Abstract
Background
Deficits in GABA neuron-related markers, including the GABA synthesizing enzyme GAD67, the calcium-binding protein parvalbumin, the neuropeptide somatostatin, and the transcription factor Lhx6, are most pronounced in a subset of schizophrenia subjects identified as having a "low GABA marker" (LGM) molecular phenotype. Furthermore, schizophrenia shares degrees of genetic liability, clinical features and cortical circuitry abnormalities with schizoaffective disorder and bipolar disorder. Therefore, we determined the extent to which a similar LGM molecular phenotype may also exist in subjects with these disorders.
Method
Transcript levels for GAD67, parvalbumin, somatostatin, and Lhx6 were quantified using quantitative PCR in prefrontal cortex area 9 of 184 subjects with a diagnosis of schizophrenia (n=39), schizoaffective disorder (n=23) or bipolar disorder (n=35), or with a confirmed absence of any psychiatric diagnoses (n=87). A blinded clustering approach was employed to determine the presence of a LGM molecular phenotype across all subjects.
Results
Approximately 49% of the subjects with schizophrenia, 48% of the subjects with schizoaffective disorder, and 29% of the subjects with bipolar disorder, but only 5% of unaffected subjects, clustered in the cortical LGM molecular phenotype.
Conclusions
These findings support the characterization of psychotic and bipolar disorders by cortical molecular phenotype which may help elucidate more pathophysiologically-informed and personalized medications.
Introduction
Disturbances in inhibitory (GABA) neurons in the prefrontal cortex (PFC) are among the most widely and consistently reported findings in postmortem brain tissue in schizophrenia (Volk and Lewis, 2014). For example, multiple research groups have reported deficits in mRNA levels for the GABA synthesizing enzyme glutamate decarboxylase (GAD67) in the PFC in schizophrenia (Akbarian et al., 1995, Curley et al., 2011, Duncan et al., 2010, Guidotti et al., 2000, Straub et al., 2007, Volk et al., 2000). Furthermore, deficits in mRNA levels for the calcium-binding protein parvalbumin and the neuropeptide somatostatin, which are expressed by non-overlapping subpopulations of GABA neurons, have also been replicated across multiple cohorts of schizophrenia subjects (Fung et al., 2010, Hashimoto et al., 2003, Mellios et al., 2009, Morris et al., 2008, Volk et al., 2012). In addition, mRNA levels for the transcription factor Lhx6, which plays a critical developmental role specifically in cortical parvalbumin and somatostatin neurons (Georgiev et al., 2012, Liodis et al., 2007, Neves et al., 2013, Zhao et al., 2008), have been reported to be lower in the PFC in schizophrenia in two subject cohorts (Volk et al., 2014, Volk et al., 2012). Evidence from human studies and pharmacological studies in monkeys suggests that the deficits in these four GABA neuron-related mRNAs are not attributable to exposure to antipsychotic medications, other factors frequently comorbid with schizophrenia or potential confounds (Hashimoto et al., 2003, Morris et al., 2008, Volk et al., 2012). Taken together, these findings suggest that certain GABA neuron-related transcriptome abnormalities are a conserved feature of the disease process of schizophrenia.
In each of these studies, the variability in expression of GABA neuron-related mRNAs was generally higher than in unaffected comparison subjects (Curley et al., 2011, Volk et al., 2012). In addition, deficits in these GABA neuron-related mRNAs were not seen in all schizophrenia subjects relative to pair-matched unaffected comparison subjects (Curley et al., 2011, Volk et al., 2012). These findings are perhaps consistent with the idea that the clinical syndrome recognized as schizophrenia is a collection of heterogeneous disorders of diverse etiologies and molecular phenotypes (Horvath and Mirnics, 2015, Volk and Lewis, 2015). Consistent with this concept, we recently reported the presence of a "low GABA marker" (LGM) molecular phenotype in which deficits in GAD67, parvalbumin, somatostatin, and Lhx6 mRNAs in the PFC distinguished approximately half of the schizophrenia subjects from the other schizophrenia subjects and the unaffected comparison subjects which did not differ from each other by these measures (Volk et al., 2012).
Schizophrenia shares some genetic risk factors (Fanous et al., 2012, Purcell et al., 2009, Ripke et al., 2011, Wang et al., 2010) and certain clinical features, such as psychosis and cognitive impairments (Zanelli et al., 2010), with schizoaffective disorder and bipolar disorder. In addition, lower mean levels of some GABA-related transcripts have been reported in the PFC from subjects with each of these disorders (Guidotti et al., 2000, Sibille et al., 2011, Woo et al., 2008). Together, these findings suggest the hypothesis that the cortical LGM molecular phenotype may be characteristic of a subset of subjects from each of these three diagnostic groups. To test this hypothesis, we quantified mRNA levels for four GABA neuron-related markers in the PFC from 184 subjects (including 84 previously studied subjects (Volk et al., 2012)) with a diagnosis of schizophrenia, schizoaffective disorder or bipolar disorder, or with a confirmed absence of any psychiatric diagnosis and employed a pre-specified blinded clustering approach to determine the presence of an LGM molecular phenotypes among all subjects.
Method
Human subjects
Brain specimens were obtained during routine autopsies conducted at the Allegheny County Office of the Medical Examiner (Pittsburgh, Pennsylvania) after consent was obtained from next-of-kin. An independent committee of experienced research clinicians made consensus DSMIV(American Psychiatric, 1994) diagnoses for each subject using structured interviews with family members and review of medical records, and the absence of a psychiatric diagnosis was confirmed in unaffected comparison subjects using the same approach (Volk et al., 2011). To control for experimental variation, each subject with schizophrenia (n=39), schizoaffective disorder (n=23) or bipolar disorder (n=35), was matched to one unaffected comparison subject (n=87) for sex and as closely as possible for age (Table 1; Supplemental Tables S1, S2, and S3). Ten unaffected subjects were previously used as comparison subjects both in published studies for bipolar disorder (Kimoto et al., 2015, Sibille et al., 2011) and studies involving a mixture of subjects with a schizophrenia or schizoaffective disorder (Volk et al., 2015a, Volk et al., 2014, Volk et al., 2012). Consequently, this pairing was retained in the present study in which ten bipolar disorder subjects share the same unaffected comparison subjects with ten subjects with either schizophrenia or schizoaffective disorder (Supplemental Tables S1, S2, S3; see Quantitative PCR below). Tissue samples from subjects in a pair were processed together throughout all stages of the study. The mean age, postmortem interval, RNA integrity number (RIN), brain pH and tissue freezer storage time did not differ between diagnostic groups and their matched unaffected comparison subjects (Table 1). All procedures were approved by the University of Pittsburgh’s Committee for Oversight of Research and Clinical Training Involving Decedents and Institutional Review Board for Biomedical Research.
Table 1.
Summary of demographic and postmortem characteristics of human subjects
| Parameter | Unaffected | Schizophrenia | Unaffected | Schizoaffective Disorder |
Unaffected | Bipolar Disorder |
|
|---|---|---|---|---|---|---|---|
| N | 39 | 39 | 23 | 23 | 35 | 35 | |
| Sex | 33M / 6F | 33M / 6F | 14M / 9F | 14M / 9F | 20M / 15F | 20M / 15F | |
| Race | 34W / 5B | 29W / 10B | 18W / 5B | 17W / 6B | 32W / 3B | 34W / 1B | |
| Age (years) | 49.8 ± 12.9 | 48.8 ± 12.9 | 47.0 ± 15.4 | 45.7 ± 12.5 | 46.4 ± 12.7 | 45.5 ± 12.2 | |
| Postmortem Interval (hours) | 18.5 ± 5.2 | 18.7 ± 8.1 | 19.3 ± 6.1 | 20.2 ± 9.4 | 19.1 ± 5.1 | 20.5 ± 7.0 | |
| Freezer Storage Time (months) | 135.5 ± 56.3 | 128.7 ± 61.6 | 125.3 ± 58.3 | 126.6 ± 62.3 | 103.0 ± 49.8 | 109.9 ± 45.1 | |
| Brain pH | 6.7 ± 0.3 | 6.6 ± 0.3 | 6.7 ± 0.2 | 6.6 ± 0.3 | 6.7 ± 0.3 | 6.6 ± 0.3 | |
| RNA Integrity Number | 8.2 ± 0.6 | 8.0 ± 0.6 | 8.1 ± 0.7 | 8.1 ± 0.7 | 8.1 ± 0.6 | 8.0 ± 0.5 | |
| Medications At Time of Death* | |||||||
| Antipsychotic | - | 35 (89.7%) | - | 19 (82.6%) | - | 12 (34.3%) | |
| Antidepressant | - | 19 (48.7%) | - | 8 (34.8%) | - | 22 (62.9%) | |
| Benzodiazepine/Anticonvulsant | - | 13 (33.3%) | - | 11 (47.8%) | - | 16 (45.7%) | |
| Lithium | - | 2 (5.1%) | - | 2 (8.7%) | - | 3 (8.6%) | |
The mean (± standard deviation) age, postmortem interval, RNA integrity number (RIN), brain pH and tissue freezer storage time did not differ between the individual diagnostic groups and their matched unaffected comparison subjects (i.e. schizophrenia: all t76 ≤1.8, all p ≥0.07; schizoaffective disorder: all t44≤1.9, all p ≥0.07, and bipolar disorder: all t68 ≤1.1, all p ≥0.29).
For medications at time of death, the number and percentage (in parentheses) of subjects in each applicable category are provided.
Quantitative PCR
Frozen tissue blocks containing the middle portion of the right superior frontal sulcus were confirmed to contain PFC area 9 using Nissl-stained, cryostat tissue sections for each subject (Volk et al., 2000). The gray-white matter boundary of PFC area 9 in a tissue block from each subject was carefully scored with a scalpel blade where the gray matter was cut perpendicular to the pia matter and had uniform thickness and the gray-white matter boundary was easily delineated. This approach ensured minimal white matter contamination (Volk et al., 2011, Volk et al., 2013). The scored gray matter region of the tissue block was then digitally photographed, and the number of tissue sections (40 µm) required to collect ~30 mm3 of gray matter was determined for each subject. The calculated number of required tissue sections for each subject was then cut by cryostat, and gray matter was separately collected into a tube containing TRIzol reagent in a manner consistent with excellent RNA preservation (Volk et al., 2011, Volk et al., 2013). Standardized dilutions of total RNA for each subject were used to synthesize cDNA. All primer pairs (Supplemental Table S4) demonstrated high amplification efficiency (>97%) across a range of four cDNA dilutions and specific single products in dissociation curve analysis. Control studies in which the cDNA template was not included in the quantitative PCR reaction resulted in a complete lack of amplification. Quantitative PCR was performed using the comparative cycle threshold (CT) method with Power SYBR Green dye and the ViiA-7 Real-Time PCR System (Applied Biosystems), as previously described (Volk et al., 2010, Volk et al., 2013). Three reference genes (beta actin, cyclophilin A, and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) that were previously reported to be stably expressed in the present cohort of schizophrenia and schizoaffective disorder subjects relative to the matched unaffected comparison subjects (Volk et al., 2015a) were used to normalize target mRNA levels (Hashimoto et al., 2008). Furthermore, the mean relative expression level for each reference gene relative to the two other reference genes did not differ between bipolar and unaffected comparison subjects, respectively (beta actin: 1.43 ± 0.19 versus 1.40 ± 0.15; cyclophilin: 0.47 ± 0.07 versus 0.48 ± 0.06; GAPDH: 1.53 ± 0.27 versus 1.53 ± 0.27). The difference in CT (dCT) for each target transcript was calculated by subtracting the geometric mean CT for the three reference genes from the CT of the target transcript (mean of four replicate measures). Because dCT represents the log2-transformed expression ratio of each target transcript to the reference genes, the relative level of the target transcript for each subject is reported as 2−dCT (Vandesompele et al., 2002, Volk et al., 2010, Volk et al., 2013). For the ten unaffected subjects used as shared comparison subjects for both schizophrenia or schizoaffective disorder subjects and bipolar disorder subjects, mRNA levels were quantified in two separate qPCR runs, once with the matched schizophrenia or schizoaffective disorder subject and once with the matched bipolar disorder subject, and the mRNA levels from these two runs were averaged for the cluster analysis described below. Although levels of these transcripts have been previously reported in some of these subject pairs, it is important to note that all data from all subject pairs reported in this paper were obtained in new assays conducted at the same time.
Statistical analysis
In order to account for individual characteristics when doing the blinded clustering analysis, a linear regression model with stepwise forward selection was first employed to examine the effects of age, sex, brain pH, RIN, and freezer storage time on GAD67, parvalbumin, somatostatin, and Lhx6 mRNA levels based upon the 184 subjects. Consequently, for the cluster analysis using these subjects, somatostatin mRNA levels were adjusted by age (p <.0001) and brain pH (p =.005); parvalbumin mRNA levels were adjusted by brain pH (p <.0001); and GAD67 mRNA levels by brain pH (p <.0001). The statistical investigators (YZ and ARS) were blinded to subject diagnosis in doing the cluster analysis. Ward's method (Ward, 1963) was used in SAS in PROC Cluster for the hierarchical detection of possible clusters. Each of the 184 subjects was exclusively assigned to exactly one of the two identified clusters. Based upon these blinded data, the distributions of the mRNAs were examined for each cluster to obtain a better understanding of the differences between clusters vis-à-vis their four mRNA values. A comparison of the four mRNA values among the found clusters was done graphically and also with two-sample t-tests statistical tests comparing the individual adjusted means among the found clusters. Based upon these analyses, one of the two clusters produced lower values for all four mRNAs and is designated the LGM cluster. The blind was broken after the two clusters (i.e., the new LGM cluster and the new non-LGM cluster) were identified. To determine whether cluster membership was related to disorder status, membership rates in the LGM and non-LGM clusters for subjects with schizophrenia, schizoaffective disorder, and/or bipolar disorder were compared to their respectively matched unaffected comparison subjects using McNemar's chi-squared test. This clustering approach using Ward’s method was then repeated after excluding the unaffected comparison subjects using a database that did not have the specific disorder identified with a subject.
Results
Cluster analysis to identify the LGM molecular phenotype across psychiatric disorders
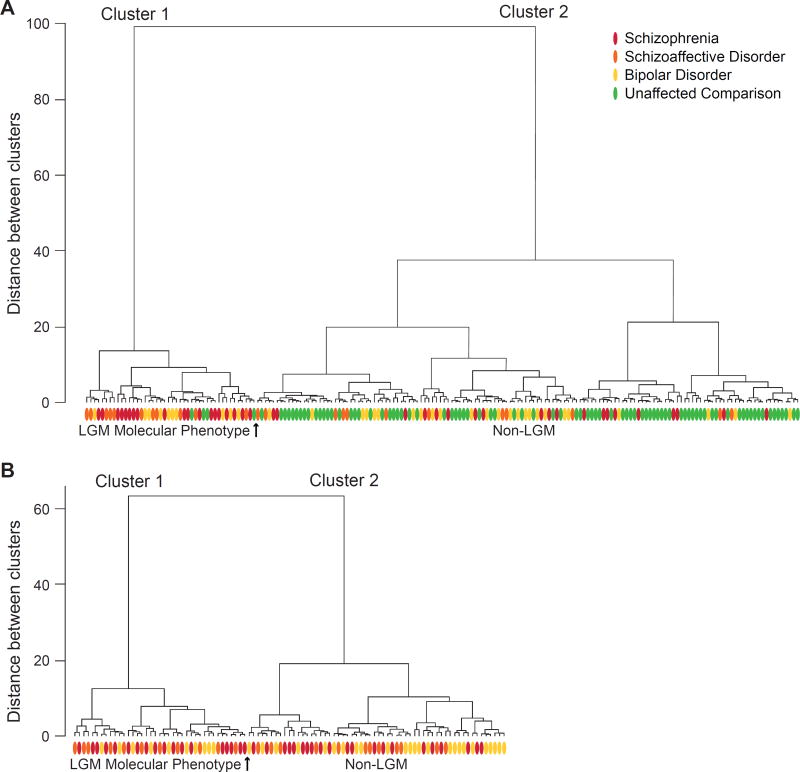
We first determined the extent to which the LGM molecular phenotype is present in a cohort of schizophrenia, schizoaffective disorder, bipolar disorder, and unaffected comparison subjects by quantifying GAD67, parvalbumin, somatostatin, and Lhx6 mRNA levels obtained for all 184 subjects in the same series of quantitative PCR assays. Two primary clusters were identified from the Clustering Tree using the blinded clustering method. Cluster 1 (Figure 1A, left) consisted of 44 subjects, and cluster 2 (Figure 1A, right) consisted of 140 subjects. In cluster 1, mean adjusted transcript levels were lower for GAD67 (−30%; t182=−14.5, p<.00001), parvalbumin (−28%; t182=−8.4, p<.00001), somatostatin (−48%; t182=−12.7, p<.00001), and Lhx6 (−23%; t182=−10.2, p<.00001) relative to cluster 2, consistent with our identification of cluster 1 as the LGM molecular phenotype.
Figure 1. Dendrograms illustrating a cluster analysis of adjusted GABA neuron-related mRNA levels in schizophrenia, schizoaffective disorder, bipolar disorder, and unaffected subjects.
A. Cluster 1 (left of arrow) consisted of 44 subjects with the LGM phenotype including 19 schizophrenia subjects (red ovals), 11 schizoaffective disorder subjects (orange ovals), 10 bipolar disorder subjects (yellow ovals) and 4 unaffected comparison subjects (green ovals), while cluster 2 (right of arrow) consisted of the remaining 140 subjects. B. Excluding all unaffected subjects (n=87) and repeating the cluster analysis including only the schizophrenia, schizoaffective disorder and bipolar disorder subjects (n=97) resulted in two distinct clusters that were highly similar to those in panel A.
Removing the blind on the diagnosis of each subject revealed that 48.7% of schizophrenia subjects (i.e. 19 of 39 subjects), 47.8% (11/23) of schizoaffective disorder subjects, 28.6% (10/35) of bipolar disorder subjects, but only 4.6% (4/87) of unaffected comparison subjects were classified as having the LGM molecular phenotype. The proportions of subjects with schizophrenia (χ2(1)=12.5, p<.0004), schizoaffective disorder (χ2(1)=6.8, p=.009), or bipolar disorder (χ2(1)=8.1, p=.004) with the LGM molecular phenotype were significantly higher relative to their matched unaffected comparison subjects. The proportion of subjects with schizophrenia with the LGM phenotype did not differ from schizoaffective disorder subjects (χ2(1)=0.005, p=.95), but the proportion of subjects with schizophrenia or schizoaffective disorder with the LGM phenotype was nearly significantly higher than in bipolar disorder subjects (χ2(1)=3.6, p=.057). We also found that 19% of bipolar I disorder subjects (4/21) and none of their matched unaffected comparison subjects were respectively classified as having the LGM molecular phenotype (χ2(1)=2.25, p=.13). In addition, 42.9% of bipolar II disorder or bipolar disorder not otherwise specified subjects (6/14) and none of their matched unaffected comparison subjects were respectively classified as having the LGM molecular phenotype (χ2(1)=4.17, p=0.04). Furthermore, the proportion of bipolar I disorder subjects with the LGM phenotype (4/21) did not differ from the proportion of bipolar II or not otherwise specified disorder subjects with the LGM phenotype (6/14; χ2(1)=2.33, p=.13). The proportion of subjects with bipolar I disorder with psychotic features having the LGM phenotype (20%; 2/10) did not differ from the proportion of bipolar I disorder subjects without psychotic features having the LGM phenotype (18.2%; 2/11; χ2(1)=0.01, p=.92).
Transcript levels of GABA neuron-related markers in psychiatric disorder subjects with and without the LGM molecular phenotype
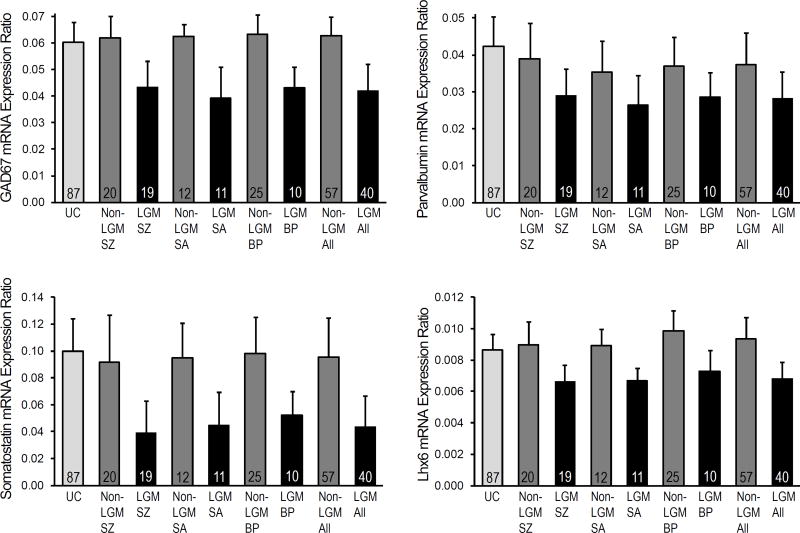
We next examined the differences in mean unadjusted mRNA levels for these GABA neuron-specific markers across the LGM and non-LGM subject diagnostic groups and the unaffected subjects (Figure 2; Supplemental Table S5). LGM subjects with schizophrenia, schizoaffective disorder or bipolar disorder each had lower observed GAD67, parvalbumin, somatostatin, and Lhx6 mRNA levels relative to all unaffected subjects (Figure 2) and had significantly lower values compared to both their pair-matched unaffected subjects and non-LGM subjects with the same diagnosis (Supplemental Table S5). In contrast, transcript levels for GAD67, parvalbumin, somatostatin, and Lhx6 did not differ between non-LGM schizophrenia subjects, schizoaffective or bipolar disorder subjects and all unaffected subjects (Figure 2) and were not significantly different compared to their pair-matched unaffected subjects (Supplemental Table S5) with two exceptions. First, parvalbumin mRNA levels for non-LGM bipolar subjects were lower (−15%, p= .001) than their matched unaffected comparison subjects, and second, somatostatin mRNA levels for non-LGM schizophrenia subjects were marginally lower (−14%, p=.05) than their matched unaffected comparison subjects (Supplemental Table S5). Finally, all LGM subjects with a diagnosis of schizophrenia, schizoaffective disorder, or bipolar disorder (n=40) had lower mRNA levels for GAD67, parvalbumin, somatostatin, and Lhx6 relative to all non-LGM subjects and unaffected comparison subjects (Figure 2) and significantly lower mRNA values both compared to their pooled pair-matched unaffected subjects and to all non-LGM schizophrenia, schizoaffective and bipolar disorder subjects (n=57) (Supplemental Table S5). Very similar results were obtained when the preceding analyses were done on the adjusted mRNA values.
Figure 2. GABA neuron-related mRNA levels in the LGM molecular phenotype of schizophrenia, schizoaffective disorder, and bipolar disorder.
Bar graphs illustrating mean and standard deviation values for unadjusted GAD67, parvalbumin, somatostatin, and Lhx6 mRNA levels in unaffected comparison (UC) subjects; "Low GABA Marker" molecular phenotype (LGM) subjects with schizophrenia (LGM SZ), schizoaffective disorder (LGM SA), bipolar disorder (LGM BP) or together (LGM All), and non-LGM schizophrenia subjects (Non-LGM SZ), schizoaffective disorder subjects (Non-LGM SA), bipolar disorder subjects (Non-LGM BP), or together (Non-LGM All).
Cluster analysis to identify the LGM molecular phenotype exclusively in psychiatric disorder subjects
To further confirm the presence of two distinctive clusters of psychiatric disorder subjects with and without the LGM molecular phenotype, we next excluded the unaffected subjects (n=87) and repeated the Ward cluster analysis including only the schizophrenia, schizoaffective disorder and bipolar disorder subjects (n=97), using the same adjusted mRNA values for each subject employed in the first cluster analysis. We again found two distinct clusters (Figure 1B). In cluster 1 (n=39), mean adjusted transcript levels were lower for GAD67 (−32%; t95=−13.0, p<.0001), parvalbumin (−24%; t95=−6.2, p<.0001), somatostatin (−46%; t95=−8.1, p<.0001), and Lhx6 (−27%; t95=−9.6, p<.0001) relative to cluster 2 (n=58), consistent with this cluster capturing the LGM molecular phenotype. Removing the blind on the diagnosis of each subject revealed that 46.2% (18/39) of schizophrenia subjects, 47.8% (11/23) of schizoaffective disorder subjects, and 28.6% (10/35) of bipolar disorder subjects were classified as having the LGM molecular phenotype. Importantly, 96 of the 97 schizophrenia, schizoaffective disorder, and bipolar disorder subjects received the same LGM classification status that was assigned in the cluster analysis containing the unaffected comparison subjects (Figure 1A).
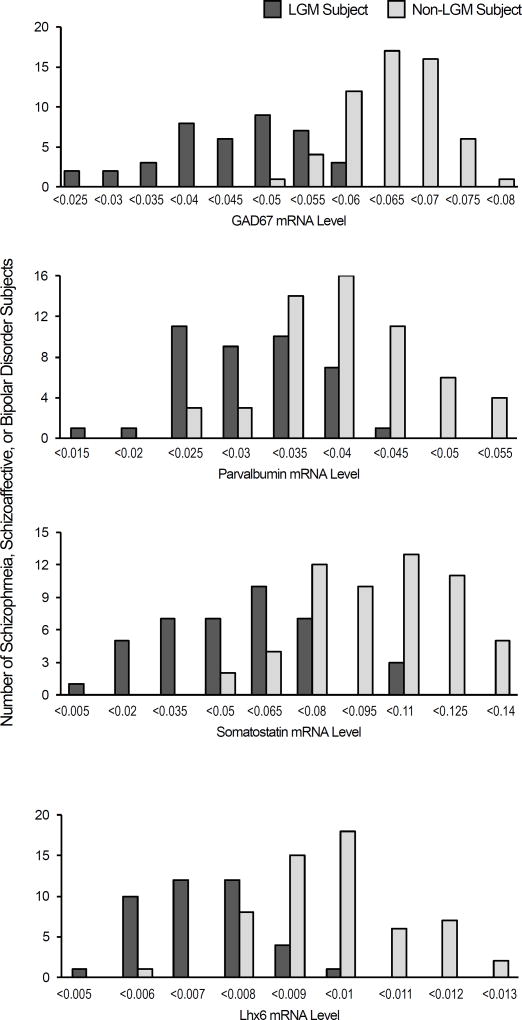
Histogram plots of adjusted mRNA values for GAD67, parvalbumin, somatostatin, and Lhx6 were then created for the schizophrenia, schizoaffective disorder and bipolar disorder subjects classified as having or not having the LGM molecular phenotype (Figure 3). These histograms revealed that the distributions of GAD67, parvalbumin, somatostatin, and Lhx6 mRNA levels for the LGM and non-LGM subjects were clearly distinct and only partially overlapping with the LGM subjects having lower mRNA levels for all four GABA neuron-related mRNA levels.
Figure 3. Distribution of GABA neuron-related mRNA levels in the LGM molecular phenotype.
Histograms showing the distribution of schizophrenia, schizoaffective disorder, and bipolar disorder subjects with (dark gray) and without (light gray) the "Low GABA Marker" classification (Y axis) within a range of adjusted GAD67, parvalbumin, somatostatin, and Lhx6 mRNA levels (X axis; values adjusted for significant covariates as described in the Statistics section).
The LGM molecular phenotype and psychotropic medications, substance use disorders, and suicide as manner of death
Among the subjects with a psychiatric diagnosis, those with the LGM molecular phenotype (n=40) were not more likely: to be exposed to antipsychotics (χ2(1)=2.8, p=.09), antidepressants (χ2(1)=0.007, p=.93), benzodiazepines/anticonvulsants (χ2(1)=0.40, p=.53), or tobacco (χ2(1)=0.82, p=.37) at time of death; to have a substance use disorder current at time of death (χ2(1)=3.1, p=.08); or to have a history of a cannabis use disorder (χ2(1)=0.11, p=.74) than subjects without the LGM molecular phenotype (n=57). Furthermore, subjects with the LGM molecular phenotype were less likely to have suicide as the manner of death (17.5%; 7/40; χ2(1)=4.3, p=.039) relative to subjects without the LGM molecular phenotype (36.8%; 21/57).
Discussion
In this study, we tested the hypothesis that the cortical LGM molecular phenotype identifies a subset of subjects with schizophrenia, schizoaffective disorder, or bipolar disorder. Using a blinded cluster analysis of mRNA levels of four GABA neuron-related markers, we found that 49% of the subjects with schizophrenia, 48% of the subjects with schizoaffective disorder, and 29% of subjects with bipolar disorder share the cortical LGM molecular phenotype. Accordingly, these GABA neuron-related markers were prominently decreased in schizophrenia, schizoaffective disorder, and bipolar disorder subjects with the LGM classification relative to unaffected comparison subjects. Even when the unaffected subjects were excluded from the analysis, the proportion and identity of schizophrenia, schizoaffective disorder, and bipolar disorder subjects with the LGM phenotype remained constant, indicating robust differences in GABA-related mRNA expression between psychiatrically ill subjects with and without this molecular phenotype. In contrast, mRNA levels of these GABA neuron-related markers were largely indistinguishable between psychiatric disorder subjects without the LGM classification and unaffected comparison subjects.
The presence of the LGM phenotype across these severe psychiatric disorders is consistent with other lines of evidence implicating genetic and clinical similarities across these diagnoses. For example, genome-wide association studies have found genetic risk variants that are common to schizophrenia, schizoaffective disorder, and bipolar disorder (Fanous et al., 2012, Purcell et al., 2009, Ripke et al., 2011, Wang et al., 2010), and subjects with these disorders may also share certain clinical features such as psychosis or cognitive impairments (Zanelli et al., 2010). The multiple prior reports of lower mean values for GAD67, parvalbumin, somatostatin, and Lhx6 mRNAs in the PFC across different cohorts of schizophrenia and/or schizoaffective subjects (Akbarian et al., 1995, Curley et al., 2011, Fung et al., 2010, Guidotti et al., 2000, Mellios et al., 2009, Morris et al., 2008, Straub et al., 2007, Volk et al., 2000, Volk et al., 2015a, Volk et al., 2012) is consistent with our findings that the LGM phenotype is present in a relatively high proportion of these subjects. In contrast, deficits in these GABA-related mRNAs have been reported less consistently in the PFC in bipolar disorder (Guidotti et al., 2000, Sibille et al., 2011, Woo et al., 2008), which may reflect a population sampling bias attributable to the relatively lower frequency (~29%) of the LGM phenotype in bipolar disorder found in the present study. Interestingly, lower mRNA levels for somatostatin, but not GAD67 or parvalbumin, have been reported in the PFC in major depressive disorder (Sibille et al., 2011), and psychosis can occur during severe major depressive episodes. Because the LGM phenotype is present to varying degrees in schizophrenia, schizoaffective disorder and bipolar disorder, these data suggest the hypothesis that the LGM molecular phenotype may also be present, but at a much lower relative frequency, in major depressive disorder. Thus, the LGM phenotype could represent a trans-diagnostic feature that provides a molecular analogue to behavioral dimensions of the Research Domain Criteria (Cuthbert, 2014).
Other pathophysiological processes may be also more prominent in subjects with the LGM molecular phenotype. For example, cortical immune activation, characterized by elevated cytokine mRNA levels and activated microglia (Bloomfield et al., 2016, Fillman et al., 2013, Volk et al., 2015b), has been reported in schizophrenia. Furthermore, levels of immune markers and GABA neuron-related markers have been reported to be negatively correlated in the PFC in schizophrenia (Siegel et al., 2014), and schizophrenia subjects with a "high inflammatory state" have been reported to have greater deficits in GABA neuron-related markers (Fillman et al., 2013). However, additional studies are needed to determine whether cortical immune activation contributes to, or is a response to, disturbances in cortical GABA neurons. These findings highlight the importance of further identification of molecular phenotypes in psychiatric disorders in order to better understand the potential interactions between different pathological findings in the same subjects.
Alterations in inhibitory neurotransmission have been reported to contribute to cognitive impairments that are present in schizophrenia, schizoaffective disorder and bipolar disorder (Volk and Lewis, 2014), and the presence of the LGM molecular phenotype may represent a substrate for cognitive impairments in subsets of these subjects. However, a substantial proportion of subjects with these disorders (and in the case of bipolar disorder, the majority of subjects) lack the LGM phenotype and, consequently, may have pathophysiological disturbances in other related components of cortical circuitry that contribute to cognitive impairments. Indeed, our initial report did not find differences in measures of illness severity, such as socioeconomic status and independent living, between subjects with and without the LGM molecular phenotype, suggesting that cognitive dysfunction is likely still present in the subjects without the LGM molecular phenotype (Volk et al., 2012). Interestingly, a lower density of pyramidal neuron dendritic spines, the primary source of excitatory synaptic input, has been commonly reported in the PFC in schizophrenia (Garey et al., 1998, Glantz and Lewis, 2000, Kolluri et al., 2005) and has also recently been reported in bipolar disorder (Konopaske et al., 2014). Experimental models of dendritic spine pathology have been reported to produce cognitive impairments similar to those seen in schizophrenia (Glausier and Lewis, 2013). Thus, perhaps the schizophrenia, schizoaffective disorder and bipolar disorder subjects identified in the present study as not having the LGM phenotype may instead have pyramidal neuron dendritic spine pathology, or perhaps another undetermined molecular phenotype, which then contributes to cognitive impairments in these subjects.
The findings of the present study suggest that independent of diagnosis, subsets of individuals with serious psychiatric illnesses share the same molecular phenotype, and thus might have the same underlying pathogenetic process(es) that contribute to PFC dysfunction. The differences in clinical phenotype, as captured by different DSM diagnoses, across these individuals may reflect differences in genetic background, environmental exposures, and/or other unspecified pathogenetic processes. Alternatively, the LGM molecular phenotype may represent a common endpoint of multiple different pathogenetic processes in individuals that converge to produce similar disruptions in cortical circuitry function. In either case, individuals with the LGM molecular phenotype may potentially benefit from pharmacological treatments that focus on GABA-related disturbances. However, the delivery of such treatment first requires the ability to identify subjects with particularly prominent GABA neuron-related disturbances using peripheral or non-invasive methods. Interestingly, parvalbumin neurons enable the synchronization of cortical neural activity at gamma frequencies (30–80Hz) (Sohal, 2012, Sohal et al., 2009) and deficits in PFC gamma oscillations have been reported in schizophrenia (Cho et al., 2006, Minzenberg et al., 2010). Thus, electroencephalogram measures of gamma oscillations, perhaps in conjunction with magnetic resonance spectroscopy measures of cortical GABA levels (Rowland et al., 2016), may represent useful biomarkers to identify the most severely affected patients, presumably those with the LGM molecular phenotype, and even monitor treatment response. Going forward, advancing the next generation of efficacious neuropharmacological treatment approaches to these disorders may benefit from the identification of the contributing molecular phenotypes.
Supplementary Material
Acknowledgments
The authors wish to acknowledge Siyu Li for her assistance with the statistical design. This study was supported by grants from the National Institutes of Health (MH100066 to Dr. Volk; MH043784 and MH051234 to Dr. Lewis).
David A. Lewis currently receives investigator-initiated research support from Pfizer. In 2013–2015, he served as a consultant in the areas of target identification and validation and new compound development to Autifony, Bristol-Myers Squibb, Concert Pharmaceuticals and Sunovion. Yun Zhang is currently a part-time employee of Janssen Pharmaceutical Research and Development under the auspices of the University of Pittsburgh’s Curricular Practical Training during her Ph.D. degree program.
Footnotes
Declaration of Interest
All other authors have nothing to disclose.
References
- Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney JWE, Jones EG. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Archives of General Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- American Psychiatric, A. DSM-IV. Diagnostic and Statistical Manual of Mental Disorders. Fourth. American Psychiatric Association; Washington, D.C.: 1994. [Google Scholar]
- Bloomfield PS, Selvaraj S, Veronese M, Rizzo G, Bertoldo A, Owen DR, Bloomfield MA, Bonoldi I, Kalk N, Turkheimer F, McGuire P, de Paola V, Howes OD. Microglial Activity in People at Ultra High Risk of Psychosis and in Schizophrenia: An [(11)C]PBR28 PET Brain Imaging Study. American Journal of Psychiatry. 2016;173:44–52. doi: 10.1176/appi.ajp.2015.14101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RY, Konecky RO, Carter CS. Impairments in frontal cortical gamma synchrony and cognitive control in schizophrenia. Proceedings of the National Academy of Sciences U S A. 2006;103:19878–19883. doi: 10.1073/pnas.0609440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curley AA, Arion D, Volk DW, Asafu-Adjei JK, Sampson AR, Fish KN, Lewis DA. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: Clinical, protein, and cell type-specific features. American Journal of Psychiatry. 2011;168:921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert BN. The RDoC framework: facilitating transition from ICD/DSM to dimensional approaches that integrate neuroscience and psychopathology. World Psychiatry. 2014;13:28–35. doi: 10.1002/wps.20087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CE, Webster MJ, Rothmond DA, Bahn S, Elashoff M, Shannon WC. Prefrontal GABA(A) receptor alpha-subunit expression in normal postnatal human development and schizophrenia. Journal of Psychiatric Research. 2010;44:673–681. doi: 10.1016/j.jpsychires.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Fanous AH, Middleton FA, Gentile K, Amdur RL, Maher BS, Zhao Z, Sun J, Medeiros H, Carvalho C, Ferreira SR, Macedo A, Knowles JA, Azevedo MH, Pato MT, Pato CN. Genetic overlap of schizophrenia and bipolar disorder in a high-density linkage survey in the Portuguese Island population. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2012;159B:383–391. doi: 10.1002/ajmg.b.32041. [DOI] [PubMed] [Google Scholar]
- Fillman SG, Cloonan N, Catts VS, Miller LC, Wong J, McCrossin T, Cairns M, Weickert CS. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Molecular Psychiatry. 2013;18:206–214. doi: 10.1038/mp.2012.110. [DOI] [PubMed] [Google Scholar]
- Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. American Journal of Psychiatry. 2010;167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- Garey LJ, Ong WY, Patel TS, Kanani M, Davis A, Mortimer AM, Barnes TRE, Hirsch SR. Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. Journal of Neurology, Neurosurgery, and Psychiatry. 1998;65:446–453. doi: 10.1136/jnnp.65.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev D, Gonzalez-Burgos G, Kikuchi M, Minabe Y, Lewis DA, Hashimoto T. Selective expression of KCNS3 potassium channel alpha-subunit in parvalbumin-ontaining GABA neurons in the human prefrontal cortex. PLoS One. 2012;7:e43904. doi: 10.1371/journal.pone.0043904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Archives of General Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Gerevini VD, Dwivedi Y, Grayson DR, Impagnatiello F, Pandey G, Pesold C, Sharma R, Uzunov D, Costa E. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder. Archives of General Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. American Journal of Psychiatry. 2008;165:479–489. doi: 10.1176/appi.ajp.2007.07081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, Sampson AR, Lewis DA. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. Journal of Neuroscience. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Mirnics K. Schizophrenia as a disorder of molecular pathways. Biological Psychiatry. 2015;77:22–8. doi: 10.1016/j.biopsych.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto S, Zaki MM, Bazmi HH, Lewis DA. Altered markers of cortical gamma-aminobutyric acid neuronal activity in schizophrenia: Role of the NARP Gene. JAMA Psychiatry. 2015;72:747–56. doi: 10.1001/jamapsychiatry.2015.0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolluri N, Sun Z, Sampson AR, Lewis DA. Lamina-specific reductions in dendritic spine density in the prefrontal cortex of subjects with schizophrenia. American Journal of Psychiatry. 2005;162:1200–1202. doi: 10.1176/appi.ajp.162.6.1200. [DOI] [PubMed] [Google Scholar]
- Konopaske GT, Lange N, Coyle JT, Benes FM. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 2014;71:1323–1331. doi: 10.1001/jamapsychiatry.2014.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. Journal of Neuroscience. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biological Psychiatry. 2009;65:1006–1014. doi: 10.1016/j.biopsych.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Minzenberg MJ, Firl AJ, Yoon JH, Gomes GC, Reinking C, Carter CS. Gamma oscillatory power is impaired during cognitive control independent of medication status in first-episode schizophrenia. Neuropsychopharmacology. 2010;35:2590–2599. doi: 10.1038/npp.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris HM, Hashimoto T, Lewis DA. Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder. Cerebral Cortex. 2008;18:1575–1587. doi: 10.1093/cercor/bhm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Shah MM, Liodis P, Achimastou A, Denaxa M, Roalfe G, Sesay A, Walker MC, Pachnis V. The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cerebral Cortex. 2013;23:1811–1823. doi: 10.1093/cercor/bhs159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, Scolnick E, Cichon S, St Clair D, Corvin A, Gurling H, Werge T, Rujescu D, Blackwood DH, Pato CN, Malhotra AK, Purcell S, Dudbridge F, Neale BM, Rossin L, Visscher PM, Posthuma D, Ruderfer DM, Fanous A, Stefansson H, Steinberg S, Mowry BJ, Golimbet V, De Hert M, Jonsson EG, Bitter I, Pietilainen OP, Collier DA, Tosato S, Agartz I, Albus M, Alexander M, Amdur RL, Amin F, Bass N, Bergen SE, Black DW, Borglum AD, Brown MA, Bruggeman R, Buccola NG, Byerley WF, Cahn W, Cantor RM, Carr VJ, Catts SV, Choudhury K, Cloninger CR, Cormican P, Craddock N, Danoy PA, Datta S, de Haan L, Demontis D, Dikeos D, Djurovic S, Donnelly P, Donohoe G, Duong L, Dwyer S, Fink-Jensen A, Freedman R, Freimer NB, Friedl M, Georgieva L, Giegling I, Gill M, Glenthoj B, Godard S, Hamshere M, Hansen M, Hansen T, Hartmann AM, Henskens FA, Hougaard DM, Hultman CM, Ingason A, Jablensky AV, Jakobsen KD, Jay M, Jurgens G, Kahn RS, Keller MC, Kenis G, Kenny E, Kim Y, Kirov GK, Konnerth H, Konte B, Krabbendam L, Krasucki R, Lasseter VK, Laurent C, Lawrence J, Lencz T, Lerer FB, Liang KY, Lichtenstein P, Lieberman JA, Linszen DH, Lonnqvist J, Loughland CM, Maclean AW, Maher BS, Maier W, Mallet J, Malloy P, Mattheisen M, Mattingsdal M, McGhee KA, McGrath JJ, McIntosh A, McLean DE, McQuillin A, Melle I, Michie PT, Milanova V, Morris DW, Mors O, Mortensen PB, Moskvina V, Muglia P, Myin-Germeys I, Nertney DA, Nestadt G, Nielsen J, Nikolov I, Nordentoft M, Norton N, Nothen MM, O'Dushlaine CT, Olincy A, Olsen L, O'Neill FA, Orntoft TF, Owen MJ, Pantelis C, Papadimitriou G, Pato MT, Peltonen L, Petursson H, Pickard B, Pimm J, Pulver AE, Puri V, Quested D, Quinn EM, Rasmussen HB, Rethelyi JM, Ribble R, Rietschel M, Riley BP, Ruggeri M, Schall U, Schulze TG, Schwab SG, Scott RJ, Shi J, Sigurdsson E, Silverman JM, Spencer CC, Stefansson K, Strange A, Strengman E, Stroup TS, Suvisaari J, Terenius L, Thirumalai S, Thygesen JH, Timm S, Toncheva D, van den OE, van Os J, van Winkel R, Veldink J, Walsh D, Wang AG, Wiersma D, Wildenauer DB, Williams HJ, Williams NM, Wormley B, Zammit S, Sullivan PF, O'Donovan MC, Daly MJ, Gejman PV. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland LM, Summerfelt A, Wijtenburg SA, Du X, Chiappelli JJ, Krishna N, West J, Muellerklein F, Kochunov P, Hong LE. Frontal Glutamate and gamma-Aminobutyric Acid Levels and Their Associations With Mismatch Negativity and Digit Sequencing Task Performance in Schizophrenia. JAMA Psychiatry. 2016;73:166–74. doi: 10.1001/jamapsychiatry.2015.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibille E, Morris HM, Kota RS, Lewis DA. GABA-related transcripts in the dorsolateral prefrontal cortex in mood disorders. International Journal of Neuropsychopharmacology. 2011;14:721–734. doi: 10.1017/S1461145710001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel BI, Sengupta EJ, Edelson JR, Lewis DA, Volk DW. Elevated viral restriction factor levels in cortical blood vessels in schizophrenia. Biological Psychiatry. 2014;76:160–167. doi: 10.1016/j.biopsych.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS. Insights into cortical oscillations arising from optogenetic studies. Biological Psychiatry. 2012;71:1039–1045. doi: 10.1016/j.biopsych.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Lipska BK, Egan MF, Goldberg TE, Callicott JH, Mayhew MB, Vakkalanka RK, Kolachana BS, Kleinman JE, Weinberger DR. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Molecular Psychiatry. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:34.1–34.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Archives of General Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- Volk DW, Chitrapu A, Edelson JR, Lewis DA. Chemokine receptors and cortical interneuron dysfunction in schizophrenia. Schizophrenia Research. 2015a;167:12–7. doi: 10.1016/j.schres.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Chitrapu A, Edelson JR, Roman KM, Moroco AE, Lewis DA. Molecular mechanisms and timing of cortical immune activation in schizophrenia. American Journal of Psychiatry. 2015b;172:1112–21. doi: 10.1176/appi.ajp.2015.15010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Edelson JR, Lewis DA. Cortical inhibitory neuron disturbances in schizophrenia: role of the ontogenetic transcription factor Lhx6. Schizophrenia Bulletin. 2014;40:1053–61. doi: 10.1093/schbul/sbu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Eggan SM, Lewis DA. Alterations in metabotropic glutamate receptor 1alpha and regulator of G protein signaling 4 in the prefrontal cortex in schizophrenia. American Journal of Psychiatry. 2010;167:1489–1498. doi: 10.1176/appi.ajp.2010.10030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Lewis DA. Early developmental disturbances of cortical inhibitory neurons: contribution to cognitive deficits in schizophrenia. Schizophrenia Bulletin. 2014;40:952–7. doi: 10.1093/schbul/sbu111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Lewis DA. Schizophrenia. In: Rosenberg RN, Pascal JM, editors. Rosenberg's Molecular and Genetic Basis of Neurological and Psychiatric Disease. Academic Press/Elsevier; 2015. pp. 1293–1299. [Google Scholar]
- Volk DW, Matsubara T, Li S, Sengupta EJ, Georgiev D, Minabe Y, Sampson A, Hashimoto T, Lewis DA. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. American Journal of Psychiatry. 2012;169:1082–1091. doi: 10.1176/appi.ajp.2012.12030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Radchenkova PV, Walker EM, Sengupta EJ, Lewis DA. Cortical opioid markers in schizophrenia and across postnatal development. Cerebral Cortex. 2011;22:1215–1223. doi: 10.1093/cercor/bhr202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Siegel BI, Verrico CD, Lewis DA. Endocannabinoid metabolism in the prefrontal cortex in schizophrenia. Schizophrenia Research. 2013;147:53–57. doi: 10.1016/j.schres.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu XF, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophrenia Research. 2010;124:192–199. doi: 10.1016/j.schres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Ward JH. Hierarchical grouping to optimize an objective function. Journal of the American Statistical Association. 1963;58:236–244. [Google Scholar]
- Woo TU, Kim AM, Viscidi E. Disease-specific alterations in glutamatergic neurotransmission on inhibitory interneurons in the prefrontal cortex in schizophrenia. Brain Research. 2008;1218:267–277. doi: 10.1016/j.brainres.2008.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli J, Reichenberg A, Morgan K, Fearon P, Kravariti E, Dazzan P, Morgan C, Zanelli C, Demjaha A, Jones PB, Doody GA, Kapur S, Murray RM. Specific and generalized neuropsychological deficits: a comparison of patients with various first-episode psychosis presentations. American Journal of Psychiatry. 2010;167:78–85. doi: 10.1176/appi.ajp.2009.09010118. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. Journal of Comparative Neurology. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.