Abstract
Long non-coding RNAs (lncRNAs) are a subgroup of non-coding RNA transcripts greater than 200 nucleotides in length with little or no protein-coding potential. Emerging evidence indicates that lncRNAs may play important regulatory roles in the pathogenesis and progression of human cancers, including hepatocellular carcinoma (HCC). Certain lncRNAs may be used as diagnostic or prognostic markers for HCC, a serious malignancy with increasing morbidity and high mortality rates worldwide. Therefore, elucidating the functional roles of lncRNAs in tumors can contribute to a better understanding of the molecular mechanisms of HCC and may help in developing novel therapeutic targets. In this review, we summarize the recent progress regarding the functional roles of lncRNAs in HCC and explore their clinical implications as diagnostic or prognostic biomarkers and molecular therapeutic targets for HCC.
Keywords: Hepatocellular carcinoma, Long non-coding RNAs, Function, Biomarker, Therapeutic target
Core tip: Emerging evidence indicates that long non-coding RNAs (lncRNAs) may play important regulatory roles in the pathogenesis and progression of human cancers, including hepatocellular carcinoma (HCC). Therefore, elucidating the functional roles of lncRNAs in tumors can contribute to a better understanding of the molecular mechanisms of HCC and may help in developing novel therapeutic targets. In this review, we summarize the recent progress regarding the functional roles of lncRNAs in HCC and explore their clinical implications as diagnostic or prognostic biomarkers and molecular therapeutic targets for HCC.
INTRODUCTION
Hepatocellular carcinoma (HCC), a major type of primary liver cancer, is the second leading cause of cancer death worldwide[1]. Unfortunately, the incidence and mortality rates of HCC have continued to increase globally. The high mortality of HCC patients is mainly due to late diagnosis, leading to limited therapeutic options. Accordingly, there is an urgent need to elucidate the molecular mechanisms involved in the initiation and progression of HCC to identify reliable biomarkers for early diagnosis and therapeutic targets to improve the survival of these patients. Recent data have demonstrated that the complexity of human carcinogenesis cannot be accounted for by genetic alterations alone and that epigenetic changes may also be involved[2]. In fact, it is becoming increasingly evident that dysregulated epigenetic regulatory processes play a central role in cancer onset and progression[3]. In human HCC, for example, epigenetic changes in various cancer-related genes are more frequently observed than genetic changes[4], suggesting the crucial impact of epigenetic alterations in hepatocarcinogenesis.
Epigenetic alterations include changes in DNA methylation, histone modifications, and non-coding RNA-mediated gene silencing[5]. Recent studies have revealed that the vast majority of the human genome is actively transcribed into non-coding RNAs (ncRNAs), only 1%-2% of which encode proteins[6,7]. As most cancer studies to date have principally focused on protein-coding genes, the function of ncRNAs in cancer remains largely unknown. Nonetheless, accumulating evidence is shedding light on the functional importance of ncRNAs in cancer biology, and these molecules are emerging as new regulators of diverse biological functions, with important roles in oncogenesis and tumor progression[8]. NcRNAs can be roughly classified into the following two groups based on length: small ncRNAs (< 30 nucleotides) and long ncRNAs (lncRNAs; > 200 nucleotides)[9]. Small ncRNAs, especially microRNAs (miRNAs), have been studied extensively. In contrast, lncRNAs are the least studied transcripts and their functions remain largely unknown, even though they constitute the majority of ncRNAs.
LncRNAs were initially regarded as “transcriptional noise” of the transcriptome. However, the recent application of next-generation sequencing, particularly RNA-sequencing (RNA-Seq), has broadened and deepened our knowledge of lncRNAs related to various types of diseases, including cancer. It is clear that lncRNAs act as critical regulators of multiple cellular processes, especially gene expression. It has been well documented that many lncRNAs are frequently aberrantly expressed in human cancers in which they may serve as oncogenes or tumor suppressors[10-12], suggesting that they may act as novel drivers of tumorigenesis. Compared with protein-coding genes, lncRNA alterations are highly tumor- and cell line-specific[13], and this characteristic of specificity makes lncRNAs promising biomarkers for diagnosis. Importantly, lncRNAs play critical regulatory roles in the pathogenesis and progression of cancers, including cell proliferation, differentiation, apoptosis, tumorigenesis, and progression[14-17]. All of these findings point to lncRNAs as promising diagnostic or prognostic biomarkers and potential therapeutic targets for cancer.
Given the critical roles of lncRNAs in the initiation and progression of cancer, it is not surprising that lncRNAs have aroused considerable interest in HCC research. To date, multiple HCC-related lncRNAs have been identified. In vitro and in vivo functional experiments have shown that in HCC cells, lncRNAs are involved in the regulation of diverse biological processes, such as proliferation, migration, apoptosis, the cell cycle, tumorigenesis, and metastasis. Moreover, increasing evidence indicates that lncRNAs may play irreplaceable roles in the initiation and progression of HCC. As lncRNAs may serve as diagnostic or prognostic biomarkers and therapeutic targets for HCC, elucidating the roles of lncRNAs in tumors can contribute to a better understanding of the molecular mechanisms of HCC and assist in the development of novel therapeutic targets. In this review, we summarize the recent progress regarding the functions of lncRNAs in HCC and explore their clinical implications as diagnostic or prognostic biomarkers and molecular therapeutic targets.
CLASSIFICATION OF LNCRNAs
As they can be categorized according to their various properties, such as transcript length, genomic location and context, sequence and structure conservation, effects on DNA sequences, functional mechanisms and targeting mechanisms, association with protein-coding genes or subcellular structures, many different classifications of lncRNAs have been proposed[18,19]. For example, according to their genomic location relative to neighboring protein-coding genes, lncRNAs have generally been categorized into five classes: sense, antisense, intronic, intergenic, and bidirectional lncRNAs[20]. LncRNAs may also be classified according to their targeting mechanisms: signal, decoy, guide, and scaffold[21].
However, there has been no systematic and unambiguous classification of lncRNAs to date, and many existing lncRNA classifications are conflicting and overlapping. Different criteria (databases, projects, and methodologies) used to classify lncRNAs may be primarily responsible for the classification overlap. In reality, lncRNAs are not a homogeneous class of molecules but rather a mixture of multiple functional classes with distinct biological mechanisms and/or roles[22]. Many lncRNAs are not easily classified into any particular category, and it is likely that the same lncRNAs may be listed in different groups in all classifications[23,24]. In addition, the vast majority of lncRNAs remain functionally uncharacterized, which hampers their functional classification.
Given their complexity, from biogenesis to function, these overlapping and conflicting classifications would inevitably add another layer of difficulty to our understanding of lncRNA biology. Interestingly, the authors of a recent review highlight the roles of large systems biology-based datasets as conceptual guidelines for lncRNA classification and functional annotation[19]. Specifically, advances in high-throughput transcriptome sequencing technologies will contribute to uncovering previously unknown functions of lncRNAs, and as such, the arbitrary classifications will need to be redefined.
SUBCELLULAR LOCALIZATION PATTERNS OF LNCRNAs
LncRNAs have diverse subcellular localization patterns, ranging from bright sub-nuclear foci to almost exclusive cytoplasmic localization; some lncRNAs are found in both compartments[25,26], with the majority preferentially localized to the nucleus and chromatin[20,27-29]. Importantly, it is becoming increasingly clear that the function of lncRNAs depends on their subcellular localization[30]. In general, nuclear lncRNAs are recognized as important transcriptional and epigenetic modulators of nuclear functions[15,31,32], whereas cytoplasmic lncRNAs have been described as modulating mRNA stability and translation[32,33]. Compared with the mostly highly abundant cellular RNAs, the vast majority of lncRNAs that are typically less abundant in a population of cells can be highly abundant in individual cells[25,34]. To more precisely locate and confirm the sub-cellular localization of lncRNAs, two recent reports have suggested that rather than using conventional RNA fluorescence in situ hybridization (FISH) techniques that have a relatively low sensitivity, it may be more effective to study lncRNAs by applying single-molecule RNA FISH[25,35].
MECHANISMS OF LNCRNA-MEDIATED GENE EXPRESSION
To date, the biological functions and molecular mechanisms of most lncRNAs remain largely elusive, with only very few being partially characterized. Nevertheless, existing evidence demonstrates that these molecules play critical roles in the regulation of specific cellular processes, specifically in protein-coding gene expression at the epigenetic, transcriptional and post-transcriptional levels[36-40].
Epigenetic regulation
Epigenetic regulatory mechanisms can act at genomic (DNA methylation or demethylation) or nucleosomal and chromatin (post-translational histone modifications and chromatin remodeling complexes) levels[41]. As stated above, the majority of lncRNAs localize preferentially to the nucleus and chromatin, and increasing evidence indicates that some nuclear lncRNAs epigenetically regulate gene expression by altering chromatin structure[42]. There are two underlying mechanisms by which lncRNAs mediate changes in chromatin and gene expression. First, they can directly interact with chromatin-modifying enzymes, functioning as guides in cis or trans by recruiting chromatin modifiers to specific genomic loci to mediate DNA methylation or histone modification, thereby modulating chromatin states and impacting gene expression[32,43-47]. Second, lncRNAs function as adaptors that link specific chromatin loci with ATP-dependent chromatin-remodeling complexes[48,49], serving as guides to target these complexes to regulate nucleosome remodeling and gene expression[47,50,51].
In addition, lncRNAs have been identified as crucial regulators of epigenetic processes such as X-chromosome inactivation[52,53], genomic imprinting[53,54], cellular differentiation determination[55,56], and cell identity maintenance[57]. Thus, lncRNAs play crucial roles in the epigenetic regulation of gene expression. In particular, investigation of the interrelationships between lncRNAs and epigenetic modifications will provide new insight into cancer diagnosis and therapy.
Transcriptional regulation
At the level of transcriptional regulation, lncRNAs regulate gene expression by (1) recruiting and guiding transcription factors to the promoter region of target genes to regulate their transcription; (2) functioning as transcriptional activators or repressors to mediate gene transcription; (3) interacting with RNA polymerase II to regulate gene transcription; (4) interfering with transcription of adjacent genes in cis; (5) forming lncRNA-DNA hybrids to repress transcription of a target; and (6) affecting protein localization to regulate gene expression[24,58-63].
Post-transcriptional regulation
LncRNAs regulate the expression of genes responsible for biological functions at the post-transcriptional level by modulating messenger RNA (mRNA) stability, translation, degradation, and pre-mRNA alternative splicing genes. These molecules also function as competing endogenous RNA (ceRNA) or endogenous microRNA (miRNA) sponges, act as precursors of miRNAs, and interact with proteins to mediate their activity or alter their localization[58,64-71]. Through these mechanisms, lncRNAs play crucial roles in the post-transcriptional regulation of gene expression.
Taken together, these distinct molecular mechanisms allow dysregulated lncRNAs to up-regulate or down-regulate gene expression, thereby determining their regulatory functions in various biological processes. Nevertheless, the complicated mechanisms underlying such regulatory behaviors need further investigation. The biological functions and molecular mechanisms of action of lncRNAs are presented in Figure 1.
Figure 1.
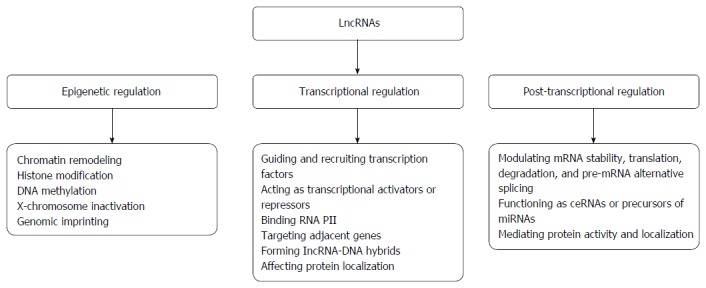
The regulatory mechanisms of long non-coding RNAs. LncRNAs: Long non-coding RNAs; RNA PII: RNA polymerase II; ceRNAs: Competing endogenous RNAs; mRNA: Messenger RNA; miRNAs: MicroRNAs.
FUNCTIONAL ROLES OF LNCRNAs AND MECHANISMS UNDERLYING LNCRNAs DYSREGULATION IN CANCER
Numerous investigations have indicated that aberrantly expressed lncRNAs play critical roles in cancer initiation and progression. However, the biological functions and mechanisms of the majority of lncRNAs in cancer remain largely unknown. In general, lncRNAs regulate gene expression in cancer at the epigenetic, transcriptional, and post-transcriptional levels. Consequently, lncRNAs affect cell proliferation, survival, migration, or genomic stability[72], thereby contributing to tumor development. Specifically, evidence to date demonstrates that lncRNAs are frequently aberrantly expressed in human cancers in which they may serve as oncogenes or tumor suppressors[73,74]. These lncRNAs can mediate several cancer-associated processes, including epigenetic regulation, the DNA damage response, cell cycle control, and miRNA silencing[75]. Furthermore, dysregulated lncRNAs can disrupt multiple cellular oncogenic pathways by exerting oncogenic and/or tumor suppressive functions. LncRNAs also drive many important cancer phenotypes through interactions with other cellular macromolecules, including DNA, protein, and RNA[76]. In brief, the role of lncRNAs in cancer initiation and progression is evident, yet the detailed mechanisms of their involvement in this process need to be clarified.
To date, researchers have elucidated genetic, epigenetic, and transcriptional regulatory mechanisms responsible for dysregulation of lncRNAs in cancer[77]. For instance, genetic regulatory factors, such as genetic instability and single-nucleotide polymorphisms, can be found in lncRNAs and might contribute to their aberrant expression in cancer[77]. Additionally, aberrant expression of lncRNAs with oncogenic properties can be caused by gene amplifications and point mutations[78]. Epigenetic regulation, such as DNA methylation or histone acetylation in the promoter region of lncRNAs, can alter their expression in cancer[79,80], and expression of some cancer-associated lncRNAs can also be initiated by some key transcription factors, such as Myc and p53[81,82], or signaling cascades such as Notch[83]. Taken together, the above-mentioned regulatory factors contribute to aberrant expression of lncRNAs in cancer, with the dysregulated lncRNAs consequently acting as important regulators of cancer initiation and progression.
DYSREGULATED EXPRESSION OF LNCRNAs IN HCC
It has been proven that aberrant lncRNA expression leads to dysregulation of downstream effectors and that lncRNAs may provide a cellular growth advantage resulting in HCC[84], suggesting that lncRNAs may serve as promising diagnostic biomarkers and potential therapeutic targets for HCC. Thus far, multiple dysregulated lncRNAs have been identified as participating in the initiation and progression of HCC. Here, we briefly summarize seven well-documented lncRNAs in HCC: H19, HOTAIR, HULC, HOTTIP, MALAT1, MVIH, and MEG3. FTX, a novel lncRNA associated with HCC, is also discussed. Up-regulated expression of lncRNAs in HCC is thought to have an oncogenic function, whereas a few lncRNAs exhibiting down-regulated expression in HCC may act as tumor suppressors (Table 1).
Table 1.
Hepatocellular carcinoma associated long non-coding RNAs in this review
| LncRNA | Chromosomal location | Dysregulation | Biological roles | Ref. |
| H19 | 11p15.5 | Up-regulated | Promotes HCC growth | Matouk et al[93] |
| Down-regulated | Inhibits migration and invasion of HCC cells | Lv et al[98] | ||
| HOTAIR | 12q13.13 | Up-regulated | Promotes HCC growth | Geng et al[107] |
| HOTTIP | 7p15.2 | Up-regulated | Promotes proliferation of HCC cells | Quagliata et al[115] |
| HULC | 6p24.3 | Up-regulated | Promotes HCC growth | Zhang et al[127] |
| MALAT1 | 11q 13.1 | Up-regulated | Promotes invasion | Lai et al[148] |
| MVIH | 10q22-q23 | Up-regulated | Promotes HCC growth, microvascular invasion, and intrahepatic metastasis | Shi et al[153] |
| MEG3 | 14q32.2 | Down-regulated | Inhibits cell growth | Zhu et al[166] |
| Lnc-FTX | Xq13.2 | Up-regulated | Promotes proliferation and cell cycle progression of HCC cells | Liu et al[175] |
| Down-regulated | Inhibits proliferation and cell cycle progression of HCC cells | Liu et al[176] |
HCC: Hepatocellular carcinoma; LncRNA: Long non-coding RNA; H19: H19, imprinted maternally expressed transcript; HOTAIR: HOX antisense intergenic RNA; HOTTIP: HOXA transcript at the distal tip; HULC: Highly up-regulated in liver cancer; MALAT1: Metastasis-associated lung adenocarcinoma transcript 1; MEG3: Maternally expressed gene 3; MVIH: Microvascular invasion in HCC; FTX: Five prime to Xist.
H19
The human H19 gene (H19) is a paternally imprinted gene located on human chromosome 11p15.5, a locus that contains several imprinted genes, such as insulin-like growth factor 2 (IGF2) and H19. Although H19 has been investigated for years, its role in tumorigenesis is still controversial. Increasing evidence suggests that H19 is highly expressed in many human cancers[73,85-88], indicating that it acts as an oncogene and that its activation may play a critical role in tumorigenesis. Nonetheless, several studies have shown that H19 functions as a tumor suppressor[89-92]. Apparently, H19 has a dual role in tumorigenesis, reflecting the complexity of H19 function. According to the literature, H19 function in HCC is seemingly much more complicated than that in other types of cancers; indeed, its function in hepatocarcinogenesis is largely debated. Numerous investigations have shown that the H19 gene behaves as an oncogene, with its activation contributing to hepatocarcinogenesis. For example, hypoxia induces H19 expression in HCC cells both in vitro and in vivo. Furthermore, silencing H19 expression attenuates tumor growth in vivo, suggesting that H19 behaves as an oncogene and enhances the tumorigenic potential of HCC cells in vivo[93]. A mechanism by which H19 exerts its oncogenic activity in hepatocarcinogenesis has been proposed. Alterations in gene expression at the H19/IGF2 locus are associated with malignancies[87]. In particular, H19 is a precursor of miR-675, and H19 and miR-675 are increasingly described as having key roles in the progression and metastasis of cancers of different tissue origins[94]. Recent data indicate that H19-derived miR-675 favors tumor progression in HCC by repressing expression of twist-related protein 1[95], and miR-675 up-regulates H19 by activating EGR1 in human liver cancer[96]. These findings suggest that the oncogenic role of H19 is mediated through miR-675. Aflatoxin B1 (AFB1) presents another mechanism related to the oncogenic function of H19. AFB1 induces expression of transcriptional factor E2F1 (E2F1), and AFB1-induced E2F1 up-regulates the expression of H19 in HCC HepG2 cells, thereby promoting cellular growth and invasion[97].
Regardless, current evidence supports a role of H19 as a tumor suppressor. A study investigating the effect and mechanism of H19 and miR-675 on HCC cell migration and invasion reported that inhibition of H19 and miR-675 expression can promote the migration and invasion of HCC cells via the AKT/GSK-3β/Cdc25A signaling pathway[98]. This finding suggests that H19 acts a tumor suppressor in HCC cells. Intriguingly, recent data indicate that H19 is down-regulated in intratumoral HCC tissues compared with peritumoral tissues[99]. Additionally, H19 plays a role in promoting tumor initiation but exerts its tumor-suppressive effect on subsequent tumor progression and metastasis in HCC[99]. These findings suggest a tumor-promoting mechanism for H19 in peritumoral HCC tissues and also indicate that H19 has distinct roles at different stages of HCC development. Given the complexity of H19 function in HCC, there is a need for further investigation to resolve the discrepancy.
In particular, a recent study found that up-regulation of H19 has a statistically significant linear correlation with AFP mRNA levels in HCC tumor samples[95], suggesting its role as a potential non-invasive diagnostic biomarker in HCC. Therefore, it should be feasible to detect both AFP and H19 simultaneously to achieve better performance in HCC management.
HOTAIR
HOX transcript antisense intergenic RNA (HOTAIR) is a human gene located on chromosome 12q13.13 that is co-expressed with HOXC genes. HOTAIR has been identified as regulating chromatin silencing of the adjacent HOX locus[100]. Recent studies have revealed that HOTAIR functions as a molecular scaffold to link polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1/REST corepressor 1/RE1-silencing transcription factor (LSD1/CoREST/REST) complexes and direct them to specific gene sites, leading to altered histone H3 lysine 27 (H3K27) methylation and H3K4 demethylation and ultimately resulting in epigenetic gene silencing[46,101]. Accumulating evidence demonstrates that HOTAIR is dysregulated in a variety of human cancers and that overexpression of HOTAIR is associated with cancer cell proliferation, apoptosis, invasion, progression, and metastasis as well as poor survival[102-105].
It has been reported that HOTAIR expression in HCC tissues is significantly higher than that in adjacent non-cancerous tissues[106,107]. In addition, the expression levels of HOTAIR in liver cancer cell lines were found to be higher than those in normal liver cell lines[106]. These findings suggest that HOTAIR exhibits oncogenic activity in HCC. Thus far, several studies have investigated the clinical implications of HOTAIR in HCC. Patients with HCC that overexpress HOTAIR have an increased risk of recurrence following hepatectomy, and there is also a correlation between HOTAIR overexpression and increased risk of lymph node metastasis[108]. A high level of HOTAIR expression has potential as a candidate biomarker for predicting HCC recurrence in liver transplantation (LT) patients[106]. Furthermore, patients with high expression of HOTAIR have a significantly shorter recurrencefree survival than patients with low expression of HOTAIR[109]. Taken together, these findings support the role of HOTAIR as a metastatic biomarker. Indeed, just as in most other types of cancer, HOTAIR is considered most valuable as a prognostic indicator in HCC, particularly as a metastatic biomarker rather than as a diagnostic biomarker[110].
Various mechanisms have been proposed for the oncogenic activity of HOTAIR in HCC. For example, a regulatory network between miR-218 and HOTAIR was elucidated, whereby HOTAIR inactivates P16 (Ink4a) and P14 (ARF) signaling by down-regulating miR-218 expression in HCC via EZH2 targeting of the miR-218-2 promoter regulatory axis and enhancing Bmi-1 expression, resulting in hepatocarcinogenesis[111]. In addition, up-regulation of HOTAIR promotes proliferation, migration, and invasion of human HCC cells by activating autophagy[112], by inhibiting RNA binding motif protein 38 (RBM38)[113], or in part by modulating miR1[114].
HOTTIP
HOXA transcript at the distal tip (HOTTIP), which is transcribed from the 5’ tip of the HOXA locus, has been observed to be up-regulated in various cancers, including HCC[115]. For example, a recent meta-analysis demonstrated that a higher expression level of HOTTIP is correlated with positive lymph node metastasis (LNM) and poor overall survival (OS) in patients with diverse cancers[116], suggesting that HOTTIP might be a potentially promising predictor of LNM and survival in human cancer.
Another recent study showed that HOTTIP expression is significantly up-regulated in HCC tissues compared with adjacent non-neoplastic tissues[115]. Patients with higher levels of HOTTIP and homeobox protein Hox-A13 (HOXA13) showed increased metastasis formation and decreased OS. Moreover, knockdown of HOTTIP inhibited the proliferation of liver cancer-derived cell lines[115]. These findings indicate that HOTTIP might serve as a potential predictor of LNM and survival in patients with HCC. Intriguingly, these authors have also observed marked up-regulation of HOXA13 in HCC, with HOTTIP and HOXA13 having a highly positive correlation. In addition, knock-down of HOTTIP expression led to a reduction in HOXA13 expression in HCC cell lines[115], suggesting that HOTTIP may serve as a transcriptional regulator of HOXA13 in HCC cells. HOTTIP is located at the 5’ end of the HoxA cluster, and can enhance expression of upstream HoxA genes, most prominently HOXA13[117]. Furthermore, HOXA13 has been shown to play a critical role in hepatocarcinogenesis. In a recent study, HOXA13 expression was found to be significantly up-regulated in HCC tissues compared with corresponding paracarcinomatous tissues, and all HOXA13-positive paracarcinomatous tissues exhibited different levels of atypical hyperplasia. Moreover, HOXA13 overexpression may be associated with tumor angiogenesis in HCC[118]. These findings indicate that HOXA13 may play a crucial role in hepatocyte carcinogenesis. Another study found that HOXA13 was the only HOX network gene to be constitutively overexpressed in all tested HCCs, independently of stage[119], suggesting its involvement in the tumorigenic process of HCC. These authors speculated that HOXA13 deregulation is involved in HCC, possibly through nuclear export of eIF4E-dependent transcripts[119]. In addition, overexpression of HOXA13 was shown to rescue the phenotype of HOTTIP knock-down HCC cells, further supporting that up-regulation of HOTTIP in HCC may enhance expression of HOXA13 and eventually mediate HCC carcinogenesis[120]. Overall, HOTTIP exerts its oncogenic functions in hepatocarcinogenesis at least partly by modulating HOXA13. Additionally, the HOTTIP/HOXA13 axis may represent a predictor of prognosis in patients with HCC and a potential therapeutic target for this fatal disease.
Increasing evidence reveals that lncRNAs can interact with miRNAs. Indeed, lncRNAs can act as miRNA sponges, reducing their regulatory effect; in turn, miRNAs may directly interact with lncRNAs and silence their expression[121,122]. MiR-125b has been shown to be a post-transcriptional regulator of HOTTIP in HCC, whereby loss of miR-125b expression might contribute to the frequent up-regulation of HOTTIP[120]. In another recent study, the authors found that both miR-192 and miR-204 function as tumor suppressors to reduce HOTTIP expression via the Argonaute2-mediated RNA interference pathway in HCC. Furthermore, glutaminase has been identified as a potential downstream target of the miR-192/-204-HOTTIP axis in HCC[123].
In summary, the afore-mentioned results suggest the existence of a complex regulatory interaction between HOTTIP and HoxA genes or miRNAs. Upregulation of HOTTIP contributes to hepatocarcinogenesis at least partly by regulating expression of HoxA genes, especially HOXA13, and interacting with miRNAs. Further studies are required to determine whether the regulatory loop between HOTTIP and HOXA13 or miRNAs may serve as potential therapeutic targets for HCC.
HULC
Expression of the highly up-regulated in liver cancer (HULC) gene, which is located on chromosome 6p24.3, is increased in HCC[124], and several recent studies have helped shed light on the factors that contribute to its aberrant up-regulation. For example, research has found that expression of HULC can be enhanced by the transcription factor CREB (cAMP response element-binding protein) through interaction with miR-372[125]. In addition, up-regulation of HULC by the hepatitis B virus (HBV) X protein promotes the proliferation of hepatoma cells through down-regulation of the tumor suppressor p18[126]. Furthermore, it has been shown that HULC might function as an miRNA sponge for miR-372 in HCC and may thereby regulate gene expression at the post-transcriptional level[125].
As an oncogene, HULC is implicated in hepatocarcinogenesis via regulation of multiple biological processes. HULC promotes the proliferation of HCC cells by regulating tumor cell proliferation-associated genes, especially cell cycle-related genes to alter the cell cycle in HCC cells[127]. HULC also contributes to HCC growth by acting mechanistically to deregulate lipid metabolism through a signaling pathway involving miR-9, peroxisome proliferator-activated receptor alpha (PPARA), and acyl-CoA synthetase long chain family member 1 (ACSL1)[128]. In addition, HULC is responsible for perturbations in the circadian rhythm by up-regulating the circadian oscillator CLOCK (clock circadian regulator) in hepatoma cells, resulting in the promotion of hepatocarcinogenesis[129]. Other biological processes, such as angiogenesis, alterations in cell metabolism, activation of a precursor cell compartment, and tissue remodeling, as well as survival, invasion and migration[124,130], may also contribute to hepatocarcinogenesis. Furthermore, HULC functions as a ceRNA to activate the epithelial-mesenchymal transition, stimulating HCC progression and metastasis through the miR-200a-3p/ZEB1 signaling pathway[130]. A recent study provides new insight into the molecular mechanisms underlying the functions of HULC in hepatocarcinogenesis. The authors demonstrate that HULC specifically binds to Y-box protein-1 (YB-1) to promote its phosphorylation through ERK kinase and in turn regulates the interaction of YB-1 with certain oncogenic mRNAs, consequently accelerating the translation of these oncogenic mRNAs in hepatocarcinogenesis[131]. All of these findings indicate that HULC might be involved in the pathogenesis and progression of HCC.
However, there are conflicting data in the literature regarding whether HULC in HCC is associated with a favorable or an unfavorable prognosis. According to a recent study from China, high HULC expression is significantly associated with higher clinical stage and probability of intrahepatic metastasis, and HCC patients with high expression of HULC had worse survival than those with low or no HULC expression[130]. Conversely, two recent studies from South Korea and Germany, propose that high HULC expression is significantly associated with a low stage and grade and less vascular invasion and that HCC patients with high HULC expression have better survival than those with low or no HULC expression[132,133]. These conflicting findings might be largely due to the inclusion of different racial and regional groups. Future studies with larger patient cohorts and various geographic and etiologic backgrounds are needed to confirm the prognostic value of HULC in HCC.
Compared with healthy controls, the plasma level of HULC was found to be dramatically increased in a large cohort of HCC patients, and higher HULC expression was significantly associated with larger tumor size, and no tumor encapsulation[134], as well as higher Edmondson grades and HBV-positive status[135]. Therefore, plasma HULC might act as a potential noninvasive biomarker for predicting the growth, progression and metastasis in HCC.
In summary, the afore-mentioned findings suggest that HULC may contribute to the carcinogenesis and progression of HCC. Therefore, HULC may act as a potential noninvasive biomarker for predicting the growth, progression, metastasis, and prognosis of HCC.
MALAT1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is also known as non-coding nuclear-enriched abundant transcript 2. The MALAT1 locus at 11q13.1 has been reported to harbor chromosomal translocation breakpoints, deletions, translocations, and point mutations linked to cancer[136,137]. These studies have suggested that patients with these phenotypes are more susceptible to cancer.
Nonetheless, the molecular mechanism of MALAT1 in cancer is currently uncertain. Previous cell culture studies have shown that MALAT1 is specifically retained in nuclear speckles to regulate alternative splicing of pre-mRNAs by modulating the functional levels of serine/arginine (SR) splicing proteins[138,139]. Moreover, a recent study suggests that MALAT1 function is only apparent in particular cell types, such as metastatic cancer cells[140]. These studies indicate that aberrant MALAT1 expression promotes tumor metastasis by modulating alternative pre-mRNA splicing. However, another study has suggested a mechanism of gene regulation[141]. Two molecular functions of MALAT1 in cell-based models, contributing to its association with tumor metastasis, have been proposed: regulation of gene expression and alternative splicing[142-144]. For example, regulation of expression of metastasis-associated genes, rather than alternative splicing, is the critical function of MALAT1 in lung cancer metastasis[145]. Although alternative splicing is critical for regulating gene expression, it may not be a major mechanism for modulating gene expression, and alternative splicing alone cannot explain the role of MALAT1 in some cancer cell lines or tissues. Overall, MALAT1 functions as a regulator of alternative splicing or gene expression, governing the hallmarks of cancer metastasis.
Increasing evidence shows that MALAT1 is frequently up-regulated in both liver cancer cell lines and human HCC tissue samples[146], suggesting that it plays an oncogenic role in HCC. A few studies to date have investigated the roles and clinical implications of MALAT1 in HCC. In one study, MALAT1 expression was found to be significantly up-regulated in HCC tumor tissues compared with corresponding non-tumor tissues. Furthermore, MALAT1 was found to act as a marker with high sensitivity for human HCCs at both early and late stages[147], suggesting that the gene can serve as a potential diagnostic tool for HCC. In another study, patients with high expression levels of MALAT1 had a significantly increased risk of tumor recurrence after LT, and silencing MALAT1 with siRNA in HepG2 cells effectively reduced cell viability, motility, and invasiveness and also increased susceptibility to apoptosis[148]. These findings suggest that MALAT1 may play a critical role in HCC progression and serve as a potential predictor of HCC recurrence after LT. Importantly, inhibition of MALAT1 may be a potential therapeutic target for treatment of HCC.
A recent study investigated the role of specificity protein 1/3 (Sp1/3) in the regulation of MALAT1 transcription in HCC cells, and the authors found that Sp1 and Sp3 play roles in up-regulating MALAT1 expression[149]. Several potential mechanisms linking MALAT1 with HCC oncogenesis have been proposed. For instance, MALAT1 was found to be up-regulated in HCC and to act as a proto-oncogene to promote HCC cell growth through Wnt pathway activation and induction of oncogenic serine/arginine-rich splicing factor 1 (SRSF1). In addition, inhibition of SRSF1 expression or mTOR activity abolished the oncogenic properties of MALAT1, and the authors concluded that MALAT1 promotes HCC development through SRSF1 up-regulation and mTOR activation[150]. Nevertheless, the molecular mechanisms underlying the biological functions of MALAT1 in HCC remain largely elusive and require further investigation.
MVIH
The lncRNA microvascular invasion in hepatocellular carcinoma (MVIH) is located in the intron of the RPS24 gene, which encodes a protein belonging to the S24E family of ribosomal proteins[151]. MVIH functions as a tumor promoter and is thus up-regulated in many human cancers. Furthermore, MVIH has been shown to activate angiogenesis[152]. Thus far, only a few studies have shown that MVIH is involved in the pathogenesis and progression of HCC, and the function and mechanism of MVIH in HCC still need to be fully investigated.
A recent study found that MVIH expression was significantly increased in HCC tissues and cells and that MVIH promoted HCC cell growth and inhibited apoptosis by inhibiting miR-199a expression in vitro and in vivo[153]. Taken together, these findings provide evidence that MVIH acts as an miR-199a sponge, linking regulation of gene expression in HCC pathogenesis. In addition to its role in HCC pathogenesis, MVIH has also been shown to activate angiogenesis. A previous study demonstrated that MVIH is generally overexpressed in HCC and plays a key role in activating angiogenesis; consequently, dysregulation of MVIH might serve as a predictor of poor recurrence-free survival of HCC patients after hepatectomy[154]. It is well-known that pathological angiogenesis is essential for oncogenesis, tumor invasion and metastasis. The above-mentioned results suggest that blocking MVIH function might inhibit tumor angiogenesis. Thus, MVIH might serve as a promising therapeutic target for HCC antiangiogenic therapy.
MEG3
Maternally expressed gene 3 (MEG3) is an imprinted gene located at chromosome 14q32.3; imprinting of this gene is controlled by the upstream intergenic differentially methylated region (IG-DMR)[155]. Although MEG3 is expressed in many normal tissues, its expression is lost in various human cancers or cancer cell lines. Numerous studies have verified the functional role of MEG3 as a tumor suppressor in many human cancers[156-158]. Therefore, loss of MEG3 expression may contribute to tumor pathogenesis in a wide range of tissues of different origin. In recent years, hypermethylation of the MEG3 promoter or the MEG-3IG-DMR has been shown to contribute to loss of MEG3 expression in human cancer cells[159-161], and increasing evidence shows that hypermethylation of the MEG3 promoter plays an important role in loss of MEG3 expression in tumors[156,158,162-165]. Overall, hypermethylation in specific MEG3 regions might result in permanent gene transcriptional silencing and the consequent loss of its antiproliferative function, thus contributing to oncogenesis[159].
MEG3 expression was found to be markedly reduced in HCC tissues and cell lines compared with that in adjacent normal liver tissues and normal hepatocytes[79,166]. Furthermore, ectopic expression of MEG3 in hepatoma cells significantly inhibits proliferation and induces apoptosis[166,167], and forced expression of MEG3 in HCC cells significantly decreases both anchorage-dependent and -independent growth and induces apoptosis[79,160]. These data therefore indicate that MEG3 functions as a tumor suppressor in hepatoma cells and plays an important role in hepatocarcinogenesis. Several studies have investigated the mechanism underlying loss of or reduction in MEG3 expression in HCC. Similar to many other cancers, it has been revealed that loss of MEG3 expression in HCC is associated with hypermethylation of its promoter region[79,160,167,168].
It has been proven that MEG3 can inhibit cell proliferation and promote apoptosis through a p53-related pathway[169]. Several studies have also confirmed that overexpression of MEG3 results in an increase in p53 protein and stimulates its transactivational activity in HCC cells[166,170,171]. Further investigation showed that MEG3 functions as a tumor suppressor in hepatoma cells by interacting with p53 to enhance p53-mediated transcriptional activity and influence the expression of partial p53 target genes[166]. In addition, dysregulated tissue-specific expression of miR-29a in HCC epigenetically modulates MEG3 expression through promoter hypermethylation[79].
Kaplan-Meier analysis demonstrated that patients with low MEG3 expression have worse overall and relapse-free survival compared with those with high expression of MEG3, and Cox proportional hazard analyses showed MEG3 expression to be an independent prognostic factor for HCC patients[171]. These findings suggest that decreased expression of MEG3 contributes to HCC development and progression. Overall, MEG3 may serve as a useful molecular diagnostic marker and a potential therapeutic target for HCC.
FTX
The gene five prime to XIST (FTX) is located upstream of XIST, within the X-inactivation center (XIC). FTX is thought to positively regulate the expression of XIST, which is essential for the initiation and spread of X-inactivation[172], and recent studies have indicated the pro-oncogenic potential of FTX in several types of cancer, including renal cell carcinoma[173] and glioma[174].
Surprisingly, there are two opposite findings regarding the role of FTX in HBV-related HCC in a Chinese population. In one study, FTX and FTX-derived miR-545 were found to be up-regulated in HCC tissues compared with matched tumor-adjacent tissues, and patients with high FTX expression exhibited poor survival[175], indicating that FTX functions as an oncogenic lncRNA in HCC. Conversely, in another study, FTX was found to be significantly down-regulated in HCC tissues compared with that in normal liver tissues, and patients with higher FTX expression exhibited longer survival, suggesting that FTX acts as a tumor suppressor in HCC[176]. There are several possible explanations for these two contradictory findings. First, FTX might play distinct roles in HCC because it can function as a precursor for miRNAs and as an endogenous miRNA sponge (also termed ceRNA). FTX can encode a related cluster of miRNAs (miR-374a and miR-545) in most mammalian species[177]. Accordingly, in HCC, FTX can function as an oncogene when it serves as the precursor of miR-545, with which it is co-transcribed, or as a tumor suppressor when it acts as a microRNA sponge for miR-374a to inhibit the binding of miR-374a to its targets. Second, in two studies, FTX was either up-regulated or down-regulated in HCC compared with non-tumor liver samples, suggesting a high FTX variability across different cohorts of patients. Third, different levels of FTX distribution at different sites of the HCC nodule may exist, and inadequate tumor sampling may also be a factor. Fourth, different methods were used to detect FTX in these two studies, with the former using quantitative reverse transcription-quantitative polymerase chain reaction, and the latter in situ hybridization.
PROBLEMS AND PERSPECTIVES
In this review, we summarize the recent progress regarding the functional roles of lncRNAs associated with HCC, including H19, HOTAIR, HULC, HOTTIP, MALAT1, MVIH, MEG3, and FTX. As potent gene regulators, these HCC-related lncRNAs are involved in diverse biological functions, such as cell proliferation, apoptosis, migration, invasion, metastasis, and angiogenesis, thereby contributing to the initiation and progression of HCC. In addition, these HCC-related lncRNAs may serve as potential diagnostic or prognostic biomarkers and also as therapeutic targets for HCC.
Intriguingly, due to their highly specific expression patterns in particular types of cancer[178], efficient detection in the bodily fluids of patients (e.g., blood, plasma, and urine) and relatively stable local secondary structures, lncRNAs have the potential to serve as novel noninvasive biomarkers[13]. For example, HULC is detected with a higher frequency in the plasma of HCC patients than in healthy controls[135], suggesting the possibility of using HULC as a potent circulating biomarker to facilitate early diagnosis of HCC. Nevertheless, further investigations in larger patient cohorts are necessary to validate the diagnostic effectiveness of circulating HULC in HCC.
Despite the importance of lncRNAs in HCC, our current understanding of HCC-related lncRNAs remains rather limited. First, the behavioral characteristics and mechanisms underlying HCC-related lncRNAs contributing to HCC remain largely unclear. Second, “driver lncRNAs” associated with tumorigenesis and progression of HCC have not yet been identified. To gain insight into lncRNA functions and mechanisms of action in HCC, several major issues need to be addressed: (1) technological advances in high-throughput RNA-Seq and high-resolution imaging of RNAs are required. In addition, computational algorithm analysis and integrated datasets are also essential; (2) rather than acting alone, the regulatory role of lncRNAs typically occurs through a large complex network that involves mRNAs, miRNAs, DNA, and proteins[179]. Therefore, it is critical to understand how lncRNAs interact with RNA, DNA, and proteins and how aberrant crosstalk may be regulated in HCC; and (3) most of the previous studies concerning lncRNAs have been retrospective single-center analyses with a relatively small sample size. Thus, a multicenter prospective cohort study with a large sample is needed to gain a deeper understanding of the explicit roles of lncRNAs in HCC in various ethnic populations[85].
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Conflict-of-interest statement: The authors declare no conflict of interests.
Peer-review started: March 21, 2017
First decision: April 26, 2017
Article in press: July 24, 2017
P- Reviewer: Chiu KW S- Editor: Gong ZM L- Editor: Wang TQ E- Editor: Zhang FF
Contributor Information
Zhao-Shan Niu, Laboratory of Micromorphology, School of Basic Medicine, Medical Department of Qingdao University, Qingdao 266071, Shandong Province, China. z.s.niu@qdu.edu.cn.
Xiao-Jun Niu, Oncology Specialty, Medical Department of Qingdao University, Qingdao 266071, Shandong Province, China.
Wen-Hong Wang, Department of Pathology, School of Basic Medicine, Medical Department of Qingdao University, Qingdao 266071, Shandong Province, China.
References
- 1.Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, McGlynn KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978-2007. Int J Cancer. 2016;139:1534–1545. doi: 10.1002/ijc.30211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 3.Morera L, Lübbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics. 2016;8:57. doi: 10.1186/s13148-016-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishida N, Kudo M. Clinical Significance of Epigenetic Alterations in Human Hepatocellular Carcinoma and Its Association with Genetic Mutations. Dig Dis. 2016;34:708–713. doi: 10.1159/000448863. [DOI] [PubMed] [Google Scholar]
- 5.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014;455:43–57. doi: 10.1016/j.bbrc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansji H, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet. 2014;5:379. doi: 10.3389/fgene.2014.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boon RA, Jaé N, Holdt L, Dimmeler S. Long Noncoding RNAs: From Clinical Genetics to Therapeutic Targets? J Am Coll Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 8.Majem B, Rigau M, Reventós J, Wong DT. Non-coding RNAs in saliva: emerging biomarkers for molecular diagnostics. Int J Mol Sci. 2015;16:8676–8698. doi: 10.3390/ijms16048676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ragusa M, Barbagallo C, Statello L, Condorelli AG, Battaglia R, Tamburello L, Barbagallo D, Di Pietro C, Purrello M. Non-coding landscapes of colorectal cancer. World J Gastroenterol. 2015;21:11709–11739. doi: 10.3748/wjg.v21.i41.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amicone L, Citarella F, Cicchini C. Epigenetic regulation in hepatocellular carcinoma requires long noncoding RNAs. Biomed Res Int. 2015;2015:473942. doi: 10.1155/2015/473942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi R, Miyagawa R, Yamashita H, Morikawa T, Okuma K, Fukayama M, Ohtomo K, Nakagawa K. Increased expression of long non-coding RNA XIST predicts favorable prognosis of cervical squamous cell carcinoma subsequent to definitive chemoradiation therapy. Oncol Lett. 2016;12:3066–3074. doi: 10.3892/ol.2016.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takenaka K, Chen BJ, Modesitt SC, Byrne FL, Hoehn KL, Janitz M. The emerging role of long non-coding RNAs in endometrial cancer. Cancer Genet. 2016;209:445–455. doi: 10.1016/j.cancergen.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isin M, Dalay N. LncRNAs and neoplasia. Clin Chim Acta. 2015;444:280–288. doi: 10.1016/j.cca.2015.02.046. [DOI] [PubMed] [Google Scholar]
- 15.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 16.Serviss JT, Johnsson P, Grandér D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet. 2014;5:234. doi: 10.3389/fgene.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liyanarachchi S, Li W, Yan P, Bundschuh R, Brock P, Senter L, Ringel MD, de la Chapelle A, He H. Genome-Wide Expression Screening Discloses Long Noncoding RNAs Involved in Thyroid Carcinogenesis. J Clin Endocrinol Metab. 2016;101:4005–4013. doi: 10.1210/jc.2016-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Shishkin AA, Zhu X, Kadri S, Maza I, Guttman M, Hanna JH, Regev A, Garber M. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol. 2016;17:19. doi: 10.1186/s13059-016-0880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Li C, Pan Y, Han S, Feng B, Gao Y, Chen J, Zhang K, Wang R, Chen L. The Emerging Role and Promise of Long Noncoding RNAs in Lung Cancer Treatment. Cell Physiol Biochem. 2016;38:2194–2206. doi: 10.1159/000445575. [DOI] [PubMed] [Google Scholar]
- 24.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 25.Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Zong X, Huang L, Tripathi V, Peralta R, Freier SM, Guo S, Prasanth KV. Knockdown of nuclear-retained long noncoding RNAs using modified DNA antisense oligonucleotides. Methods Mol Biol. 2015;1262:321–331. doi: 10.1007/978-1-4939-2253-6_20. [DOI] [PubMed] [Google Scholar]
- 29.Singh DK, Prasanth KV. Functional insights into the role of nuclear-retained long noncoding RNAs in gene expression control in mammalian cells. Chromosome Res. 2013;21:695–711. doi: 10.1007/s10577-013-9391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Degirmenci U, Lei S. Role of lncRNAs in Cellular Aging. Front Endocrinol (Lausanne) 2016;7:151. doi: 10.3389/fendo.2016.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 33.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz AA, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunagin M, Cabili MN, Rinn J, Raj A. Visualization of lncRNA by single-molecule fluorescence in situ hybridization. Methods Mol Biol. 2015;1262:3–19. doi: 10.1007/978-1-4939-2253-6_1. [DOI] [PubMed] [Google Scholar]
- 36.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saayman S, Ackley A, Turner AM, Famiglietti M, Bosque A, Clemson M, Planelles V, Morris KV. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther. 2014;22:1164–1175. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez SR, Gay MS, Zhang L. Epigenetic mechanisms in heart development and disease. Drug Discov Today. 2015;20:799–811. doi: 10.1016/j.drudis.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R, Xia LQ, Lu WW, Zhang J, Zhu JS. LncRNAs and cancer. Oncol Lett. 2016;12:1233–1239. doi: 10.3892/ol.2016.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12:1094–1098. doi: 10.1080/15476286.2015.1063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinn JL. lncRNAs: linking RNA to chromatin. Cold Spring Harb Perspect Biol. 2014;6:pii: a018614. doi: 10.1101/cshperspect.a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Böhmdorfer G, Wierzbicki AT. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nainar S, Feng C, Spitale RC. Chemical Tools for Dissecting the Role of lncRNAs in Epigenetic Regulation. ACS Chem Biol. 2016;11:2091–2100. doi: 10.1021/acschembio.6b00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts TC, Morris KV, Weinberg MS. Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics. 2014;9:13–20. doi: 10.4161/epi.26700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montes M, Lund AH. Emerging roles of lncRNAs in senescence. FEBS J. 2016;283:2414–2426. doi: 10.1111/febs.13679. [DOI] [PubMed] [Google Scholar]
- 52.Autuoro JM, Pirnie SP, Carmichael GG. Long noncoding RNAs in imprinting and X chromosome inactivation. Biomolecules. 2014;4:76–100. doi: 10.3390/biom4010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furlan G, Rougeulle C. Function and evolution of the long noncoding RNA circuitry orchestrating X-chromosome inactivation in mammals. Wiley Interdiscip Rev RNA. 2016;7:702–722. doi: 10.1002/wrna.1359. [DOI] [PubMed] [Google Scholar]
- 54.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 55.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopez-Pajares V. Long non-coding RNA regulation of gene expression during differentiation. Pflugers Arch. 2016;468:971–981. doi: 10.1007/s00424-016-1809-6. [DOI] [PubMed] [Google Scholar]
- 57.Jain AK, Xi Y, McCarthy R, Allton K, Akdemir KC, Patel LR, Aronow B, Lin C, Li W, Yang L, et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol Cell. 2016;64:967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Zhang S. Long noncoding RNAs in cell differentiation and pluripotency. Cell Tissue Res. 2016;366:509–521. doi: 10.1007/s00441-016-2451-5. [DOI] [PubMed] [Google Scholar]
- 59.Quan M, Chen J, Zhang D. Exploring the secrets of long noncoding RNAs. Int J Mol Sci. 2015;16:5467–5496. doi: 10.3390/ijms16035467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–18. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tian X, Tian J, Tang X, Ma J, Wang S. Long non-coding RNAs in the regulation of myeloid cells. J Hematol Oncol. 2016;9:99. doi: 10.1186/s13045-016-0333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi X, Sun M, Wu Y, Yao Y, Liu H, Wu G, Yuan D, Song Y. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumour Biol. 2015;36:503–513. doi: 10.1007/s13277-015-3106-y. [DOI] [PubMed] [Google Scholar]
- 65.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rashid F, Shah A, Shan G. Long Non-coding RNAs in the Cytoplasm. Genomics Proteomics Bioinformatics. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX, Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547:1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 68.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14:361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 69.Ankö ML, Neugebauer KM. Long noncoding RNAs add another layer to pre-mRNA splicing regulation. Mol Cell. 2010;39:833–834. doi: 10.1016/j.molcel.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 70.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 73.Soudyab M, Iranpour M, Ghafouri-Fard S. The Role of Long Non-Coding RNAs in Breast Cancer. Arch Iran Med. 2016;19:508–517. [PubMed] [Google Scholar]
- 74.Nikpayam E, Tasharrofi B, Sarrafzadeh S, Ghafouri-Fard S. The Role of Long Non-Coding RNAs in Ovarian Cancer. Iran Biomed J. 2017;21:3–15. doi: 10.6091/.21.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benetatos L, Voulgaris E, Vartholomatos G. The crosstalk between long non-coding RNAs and PI3K in cancer. Med Oncol. 2017;34:39. doi: 10.1007/s12032-017-0897-2. [DOI] [PubMed] [Google Scholar]
- 76.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 78.Babaian A, Mager DL. Endogenous retroviral promoter exaptation in human cancer. Mob DNA. 2016;7:24. doi: 10.1186/s13100-016-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 81.Hart JR, Roberts TC, Weinberg MS, Morris KV, Vogt PK. MYC regulates the non-coding transcriptome. Oncotarget. 2014;5:12543–12554. doi: 10.18632/oncotarget.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grossi E, Sánchez Y, Huarte M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim Biophys Acta. 2016;1859:200–208. doi: 10.1016/j.bbagrm.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 83.Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang D, Cao C, Liu L, Wu D. Up-regulation of LncRNA SNHG20 Predicts Poor Prognosis in Hepatocellular Carcinoma. J Cancer. 2016;7:608–617. doi: 10.7150/jca.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bikle DD, Jiang Y, Nguyen T, Oda Y, Tu CL. Disruption of Vitamin D and Calcium Signaling in Keratinocytes Predisposes to Skin Cancer. Front Physiol. 2016;7:296. doi: 10.3389/fphys.2016.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guan GF, Zhang DJ, Wen LJ, Xin D, Liu Y, Yu DJ, Su K, Zhu L, Guo YY, Wang K. Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. Int J Med Sci. 2016;13:914–922. doi: 10.7150/ijms.16571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Angrand PO, Vennin C, Le Bourhis X, Adriaenssens E. The role of long non-coding RNAs in genome formatting and expression. Front Genet. 2015;6:165. doi: 10.3389/fgene.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao H, Peng R, Liu Q, Liu D, Du P, Yuan J, Peng G, Liao Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys. 2016;610:1–7. doi: 10.1016/j.abb.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 89.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 90.Cui H, Hedborg F, He L, Nordenskjöld A, Sandstedt B, Pfeifer-Ohlsson S, Ohlsson R. Inactivation of H19, an imprinted and putative tumor repressor gene, is a preneoplastic event during Wilms’ tumorigenesis. Cancer Res. 1997;57:4469–4473. [PubMed] [Google Scholar]
- 91.Fukuzawa R, Umezawa A, Ochi K, Urano F, Ikeda H, Hata J. High frequency of inactivation of the imprinted H19 gene in “sporadic” hepatoblastoma. Int J Cancer. 1999;82:490–497. doi: 10.1002/(sici)1097-0215(19990812)82:4<490::aid-ijc4>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 92.Wang L, Sun Y, Yi J, Wang X, Liang J, Pan Z, Li L, Jiang G. Targeting H19 by lentivirus-mediated RNA interference increases A549 cell migration and invasion. Exp Lung Res. 2016;42:346–353. doi: 10.1080/01902148.2016.1223229. [DOI] [PubMed] [Google Scholar]
- 93.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X, Adriaenssens E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6:29209–29223. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hernandez JM, Elahi A, Clark CW, Wang J, Humphries LA, Centeno B, Bloom G, Fuchs BC, Yeatman T, Shibata D. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Ann Surg Oncol. 2013;20 Suppl 3:S625–S635. doi: 10.1245/s10434-013-3106-3. [DOI] [PubMed] [Google Scholar]
- 96.Li H, Li J, Jia S, Wu M, An J, Zheng Q, Zhang W, Lu D. miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget. 2015;6:31958–31984. doi: 10.18632/oncotarget.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lv J, Yu YQ, Li SQ, Luo L, Wang Q. Aflatoxin B1 promotes cell growth and invasion in hepatocellular carcinoma HepG2 cells through H19 and E2F1. Asian Pac J Cancer Prev. 2014;15:2565–2570. doi: 10.7314/apjcp.2014.15.6.2565. [DOI] [PubMed] [Google Scholar]
- 98.Lv J, Ma L, Chen XL, Huang XH, Wang Q. Downregulation of LncRNAH19 and MiR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3β/Cdc25A signaling pathway. J Huazhong Univ Sci Technolog Med Sci. 2014;34:363–369. doi: 10.1007/s11596-014-1284-2. [DOI] [PubMed] [Google Scholar]
- 99.Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 100.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bayram S, Sümbül AT, Batmacı CY, Genç A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour Biol. 2015;36:3863–3870. doi: 10.1007/s13277-014-3028-0. [DOI] [PubMed] [Google Scholar]
- 103.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou X, Chen J, Tang W. The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance. Acta Biochim Biophys Sin (Shanghai) 2014;46:1011–1015. doi: 10.1093/abbs/gmu104. [DOI] [PubMed] [Google Scholar]
- 105.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 106.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 107.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 108.Gao JZ, Li J, DU JL, Li XL. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11:1791–1798. doi: 10.3892/ol.2016.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cai B, Wu Z, Liao K, Zhang S. Long noncoding RNA HOTAIR can serve as a common molecular marker for lymph node metastasis: a meta-analysis. Tumour Biol. 2014;35:8445–8450. doi: 10.1007/s13277-014-2311-4. [DOI] [PubMed] [Google Scholar]
- 111.Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, Liang WC, Wang SS, Ko CH, Waye MM, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63:886–895. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 112.Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12:2605–2612. doi: 10.1039/c6mb00114a. [DOI] [PubMed] [Google Scholar]
- 113.Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C, Peng C, Xie H, Zhou L, Wu J, et al. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15:4060–4076. doi: 10.3390/ijms15034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Su DN, Wu SP, Chen HT, He JH. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol Lett. 2016;12:4061–4067. doi: 10.3892/ol.2016.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen Z, He A, Wang D, Liu Y, Huang W. -Long noncoding RNA HOTTIP as a novel predictor of lymph node metastasis and survival in human cancer: a systematic review and meta-analysis. Oncotarget. 2017;8:14126–14132. doi: 10.18632/oncotarget.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pan TT, Jia WD, Yao QY, Sun QK, Ren WH, Huang M, Ma J, Li JS, Ma JL, Yu JH, et al. Overexpression of HOXA13 as a potential marker for diagnosis and poor prognosis of hepatocellular carcinoma. Tohoku J Exp Med. 2014;234:209–219. doi: 10.1620/tjem.234.209. [DOI] [PubMed] [Google Scholar]
- 119.Cillo C, Schiavo G, Cantile M, Bihl MP, Sorrentino P, Carafa V, D’ Armiento M, Roncalli M, Sansano S, Vecchione R, et al. The HOX gene network in hepatocellular carcinoma. Int J Cancer. 2011;129:2577–2587. doi: 10.1002/ijc.25941. [DOI] [PubMed] [Google Scholar]
- 120.Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, Ng IO, Wong CM. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35:1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 121.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 123.Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L, Han S, Yuan Q, Yang M. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 125.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Y, Li Z, Zhang Y, Zhong Q, Chen Q, Zhang L. Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. Int J Clin Exp Med. 2015;8:12956–12962. [PMC free article] [PubMed] [Google Scholar]
- 128.Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 129.Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia. 2015;17:79–88. doi: 10.1016/j.neo.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J, Qiao Y, Zhan Q. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2017;65:1612–1627. doi: 10.1002/hep.29010. [DOI] [PubMed] [Google Scholar]
- 132.Yang Z, Lu Y, Xu Q, Tang B, Park CK, Chen X. HULC and H19 Played Different Roles in Overall and Disease-Free Survival from Hepatocellular Carcinoma after Curative Hepatectomy: A Preliminary Analysis from Gene Expression Omnibus. Dis Markers. 2015;2015:191029. doi: 10.1155/2015/191029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hämmerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K, et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1) Hepatology. 2013;58:1703–1712. doi: 10.1002/hep.26537. [DOI] [PubMed] [Google Scholar]
- 134.Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, Sun B. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687–696. doi: 10.1159/000430387. [DOI] [PubMed] [Google Scholar]
- 135.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rajaram V, Knezevich S, Bove KE, Perry A, Pfeifer JD. DNA sequence of the translocation breakpoints in undifferentiated embryonal sarcoma arising in mesenchymal hamartoma of the liver harboring the t(11;19)(q11;q13.4) translocation. Genes Chromosomes Cancer. 2007;46:508–513. doi: 10.1002/gcc.20437. [DOI] [PubMed] [Google Scholar]
- 137.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eißmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Groß M, Schirmacher P, Rippe K, Braun T, Zörnig M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 144.Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859:192–199. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 145.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guerrieri F. Long non-coding RNAs era in liver cancer. World J Hepatol. 2015;7:1971–1973. doi: 10.4254/wjh.v7.i16.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 148.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 149.Huang Z, Huang L, Shen S, Li J, Lu H, Mo W, Dang Y, Luo D, Chen G, Feng Z. Sp1 cooperates with Sp3 to upregulate MALAT1 expression in human hepatocellular carcinoma. Oncol Rep. 2015;34:2403–2412. doi: 10.3892/or.2015.4259. [DOI] [PubMed] [Google Scholar]
- 150.Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017;77:1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 152.Kumar MM, Goyal R. LncRNA as a Therapeutic Target for Angiogenesis. Curr Top Med Chem. 2017;17:1750–1757. doi: 10.2174/156802661766616111644744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shi Y, Song Q, Yu S, Hu D, Zhuang X. Microvascular invasion in hepatocellular carcinoma overexpression promotes cell proliferation and inhibits cell apoptosis of hepatocellular carcinoma via inhibiting miR-199a expression. Onco Targets Ther. 2015;8:2303–2310. doi: 10.2147/OTT.S86807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 155.Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 156.Zhang J, Lin Z, Gao Y, Yao T. Downregulation of long noncoding RNA MEG3 is associated with poor prognosis and promoter hypermethylation in cervical cancer. J Exp Clin Cancer Res. 2017;36:5. doi: 10.1186/s13046-016-0472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kruer TL, Dougherty SM, Reynolds L, Long E, de Silva T, Lockwood WW, Clem BF. Expression of the lncRNA Maternally Expressed Gene 3 (MEG3) Contributes to the Control of Lung Cancer Cell Proliferation by the Rb Pathway. PLoS One. 2016;11:e0166363. doi: 10.1371/journal.pone.0166363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Modali SD, Parekh VI, Kebebew E, Agarwal SK. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol. 2015;29:224–237. doi: 10.1210/me.2014-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 160.Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Lehmann U. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One. 2012;7:e49462. doi: 10.1371/journal.pone.0049462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gejman R, Batista DL, Zhong Y, Zhou Y, Zhang X, Swearingen B, Stratakis CA, Hedley-Whyte ET, Klibanski A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocrinol Metab. 2008;93:4119–4125. doi: 10.1210/jc.2007-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chak WP, Lung RW, Tong JH, Chan SY, Lun SW, Tsao SW, Lo KW, To KF. Downregulation of long non-coding RNA MEG3 in nasopharyngeal carcinoma. Mol Carcinog. 2017;56:1041–1054. doi: 10.1002/mc.22569. [DOI] [PubMed] [Google Scholar]
- 163.Sheng X, Li J, Yang L, Chen Z, Zhao Q, Tan L, Zhou Y, Li J. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol Rep. 2014;32:277–285. doi: 10.3892/or.2014.3208. [DOI] [PubMed] [Google Scholar]
- 164.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 165.Gao Y, Lu X. Decreased expression of MEG3 contributes to retinoblastoma progression and affects retinoblastoma cell growth by regulating the activity of Wnt/β-catenin pathway. Tumour Biol. 2016;37:1461–1469. doi: 10.1007/s13277-015-4564-y. [DOI] [PubMed] [Google Scholar]
- 166.Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, Wei L, Jin Y, Fu H, Wu Y, et al. Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells. PLoS One. 2015;10:e0139790. doi: 10.1371/journal.pone.0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu LX, Deng W, Zhou XT, Chen RP, Xiang MQ, Guo YT, Pu ZJ, Li R, Wang GF, Wu LF. The mechanism of adenosine-mediated activation of lncRNA MEG3 and its antitumor effects in human hepatoma cells. Int J Oncol. 2016;48:421–429. doi: 10.3892/ijo.2015.3248. [DOI] [PubMed] [Google Scholar]
- 168.Zamani M, Sadeghizadeh M, Behmanesh M, Najafi F. Dendrosomal curcumin increases expression of the long non-coding RNA gene MEG3 via up-regulation of epi-miRs in hepatocellular cancer. Phytomedicine. 2015;22:961–967. doi: 10.1016/j.phymed.2015.05.071. [DOI] [PubMed] [Google Scholar]
- 169.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 170.Chang L, Wang G, Jia T, Zhang L, Li Y, Han Y, Zhang K, Lin G, Zhang R, Li J, et al. Armored long non-coding RNA MEG3 targeting EGFR based on recombinant MS2 bacteriophage virus-like particles against hepatocellular carcinoma. Oncotarget. 2016;7:23988–24004. doi: 10.18632/oncotarget.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, Wang P, Sun B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol Carcinog. 2016;55:209–219. doi: 10.1002/mc.22270. [DOI] [PubMed] [Google Scholar]
- 172.Chureau C, Chantalat S, Romito A, Galvani A, Duret L, Avner P, Rougeulle C. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet. 2011;20:705–718. doi: 10.1093/hmg/ddq516. [DOI] [PubMed] [Google Scholar]
- 173.Zhang W, Bi Y, Li J, Peng F, Li H, Li C, Wang L, Ren F, Xie C, Wang P, et al. Long noncoding RNA FTX is upregulated in gliomas and promotes proliferation and invasion of glioma cells by negatively regulating miR-342-3p. Lab Invest. 2017;97:447–457. doi: 10.1038/labinvest.2016.152. [DOI] [PubMed] [Google Scholar]
- 174.He X, Sun F, Guo F, Wang K, Gao Y, Feng Y, Song B, Li W, Li Y. Knockdown of Long Noncoding RNA FTX Inhibits Proliferation, Migration, and Invasion in Renal Cell Carcinoma Cells. Oncol Res. 2017;25:157–166. doi: 10.3727/096504016X14719078133203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y, Jia Y, Li Q, Zhang H, Tu K, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350–25365. doi: 10.18632/oncotarget.8129. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 176.Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–5434. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 177.Romito A, Rougeulle C. Origin and evolution of the long non-coding genes in the X-inactivation center. Biochimie. 2011;93:1935–1942. doi: 10.1016/j.biochi.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 178.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ernst C, Morton CC. Identification and function of long non-coding RNA. Front Cell Neurosci. 2013;7:168. doi: 10.3389/fncel.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]


