ABSTRACT
Candida glabrata is a promising producer of organic acids. To elucidate the physiological function of the Mediator tail subunit Med15B in the response to low-pH stress, we constructed a deletion strain, C. glabrata med15BΔ, and an overexpression strain, C. glabrata HTUΔ/CgMED15B. Deletion of MED15B caused biomass production, glucose consumption rate, and cell viability to decrease by 28.3%, 31.7%, and 26.5%, respectively, compared with those of the parent (HTUΔ) strain at pH 2.0. Expression of lipid metabolism-related genes was significantly downregulated in the med15BΔ strain, whereas key genes of ergosterol biosynthesis showed abnormal upregulation. This caused the proportion of C18:1 fatty acids, the ratio of unsaturated to saturated fatty acids (UFA/SFA), and the total phospholipid content to decrease by 11.6%, 27.4%, and 37.6%, respectively. Cells failed to synthesize fecosterol and ergosterol, leading to the accumulation and a 60.3-fold increase in the concentration of zymosterol. Additionally, cells showed reductions of 69.2%, 11.6%, and 21.8% in membrane integrity, fluidity, and H+-ATPase activity, respectively. In contrast, overexpression of Med15B increased the C18:1 levels, total phospholipids, ergosterol content, and UFA/SFA by 18.6%, 143.5%, 94.5%, and 18.7%, respectively. Membrane integrity, fluidity, and H+-ATPase activity also increased by 30.2%, 6.9%, and 51.8%, respectively. Furthermore, in the absence of pH buffering, dry weight of cells and pyruvate concentrations were 29.3% and 61.2% higher, respectively, than those of the parent strain. These results indicated that in C. glabrata, Med15B regulates tolerance toward low pH via transcriptional regulation of acid stress response genes and alteration in lipid composition.
IMPORTANCE This study explored the role of the Mediator tail subunit Med15B in the metabolism of Candida glabrata under acidic conditions. Overexpression of MED15B enhanced yeast tolerance to low pH and improved biomass production, cell viability, and pyruvate yield. Membrane lipid composition data indicated that Med15B might play a critical role in membrane integrity, fluidity, and H+-ATPase activity homeostasis at low pH. Thus, controlling membrane composition may serve to increase C. glabrata productivity at low pH.
KEYWORDS: Candida glabrata, Mediator subunit Med15B, low-pH stress, transcriptomics, membrane lipid
INTRODUCTION
The Mediator coactivator complex is required for transcription initiation in eukaryotes (1, 2). It is recruited by transcription activators and conveys regulatory information from gene-specific regulators to promoters (3, 4). Mediator can influence almost all stages of transcription and coordinated processes such as chromatin remodeling, transcription elongation, and posttranslational modifications (5, 6). Mediator is a multisubunit assembly comprising four modules: head, middle, tail, and cyclin-dependent kinase 8 (CDK8) (7). The head and middle modules are highly conserved and essential for viability, whereas the tail and CDK8 modules are less conserved and nonessential (8). The head module can bind to the carboxy-terminal domain of RNA polymerase II (9). The tail module serves to recruit Mediator to upstream activating sequence (UAS) and directly interacts with a number of transcription activators, including the general control protein (Gcn4), oleate-activated transcription factor (Oaf1), pleiotropic drug response factor (Pdr1), and others (10–13). The kinase module is often implicated in transcriptional repression because deletion of its subunits leads to global gene upregulation and facilitates Mediator-UAS interactions (14, 15).
Mediator is required for gene expression and is involved in many transcriptional regulatory pathways that affect the cell response to environmental stressors (3, 16). In Saccharomyces cerevisiae, Mediator subunits Med18, Med19, and Med20, which activate the cytochrome c1 (CYC1) gene in response to oxidative stress, are required for cell viability (17). Med17 links transcription and DNA repair by facilitating the recruitment of the Rad2/XPG endonuclease to transcribed genes during exogenous genotoxic stress (18). The tail module subunits Med2, Med3, and Med15 can form a subcomplex and are involved in a heat shock response through recruitment by activated heat shock transcription factor 1 (Hsf1) to its target genes (19). By changing the Mediator subunit composition, the tail subcomplex affects the response to oxidative stress (20). The tail module subunits are also required for mitogen-activated protein kinase (MAPK) Hog1-mediated gene expression, which is essential for proper cell adaptation to osmotic stress (21). Manipulation of the Mediator complex, involved in acid stress tolerance, might serve as a new strategy to expand the use of Candida glabrata in organic acid production.
C. glabrata is an asexual, facultative aerobic, haploid yeast widely used for the production of organic acids such as malic, fumaric, and pyruvic acids (22–25). However, accumulation of organic acids causes acidification of the fermentation broth and results in intracellular acidification and oxidative stress, leading to a decrease in metabolite production (26, 27). In S. cerevisiae, the tail module subunit Med15 can be involved in glucocorticoid metabolism (28) and various external stress responses as described above. Based on BLAST data, C. glabrata Med15B (CAGL0H06215g) shares 90% nucleotide sequence identity with Med15 from S. cerevisiae. In C. glabrata, Med15B is required for mediation of multidrug resistance (10) and in response to starvation. In this study, we assessed the function of Med15B in acid stress tolerance of C. glabrata and investigated whether its overexpression could enhance cell growth at a low pH. To this end, we constructed and characterized med15BΔ deletion and HTUΔ/CgMED15B overexpression strains.
RESULTS
Med15B is necessary for growth at pH 2.0.
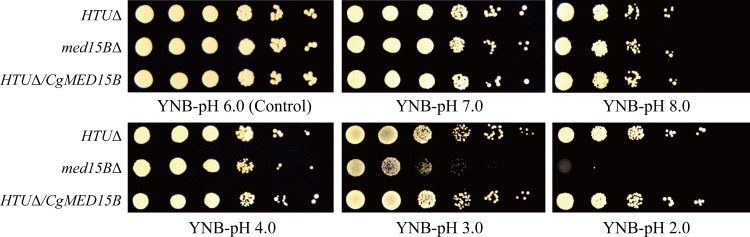
First, we determined whether Med15B was required for the growth of C. glabrata at low pH. The parent strain (C. glabrata HTUΔ) and the med15BΔ and HTUΔ/CgMED15B mutants were spotted onto YNB (0.67% yeast nitrogen base, 2% glucose [pH 6.0]) medium at pH values ranging from 2.0 to 8.0 (Fig. 1). Cells lacking the MED15B gene showed significantly reduced growth at pH 2.0, whereas no significant difference in cell growth was observed between Med15B-overexpressing and parent strains (Fig. 1).
FIG 1.
Spot assays show that Med15B is required for C. glabrata growth at low pH. Growth profiles of the parent (HTUΔ), med15AΔ, and HTUΔ/CgMED15B strains grown on YNB medium at different pHs.
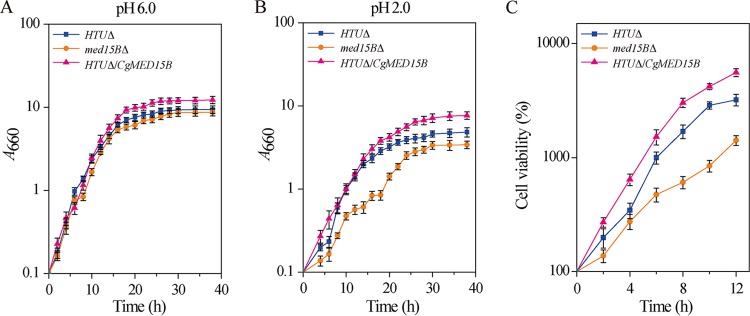
The growth curves of the three strains were compared at pH 6.0 and pH 2.0 (Fig. 2). At pH 6.0, growth of the med15BΔ strain was similar to that of the parent strain, whereas the final biomass and glucose consumption rate of the HTUΔ/CgMED15B strain were 28.6% and 20.1% higher, respectively, than those of the parent strain (Fig. 2A). At pH 2.0, the final biomass and glucose consumption rate of the med15BΔ strain were 28.3% and 31.7% lower, respectively, than those of the parent strain. In contrast, in the HTUΔ/CgMED15B strain, the values were 36.5% and 32.5% higher, respectively, than those of the parent strain (Fig. 2B). Consistent with the results of the spot assay shown in Fig. 1, growth of the med15BΔ strain was much slower than that of the parent strain. The observed differences could be due to differences in viability between the cell populations of C. glabrata at pH 2.0. To test this hypothesis, viability was determined by counting CFU. After 12 h of incubation at pH 2.0, viability of the med15BΔ strain was reduced by 26.5%, whereas that of the HTUΔ/CgMED15B strain was 2.3-fold higher (Fig. 2C; see also Table S1 in the supplemental material). These results strongly suggested that Med15B plays a vital role in the growth of C. glabrata at pH 2.0 and that growth defects in cells lacking Med15B correlate with decreased viability at low pH.
FIG 2.
Deletion of Med15B decreased growth and cell viability. Growth curves of the parent (HTUΔ), med15BΔ, and HTUΔ/CgMED15B strains at 660 nm at pH 6.0 (A) and pH 2.0 (B). (C) Cell viability of all three strains at pH 2.0. The experiments were performed in biological triplicates. Error bars indicate the standard deviations.
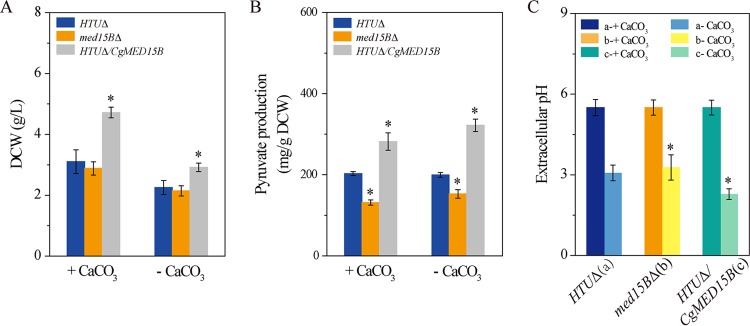
Given that low pH values reduce drastically cell growth, we added CaCO3 as a buffering agent. CaCO3 reacts with the pyruvic acid produced by C. glabrata to form calcium malate and thus maintains the pH at 5.5. We also analyzed pyruvate production without pH buffering by CaCO3 (Fig. 3). When the med15BΔ strain was grown in medium with CaCO3, dry weight of cells and pyruvate production were 7.2% and 35.2% lower, respectively, than those of the parent strain. In contrast, dry weight of cells and pyruvate production by the HTUΔ/CgMED15B strain were 60.0% and 38.7% higher, respectively, than those of the parent strain (Fig. 3A and B). In medium without CaCO3, dry weight of cells and pyruvate production were 6.4% and 23.4% lower, respectively, in the med15BΔ strain but 29.3% and 61.2% higher, respectively, in the HTUΔ/CgMED15B strain than those of the parent strain (Fig. 3A and B). To evaluate the influence of pyruvate accumulation on the pH of the fermentation broth, we measured pH in all three strain cultures. pH was significantly higher, by 0.3 units, in the med15BΔ culture but significantly lower, by 0.9 units, in the HTUΔ/CgMED15B culture, than in the parent strain (Fig. 3C). Consistent with this observation, dry weight of cells was lower whereas pyruvate production was not affected or increased as pH decreased. These results indicate that low pH reduced pyruvate accumulation as a result of growth inhibition and that overexpression of the MED15B gene could enhance cell growth at low pH.
FIG 3.
Med15B is required for pyruvate production. Cell growth (A), pyruvate production (B), and extracellular pH (C) by the parent (HTUΔ, a), med15BΔ (b), and HTUΔ/CgMED15B (c) strains were carried out after 52 h of culturing. The experiments were performed in biological triplicates. Error bars indicate the standard deviations. (*, P < 0.05 compared with the parent stain, as determined by the t test).
Global transcriptome analysis of the parent and med15BΔ strains.
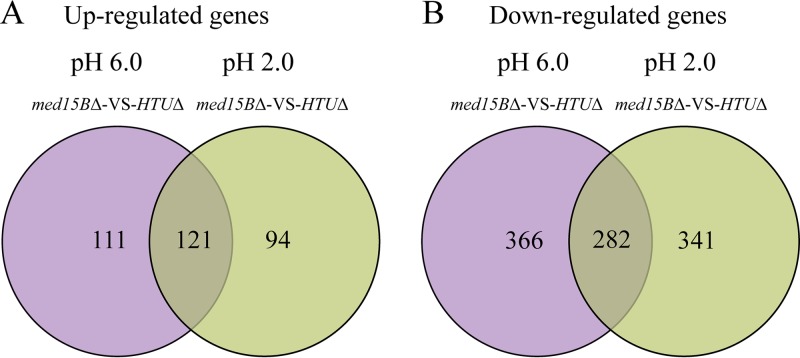
To identify genes that were differentially regulated and may therefore contribute to the growth defects of cells lacking Med15 at low pH, we used transcriptome sequencing (RNA-seq) to compare global gene expression in the parent and med15BΔ strains at pH 6.0 and 2.0. We extracted RNA from each strain 6 h after incubation at pH 2.0 and from three independent experiments. This time point was chosen based on the growth curves because it corresponded to early log phase. The numbers of genes that were differentially expressed (fold change > 1.5, P ≤ 0.01) in the med15BΔ strain compared to their expression in the parent strain are depicted by Venn diagrams in Fig. 4A and B and in Data Sets S1 and S2 in the supplemental material. Transcriptional profiling revealed 232 upregulated and 648 downregulated genes in the med15BΔ strain compared to the parent strain at pH 6.0. At pH 2.0, there were 215 upregulated and 623 downregulated genes in the med15BΔ strain. Interestingly, 121 upregulated and 282 downregulated genes were uniquely differentially expressed in the med15BΔ strain at low pH. Based on gene ontology analysis, genes commonly upregulated in the two strains were involved in many cell processes, such as sterol biosynthesis (0006694), fatty acid metabolism (0006631), sphingolipid metabolism (0006631), gene expression (0010467), ribosome biogenesis (0042254), response to stimuli (0050896), and glucose metabolism (0006007). Meanwhile, the downregulated genes were involved in the cell cycle (0000087), DNA repair (0006281), glucose metabolism (0006007), cellular protein modification process (0006464), fatty acid metabolism (0006631), phospholipid metabolism (0006644), ATP biosynthesis (0006754), cellular homeostasis (0019725), transport (0006810), and signal transduction (0007165), among others (Data Sets S1 and S2).
FIG 4.
Differently regulated genes in the med15BΔ mutant. Transcription profile analysis in response to acid stress is shown in Venn diagrams, depicting the overlap between upregulated (A) and downregulated (B) genes in the med15BΔ strain compared with their expression in the parent (HTUΔ) strain. The results of RNA-seq represent biological triplicates (fold change > 1.5, P ≤ 0.01).
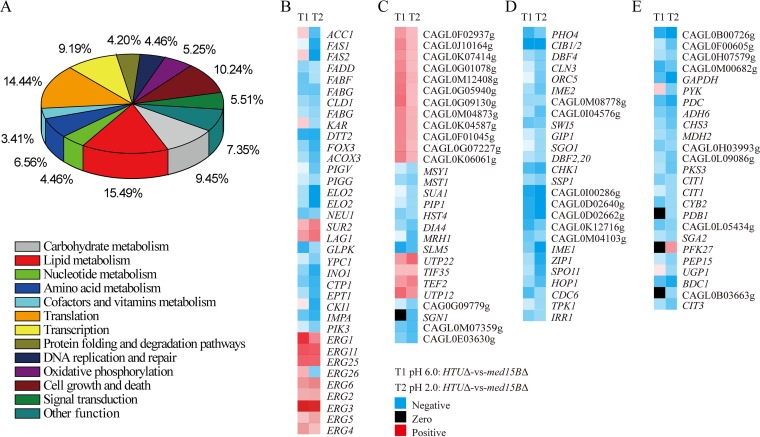
In addition, statistical analysis of the metabolic pathways involving the identified differentially regulated genes was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping. We concluded that membrane lipid metabolism, translation, cell growth and death, and carbohydrate metabolism were the four most notable differentially regulated pathways, accounting for 15.49%, 14.44%, 10.24%, and 9.45%, respectively, of all genes distinctively regulated in the med15BΔ strain at pH 2.0 (Fig. 5A). The next set corresponded to transcription, amino acid metabolism, oxidative phosphorylation, and nucleotide metabolism pathways. Signal transduction, DNA replication and repair, and protein folding and degradation pathways responding to stress were also affected (Fig. 5A; see also Data Set S3 in the supplemental material). The differentially regulated genes associated with the four most notable differentially regulated pathways showed downregulation in heat maps (Fig. 5B to E). These results suggested that deletion of the MED15B gene strongly affects membrane lipid biosynthesis and metabolism in response to an acidic environment.
FIG 5.
Statistical analysis of the metabolic pathways in which the identified significant differentially expressed genes in the med15BΔ strain compared with the parent strain at pH 2.0 are involved. (A) Top 12 statistics of pathway enrichment based on KEGG databases. Heat maps of differentially regulated genes involved in the four most notable differentially regulated pathways, i.e., lipid metabolism (B), translation (C), cell growth and death (D), and carbohydrate metabolism (E) are shown. RNA-seq was performed in biological triplicates.
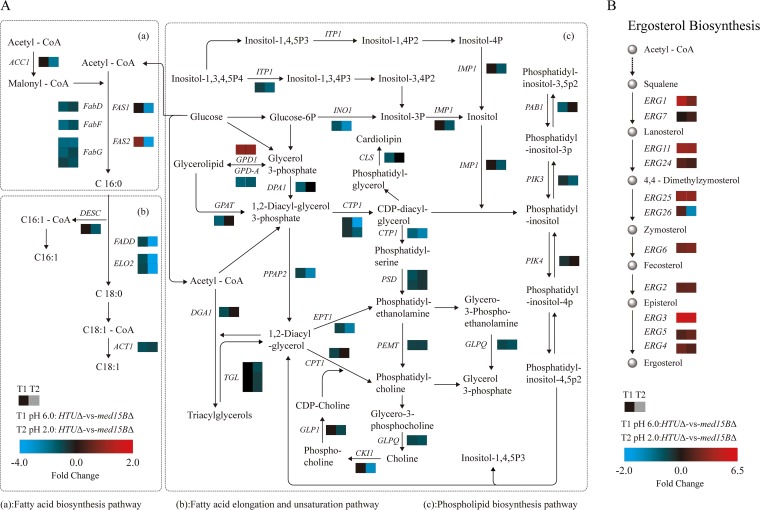
Next, we compared the expression levels in the med15BΔ and parent strains of the genes associated with membrane lipid metabolism (Fig. 6A and B; see also Data Set S4 in the supplemental material). We observed selective downregulation of genes involved in (i) fatty acid biosynthesis, such as the genes encoding acetyl coenzyme A (acetyl-CoA)/propionyl-CoA carboxylase (ACC1), tetrafunctional fatty acid synthase (FAS1), fatty acid synthase subunit alpha (FAS2), 3-oxoacyl-[acyl-carrier-protein] synthase II (FABF), and 3-oxoacyl-[acyl-carrier-protein] reductase (FABG), which were downregulated by 2.7-, 3.5-, 3.9-, 1.7-, and 2.4-fold, respectively; (ii) fatty acid elongation, such as the genes encoding fatty acid elongase 2 (ELO2) and very-long-chain 3-oxoacyl-CoA reductase (KAR), which were downregulated by 6.7- and 1.5-fold, respectively; (iii) fatty acid unsaturation such as the gene encoding fatty acid desaturase (DESC), which was downregulated by 1.5-fold; and (iv) fatty acid metabolism, such as the genes encoding acetyl-CoA oxidase (ACOX3), acetyl-CoA acyltransferase (FADA), and long-chain acyl-CoA synthetase (FADD), which were downregulated by 2.7-, 3.0-, and 2.2-fold, respectively. Additional downregulated genes included those involved in (i) glycerolipid metabolism, such as the genes encoding glycerate 2-kinase (GLXK), glycerol kinase (GLPK), and triacylglycerol lipase (TGL3), which were downregulated by 1.9-, 1.7-, and 1.5-fold, respectively; (ii) glycerophospholipid metabolism, such as the genes encoding mitochondrial citrate transport protein 1 (CTP1), casein kinase-1 (CKI1), ethanolaminephosphotransferase (EPT1), diacylglycerol cholinephosphotransferase (CPT1), CDP-diacylglycerol-serine O-phosphatidyltransferase (PSSA), glycerol-3-phosphate dehydrogenase (GLPA), and cardiolipin synthase (CMP-forming) (CRD1), which were downregulated by 3.9-, 3.4-, 3.0-, 3.0-, 2.9-, 1.9-, and 1.8-fold, respectively; (iii) sphingolipid metabolism, such as the genes encoding sialidase-1 (NEU1) and phytoceramidase (YPC1), which were downregulated by 2.2- and 2.1-fold, respectively; and (iv) inositol phosphate metabolism, such as the genes encoding inositol-3-phosphate synthase (INO1), itaconate transport protein (ITP1), myo-inositol-1(or 4)-monophosphatase (IMPA), and phosphatidylinositol 3-kinase (PIK3), which were downregulated by 6.3-, 2.2-, 3.4-, and 1.6-fold, respectively (Data Set S4). We also observed a set of upregulated genes involved in sterol biosynthesis, such as those encoding lanosterol 14-alpha-demethylase (ERG11), methylsterol monooxygenase (ERG25), sterol methyltransferase (ERG6), Δ8-Δ7-sterol isomerase (ERG2), C-5 sterol desaturase (ERG3), C-22 sterol desaturase (ERG5), and C-24 sterol reductase (ERG4) (Fig. 6B). These genes, playing an important role in sterol biosynthesis, from acetyl-CoA to ergosterol, were overexpressed in the med15BΔ strain at both pH 6.0 and pH 2.0 (Fig. 6B). However, the expression of the ERG26 gene, encoding C-3 sterol dehydrogenase (C-4 decarboxylase), was increased in the med15BΔ strain at pH 6.0 but decreased at pH 2.0. These results demonstrated that pathways influencing membrane composition, such as those for fatty acid synthesis and phospholipid and inositol metabolism, as well as ergosterol biosynthesis, were affected in the deletion mutant. This suggested that Med15B might regulate the cell membrane’s lipid composition under acid stress conditions.
FIG 6.
Effect of deletion of Med15B on the lipid membrane at low pH. Shown are measured changes in the expression of genes involved in fatty acid (panel A, subpanels a and b [left]) and phospholipid (panel A, subpanel c [right]) metabolism and sterol biosynthesis (panel B) in the med15BΔ strain compared to those in the parent strain at pH 6.0 and at pH 2.0. RNA-seq was performed in biological triplicates.
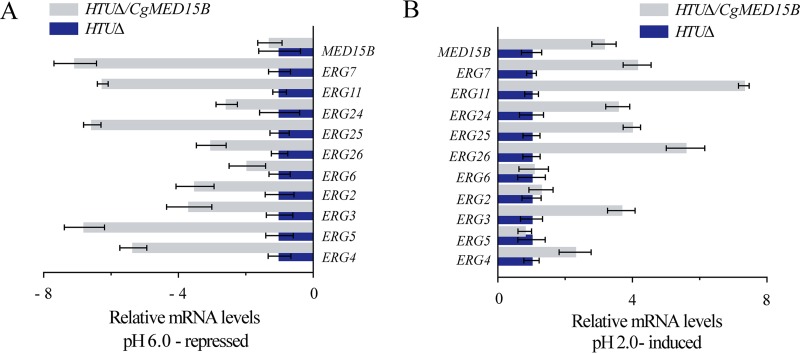
Given the abnormal upregulation of ERG genes in the med15BΔ strain, we analyzed the expression of ERG genes in the HTUΔ/CgMED15B and parent strains at pH 6.0 and pH 2.0 (Fig. 7A and B). The levels of expression of MED15B and ERG genes decreased significantly in the HTUΔ/CgMED15B strain at pH 6.0 (Fig. 7A) but increased significantly at pH 2.0 (Fig. 7B). Thus, these results suggested that the expression of genes involved in sterol biosynthesis is highly dependent on Med15B.
FIG 7.
Effect of overexpression of Med15B on the sterol biosynthesis at low pH. Changes in ERG gene expression in the HTUΔ/CgMED15B strain compared with the parent strain at pH 6.0 (A) and at pH 2.0 (B). The experiments were performed in biological triplicates. Error bars indicate the standard deviations.
MED15B regulates membrane lipid composition.
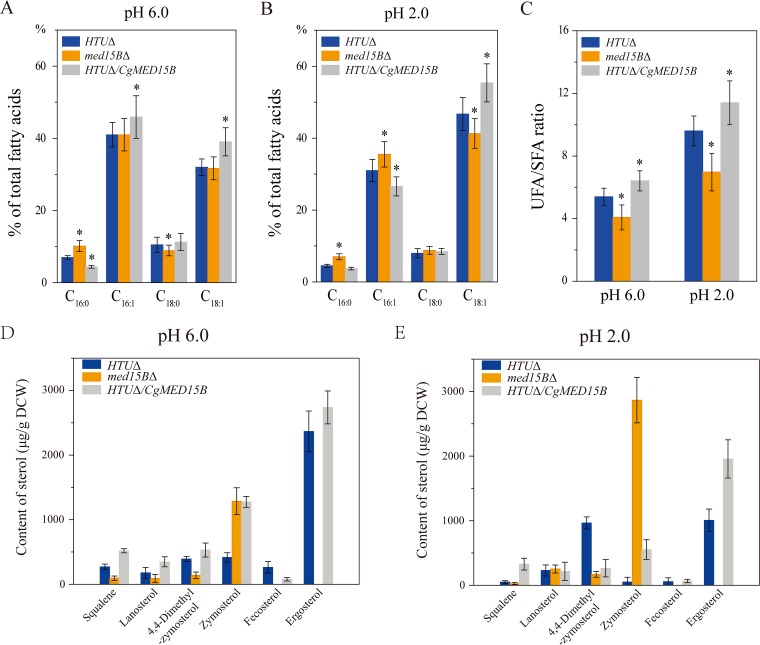
Next, we determined membrane compositions of the parent, med15BΔ, and HTUΔ/CgMED15B strains at pH 6.0 and 2.0 (Fig. 8). At pH 6.0, C16:0 and C18:0 levels in the med15BΔ strain were 45.2% higher and 14.9% lower, respectively, than those in the parent strain, whereas C16:1 and C18:1 levels were unchanged. In the HTUΔ/CgMED15B strain, the C16:0 level was 37.2% lower, whereas those of C18:0, C16:1, and C18:1 were 7.4%, 12.1%, and 21.6% higher, respectively (Fig. 8A). At pH 2.0, the levels of C16:0, C18:0, and C16:1 were 56.3%, 9.7%, and 14.5% higher, respectively, while that of C18:1 was 11.6% lower in the med15BΔ strain than those in the parent strain. In the HTUΔ/CgMED15B strain, C16:0 and C16:1 were 17.4% and 14.3% lower, respectively, whereas C18:0 and C18:1 were 6.5% and 18.6% higher, respectively, than those in the parent strain (Fig. 8B). Overall, the ratio of unsaturated to saturated fatty acids (UFA/SFA) decreased by 24.5% in the med15BΔ strain and increased by 8.1% in the HTUΔ/CgMED15B strain with respect to the parent strain at pH 6.0 (Fig. 8C). However, at pH 2.0, UFA/SFA exhibited a decrease of 27.4% in the med15BΔ strain and an increase of 18.7% in the HTUΔ/CgMED15B strain.
FIG 8.
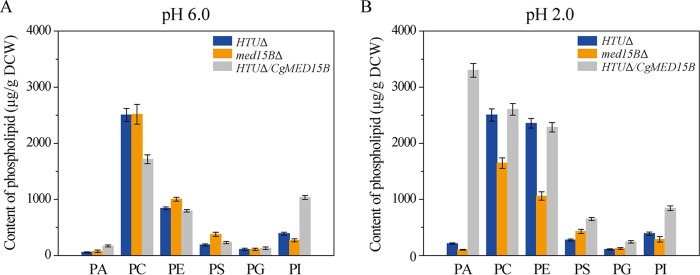
Med15B affects the membrane fatty acids and sterol of C. glabrata. Changes in the percentage of fatty acids in the parent (HTUΔ), med15BΔ, and HTUΔ/CgMED15B strains at pH 6.0 (A) and pH 2.0 (B). (C) Changes in UFA/SFA at pH 6.0 and pH 2.0. Sterol analysis was carried out on strains cultivated in shake-flasks at pH 6.0 (D) and pH 2.0 (E). The experiments were performed in biological triplicates. Error bars indicate the standard deviations. (*, P < 0.05 compared with the parent strain, as determined by the t test).
Membrane sterol content was measured to determine whether Med15B could affect ergosterol biosynthesis (Fig. 8D and E). We found that cells lacking the MED15B gene failed to catalyze the conversion of zymosterol to fecosterol and ergosterol. This caused zymosterol accumulation; its concentration was 1,285.8 μg/g cells (dry weight) at pH 6.0 and 2,866.1 μg/g cells (dry weight) at pH 2.0. At pH 6.0, the amount of total sterols decreased by 43.5% in the med15BΔ strain, whereas the concentration of zymosterol increased 2.1-fold compared to that in the parent strain. In the HTUΔ/CgMED15B strain, the amounts of total sterols, zymosterol, and ergosterol increased by 65.1%, 206.3%, and 15.7%, respectively, whereas that of fecosterol decreased by 70.4% (Fig. 8D). At pH 2.0, deletion of the MED15B gene resulted in a 91.4% decrease in the amount of total sterols, whereas zymosterol increased about 60.3-fold versus the amounts in the parent strain. Overexpression of Med15B improved the biosynthesis of total sterols, zymosterol, fecosterol, and ergosterol at pH 2.0. Their concentrations increased by 60.8%, 971.5%, 16.6%, and 94.5%, respectively, with respect to the levels in the parent strain (Fig. 8E). In the HTUΔ/CgMED15B strain, the concentrations of ergosterol precursors, such as squalene, lanosterol, and 4,4-dimethylzymosterol, which play an important role in the response to environmental variations, were 1.1-fold higher at pH 6.0 and 40.2% higher at pH 2.0 than those in the parent strain. Taken together, the data indicated that the Med15B protein is critical for sterol biosynthesis from zymosterol to fecosterol and ergosterol and is important for regulating the type and amount of cell membrane sterols.
We found that the total content of phospholipids increased by 43.0% and 143.5% in the parent and HTUΔ/CgMED15B strains, respectively, and decreased by 16.2% in the med15BΔ strain when pH was lowered from 6.0 to 2.0. At pH 6.0, the contents of phosphatidic acid (PA), phosphatidylethanolamine (PE), and phosphatidylserine (PS) increased by 33.3%, 19.3%, and 102.0%, respectively, whereas the content of phosphatidylinositol (PI) decreased by 30.8% in the med15BΔ strain compared to those in the parent strain. The contents of phosphocholine (PC) and phosphatidylglycerol (PG) were unchanged. In the HTUΔ/CgMED15B strain, PA, PS, PG, and PI contents increased by 205.3%, 23.5%, 17.0%, and 165.2%, respectively; PC content decreased by 31.5%; and that of PE was unchanged (Fig. 9A). In the med15BΔ strain at pH 2.0, total phospholipid, PA, PC, PE, and PI contents decreased by 37.6%, 52.5%, 34.3%, 55.0%, and 26.5%, respectively, whereas PS and PG contents increased by 55.5% and 14.5%, respectively, compared to those in the parent strain. In the HTUΔ/CgMED15B strain, total phospholipid, PA, PS, PG, and PI contents increased 1.4-, 14.6-, 1.4-, 1.2-, and 1.2-fold, respectively, whereas PC and PE content did not differ significantly from that of the parent strain. At low pH, Med15B appears to contribute to the biosynthesis of PA, thereby possibly enhancing the biosynthesis of other phospholipids.
FIG 9.
Med15B affects the membrane phospholipids of C. glabrata. Changes in the content of phospholipids in the parent (HTUΔ), med15BΔ, and HTUΔ/CgMED15B strains at pH 6.0 (A) and pH 2.0 (B). Abbreviations: PA, phosphatidic acid; PC, phosphocholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PG, phosphatidylglycerol; PI, phosphatidylinositol. The experiments were performed in biological triplicates. Error bars indicate the standard deviations.
Med15B is necessary for membrane integrity and fluidity.
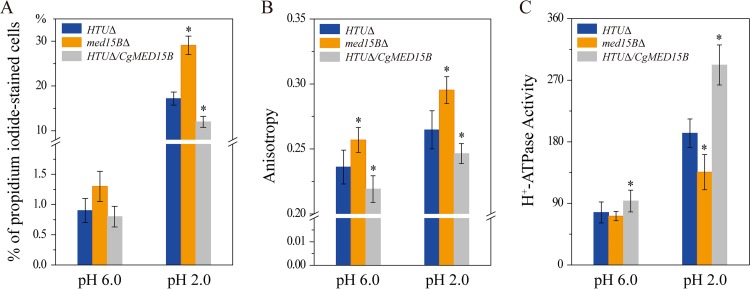
To investigate the role of Med15B in membrane integrity and fluidity, cells of the parent, med15BΔ, and HTUΔ/CgMED15B strains were subjected to acidic (pH 2.0) conditions for 8 h. We observed no significant differences in the percentages of propidium iodide-stained cells at pH 6.0. Instead, at pH 2.0, the percentage of propidium iodide-positive staining increased significantly, by 69.2%, among med15BΔ cells but decreased significantly, by 30.2%, among HTUΔ/CgMED15B cells compared to the parent strain (Fig. 10A).
FIG 10.
Med15B affects the membrane integrity, fluidity, and H+-ATPase activity. (A) Flow cytometry analyses of membrane integrity in the parent (HTUΔ), med15BΔ, and HTUΔ/CgMED15B strains at pH 6.0 and pH 2.0. Cells were stained with propidium iodide. (B) Fluidity assays in the same strains as for panel A at pH 6.0 and pH 2.0. (C) H+-ATPase activity assays in the same strains as for panel A at pH 6.0 and pH 2.0. The experiments were performed in biological triplicates. Error bars indicate the standard deviations. (*, P < 0.05 compared with the parent strain, as determined by the t test).
Next, we evaluated membrane fluidity and found that at pH 6.0, it decreased by 8.8% in the med15BΔ strain and increased by 7.2% in the HTUΔ/CgMED15B strain compared with that in the parent strain. At pH 2.0, in response to low-pH stress, membrane fluidity was 11.6% lower and 6.9% higher in the med15BΔ and HTUΔ/CgMED15B strains, respectively, than that in the parent strain (Fig. 10B).
At pH 6.0, the activity of the H+-ATPase was 7.2% lower in the med15BΔ strain but 21.8% higher in the HTUΔ/CgMED15B strain than that in the parent strain. At pH 2.0, the activity of the H+-ATPase decreased by 29.5% in the med15BΔ strain and increased by 51.8% in the HTUΔ/CgMED15B strain (Fig. 10C). In conclusion, Med15B is essential for membrane function at low-pH-stress-inducing conditions. Overall, cells lacking the MED15B gene were defective in the integrity and fluidity of the membrane and in the activity of the H+-ATPase, especially under acidic conditions.
DISCUSSION
In this work, we investigated the response of the Mediator subunit Med15B to low pH. We report that Med15B plays an important role in regulating the expression of genes related to lipid metabolism, thereby altering membrane lipid composition (fatty acids, sterols, phospholipids) and influencing membrane integrity, fluidity, and H+-ATPase activity. This function of C. glabrata Med15B is critical for cell growth and pyruvate accumulation at low pH.
Various approaches can be used to enhance pyruvate production of C. glabrata, such as random mutagenesis, optimization of the fermentation medium, biphasic fermentation, and lowering by-product production through removal of certain factors from the metabolic network. Nevertheless, such approaches have some serious drawbacks, such as a heavy workload, uncertainty, and growth defects (23, 29–31). Here, we propose a strategy that could overcome the above-mentioned problems by enhancing pyruvate production through overexpression of Med15B and increased dry weight of cells. In this study, we found that deletion of MED15B caused a significant growth defect at pH 2.0 whereas overexpression of MED15B enhanced cell tolerance and growth at low pH. Additionally, we tested the phenotypes of the med15BΔ and HTUΔ/CgMED15B strains under different biological stimuli and found that Med15B was essential for cell tolerance under oxidative and osmotic stress conditions (data not shown). These data are consistent with Med15 from S. cerevisiae having a critical role in the stress response (20, 21, 32). Our results indicated that Med15B has a vital modulatory role in pH homeostasis in C. glabrata, so it will be important to examine this function in other eukaryotes.
Deletion of MED15B caused significant changes at the global transcriptome level at pH 2.0. Differentially expressed genes were involved in various metabolic pathways, most notably lipid metabolism. A set of genes involved in fatty acid biosynthesis, elongation, unsaturation, and metabolism were downregulated. Many of these genes were reported to be involved in the length and number of double bonds in fatty acids and in the response to various forms of stress (33, 34), which may influence the species of phospholipid produced (35). Meanwhile, key genes involved in phospholipid and sphingolipid metabolism were also downregulated, thus contributing to a decreased phospholipid and sphingolipid biosynthesis in the med15BΔ strain. Several reports have shown that the type and proportion of phospholipids and sphingolipids can affect acid tolerance (36–38). In particular, previous studies have shown that ERG25 and ERG3 deletion or inactivation decreased the biosynthesis of ergosterol under various stress conditions (39, 40). Our study showed that deletion or overexpression of MED15B increased expression of genes related to sterol biosynthesis at pH 2.0. Quantification of sterol content showed significant differences in sterol composition between med15BΔ, HTUΔ/CgMED15B, and parent strains. Deletion of MED15 has been shown to lead to the selective repression of genes involved in ribosomal biogenesis (41). However, we found that the expression of genes encoding large- or small-subunit ribosomal proteins increased (Fig. 5C). Expression of the battenin gene (CLN3), involved in the cell cycle, was downregulated by 2.0-fold. This result is consistent with the observation that inactivation of Med15 causes the cell cycle-related genes to be downregulated (42). Med15 supports the expression levels of genes involved in energy homeostasis, including glucose and lipid metabolism, in S. cerevisiae and Caenorhabditis elegans (13, 43). Here, key genes of glycolysis, of the tricarboxylic acid cycle, and of oxidative phosphorylation showed downregulation in the med15BΔ strain at pH 2.0. These results may explain the defective pyruvate production and the lower membrane H+-ATPase activity of the med15BΔ strain. Additionally, DNA damage may affect cell growth under various stress conditions (44). Genes involved in DNA repair and the MAPK signaling pathway, which are stress response pathways, were also significantly affected. These results are in agreement with previous reports that showed that these Med15B-dependent genes were related to various environmental stress responses (45).
Alteration of membrane composition serves as an adaptive response to variations in pH (33). Our genome-wide analyses confirmed the critical role played by the composition and structure of the membrane lipid and its physiological function in acid tolerance (46–48). Deletion of MED15B decreased the long-chain fatty acid and phospholipid proportion and the UFA/SFA ratio and blocked the biosynthesis of fecosterol and ergosterol, thereby increasing the zymosterol concentration to 2,866.1 μg/g cells (dry weight) at pH 2.0. At the same time, these parameters increased upon MED15B overexpression. Sphingolipid is one of the important components of membrane. Our work has revealed that Med15B influences the expression of genes involved in sphingolipid biosynthesis at pH 2.0. Further studies will be required to determine the role played by Med15B in sphingolipid content change at low pH. Consistent with the alteration of lipid composition, membrane integrity, fluidity, and H+-ATPase activity were significantly changed. These results are in agreement with previous reports that have demonstrated that to adapt to environmental acidification, acid-resistant microorganisms developed a number of strategies. These include (i) increasing the content of long-chain fatty acids and UFAs, which leads to greater membrane fluidity and integrity (47, 48); (ii) augmenting sterol content, which has an effect on membrane fluidity, increasing its thickness and sturdiness (49); and (iii) enhancing H+-ATPase activity, which promotes proton efflux (35). Changes in the lipid composition of the med15BΔ strain might affect the activity of membrane proteins, which are essential for barrier function (50, 51). The transcription factor Crz1 was shown to function in low-pH tolerance in C. glabrata (33). In this study, we explored the role of C. glabrata Mediator subunit Med15B in providing tolerance to low-pH conditions. crz1Δ and med15BΔ strains were observed to possess a similar phenotype. Moreover, we identified a shared function of Crz1 and Med15B mutants, characterized by downregulation of genes involved in lipid metabolism and a growth defect at low pH. This finding suggests that the transcriptional activator Crz1 could be modulating Med15B-dependent gene expression. It will be interesting to examine how such an interaction may occur. In summary, our results demonstrated that (i) it is possible to enhance pyruvate production by overexpressing MED15B; (ii) increased tolerance at low pH is associated with altered membrane composition and function; and (iii) acquiring tolerance to low pH may lead to an increased tolerance to the membrane-damaging stress caused by other organic acids produced by C. glabrata.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Unless stated otherwise, YNB (0.67% yeast nitrogen base, 2% glucose [pH 6.0]) medium containing essential nutrients was used to culture C. glabrata. Indicated media were brought to the desired pH with HCl or NaOH. Strains were incubated at 30°C with shaking at 200 rpm.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| C. glabrata ATCC 2001 | Wild-type strain | 45 |
| C. glabrata ATCC 55 (HTUΔ) | his3Δ trp1Δ ura3Δ | 45 |
| C. glabrata med15BΔ | his3Δ trp1Δ ura3Δ med15BΔ::CgHIS3 | This work |
| C. glabrata HTUΔ/pY26 | his3Δ trp1Δ ura3Δ | This work |
| C. glabrata HTUΔ/CgMED15B | his3Δ trp1Δ ura3Δ::URA3(pY26) CgMED15B::URA3(pY26/NotI-SacII) | This work |
| Plasmids | ||
| pY26 | 2μm, Ampr,a URA3, PGPD, PTEF | 24 |
| pY26-CgMED15B | 2μm, Ampr, URA3, PGPD, PTEF-CgMED15B | This work |
Ampr, ampicillin resistance.
All mutant strains were generated from C. glabrata ATCC 55 (HTUΔ::his3Δ trp1Δ ura3Δ; a gift from Karl Kuchler). The med15BΔ mutant was generated by genomic integration of a HIS3 maker in the MED15B locus. The maker gene and flanking regions were amplified from the genome of C. glabrata ATCC 2001 (wild type; a gift from Karl Kuchler) by PCR. The fusion PCR fragments were transformed into the parent strain by electroporation as described previously (27). The deletion strain was verified by genomic PCR and DNA sequencing.
The PCR fragments of gene CgMED15B were amplified from the genome of the wild-type strain, cut at the introduced NotI and SacII sites, and ligated to the shuttle plasmid pY26, which was digested with NotI and SacII, resulting in pY26-CgMED15B. Plasmids pY26 and pY26-CgMED15B were transformed into the parent strain by electroporation and yielded overexpression HTUΔ/CgMED15B strain. The primer sequences used in this work are shown in Table 2. The detailed protocols for strain construction are described in the supplemental material.
TABLE 2.
Primers used for the strains construction
| Target | Primer name | Sequencea |
|---|---|---|
| Deletion | ||
| MED15B-Up | US-MED15Bdel | CCTGTGACAGCTAAATGGAAG |
| UA-MED15Bdel | ACCCTCTTAACAAACGCCATTGTTTCATTGTCAGTTGTGTATTT | |
| Marker HIS3 | MS-HIS3del | ACACAACTGACAATGAAACAATGGCGTTTGTTAAGAGGGTT |
| MA-HIS3del | TCATCCCTGTCATTTACAGTCTATGCTAGGACACCCTTAGTGG | |
| MED15B-Down | DS-MED15Bdel | CCACTAAGGGTGTCCTAGCATAGACTGTAAATGACAGGGATGACTT |
| DA-MED15Bdel | CGAAATTAAAAACGACACAACTC | |
| Deletion check | ||
| med15BΔ | S-MED15Bdel-ch | CCATGCTACCACTACAATAACG |
| A-MED15Bdel-ch | TGGATTCCTTCCCATTCTTA | |
| Overexpression | ||
| pY26-CgMED15B | S-MED15Bover | ATAAGAATGCGGCCGCATGTCTAGCAAAGAGACCATTCC (NotI) |
| A-MED15Bover | TCCCCGCGGTTACTCTATACTTGTCCAGAAATTCC (SacII) | |
| Overexpression check | ||
| HTUΔ/CgMED15B | S-MED15Bover-ch | AAGTTTTCTAGAACTAGCGCG |
| A-MED15Bover-ch | GTGTCAACAACGTATCTACCAAC |
Regions flanking the target gene and restriction site sequences are underlined.
Phenotypic analysis.
Log-phase cells were diluted in sterile water to an absorbance at 660 nm (A660) of 1.0. Serial dilutions (10-fold) of 4 μl were spotted onto plates at different pH values and incubated at 30°C for 3 to 5 days.
Growth and viability analysis.
Log-phase cells were inoculated into fresh YNB medium at pH 6.0 and pH 2.0 with an A660 of 1.0. Cultures were taken at regular time intervals, and the A660 values were recorded. For viability analysis, cultures grown in YNB at pH 2.0 were appropriately diluted and plated onto YNB plates at various time points. Total CFU were calculated by counting viable colonies that appeared after incubation at 30°C for 2 days. A histogram was prepared to illustrate cell survival over time.
Pyruvate production and analysis.
The strains were cultivated at 30°C in 250-ml flasks containing 25 ml medium A, consisting of, per liter, 30 g glucose, 10 g peptone, 1.0 g KH2PO4, and 0.5 g MgSO4·7H2O, for 24 h. Cells were centrifuged, washed with distilled water, and inoculated into 500-ml flasks containing 50 ml medium B, consisting of, per liter, 100 g glucose, 3 g KH2PO4, 0.8 g MgSO4·7H2O, 3 g sodium acetate, 18 μg thiamine-HCl, 4.0 μg biotin, 40 μg pyridoxine-HCl, and 0.8 μg nicotinic acid, with an initial biomass (dry weight) of 0.5 g/liter. Fermentation was performed at 30°C over 48 h in medium B alone or buffered with 40 g/liter CaCO3.
Analytical methods.
The A660 was calibrated against the dry weight of cells on the basis of a standard curve (52): A660/DCW = 1/0.23 g/liter, where DCW is the dry weight of cells.
The pyruvate concentrations were determined by high-performance liquid chromatography (HPLC; Dionex, Shanghai, China), using an Aminex HPX-87H column eluted at a flow rate of 0.6 ml min−1 (53). The extracellular pH was measured by a pH meter (Dionex).
Total RNA extraction and transcriptome sequencing (RNA-seq).
Log-phase cells were inoculated into fresh YNB medium at pH 6.0 and pH 2.0 with an A660 of 1.0. After incubation for 6 h, cultures were centrifuged and washed twice with phosphate-buffered saline (PBS; pH 7.4). Samples were frozen with liquid nitrogen, and total RNA was extracted using the Mini BEST Universal RNA extraction kit (TaKaRa, Japan) and sent to the Beijing Genomics Institute (Beijing, China) (http://www.genomics.cn/index), which provides global gene analysis services. rRNA was removed, and mRNA was enriched using the NEBNext poly(A) mRNA magnetic isolation module (E7490; New England BioLabs [NEB], Ipswich, MA, USA). The NEBNext mRNA library prep master mix set for Illumina (E6110; NEB) and NEBNext multi-oligos for Illumina (E7500; NEB) were used to construct the libraries. Clusters were obtained from the qualified libraries on an Illumina cBot (Illumina, San Diego, CA, USA), and Illumina HiSeq 2500 was used to perform the sequencing. C. glabrata genes were annotated according to the similarity of their sequence to putative orthologs in other yeast species.
qRT-PCR.
Total RNA extraction was carried out as described above. Total RNA (1 μg) was taken to synthesize cDNA using the PrimeScript II 1st-strand cDNA synthesis kit (TaKaRa, Japan). The cDNA mixture was diluted to about 100 ng/μl and used as the template for the gene expression level analysis by quantitative reverse transcription-PCR (qRT-PCR). qRT-PCR was performed with SYBR Premix Ex Taq (TaKaRa, Japan) using an iQ5 continuous fluorescence detector system (Bio-Rad, Hercules, CA). Data were normalized to that of β-actin gene ACT1. The primer sequences used in qRT-PCR are listed in Table 3.
TABLE 3.
Primers used for the qRT-PCR
| Target | Primer name | Sequence |
|---|---|---|
| MED15B | MED15B-S | GGTCCAGTAATGAACGATGC |
| MED15B-A | GGTTTGCTCCTTTGGTCTATT | |
| ERG2 | ERG2-S | CCGGTGTCGGTTACCTTT |
| ERG2-A | GTGCTTAGCCAATGCGTCT | |
| ERG3 | ERG3-S | CACACCGTCCACCACTTG |
| ERG3-A | CGTCACCTTCCACCTCTTG | |
| ERG4 | ERG4-S | TGGGTATGCTAATTGGCTTCC |
| ERG4-A | CAGTAGTAAGGTAGTTGCTTGTTC | |
| ERG5 | ERG5-S | TTGTGGTGTGCTGGTGTG |
| ERG5-A | TGACGAACTTGTGGAAGATGG | |
| ERG6 | ERG6-S | ATCTTGCTGACGAGGATGAC |
| ERG6-A | GCGACGAATAGGAACATTGG | |
| ERG7 | ERG7-S | ATACATCGTCAATACAGCACATC |
| ERG7-A | CCAATAGCACCGCCTAACC | |
| ERG11 | ERG11-S | GGACACCGACTTCGCTTAC |
| ERG11-A | CCATCAAGACACCAATCAATAGG | |
| ERG24 | ERG24-S | GGCGTCGGCACTAATGAG |
| ERG24-A | CAATAACCACAGCAGCATACC | |
| ERG25 | ERG25-S | ACCTTATGTGTATGGATTACCTTG |
| ERG25-A | CTTCTCTGGCGACCTTAGC | |
| ERG26 | ERG26-S | TTCTGGCAAGTAAGTGTAATGTAG |
| ERG26-A | GGACCATATCTTCAGCAATAGC | |
| β-ACTIN | β-ACTIN -S | ACCGCTGCTCAATCTTCC |
| β-ACTIN -A | GGTTTGCTCCTTTGGTCTATT |
Fatty acids extraction and measurement.
Log-phase cells were inoculated into fresh YNB medium at pH 6.0 and pH 2.0 with an A660 of 1.0 for 6 to 8 h. Cells were harvested, washed twice with PBS, and freeze-dried. Fifty milligrams of dried cells was resuspended in an NaOH-methanol-distilled water solution (3:10:10, wt/vol/vol). The sample saponification and methylation and extraction of total fatty acids were carried out as described previously (54). Samples were analyzed by gas chromatography (GC) with a polyethylene glycol capillary column eluted at a flow rate of 29.6 ml/min as described previously (55).
Sterol extraction and measurement.
Freeze-dried samples were obtained as described above, and 50 mg dried cells was used for saponification. The saponified samples were resuspended in hexane for total sterol extraction as described previously (33). Samples of total sterol were analyzed by gas chromatography-mass spectrometry (GC-MS) (Waters, Santa Clara, MA) with a fused-silica capillary column (30 m by 0.25 mm by 0.25 μm DB-5MS stationary-phase column; J&W Scientific, Folsom, CA, USA) (56). A 10-μl solution was injected using an autosampler, and the injector temperature was 280°C. The data mass spectrometry was operated at a range of m/z 50 to 800.
Phospholipid extraction and measurement.
Freeze-dried samples were obtained as described above, and 50 mg dried cells was used for phospholipid analyses. Phospholipid extraction was performed in four steps with different ratios of chloroform to methanol as described previously (57). The extracted phospholipids were dried under a nitrogen stream and dissolved in chloroform-methanol (1:1, vol/vol). Samples of phospholipids were analyzed by electrospray ionization mass spectrometry (ESI-MS; Shimadzu, Japan) (58).
Cell membrane integrity analysis.
Log-phase cells were inoculated into fresh YNB medium at pH 6.0 and pH 2.0 with an A660 of 1.0 for 6 to 8 h. Samples were centrifuged, washed twice with PBS, and diluted to an A660 of 0.5. Diluted samples (0.5 ml) were incubated with 3 μl of 1 mg/ml propidium iodide (Sigma, Shanghai City, China), used to monitor the cell membrane integrity. The mixtures were quickly homogenized, kept in the dark for 5 min at room temperature, and then used for flow cytometry analysis.
A FACSCalibur apparatus (BD Biosciences, Shanghai City, China) was used for flow cytometry analysis. The fluorescence emission of propidium iodide-labeled cells was determined through a 660/16-nm bandpass filter. More than 20,000 events were analyzed for each sample and at a rate of 600 to 1,000 events/s. The data were acquired and analyzed using CellQuest software (59).
Cell membrane fluidity analysis.
C. glabrata cells were collected and prepared as described above. Diluted samples (0.5 ml) were incubated with 1 μl of 1-mmol/liter 1,6-diphenyl-1,3,5-hexatriene, used as a probe to monitor changes in membrane dynamics. A spectrofluorimeter (Photon Technology International, Princeton, NJ, USA) was used to determine the steady-state fluorescence anisotropy as described previously (54), with excitation at 360 nm and emission at 450 nm. The degree of fluorescence polarization (p) was calculated as follows: p = (IVV − GIVH)/(IVV + GIVH), where G (G = IHV/IHH) is the correlation fluorescence, I is the fluorescence intensity, and subscripts V and H indicate vertical orientation and horizontal orientation, respectively, of the excitation and analyzer polarizer. The fluorescence anisotropy value (r) was calculated as follows: r = 2p/(3 − p).
Plasma membrane H+-ATPase activity analysis.
C. glabrata cells were collected as described above. Samples were disrupted with an MP FastPrep-homogenizer, and the membrane fractions were isolated from the cell-free supernatant by centrifugation as described previously (60). Membrane H+-ATPase (H+-ATPase) activity was determined according to the amount of inorganic phosphate (Pi) released from ATP in the reaction mixture as described previously (33). A 5-μg membrane fraction was incubated in 120 μl assay mixture A (5 mM ATP, 10 mM MgSO4, 50 mM KCl, 50 mM MES, 50 mM KNO3, 50 mM NaN3, 0.2 mM ammonium molybdate [pH 6.0]), where MES is 2-(N-morpholino)ethanesulfonic acid, at 30°C for 30 min. The reaction was stopped by adding 130 μl assay mixture B (0.6 M H2SO4 with 1% SDS, 1.2% ammonium molybdate, and 1.6% ascorbic acid [wt/vol]). The amount of Pi released was measured at 750 nm after 10 min of incubation at room temperature. The H+-ATPase activity was expressed in micromoles of Pi released per minute per milligram of the total membrane protein.
Accession number(s).
The RNA-seq raw reads were submitted to NCBI under BioProject number PRJNA383945 and SRA study number SRP106931. The Sequence Read Archive [SRA] entries are SRX2802836, SRX2802992, SRX2802993, and SRX2802994.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karl Kuchler for the generous gift of the C. glabrata ATCC 2001 and ATCC 55 strains.
This work was supported by the National Natural Science Foundation of China (grants 31270079 and 21422602).
Author contributions: Y.Q., X.C., and L.L. designed the research; Y.Q., H.L., and J.Y. performed the research; H.L. contributed new reagents; Y.Q. and X.C. analyzed the data; and Y.Q. and L.L. wrote the paper.
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01128-17.
REFERENCES
- 1.Allen BL, Taatjes DJ. 2015. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD. 2016. Structure of a complete Mediator-RNA polymerase II pre-initiation complex. Cell 166:1411–1422. doi: 10.1016/j.cell.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poss ZC, Ebmeier CC, Taatjes DJ. 2013. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plaschka C, Lariviere L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F, Villa E, Cramer P. 2015. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 5.Conaway RC, Conaway JW. 2013. The Mediator complex and transcription elongation. Biochim Biophys Acta 1829:69–75. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kremer SB, Kim S, Jeon JO, Moustafa YW, Chen A, Zhao J, Gross DS. 2012. Role of Mediator in regulating Pol II elongation and nucleosome displacement in Saccharomyces cerevisiae. Genetics 191:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorklund S, Gustafsson CM. 2005. The yeast Mediator complex and its regulation. Trends Biochem Sci 30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Lariviere L, Seizl M, Cramer P. 2012. A structural perspective on Mediator function. Curr Opin Cell Biol 24:305–313. doi: 10.1016/j.ceb.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Robinson PJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. 2012. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc Natl Acad Sci U S A 109:17931–17935. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thakur JK, Arthanari H, Yang FJ, Pan SJ, Fan XC, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Naar AM. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604–612. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 11.Thakur JK, Arthanari H, Yang FJ, Chau KH, Wagner G, Naar AM. 2009. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem 284:4422–4428. doi: 10.1074/jbc.M808263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE, Hahn S. 2011. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell 44:942–953. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ansari SA, Ganapathi M, Benschop JJ, Holstege FCP, Wade JT, Morse RH. 2012. Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J 31:44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. 2013. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol 20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Wang J, Ding Z, Ji J, Sun Q, Cai G. 2013. Structural flexibility and functional interaction of Mediator Cdk8 module. Protein Cell 4:911–920. doi: 10.1007/s13238-013-3069-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul E, Zhu ZI, Landsman D, Morse RH. 2015. Genome-wide association of Mediator and RNA polymerase II in wild-type and Mediator mutant yeast. Mol Cell Biol 35:331–342. doi: 10.1128/MCB.00991-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SK, Chen X, Huang L, Stargell LA. 2013. The head module of Mediator directs activation of preloaded RNAPII in vivo. Nucleic Acids Res 41:10124–10134. doi: 10.1093/nar/gkt796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eyboulet F, Cibot C, Eychenne T, Neil H, Alibert O, Werner M, Soutourina J. 2013. Mediator links transcription and DNA repair by facilitating Rad2/XPG recruitment. Gene Dev 27:2549–2562. doi: 10.1101/gad.225813.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anandhakumar J, Moustafa YW, Chowdhary S, Kainth AS, Gross DS. 2016. Evidence for multiple Mediator complexes in yeast independently recruited by activated heat shock factor. Mol Cell Biol 36:1943–1960. doi: 10.1128/MCB.00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu XF, Chen LH, Carlsten JOP, Liu Q, Yang JS, Liu BD, Gustafsson CM. 2015. Mediator tail subunits can form amyloid-like aggregates in vivo and affect stress response in yeast. Nucleic Acids Res 43:7306–7314. doi: 10.1093/nar/gkv629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E. 2007. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol 27:3900–3910. doi: 10.1128/MCB.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyata R, Yonehara T. 2000. Breeding of high-pyruvate-producing Torulopsis glabrata and amino acid auxotrophic mutants. J Biosci Bioeng 90:137–141. doi: 10.1016/S1389-1723(00)80100-0. [DOI] [PubMed] [Google Scholar]
- 23.Wang QH, He P, Lu DJ, Shen A, Jiang N. 2005. Metabolic engineering of Torulopsis glabrata for improved pyruvate production. Enzyme Microb Technol 36:832–839. doi: 10.1016/j.enzmictec.2005.01.015. [DOI] [Google Scholar]
- 24.Chen X, Xu G, Xu N, Zou W, Zhu P, Liu L, Chen J. 2013. Metabolic engineering of Torulopsis glabrata for malate production. Metab Eng 19:10–16. doi: 10.1016/j.ymben.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Wu J, Song W, Zhang L, Wang H, Liu L. 2015. Fumaric acid production by Torulopsis glabrata: engineering the urea cycle and the purine nucleotide cycle. Biotechnol Bioeng 112:156–167. doi: 10.1002/bit.25334. [DOI] [PubMed] [Google Scholar]
- 26.Patnaik R, Louie S, Gavrilovic V, Perry K, Stemmer WP, Ryan CM, del Cardayre S. 2002. Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol 20:707–712. doi: 10.1038/nbt0702-707. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Chen XL, Cai LJ, Tang L, Liu LM. 2015. Transcription factors Asg1p and Hal9p regulate pH homeostasis in Candida glabrata. Front Microbiol 6:843. doi: 10.3389/fmicb.2015.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DH, Kim GS, Yun CH, Lee YC. 2008. Functional conservation of the glutamine-rich domains of yeast Gal11 and human SRC-1 in the transactivation of glucocorticoid receptor Tau 1 in Saccharomyces cerevisiae. Mol Cell Biol 28:913–925. doi: 10.1128/MCB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua Q, Araki M, Koide Y, Shimizu K. 2001. Effects of glucose, vitamins, and DO concentrations on pyruvate fermentation using Torulopsis glabrata IFO 0005 with metabolic flux analysis. Biotechnol Progr 17:62–68. doi: 10.1021/bp000138l. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Gao NF. 2007. Application of response surface methodology in medium optimization for pyruvic acid production of Torulopsis glabrata TP19 in batch fermentation. J Zhejiang Univ Sci B 8:98–104. doi: 10.1631/jzus.2007.B0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Z, Zeng W, Du G, Liu S, Fang F, Zhou J, Chen J. 2017. A high-throughput screening procedure for enhancing pyruvate production in Candida glabrata by random mutagenesis. Bioprocess Biosyst Eng 40:693–701. doi: 10.1007/s00449-017-1734-x. [DOI] [PubMed] [Google Scholar]
- 32.Kim S, Gross DS. 2013. Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and Mediator tail subunits Med15 and Med16. J Biol Chem 288:12197–12213. doi: 10.1074/jbc.M112.449553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan DN, Lin XB, Qi YL, Liu H, Chen XL, Liu LM, Chen J. 2016. Crz1p regulates pH homeostasis in Candida glabrata by altering membrane lipid composition. Appl Environ Microbiol 82:6920–6929. doi: 10.1128/AEM.02186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson DK, Frohlich F, Christiano R, Hannibal-Bach HK, Ejsing CS, Walther TC. 2015. Rom2-dependent phosphorylation of Elo2 controls the abundance of very long-chain fatty acids. J Biol Chem 290:4238–4247. doi: 10.1074/jbc.M114.629279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasution O, Lee YM, Kim EJ, Lee YJ, Kim WK, Choi WJ. 2017. Overexpression of OLE1 enhances stress tolerance and constitutively activates the MAPK HOG pathway in Saccharomyces cerevisiae. Biotechnol Bioeng 114:620–631. doi: 10.1002/bit.26093. [DOI] [PubMed] [Google Scholar]
- 36.Young BP, Shin JJH, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, Smits GJ, Loewen CJR. 2010. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- 37.MacGilvray ME, Lapek JD, Friedman AE, Quivey RG. 2012. Cardiolipin biosynthesis in Streptococcus mutans is regulated in response to external pH. Microbiology 158:2133–2143. doi: 10.1099/mic.0.057273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerreiro JF, Muir A, Ramachandran S, Thorner J, Sa-Correia I. 2016. Sphingolipid biosynthesis upregulation by TOR complex 2-Ypk1 signaling during yeast adaptive response to acetic acid stress. Biochem J 473:4311–4325. doi: 10.1042/BCJ20160565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blosser SJ, Merriman B, Grahl N, Chung D, Cramer RA. 2014. Two C4-sterol methyl oxidases (Erg25) catalyse ergosterol intermediate demethylation and impact environmental stress adaptation in Aspergillus fumigatus. Microbiology 160:2492–2506. doi: 10.1099/mic.0.080440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallstrom BM, Petranovic D, Nielsen J. 2014. Altered sterol composition renders yeast thermotolerant. Science 346:75–78. doi: 10.1126/science.1258137. [DOI] [PubMed] [Google Scholar]
- 41.Miller C, Matic I, Maier KC, Schwalb B, Roether S, Strasser K, Tresch A, Mann M, Cramer P. 2012. Mediator phosphorylation prevents stress response transcription during non-stress conditions. J Biol Chem 287:44017–44026. doi: 10.1074/jbc.M112.430140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsson M, Uvell H, Sandstrom J, Ryden P, Selth LA, Bjorklund S. 2013. Functional studies of the yeast Med5, Med15 and Med16 Mediator tail subunits. PLoS One 8:e73137. doi: 10.1371/journal.pone.0073137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taubert S, Hansen M, Van Gilst MR, Cooper SB, Yamamoto KR. 2008. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet 4:e1000021. doi: 10.1371/journal.pgen.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou JW, Moffat J, Boone C, Davis TN, Nislow C, Brown GW. 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol 14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roetzer A, Gregori C, Jennings AM, Quintin J, Ferrandon D, Butler G, Kuchler K, Ammerer G, Schuller C. 2008. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol Microbiol 69:603–620. doi: 10.1111/j.1365-2958.2008.06301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P, Chernyshov A, Najdi T, Fu Y, Dickerson J, Sandmeyer S, Jarboe L. 2013. Membrane stress caused by octanoic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:3239–3251. doi: 10.1007/s00253-013-4773-5. [DOI] [PubMed] [Google Scholar]
- 47.Royce LA, Yoon JM, Chen YX, Rickenbach E, Shanks JV, Jarboe LR. 2015. Evolution for exogenous octanoic acid tolerance improves carboxylic acid production and membrane integrity. Metab Eng 29:180–188. doi: 10.1016/j.ymben.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 48.Yang X, Hang XM, Zhang M, Liu XL, Yang H. 2015. Relationship between acid tolerance and cell membrane in Bifidobacterium, revealed by comparative analysis of acid-resistant derivatives and their parental strains grown in medium with and without Tween 80. Appl Microbiol Biotechnol 99:5227–5236. doi: 10.1007/s00253-015-6447-y. [DOI] [PubMed] [Google Scholar]
- 49.de Kroon AIPM, Rijken PJ, De Smet CH. 2013. Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog Lipid Res 52:374–394. doi: 10.1016/j.plipres.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Mollinedo F. 2012. Lipid raft involvement in yeast cell growth and death. Front Oncol 2:140. doi: 10.3389/fonc.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kodedova M, Sychrova H. 2015. Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10:e0139306. doi: 10.1371/journal.pone.0139306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu LM, Li Y, Li HZ, Chen J. 2004. Manipulating the pyruvate dehydrogenase bypass of a multi-vitamin auxotrophic yeast Torulopsis glabrata enhanced pyruvate production. Lett Appl Microbiol 39:199–206. doi: 10.1111/j.1472-765X.2004.01563.x. [DOI] [PubMed] [Google Scholar]
- 53.Xu G, Liu L, Chen J. 2012. Reconstruction of cytosolic fumaric acid biosynthetic pathways in Saccharomyces cerevisiae. Microb Cell Fact 11:24. doi: 10.1186/1475-2859-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu CD, Zhang J, Wang M, Du GC, Chen J. 2012. Lactobacillus casei combats acid stress by maintaining cell membrane functionality. J Ind Microbiol Biotechnol 39:1031–1039. doi: 10.1007/s10295-012-1104-2. [DOI] [PubMed] [Google Scholar]
- 55.Luo QL, Wu J, Wu MC. 2014. Enhanced acetoin production by Bacillus amyloliquefaciens through improved acetoin tolerance. Process Biochem 49:1223–1230. doi: 10.1016/j.procbio.2014.05.005. [DOI] [Google Scholar]
- 56.Tian HC, Zhou J, Qiao B, Liu Y, Xia JM, Yuan YJ. 2010. Lipidome profiling of Saccharomyces cerevisiae reveals pitching rate-dependent fermentative performance. Appl Microbiol Biotechnol 87:1507–1516. doi: 10.1007/s00253-010-2615-2. [DOI] [PubMed] [Google Scholar]
- 57.Lopez-Malo M, Chiva R, Rozes N, Manuel Guillamon J. 2013. Phenotypic analysis of mutant and overexpressing strains of lipid metabolism genes in Saccharomyces cerevisiae: implication in growth at low temperatures. Int J Food Microbiol 162:26–36. doi: 10.1016/j.ijfoodmicro.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 58.Guan XL, Wenk MR. 2006. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast 23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- 59.McKenna SL, Cotter TG. 2000. Inhibition of caspase activity delays apoptosis in a transfected NS/0 myeloma cell line. Biotechnol Bioeng 67:165–176. [PubMed] [Google Scholar]
- 60.Nakamura K, Niimi M, Niimi K, Holmes AR, Yates JE, Decottignies A, Monk BC, Goffeau A, Cannon RD. 2001. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob Agents Chemother 45:3366–3374. doi: 10.1128/AAC.45.12.3366-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.