ABSTRACT
Shellfish-transmitted Vibrio parahaemolyticus infections have recently increased from locations with historically low disease incidence, such as the Northeast United States. This change coincided with a bacterial population shift toward human-pathogenic variants occurring in part through the introduction of several Pacific native lineages (ST36, ST43, and ST636) to nearshore areas off the Atlantic coast of the Northeast United States. Concomitantly, ST631 emerged as a major endemic pathogen. Phylogenetic trees of clinical and environmental isolates indicated that two clades diverged from a common ST631 ancestor, and in each of these clades, a human-pathogenic variant evolved independently through acquisition of distinct Vibrio pathogenicity islands (VPaI). These VPaI differ from each other and bear little resemblance to hemolysin-containing VPaI from isolates of the pandemic clonal complex. Clade I ST631 isolates either harbored no hemolysins or contained a chromosome I-inserted island we call VPaIβ that encodes a type 3 secretion system (T3SS2β) typical of Trh hemolysin producers. The more clinically prevalent and clonal ST631 clade II had an island we call VPaIγ that encodes both tdh and trh and that was inserted in chromosome II. VPaIγ was derived from VPaIβ but with some additional acquired elements in common with VPaI carried by pandemic isolates, exemplifying the mosaic nature of pathogenicity islands. Genomics comparisons and amplicon assays identified VPaIγ-type islands containing tdh inserted adjacent to the ure cluster in the three introduced Pacific and most other emergent lineages that collectively cause 67% of infections in the Northeast United States as of 2016.
IMPORTANCE The availability of three different hemolysin genotypes in the ST631 lineage provided a unique opportunity to employ genome comparisons to further our understanding of the processes underlying pathogen evolution. The fact that two different pathogenic clades arose in parallel from the same potentially benign lineage by independent VPaI acquisition is surprising considering the historically low prevalence of community members harboring VPaI in waters along the Northeast U.S. coast that could serve as the source of this material. This illustrates a possible predisposition of some lineages to not only acquire foreign DNA but also become human pathogens. Whereas the underlying cause for the expansion of V. parahaemolyticus lineages harboring VPaIγ along the U.S. Atlantic coast and spread of this element to multiple lineages that underlies disease emergence is not known, this work underscores the need to define the environment factors that favor bacteria harboring VPaI in locations of emergent disease.
KEYWORDS: emerging pathogen, genomics, HGT, pathogen evolution, pathogenicity islands, type III secretion systems, vibrio, whole-genome phylogeny, molecular epidemiology
INTRODUCTION
Vibrio parahaemolyticus is an emergent pathogen capable of causing human gastric infections when consumed, most often with contaminated shellfish (1, 2). Some human-pathogenic V. parahaemolyticus variants evolve from diverse nonpathogenic communities through horizontal acquisition of Vibrio pathogenicity islands (VPaI) (3–5). Enteropathogenic V. parahaemolyticus typically harbor islands with at least one of two types of horizontally acquired hemolysin genes (tdh and trh) that are routinely used for pathogen discrimination even though their role in disease appears modest (6–11). Most pathogenic V. parahaemolyticus isolates also carry accessory type 3 secretion systems (T3SS) that translocate effector proteins that contribute to host interaction (12–14). Two evolutionarily divergent horizontally acquired accessory systems (T3SS2α and T3SS2β) contribute to human disease and are genetically linked to hemolysin genes (two tdh genes with T3SS2α, and trh with T3SS2β) in contiguous but distinct islands (4, 15–17). The first described tdh-harboring island (called by several different names including Vp-PAI [15], VPaI-7 [4], and tdhVPA [17]) from an Asian pandemic strain called RIMD 2210366 is fairly well characterized (4, 5, 13, 18, 19). In contrast, islands containing T3SS2β linked to trh and a urease (ure) cluster, which confers a useful diagnostic phenotype, (where similar islands are described by others as Vp-PAITH3966 [16] or trhVPA [17, 20]) have received only modest attention. Pathogenic variants harboring both tdh and trh are increasingly associated with disease in North America (21–26), and yet, to our knowledge, the exact configuration of hemolysin-associated VPaI in isolates that contain both tdh and trh has not yet been described (see also reference 20). Thus, it is unclear how virulence loci and islands in these emergent pathogen lineages carrying both hemolysins evolved and spread.
The expanding populations of V. parahaemolyticus have increased infections even in temperate regions previously only rarely impacted by this pathogen and where most environmental isolates harbor no known virulence determinants (27). A related complex of Asia-derived pandemic strains, most often identified as serotype O3:K6 and also known as sequence type 3 (ST3; based on allele combinations of seven housekeeping genes) causes the most disease globally (28). An unrelated Pacific native lineage called ST36 (also described as serotype O4:K12) currently dominates infections in North America, including from the Northeast United States (21, 26, 29). The introduction of ST36 into the Atlantic Ocean by an unknown route precipitated a series of outbreaks from Atlantic shellfish starting in 2012 (29, 30). Prior to 2012, resident lineages contributed to low but increasing sporadic infection rates on the Northeast U.S. coast (https://www.cdc.gov/vibrio/surveillance.html, 2017; see also reference 21), with ST631 emerging as the major lineage that is endemic to nearshore areas of the Atlantic Ocean bordering North America (the northwest Atlantic Ocean) (31). However, we previously identified a single ST631 isolate lacking hemolysins (21, 27), suggesting this pathogen lineage may have recently evolved through VPaI acquisition.
The goal of our study was to understand the genetic events and changing population context for the evolution of the ST631 pathogenic lineage. We conducted whole and core genome phylogenetic analyses of 3 environmental and 39 clinical ST631 isolates, along with isolates from other emergent lineages from the region, which revealed two ST631 clades of common ancestry, from which human pathogens evolved in parallel. The single clade I clinical isolate acquired a recA gene insertion previously seen associated with Asian lineages and had a VPaI that is typical of isolates harboring trh in the absence of tdh. In contrast, isolates from the clonal ST631 clade II that dominates Atlantic-derived ST631 infections (31) had a related but distinct VPaI. This VPaI contained a tdh gene inserted within, not next to, an existing ure-trh-T3SS2β island in close proximity to the ure cluster. Nearly all emergent resident and invasive lineages, including all three Pacific lineages (ST36, ST636, and ST43) contained islands that similarly had a tdh gene inserted within the VPaI in an identical location adjacent to the ure cluster providing a mechanism for simultaneous acquisition of both hemolysins with T3SS2β.
RESULTS
Atlantic endemic ST631 and several invasive lineages harboring both the tdh and trh hemolysin genes are clinically prevalent in four reporting Northeast U.S. states.
Ongoing analysis of clinical isolates revealed that even as the Pacific-derived ST36 lineage continued to dominate infections (50%), the endemic (autochthonous) ST631 lineage accounted for 14% of infections (Table 1). Concurrently, a limited number of other lineages contributed individually to fewer infections (≤3% each), among which were two lineages that have caused infections in the Pacific Northwest in prior decades: ST43 and ST636 (22, 23). ST43 and ST636 only recently (2013 and 2011, respectively) (21) have been linked to product harvested from waters along the Northeast U.S. coast and also caused infections in subsequent years. As is common among U.S. clinical isolates, pathogenic isolates of all the aforementioned lineages harbor both tdh and trh hemolysin genes (Table 1). Among environmental isolates, ST34 and ST674 are the most frequently recovered pathogen lineages, but these caused comparatively few infections (Table 1). ST34 was first reported from the environment in 1998, both from the Gulf of Mexico and nearshore of Massachusetts, and it was also recovered in New Hampshire in 2012 (21), suggesting it is an established resident in the region. ST674, which was first reported from an infection in Virginia in 2007 (32), was first recovered from the local environment in 2012 (https://pubmlst.org/vparahaemolyticus/; see also reference 21). Notably, even though all four ST674 environmental isolates, like ST34, harbored both hemolysin genes, the single ST674 clinical isolate (MAVP-21) lacked the Tdh and Trh hemolysins (Table 1) (21). The decrease in clinical prevalence of trh-harboring Atlantic-endemic ST1127, which caused no infections in the last 3 years, coincided with the increase in clinical prevalence of all three Pacific-derived lineages which harbor both hemolysins. Notably, very few other clinical isolates harbored trh in the absence of tdh and clinical isolates containing only tdh (i.e., ST1725) were extremely rare (Table 1). Concurrent with this shift in the composition of clinical lineages that includes multiple Pacific-derived lineages, hemolysin producers have increased in relative abundance in nearshore areas of the region, where historically these represented ∼1% of all isolates (27). Since 2012, hemolysin producers have been recovered more frequently and in the last 2 years their proportion has increased by up to an order of magnitude (comprising as much as 10%) in some regional shellfish-associated populations (data not shown).
TABLE 1.
Clinical and environmental prevalence of emergent Northeast U.S. V. parahaemolyticus lineages with associated virulence features
| Sequence typea | No. of isolates |
Hemolysin genotype | VPaI typed | |||
|---|---|---|---|---|---|---|
| Northeast United Statesb |
MLST databasec |
|||||
| Clinical | Environmental | Clinical | Environmental | |||
| 3 | 2 | 0 | 217 | 33 | tdh+ | α |
| 36 | 91 | 1 | 58 | 5 | tdh+ trh+ | γ |
| 631 | 24 | 0 | 12 | 0 | tdh+ trh+ | γ |
| 1e | 2 | 0 | 0 | trh+ | β | |
| 0 | 1 | 0 | 0 | Neither | Absent | |
| 43 | 5 | 0 | 17 | 4 | tdh+ trh+ | γ |
| 636 | 4 | 0 | 2 | 0 | tdh+ trh+ | γ |
| 1127 | 4 | 0 | 0 | 0 | trh+ | β |
| 110 | 3 | 0 | 0 | 1 | tdh+ trh+ | γ |
| 34/324 | 2 | 2 | 4 | 19 | tdh+ trh+ | γ |
| 674 | 0 | 4 | 1 | 20 | tdh+ trh+ | γ |
| 1 | 0 | 0 | 0 | Neither | Absent | |
| 308 | 2 | 0 | 0 | 2 | tdh+ trh+ | γ |
| 12 | 2 | 0 | 0 | 4 | trh+ | β |
| 162 | 2 | 0 | 1 | 1 | Neither | Absent |
| 194 | 2 | 0 | 1 | 0 | Neither | Absent |
| 809 | 2 | 0 | 0 | 1 | trh+ | β |
| 1716 | 2 | 0 | 0 | 0 | trh+ | β |
| 1123 | 1 | 1 | 0 | 0 | trh+ | β |
| 8 | 1 | 0 | 13 | 5 | trh+ | β |
| 23 | 1 | 0 | 0 | 3 | tdh+ trh+ | γ |
| 749 | 1 | 0 | 1 | 0 | tdh+ trh+ | γ |
| 1295 | 1 | 0 | 0 | 1 | Neither | Absent |
| 134 | 1 | 0 | 1 | 0 | Neither | Absent |
| 741 | 1 | 0 | 0 | 1 | Neither | Absent |
| 98 | 1 | 0 | 0 | 1 | trh+ | β |
| 1205 | 1 | 0 | 0 | 1 | Neither | Absent |
| 1561 | 1 | 0 | 0 | 0 | Neither | Absent |
| 1717 | 1 | 0 | 0 | 0 | Neither | Absent |
| 1725 | 1 | 0 | 0 | 0 | tdh+ | α |
Some clinical isolates had insufficient sequencing coverage to determine sequence type and included 8 tdh+ trh+ isolates, 1 tdh+ isolate, 4 trh+ isolates, and 11 isolates without hemolysins, some of which were from wound infections. Two wound infection isolates lacking hemolysins were of known sequence types and are not listed above.
Data generated from all available gastric infection clinical and environmental isolates from four reporting Northeast U.S. states, including ME, NH, MA, and CT, between 2010 and 2016.
Source: http://pubmlst.org/vparahaemolyticus, 2017 (36, 58).
The presence of the VPaIγ architecture was determined by PacBio genome sequencing of isolate MAVP-Q and MAVP-26, whereas for other isolates identification of VPaI type was determined through Illumina genome sequencing, PCR amplification, and Sanger sequencing.
This single isolate harbors a recA allele (allele 21) typical of ST631 with an inserted allele (allele 107) previously described (33).
A single clinical ST631 lineage isolate with an unusual recA allele harbors trh in the absence of tdh.
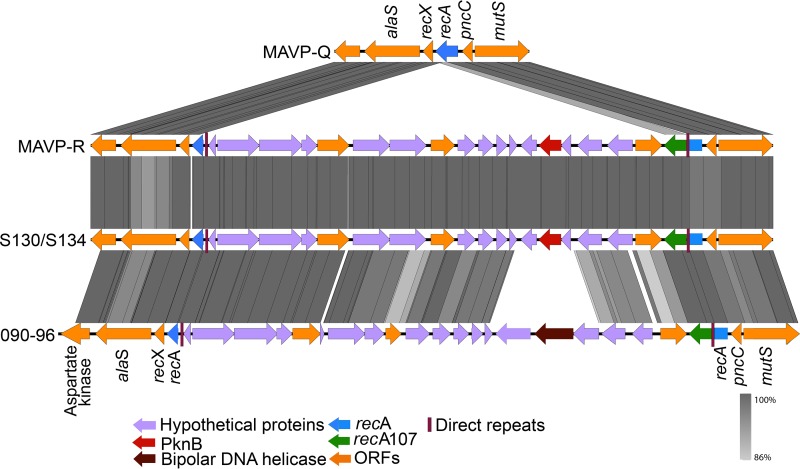
Employing ST631-specific marker-based assays (see Materials and Methods), we identified two additional 2015 environmental isolates (one from New Hampshire and one from Massachusetts) and one additional 2011 local-source clinical isolate (MAVP-R) (21) with a hemolysin profile (trh+ without tdh) that is atypical of the ST631 lineage (Table 1). Although analysis of the seven-housekeeping-gene allele combination confirmed that the environmental isolates were indeed ST631, MAVP-R was not ST631 based on only one locus: recA. Examination of the recA locus of MAVP-R uncovered a large insertion within the ancestral ST631 recA gene (allele recA21 [https://pubmlst.org/vparahaemolyticus/]) incorporating an intact but different recA gene into the locus (allele recA107 [33]) and fragmenting the ancestral gene (Fig. 1). The insertion in the ancestral recA gene in MAVP-R is identical to one observed in the recA locus of two Hong Kong isolates (isolates S130 and S134) and similar to the one in isolate 090-96 (ST189a) isolated in Peru but believed to have originated in Asia (33).
FIG 1.
Schematic of a horizontally acquired insertion in the recA-encoding region of MAVP-R. Sequences of the recA gene and flanking region from MAVP-Q (reference ST631 genome), MAVP-R, Asia-derived isolates S130/S134, and Peru-derived isolate 090-96 were extracted and aligned. ORFs designated by arrows and illustrated by representative colors highlight homologous and unique genes. The percent similarity between homologs is illustrated by gray bars.
ST631 forms two divergent clades.
The existence of three different hemolysin profiles (Table 1) among all available ST631 draft genomes suggested there could be more than one ST631 lineage. Therefore, we evaluated whole-genome maximum-likelihood (ML) phylogenies of select ST631 isolates and all other lineages causing two or more infections reported in four states in Northeast United States to evaluate whether there was more than one ST631 lineage (Table 1 and Fig. 2). The phylogenetic tree showed that ST631 isolates, regardless of their hemolysin genotype, clustered together, but they formed two distinct clades, a finding indicative of common ancestry (Fig. 2). Clade I harbored either trh or no hemolysins and consisted of all three environmental isolates which were from Massachusetts and New Hampshire, and the single clinical isolate MAVP-R, whereas clade II consisted of all other isolates, all of which harbor both hemolysins. The two distinct ST631 clades shared 85% of their DNA in common and displayed polymorphisms in ≤12% of the shared DNA content. The most closely related sister lineage to ST631 was formed by trh-harboring ST1127 isolates that have been exclusively reported from clinical sources in the Northeast United States (21).
FIG 2.
Phylogenetic relationships of V. parahaemolyticus lineages and identification of distinct ST631 clades. An ML phylogeny of representative V. parahaemolyticus genomes of clinical isolates causing two or more infections was built on whole-genome SNPs identified by reference-free comparisons, as described in Materials and Methods. The branch length represents the number of nucleotide substitutions per site. Numbers at the nodes represent percent bootstrap support where unlabeled nodes had bootstraps of <70%.
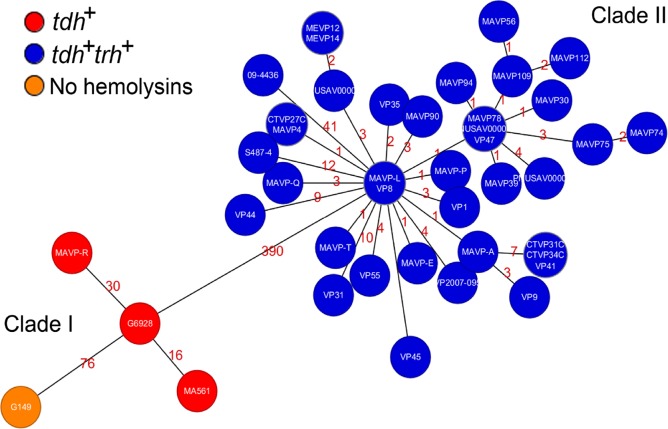
We next evaluated the relationships of all available ST631 isolate genomes at NCBI and sequenced by us (Table 2) using a custom core genome multilocus sequence typing (cgMLST) method as previously described (31). Minimum spanning trees built from core genome loci from 42 ST631 isolates indicated that only 390 loci varied between the most closely related isolate of clade I (MAVP-L) and clade II (G6928) (Fig. 3). The most distantly related isolates within clade I (G149 and MAVP-R) exhibited 80 core genome locus differences, whereas clade II is clonal with only 51 variant loci between the most divergent isolates: clinical isolate 09-4436 and environmental isolate S487-4, both reported from Prince Edward Island, Canada (Fig. 3) (31).
TABLE 2.
Draft genomes utilized for phylogenetic analysis
| Isolate | NCBI accession or SRA no. | VPaI type | Sequence type | Reference |
|---|---|---|---|---|
| VP2007-095 | AVOI01000000 | γ | ST631 clade II | 31 |
| 09-4436 | LRAJ01000000 | γ | ST631 clade II | 31 |
| G149 | MPPO00000000 | None | ST631 clade I | This study |
| MAVP-A | MDWP00000000 | γ | ST631 clade II | 31 |
| MAVP-E | LBHP00000000 | γ | ST631 clade II | 31 |
| MAVP-P | MDWQ00000000 | γ | ST631 clade II | 31 |
| MAVP-T | MDWR00000000 | γ | ST631 clade II | 31 |
| MAVP-L | MDWS00000000 | γ | ST631 clade II | 31 |
| MAVP-Q | MDWT00000000 | γ | ST631 clade II | 31 |
| MAVP-R | MPPP00000000 | β | ST631 clade I | This study |
| VP1 | JNSM00000000 | γ | ST631 clade II | 31 |
| VP8 | JNSN01000000 | γ | ST631 clade II | 31 |
| VP9 | JNSO00000000 | γ | ST631 clade II | 31 |
| CTVP27C | NJHM00000000 | γ | ST631 clade II | 31 |
| CTVP31C | NJHL00000000 | γ | ST631 clade II | 31 |
| CTVP34C | NJHK00000000 | γ | ST631 clade II | 31 |
| MAVP-4 | MDWU00000000 | γ | ST631 clade II | 31 |
| MAVP-30 | MDWW00000000 | γ | ST631 clade II | 31 |
| MAVP-39 | MDWV00000000 | γ | ST631 clade II | 31 |
| MAVP-56 | MDWX00000000 | γ | ST631 clade II | 31 |
| S487-4 | LFZE01000000 | γ | ST631 clade II | 31 |
| VP31 | JNSP00000000 | γ | ST631 clade II | 31 |
| VP35 | JNSQ00000000 | γ | ST631 clade II | 31 |
| VP41 | JNSR00000000 | γ | ST631 clade II | 31 |
| VP44 | JNSS00000000 | γ | ST631 clade II | 31 |
| VP45 | JNST00000000 | γ | ST631 clade II | 31 |
| VP47 | SRR4032360 | γ | ST631 clade II | 31 |
| MAVP-74 | MDWY00000000 | γ | ST631 clade II | 31 |
| MAVP-75 | MDWZ00000000 | γ | ST631 clade II | 31 |
| MAVP-78 | MDXA00000000 | γ | ST631 clade II | 31 |
| VP55 | SRR4032361 | γ | ST631 clade II | 31 |
| G6928 | MPPN00000000 | β | ST631 clade I | This study |
| MA561 | MPPM00000000 | β | ST631 clade I | This study |
| MAVP-90 | MPPQ00000000 | γ | ST631 clade II | 31 |
| MAVP-94 | MPPR00000000 | γ | ST631 clade II | 31 |
| MAVP-109 | MPPS00000000 | γ | ST631 clade II | 31 |
| MAVP-112 | MPPT00000000 | γ | ST631 clade II | 31 |
| MEVP-12 | NJHJ00000000 | γ | ST631 clade II | 31 |
| MEVP-14 | NJHI00000000 | γ | ST631 clade II | 31 |
| PNUSAV000012 | SRR4016797 | γ | ST631 clade II | 31 |
| PNUSAV000015 | SRR4016801 | γ | ST631 clade II | 31 |
| PNUSAV00021 | SRR4018053 | γ | ST631 clade II | 31 |
| G4186 | NIYP00000000 | γ | ST34 | This study |
| MAVP-14 | NJAP00000000 | γ | ST324 | This study |
| MAVP-71 | NIXZ00000000 | γ | ST43 | This study |
| MAVP-46 | NJAO00000000 | γ | ST110 | This study |
| MAVP-50 | NIXY00000000 | γ | ST636 | This study |
| MAVP-67 | NIXX00000000 | γ | ST308 | This study |
| MAVP-99 | NIXW00000000 | β | ST12 | This study |
| G3578 | NIYO00000000 | γ | ST674 | This study |
| MAVP-21 | NIXV00000000 | None | ST674 | This study |
| CTVP43C | NIXU00000000 | None | ST162 | This study |
| MAVP-3 | NIXT00000000 | α | ST3 | This study |
| MAVP-25 | NJAN00000000 | β | ST1127 | This study |
| CTVP35C | NIXS00000000 | None | ST194 | This study |
| MAVP-G | NIXR00000000 | β | ST809 | This study |
| MAVP-108 | NIXQ00000000 | β | ST1716 | This study |
FIG 3.
Minimum spanning tree relationships among clade I and clade II ST631. A cgMLST core gene-by-gene analysis (excluding accessory genes) was performed, and SNPs were identified within different alleles. The numbers above the connected lines (not drawn to scale) represent SNP differences identified within loci with different alleles. The isolates are colored based on different hemolysin genotypes as labeled.
Each ST631 clade independently acquired a distinct pathogenicity island positioned on different chromosomes.
Given the variation in ST631, comparisons between these isolates could not only elucidate the events that led to the evolution of two pathogenic clades but also address unresolved questions about the unique configurations and contents of pathogenicity islands in western Atlantic Ocean emergent lineages. The physical proximity of tdh with the ure cluster and trh, and the cooccurrence of tdh with T3SS2β reported in many tdh+ trh+ clinical isolates, suggested tdh could be located within or next to the same pathogenicity island harboring trh in at least some lineages as was previously suggested (20, 24, 34).
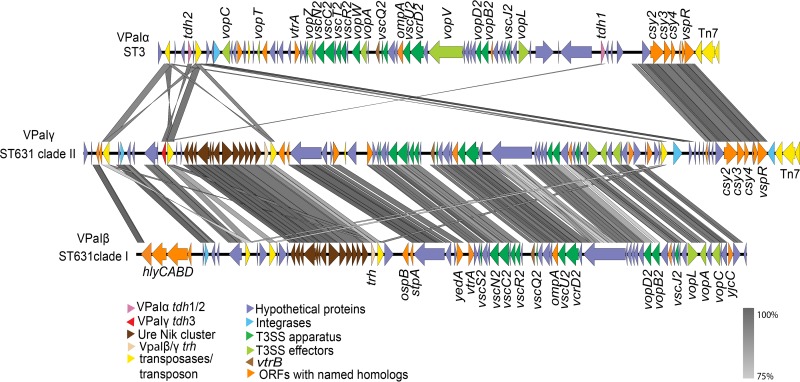
To identify the location and determine the architecture of the pathogenicity elements harboring hemolysin genes, we generated high-quality annotated genomes for the clade I ST631 isolate MAVP-R and clade II ST631 isolate MAVP-Q (both reported in 2011 from Massachusetts) employing PacBio sequencing. The pathogenicity island regions in these isolates genomes were extracted and aligned, and the contents were compared with pathogenicity islands harboring two tdh genes (previously called Vp-PAI [15], VPaI-7 [4], and tdhVPA [17]) from RIMD 2210366 and Vp-PAITH3996 (16; also called trhVPI [17]) harboring trh (see Table S1 in the supplemental material). This comparison revealed that MAVP-R harbored a pathogenicity island typical of trh-containing isolates that includes a linked ure cluster and T3SS2β that is orthologous, with the exception of a few unique regions, with Vp-PAITH3996 (16) (see Table S1 in the supplemental material and Fig. 4). Because the lack of convention in uniformly naming syntenic islands that distinguish them from distinctive and yet functionally analogous islands can impede communication, we will consistently reference the same island by a common descriptive name regardless of isolate lineage. We refer to islands sharing the same general configuration to that in MAVP-R by the name VPaIβ and refer to tdh-containing islands similar to that described in strain RIMD 2210366 by the name VPaIα, regardless of bacterial isolate background. We adopted this simplified nomenclature in reference to the version of the key virulence determinant carried in the islands (T3SS2α and T3SS2β) in the two already-described island types. This scheme importantly accommodates naming of additional uniquely configured islands as they are identified. As noted previously (16, 17, 20), VPaIβ is dissimilar to VPaIα in most gene content with ∼78 open reading frames (ORFs) unique to VPaIβ (where the number of identified ORFs used for comparison can differ slightly depending on which annotation program is applied) (see Table S1 in the supplemental material and Fig. 4). Even so, VPaIβ had many homologous genes of varying sequence identity (n = ∼38 ORFs, excluding tdh homology with trh) compared to VPaIα (Table S1 and Fig. 4) (4, 5, 16). Identification of some homologs required that we relax matching to 50%, such as for the divergent, but homologous T3SS2α and T3SS2β genes encoding the apparatus, chaperones, and some shared effectors (see Table S1 in the supplemental material). No homolog of the T3SS2α effector gene vopZ was identified, but a single ORF whose deduced protein sequence bears only 27% identity with VopZ is located in its place (Fig. 2 and Table S1 in the supplemental material). VPaIβ from strain TH3996 and VPaIα from pandemic strain RIMD 2210633 are inserted in an identical location in chromosome II adjacent to an acyl coenzyme A (acyl-CoA) hydrolase-encoding gene. In contrast, the VPaIβs in MAVP-R, ST1127 isolate MAVP-25, and Asia-derived AQ4037 are in chromosome I, in each case in the same insertion location identified for strain AQ4037 (17).
FIG 4.
Comparisons of the pathogenicity islands containing hemolysins and T3SS2. Sequences of VPaI were extracted from select genomes and aligned. VPaIα was derived from ST3 strain RIMD2210633, VPaIγ was derived from ST631 clade II isolate MAVP-Q, and VPaIβ was derived from ST631 clade I isolate MAVP-R. ORFs are depicted in defined colors, and similarities (≥75%) among ORFs are illustrated in gray blocks. Homologs between VPaIα and VPaIβ/γ (50 to >75% identity) are named and listed in Table S1 in the supplemental material.
MAVP-Q contained both tdh and trh within the same contiguous unique VPaI (here called VPaIγ) that shared features with both VPaIα and VPaIβ (Fig. 4; see also Table S1 in the supplemental material). Specifically, VPaIγ had a core that with few exceptions was orthologous in content and syntenous with VPaIβ from MAVP-R (Fig. 4). VPaIγ displays high conservation with VPaIα near its 3′ end, as has been described in other draft tdh+ trh+ harboring genomes (20), as well as in the VPaIβ of strain TH3996, although the presence of this element may not be typical of VPaIβ (e.g., it is absent in the islands from AQ4037 [17], MAVP-R, and MAVP-25). The VPaIγ also contained a tdh gene homologous to tdh2 (also called tdhA) from VPaIα (98.6%) near its 5′ end but not at the 5′ terminus of the island (Fig. 4). Rather, the DNA flanking both sides of the tdh gene in VPaIγ was conserved in VPaIβ of MAVP-R and absent from VPaIα (Fig. 4). Analysis of 300 genomes of V. parahaemolyticus (representing a minimum of 28 distinct sequence types) of sufficient quality for analysis confirmed that the module of four hypothetical proteins preceding the tdh2 homolog was present only in trh-harboring genomes but not in genomes harboring tdh in the absence of trh (i.e., VPaIα-containing genomes), providing evidence that the tdh gene was acquired horizontally by insertion into, not next to, an existing VPaIβ, perhaps through activity of the adjacent transposase gene (11) (see Table S2 and Fig. S1 in the supplemental material; also, data not shown). As with VPaIα from RIMD 2210633 and VPaIβ of TH3996, VPaIγ of clade II ST631 is located in a conserved location of chromosome II, adjacent to an acyl-CoA hydrolase-encoding gene.
The final environmental ST631 clade I isolate that lacked hemolysins, G149, had no VPaIα, -β, or -γ elements in its genome. Close examination of the DNA corresponding to the VPaI insertion sites in either chromosome revealed no remnants of these islands in either chromosomal location, indicating this isolate likely never acquired a VPaIα, -β, or -γ (see Fig. S2 in the supplemental material; also, data not shown). Because clade I isolate G149 lacked these islands, this could be the ancestral state of the ST631 lineage (21).
Most clinically prevalent isolates from the Northeast United States harbor similar contiguous pathogenicity islands containing tdh inserted in the same island location.
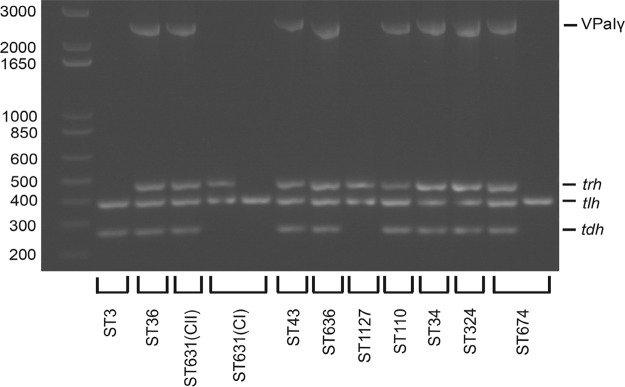
We next sought to determine whether isolates from other lineages likely residing within the mixed population with ST631 in nearshore areas of the Northeast United States harbored islands with structures similar to VPaIγ that contain both hemolysin genes. Assembly of short-read sequences into contigs that cover the full length of VPaI, which is necessary for comparative analysis of entire island configuration, was impeded by the fact that homologous transposase genes and other sequences were repeated multiple times throughout the island. Therefore, we determined whether other lineages harboring both hemolysin genes harbor tdh in the same island location, between the conserved VPaIβ/γ module of four hypothetical protein-encoding genes (to the left or 5′ of tdh) and the ure cluster (to the right or 3′ of tdh) (Fig. 4) by combining bioinformatics analysis of sequenced genomes with amplicon assays (see Fig. S1 in the supplemental material). First, we analyzed assembled draft genomes for tdh cooccurrence and proximity with the four adjacent hypothetical protein-encoding genes that are absent in VPaIα but present in VPaIβ/γ (see Materials and Methods). Every emergent pathogenic lineage of the Northeast United States (Table 1) harboring both tdh and trh carried homologous DNA corresponding to all four hypothetical proteins adjacent to the tdh gene in a contiguous segment (see Table S2 in the supplemental material). To determine whether tdh was also adjacent to the ure cluster in these same isolates, we next designed specific flanking primers and amplified the unique juncture between the tdh-containing transposon-associated module and the ure cluster for all clinical isolates harboring both tdh and trh (see Materials and Methods; see also Fig. S1 in the supplemental material). The results were congruent with our bioinformatics assessment (see Table S2 in the supplemental material) and demonstrated that isolates from all emergent pathogenic lineages harboring both hemolysins have tdh inserted in close proximity to an ure cluster in a configuration similar to VPaIγ from MAVP-Q (Fig. 5 and Table 1). This confirmed that these isolates harboring both hemolysins harbor tdh within, and not next to, the same VPaI, thereby facilitating simultaneous acquisition of both hemolysin genes.
FIG 5.
Distribution of VPaIγ in emergent pathogen lineages. The presence of tdh, trh, and VPaIγ, along with positive-control tlh, was determined by PCR amplification using gene-specific primers and visualized on a 1.2% agarose gel. The order from left to right is a >1-kb ladder, ST3 (MAVP-C), ST36 (MAVP-26), ST631 CII (clade II isolate MAVP-Q), ST631 CI (clade I isolates MAVP-R and G149), ST43 (MAVP-71), ST636 (MAVP-50), ST1127 (MAVP-M), ST110 (MAVP-46), ST34 (CTVP19C), ST324 (MAVP-14), and ST674 (CT4291 and MAVP21). The corresponding sizes of the ladder fragments (bp) are labeled on the left, and the identities of the amplicons are listed to the right of the gel image.
DISCUSSION
Even preceding the increased illnesses from Pacific-invasive lineages, two different clades of the predominant endemic Atlantic lineage of pathogenic V. parahaemolyticus, ST631 (31) evolved and contributed to a rise in sporadic illnesses in four reporting Northeast U.S. states (Table 1 and Fig. 2 and 3). Several lines of evidence support the interpretation of parallel pathogen evolution. The two lineages exhibit differences in both clinical and environmental prevalence, suggesting the pathogenic variants of each clade have not evolved the same degree of virulence (Table 1). Pathogenic members in each lineage also acquired different pathogenicity islands with different hemolysin gene contents (Fig. 2 and 3). Although it was a formal possibility that ST631 clade II evolved from clade I by independent horizontal acquisition of tdh into its existing VPaIβ, it is notable that other resident and even invasive lineages now in the Atlantic harbor VPaIγ with tdh inserted into the same location of the island, suggesting a common evolutionary origin of this hybrid-type island (Fig. 4 and see Fig. S1 in the supplemental material). Finally, each of the two clades harbor VPaI insertions on different chromosomes: the less clinically prevalent ST631 clade I contains three isolates that harbor VPaIβ in chromosome I (Fig. 3) and a single environmental isolate lacking any island (Table 1; see also Fig. S2 in the supplemental material), whereas the clonal ST631 clade II isolates all harbor VPaIγ in chromosome II.
Given that several other resident lineages harbor similar β- and γ-type VPaI, pathogens in each clade could have acquired their islands from the reservoir of resident bacteria already circulating in the Atlantic even before the presumed arrival of invasive Pacific lineages. Several well-documented members of the Gulf of Mexico V. parahaemolyticus population (35–37) may also have expanded their range through movement of ocean currents and could be the source for these VPaI (Table 1 and Fig. 5). However, historically, hemolysin producers are extremely rare in nearshore areas of the Atlantic U.S. coast (25) and represented only about ∼1% of isolates in an estuary of New Hampshire as of a decade ago (27), limiting the potential for interacting partners or sources for acquired VPaI. Given this historical context, it is remarkable that two different clades from the same lineage independently acquired different VPaI, which for clade II ST631 occurred prior to 2007, well before the recent shift in abundance of hemolysin producers.
The parallel evolution of two different lineages through lateral DNA acquisition alludes to the possibility that as-yet-undefined attributes may increase the chances of acquisition or prime some bacterial lineages (such as ST631) to more readily acquire and maintain genetic material or become pathogenic upon island acquisition. Even though the ecological niche in which horizontal island acquisition took place is unknown, it is conceivable that cocolonization of hosts or substrates favorable to the growth of ST631 and hemolysin producers may have facilitated island movement. Certainly, an association of bacteria with specific marine substrates such as chitinous surfaces of plankton that also induce a natural state of competence could promote lateral transfer through close contact between the progenitors of the pathogenic subpopulation of each clade and island donors (3, 38, 39). Alternatively, conjugative plasmids or transducing phage could have been the agents of island delivery. The finding that the only clinical clade I isolate, MAVP-R, also harbors a second horizontal insertion in its recA locus that matched one previously found in Asia-derived strains (33) indicates it acquired more than one segment of foreign DNA during its evolution as a pathogen (Fig. 1), further illustrating that mechanisms that facilitate DNA transfer and acquisition may both have been at play. This also suggests that horizontal transfer of DNA from introduced bacteria not yet detected in the Atlantic could add to the genetic material available for pathogen evolution from Atlantic Ocean populations. More detailed molecular epidemiological, comparative genomics, and functional analyses necessary to assess the impact of introduced pathogens on resident Atlantic lineages are warranted given this evidence and the documented introduction of multiple Pacific-derived lineages in the region (Table 1).
There has been some consideration of the roles of human virulence determinants in ecological fitness, but the natural context of pathogenic V. parahaemolyticus evolution is still unknown (40–42). Whereas tdh and T3SS2α each may promote growth when bacteria are under predation, isolates that carry trh-containing islands (which likely also have T3SS2β) do not derive similar benefits from their islands (43). This is surprising considering the islands encode several homologous effectors (Fig. 4 and see Table S1 in the supplemental material) that do not have an established role in enteric disease, but they could alternatively or additionally mediate eukaryotic cell interactions with natural hosts, thereby promoting environmental fitness (13, 14). However, these islands also lack a homologous gene for the VPaIα effector that is most closely associated with enteric disease: vopZ (11) (Fig. 4 and see Table S1 in the supplemental material). The general lack of knowledge of unique T3SS2β effectors and other gene function in these islands (Fig. 4 and Table S1 in the supplemental material), even with regard to enteric disease, limits comparative analysis with the well-studied and functionally defined VPaIα, which could elucidate the bases for pathogen evolution. The higher clinical prevalence of clade II ST631 than clade I, which has also been recovered on more than one occasion from the environment (Table 1), could indicate that VPaIγ confers greater virulence potential than VPaIβ, perhaps owing to the presence of tdh, a known virulence factor (1, 7, 44). However, the resident community members in both the Pacific and the Atlantic Ocean that harbor tdh and T3SS2α comparatively rarely cause human infections (21–23). The unique environmental conditions that underlie pathogen success from northern latitudes that favors bacteria with VPaIβ and VPaIγ, including two different ST631 lineages, suggest that the shared content of these islands could confer abilities that are distinct from VPaIα which could underlie the repeated acquisition and maintenance of these related islands by so many different lineages now present in nearshore areas of the Northeast United States.
MATERIALS AND METHODS
Bacterial isolates, media, and growth conditions.
V. parahaemolyticus clinical isolates for this study were provided by cooperating public health laboratories in Massachusetts, New Hampshire, Maine, and Connecticut, whereas a select number of environmental isolates were enriched from estuarine substrates as described previously (21). Detailed information about these isolates was described previously (31) and is listed in Table 2. Isolates were routinely cultured in heart infusion media supplemented with NaCl and grown at 37°C as described previously (21).
Whole-genome sequencing, assembly, annotation, and sequence type identification.
Genomic DNA was extracted by using a Wizard Genomic DNA purification kit (Promega, Madison, WI) or by organic extraction (21). The quality of genomic DNA was determined by spectrophotometric measurements by NanoDrop (Thermal Fisher, Waltham, MA). Libraries for DNA sequencing were prepared using a high-throughput Nextera DNA preparation protocol (45) using an optimal DNA concentration of 2 ng/μl. Genomic DNA was sequenced using an Illumina HiSeq2500 device at the Hubbard Center for Genome Studies at the University of New Hampshire with a 150-bp paired-end library. De novo assembly was performed using the A5 pipeline (46), and the assemblies were annotated with Prokka1.9 using the “genus” option and selecting “Vibrio” for the reference database (47). The sequence types were subsequently determined using the SRST2 pipeline (48). The sequence type of each genome was determined when using the V. parahaemolyticus database (https://pubmlst.org/vparahaemolyticus/). For most isolates where the combination of each allele was not found in the database representing novel sequence types, the genome was submitted for a new sequence type designation (https://pubmlst.org/vparahaemolyticus/).
Isolates MAVP-Q and MAVP-R were sequenced using the Pacific Biosciences RSII technology. Using between 3.7 and 5.3 μg of DNA, the library preparation and sequencing was performed according to the manufacturer's instructions (Pacific Biosciences, Menlo Park, CA) and reflects the P5-C3 sequencing enzyme and chemistry for the MAVP-Q isolate and the P6-C4 configuration for MAVP-R. The mass of double-stranded DNA was determined by Qubit (Waltham, MA), and the sample was diluted to a final concentration of 33 μg/μl in a volume of 150 μl of elution buffer (Qiagen, Germantown, MD). The DNA was sheared for 60 s at 4,500 rpm in a G-tube spin column (Covaris, Woburn, MA) that was subsequently flipped and respun for another 60 s at 4,500 rpm, resulting in an ∼20,000-bp DNA verified using a DNA 12000 Bioanalyzer gel chip (Agilent, Santa Clara, CA). The sheared DNA isolate was then repurified using a 0.45× AMPure XP purification step (Beckman Coulter, Indianapolis, IN). The DNA was repaired by incubation in DNA Damage Repair solution. The library was again purified using 0.45× Ampure XP and SMRTbell adapters ligated to the ends of the DNA at 25°C overnight. The library was treated with an exonuclease cocktail (1.81 U/μl Exo III 18 and 0.18 U/μl Exo VII) at 37°C for 1 h to remove unligated DNA fragments. Two additional 0.45× Ampure XP purifications steps were performed to remove <2,000-bp molecular weight DNA and organic contaminant.
Upon completion of library construction, samples were validated using an Agilent DNA 12000 gel chip. The isolate library was subjected to additional size selection to the range of 7,000 to 50,000 bp to remove any SMRTbells less than 5,000 bp using Sage Science Blue Pippin 0.75% agarose cassettes to maximize the SMRTbell sub-read length for optimal de novo assembly. Size selection was confirmed by Bio-Analysis, and the mass was quantified using the Qubit assay. Primer was then annealed to the library (80°C for 2.5 min, followed by decreasing the temperature by 0.1°/s to 25°C). The polymerase-template complex was then bound to the P5 or P6 enzyme using a 10:1 ratio of polymerase to SMRTbell at 0.5 nM for 4 h at 30°C and then held at 4°C until ready for MagBead loading prior to sequencing. The magnetic bead-loading step was conducted at 4°C for 60 min according to the manufacturer's guidelines. The MagBead-loaded, polymerase-bound, SMRTbell libraries were placed onto the RSII machine at a sequencing concentration of 110 to 150 pM and configured for a 180-min continuous sequencing run. Long-read assemblies were constructed using HGAP version 2.3.0 for de novo assembly generation. Further, hybrid assemblies were generated and error corrected with Illumina raw reads using Pilon v1.20 (49).
Lineage-specific marker-based assays.
To more rapidly identify ST631 isolates from clinical and environmental collections, we developed PCR-amplicon assays to unique gene content in ST631. Whole-genome comparisons were performed on MAVP-Q (a ST631 clinical isolate), G149 (a ST631 environmental isolate), MAVP-26 (ST36), RIMD2210633 (ST3), and AQ4037 (ST96) (see Fig. S3 in the supplemental material). A total of 26 distinct genomic regions, each >1 kb in size, were present in MAVP-Q but absent in other comparator genomes, including environmental ST631 that lacks hemolysins (G149) (see Fig. S3 in the supplemental material). Within a large genomic island ∼37.6 kb in length with an integrase at one terminus and an overall lower GC content (40.6% compared to 45.8% for the genome), a single ORF homologous to restriction endonucleases (AB831_06355) that was restricted to clinical ST631 isolates in our collection and publicly available draft genomes (n = 693; http://www.ncbi.nlm.nih.gov/genome/691; 2017) was selected as a suitable amplicon target. The distribution of this locus was further analyzed using the BLAST algorithm by a query against the nucleotide collection, the nonredundant protein sequences, and against the genus Vibrio (taxid 662), excluding V. parahaemolyticus (taxid 691), using the default settings for BLASTn (50). Similar approaches were applied to identify ST631 diagnostic loci inclusive of the single environmental isolate (G149), which identified a hypothetical protein encoding region (AB831_06535) (ST631env). Oligonucleotide primers were designed to amplify the diagnostic regions, including AB831_06355, using the primers ST631endF (5′-AGTTCATCAGGTAGAGAGTTAGAGGA-3′) and ST631endR (5′-TCTTCGTTACCATAGTATGAGCCA-3′), which produce an amplicon of ca. 494 bp, and AB831_06535, using the primers ST631envF (5′-TGGGCGTTAGGCTTTGC-3′) and ST631-envR (5′-GGGCTTCTACGACTTTCTGCT-3′), which produce an amplicon of 497 bp.
Amplification of diagnostic loci was evaluated in individual assays using genomic DNA from positive and negative controls: MAVP-Q and G149 (ST631), G4186 (ST34), G3578 (ST674), MAVP-M (ST1127), MAVP-26 (ST36), and G61 (ST1125). Amplification of specific sequence types were performed with Accustart enzyme mix on purified DNA. Cycling was performed with an initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and amplification at 72°C for 30 s, with a final elongation at 72°C for 5 min. The primer pairs only produced amplicons from template DNA from ST631, and each was the expected size (data not shown; see Fig. S3 in the supplemental material). Amplicon assays were applied to 208 clinical isolates from Northeast U.S. states (Maine, New Hampshire, Massachusetts, and Connecticut) and 1,140 environmental isolates collected from 2015 to 2016 from New Hampshire and Massachusetts. These assays identified all known ST631 clinical isolates with 100% specificity and also identified an additional seven tdh+ trh+ clinical isolates (ST631end and ST631env positive) and two environmental isolates (ST631end negative and ST631env positive) from our archived collection. Each, with the exception of MAVP-R, was subsequently confirmed to be ST631 by seven-locus MLST (www.pubmlst.org).
Examination of recA allele and adjacent sequences.
The PacBio sequenced genome of MAVP-R, contig 000001 (accession no. MPPP00000000) that contained the recA gene, was annotated using PROKKA1.9 (47). The sequences of recA and its surrounding DNA were then compared to the contig-containing recA region from isolate S130 (AWIW01000000), S134 (AWIS01000000), 090-96 (JFFP01000036) (33), and MAVP-Q (accession no. MDWT00000000). The map of recA region of the five isolates was illustrated using Easyfig (51).
Core genome SNP determination and phylogenetic analysis.
Whole-genome phylogenies were constructed with single-nucleotide polymorphisms (SNPs) identified from draft genomes using kSNP3 to produce aligned SNPs in FASTA format (52). A maximum-likelihood (ML) tree was then built from the FASTA file using raxMLHPC with model GTRGAMMA and the “-f option,” as well as 100 bootstraps (53). Since there were no differences among the clade II ST631 isolates, we used a subset representing geographic and temporal span of isolation.
Minimum-spanning tree analysis was built based on core gene SNPs produced from a cluster analysis. The cluster analysis of ST631 was performed using a custom core genome multilocus sequence type (cgMLST) analysis using RidomSeqSphere+software v3.2.1 (Ridom GmbH, Münster, Germany) as previously described (31). Briefly, the software first defines a cgMLST scheme using the target definer tool with default settings using the PacBio generated MAVP-Q genome as the reference. Then, five other V. parahaemolyticus genomes (BB22OP, CDC_K4557, FDA_R31, RIMD2210633, and UCM-V493) were used for comparison with the reference genome to establish the core and accessory genome genes. Genes that are repeated in more than one copy in any of the six genomes were removed from the analysis. Subsequently, a task template was created that contains both core and accessory genes. Each individual gene locus from MAVP-Q was assigned allele number 1. Each ST631 isolate genome assembly was then queried against the task template, where any locus that differed from the reference genome or any other queried genome was assigned a new allele number. The cgMLST performed a gene-by-gene analysis of all core genes (excluding accessory genes) and identified SNPs within different alleles to establish genetic distance calculations.
Configuration and distribution of VPaI.
The VPaI sequence from the PacBio sequenced genomes of MAVP-Q and MAVP-R were identified by comparison with the published RIMD2210633 VPaI-7 (NC_004605 region between VPA1312 and VPA1395) and VPaITH3996 (AB455531) (16). Identification of the complete MAVP-Q VPaIγ and genomic junctures in chromosome II was done by comparison to the same region of chromosome II in MAVP-R and G149 (which lack an island in this location) using Mauve (54). In a reciprocal manner, the absence of an island in chromosome I in MAVP-Q and G149 was assessed by comparison with chromosome I of MAVP-R. MAVP-Q VPaIγ (MF066646) and MAVP-R VPaIβ (MF066647) were then extracted as a single contiguous sequence and annotated using Prokka 1.9. Gene content and order of the VPaI elements in MAVP-Q, MAVP-R, and RIMD2210633 were then illustrated by Easyfig (51). Roary (55) was then used to determine homologs among VPaI based on each island's annotated sequences with identity set at 50%. Identification of the genome locations of VPaIβ in ST1127 isolate MAVP-M (accession number GCA_001023155) and of VPaIγ in AQ4037 (accession number GCA_000182365) (17) was also done using Mauve (54).
To examine the distribution of the VPaIγ in all publicly available draft genomes (https://www.ncbi.nlm.nih.gov/genome/genomes/691; 2016) and genomes from archived regional isolates, whole draft genome sequences were aligned to a 6,118-bp subsequence of the MAVP-Q VPaI with NASP version 1.0.2 (56) (https://pypi.python.org/pypi/nasp/1.0.2; 2017). This subsequence spanned the unique juncture of the four conserved hypothetical proteins (AB831_22090, AB831_22095, AB831_22100, and AB831_22105) with the adjacent inserted tdh (AB831_22110, ca. 2,549 bp upstream of ure cluster) (see Fig. S1 in the supplemental material). The percent coverage of the reference sequence was used to determine whether each genome harbored only the four hypothetical protein-encoding genes, only a tdh gene, or the entire module including the fusion of the four genes with tdh (see Fig. S1 and Table S2 in the supplemental material). The sequence type of each genome harboring the fused element characteristic of VPaIγ was then determined using the SRST2 pipeline (48). Where sequencing reads were not available as the input for SRST2, they were simulated from assemblies using an in-house Python script (https://github.com/kpdrees/fasta2reads).
A PCR amplification approach was developed and applied to survey the presence of tdh adjacent to the ure gene cluster. Primers were designed to anneal to conserved sequences at the 3′ end of tdh (PIHybF8, 5′-GCCAACATGGATATAAATAAAAATGA-3′) and the 5′ end of ureG (tdhUreGrev5, 5′-GACAAAGGTATGCTGCCAAAAGTG-3′) as determined by gene alignments, which when used together produced a 2,631-bp amplicon of the insertion juncture when used with MAVP-Q as a template (see Fig. S4 in the supplemental material). Amplification was performed on purified DNA with Accustart enzyme mix, with an initial denaturation at 94°C for 3 min, followed by 30 cycles of a denaturation at 94°C for 1 min, annealing at 61°C for 1 min, and amplification at 72°C for 2.5 min, with a final elongation at 72°C for 5 min. This amplification was performed in parallel with a diagnostic multiplex PCR amplification of tdh, trh, and tlh according to published methods (10, 57) to investigate the cooccurrence of VPaIγ with both hemolysin-encoding genes in representative isolates of various clinically prevalent sequence types. Amplicons were visualized using a 1.2% agarose gel in Tris-acetate-EDTA (TAE) buffer (see Fig. S4 in the supplemental material).
Accession number(s).
The accession numbers of the Pacific Biosciences sequenced genome for MAVP-Q are CP022473 for chromosome I and CP022472 for chromosome II, and those for MAVP-R are CP022552 for chromosome I, CP022553 for chromosome II, and CP022554 and CP022555 for the extrachromosomal (plasmid) sequence. Detailed information about all clade II ST631 strain genomes were described previously (31), and these and the accession numbers for the remaining sequenced genomes are listed in Table 2. The accession number of VPaIβ from MAVP-R is MF066647, and the accession number of VPaIγ from MAVP-Q is MF066646.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for clinical isolates and thank Jana Ferguson and Tracy Stiles of the Massachusetts Department of Public Health and M. Hickey and C. Schillaci from the Massachusetts Department of Marine Fisheries, J. K. Kanwit of the Maine Department of Marine Resources and A. Robbins from the Maine Department of Health and Human Services, and Laurn Mank from the Connecticut Department of Public Health Laboratory and K. DeRosia-Banick, Connecticut Department of Agriculture, Bureau of Aquaculture. Assistance with genome sequencing was provided by W. K. Thomas, helpful comments on the manuscript were provided by J. Foster, and technical assistance was provided by J. Lemaire, K. Hartman, C. Hallee, M. Malanga, S. Ilyas, J. Hall, J. Sevigny, M. Dillon, K. Flynn, A. Goupil, J. Means, Sarah Eggert, Ashley Marcinkiewicz, R. Foxall, E. DaSilva, and M. S. Pankey.
Partial funding for this work was provided by the U.S. Department of Agriculture National Institute of Food and Agriculture (Hatch projects NH00574, NH00609 [accession number 233555], and NH00625 [accession number 1004199]). This is Scientific Contribution number 2722. Additional funding was provided by the National Oceanic and Atmospheric Administration College Sea Grant program and grants R/CE-137, R/SSS-2, and R/HCE-3. Support was also provided through the National Institutes of Health (1R03AI081102-01), the National Science Foundation (EPSCoR IIA-1330641), and the National Science Foundation (DBI 1229361 NSF MRI). N.G.-E. was funded through the FDA Foods Science and Research Intramural Program. F. Xu and C. A. Whistler declare a potential conflict of interest in the form of a pending patent application (U.S. patent application 62/128,764).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01168-17.
REFERENCES
- 1.Hiyoshi H, Kodama T, Iida T, Honda T. 2010. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect Immun 78:1772–1780. doi: 10.1128/IAI.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazen TH, Pan L, Gu J-D, Sobecky PA. 2010. The contribution of mobile genetic elements to the evolution and ecology of vibrios. FEMS Microbiol Ecol 74:485–499. doi: 10.1111/j.1574-6941.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 4.Hurley CC, Quirke A, Reen FJ, Boyd EF. 2006. Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7:104. doi: 10.1186/1471-2164-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd EF, Cohen AL, Naughton LM, Ussery DW, Binnewies TT, Stine OC, Parent MA. 2008. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol 8:110. doi: 10.1186/1471-2180-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kishishita M, Matsuoka N, Kumagai K, Yamasaki S, Takeda Y, Nishibuchi M. 1992. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol 58:2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honda T, Ni Y, Miwatani T, Adachi T, Kim J. 1992. The thermostable direct hemolysin of Vibrio parahaemolyticus is a pore-forming toxin. Can J Microbiol 38:1175–1180. doi: 10.1139/m92-192. [DOI] [PubMed] [Google Scholar]
- 8.Park K-S, Ono T, Rokuda M, Jang M-H, Iida T, Honda T. 2004. Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol Immunol 48:313–318. doi: 10.1111/j.1348-0421.2004.tb03512.x. [DOI] [PubMed] [Google Scholar]
- 9.Shirai H, Ito H, Hirayama T, Nakamoto Y, Nakabayashi N, Kumagai K, Takeda Y, Nishibuchi M. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun 58:3568–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panicker G, Call DR, Krug MJ, Bej AK. 2004. Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA microarrays. Appl Environ Microbiol 70:7436–7444. doi: 10.1128/AEM.70.12.7436-7444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishibuchi M, Kaper JB. 1995. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect Immun 63:2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park K-S, Ono T, Rokuda M, Jang M-H, Okada K, Iida T, Honda T. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun 72:6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broberg CA, Calder TJ, Orth K. 2011. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microb Infect 13:992–1001. doi: 10.1016/j.micinf.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Orth K. 2013. Virulence determinants for Vibrio parahaemolyticus infection. Curr Opin Microbiol 16:70–77. doi: 10.1016/j.mib.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 16.Okada N, Iida T, Park K-S, Goto N, Yasunaga T, Hiyoshi H, Matsuda S, Kodama T, Honda T. 2009. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun 77:904–913. doi: 10.1128/IAI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Stine OC, Badger JH, Gil AI, Nair GB, Nishibuchi M, Fouts DE. 2011. Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics 12:1. doi: 10.1186/1471-2164-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Gewurz BE, Ritchie JM, Takasaki K, Greenfeld H, Kieff E, Davis BM, Waldor MK. 2013. A Vibrio parahaemolyticus T3SS effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting TAK1 activation. Cell Rep 3:1690–1702. doi: 10.1016/j.celrep.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard TP, Chao MC, Abel S, Blondel CJ, zur Wiesch PA, Zhou X, Davis BM, Waldor MK. 2016. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc Nat Acad Sci U S A 113:6283–6288. doi: 10.1073/pnas.1601718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronholm J, Petronella N, Leung CC, Pightling A, Banerjee S. 2016. Genomic features of environmental and clinical Vibrio parahaemolyticus isolates lacking recognized virulence factors are dissimilar. Appl Environ Microbiol 82:1102–1113. doi: 10.1128/AEM.03465-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu F, Ilyas S, Hall JA, Jones SH, Cooper VS, Whistler CA. 2015. Genetic characterization of clinical and environmental Vibrio parahaemolyticus from the Northeast USA reveals emerging resident and non-indigenous pathogen lineages. Front Microbiol 6:272. doi: 10.3389/fmicb.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee SK, Kearney AK, Nadon CA, Peterson C-L, Tyler K, Bakouche L, Clark CG, Hoang L, Gilmour MW, Farber JM. 2014. Phenotypic and genotypic characterization of Canadian clinical isolates of Vibrio parahaemolyticus collected from 2000 to 2009. J Clin Microbiol 52:1081–1088. doi: 10.1128/JCM.03047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner JW, Paranjpye RN, Landis ED, Biryukov SV, González-Escalona N, Nilsson WB, Strom MS. 2013. Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest coast of the United States. PLoS One 8:e55726. doi: 10.1371/journal.pone.0055726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones JL, Lüdeke CH, Bowers JC, Garrett N, Fischer M, Parsons MB, Bopp CA, DePaola A. 2012. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J Clin Microbiol 50:2343–2352. doi: 10.1128/JCM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePaola A, Ulaszek J, Kaysner CA, Tenge BJ, Nordstrom JL, Wells J, Puhr N, Gendel SM. 2003. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl Environ Microbiol 69:3999–4005. doi: 10.1128/AEM.69.7.3999-4005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haendiges J, Timme R, Allard MW, Myers RA, Brown EW, Gonzalez-Escalona N. 2015. Characterization of Vibrio parahaemolyticus clinical strains from Maryland (2012-2013) and comparisons to a locally and globally diverse V. parahaemolyticus strains by whole-genome sequence analysis. Front Microbiol 6:125. doi: 10.3389/fmicb.2015.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis CN, Schuster BM, Striplin MJ, Jones SH, Whistler CA, Cooper VS. 2012. Influence of seasonality on the genetic diversity of Vibrio parahaemolyticus in New Hampshire shellfish waters as determined by multilocus sequence analysis. Appl Environ Microbiol 78:3778–3782. doi: 10.1128/AEM.07794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3: K6 and its serovariants. Clin Microbiol Rev 20:39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Urtaza J, Baker-Austin C, Jones JL, Newton AE, Gonzalez-Aviles GD, DePaola A. 2013. Spread of Pacific Northwest Vibrio parahaemolyticus strain. N Engl J Med 369:1573–1574. doi: 10.1056/NEJMc1305535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newton AE, Garrett N, Stroika SG, Halpin JL, Turnsek M, Mody RK, Division of Foodborne W, Environmental D . 2014. Notes from the field: Increase in Vibrio parahaemolyticus infections associated with consumption of Atlantic coast shellfish—2013. MMWR Morb Mortal Wkly Rep 63:335–336. [PMC free article] [PubMed] [Google Scholar]
- 31.Xu F, Gonzalez-Escalona N, Haendiges J, Myers RA, Ferguson J, Stiles T, Hickey E, Moore M, Hickey JM, Schillaci C. 2017. Sequence type 631 Vibrio parahaemolyticus, an emerging foodborne pathogen in North America. J Clin Microbiol 55:645–648. doi: 10.1128/JCM.02162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lüdeke CH, Gonzalez-Escalona N, Fischer M, Jones JL. 2015. Examination of clinical and environmental Vibrio parahaemolyticus isolates by multi-locus sequence typing (MLST) and multiple-locus variable-number tandem-repeat analysis (MLVA). Front Microbiol 6:564. doi: 10.3389/fmicb.2015.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González-Escalona N, Gavilan RG, Brown EW, Martinez-Urtaza J. 2015. Transoceanic spreading of pathogenic strains of Vibrio parahaemolyticus with distinctive genetic signatures in the recA gene. PLoS One 10:e0117485. doi: 10.1371/journal.pone.0117485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park K-S, Suthienkul O, Kozawa J, Yamaichi Y, Yamamoto K, Honda T. 1998. Close proximity of the tdh, trh, and ure genes on the chromosome of Vibrio parahaemolyticus. Microbiology 144:2517–2523. doi: 10.1099/00221287-144-9-2517. [DOI] [PubMed] [Google Scholar]
- 35.Johnson C, Flowers A, Young V, Gonzalez-Escalona N, DePaola A, Noriea N III, Grimes D. 2009. Genetic relatedness among tdh+ and trh+ Vibrio parahaemolyticus cultured from Gulf of Mexico oysters (Crassostrea virginica) and surrounding water and sediment. Microb Ecol 57:437–443. doi: 10.1007/s00248-008-9418-3. [DOI] [PubMed] [Google Scholar]
- 36.González-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus L-A, DePaola A. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol 190:2831–2840. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellingsen BA, Olsen JS, Granum PE, Rorvik LM, González-Escalona N. 2013. Genetic characterization of trh positive Vibrio spp. isolated from Norway. Front Cell Infect Microbiol 3:107. doi: 10.3389/fcimb.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Dai J, Morris JG, Johnson JA. 2010. Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3: K6. BMC Microbiol 10:1. doi: 10.1186/1471-2180-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 40.Takemura AF, Chien DM, Polz MF. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovell CR. 2017. Ecological fitness and virulence features of Vibrio parahaemolyticus in estuarine environments. Appl Microbiol Biotechnol 101:1781–1794. doi: 10.1007/s00253-017-8096-9. [DOI] [PubMed] [Google Scholar]
- 42.Johnson CN. 2013. Fitness factors in vibrios: a minireview. Microb Ecol 65:826–851. doi: 10.1007/s00248-012-0168-x. [DOI] [PubMed] [Google Scholar]
- 43.Matz C, Nouri B, McCarter L, Martinez-Urtaza J. 2011. Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS One 6:e20275. doi: 10.1371/journal.pone.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishibuchi M, Kaper JB. 1985. Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J Bacteriol 162:558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tritt A, Eisen JA, Facciotti MT, Darling AE. 2012. An integrated pipeline for de novo assembly of microbial genomes. PLoS One 7:e42304. doi: 10.1371/journal.pone.0042304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 48.Inouye M, Conway TC, Zobel J, Holt KE. 2012. Short read sequence typing (SRST): multi-locus sequence types from short reads. BMC Genomics 13:338. doi: 10.1186/1471-2164-13-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 53.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 54.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahl JW, Lemmer D, Travis J, Schupp J, Gillece J, Aziz M, Driebe E, Drees K, Hicks N, Williamson C. 2016. The Northern Arizona SNP Pipeline (NASP): accurate, flexible, and rapid identification of SNPs in WGS datasets. Microb Genom 2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whistler CA, Hall JA, Xu F, Ilyas S, Siwakoti P, Cooper VS, Jones SH. 2015. Use of whole-genome phylogeny and comparisons for development of a multiplex PCR assay to identify sequence type 36 Vibrio parahaemolyticus. J Clin Microbiol 53:1864–1872. doi: 10.1128/JCM.00034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jolley KA, Chan M-S, Maiden MC. 2004. mlstdbNet-distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.