Abstract
Studies of single cells have previously shown intracellular clonal expansion of mitochondrial DNA (mtDNA) mutations to levels that can cause a focal cytochrome c oxidase (COX) defect. Whilst techniques are available to study mtDNA rearrangements at the level of the single cell, recent interest has focused on the possible role of somatic mtDNA point mutations in ageing, neurodegenerative disease and cancer. We have therefore developed a method that permits the reliable determination of the entire mtDNA sequence from single cells without amplifying contaminating, nuclear-embedded pseudogenes. Sequencing and PCR–RFLP analyses of individual COX-negative muscle fibres from a patient with a previously described heteroplasmic COX II (T7587C) mutation indicate that mutant loads as low as 30% can be reliably detected by sequencing. This technique will be particularly useful in identifying the mtDNA mutational spectra in age-related COX-negative cells and will increase our understanding of the pathogenetic mechanisms by which they occur.
INTRODUCTION
Human cells contain multiple copies of mitochondrial DNA (mtDNA) located in the mitochondrial matrix. The mitochondrial genome is highly compact and exhibits little redundancy throughout its sequence of 16 569 bp, which encodes 37 genes: two genes encode ribosomal RNAs, 22 encode transfer RNAs (tRNA) and 13 encode polypeptides, all of which are essential proteins of the mitochondrial respiratory chain (1). The only non-coding region is the 1.1 kb mtDNA control region, which contains elements crucial for mtDNA replication and transcription (2).
Mutations of the mitochondrial genome are widely recognised as important causes of disease (3). These cover a broad spectrum of clinical manifestations affecting many different tissues, although many patients typically present with a progressive neurological syndrome (4). The genetic lesion involves either single, large-scale rearrangements which tend to be sporadic or maternally inherited point mutations in protein encoding or RNA genes. Pathogenic mtDNA mutations frequently coexist with wild-type mtDNA, a phenomenon termed heteroplasmy, with higher levels of mutation accumulating in post-mitotic tissues such as skeletal muscle, heart and the central nervous system (5). Single cell and tissue culture studies of pathogenic tRNA mutations have shown that this ratio of mutant to wild-type mtDNA critically determines expression of the genetic defect, and hence the clinical phenotype, with mutant loads in excess of 90% required to cause respiratory chain dysfunction (6–8). Since this proportion varies between different tissues, the identification of causative mtDNA mutations may prove difficult.
Less clear is the role of somatic mtDNA mutations in human pathology. Lacking protective histones and many of the repair mechanisms associated with nuclear DNA, the mitochondrial genome is highly susceptible to oxidative damage (9,10), and hence mutation. The age-dependent accumulation of somatic mtDNA deletions to levels that affect mitochondrial function is well documented in neurological tissue (11,12), with clonal expansion of individual mutational events restricted to single cells. As the level of deletion exceeds a critical threshold, this causes a biochemical defect, as shown by a selective decrease in the histochemical activity of cytochrome c oxidase (COX), a component of the respiratory chain whose three major catalytic subunits are coded by mtDNA (13,14). Similar patterns of focal COX deficiency in individual neurones have been described in spinal cord from ALS patients (15) and choroid plexus and hippocampus in patients with Alzheimer’s disease (16), although the precise molecular defect in these cells is as yet unknown. Whilst various PCR-based technologies exist for the study of mtDNA deletions in single cells (13,14,17,18), sensitive techniques that can identify mtDNA sequence variants in single cells are nevertheless required to investigate whether acquired mtDNA point mutations might be responsible for the biochemical defect in these COX-negative cells.
Our method for determining the complete mtDNA sequence of a sample uses a series of 28 overlapping PCR-amplified fragments that span the entire length of the mitochondrial genome (19). As each PCR reaction requires ∼100 ng total DNA template to generate sufficient product to sequence, ∼3 µg total DNA template is needed for a complete genome analysis. Absolute amounts of total mtDNA, estimated at ∼0.1% of the total genetic complement of a cell, have been calculated in the femtogram range in single muscle fibres (20). In order to generate sufficient template to sequence, we have therefore devised a two-stage PCR strategy that permits the accurate amplification and sequencing of the entire (16.6 kb) mitochondrial genome from single cells. Here we describe the identification of homoplasmic, neutral polymorphic variants to prove the fidelity of this protocol, and the sequencing of single muscle fibres from a patient with a heteroplasmic T7587C COX II mutation to demonstrate its capability of detecting levels of mtDNA heteroplasmy sufficient to cause a biochemical defect.
MATERIALS AND METHODS
Patients
Control muscle was obtained from a young individual (12 years old) who underwent muscle biopsy for the investigation of exercise-induced muscle pain. The biopsy findings were unremarkable, with no evidence of COX-negative fibres or signs consistent with mitochondrial myopathy. The patient with the heteroplasmic T7587C COX II mutation presented with an extensive history of fatigue and unsteadiness and has been reported previously (21). The mutation changes the initiating methionine to threonine, consequently impairing the initiation of protein translation. The biochemical threshold of the T7587C mutation has been shown to be low, with levels of 55–65% mutation required to cause a defect in COX activity (21).
Cell culture
Human osteosarcoma-derived cell lines either containing mtDNA (143B.TK–, ρ+) or lacking mtDNA (143B206 ρ0) (kindly provided by Dr M. Davidson, Department of Neurology, Columbia University, New York, NY) were grown as previously described (22) and total DNA was extracted using standard protocols. The absence of mtDNA in the 143B206 ρ0 cells had previously been confirmed by Southern blot analysis.
Histochemical analysis of muscle tissue
Transversely orientated blocks were frozen in isopentane and cooled to –190°C in liquid nitrogen. Cryostat sections (20 µm) of skeletal muscle were assayed for COX activities or, in some cases, both COX and succinate dehydrogenase activities (23), permitting the identification of COX-negative fibres.
Isolation of DNA from single cells
Individual muscle fibres were microdissected using fine glass capillaries and placed in a sterile microfuge tube containing 12 µl of lysis buffer (50 mM Tris–HCl, pH 8.5, 1 mM EDTA, 0.5% Tween-20, 200 µg ml–1 proteinase K) (24). Cells were incubated at 55°C for 2 h, followed by heat inactivation of the proteinase K at 95°C for 10 min.
Mitochondrial DNA sequencing
Primary PCR reactions. A series of overlapping primer pairs (SC-A–SC-I; Table 1) were designed to amplify the human mitochondrial genome in nine fragments of ∼2 kb from the single cell lysate. All PCR amplifications were performed in a 50 µl volume containing 1× PCR buffer (10 mM Tris–HCl, pH 8.3, 1.5 mM MgCl2, 50 mM KCl, 0.001% w/v gelatin), 0.2 mM dNTPs, 0.6 µM primers, 1 U AmpliTaq Gold DNA polymerase (Applied Biosystems) and 1 µl single cell lysate. Reaction conditions were 94°C for 12 min and 35 cycles of 94°C for 1 min, 58°C for 1 min and 72°C for 1.5 min. The final extension proceeded for 8 min. The capacity of these primer pairs to amplify nucleus-embedded mtDNA pseudogenes (25,26) was investigated by performing each of the nine primary PCR reactions using either 500 ng ρ+ or ρ0 cell DNA as template.
Table 1. Oligonucleotide primer pairs used for the primary PCR amplification of mtDNA from single cell lysates.
| Forward primer |
Nucleotide positions |
Reverse primer |
Nucleotide positions |
Product size (bp) |
Template for stage 2 PCRa |
| SC-A-for | 503–520 | SC-A-rev | 2484–2463 | 1982 | 1–3 |
| SC-B-for | 2364–2386 | SC-B-rev | 4249–4228 | 1886 | 4–6 |
| SC-C-for | 4155–4175 | SC-C-rev | 6220–6199 | 2066 | 7–9 |
| SC-D-for | 6113–6133 | SC-D-rev | 8017–7996 | 1905 | 10–12 |
| SC-E-for | 7925–7944 | SC-E-rev | 9884–9863 | 1960 | 13–15 |
| SC-F-for | 9767–9784 | SC-F-rev | 11748–11727 | 1982 | 16–18 |
| SC-G-for | 11614–11635 | SC-G-rev | 13638–13617 | 2025 | 19–21 |
| SC-H-for | 13539–13559 | SC-H-rev | 15431–15409 | 1893 | 22–24 |
| SC-I-for | 15331–15350 | SC-I-rev | 836–815 | 2075 | 25, D1–D3 |
All nucleotide positions shown refer to the original Cambridge Reference Sequence report (1).
aSee Table 2 for details of the primers used in the secondary PCR.
Secondary PCR reactions. During the course of extensive sequencing investigations of patients suspected of having pathogenic mtDNA mutations, we have designed 28 pairs of primers to generate overlapping fragments of between 600 and 700 bp that span the entire sequence of the human mitochondrial genome. To facilitate the direct sequencing of PCR-amplified products, all primers are tagged with M13 sequence. Forward primers are tagged with 18 nt of –21 M13 forward sequence (5′-TGTAAAACGACGGCCAGT-3′) and reverse primers tagged with 18 nt of the reverse M13 sequence (5′-CAGGAAACAGCTATGACC-3′). Each primer pair is designed to anneal optimally at 58°C, thereby permitting simultaneous amplification of the 28 PCR reactions required to amplify a complete genome. For amplification of templates generated in the primary PCR of single cell lysates, 2 µl of this initial PCR product was used as template in the secondary PCR amplification. Reaction conditions (30 cycles) were exactly as described above, apart from the inclusion of 10% DMSO in all reactions with the exception of 6F/6R, 8F/8R, 16F/16R, 17F/17R and 23F/23R (Table 2) and a shortening of the extension time to 1 min. Inclusion of DMSO in these amplification reactions has been noted to markedly decrease the efficiency of amplification (unpublished observations). PCR-amplified products were subsequently purified (QIAquick PCR purification kit; QIAGEN) to remove unincorporated primer and sequenced directly using BigDye terminator cycle sequencing chemistry (PE Biosystems) on an ABI 377 automated DNA sequencer. The sequences generated were compared to the revised Cambridge Reference Sequence (rCRS) (19) using Sequence Navigator and Factura software (PE Biosystems).
Table 2. M13-tagged oligonucleotide primer pairs used for the secondary PCR amplification of mtDNA templates.
| Forward primer |
Nucleotide positions |
Reverse primer |
Nucleotide positions |
Product size (bp) |
| 1F | 516–534 | 1R | 1190–1172 | 675 |
| 2F | 1138–1156 | 2R | 1801–1782 | 664 |
| 3F | 1756–1776 | 3R | 2444–2426 | 689 |
| 4F | 2395–2415 | 4R | 3074–3054 | 680 |
| 5F | 2995–3013 | 5R | 3645–3627 | 651 |
| 6F | 3536–3553 | 6R | 4239–4219 | 704 |
| 7F | 4184–4202 | 7R | 4869–4852 | 686 |
| 8F | 4832–4849 | 8R | 5570–5551 | 739 |
| 9F | 5526–5545 | 9R | 6188–6171 | 663 |
| 10F | 6115–6134 | 10R | 6781–6761 | 667 |
| 11F | 6730–6750 | 11R | 7398–7379 | 669 |
| 12F | 7349–7369 | 12R | 8009–7990 | 661 |
| 13F | 7960–7979 | 13R | 8641–8621 | 682 |
| 14F | 8563–8581 | 14R | 9231–9212 | 669 |
| 15F | 9181–9198 | 15R | 9867–9848 | 687 |
| 16F | 9821–9841 | 16R | 10516–10497 | 696 |
| 17F | 10394–10414 | 17R | 11032–11013 | 639 |
| 18F | 10985–11004 | 18R | 11708–11689 | 724 |
| 19F | 11633–11651 | 19R | 12361–12341 | 729 |
| 20F | 12284–12302 | 20R | 13005–12987 | 722 |
| 21F | 12951–12969 | 21R | 13614–13595 | 664 |
| 22F | 13568–13587 | 22R | 14276–14258 | 709 |
| 23F | 14227–14246 | 23R | 14928–14911 | 702 |
| 24F | 14732–14752 | 24R | 15419–15400 | 688 |
| 25F | 15372–15391 | 25R | 16067–16048 | 696 |
| D1F | 15879–15897 | D1R | 16545–16526 | 667 |
| D2F | 16495–16514 | D2R | 389–370 | 446 |
| D3F | 315–332 | D3R | 803–786 | 489 |
Quantification of T7587C mutation in single muscle fibres
The level of mutated mtDNA was determined by PCR–RFLP analysis essentially as described elsewhere (21). A 455-bp fragment encompassing the mutation site was amplified with the M13-tailed mismatch forward primer (M13 sequence shown in lower case, mismatch base in bold) L7565 (nucleotide positions 7565–7585, 5′-tgtaaaacgacggccagtGGCTAAATCCTATATATGTTA-3′) and the reverse primer H8001 (nucleotide positions 8001–7984, 5′-TCAACGTCAAGGAGTCGC-3′). Following 30 cycles of amplification, an additional cycle was performed in the presence of 5 µCi [α-32P]dCTP (3000 Ci/mmol). Labelled products were precipitated and equal amounts (1000–2000 c.p.m.) digested with 8 U HindII (Roche Molecular Biochemicals). Restriction fragments were separated through an 8% non-denaturing polyacrylamide gel, dried onto a support and analysed with ImageQuant software (Molecular Dynamics) following exposure to a PhosphorImager. A single HindII recognition site in the wild-type product generates fragments of 309 and 146 bp. The T7587C transition, in concert with the mismatch base in the forward primer, generates an additional recognition site cutting the 309-bp fragment into two smaller products of 272 and 37 bp.
RESULTS
Two-step PCR amplification of mtDNA
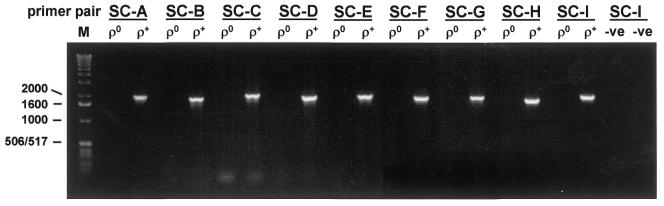
In order to generate sufficient template to sequence whole mtDNA genomes from individual cells, we designed nine pairs of PCR primers to amplify fragments of ∼2 kb that encompassed the entire mitochondrial genome (Table 1). Each primer pair was tested using total homogenate DNA extracted from ρ0 (mtDNA-less) and ρ+ cells and shown to amplify true mtDNA sequences (Fig. 1).
Figure 1.
PCR amplification of ρ+ and ρ0 cell DNA using the nine primer pairs designed to amplify mtDNA from single cell lysates in the first round amplification. The predicted sizes of each of the products are given in Table 1. M, DNA size standard with sizes (bp) on the left; –ve, template omitted from PCR reaction.
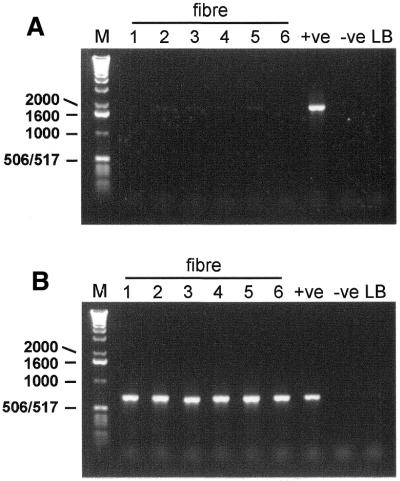
Subsequently, these primers were used to amplify mtDNA using single cell lysate as template. Figure 2 shows the primary PCR reaction products for one pair of primers using DNA extracted from six individual muscle fibres as template. PCR products from single fibres are barely visible on the gel (Fig. 2A, fibre 5), whereas a clear, visible product is generated using a control homogenate (ρ+ cell) DNA. Using 2 µl of the 50 µl first round PCR product as template for the subsequent PCR reactions with the sequencing primers (Table 2), products of the expected size were amplified from each of the single fibre lysates (Fig. 2B). Importantly, no visible PCR products are evident in either of the two negative control lanes, indicating that these PCR products must have been amplified from the single cell template DNA.
Figure 2.
Two-step PCR amplification of mtDNA isolated from six individual muscle fibres. Representative gels of each round of amplification are depicted. (A) An aliquot of 1 µl total DNA template released by lysis of six individual fibres (1–6) was amplified (50 µl PCR reaction) by primer pair SC-D (Table 1). The expected size of this product was 1905 bp. (B) Amplified product (2 µl) from the first round of amplification was used as template in a second round of PCR with primer pair 12F/12R (Table 2). The expected size of this product was 661 bp. Appropriate controls are shown for each round of amplification. +ve, 500 ng ρ+ DNA; –ve, template omitted from PCR reaction; LB, 2 µl aliquot of the single cell lysis buffer used in the DNA extraction; M, DNA size standard with sizes (bp) on the left.
Whole genome sequencing from individual muscle fibres
We next investigated the quality of mtDNA sequence derived from single cell amplifications. Eighteen individual muscle fibres were picked from a 20 µm section of control muscle and the DNA isolated was subjected to two rounds of amplification using primer pair SC-I (forward and reverse) for the first round of amplification and primer pairs D1, D2 and D3 (forward and reverse) for the second round. Together, these primers amplify across the polymorphic, non-coding control region or D-loop. All 18 fibres yielded excellent sequence, revealing 13 changes from the rCRS (Table 3) which were identical to the sequence derived from sequencing a DNA homogenate of this muscle (data not shown). No other sequence variations were observed that distinguished any fibre from the others and each change was represented in the MITOMAP database of sequence polymorphisms (27).
Table 3. mtDNA D-loop sequence changes from the rCRS in individual muscle fibres (n = 18).
| Nucleotide position |
Nucleotide change |
Comment |
| 73 | A→G | Common polymorphisma |
| 143 | G→A | Common polymorphism |
| 189 | A→G | Common polymorphism |
| 192 | T→C | Common polymorphism |
| 194 | C→T | Common polymorphism |
| 195 | T→C | Common polymorphism |
| 196 | T→C | Common polymorphism |
| 204 | T→C | Common polymorphism |
| 207 | G→A | Common polymorphism |
| 263 | A→G | Common polymorphism |
| 311 | C→CC | Common polymorphism |
| 16223 | C→T | Common polymorphism |
| 16519 | T→C | Common polymorphism |
aSequence variant documented in the MITOMAP database (27) as polymorphic.
The DNA extracted from 9 of the 18 individual muscle fibres was subsequently subjected to further amplification with the remaining eight pairs of primary PCR primers and the entire coding region of mtDNA sequence deduced. The quality of the sequence information was excellent for every fibre, revealing a further 24 polymorphic variants from the rCRS (Table 4). These changes were present in all of the nine single fibres that were sequenced and were also present in the sequence from the muscle homogenate (data not shown). Most of these coding sequence changes had been previously described, whilst we have observed others (T1243C, A3505G, G5046A, C11674T, A11947G, T12414C and G15884C) in our own database of complete human mtDNA sequences (28). One silent change, G3531A in ND1, appeared to be novel.
Table 4. mtDNA coding sequence changes from the rCRS in individual muscle fibres (n = 9).
| Nucleotide position |
Gene |
Nucleotide change |
Amino acid change |
Comment |
| 709 | 12S rRNA | G→A | – | Common polymorphism |
| 750 | 12S rRNA | A→G | – | Common polymorphism |
| 1243 | 12S rRNA | T→C | – | Rare polymorphisma |
| 1438 | 12S rRNA | A→G | – | Common polymorphism |
| 2706 | 16S rRNA | A→G | – | Common polymorphism |
| 3106 | 16S rRNA | C→CC | – | Common polymorphism |
| 3505 | ND1 | A→G | T→A | Rare polymorphism |
| 3531 | ND1 | G→A | Silent/P | Unreported polymorphismb |
| 4769 | ND2 | A→G | Silent/M | Common polymorphism |
| 5046 | ND2 | G→A | V→I | Rare polymorphism |
| 5460 | ND2 | G→A | A→T | Common polymorphism |
| 7028 | COXI | C→T | Silent/A | Common polymorphism |
| 8251 | COXII | G→A | Silent/G | Common polymorphism |
| 8860 | ATPase 6 | A→G | T→A | Common polymorphism |
| 8994 | ATPase 6 | G→A | Silent/L | Common polymorphism |
| 11674 | ND4 | C→T | Silent/T | Rare polymorphism |
| 11719 | ND4 | G→A | Silent/G | Common polymorphism |
| 11947 | ND4 | A→G | Silent/T | Rare polymorphism |
| 12414 | ND5 | T→C | Silent/P | Rare polymorphism |
| 12705 | ND5 | C→T | Silent/I | Common polymorphism |
| 14766 | Cyt b | C→T | I→T | Common polymorphism |
| 15326 | Cyt b | A→G | T→A | Common polymorphism |
| 15884 | Cyt b | G→C | A→P | Rare polymorphism |
| 15924 | tRNAThr | A→G | – | Common polymorphism |
aSequence variant documented in our own database of 66 complete mtDNA sequences (28).
bSequence variant not previously reported.
Detection of mtDNA heteroplasmy in single cells
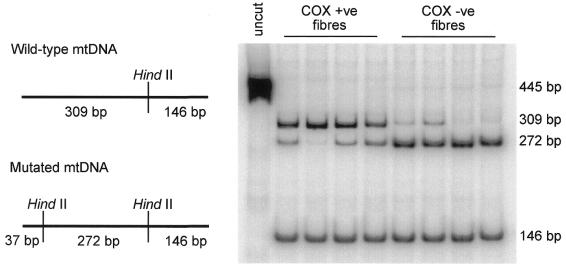
To establish the limits of detection of mtDNA heteroplasmy by single cell mtDNA sequencing, we performed experiments on individual muscle fibres from a patient with a well-characterised heteroplasmic T7587C mutation within the COX II gene (21). Single fibre PCR–RFLP analysis confirmed that higher levels of the mutant 7587C allele were present in COX-negative fibres than COX-positive fibres (Fig. 3).
Figure 3.
Quantification of the 7587C mutant allele in single muscle fibres expressing either a COX-positive or COX-negative phenotype. PCR and RFLP were performed as described in Materials and Methods and labelled products were separated through an 8% non-denaturing polyacrylamide gel. A schematic of the HindII digestion pattern expected in both wild-type and mutant mtDNA is shown on the left. Sizes of expected fragments (bp) are shown on the right of the gel.
Next, by amplifying the DNA isolated from these individual fibres by the two-stage strategy, we were then able to correlate the quantifiable levels of the T7587C mutation with the sequence chromatogram. Using the primary PCR primers SC-D-for and SC-D-rev (Table 1) to amplify across the mutation site, the products obtained from both COX-positive and COX-negative fibres were used simultaneously as (i) template in the PCR–RFLP analysis of T7587C mutation load and (ii) template for PCR amplification with sequencing primer pair 12F/12R (Table 2). Representative data for this experiment are shown in Figure 4. Even in COX-positive fibres with loads of the 7587C allele in the region of 25–40%, there was an observable mutant C peak on the sequence chromatogram that was recognised by the software as different from the wild-type sequence. With higher levels of mutation, the relative proportion of C/T increased such that fibres with >65% mutant loads ‘appeared’ homoplasmic for the 7587C allele on the sequence chromatogram. A total of 26 individual muscle fibres (20 COX-negative, 6 COX-positive) were sequenced and every COX-negative fibre [mean level of mutant DNA 85.3 ± 11.8% (n = 20), range 54–98%] read a C on the sequence chromatogram. Together, these observations suggest that heteroplasmic mtDNA variants representing as low as 30% of the mtDNA genotype of a single cell can be detected by this sequencing method.
Figure 4.
Detection of T7587C mtDNA heteroplasmy by sequencing individual muscle fibres. Products (2 µl) from the first round amplification of single cell DNA isolates by primer pair SC-D (Table 1) were used as template in both the PCR–RFLP analysis of T7587C mutation load and amplification with sequencing primer pair 12F/12R (Table 2). Comparative sequence chromatograms and RFLP gels are shown together. The mutated base is shown (*).
DISCUSSION
We have devised a two-stage PCR amplification strategy to determine the complete sequence of the human mitochondrial genome from single cells. The sequencing of 18 control regions and nine complete genomes equates to nearly 160 000 bases sequenced and every change from the rCRS observed was in agreement with the findings of other, independent experiments between different fibres. Furthermore, we were able to generate sufficient template to permit not only determination of the nucleotide sequence, but to subsequently investigate any potential pathogenic changes by other PCR-based technologies such as PCR–RFLP analysis (see Fig. 4).
The accumulation over time of somatic mtDNA mutations via clonal expansion of specific mutational events is a recognised phenomenon in non-dividing tissues. A broad spectrum of mtDNA deletions has been detected in various post-mitotic tissues of older individuals that are not observed in young control subjects (11,12). Despite these mutant mtDNAs accounting for <0.1% of total mtDNA in a tissue, individual cells within this tissue may contain very high levels of a single mutant mtDNA species (13). When the proportion of mutant mtDNA exceeds a critical threshold level, this results in an observable defect of respiratory chain function, which may eventually compromise organ function. In order to further our understanding of the role of somatic mtDNA mutations in ageing and other neurodegenerative disorders, it is essential to develop sensitive techniques that can detect and identify heteroplasmic mtDNA abnormalities in individual cells. Various PCR-based technologies exist for the study of mtDNA deletions (13,14,17,18), mtDNA copy number (20,29) and discrete mtDNA control region point mutations that have been implicated in the process of ageing (30,31). Nevertheless, the identification of novel point mutations is fraught with difficulties. Human mtDNA exhibits little redundancy of its coding sequence and, as such, a single base mutation in any region of the 16.6 kb genome could theoretically lead to a defect in mitochondrial protein synthesis and translation and hence oxidative phosphorylation. The ability to accurately determine the sequence of mitochondrial genomes from individual cells would therefore provide a powerful tool to investigate the pathophysiology of somatic mtDNA sequence variants.
Perhaps the biggest obstacle we faced in developing this technique was the generation of sufficient product to sequence the entire mitochondrial genome from a single cell containing femtogram amounts of template. Based on our findings, however, we are confident that even with such low amounts of starting template DNA from a single cell, the two-stage PCR faithfully amplifies only mtDNA sequence and does not result in the artefactual PCR amplification of mtDNA pseudogenes (25,26,32). By designing the nine primer pairs that generate template for the secondary, sequencing PCR reactions sufficiently far enough apart, co-amplification of mtDNA pseudogenes is minimised. Since these primary PCR primers failed to generate a product when the mtDNA-deficient ρ0 cell DNA was used as template, we conclude that only human mtDNA is amplified.
Somatic mtDNA mutations must accumulate to high levels before a biochemical defect is apparent and, as such, the identification of any unknown pathogenic mutation by sequencing is likely to be complicated by the coexistence of wild-type mtDNA in these cells. Many pathogenic mtDNA mutations are highly recessive, with expression of the biochemical abnormality exquisitely dependent on the ratios of the mutant and wild-type molecule. Experiments using primary cultures and transmitochondrial cybrids harbouring the A3243G tRNALeu(UUR) and A8344G tRNALys point mutations have clearly demonstrated that mitochondrial function only becomes impaired with mutation loads in excess of 90% (6–8). The mutant load of these point mutations in COX-negative fibres from patients would be at a comparable level and would certainly be detected by sequencing through the relevant region of the genome. By studying a mutation (T7587C) with a low biochemical threshold for disease expression, an increasingly recognised phenomenon amongst mtDNA-encoded structural gene mutations (21,33–36), we confirmed that even the lowest level of mutant mtDNA observed in a biochemically affected cell (i.e. COX-negative) would be detected by this technique.
Recent studies have documented the accumulation of point mutations in the non-coding control region of mtDNA in fibroblast cell lines from old individuals, suggesting that they may play a role in ageing (30). One particular mutation (T414G), which resides adjacent to transcriptional start sites, was found in more than half of the cell lines investigated and reached levels of heteroplasmy nearing 50%. This mutation also accumulates in skeletal muscle (31), although it is absent in brain (37). The relevance of these findings to the role of mtDNA mutations in ageing remains contentious, but it highlights the need to search for these mutations both in coding and non-coding regions of the genome. We believe that the development of sensitive sequencing protocols as reported here that accurately and reliably determine the complete human mtDNA sequences in single cells will provide the methodological basis for investigating the role of somatic mtDNA mutations in ageing and neurodegenerative disease and begin to answer these questions. Furthermore, its application at the level of individual cells will help to further clarify the role of somatic mtDNA point mutations in cancer (38,39) and at what stage of the malignant process these mutations are fixed.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Emma Blakely for her help in the development of these techniques. This work was supported by the Medical Research Council and the Wellcome Trust.
References
- 1.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H., Smith,A.J., Staden,R. and Young,I.G. (1981) Sequence and organisation of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 2.Clayton D.A. (1991) Nuclear gadgets in mitochondrial DNA replication and transcription. Trends Biochem. Sci., 16, 107–111. [DOI] [PubMed] [Google Scholar]
- 3.Schon E.A., Bonilla,E. and DiMauro,S. (1997) Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomembr., 29, 131–149. [DOI] [PubMed] [Google Scholar]
- 4.Chinnery P.F. and Turnbull,D.M. (1997) Clinical features, investigation and management of patients with defects of mitochondrial DNA. J. Neurol. Neurosurg. Psychiatry, 63, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lightowlers R.N., Chinnery,P.F., Turnbull,D.M. and Howell,N. (1997) Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet., 13, 450–455. [DOI] [PubMed] [Google Scholar]
- 6.Chomyn A., Martinuzzi,A., Yoneda,M., Daga,A., Hurko,O., Johns,D., Lai,S.T., Nonaka,I., Angelini,C. and Attardi,G. (1992) MELAS mutation in mtDNA binding site for transcription termination factor causes defect in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc. Natl Acad. Sci. USA, 89, 4221–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attardi G., Yoneda,M. and Chomyn,A. (1995) Complementation and segregation behavior of disease causing mitochondrial DNA mutations in cellular model systems. Biochim. Biophys. Acta, 1271, 241–248. [DOI] [PubMed] [Google Scholar]
- 8.Boulet L., Karpati,G. and Shoubridge,E.A. (1992) Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am. J. Hum. Genet., 51, 1187–1200. [PMC free article] [PubMed] [Google Scholar]
- 9.Richter C., Park,J.W. and Ames,B. (1988) Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl Acad. Sci. USA, 85, 6465–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mecocci P., MacGarvey,U., Kaufman,A.E., Koontz,D., Shoffner,J.M., Wallace,D.C. and Beal,M.F. (1993) Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann. Neurol., 34, 609–616. [DOI] [PubMed] [Google Scholar]
- 11.Cortopassi G.A. and Arnheim,N. (1990) Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res., 18, 6927–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corral-Debrinski M., Horton,T., Lott,M.T., Shoffner,J.M., Beal,M.F. and Wallace,D.C. (1992) Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nature Genet., 2, 324–329. [DOI] [PubMed] [Google Scholar]
- 13.Brierley E.J., Johnson,M.A., Lightowlers,R.N., James,O.F.W. and Turnbull,D.M. (1998) Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann. Neurol., 43, 217–223. [DOI] [PubMed] [Google Scholar]
- 14.Bodyak N.D., Nekhaeva,E., Wei,J.Y. and Khrapko,K. (2001) Quantification and sequencing of somatic deleted mtDNA in single cells: evidence for partially duplicated mtDNA in aged human tissues. Hum. Mol. Genet., 10, 17–24. [DOI] [PubMed] [Google Scholar]
- 15.Borthwick G.M., Johnson,M.A., Ince,P.G., Shaw,P.J. and Turnbull D.M. (1999) Mitochondrial enzyme activity in amyotrophic lateral sclerosis: implications for the role of mitochondria in neuronal cell death. Ann. Neurol., 46, 787–790. [DOI] [PubMed] [Google Scholar]
- 16.Cottrell D.A., Blakely,E.L., Johnson,M.A., Ince,P.G., Borthwick,G.M. and Turnbull,D.M. (2001) Cytochrome c oxidase deficient cells accumulate in the hippocampus and choroid plexus with age. Neurobiol. Aging, 22, 265–272. [DOI] [PubMed] [Google Scholar]
- 17.Sciacco M., Bonilla,E., Schon,E.A., DiMauro,S. and Moraes,C.T. (1994) Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from pateints with mitochondrial myopathy. Hum. Mol. Genet., 3, 13–19. [DOI] [PubMed] [Google Scholar]
- 18.Moslemi A.-R., Melberg,A., Holme,E. and Oldfors,A. (1996) Clonal expansion of mitochondrial DNA with multiple deletions in autosomal dominant progressive external ophthalmoplegia. Ann. Neurol., 40, 707–713. [DOI] [PubMed] [Google Scholar]
- 19.Andrews R.M., Kubacka,I., Chinnery,P.F., Lightowlers,R.N., Turnbull,D.M. and Howell,N. (1999) Reanalysis and revision of the Cambridge Reference Sequence for human mitochondrial DNA [Letter]. Nature Genet., 23, 147. [DOI] [PubMed] [Google Scholar]
- 20.Sciacco M., Gasparo-Rippa,P., Vu,T.H., Tanji,K., Shanske,S., Mendell,J.R., Schon,E.A., DiMauro,S. and Bonilla,E. (1998) Study of mitochondrial DNA depletion in muscle by single-fiber polymerase chain reaction. Muscle Nerve, 21, 1374–1381. [DOI] [PubMed] [Google Scholar]
- 21.Clark K.M., Taylor,R.W., Johnson,M.A., Chinnery,P.F., Chrzanowska-Lightowlers,Z.M.A., Andrews,R.M., Nelson,I.P., Wood,N.W., Lamont,P.J., Hanna,M.G., Lightowlers,R.N. and Turnbull,D.M. (1999) An mtDNA mutation in the initiation codon of the cytochrome c oxidase subunit II gene results in lower levels of the protein and a mitochondrial encephalomyopathy. Am. J. Hum. Genet., 64, 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King M.P. and Attardi,G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science, 246, 500–503. [DOI] [PubMed] [Google Scholar]
- 23.Old S.L. and Johnson,M.A. (1989) Methods of microphotometric assay of succinate dehydrogenase and cytochrome c oxidase activities for use on human skeletal muscle. Histochem. J., 21, 545–556. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L., Chomyn,A., Attardi,G. and Miller,C.A. (1997) Myoclonic epilepsy and ragged red fibers (MERRF) syndrome: selective vulnerability of CNS neurons does not correlate with the level of mitochondrial tRNAlys mutation in individual neuronal isolates. J. Neurosci., 17, 7746–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano M., Shtilbans,A., Mayeux,R., Davidson,M., DiMauro,S., Knowles,J.A. and Schon,E.A. (1997) Apparent mtDNA heteroplasmy in Alzheimer’s disease patients and in normals due to PCR amplification of nucleus-embedded mtDNA pseudogenes. Proc. Natl Acad. Sci. USA, 94, 14894–14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor R.W., Taylor,G.A., Morris,C.M., Edwardson,J.A. and Turnbull,D.M. (1998). Diagnosis of mitochondrial disease: assessment of mitochondrial DNA heteroplasmy in blood. Biochem. Biophys. Res. Commun., 251, 883–887. [DOI] [PubMed] [Google Scholar]
- 27. MITOMAP: A Human Mitochondrial Genome Database. Center for Molecular Medicine, Emory University, Atlanta, GA, USA. http://www.gen.emory.edu/mitomap.html, 2001.
- 28.Elson J.L., Andrews,R.M., Chinnery,P.F., Lightowlers,R.N., Turnbull,D.M. and Howell,N. (2001) Analysis of European mtDNAs for recombination. Am. J. Hum. Genet., 68, 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szuhai K., van den Ouweland,J.M., Dirks,R.W., Lemaitre,M., Truffert,J.-C., Janssen,G.M., Tanke,H.J., Holme,E., Maassen,J.A. and Raap,A.K. (2001) Simultaneous A8344G heteroplasmy and mitochondrial DNA copy number quantification in Myoclonus Epilepsy and Ragged-Red Fibers (MERRF) syndrome by a multiplex Molecular Beacon based real-time fluorescence PCR. Nucleic Acids Res., 29, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michikawa Y., Mazzucchelli,F., Bresolin,N., Scarlato,G. and Attardi,G. (1999) Aging-dependent accumulation of point mutations in human mtDNA control region for replication. Science, 286, 774–779. [DOI] [PubMed] [Google Scholar]
- 31.Murdock D.G., Christacos,N.C. and Wallace,D.C. (2000) The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Res., 28, 4350–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis R.E., Miller,S., Herrnstadt,C., Ghosh,S.S., Fahy,E., Shinobu,L.A., Galasko,D., Thal,L.J., Beal,M.F., Howell,N. and Parker,W.D. (1997) Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease. Proc. Natl Acad. Sci. USA, 94, 4526–4531. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Hanna M.G., Nelson,I.P., Rahman,S., Lane,R.J.M., Land,J., Heales,S., Cooper,M.J., Schapira,A.H.V., Morgan-Hughes,J.A. and Wood,N.W. (1998) Cytochrome c oxidase deficiency associated with the first stop-codon point mutation in human mtDNA. Am. J. Hum. Genet., 63, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pulkes T., Eunson,L., Patterson,V., Siddiqui,A., Wood,N.W., Nelson,I.P., Morgan-Hughes,J.A. and Hanna,M.G. (1999) The mitochondrial DNA G13513A transition in ND5 is associated with a LHON/MELAS overlap syndrome and may be a frequent cause of MELAS. Ann. Neurol., 46, 916–919. [DOI] [PubMed] [Google Scholar]
- 35.Andreu A.L., Tanji,K., Bruno,C., Hadjigeorgiou,G.M., Sue,C.M., Jay,C., Ohnishi,T., Shanske,S., Bonilla,E. and DiMauro,S. (1999) Exercise intolerance due to a nonsense mutation in the mtDNA ND4 gene. Ann. Neurol., 45, 820–823. [DOI] [PubMed] [Google Scholar]
- 36.Taylor R.W., Singh-Kler,R., Hayes,C.M., Smith,P.E.M. and Turnbull,D.M. (2001) Progressive mitochondrial disease due to a novel missense mutation in the mitochondrial DNA ND3 gene. Ann. Neurol., 50, 104–107. [DOI] [PubMed] [Google Scholar]
- 37.Chinnery P.F., Taylor,G.A., Howell,N., Brown,D.T., Parsons,T.J. and Turnbull,D.M. (2001) Point mutations of the mtDNA control region in normal and neurodegenerative human brains. Am. J. Hum. Genet., 68, 529–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polyak K., Li,Y., Zhu,H., Lengauer,C., Willson,J.K.V., Markowitz,S.D., Trush,M.A., Kinzler,K.W. and Vogelstein,B. (1998) Somatic mutations of the mitochondrial genome in human colorectal cancers. Nature Genet., 20, 291–293. [DOI] [PubMed] [Google Scholar]
- 39.Fliss M.S., Usadel,H., Caballero,O.L., Wu,L., Buta,M.R., Eleff,S.M., Jen,J. and Sidransky,D. (2000) Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science, 287, 2017–2019. [DOI] [PubMed] [Google Scholar]