Abstract
Activation of prefrontal cortical (PFC), striatal, and hippocampal dopamine 1-class receptors (D1R and D5R) is necessary for normal spatial information processing. Yet the precise role of the D1R versus the D5R in the aforementioned structures, and their specific contribution to the water-maze spatial learning task remains unknown. D1R- and D5R- specific in situ hybridization probes showed that forebrain restricted D1R and D5R KO mice (F-D1R/D5R KO) displayed D1R mRNA deletion in the medial (m)PFC, dorsal and ventral striatum, and the dentate gyrus (DG) of the hippocampus. D5R mRNA deletion was limited to the mPFC, the CA1 and DG hippocampal subregions. F-D1R/D5R KO mice were given water-maze training and displayed subtle spatial latency differences between genotypes and spatial memory deficits during both regular and reversal training. To differentiate forebrain D1R from D5R activation, forebrain restricted D1R KO (F-D1R KO) and D5R KO (F-D5R KO) mice were trained on the water-maze task. F-D1R KO animals exhibited escape latency deficits throughout regular and reversal training as well as spatial memory deficits during reversal training. F-D1R KO mice also showed perseverative behavior during the reversal spatial memory probe test. In contrast, F-D5R KO animals did not present observable deficits on the water-maze task. Because F-D1R KO mice showed water-maze deficits we tested the necessity of hippocampal D1R activation for spatial learning and memory. We trained DG restricted D1R KO (DG-D1R KO) mice on the water-maze task. DG-D1R KO mice did not present detectable spatial memory deficit, but did show subtle deficits during specific days of training. Our data provides evidence that forebrain D5R activation plays a unique role in spatial learning and memory in conjunction with D1R activation. Moreover, these data suggest that mPFC and striatal, but not DG D1R activation are essential for spatial learning and memory.
Keywords: Dopamine-1 Receptor, Dopamine-5 Receptor, Water-maze, Conditional KO, Neuromodulator
Introduction
The water-maze task is a spatial learning and memory paradigm whereby an animal randomly swims to find a hidden escape platform. Successful water-maze spatial learning requires the hippocampus (Morris et al., 1982). The hippocampus is hypothesized to form an allocentric representation of location whereby external cues provide spatial information regarding position of an escape platform (Eichenbaum et al., 1990; Nakazawa et al., 2002). Relatedly, the dorsal striatum and ventral striatum contribute to spatial processing of early water-maze learning (Annett et al., 1989; Groenewegen et al., 1987; Lee et al., 2014; Woolley et al., 2013). Moreover, the PFC is necessary for water-maze reversal training, which requires extinguishing previous spatial associations and updating newly acquired information into pre-existing spatial schemas (Bartlett, 1932; Lacroix et al., 2002; Piaget, 1926). The hippocampus is also required for updating spatial schemas (Bethus et al., 2010; Dragoi and Tonegawa, 2013; McKenzie et al., 2013; Tse et al., 2007). Although the brain structures required for processing spatial information are well known, the mechanisms underlying spatial learning and memory in these structures remain unclear.
Dopamine 1- and 5-receptor (D1R and D5R) activation is linked to spatial learning and memory processing (Bethus et al., 2010; O’Carroll et al., 2006; Sawaguchi and Goldman-Rakic, 1991; Silva et al., 2012). Constitutive deletion of the D1R significantly impairs regular and reversal learning on the water-maze task (El-Ghundi et al., 1999; Granado et al., 2008; Holmes et al., 2001; Karasinska et al., 2000; Smith et al., 1998). However, constitutive D5R deletion has not shown deficits in water-maze spatial learning and memory (Holmes et al., 2001). Direct injection of D1R/D5R antagonists into the hippocampus impairs regular and reversal water-maze learning, while injection into the striatum impairs water-maze spatial learning and memory (Mele et al., 2004; Silva et al., 2012). However, constitutive KO and pharmacological studies cannot differentiate the specific contribution of region-specific D1R and D5R activation in spatial learning and memory. Constitutive D1R deletion is not region specific, while D1R/D5R antagonists cannot discriminate between D1Rs and D5Rs (Missale et al., 1998). Moreover, the D1R and D5R receptor are functionally distinct (Lee et al., 2002; Liu et al., 2000; Sariñana et al., 2014). Thus, constitutive KO and pharmacological techniques are unable to differentiate the functional role of subregion-specific PFC, striatal, and hippocampal D1Rs and D5Rs in spatial learning and memory.
In this study, we overcome the aforementioned limitations of pharmacological and constitutive KO studies by utilizing region-specific D1R/D5R KO, D1R KO, and D5R KO mice. We found that deletion of both forebrain D1Rs and D5Rs resulted in spatial learning and memory deficits. When comparing single forebrain D1R deletion from forebrain D5R deletion, D1R deletion produced spatial learning and memory deficits while forebrain D5R deletion did not. Given the deficits in forebrain D1R, but not D5R, KO animals we tested the necessity of hippocampal D1Rs in spatial learning and memory. Dentate gyrus (DG) D1R deletion resulted in subtle latency deficits but no statistical differences were found on spatial memory performance. Although forebrain D5R deletion did not impact spatial learning and memory, the double D1R and D5R KO animals showed qualitative phenotypic differences when compared to forebrain D1R KO mice. Thus, D5R activation in conjunction with D1R activation play a vital role in spatial information processing. In using our subregion specific KO mice, we further the understanding of D1R and D5R function in spatial learning and memory.
Materials and Methods
Animals
All experiments were carried out on homozygous flx D1R, D5R, or D1R/D5R male mice (C57/BL6 background) that were either positive (KO mice) or negative carriers (flx control mice) of the cre-recombinase transgene. DG-D1R KO animals were trained between 16 and 24 weeks of age, while forebrain animals were between 28 and 40 weeks of age. The original characterization of DG-D1R and F-D1R/D5R KO animals showed that deletion of the D1R or D5R was complete and spatially restricted during the aforementioned ages (Sariñana et al., 2014). Each cage contained 2 to 4 mice with ad libitum access to food and water. Experiments and analyses were conducted blind to mouse genotypes. All mice were on a 12 hour light/dark cycle. All procedures conformed to the Institutional and NIH guidelines.
Water-Maze Training and Testing
The Morris water-maze task was used to assess spatial learning and memory. All mice were given four training trials per day (45 min. inter-trial interval) for ten consecutive days. Mice were given 90 seconds to find a 12 cm platform submerged in opaque water in a 1.6 m diameter pool. The starting point to find the platform was pseudo-randomized for each training trial. If mice did not find the platform within 90 seconds, they were placed onto the platform for 15 seconds. High contrast external cues were prominently displayed on each wall of the training room surrounding the pool. On days 6 and 11, mice received a 60 second probe trial. During the probe, the submerged platform was removed, and the animal was placed into the pool center at the start of the probe. During reversal training and probe trials, the animal received the same protocol as described above. However, during reversal training, the platform was moved to the adjacent quadrant. If a mouse exhibited floating behavior during a single probe trial, they were removed from analysis. If the mouse floated during both probe trials (e.g., reverse probe 1 and 2), they were removed from the study. All water maze data was collected using Image Water 2020 software.
In Situ Hybridization and Receptor Quantification
See references (McHugh et al., 2007; Sariñana et al., 2014) for in situ hybridization probes and general methods. See references (Lazic, 2009; Sariñana et al., 2014) for receptor quantification.
Behavioral Batteries
Open field activity - Mice were handled for 2-min each day for three consecutive days before the start of the open field test. Activity was measured by infrared beam interruption and recorded in 1-min intervals over a 10-min period (F-D5R flx and KO) or 15-min period (F-D1R flx and KO) (Digiscan apparatus, Accuscan Instruments). DG-D1R KO and F-D1R/D5R KO mice open field and rotarod behavior have previously been reported and do not show abnormal gross motor activity (Sariñana et al., 2014).
Rotarod – Mice were placed on a rotating platform that increases in the rate of rotation over the period of 300 seconds. The time from placement onto the apparatus to the time to fall was recorded.
Data analysis
Data analysis was performed with GraphPad PRISM software (GraphPad, San Diego, CA) and all reported values are as SEM. Statistical significance was determined by 2 × ANOVA with multiple comparisons performed with Bonferroni, Fisher least significant differences, posttests or by two-tailed unpaired Student’s t-tests; P < 0.05 was considered significant.
Results
D1R and D5R Forebrain Expression and Deletion Patterns
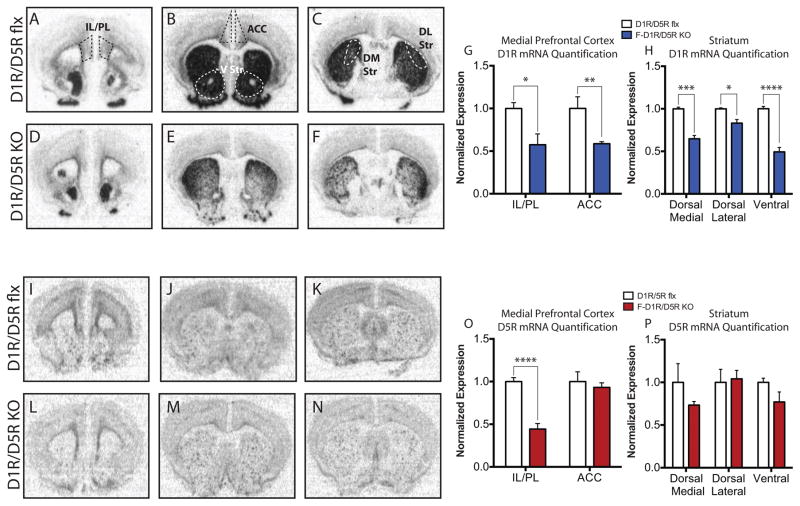
Forebrain KO mice were developed by crossing CaMKII-Cre mice (Tsien et al., 1996a) with floxed (flx) D1R/D5R, D1R, and D5R mice (Sariñana et al., 2014). Using a D1R and D5R specific riboprobe, we show that the D1R mRNA is expressed throughout the dorsal and ventral (V) striatum and mPFC, which consists of the infralimbic (IL), prelimbic (PC), and anterior cingulate cortices (ACC) (Fig. 1A–C). D1R mRNA expression was also observed in the DG of hippocampus, consistent with our previous report (Sariñana et al., 2014) (Supp. Fig. 2A). D5R mRNA signal was primarily observed in the IL/PL region, with minimal expression in the ACC, and sparse signal throughout the striatum (Fig. 1I–K). D5R mRNA expression was also observed in CA1, CA3, and DG subregions of the hippocampus, consistent with our previous report (Sariñana et al., 2014) (Supp. Fig. 2C).
Figure 1. Pattern and Quantification of D1R and D5R Expression.
(A) IL/PL D1R mRNA expression. (B) ACC and ventral (V) striatum D1R mRNA expression (C) dorsal medial (DM) and dorsal lateral (DL) striatum D1R mRNA expression. (A–C) Black or white dashed lines indicate regions of interest used to quantify D1R and D5R mRNA signal. (D) IL/PL D1R mRNA expression in F-D1R/D5R KO mice. (E) ACC and V striatum D1R mRNA expression in F-D1R/D5R KO mice. (F) DM and DL striatum D1R mRNA expression in F-D1R/D5R KO mice. (G) Quantification of IL/PL and ACC D1R mRNA signal (D1R/D5R flx IL/PL, n = 7 and F-D1R/D5R KO IL/PL, n = 6; D1R/D5R flx ACC, n = 4 and F-D1R/D5R KO ACC = 6). (H) Quantification of DM, DL, and V striatum D1R mRNA signal (D1R/D5R flx DM/DL, n = 3 and F-D1R/D5R KO DM/DL, n = 6; D1R/D5R flx V striatum, n = 6 and F-D1R/D5R KO V striatum = 3). (I) IL/PL D5R mRNA expression. (J) ACC and V striatum D5R mRNA expression (K) DM and DL striatum D5R mRNA expression. (L) IL/PL D5R mRNA expression in F-D1R/D5R KO mice. (M) ACC and V striatum D5R mRNA expression in F-D1R/D5R KO mice (N) DM and DL striatum D5R mRNA expression in F-D1R/D5R KO mice. (O) Quantification of IL/PL and ACC D5R mRNA signal (D1R/D5R flx IL/PL, n = 5 and F-D1R/D5R KO IL/PL, n = 5; D1R/D5R flx ACC, n = 3 and F-D1R/D5R KO ACC = 5). (P) Quantification of DM, DL, and V striatum D5R mRNA signal in F-D1R/D5R KO (D1R/D5R flx DM/DL/V striatum, n = 3 and F-D1R/D5R KO DM/DL/V striatum, n = 4). * denotes P value < 0.05, ** P value < 0.01, *** P value < 0.001, **** P value < 0.0001.
D1R and D5R deletion in F-D1R/D5R KO animals was quantified (Fig. 1G, H, and O,P). Significant reduction of D1R mRNA in the infralimbic (IL), prelimbic (PC), and anterior cingulate cortices (ACC) (IL/PL - unpaired t test, P < 0.05; ACC - unpaired t test, P < 0.01) was observed in F-D1R/D5R KO mice (Fig. D–F and G). Significant deletion was also observed in the dorsal medial (DM), dorsal lateral (DL), and ventral striatum (VS) (DM - unpaired t test, P < 0.001; DL- unpaired t test, P < 0.05; VS - unpaired t test, P < 0.0001) (Fig. 1D–F and H). Consistent with our previous findings, D1R deletion also occurred in the DG of the hippocampus (Sariñana et al., 2014) in KO animals (Supp. Fig. 2B). The D5R mRNA signal was significantly reduced in the IL/PL subregions of the mPFC (unpaired t test, P < 0.0001) (Fig. 1L and O). Although some reduction of the D5R mRNA signal in the DM striatum seems to occur, there is no significant difference in the D5R mRNA signal between KO and control animals (P = 0.2216). There was not significant differences between KO and control animals when comparing DL or V striatum D5R mRNA levels (Fig. 1M,N, and P). Consistent with our previous findings, D5R deletion also occurred in the DG and CA1 hippocampal subregions (Supp. Fig. 2D) (Sariñana et al., 2014).
Forebrain D1R and D5R Deletion Impairs Spatial Learning and Memory
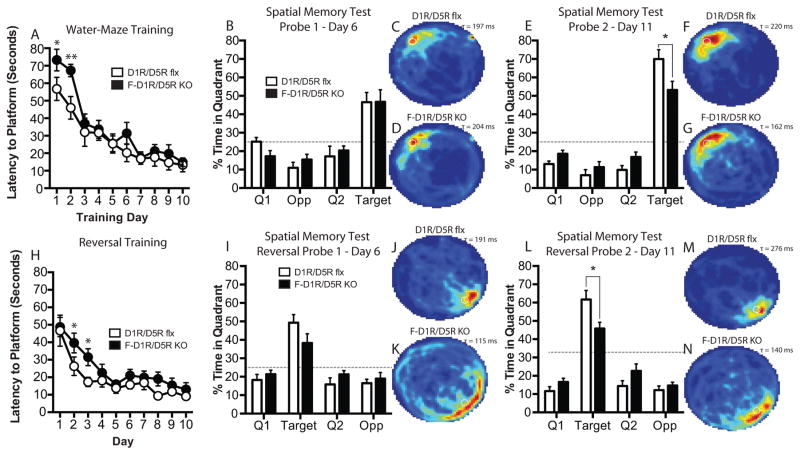
D1R/D5R flx control and forebrain D1R/D5R KO (F-D1R/D5R KO) animals received water-maze training of four trials per day for 10 consecutive days. During each training trial, mice were given up to 90 seconds to find a submerged and hidden escape platform. F-D1R/D5R KO animals did not show a significant difference between genotypes in their latency to platform across regular training [two-way ANOVA, F(1, 14)genotype = 3.846, P = 0.0701] (Fig. 2A). However, Bonferroni post-test reveals a significant difference between genotypes on day-2 (P < 0.05) of water-maze training. When multiple comparisons are not corrected for, using Fishers least significant differences (LSD) post-test, a significant difference in latency to platform between genotypes on day 1 (P = 0.0235) and 2 (P = 0.0036) (Fig. 2A) is revealed. Memory consolidation during regular water-maze training did not significantly differ between genotypes (unpaired t test, P > 0.05) (Supp. Fig. 1K). On days 6 and 11, the escape platform was removed and animals were given a 60 second spatial memory probe test. During the probe-1 trial, F-D1R/D5R KO animals did not exhibit a significant difference in time spent in the target quadrant as compared to control mice (unpaired t test, P > 0.05) (Fig. 2B–D). In contrast, F-D1R/D5R KO mice exhibited a significant difference in time spent in the target quadrant during the probe-2 trial (unpaired t test, P < 0.05) (Fig. 2E–G). To test the animal’s ability to extinguish the original platform location and update new spatial information into a pre-existing spatial schema, mice were given reversal training (Fig. 2H). In reversal training, the escape platform was moved to the opposite quadrant and mice were given the same training schedule as during regular training. The latency curves during reversal training did not significantly differ between F-D1R/D5R KO animals and control mice [two-way ANOVA, F(9, 126)Interaction = 0.6857, P = 0.7208] (Fig. 2H). Bonferroni post-test did not reveal a significant difference between genotypes on any single training day. Using Fishers LSD post-test, we found that F-D1R/D5R KO animals were significantly slower to reach the platform on days 2 (P < 0.0279) and 3 (P < 0.0195) in comparison to control mice during reversal training (Fig. 2H). Memory consolidation during reversal training was not significantly different between F-D1R/D5R KO and control animals (unpaired t test, P > 0.05) (Supp. Fig. 4K). KO and control animals did not significantly differ in time spent in the target quadrant (unpaired t test, P > 0.05) during reversal probe-1 (Fig. 2I–K). On the second reversal probe trial, F-D1R/D5R KO mice spent significantly less time in the target quadrant compared to control mice (unpaired t test, P < 0.05) (Fig. 2L–N).
Figure 2. Performance on watermaze training and spatial memory in F-D1R/D5R KO animals.
(A) Escape latency (D1R/D5R flx, n = 8; F-D1R/D5R KO, n = 8). (B) Spatial memory probe-1 (D1R/D5R flx, n = 7; F-D1R/D5R KO, n = 8). (C) D1R/D5R flx heat map of average search time during probe-1 (τ = 197 ms). (D) F-D1R/D5R KO heat map of average search time during probe-1 (τ = 204 ms). (E) Spatial memory probe-2 (D1R/D5R flx, n = 8; F-D1R/D5R KO, n = 8). (F) D1R/D5R flx heat map of average search time during probe-2 (τ = 220 ms). (G) F-D1R/D5R KO heat map of average search time during probe-2 (τ = 162 ms). (H) Reversal escape latency (D1R/D5R flx, n = 8; F-D1R/D5R KO, n = 8). (I) Reversal spatial memory probe-1 (D1R/D5R flx, n = 7; F-D1R/D5R KO, n = 7). (J) D1R/D5R flx heat map of average search time during probe-1 (τ = 191 ms). (K) F-D1R/D5R KO heat map of average search time during probe-1 (τ = 115 ms). (L) Spatial memory probe-2 (D1R/D5R flx, n = 7; F-D1R/D5R KO, n = 8. (M) D1R/D5R flx heat map of average search time during probe-2 (τ = 276 ms). (N) F-D1R/D5R KO heat map of average search time during probe-2 (τ = 140 ms). * denotes P value < 0.05. Fisher’s LSD test was used to determine significance between genotypes for daily platform latency. ms – milliseconds.
F-D1R/D5R KO animals displayed similar swim speed during probe-1 (unpaired t test, P > 0.05), but enhanced swim speeds during probe-2 in comparison to flx control animals (unpaired t test, P < 0.05) (Supp. Fig. 1L). Swim speeds did not significantly differ in F-D1R/D5R KO mice for reversal probes-1 and -2 (probe-1 – unpaired t test; P > 0.05; probe-2 – unpaired t test; P > 0.05) (Supp. Fig. 4L). No significant differences were found between F-D1R/D5R KO animals and control mice in thigmotaxis during regular and reversal spatial memory probes (Supp. Fig. 1J and 4J). The rotarod task is an important behavioral protocol to test gross motor ability in rodents (Crawley, 1999). Gross motor impairments could affect swimming ability during water-maze training. We previously reported that F-D1R/D5R KO mice did no show impaired motor behavior as measured on the rotarod task (Sariñana et al., 2014).
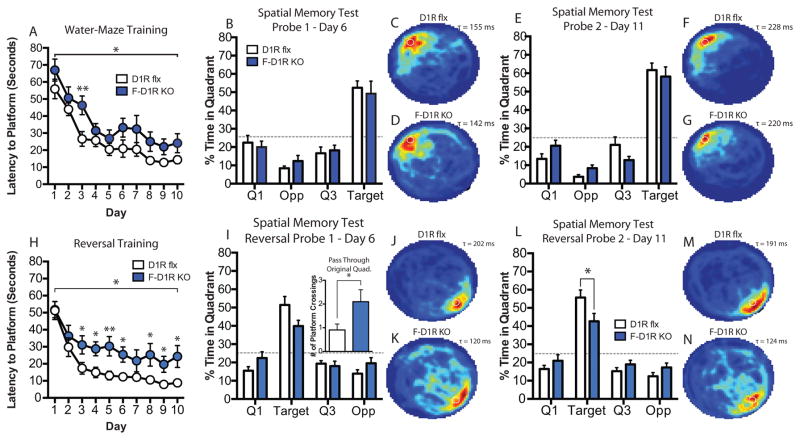
Forebrain D1R Deletion Impairs Spatial Learning, Memory, and Results in Perseverative Behavior
F-D1R KO mice exhibited deficits in spatial learning. F-D1R KOs showed deficits in their latency to reach the platform during training when compared to control animals [two-way ANOVA, F(1, 19)genotype = 8.076, P = 0.0104] (Fig. 3A). Bonferroni post-test revealed a significant difference between genotypes on day 3 of training. Fisher’s LSD post-test also showed a significant difference between F-D1R KO and control animals on Day 3 of training (P = 0.0041) (Fig. 3A). No significant differences in memory consolidation during training was observed between KO animals and control mice (unpaired t test, P > 0.05) (Supp. Fig. 1E). F-D1R KO animals exhibited similar performance during probe-1 (unpaired t test, P > 0.05) (Fig. 3B–D) and probe-2 (unpaired t test, P > 0.05) (Fig. 3E–G) trials as compared to control animals. F-D1R KO mice exhibited significant latency deficits across reversal training [two-way ANOVA, F(1, 19)genotype = 7.763, P = 0.0118]. Although Bonferroni post-test did not reveal any significant differences during any day of training, Fishers LSD post-test showed significant latency differences at several days across training (Fig. 3H). F-D1R KO animals also displayed consolidation deficits during reversal training (unpaired t test, P < 0.05) (Supp. Fig. 4E) as compared to controls. During reversal spatial memory probe-1 (Fig. 3I–K), F-D1R KO mice displayed perseverative behaviors as shown by their increased return to the original platform location during regular water-maze training (unpaired t test, P < 0.05) (Fig. 3I inset). On the reversal probe-2 trial, D1R-KO mice showed significant deficits in spatial memory performance as compared to control animals (unpaired t test, P < 0.05) (Fig. 3L–N).
Figure 3. Performance on watermaze spatial learning and memory in F-D1R KO mice.
(A) Escape latency (D1R flx, n = 11; F-D1R KO, n = 10). (B) Spatial memory probe-1 (D1R flx, n = 11; F-D1R KO, n = 10). (C) D1R flx heat map of average search time during probe-1 (τ = 155 ms). (D) F-D1R KO heat map of average search times during probe-1 (τ = 142 ms). (E) Spatial memory probe-2 (D1R flx, n = 11; F-D1R KO, n = 10). (F) D1R flx heat map of average search time during probe-2 (τ = 228 ms). (G) F-D1R KO heat map of average search time during probe-2 (τ = 220 ms). (H) Reversal escape latency (D1R flx, n = 11; F-D1R KO, n = 10). (I) Reversal spatial memory probe-1 (D1R flx, n = 11; F-D1R KO, n = 10); Inset – crossings of original escape platform location. (J) D1R flx heat map of average search time during probe-1 (τ = 202 ms). (K) F-D1R KO heat map of average search time during probe-1 (τ = 120 ms). (L) Spatial memory probe-2 (D1R flx, n = 11; F-D1R KO, n = 10). (M) D1R flx heat map of average search time during probe-2 (τ = 191 ms). (N) F-D1R KO heat map of average search time during probe-2 (τ = 124 ms). * denotes P value < 0.05. Fisher’s LSD test was used to determine significance between genotypes for daily platform latency. ms – milliseconds.
The striatum is essential for the development of motor skills (Jueptner et al., 1997). F-D1R KO mice showed no difference in swim speed during probe-1 (unpaired t test; P > 0.05) and probe-2 trials (unpaired t test; P > 0.05) (Supp. Fig. 1F). Similarly, F-D1R KO animals did not show a significant difference in swim speed during reversal probes (probe-1 and -2, P > 0.05) (Supp. Fig. 4F). F-D1R KO animals mice did not significantly differ in thigmotaxis behavior during probe-1 (unpaired t test, P > 0.05), but significantly reduced levels of thigmotaxis during probe-2 (unpaired t test, P < 0.05) (Supp. Fig. 1D). During reversal probes, F-D1R KO animals did not show significant differences in thigmotaxis during probe-1 (unpaired t test, P > 0.05) but significant reduction during probe-2 (unpaired t test, < 0.01) (Supp. Fig. 4D). F-D1R KO animals did not significantly differ in total distance traveled in the open field task when compared to control mice [two-way ANOVA, F(1, 14)genotype = 2.836, P = 0.1143] (Supp. Fig. 3A). Moreover, F-D1R KO present similar motor activity on the rotarod task in comparison to flx control animals [two-way ANOVA, F(1, 14)genotype = 0.2266, P = 0.6414] (Supp. Fig. 3C)..
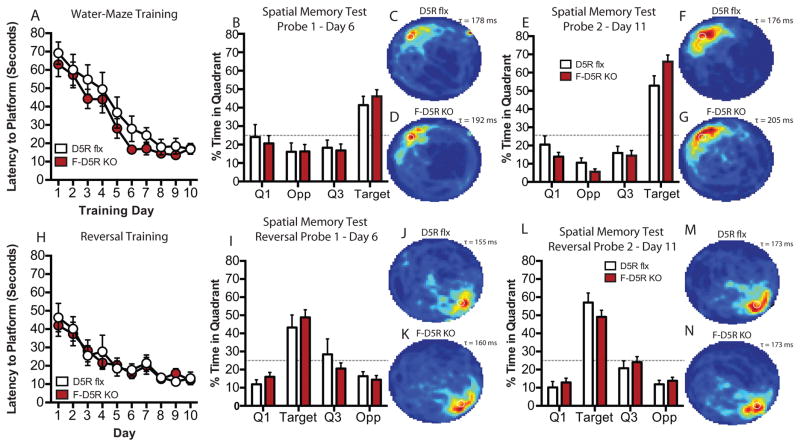
Forebrain D5R Deletion Does not Result in Observable Spatial Learning and Memory Deficits
F-D5R KO mice did not significantly differ in latency during training [two-way ANOVA, F(1, 17)genotype = 2.495, P = 0.1326] (Fig. 4A), memory consolidation (unpaired t test, P > 0.05) (Supp. Fig. 1H), and time spent in the target quadrant during the spatial memory probe trials as compared to flx controls (probe -1 and -2, unpaired t test, P > 0.05) (Fig. 4B–D and E–G). F-D5R KO mice did not exhibit observable deficits in reversal escape latency [two-way ANOVA F(1,16)genotype = 0.05639, P = 0.8153] (Fig. 4H) or memory consolidation (unpaired t test, P > 0.05) (Supp. Fig. 4H) during reversal training. F-D5R KO mice did not exhibit observable reversal spatial learning and memory deficits on reversal probe-1 or reversal probe 2 (Probe-1 and -2, unpaired t test, P > 0.05) as compared to control mice (Fig. 4I–K and L–N).
Figure 4. Performance on watermaze spatial learning and memory in F-D5R KO mice.
(A) Escape latency (D5R flx, n = 8; F-D5R KO, n = 10). (B) Spatial memory probe-1 (D5R flx, n = 8; F-D5R KO, n = 10). (C) D5R flx heat map of average search time during probe-1 (τ = 178 ms). (D) F-D5R KO heat map of average search time during probe-1 (τ = 192 ms). (E) Spatial memory probe-2 (D5R flx, n = 8; F-D5R KO, n = 10). (F) D5R flx heat map of average search time during probe-2 (τ = 176 ms). (G) F-D5R KO heat map of average search time during probe-2 (τ = 205 ms). (H) Reversal escape latency (D5R flx, n = 8; F-D5R KO, n = 10). (I) Reversal spatial memory probe-1 (D5R flx, n = 8; F-D5R KO, n = 10. (J) D5R flx heat map of average search time during probe-1 (τ = 155 ms). (K) F-D5R KO heat map of average search time during probe-1 (τ = 160 ms). (L) Reversal spatial memory probe-2 (D5R flx, n = 8; F-D5R KO, n = 10). (M) D5R flx heat map of average search time during probe-2 (τ = 173 ms). (N) F-D5R KO heat map of average search time during probe-2 (τ = 173 ms). ms – milliseconds.
F-D5R KO mice showed no significant difference in swim speed during probe-1 (unpaired t test, P > 0.05) and probe-2 trials (unpaired t test, P > 0.05) (Supp. Fig. 1I). During reversal probes-1 and -2 F-D5R KO animals did not show significant differences in swim speed (reversal probe-1 and -2, unpaired t test, P > 0.05) (Supp. Fig 4I). F-D5R KO animals displayed similar thigmotaxis behavior during probe-1 (unpaired t test, P > 0.05), but significantly reduced levels of thigmotaxis during probe-2 (unpaired t test, P < 0.05) (Supp. Fig. 1G). During reversal probes -1 and -2, F-D5R KOs did not significantly differ in thigmotaxic behavior (probe -1 and -2, unpaired t test, P > 0.05) (Supp. Fig. 4G). F-D5R KO animals did not significantly differ in total distance traveled in the open field task when compared to control mice [two-way ANOVA, F(1, 30)genotype = 0.3935, P = 0.5352] (Supp. Fig. 3B). F-D5R KO animals did not present significant differences in motor activity on the rotarod task [two-way ANOVA, F(1, 14)genotype = 1.013, P = 0.3313] (Supp. Fig. 3D).
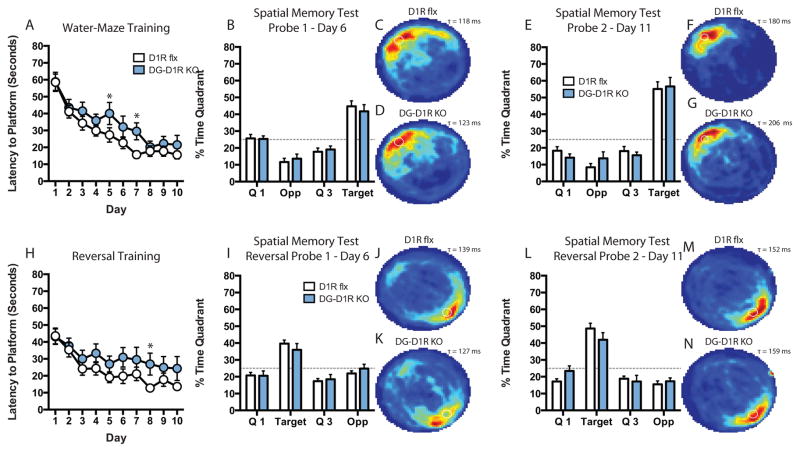
DG D1R Deletion Does not Impair Spatial Memory
We recently reported on the generation and characterization of a DG-restricted D1R KO mouse line (DG-D1R KO). We showed that D1Rs are primarily expressed in the DG of the hippocampus. In order to spatially restrict and isolate D1R function in hippocampal-dependent spatial learning and memory, we trained DG-D1R KO mice on the water-maze task. Control D1R flx and DG-D1R KO mice Control and DG-D1R KOs exhibited similar escape latencies during training [two-way ANOVA, F(1, 29)genotype = 2.896, P = 0.0995] (Fig 5A). Bonferroni post-test did not reveal a difference between genotypes on any single day during training. However, a Fisher’s LSD post-test revealed significant differences on days 5 (unpaired t test, P < 0.05) and 7 (unpaired t test, P < 0.05) (Fig 5A). Memory consolidation during regular training was similar between DG-D1R KO animals and control mice (Supp. Fig. 1B). There was no difference between control and DG-D1R KO mice on the time spent in the target quadrant on probe-1 (unpaired t test, P >0.05) and on probe-2 (unpaired t test, P >0.05) (Fig. 5B–D and E–G). We observed no significant difference between control D1R flx and DG-D1R KO mice on reversal training latency [two-way ANOVA, F(1, 29)genotype = 2.103, P = 0.1578] (Fig. 5H). Bonferroni post-test did not reveal significant differences between genotypes on any single day of training. However, Fisher’s LSD post-test revealed significant differences between DG-D1R KO and flx controls on day 8 (unpaired t test, P < 0.05) (Fig. 5H). Memory consolidation was similar between DG-D1R KO animals and control mice (Supp. Fig. 4B). Time spent in the target quadrant during reversal probe-1 (unpaired t test, P > 0.05) (Fig. 5I–K) and -2 (unpaired t test, P > 0.05) (Fig. 5L–N) were not significantly different between genotypes. Both flx and DG-D1R KO animals displayed similar time spent using non-spatial strategies to search for the escape platform, that is, thigmotaxis (Supp. Fig. 1A)
Figure 5. Performance on water-maze training and spatial memory in DG-D1R KO Mice.
(A) Escape latency (D1R flx, n = 16; DG-D1R KO, n = 15) (B) Spatial memory probe-1 (D1R flx, n = 16; DG-D1R KO, n = 15). (C) D1R flx heat map of average search time during probe-1 (τ = 118 ms). (D) DG-D1R KO heat map of average search time during probe-1 (τ = 123 ms). (E) Spatial memory probe-2 (D1R flx, n = 16; DG-D1R KO, n = 15). (F) D1R flx heat map of average search time during probe-2 (τ = 180 ms). (G) DG-D1R KO heat map of average search time during probe-2 (τ = 206 ms). (H) Reversal escape latency (D1R flx, n = 16; DG-D1R KO, n = 15). (I) Spatial memory probe-1 (D1R flx, n = 16; DG-D1R KO, n = 15). (J) D1R flx heat map of average search time during probe-1 (τ = 139 ms). (K) DG-D1R KO heat map of average search time during probe-1 (τ = 127 ms). (L) Spatial memory probe-2 (D1R flx, n = 16; DG-D1R KO, n = 15). (M) D1R flx heat map of average search time during probe-2 (τ = 152 ms). (N) DG-D1R KO heat map of average search time during probe-2 (τ = 159 ms). * denotes P value < 0.05. Fisher’s LSD test was used to determine significance between genotypes for daily platform latency. ms – milliseconds.
DG-D1R KO mice showed no significant differences in swim speed during probe-1 (unpaired t test, P > 0.05) and probe-2 trials (unpaired t test, P > 0.05) (Supp. Fig. 1C). During reversal probes-1 and -2 DG-D1R KO animals did not show a significant difference in swim speed (probe-1 and -2, unpaired t test, P > 0.05) (Supp. Fig 4C). DG-D1R KO animals did not show a significant difference in thigmotaxis during regular and reversal probes (probe-1 and -2, unpaired t test, P > 0.05, reversal probe-1 and -2, unpaired t test, P > 0.05) (Supp. Fig. 1A and 4A). We previously reported that DG-D1R KO mice did exhibit significant impairment in motor behavior as measured on the rotarod task or significant differences traveled in the open field task (Sariñana et al., 2014).
Discussion
In this study, we found that forebrain D1R, but not D5R, activation is crucial for spatial learning and memory, and DG D1Rs are not necessary for spatial memory. Our in situ data shows that forebrain D1R mRNA signal is reduced in the IL/PL, ACC, and MD, ML, and V striatum in F-D1R/D5R KO mice (Fig. 1D–H). In contrast, the D5R mRNA signal was significantly reduced only in the IL/PL region (Fig. 1L and O). During water-maze training, F-D1R/D5R KO animals showed slight deficits in latency to platform during regular and reversal learning (Fig. 2A and H). Still, F-D1R/D5R KO mice reach the same criterion as their flx control counterparts. F-D1R/D5R KO animals showed spatial memory deficits during both regular and reversal training (Fig. 2E and L). To differentiate the contribution of forebrain D1R from D5R activation in spatial learning and memory processing, we utilized forebrain restricted D1R and D5R KO animals. F-D1R KO mice displayed deficits in spatial learning and memory. Specifically, F-D1R KO animals displayed deficits in escape latency during regular water-maze training (Fig. 3A). However, F-D1R KO animals learned the spatial location of the escape platform well enough to show spatial memory that’s similar to that of flx control animals (Fig. 3B and E). In contrast, F-D1R KO mice presented greater latency deficits during reversal training (Fig. 3H), as well as deficits in memory consolidation (Supp. Fig. 4E) and spatial memory (Fig. 3L). Moreover, during reversal probe-1, F-D1R KOs swam back to the original water-maze platform location significantly more than the control animals, suggesting deficits in extinguishing the prior platform location (Fig. 3I inset). Although F-D5R KO animals did not exhibit any observable deficits in spatial learning and memory (Fig. 4 and Table 1), double F-D1R/D5R KO mice presented deficits that differed from F-D1R KOs. Moreover, unlike F-D1R KO mice, F-D1R/D5R KO animals were able to extinguish the prior escape platform location to update the former spatial schema learned during regular water-maze training. Unexpectedly, DG-D1R KO mice only showed a slight impairment during water-maze regular and reversal training, but no significant difference in spatial memory performance in comparison to control mice (Fig. 5 and Table 1). Taken together, our data shows for the first time that forebrain D5R activation must contribute to spatial information processing. We also demonstrate that mPFC and striatal, but not DG, D1Rs are essential for spatial learning and memory.
Table 1.
| Regular Training | Reversal Training | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Area of Gene Deletion | Latency to Platform | Probe 1 | Probe 2 | Latency to Platform | Probe 1 | Probe 2 | |
| F-D1R/D5R KO | See single gene deletion below | *Deficit | No Deficit | *Deficit | *Deficit | No Deficit | *Deficit |
| F-D1R KO | DG IL/PL ACC Striatum |
*Deficit | No Deficit | No Deficit | *Deficit | No Deficit | *Deficit |
| F-D5R KO | DG/CA3 IL/PL |
No Deficit | No Deficit | No Deficit | No Deficit | No Deficit | No Deficit |
| DG-D1R KO | DG | *Deficit | No Deficit | No Deficit | *Deficit | No Deficit | No Deficit |
Dentate Gyrus D1Rs Are Not Required for the Morris-Water Maze Task
It is well established that the hippocampus is necessary for spatial learning and memory (Eichenbaum et al., 1990; Morris et al., 1982; Tsien et al., 1996b). Although, constitutive D1R deletion results in spatial learning and memory deficits (Granado et al., 2008), constitutive D5R deletion does not impair spatial learning and memory (Holmes et al., 2001). Our data shows that F-D5R KO animals do not exhibit significant changes in spatial learning and memory (Fig. 4). It is therefore possible that D1Rs, but not D5Rs, are necessary for spatial learning and memory. Additionally D1R/D5R antagonists infusion into the hippocampus results in water-maze deficits (Silva et al., 2012). When taken together, hippocampal D1Rs may be required for spatial learning and memory. However, D1R/D5R antagonists block norepinephrine and serotonergic receptors (5HT-2) as well as the serotonin transporter, which expresses throughout the hippocampus (Hicks et al., 1984; Ohlstein and Berkowitz, 1985; Zarrindast et al., 2011), obscuring a strong conclusion as to whether hippocampal D1R activation is truly required for spatial learning and memory. Still, hippocampal D1R activation could underlie spatial learning and memory. Our data, shows that the D1R primarily expresses in the DG of the hippocampus, but not CA3 or CA1 (Supp. Fig. 2), which is in agreement with previous findings (Fremeau et al., 1991; Mansour et al., 1992; Mu et al., 2011; Sariñana et al., 2014) However, a recent publication suggest that there is sparse D1R expression in the CA3 and CA1 hippocampal subfields, particularly in interneurons (Gangarossa et al., 2012). Based on the evidence above, we conclude that DG D1Rs are not required for spatial memory as assayed by our water-maze task. Further research studying sparse D1R expression throughout the hippocampus is required to further understand the role of hippocampal D1R activation on spatial learning and memory.
PFC D1R and D5R Activation Work in Conjunction for Spatial Memory Updating
Although D5R expression occurs throughout all hippocampal subregions, constitutive D5R KOs and forebrain wide D5R deletion does not appear to significantly affect spatial learning and memory (Holmes et al., 2001) (Fig. 4). Although D5Rs may not be required for spatial learning and memory, D5R activation could still be important for spatial memory updating. D1R and D5R mRNA co-express in the IL/PL cortex (Fig. 1A and I) and both F-D1R and F-D5R animals display significant receptor deletion of D1Rs and D5Rs, respectively, in the IL/PL cortex. F-D1R KO animals differ in phenotype in comparison to F-D1R/D5RKO animals, which suggests that D5R activation affects spatial learning and memory processing. More specifically, F-D1R KO animals display perseverative behaviors (Fig 3I insert), while F-D1R/D5R KO mice do not. Given the importance of the PFC in reversal learning (Lacroix et al., 2002), it is feasible that D5R deletion in the IL/PL cortex results in the phenotypic difference between F-D1R and F-D1R/D5R KO mice with regard to perseverative behaviors. A possible explanation for the observed differences between F-D1R/D5R KO and F-D1R KO animals is that D1R and D5R activation results in downstream processing that impairs neuronal computational power. For example, the D1R directly couples to the GluN1 and GluN2A subunits of the n-methyl-d-aspartate receptor (NMDAR) and modulates the NMDAR ionic currents (Lee et al., 2002; Pei et al., 2004). Similarly, the D5R directly couples to the γ2 subunit of the γ-aminobutyric acid subtype-A receptor (GABAAR), modulating the inhibitory current (Liu et al., 2000). Given the differences in D1R and D5R protein coupling, D1R would likely express at excitatory synapses, while D5R expression would receive inhibitory inputs along the dendritic shafts (Megias et al., 2001; Wierenga et al., 2008). A difference in D1R and D5R distribution would support enhanced computational power of single neurons to maximize memory processing and storage (Govindarajan et al., 2011; Govindarajan et al., 2006). Thus, deletion of both receptors could significantly impair neuronal computation resulting in spatial processing deficits that underlie perseverative behavior. The difference in spatial learning and memory phenotypes between F-D1R/D5R KO and F-D1R KO mice shows that forebrain D5R activation is certainly contributing to spatial information processing.
Striatal D1R Contribution to Spatial Processing
Lesions to either the medial dorsal or ventral striatum significantly impair early water-maze training and spatial memory (Annett et al., 1989; Devan and White, 1999). The dorsal medial DM striatum has been shown to process spatial information, while the DL striatum is associated with procedural learning (Voorn et al., 2004). We observed a strong deletion in the ventral striatum, which links spatial information to reinforcing events (van der Meer and Redish, 2011). The strong deletion of the D1R in the DM and ventral striatum in the D1R-KO and D1R/D5R KO animals suggests that spatial information would be impaired, while weaker deletion in the DL striatum suggests impairments in procedural learning. Given the spatial memory deficits observed in F-D1R KO and F-D1R/D5R KO, but not DG-D1R KOs, suggests that D1R deletion in the DM and ventral striatum underlies the observed spatial memory deficits (Fig. 2E,L, and 3L). Moreover, the deletion of D1Rs in the DL striatum might underlie the deficits observed during spatial learning in F-D1R KO and F-D1R/D5R KO animals given that procedural nature of water-maze training (Fig. 2A, H and 3A, H). However, neither F-D1R KO nor F-D1R/D5R KO animals exhibit gross motor deficits as measured on the rotarod task, suggesting that general motor activity does not underlie the deficits observed during water-maze training. DG-D1R KO animals also show subtle deficits during water-maze training, which suggests that DG D1R activation could also contribute to the spatial component during water-maze training.
The genetic tools used in our current study have provided a unique advantage over pharmacological and global KO studies by distinguishing the D1R from the D5R with gene-specific and spatially restricted KOs. Our findings that the primary site of D1R expression is the DG, and not CA1, are consistent with some previous findings (Fremeau et al., 1991; Mansour et al., 1992; Mu et al., 2011). However, other previous studies have focused on hippocampal CA1 D1R activation as the driver of synaptic plasticity, learning, and memory (Huang and Kandel, 1995; Lemon and Manahan-Vaughan, 2006; Li et al., 2003; Ortiz et al., 2010; Smith et al., 1998). Some of these previous studies attributed the observed deficits to CA1 D1Rs, which may be due to sparse expression of CA1 D1Rs (Gangarossa et al., 2012). Still, deficits presumed to be due to CA1 D1Rs may instead be due to disruption of D5R function. Therefore, future research on D1R versus D5R function in hippocampal memory processing as well as physiological changes within the hippocampus should be studied utilizing genetic tools given the limitations of current pharmacological reagents.
Supplementary Material
Supplemental Figure 1. Regular training probes 1 and 2 thigmotaxis, memory consolidation, and swim speed for all mouse lines. (A, D, G, J) Thigmotaxis. (B, E, H, K) Memory consolidation. (C, F, I, L) Swim speed. Probe 1 - (D1R flx, n = 16, DG-D1R KO = 15; D1R flx, n = 11; F-D1R KO = 10; D5R flx, n = 8, F-D5R KO = 10; D1R/D5R flx, n = 7; F-D1R/D5R KO = 8). Probe 2 – (D1R flx, n = 16, DG-D1R KO = 15; D1R flx, n = 11; F-D1R KO = 10; D5R flx, n = 8, F-D5R KO = 10; D1R/D5R flx, n = 8; F-D1R/D5R KO = 8).
Supplemental Figure 2. In Situ Hybridization. (A and B) D1R mRNA Probe for D1R/D5R flx and F-D1R/D5R KO mouse, respectively. (C and D) D5R mRNA Probe for D1R/D5R flx and F-D1R/D5R KO mouse, respectively.
Supplemental Figure 3. Gross motor activity in flx (D1R or D5R) controls and forebrain KO (D1R or D5R) animals. (A and B) Open field total distance (D1R flx, n = 7; F-D1R KO = 9; D5R flx, n = 15, F-D5R KO = 17). (C and D) Rotarod motor test (D1R flx, n = 7; F-D1R KO = 9; D5R flx, n = 7, F-D5R KO =9).
Supplemental Figure 4. Reversal probes 1 and 2 thigmotaxis, memory consolidation, and swim speed for all mouse lines. (A, D, G, J) Thigmotaxis. (B, E, H, K) Memory consolidation. (C, F, I, L) Swim speed. Probe 1 - (D1R flx, n = 16, DG-D1R KO = 15; D1R flx, n = 11; F-D1R KO = 10; D5R flx, n = 8, F-D5R KO = 10; D1R/D5R flx, n = 7; F-D1R/D5R KO = 7). Probe 2 – (D1R flx, n = 16, DG-D1R KO = 15; D1R flx, n = 11; F-D1R KO = 10; D5R flx, n = 8, F-D5R KO = 10; D1R/D5R flx, n = 7; F-D1R/D5R KO = 8).
Acknowledgments
The authors would like to thank Derek Buhl for water-maze heat-map scripts for analysis, Julie Moyer for experimental support, and Takashi Kitamura for comments on the manuscript.
This work was supported by the RIKEN Brain Science Institute (Grant #) and the Howard Hughes Medical Institute (Grant #).
References
- Annett LE, McGregor A, Robbins TW. The effects of ibotenic acid lesions of the nucleus accumbens on spatial learning and extinction in the rat. Behav Brain Res. 1989;31(3):231–42. doi: 10.1016/0166-4328(89)90005-3. [DOI] [PubMed] [Google Scholar]
- Bartlett FC. Remembering. Cambridge, UK: Cambridge UP; 1932. [Google Scholar]
- Bethus I, Tse D, Morris RG. Dopamine and memory: modulation of the persistence of memory for novel hippocampal NMDA receptor-dependent paired associates. J Neurosci. 2010;30(5):1610–8. doi: 10.1523/JNEUROSCI.2721-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests1. Brain Research. 1999;835(1):18–26. doi: 10.1016/s0006-8993(98)01258-x. [DOI] [PubMed] [Google Scholar]
- Devan BD, White NM. Parallel information processing in the dorsal striatum: relation to hippocampal function. J Neurosci. 1999;19(7):2789–98. doi: 10.1523/JNEUROSCI.19-07-02789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragoi G, Tonegawa S. In: Development of schemas revealed by prior experience and NMDA receptor knock-out. Eichenbaum H, editor. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H, Stewart C, Morris RG. Hippocampal representation in place learning. J Neurosci. 1990;10(11):3531–42. doi: 10.1523/JNEUROSCI.10-11-03531.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ghundi M, Fletcher PJ, Drago J, Sibley DR, O’Dowd BF, George SR. Spatial learning deficit in dopamine D(1) receptor knockout mice. Eur J Pharmacol. 1999;383(2):95–106. doi: 10.1016/s0014-2999(99)00573-7. [DOI] [PubMed] [Google Scholar]
- Fremeau RT, Duncan GE, Fornaretto M-G, Dearry A, Gingrich JA, Breese GR, Caron MG. Localization of D1 dopamine receptor mrRNA in brain supports a role in cognitive, affective, and neuroendocrine aspects of dopaminergic neurotransmission. PNAS. 1991;88:3772–3776. doi: 10.1073/pnas.88.9.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangarossa G, Longueville S, De Bundel D, Perroy J, Herve D, Girault JA, Valjent E. Characterization of dopamine D1 and D2 receptor-expressing neurons in the mouse hippocampus. Hippocampus. 2012;22(12):2199–207. doi: 10.1002/hipo.22044. [DOI] [PubMed] [Google Scholar]
- Govindarajan A, Israely I, Huang S-Y, Tonegawa S. The Dendritic Branch Is the Preferred Integrative Unit for Protein Synthesis-Dependent LTP. Neuron. 2011;69(1):132–146. doi: 10.1016/j.neuron.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan A, Kelleher RJ, Tonegawa S. A clustered plasticity model of long-term memory engrams. Nature Reviews: Neuroscience. 2006;7:575–583. doi: 10.1038/nrn1937. [DOI] [PubMed] [Google Scholar]
- Granado N, Ortiz O, Suarez LM, Martin ED, Cena V, Solis JM, Moratalla R. D1 but not D5 dopamine receptors are critical for LTP, spatial learning, and LTP-Induced arc and zif268 expression in the hippocampus. Cereb Cortex. 2008;18(1):1–12. doi: 10.1093/cercor/bhm026. [DOI] [PubMed] [Google Scholar]
- Groenewegen HJ, Vermeulen-Van der Zee E, te Kortschot A, Witter MP. Organization of the projections from the subiculum to the ventral striatum in the rat. A study using anterograde transport of Phaseolus vulgaris leucoagglutinin. Neuroscience. 1987;23(1):103–20. doi: 10.1016/0306-4522(87)90275-2. [DOI] [PubMed] [Google Scholar]
- Hicks PE, Schoemaker H, Langer SZ. 5HT-receptor antagonist properties of SCH 23390 in vascular smooth muscle and brain. European Journal of Pharmacology. 1984;105(3–4):339–342. doi: 10.1016/0014-2999(84)90628-9. [DOI] [PubMed] [Google Scholar]
- Holmes A, Hollon TR, Gleason TC, Liu Z, Dreiling J, Sibley DR, Crawley JN. Behavioral Characterization of Dopamine D5 Receptor Null Mutant Mice. Behavioral Neuroscience. 2001;115(5):1129–1144. [PubMed] [Google Scholar]
- Huang Y-Y, Kandel ER. D1/D5 receptor agonists induce a protein synthesis-dependent late potentiation in the CAl region of the hippocampus. PNAS. 1995;92:2446–2450. doi: 10.1073/pnas.92.7.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jueptner M, Frith CD, Brooks DJ, Frackowiak RS, Passingham RE. Anatomy of motor learning. II. Subcortical structures and learning by trial and error. J Neurophysiol. 1997;77(3):1325–37. doi: 10.1152/jn.1997.77.3.1325. [DOI] [PubMed] [Google Scholar]
- Karasinska JM, George SR, El-Ghundi M, Fletcher PJ, O’Dowd BF. Modification of dopamine D(1) receptor knockout phenotype in mice lacking both dopamine D(1) and D(3) receptors. Eur J Pharmacol. 2000;399(2–3):171–81. doi: 10.1016/s0014-2999(00)00347-2. [DOI] [PubMed] [Google Scholar]
- Lacroix L, White I, Feldon J. Effect of excitotoxic lesions of rat medial prefrontal cortex on spatial memory. Behavioural Brain Research. 2002;133(1):69–81. doi: 10.1016/s0166-4328(01)00442-9. [DOI] [PubMed] [Google Scholar]
- Lazic SE. Statistical evaluation of methods for quantifying gene expression by autoradiography in histological sections. BMC Neurosci. 2009;10:5. doi: 10.1186/1471-2202-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Andre JM, Pittenger C. Lesions of the dorsomedial striatum delay spatial learning and render cue-based navigation inflexible in a water maze task in mice. Front Behav Neurosci. 2014;8:42. doi: 10.3389/fnbeh.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FJ, Xue S, Pei L, Vukusic B, Chery N, Wang Y, Wang YT, Niznik HB, Yu XM, Liu F. Dual regulation of NMDA receptor functions by direct protein-protein interactions with the dopamine D1 receptor. Cell. 2002;111(2):219–30. doi: 10.1016/s0092-8674(02)00962-5. [DOI] [PubMed] [Google Scholar]
- Lemon N, Manahan-Vaughan D. Dopamine D1/D5 Receptors Gate the Acquisition of Novel Information through Hippocampal Long-Term Potentiation and Long-Term Depression. The Journal of Neuroscience. 2006;26(29):7723–7729. doi: 10.1523/JNEUROSCI.1454-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Cullen WK, Anwyl R, Rowan MJ. Dopamine-dependent facilitation of LTP induction in hippocampal CA1 by exposure to spatial novelty. Nature Neuroscience. 2003;6(5):526–531. doi: 10.1038/nn1049. [DOI] [PubMed] [Google Scholar]
- Liu F, Wan Q, Pristupa ZB, Yu X-M, Wang YT, Niznik HB. Direct protein±protein coupling enables cross-talk between dopamine D5 and g-aminobutyric acid A receptors. Nature. 2000;403:274–280. doi: 10.1038/35002014. [DOI] [PubMed] [Google Scholar]
- Mansour A, Meador-Woodruff JH, Zhou Q, Civelli O, Akil H, Watson SJ. A comparison of D1 receptor binding and mRNA in rat brain using receptor autoradiographic and in situ hybridization techniques. Neuroscience. 1992;46(4):959–971. doi: 10.1016/0306-4522(92)90197-a. [DOI] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, Tonegawa S. Dentate Gyrus NMDA Receptors Mediate Rapid Pattern Separation in the Hippocampal Network. Science. 2007;317:94–99. doi: 10.1126/science.1140263. [DOI] [PubMed] [Google Scholar]
- McKenzie S, Robinson NT, Herrera L, Churchill JC, Eichenbaum H. Learning causes reorganization of neuronal firing patterns to represent related experiences within a hippocampal schema. J Neurosci. 2013;33(25):10243–56. doi: 10.1523/JNEUROSCI.0879-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megias M, Emri Z, Freund TF, Gulyas AI. Total number and distribution of inhibitory and excitatory synapses on hippocampal CA1 pyramidal cells. Neuroscience. 2001;102(3):527–40. doi: 10.1016/s0306-4522(00)00496-6. [DOI] [PubMed] [Google Scholar]
- Mele A, Avena M, Roullet P, De Leonibus E, Mandillo S, Sargolini F, Coccurello R, Oliverio A. Nucleus accumbens dopamine receptors in the consolidation of spatial memory. Behav Pharmacol. 2004;15(5–6):423–31. doi: 10.1097/00008877-200409000-00017. [DOI] [PubMed] [Google Scholar]
- Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine Receptors: From Structure to Function. Physiological Reviews. 1998;78(1):189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- Morris RGM, Garrud P, Rawlins JNP, O’Keefe J. Place Navigation Impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Mu Y, Zhao C, Gage FH. Dopaminergic Modulation of Cortical Inputs during Maturation of Adult-Born Dentate Granule Cells. The Journal of Neuroscience. 2011;31(11):4113–4123. doi: 10.1523/JNEUROSCI.4913-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa K, Quirk MC, Chitwood RA, Watanabe M, Yeckel MF, Sun LD, Kato A, Carr CA, Johnston D, Wilson MA, et al. Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science. 2002;297(5579):211–8. doi: 10.1126/science.1071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll CM, Martin SJ, Sandin J, Frenguelli B, Morris RGM. Dopaminergic modulation of the persistence of one-trial hippocampus-dependent memory. Learning and Memory. 2006;13:760–769. doi: 10.1101/lm.321006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein EH, Berkowitz BA. SCH 23390 and SK&F 83566 are antagonists at vascular dopamine and serotonin receptors. Eur J Pharmacol. 1985;108(2):205–8. doi: 10.1016/0014-2999(85)90728-9. [DOI] [PubMed] [Google Scholar]
- Ortiz O, Delgado-Garcia JM, Espadas I, Bahi A, Trullas R, Dreyer J-L, Gruart A, Moratalla R. Associative Learning and CA3-CA1 Synaptic Plasticity Are Impaired in D1R Null, Drd1a-/- Mice and in Hippocampal siRNA Silenced Drd1a Mice. J Neurosci. 2010;30(37):12288–12300. doi: 10.1523/JNEUROSCI.2655-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Lee FJ, Moszczynska A, Vukusic B, Liu F. Regulation of dopamine D1 receptor function by physical interaction with the NMDA receptors. J Neurosci. 2004;24(5):1149–58. doi: 10.1523/JNEUROSCI.3922-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaget J. In: The child’s conception of the world. Tomlinson J, Tomlinson A, translators. New York: Harcourt, Brace; 1926. [Google Scholar]
- Sariñana J, Kitamura T, Künzler P, Sultzman L, Tonegawa S. Differential roles of the dopamine 1-class receptors, D1R and D5R, in hippocampal dependent memory. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1407395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaguchi T, Goldman-Rakic PS. D1 dopamine receptors in prefrontal cortex: involvement in working memory. Science. 1991;251(4996):947–50. doi: 10.1126/science.1825731. [DOI] [PubMed] [Google Scholar]
- Silva WC, Kohler CA, Radiske A, Cammarota M. D(1)/D(5) dopamine receptors modulate spatial memory formation. Neurobiol Learn Mem. 2012;14:14. doi: 10.1016/j.nlm.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Smith DR, Striplin CD, Geller AM, Mailman RB, Drago J, Lawler CP, Gallagher M. Behavioural Assessment of Mice Lacking D1A Dopamine Receptors. Neuroscience. 1998;86(1):135–146. doi: 10.1016/s0306-4522(97)00608-8. [DOI] [PubMed] [Google Scholar]
- Tse D, Langston RF, Kakeyama M, Bethus I, Spooner PA, Wood ER, Witter MP, Morris RG. Schemas and memory consolidation. Science. 2007;316(5821):76–82. doi: 10.1126/science.1135935. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996a;87(7):1317–26. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Huerta PT, Tonegawa S. The Essential Role of Hippocampal CA1 NMDA Receptor-Dependent Synaptic Plasticity in Spatial Memory. Cell. 1996b;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- van der Meer MAA, Redish AD. Theta Phase Precession in Rat Ventral Striatum Links Place and Reward Information. The Journal of Neuroscience. 2011;31(8):2843–2854. doi: 10.1523/JNEUROSCI.4869-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorn P, Vanderschuren LJ, Groenewegen HJ, Robbins TW, Pennartz CM. Putting a spin on the dorsal-ventral divide of the striatum. Trends Neurosci. 2004;27(8):468–74. doi: 10.1016/j.tins.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Wierenga CJ, Becker N, Bonhoeffer T. GABAergic synapses are formed without the involvement of dendritic protrusions. Nat Neurosci. 2008;11(9):1044–52. doi: 10.1038/nn.2180. [DOI] [PubMed] [Google Scholar]
- Woolley DG, Laeremans A, Gantois I, Mantini D, Vermaercke B, Op de Beeck HP, Swinnen SP, Wenderoth N, Arckens L, D’Hooge R. Homologous involvement of striatum and prefrontal cortex in rodent and human water maze learning. Proceedings of the National Academy of Sciences. 2013;110(8):3131–3136. doi: 10.1073/pnas.1217832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrindast MR, Honardar Z, Sanea F, Owji AA. SKF 38393 and SCH 23390 Inhibit Reuptake of Serotonin by Rat Hypothalamic Synaptosomes. Pharmacology. 2011;87(1–2):1–2. doi: 10.1159/000323232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Regular training probes 1 and 2 thigmotaxis, memory consolidation, and swim speed for all mouse lines. (A, D, G, J) Thigmotaxis. (B, E, H, K) Memory consolidation. (C, F, I, L) Swim speed. Probe 1 - (D1R flx, n = 16, DG-D1R KO = 15; D1R flx, n = 11; F-D1R KO = 10; D5R flx, n = 8, F-D5R KO = 10; D1R/D5R flx, n = 7; F-D1R/D5R KO = 8). Probe 2 – (D1R flx, n = 16, DG-D1R KO = 15; D1R flx, n = 11; F-D1R KO = 10; D5R flx, n = 8, F-D5R KO = 10; D1R/D5R flx, n = 8; F-D1R/D5R KO = 8).
Supplemental Figure 2. In Situ Hybridization. (A and B) D1R mRNA Probe for D1R/D5R flx and F-D1R/D5R KO mouse, respectively. (C and D) D5R mRNA Probe for D1R/D5R flx and F-D1R/D5R KO mouse, respectively.
Supplemental Figure 3. Gross motor activity in flx (D1R or D5R) controls and forebrain KO (D1R or D5R) animals. (A and B) Open field total distance (D1R flx, n = 7; F-D1R KO = 9; D5R flx, n = 15, F-D5R KO = 17). (C and D) Rotarod motor test (D1R flx, n = 7; F-D1R KO = 9; D5R flx, n = 7, F-D5R KO =9).
Supplemental Figure 4. Reversal probes 1 and 2 thigmotaxis, memory consolidation, and swim speed for all mouse lines. (A, D, G, J) Thigmotaxis. (B, E, H, K) Memory consolidation. (C, F, I, L) Swim speed. Probe 1 - (D1R flx, n = 16, DG-D1R KO = 15; D1R flx, n = 11; F-D1R KO = 10; D5R flx, n = 8, F-D5R KO = 10; D1R/D5R flx, n = 7; F-D1R/D5R KO = 7). Probe 2 – (D1R flx, n = 16, DG-D1R KO = 15; D1R flx, n = 11; F-D1R KO = 10; D5R flx, n = 8, F-D5R KO = 10; D1R/D5R flx, n = 7; F-D1R/D5R KO = 8).