ABSTRACT
Thermotoga maritima is a hyperthermophilic anaerobic bacterium that produces molecular hydrogen (H2) by fermentation. It catabolizes a broad range of carbohydrates through the action of diverse ABC transporters. However, in T. maritima and related species, highly similar genes with ambiguous annotation obscure a precise understanding of genome function. In T. maritima, three putative malK genes, all annotated as ATPase subunits, exhibited high identity to each other. To distinguish between these genes, malK disruption mutants were constructed by gene replacement, and the resulting mutant cell lines were characterized. Only a disruption of malK3 produced a defect in maltose catabolism. To verify that the mutant phenotype arose specifically from malK3 inactivation, the malK3 mutation was repaired by recombination, and maltose catabolism was restored. This study demonstrates the importance of a maltose ABC-type transporter and its relationship to sugar metabolism in T. maritima.
IMPORTANCE The application and further development of a genetic system was used here to investigate gene paralogs in the hyperthermophile Thermotoga maritima. The occurrence of three ABC transporter ATPase subunits all annotated as malK was evaluated using a combination of genetic and bioinformatic approaches. The results clarify the role of only one malK gene in maltose catabolism in a nonmodel organism noted for fermentative hydrogen production.
KEYWORDS: genetic systems, ABC transporter, maltose, hyperthermophile, homologous recombination, hydrogen
INTRODUCTION
The ATP-binding cassette (ABC) transporter gene family forms a large group. These transporters support the efficient transport of a wide variety of substrates in an ATP-dependent manner. Maltose transporters are intensively studied and usually belong to the ABC transporter family. They are composed of a substrate binding protein (encoded by malE), transmembrane proteins (encoded by malF and malG), and an ATP-hydrolyzing enzyme (encoded by malK) present as a homodimer (1, 2). Subunits of ABC transporters are frequently identified because they are contiguous with related subunits encoded within operons, and because of their pattern of regulated gene expression responding to substrate availability. However, in the case of malK, the ATPase subunit is not typically contiguous with its other subunits (malE, malF, and malG) (3–5). In addition, its transcriptional expression is not responsive to maltose availability (3, 6). This makes the identification of malK dependent on other approaches. The malK gene has been studied in model organisms, including Escherichia coli and Salmonella enterica, where only one copy is present (5–7). However, in hyperthermophilic bacteria and archaea, malK occurs in multiple copies (8–10).
T. maritima is a well-studied hyperthermophilic bacterium that belongs to the order Thermotogales. It uses simple and complex carbohydrate substrates for growth without catabolite repression (11–14). Maltose is a preferred carbon source for T. maritima (15), and its transport is mediated by the maltose ABC transporter rather than the phosphoenolpyruvate phosphotransferase system (12, 16). T. maritima has three mal operons (mal1, mal2, and mal3) (12, 17). The maltose-inducible nature and protein abundance of the subunits of the mal2 operon (malE2, malF2, and malG2) support their role as maltose transporters (9, 18). T. maritima also has three annotated malK sequences, including malK1 (THMA_0427), malK2 (THMA_1258), and malK3 (THMA_1301) (12, 19). None of the annotated malK genes were upregulated when maltose was employed as an carbon-inducing source (9, 20).
The historic lack of a T. maritima genetic system has limited the understanding of maltose transport. In this study, a reverse genetics approach guided by bioinformatic analysis was used to investigate the relative importance of putative malK genes. A similar approach may be used to study the metabolic function of other genes with identical annotations.
RESULTS
Occurrence and comparisons of MalK-like proteins from T. maritima.
Some bacteria have evolved a single copy of malK (21, 22). However, prior studies (9, 19) and four individual wild-type genome sequences of T. maritima (accession numbers AE000512, CP007013.1, CP004077, and CP011107.1) indicated the presence of three malK-like genes in this organism. In contrast to T. maritima, other thermophilic and hyperthermophilic organisms had various numbers of malK genes, as shown in Table S1 in the supplemental material. While the majority of the hyperthermophilic members of the class Thermotogae have a total of three malK-like genes, one or two malK-like genes are present in mesophilic members of the Thermotogales (Table S1).
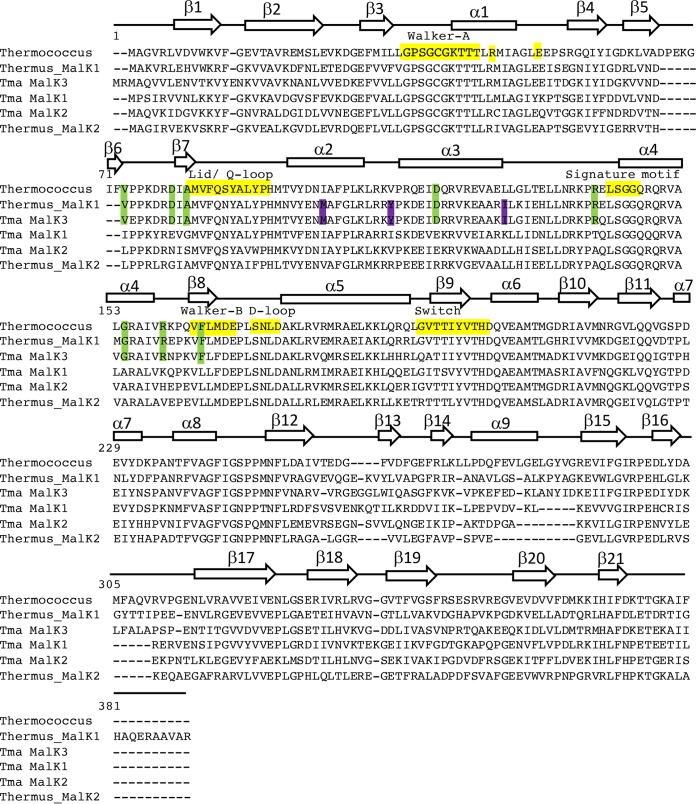
To clarify the significance of the T. maritima malK genes, genetic experimental strategies were pursued. To simplify their designation, they were renamed here malK1 (THMA_0427), malK2 (THMA_1258), and malK3 (THMA_1301). Since the N-terminal region of MalK interacts with transmembrane ABC protein subunits and plays an important role in ATP hydrolysis, this region is highly conserved (23). Previously characterized MalK proteins from Thermococcus litoralis (24) and Thermus thermophilus (10) were used for comparison to the putative MalK of T. maritima and the nonfunctional MalK2 of T. thermophilus. While the N termini of all these MalK proteins exhibited several common motifs, including Walker A and B sequences, a “signature” motif, and Q and H loops (25), the intervening region between Walker A and Walker B was divergent for MalK1, MalK2, and MalK2 of T. thermophilus (Fig. 1). In addition, MalK3 of T. maritima possessed several conserved residues that were common in the active MalK of T. litoralis and T. thermophilus. Although the sequence divergence in MalK1 and MalK2 might make them unlikely to interact with MalF and MalG, the presence of conserved motifs would allow them to act as ATPase subunits for other ABC-type transporters (26–28).
FIG 1.
Structural alignment of MalK from Thermococcus litoralis (24) and Thermus thermophilus (10). Secondary structure elements of T. litoralis MalK are indicated and numbered. Yellow-shaded sequences represent conserved motifs, and green-shaded areas show conserved residues in MalK3 and MalK of T. litoralis and T. thermophilus. The purple shading indicates conserved residues common between MalK3 and MalK of T. thermophilus. Residues of MalK1 and MalK2 of T. maritima and MalK2 of T. thermophilus that were common to the MalK of T. litoralis and T. thermophilus and MalK3 of T. maritima are not shaded.
Disruption of malK by homologous recombination.
Analysis of the T. maritima transcriptome indicated that malK3 expression was not responsive to the addition of maltose (20). Moreover, studies employing 14 monosaccharides and polysaccharides (arabinose, glucose, mannose, rhamnose, ribose, xylose, galactomannan, glucomannan, β-1,3–β-1,4-glucan, laminarin, pustulan, starch, β-xylose, and chitin) did not lead to upregulation of either of the malK genes of T. maritima (9).
In addition, malK3 was not contiguous with other ABC transporter subunits, including malE, malF, and malG. To test which, if any, of the malK genes mediated maltose transport, targeted gene disruptions were constructed using the PgroES::pyrETAF-selectable marker and a uracil auxotroph (PBL3004) deleted for pyrE, as described previously (29). To avoid counterselection for maltose catabolism, consequently, cells were cultivated in complex medium (CM) using cellobiose. This was followed by subculturing cells in defined medium (DM) containing cellobiose without uracil to enable enrichment of Pyr+ recombinants. The clonal isolates were recovered using solid DM containing cellobiose. Five isolates were genotyped from each transformation using PCR and DNA sequencing (Fig. 2). In all cases, a larger PCR product of the target malK gene was identified resulting from insertion of the selectable marker that increased the amplicon size by 763 bp relative to the size of the undisrupted allele. The possibility of homologous recombination at the PgroES locus due to homology encoded within the selectable marker was excluded by checking the size of the PgroES locus; all isolates retained an intact PgroES sequence (Fig. S1D). In addition, the pyrE deletion allele was retained in all recombinants, as indicated by PCR and DNA sequencing of this region (Fig. S1B).
FIG 2.
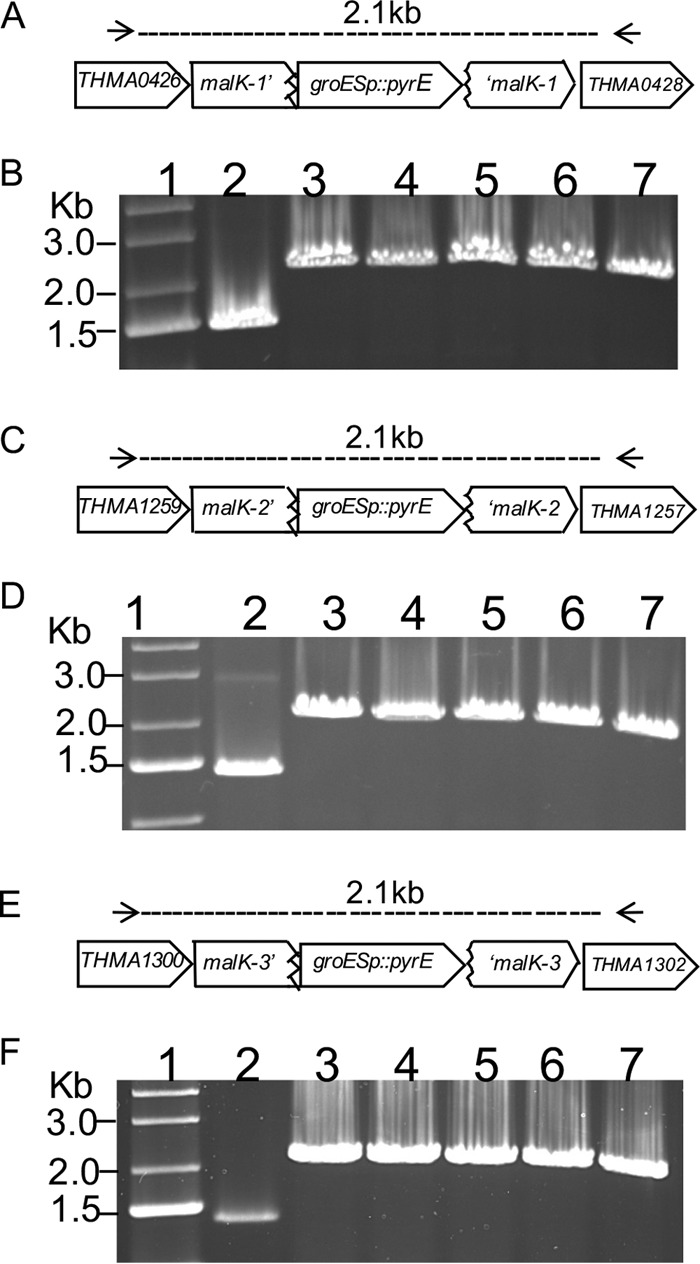
Disruption of the malK loci. (A, C, and E) Schematic representation showing a disrupted copy of malK1 (A), malK2 (C), and malK3 (E). Primers outside each malK locus are indicated by horizontal arrows. (B, D, and F) Agarose gels depicting PCR amplicons of the malK1 (B), malK2 (D), and malK3 (F) genomic regions for five isolates (lanes 3 to 7) compared to the wild-type strain (lane 2). Lane 1 shows a DNA molecular marker.
Identification of the primary maltose transporter ATPase subunit.
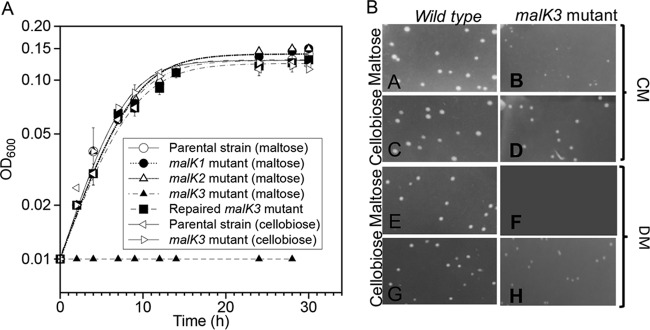
If any of the malK genes were required for maltose transport, mutant derivatives would be unable to grow using maltose as the sole source of carbon and energy. Therefore, growth of all three mutants was examined in DM with the addition of either maltose or cellobiose. Only the malK3 mutant failed to grow using maltose. In contrast, all mutants grew normally using cellobiose (Fig. 3A). Colony formation of the malK3 mutant was then examined relative to the parental wild type using CM or DM and either maltose or cellobiose. After 3 days of incubation at 80°C, the mutant failed to form colonies on maltose plates using either DM, while colonies were apparent in both media containing cellobiose (Fig. 3B). While these results indicated that malK3 was required for maltose catabolism, they also suggested that malK1 and malK2 were not and were therefore incorrectly annotated.
FIG 3.
Growth phenotypes. Growth in tubes of the wild type, malK1, malK2, and malK3 mutants, and the repaired malK3 mutant using DM (A), or on plates containing CM and DM (B) with added maltose or cellobiose, as indicated. Error bars represent results from two biological replicates.
To better understand the possible functions of malK1 and malK2, additional tests were conducted. The malK1 gene is located within an inositol operon and upstream of a xylose isomerase gene, so the malK1 mutant was tested for phenotypes related to inositol and xylose catabolism. No growth defect was observed on xylose (Fig. S2) and, in our hands, none of the strains, including the parental strain, exhibited growth using inositol as reported previously (30). The presence of an α-mannosidase located in the 5′ direction of malK2 suggested that this malK could instead be involved in mannose transport. However, no growth defect was apparent using this sugar. To test whether MalK1 or MalK2 might be involved in transporting maltose oligosaccharides, no growth defect was observed when the malK1 and malK2 mutants were grown on these oligosaccharides. To further test if MalK1 and MalK2 can transport α-linked glucose polysaccharide (starch) or β-linked glucose polysaccharide (carboxymethylcellulose [CMC]), the malK1 and malK2 mutants were tested on these polysaccharides, but no growth defect was observed (data not shown). These results suggest that the malK1 and malK2 genes are not required for maltose transport or the other tested sugars and that malK3 is specific for maltose catabolism (Fig. 4). This is consistent with transcriptomics studies where none of the malK-like genes showed upregulation in the presence of maltose (20).
FIG 4.
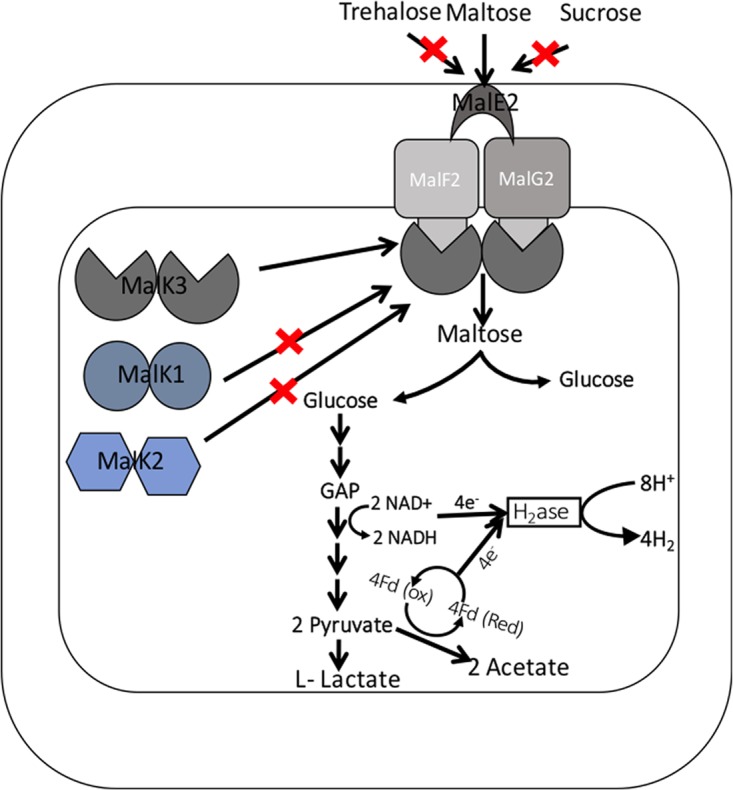
A proposed schematic of maltose transport and its relationship to metabolism in T. maritima. Maltose is transported via the maltose ABC transporter, and no other similar sugars are transported through the maltose transporter. GAP, glyceraldehyde-3-phosphate; H2ase, hydrogenase; Fd, ferredoxin.
Homologous repair of malK3.
To confirm that the maltose defect in the malK3 mutant was associated with malK3 disruption, the strain was complemented with the wild-type malK3 allele. Prior to repair, the mutant was evaluated on maltose CM plates and formed microcolonies relative to the parental wild-type strain, presumably due to the presence of trace amounts of other carbon sources. The mutant small colony size was then used as a screen for repair of the malK3 disruption mutation. To accomplish this, natural transformation was conducted as described previously (29) using a wild-type allele of malK3 and enrichment in DM supplemented with maltose and uracil. The enrichment samples were then plated on CM-maltose plates and resulted in the formation of two colony sizes (Fig. 5A). With additional incubation, the larger colonies continued to increase in size, but the small colonies did not change size. PCR and DNA sequencing demonstrated that large colonies encoded the wild-type allele, while small colonies retained the still-disrupted malK3 allele (Fig. 5B). One of the three isolates (PBL3027) encoding a wild-type allele was examined in DM containing maltose and grew normally relative to the wild-type strain (Fig. 3A). This result confirmed that the isolate had regained the capacity to catabolize maltose as a result of the repair of the malK3 disruption mutation.
FIG 5.
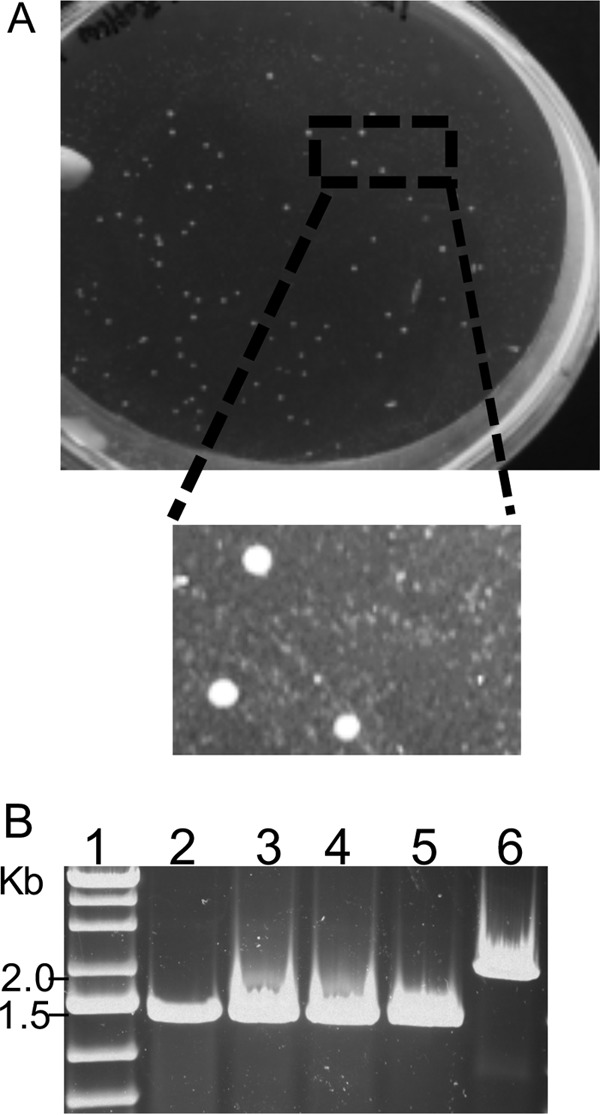
Repair of the malK3 mutation. (A) A CM maltose petri plate showing colonies of the malK3 mutant repaired with the wild-type malK3 allele. (B) An agarose gel depicting PCR amplification of the malK3 locus from three large colonies (lanes 3 to 5), one small colony (lane 6), and the wild type (lane 2). Lane 1 shows a DNA molecular marker.
DISCUSSION
The identification of genes involved in carbon catabolism is important for efforts to develop T. maritima as a host for synthesis of improved biohydrogen. The binding protein-dependent ATP-binding cassette (ABC) transporters are the major route for the assimilation of carbon and energy sources in other hyperthermophilic microbes, including the archaea Sulfolobus solfataricus (31) and Thermococcus litoralis (32) and the bacteria Thermoanaerobacter thermosulfurigenes (33), Thermoanaerobacter ethanolicus (34, 35), Alicyclobacillus acidocaldarius (36), and Thermus thermophilus (10). Interestingly, among these organisms, T. maritima possesses the second highest number of transporter genes that belong to ABC transporters (12, 37, 38). Although various transcriptomics studies have been done in T. maritima to clarify the identity of carbon transporters (9, 13), the malK gene of the maltose transporter was not clear in part due to the lack of a genetic system to identify the required genes.
T. maritima produces 4 mol H2 from per mol glucose through an electron-confurcating hydrogenase (39) and utilizes maltose efficiently (15). However, the identity of the ATPase subunit required for maltose transport was complicated by the presence of three malK-like genes (9, 19) and four wild-type genome sequences (accession numbers AE000512, CP007013.1, CP004077, and CP011107.1). In E. coli, maltose transport involves a complex signal transduction cascade triggered by maltose association with MalE that then binds MalF/MalG, followed by ATP hydrolysis catalyzed by MalK (40, 41). Therefore, a functional MalK must have highly conserved residues to achieve interactions with MalF and MalG (7, 42, 43). In this regard, the Q loop of MalK that remains in close proximity to MalF and MalG (44, 45) and the sequence between the Q loop and the signature motif help distinguish MalK from its homologs (2, 7, 42). A protein sequence alignment of malK3 with the experimentally validated MalK protein sequence of a thermophilic bacterium, Thermus thermophilus (32), and a hyperthermophilic archaeon, Thermococcus litoralis (24), suggests that positions V73, D79, A81, D114, R135, and G148 are conserved and were evident in malK3 of T. maritima. Furthermore, malK2 of Thermus thermophilus that was not involved in maltose transport (32) and both malK1 and malK2 of T. maritima lack these conserved residues. A mutation of L86F in the malK gene of Salmonella enterica serovar Typhimurium that maps inside the Q loop has been shown to exert maltose-defective growth (7), and this conserved residue is variant in malK2 in both T. maritima (L86V) and T. thermophilus (L86I), suggesting that these MalK-like proteins might not interact with MalF and MalG.
A genetic approach to clarify the functional malK from malK1 and malK2 in Thermus thermophilus HB27 (10) provided insight. The availability of a homologous recombination method for T. maritima (29) allowed for the use of a similar approach. T. maritima possesses three mal operons (mal1, mal2, and mal3) (12, 17), but only the mal2 operon is induced at the transcriptional level by the addition of maltose (9). The mal2 operon is also missing a malK subunit. Perhaps, the noncontiguous MalK3 interacts with the ABC transporter subunits of the mal2 operon. However, MalK1 and MalK2 cannot interact with the maltose ABC transporter subunit, because the malK3 mutant remained unable to catabolize maltose despite the presence of wild-type MalK1 or MalK2. While the mal1 and mal3 operons might use either MalK1 or MalK2 to transport a different sugar, no growth defect was observed when these malK mutants were cultivated using various sugars. Predicting the function of a transporter based on the affinity of the substrate binding subunit to various substrates may be deceptive. However, gene knockout studies can clarify the actual substrate that can be transported by the transporter through the manifestation of a defective phenotype, such as a loss of sugar catabolism. Although MalE2 of T. maritima was shown to bind maltose and trehalose (8), the gene disruption data presented here do not support the uptake of trehalose by the maltose transporter of T. maritima. Instead, these findings are consistent with Alicyclobacillus acidocaldarius, where MalE (SBP) bound trehalose, sucrose, lactose, and glucose efficiently, but the complete transporter was found to transport primarily maltose (36).
In T. maritima, H2 synthesis is a function of maltose transported by the maltose transporter (15). Manipulating the amount of transported maltose through the maltose transporter employing genetic engineering is likely to change the maltose metabolism and the amount of formed H2. The continued use of genetics to investigate gene function in T. maritima will promote the development of this organism as a model hyperthermophile, and specifically for the production of biohydrogen with this species.
MATERIALS AND METHODS
Strains and cultivation.
The strains, plasmids, and primers used in this study are shown in Tables 1 and 2. Thermotoga maritima MSB8 (GenBank accession number CP011107.1) and its derivatives were cultured under anaerobic conditions at 80°C in either complex medium (CM) or defined medium (DM), as described previously (29). CM was prepared as described previously (46). DM also was prepared as described previously (47) but lacked yeast extract and tryptone. T. maritima and its mutants were cultivated in Hungate tubes or serum bottles supplemented with 0.5% (15 mM) maltose or cellobiose, unless otherwise indicated. The uracil auxotroph (PB3004) was grown in DM containing 5 μg/ml uracil. Tubes were sealed with butyl rubber stoppers (Bellco Biotechnology) and crimped with metal collars, and the headspace was exchanged with N2. Growth was monitored spectrophotometrically at a wavelength of 600 nm. Sterile 1-ml syringes attached to 20.5-G needles were used for inoculation at an initial cell density of OD600 of 0.02, unless otherwise indicated. All tubes were incubated anaerobically at 80°C overnight, unless otherwise specified.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| PBL3004 | T. maritima pyrE-129 | 29 |
| PBL3023 | PBL3004 malK1′::PgroES::pyrETAF::′malK1 | Current study |
| PBL3024 | PBL3004 malK2′::PgroES::pyrETAF::′malK2 | Current study |
| PBL3025 | PBL3004 malK3′::PgroES::pyrETAF::′malK3 | Current study |
| PBL3027 | PBL3025, wild-type malK3 | Current study |
| Plasmids | ||
| pBN1312 | pUC19 PgroES::pyrETAF | 29 |
| pBN1324 | pUC19 malK1′::PgroES::pyrETAF::′malK1 | Current study |
| pBN1325 | pUC19 malK2′::PgroES::pyrETAF::′malK2 | Current study |
| pBN1326 | pUC19 malK3′::PgroES::pyrETAF::′malK3 | Current study |
| pBN1327 | pUC19 wild-type malK3 | Current study |
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Restriction sitea |
|---|---|---|
| P1 (PgroES F) | CAGAAAGAGGGCGCTTCAA | BamHI |
| P2 (pyrE R) | TTATTTTTTAATGAATCTACTTCCT | BamHI |
| P3 (THMA0427, malK1′) | ATGCCCAGTATCAGGGTTGTG | EcoRI |
| P4 (THMA0427, malK1′) | CTGAAGGTGTTTTATTTCCGCC | SacI |
| P5 (THMA0427, ′malK1) | CAGGAACTTGGAATCACCTCTGT | SalI |
| P6 (THMA0427, ′malK1) | TTACAGAATCGTTTCCTCCGTTT | SphI |
| P7 (THMA1258, malK2′) | TTGGCGCAGGTGAAAATAGA | EcoRI |
| P8 (THMA1258, malK2′) | CTTTTTCAGCTCGCTTCTCATC | SacI |
| P9 (THMA1258, ′malK2) | CTTCAGGAAAGAATTGGAGTCACC | SalI |
| P10 (THMA1258, ′malK2) | TCATGAGATTCTCTCTCCCGTCT | SphI |
| P11 (malK3′ F) | GTGAGAATGGCTCAGGTTGTCC | SacI |
| P12 (malK3′ R) | GCTTCTCATCTGCACTCTGAGTTT | KpnI |
| P13 (′mal3 F) | GAGCTCAAGAAGCTCCACCA | PstI |
| P14 (′mal3 R) | TCAGATAATAGCCTTCTCCGTTTCT | SphI |
| P15 (wt malK3 F) | GTGAGAATGGCTCAGGTTGTCC | EcoRI |
| P16 (wt malK3 R) | TCAGATAATAGCCTTCTCCGTTTCT | KpnI |
| P17 (THMA426F) | GACTCTTTCCAAGTACGTGAAAGG | NA |
| P18 (THMA428 R) | TGCCCCGTGCTATCACA | NA |
| P19 (THMA1257) | TTCAACCGCAGCTGTTTGG | NA |
| P20 (THMA1259) | TTCCAGCACACATTCTGAACAC | NA |
| P21 (THMA1300 F) | TGGTCGTGACAAAGAACGGT | NA |
| P22 (THMA1302 F) | GGACGGACTCAAGGACTACG | NA |
| P23 | CTGTGGGAGAGGACACCCT | NA |
| P24 | AGAACGATTCCTCCCTCTGTC | NA |
| P25 (pyrE F) | GTGATAAAGGAAATCCTCGAGAAAA | NA |
| P26 (pyrE R) | TCATTTCAATCCCCTGCTTCCCGGT | NA |
NA, not applicable.
Bioinformatic analysis of T. maritima MalK.
The deduced protein sequence of malK3 of T. maritima was used to retrieve paralogs from T. maritima using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. More than 50% identity was used as a criterion to predict reliable MalK-like proteins. Additionally, these sequences were used as queries to retrieve orthologs from the genomes of archaea and other bacteria. The malK sequences annotated as spermidine/putrescine transporter subunits were not considered in this study to ensure functional clarity. The region surrounded by Walker A and Walker B of the MalK protein has been reported to be important for transmembrane subunit interactions and ATP binding (23). Therefore, analysis of this region was used to characterize all three MalK proteins of T. maritima.
Construction and repair of malK mutations.
Mutant construction employed the pyrE-selectable marker that was derived from Thermosipho africanus and fused to PgroESL (PgroES::pyrETAF), as described previously (29). This cassette was cloned at the BamHI site of the pUC19 (using P1 and P2 primers) and was named pBN1312. A malK1 disruption cassette (pBN1324) was constructed by amplifying the N-terminal region of the coding region of malK1 (540 bp) (using P3/P4) and the C-terminal region (540 bp) (using P5/P6) and cloning these fragments at the EcoRI/SacI and SalI/SphI sites of pBN1312, respectively. Similarly, the malK2 (pBN1325) and malK3 (pBN1326) disruption cassettes were constructed by amplifying the N-terminal regions of either malK2 (534 bp) (using P7/P8) or malK3 (531 bp) (using P11/P12) and cloning them at the EcoRI/SacI and SacI/KpnI sites of pBN1312, respectively, followed by insertion of the amplified C-terminal regions of malK2 (534 bp) (using P9/P10) and malK3 (579 bp) (using P13/P14), respectively, at the SalI/SphI and SalI/SphI sites of pBN1312 (Table 1). The malK3 repair construct (pBN1327) resulted from cloning the open reading frame of malK3 of the wild-type strain (using P15 and P16 primers) at the EcoRI/KpnI site of pUC19. Ligation, transformation, and selection of E. coli DH5α transformants were performed as described previously (48).
The T. maritima uracil auxotroph (PBL3004) (29) and the malK3 mutant (PBL3025) were used as recipients for natural transformation, as described previously (29). Disruption constructs for malK1, malK2, and malK3 were used to transform PBL3004. Transformed cells of T. maritima were cultivated initially at 80°C for 18 to 24 h under nonselective conditions by inoculating cells into tubes containing 10 ml of CM supplemented with 0.5% cellobiose. Genetic selection was performed subsequently in DM containing 0.5% cellobiose lacking uracil. Liquid enrichments were used to isolate clonal populations by plating on DM plates supplemented with cellobiose. Five colonies from each successful transformation carried out with malK1, malK2, and malK3 disruption constructs were pursued for genotypic analysis. Selected colonies were grown in tubes containing CM, and genomic DNA was prepared as described previously (49, 50). Putative disruption mutations located at the malK1, malK2, and malK3 loci were screened by PCR using outside primers P17/P18, P19/P20, and P21/P22 to detect the PgroES::pyrETAF-disrupted alleles of malK1, malK2, and malK3. Primers outside the malK1, malK2, and malK3 loci were used to exclude the possibility of single-crossover intermediates at the malK loci. To rule out the occurrence of recombination at the native PgroES locus, the PgroES locus in all disrupted mutants was amplified by PCR using primers flanking PgroES (P23/P24). The malK3 disruption mutation was repaired using a two-step enrichment process. Transformed cells were propagated in CM supplemented with cellobiose and then in DM supplemented with maltose. Selection for maltose utilization was then performed using CM supplemented with uracil and maltose to enrich for cells containing the repaired malK3 allele. Since the malK3 mutant formed microcolonies only on CM plates with added maltose, and the repair of the malK3 mutation was anticipated to result in larger colonies, the population was examined for variations in colony size. A mixture of colony sizes was apparent, and three large colonies and one small colony were pursued for genotypic analysis using PCR. Using primers complementary to the flanking genes, the repaired wild-type and parental disrupted alleles were distinguished. Small colonies encoded the disrupted “large” malK3 allele, while large colonies encoded the small undisrupted malK3 allele. The presence or absence of a maltose-specific growth phenotype in the malK1 (PBL3023), malK2 (PBL3024), and malK3 (PBL3025) mutants was determined by cultivating these mutants in DM supplemented with maltose. Growth was measured periodically by measuring the optical density at 600 nm (OD600) of each tube. To observe the maltose-specific colony phenotype, these cells were also plated on CM and DM plates containing maltose. To test if the phenotype of the PBL3025 was solely dependent on maltose, cellobiose was used as an alternative carbon source in the tubes; the parental strain (PBL3004) was used as a control.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this study was provided by the Department of Energy (grant DE-PS02-08ER08-12) and the UNL Cell Development Facility.
We thank Julien Gradnigo for help with genome analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00930-17.
REFERENCES
- 1.Kerr ID. 2002. Structure and association of ATP-binding cassette transporter nucleotide-binding domains. Biochim Biophys Acta 1561:47–64. doi: 10.1016/S0304-4157(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 2.Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boos W, Shuman H. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev 62:204–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuman HA, Silhavy TJ. 1981. Identification of the malK gene product. A peripheral membrane component of the Escherichia coli maltose transport system. J Biol Chem 256:560–562. [PubMed] [Google Scholar]
- 5.Gilson E, Nikaido H, Hofnung M. 1982. Sequence of the malK gene in E.coli K12. Nucleic Acids Res 10:7449–7458. doi: 10.1093/nar/10.22.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bavoil P, Hofnung M, Nikaido H. 1980. Identification of a cytoplasmic membrane-associated component of the maltose transport system of Escherichia coli. J Biol Chem 255:8366–8369. [PubMed] [Google Scholar]
- 7.Hunke S, Landmesser H, Schneider E. 2000. Novel missense mutations that affect the transport function of MalK, the ATP-binding-cassette subunit of the Salmonella enterica serovar Typhimurium maltose transport system. J Bacteriol 182:1432–1436. doi: 10.1128/JB.182.5.1432-1436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nanavati DM, Nguyen TN, Noll KM. 2005. Substrate specificities and expression patterns reflect the evolutionary divergence of maltose ABC transporters in Thermotoga maritima. J Bacteriol 187:2002–2009. doi: 10.1128/JB.187.6.2002-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conners SB, Montero CI, Comfort DA, Shockley KR, Johnson MR, Chhabra SR, Kelly RM. 2005. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol 187:7267–7282. doi: 10.1128/JB.187.21.7267-7282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva Z, Sampaio MM, Henne A, Bohm A, Gutzat R, Boos W, da Costa MS, Santos H. 2005. The high-affinity maltose/trehalose ABC transporter in the extremely thermophilic bacterium Thermus thermophilus HB27 also recognizes sucrose and palatinose. J Bacteriol 187:1210–1218. doi: 10.1128/JB.187.4.1210-1218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber R, Langworthy T, König H, Thomm M, Woese C, Sleytr U, Stetter K. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch Microbiol 144:324–333. doi: 10.1007/BF00409880. [DOI] [Google Scholar]
- 12.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleischmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 13.Chhabra SR, Shockley KR, Conners SB, Scott KL, Wolfinger RD, Kelly RM. 2003. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J Biol Chem 278:7540–7552. doi: 10.1074/jbc.M211748200. [DOI] [PubMed] [Google Scholar]
- 14.Vargas M, Noll KM. 1996. Catabolite repression in the hyperthermophilic bacterium Thermotoga neapolitana is independent of cAMP. Microbiology 142:139–144. doi: 10.1099/13500872-142-1-139. [DOI] [PubMed] [Google Scholar]
- 15.Rinker KD, Kelly RM. 2000. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Biotechnol Bioeng 69:537–547. doi:. [DOI] [PubMed] [Google Scholar]
- 16.Galperin MY, Noll KM, Romano AH. 1996. The glucose transport system of the hyperthermophilic anaerobic bacterium Thermotoga neapolitana. Appl Environ Microbiol 62:2915–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noll KM, Lapierre P, Gogarten JP, Nanavati DM. 2008. Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales. BMC Evol Biol 8:7. doi: 10.1186/1471-2148-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latif H, Lerman JA, Portnoy VA, Tarasova Y, Nagarajan H, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Lee DH, Qiu Y, Zengler K. 2013. The genome organization of Thermotoga maritima reflects its lifestyle. PLoS Genet 9:e1003485. doi: 10.1371/journal.pgen.1003485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nanavati DM, Thirangoon K, Noll KM. 2006. Several archaeal homologs of putative oligopeptide-binding proteins encoded by Thermotoga maritima bind sugars. Appl Environ Microbiol 72:1336–1345. doi: 10.1128/AEM.72.2.1336-1345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen TN, Ejaz AD, Brancieri MA, Mikula AM, Nelson KE, Gill SR, Noll KM. 2004. Whole-genome expression profiling of Thermotoga maritima in response to growth on sugars in a chemostat. J Bacteriol 186:4824–4828. doi: 10.1128/JB.186.14.4824-4828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 22.Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 23.Walter C, Wilken S, Schneider E. 1992. Characterization of side-directed mutations in conserved domains of MalK, a bacterial member of the ATP-binding cassette (ABC) family. FEBS Lett 303:41–44. doi: 10.1016/0014-5793(92)80473-T. [DOI] [PubMed] [Google Scholar]
- 24.Diederichs K, Diez J, Greller G, Muller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. 2000. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J 19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider E, Hunke S. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev 22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 26.Gilson E, Higgins CF, Hofnung M, Ferro-Luzzi Ames G, Nikaido H. 1982. Extensive homology between membrane-associated components of histidine and maltose transport systems of Salmonella Typhimurium and Escherichia coli. J Biol Chem 257:9915–9918. [PubMed] [Google Scholar]
- 27.Higgins CF, Hiles ID, Whalley K, Jamieson DJ. 1985. Nucleotide binding by membrane components of bacterial periplasmic binding protein-dependent transport systems. EMBO J 4:1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White D, Singh R, Rudrappa D, Mateo J, Kramer L, Freese L, Blum P. 2016. Examination of contribution of pentose catabolism to molecular hydrogen formation by targeted disruption of arabinose isomerase (araA) in the hyperthermophilic bacterium Thermotoga maritima. Appl Environ Microbiol 83:e0631-16. doi: 10.1128/AEM.02631-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodionova IA, Leyn SA, Burkart MD, Boucher N, Noll KM, Osterman AL, Rodionov DA. 2013. Novel inositol catabolic pathway in Thermotoga maritima. Environ Microbiol 15:2254–2266. doi: 10.1111/1462-2920.12096. [DOI] [PubMed] [Google Scholar]
- 31.Albers S-V, Elferink MGL, Charlebois RL, Sensen CW, Driessen AJM, Konings WN. 1999. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J Bacteriol 181:4285–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xavier KB, Martins LO, Peist R, Kossmann M, Boos W, Santos H. 1996. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol 178:4773–4777. doi: 10.1128/jb.178.16.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahm K, Matuschek M, Muller H, Mitchell WJ, Bahl H. 1996. Molecular analysis of the amy gene locus of Thermoanaerobacterium thermosulfurigenes EM1 encoding starch-degrading enzymes and a binding protein-dependent maltose transport system. J Bacteriol 178:1039–1046. doi: 10.1128/jb.178.4.1039-1046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones CR, Ray M, Dawson KA, Strobel HJ. 2000. High-affinity maltose binding and transport by the thermophilic anaerobe Thermoanaerobacter ethanolicus 39E. Appl Environ Microbiol 66:995–1000. doi: 10.1128/AEM.66.3.995-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones CR, Ray M, Strobel HJ. 2002. Cloning and transcriptional analysis of the Thermoanaerobacter ethanolicus strain 39E maltose ABC transport system. Extremophiles 6:291–299. doi: 10.1007/s00792-001-0256-1. [DOI] [PubMed] [Google Scholar]
- 36.Hülsmann A, Lurz R, Scheffel F, Schneider E. 2000. Maltose and maltodextrin transport in the thermoacidophilic Gram-positive bacterium Alicyclobacillus acidocaldarius is mediated by a high-affinity transport system that includes a maltose binding protein tolerant to low pH. J Bacteriol 182:6292–6301. doi: 10.1128/JB.182.22.6292-6301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanFossen AL, Verhaart MR, Kengen SM, Kelly RM. 2009. Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl Environ Microbiol 75:7718–7724. doi: 10.1128/AEM.01959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen IT. 2017. TransportDB. http://www.membranetransport.org/transportDB2/index.html.
- 39.Schut GJ, Adams MW. 2009. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 191:4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Böhm A, Diez J, Diederichs K, Welte W, Boos W. 2002. Structural model of MalK, the ABC subunit of the maltose transporter of Escherichia coli: implications for mal gene regulation, inducer exclusion, and subunit assembly. J Biol Chem 277:3708–3717. doi: 10.1074/jbc.M107905200. [DOI] [PubMed] [Google Scholar]
- 41.Covitz KM, Panagiotidis CH, Hor LI, Reyes M, Treptow NA, Shuman HA. 1994. Mutations that alter the transmembrane signalling pathway in an ATP binding cassette (ABC) transporter. EMBO J 13:1752–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mourez M, Hofnung M, Dassa E. 1997. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J 16:3066–3077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ames GF, Mimura CS, Shyamala V. 1990. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia coli to human: traffic ATPases. FEMS Microbiol Rev 6:429–446. doi: 10.1111/j.1574-6968.1990.tb04110.x. [DOI] [PubMed] [Google Scholar]
- 44.Hollenstein K, Frei DC, Locher KP. 2007. Structure of an ABC transporter in complex with its binding protein. Nature 446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 45.Dawson RJ, Locher KP. 2006. Structure of a bacterial multidrug ABC transporter. Nature 443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 46.Chhabra SR, Shockley KR, Ward DE, Kelly RM. 2002. Regulation of endo-acting glycosyl hydrolases in the hyperthermophilic bacterium Thermotoga maritima grown on glucan- and mannan-based polysaccharides. Appl Environ Microbiol 68:545–554. doi: 10.1128/AEM.68.2.545-554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rinker KD, Kelly RM. 1996. Growth physiology of the hyperthermophilic archaeon Thermococcus litoralis: development of a sulfur-free defined medium, characterization of an exopolysaccharide, and evidence of biofilm formation. Appl Environ Microbiol 62:4478–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. 1989. Molecular cloning: a laboratory manual, vol 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Maezato Y, Dana K, Blum P. 2011. Engineering thermoacidophilic archaea using linear DNA recombination, p 435–445. In Williams JA. (ed), Methods in molecular biology, vol 765 Springer Science + Business Media, New York, NY. [DOI] [PubMed] [Google Scholar]
- 50.Singh R, Gradnigo J, White D, Lipzen A, Martin J, Schackwitz W, Moriyama E, Blum P. 2015. Complete genome sequence of an evolved Thermotoga maritima isolate. Genome Announc 3(3):e00557-15. doi: 10.1128/genomeA.00557-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.