3′ |
| 3′-GGCACGGATGGACTTGTCGATATGCGTGATTACGGATGCT-5′ | |
| TG | 5′-TCCTACCGTGCCTACCTGAACAGCTGGTCACACTGATGCCTACGAGTACG-3′ |
| TA | 5′-TCCTACCGTGCCTACCTGAACAGCTGGTCACACTAATGCCTACGAGTACG-3′ |
| TC | 5′-TCCTACCGTGCCTACCTGAACAGCTGGTCACACTCATGCCTACGAGTACG-3′ |
| TT | 5′-TCCTACCGTGCCTACCTGAACAGCTGGTCACACTTATGCCTACGAGTACG-3′ |
| TG*b | 5′-TCCTACCGTGCCTACCTGAACAGCTGGTCACACTG*ATGCCTACGAGTACG-3′ |
In sequences, the boldface letters show the template differences in the same experiment (“C” and “A” in 16-mer primers for steady-state kinetic analysis of the next base insertion after 8-oxoG:C or 8-oxoG:A, respectively; “A” and “a” for normal and modified bases for the binding affinity determination, respectively; “G”, “A”, “C”, “T”, and “G*” in the different templates for the fidelity analysis and determination of nucleotide incorporation efficiency).
TG*, template with damaged 8-oxoG base.
Determination of binding affinity to DNA for wild-type and mutated Dpo4.
Fluorescence titrations were performed to determine the equilibrium dissociation constant (Kd) of Dpo4 and its mutants as previously described (56, 57). A constant amount of 2-aminopurine DNA (2AP-DNA) (10 nM) was titrated against an increasing concentration of wtDpo4 or mutated Dpo4 (0 to 200 nM) in the reaction buffer [50 mM Tris acetate, pH 7.5, 50 mM sodium acetate (NaOAc), 10 mM Mg(OAc)2, 5 mM DTT, 0.05% Tween 20] at 25°C. A control experiment was carried out under identical conditions with unlabeled DNA (Table 5). Changes in fluorescence corresponding to the control experiments were subtracted from the data obtained with 2AP-DNA, and the corrected values were plotted against the corresponding polymerase concentration. The dissociation constant was calculated using the equation F = (Fmax × [polymerase])/(Kd + [polymerase]), where F is the relative fluorescence intensity and Fmax is the maximum value.
Processivity assay.
The processivity assay was performed in the presence of a trap to prevent rebinding (16, 58). When the concentration of enzyme was lower than that of the substrate, no extension band could be detected. Hence, constant doses (10 nM) of Cy3-labeled primer/template (36-mer/M13mp18) and Dpo4 (100 nM) were added to the reaction buffer [25 mM Tris-HCl, pH 8.0, 1 mM DTT, 0.1 mM (NH4)2SO2, 2.5 mM MgCl2]. Constant amounts of dNTPs (1 mM) and trap (200-fold excess of ssDNA) were added to initiate DNA synthesis at 37°C for 5 min (11); reactions were terminated by addition of 10 μl stop solution (80% formamide, 1 mg/ml xylene C, 1 mg/ml bromophenol blue, and 20 mM EDTA) followed by incubation at 95°C for 2 min. The mixture was immediately transferred on ice for 10 min and was subsequently loaded onto a 20% polyacrylamide gel containing 8 M urea and 1× Tris-borate-EDTA buffer; gels were visualized on a Typhoon Trio scanner (GE Healthcare) (59).
Fidelity analysis.
Primer extension assays to detect fidelity of wild-type and mutated enzymes were carried out as described previously (59, 60). DNA substrates were prepared using a 15-mer primer (Table 5) with each of the four templates (TA, TT, TG, and TC); these were designed to present four different nucleotides, A, T, G, and C, respectively, at the same position. The reaction mixture (20 μl) consisted of 5 mM dNTPs, 20 nM DNA substrate, 0.1 mM (NH4)2SO2, 2.5 mM MgCl2, assay buffer (25 mM Tris-HCl, pH 8.0, 1 mM DTT), and 10 nM Dpo4 (wild type or mutated). The mixture was incubated at 37°C for 2 h, after which the reaction was terminated and loaded onto a 20% polyacrylamide gel as mentioned above. The bands were visualized and recorded using a Typhoon Trio, and their intensities were quantified using ImageQuant TL 1D analysis software (GE Healthcare). Nucleotide incorporation was calculated using the equation percent incorporation = Is/(Is + Ip), where Is is the intensity of the extension band and Ip is the intensity of the primer band in the same lane. All assays were repeated thrice, and the results were reproducible.
Determination of nucleotide incorporation efficiency.
Wild-type or mutated (120 nM) Dpo4 and DNA substrate (15-mer/TG*, 30 nM) in the reaction buffer mentioned above were mixed with increasing concentrations of dCTP (0 to 400 μM) (61). The reactions were terminated after various times (0 to 720 s) using 10 μl stop solution. Reaction products were analyzed by denaturing polyacrylamide gel electrophoresis and quantified with a Typhoon Trio. The time course of product formation at each dCTP concentration was fit to a single exponential equation (equation 1).
| (1) |
Here, kobs is the observed reaction rate constant, A is the reaction amplitude, and t is time. The plot of kobs versus dCTP concentration was fit to a hyperbolic equation (equation 2).
| (2) |
Here, kp is the maximum dNTP incorporation rate and Kd,dNTP is the equilibrium dissociation constant for the ternary complex (Dpo4-DNA-dNTP). Nucleotide incorporation efficiency (kp/Kd) then was calculated.
Steady-state kinetic analysis of bypassing 8-oxoG.
Steady-state experiments of single-base incorporation and one-base extension were performed by adding a single dNTP at various concentrations. The molar ratio of Dpo4 to DNA was 1:10 (2 nM polymerase and 20 nM DNA) (62, 63). For steady-state single-base incorporation, wild-type or A181D mutated Dpo4 and DNA were mixed in the buffer mentioned for the fidelity assay. dCTP (0 to 20 μM) or dATP (0 to 200 μM) was added to initiate the reaction for 5 min at 37°C, and primer conversion to product was maintained at less than 20%. The one-base extension reaction was analogous to the single-base incorporation one, except that dNTP consisted of only dATP (0 to 200 μM). Reactions were quenched and products were analyzed and quantified. Plots of incorporation rates versus dNTP concentration were fit to a hyperbolic Michaelis-Menten equation (equation 3) to yield kcat and Km values using nonlinear regression in GraphPad Prism version 3.0 (GraphPad, San Diego, CA).
| (3) |
Here, ν is the initial rate, [D] is the dNTP concentration, [E] is the enzyme concentration, kcat is the turnover rate, and Km is the primer-template concentration at which half of the maximum activity is achieved. kcat/Km is the incorporation frequency.
Supplementary Material
ACKNOWLEDGMENTS
We have no conflicts of interest to declare.
This work was supported by the National Natural Science Foundation of China (grant number 21576117) and Natural Science Foundation of Jiangsu Province (grant number BK 20131103).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01013-17.
REFERENCES
- 1.Raper AT, Gadkari VV, Maxwell BA, Suo ZC. 2016. Single-molecule investigation of response to oxidative DNA damage by a Y-family DNA polymerase. Biochemistry 55:2187–2196. doi: 10.1021/acs.biochem.6b00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. 2001. The Y-family of DNA polymerases. Mol Cell 8:7–8. doi: 10.1016/S1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 3.Su Y, Patra A, Harp JM, Egli M, Guengerich FP. 2015. Roles of residues Arg-61 and Gln-38 of human DNA polymerase η in bypass of deoxyguanosine and 7,8-dihydro-8-oxo-2′-deoxyguanosine. J Biol Chem 290:15921–15933. doi: 10.1074/jbc.M115.653691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman MF. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem 71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 5.Zhou BL, Pata JD, Steitz TA. 2001. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell 8:427–437. doi: 10.1016/S1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 6.Ling H, Boudsocq F, Woodgate R, Yang W. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107:91–102. doi: 10.1016/S0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 7.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange SS, Takata K, Wood RD. 2011. DNA polymerases and cancer. Nat Rev Cancer 11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman MF, Woodgate R. 2013. Translesion DNA polymerases. Cold Spring Harb Perspect Biol 5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sale JE, Lehmann AR, Roger W. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. 2001. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic pol eta. Nucleic Acids Res 29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiala KA, Suo Z. 2004. Mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 43:2116–2125. doi: 10.1021/bi035746z. [DOI] [PubMed] [Google Scholar]
- 13.Glaser JI, Zamft BM, Marblestone AH, Moffitt JR, Tyo K, Boyden ES, Church G, Kording KP. 2013. Statistical analysis of molecular signal recording. PLoS Comp Biol 9:e1003145. doi: 10.1371/journal.pcbi.1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breyer WA, Matthews BW. 2001. A structural basis for processivity. Protein Sci 10:1699–1711. doi: 10.1110/ps.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Prosen DE, Mei L, Sullivan JC, Finney M, Vander Horn PB. 2004. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res 32:1197–1207. doi: 10.1093/nar/gkh271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RPP. 2000. The β clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep 1:484–488. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedford E, Tabor S, Richardson CC. 1997. The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc Natl Acad Sci U S A 94:479–484. doi: 10.1073/pnas.94.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akabayov B, Akabayov SR, Lee SJ, Tabor S, Kulczyk AW, Richardson CC. 2010. Conformational dynamics of bacteriophage T7 DNA polymerase and its processivity factor, Escherichia coli thioredoxin. Proc Natl Acad Sci U S A 107:15033–15038. doi: 10.1073/pnas.1010141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing G, Kirouac K, Shin YJ, Bell SD, Ling H. 2009. Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. Mol Microbiol 71:678–691. doi: 10.1111/j.1365-2958.2008.06553.x. [DOI] [PubMed] [Google Scholar]
- 20.Grúz P, Pisani FM, Shimizu M, Yamada M, Hayashi I, Morikawa K, Nohmi T. 2001. Synthetic activity of Sso DNA polymerase Y1, an archaeal DinB-like DNA polymerase, is stimulated by processivity factors proliferating cell nuclear antigen and replication factor C. J Biol Chem 276:47394–47401. doi: 10.1074/jbc.M107213200. [DOI] [PubMed] [Google Scholar]
- 21.Bryan TM, Goodrich KJ, Cech TR. 2000. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J Biol Chem 275:24199–24207. doi: 10.1074/jbc.M003246200. [DOI] [PubMed] [Google Scholar]
- 22.Peng Y, Mian IS, Lue NF. 2001. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol Cell 7:1201–1211. doi: 10.1016/S1097-2765(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 23.Raper AT, Suo Z. 2016. Investigation of intradomain motions of a Y-family DNA polymerase during substrate binding and catalysis. Biochemistry 55:5832–5844. doi: 10.1021/acs.biochem.6b00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh JM, Parasuram R, Rajput PR, Rozners E, Ondrechen MJ, Beuning PJ. 2012. Effects of non-catalytic, distal amino acid residues on activity of E. coli DinB (DNA polymerase IV). Environ Mol Mutag 53:766–776. doi: 10.1002/em.21730. [DOI] [PubMed] [Google Scholar]
- 25.Jacewicz A, Trzemecka A, Guja KE, Plochocka D, Yakubovskaya E, Bebenek A, Garcia-Diaz M. 2013. A remote palm domain residue of RB69 DNA polymerase is critical for enzyme activity and influences the conformation of the active site. PLoS One 8:e76700. doi: 10.1371/journal.pone.0076700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson RC, Jackson MA, Pata JD. 2013. Y-family polymerase conformation is a major determinant of fidelity and translesion specificity. Structure 21:20–31. doi: 10.1016/j.str.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherrer SM, Maxwell BA, Pack LR, Fiala KA, Fowler JD, Zhang J, Suo Z. 2012. Identification of an unfolding intermediate for a DNA lesion bypass polymerase. Chem Res Toxicol 25:1531–1540. doi: 10.1021/tx3002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polesky AH, Steitz TA, Grindley ND, Joyce CM. 1990. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem 265:14579–14591. [PubMed] [Google Scholar]
- 29.Polesky AH, Dahlberg ME, Benkovic SJ, Grindley ND, Joyce CM. 1992. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J Biol Chem 267:8417–8428. [PubMed] [Google Scholar]
- 30.Patra A, Nagy LD, Zhang QQ, Su Y, Muller L, Guengerich FP, Egli M. 2014. Kinetics, structure, and mechanism of 8-oxo-7,8-dihydro-2′-deoxyguanosine bypass by human DNA polymerase η. J Biol Chem 289:16867–16882. doi: 10.1074/jbc.M114.551820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eoff RL, Irimia A, Angel KC, Egli M, Guengerich FP. 2007. Hydrogen bonding of 7,8-dihydro-8-oxodeoxyguanosine with a charged residue in the little finger domain determines miscoding events in Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem 282:19831–19843. doi: 10.1074/jbc.M702290200. [DOI] [PubMed] [Google Scholar]
- 32.Homeyer N, Gohlke H. 2012. Free energy calculations by the molecular mechanics Poisson-Boltzmann surface area method. Mol Inform 31:114–122. doi: 10.1002/minf.201100135. [DOI] [PubMed] [Google Scholar]
- 33.Walstrom KM, Dozono JM, von Hippel PH. 1997. Kinetics of the RNA-DNA helicase activity of Escherichia coli transcription termination factor rho. 2. Processivity, ATP consumption, and RNA binding. Biochemistry 36:7993–8004. [DOI] [PubMed] [Google Scholar]
- 34.Minnick DT, Astatke M, Joyce CM, Kunkel TA. 1996. A thumb subdomain mutant of the large fragment of Escherichia coli DNA polymerase I with reduced DNA binding affinity, processivity, and frameshift fidelity. J Biol Chem 271:24954–24961. doi: 10.1074/jbc.271.40.24954. [DOI] [PubMed] [Google Scholar]
- 35.Bebenek K, Beard WA, Casasfinet JR, Kim HR, Darden TA, Wilson SH, Kunkel TA. 1995. Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J Biol Chem 270:19516–19523. doi: 10.1074/jbc.270.33.19516. [DOI] [PubMed] [Google Scholar]
- 36.Tomlinson CG, Holien JK, Mathias JAT, Parker MW, Bryan TM. 2016. The C-terminal extension of human telomerase reverse transcriptase is necessary for high affinity binding to telomeric DNA. Biochimie 128:114–121. doi: 10.1016/j.biochi.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Boudsocq F, Hong L, Yang W, Woodgate R. 2002. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair 1:343–358. doi: 10.1016/S1568-7864(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 38.Glick E, Vigna KL, Loeb LA. 2001. Mutations in human DNA polymerase η motif II alter bypass of DNA lesions. EMBO J 20:7303–7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. 2004. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J Biol Chem 279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 40.Velazquez G, Sousa R, Brieba LG. 2015. The thumb subdomain of yeast mitochondrial RNA polymerase is involved in processivity, transcript fidelity and mitochondrial transcription factor binding. RNA Biol 12:514–524. doi: 10.1080/15476286.2015.1014283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cannistraro VJ, Taylor JS. 2004. DNA-thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase eta. J Biol Chem 279:18288–18295. doi: 10.1074/jbc.M400282200. [DOI] [PubMed] [Google Scholar]
- 42.Brenlla A, Markiewicz RP, Rueda D, Romano LJ. 2014. Nucleotide selection by the Y-family DNA polymerase Dpo4 involves template translocation and misalignment. Nucleic Acids Res 42:2555–2563. doi: 10.1093/nar/gkt1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher TS, Darden T, Prasad VR. 2003. Substitutions at Phe61 in the β3-β4 hairpin of HIV-1 reverse transcriptase reveal a role for the fingers subdomain in strand displacement DNA synthesis. J Mol Biol 325:443–459. doi: 10.1016/S0022-2836(02)01225-1. [DOI] [PubMed] [Google Scholar]
- 44.Bonner G, Lafer EM, Sousa R. 1994. The thumb subdomain of T7 RNA polymerase functions to stabilize the ternary complex during processive transcription. J Biol Chem 269:25129–25136. [PubMed] [Google Scholar]
- 45.Zamft BM, Marblestone AH, Kording K, Schmidt D, Martin-Alarcon D, Tyo K, Boyden ES, Church G. 2012. Measuring cation dependent DNA polymerase fidelity landscapes by deep sequencing. PLoS One 7:e43876. doi: 10.1371/journal.pone.0043876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ibarra B, Chemla YR, Plyasunov S, Smith SB, Lázaro JM, Salas M, Bustamante C. 2009. Proofreading dynamics of a processive DNA polymerase. EMBO J 28:2794–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelman Z, O'Donnell M. 1995. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem 64:171–201. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz JJ, Quake SR. 2009. Single molecule measurement of the “speed limit” of DNA polymerase. Proc Natl Acad Sci U S A 106:20294–20300. doi: 10.1073/pnas.0907404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viguera E, Canceill D, Ehrlich SD. 2001. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J 20:2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RPP. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J 19:6259–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaisman A, Ling H, Woodgate R, Yang W. 2005. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J 24:2957–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. 2005. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J Biol Chem 280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 53.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. 2013. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Pan D, Shen Y, Jin N, Liu H, Yao X. 2012. Understanding the molecular mechanism of the broad and potent neutralization of HIV-1 by antibody VRC01 from the perspective of molecular dynamics simulation and binding free energy calculations. J Mol Model 18:4517–4527. doi: 10.1007/s00894-012-1450-z. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen TT, Mai BK, Li MS. 2011. Study of Tamiflu sensitivity to variants of A/H5N1 virus using different force fields. J Chem Infect Model 51:2266–2276. doi: 10.1021/ci2000743. [DOI] [PubMed] [Google Scholar]
- 56.Jia Y, Kumar A, Patel SS. 1996. Equilibrium and stopped-flow kinetic studies of interaction between T7 RNA polymerase and its promoters measured by protein and 2-aminopurine fluorescence changes. J Biol Chem 271:30451–30458. doi: 10.1074/jbc.271.48.30451. [DOI] [PubMed] [Google Scholar]
- 57.Beechem JM, Otto MR, Bloom LB, Eritja R, Reha-Krantz LJ, Goodman MF. 1998. Exonuclease-polymerase active site partitioning of primer-template DNA strands and equilibrium Mg2+ binding properties of bacteriophage T4 DNA polymerase. Biochemistry 37:10144–10155. doi: 10.1021/bi980074b. [DOI] [PubMed] [Google Scholar]
- 58.Mikheikin AL, Lin HK, Mehta P, Jen-Jacobson L, Trakselis MA. 2009. A trimeric DNA polymerase complex increases the native replication processivity. Nucleic Acids Res 37:7194–7205. doi: 10.1093/nar/gkp767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma A, Nair DT. 2012. MsDpo4–a DinB homolog from Mycobacterium smegmatis–is an error-prone DNA polymerase that can promote G:T and T:G mismatches. J Nucleic Acids 2012:285481–285490. doi: 10.1155/2012/285481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma A, Kottur J, Narayanan N, Nair DT. 2013. A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli. Nucleic Acids Res 41:5104–5114. doi: 10.1093/nar/gkt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vyas R, Efthimiopoulos G, Tokarsky EJ, Malik CK, Basu AK, Suo Z. 2015. Mechanistic basis for the bypass of a bulky DNA adduct catalyzed by a Y-family DNA polymerase. J Am Chem Soc 137:12131–12142. doi: 10.1021/jacs.5b08027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue Q, Zhong M, Liu B, Yong T, Wei Z, Guengerich FP, Zhang H. 2016. Kinetic analysis of bypass of 7,8-dihydro-8-oxo-2′-deoxyguanosine by the catalytic core of yeast DNA polymerase η. Biochimie 121:161–169. doi: 10.1016/j.biochi.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu B, Xue Q, Gu S, Wang W, Chen J, Li Y, Wang C, Zhang H. 2016. Kinetic analysis of bypass of O6-methylguanine by the catalytic core of yeast DNA polymerase eta. Arch Biochem Biophys 596:99–107. doi: 10.1016/j.abb.2016.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.