Abstract
Type 1 (T1D) and type 2 (T2D) diabetes share pathophysiological characteristics, yet mechanistic links have remained elusive. T1D results from autoimmune destruction of pancreatic beta cells, whereas beta cell failure inT2D is delayed and progressive. Here we find a new genetic component of diabetes susceptibility in T1D non-obese diabetic (NOD) mice, identifying immune-independent beta cell fragility. Genetic variation in Xrcc4 and Glis3 alters the response of NOD beta cells to unfolded protein stress, enhancing the apoptotic and senescent fates. The same transcriptional relationships were observed in human islets, demonstrating the role of beta cell fragility in genetic predisposition to diabetes.
Diabetes mellitus is a group of diseases in which defective insulin production or function results in the dysregulation of blood glucose levels. The most common forms of diabetes, T1D and T2D, can be considered to be at extreme ends of the etiology spectrum, with T1D caused by autoimmunity against pancreatic beta cells, resulting in insulin deficiency, and T2D initiated by metabolic changes that render target tissues resistant to insulin. The divergence in etiology is reflected in the animal models used, with the primary T1D model being the autoimmune-prone NOD mouse1, whereas T2D models typically use severe obesity to generate metabolic dysfunction2.
Despite this academic division, clinical convergence is observed for T1D and T2D, with patients with T2D commonly developing a progressive decline in total beta cell mass3. This loss of beta cells is a critical component of disease pathogenesis and has led to prolonged efforts to find shared genetic risks. Genome-wide association studies (GWAS) have demonstrated a complex genetic landscape for diabetes, with a stark separation between immunological genes in T1D4 and metabolic genes in T2D5. One of the few genes linked to both diseases is GLIS3 (refs. 5,6), which is also notable for mutations causing a form of congenital diabetes7. Another gene with mutations capable of causing congenital diabetes is EIF2AK3 (also known as PERK)8, an initiator of the unfolded protein stress response (UPR). This pathway is initiated when protein biosynthesis exceeds folding capacity, resulting in the accumulation of unfolded proteins, and is responsible for the bifurcated outcomes of stress alleviation and apoptosis. The UPR has been proposed as a major regulator of inflammation9, with potential involvement in both T1D10,11 and T2D12.
Complex longitudinal changes in beta cells during T1D progression are well established13; however, separating potential genetic beta cell defects from induced secondary changes has not been possible in human samples. The genetics of T1D susceptibility in NOD mice have been well characterized and show striking parallels with human data, making these mice an ideal testing ground for the hypothesis that genetic abnormalities in beta cells contribute to pathogenic outcome1. NOD mice have pre-autoimmune defects in glucose regulation that have been proposed to act in synergy with immune defects14. Here we directly investigate the contribution of genetic defects in beta cell robustness to the development of diabetes. Using a transgenic model that increases the basal load of unfolded protein stress in islets, we found that the NOD genetic background encodes strong genetic susceptibility to primary beta cell failure, which is independent of immune tolerance defects. This unique susceptibility is controlled by loci that include missense Xrcc4 polymorphisms and result in defective Glis3 expression, which in turn drive increased levels of beta cell apoptosis and senescence. Genetic susceptibility could be replicated by elevated levels of dietary fat. Transcriptional analysis of human islets identified the same genetic networks at play. Together, these findings demonstrate both the important role of genetic variation in beta cells for diabetes susceptibility and a mechanism by which the ‘Western diet’ may contribute to the growing diabetes epidemic.
Results
Genetic defects in beta cell robustness in NOD mice
NODk mice, with congenic replacement of the potent Idd1 major histocompatibility complex (MHC) susceptibility locus, allow for dissection of the multiple genetic components of diabetes susceptibility without spontaneous autoimmunity against pancreatic islets. In the course of constructing a matched set of transgenic mice on the B10k and NODk backgrounds15, we were surprised to observe spontaneous diabetes selectively in male NODk mice hemizygous for the insulin (Ins2) promoter–driven hen egg lysozyme (insHEL) transgene. This phenomenon was not observed in B10k.insHEL transgenic mice, nor was it observed on the B6, CBA/H or 129/Sv strain background (data not shown), suggesting that insHEL transgenic mice have a unique propensity to develop diabetes on the NODk background. To further investigate this phenomenon, we aged a cohort of B10k and NODk transgenic and non-transgenic siblings. Only NODk.insHEL and (B10k × NODk)F1.insHEL males developed diabetes (Fig. 1a). This effect was not caused by differential expression of the insHEL transgene (Supplementary Fig. 1).
Figure 1.
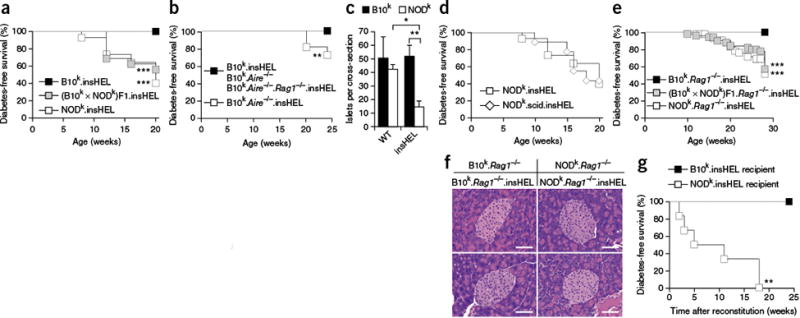
NOD mouse susceptibility to immune-independent diabetes demonstrated through a sensitized transgenic model. (a) Incidence of diabetes in male insHEL transgenic mice on the B10k (n = 22), NODk (n = 43) and (B10k × NODk)F1 (n = 16) backgrounds. No diabetes was observed in non-transgenic male littermates. (b) Incidence of diabetes in male insHEL transgenic mice on the B10k (n = 22), B10k.Aire−/− (n = 11) and B10k.Aire−/−.Rag1−/− (n = 23) backgrounds. B10k.Aire−/− mice without the insHEL transgene did not develop diabetes (n = 22). (c) Average number of islets per pancreatic section in B10k, B10k.insHEL, NODk and NODk.insHEL mice at 28 weeks of age (n = 4–5 mice/group; WT, wild type). Data are shown as means ± s.e.m. (d) Incidence of diabetes in male insHEL transgenic mice on the NODk (n = 43) and NODk.scid (n = 9) backgrounds. No diabetes was observed in nontransgenic male littermates. (e) Diabetes incidence in male insHEL transgenic mice on the B10k.Rag1−/− (n = 58), NODk. Rag1−/− (n = 44) and (B10k × NODk)F1. Rag1−/− (n = 51) backgrounds. (f) Hematoxylin and eosin histology of pancreatic islets at 28 weeks of age (representative of 7–15 mice/group). Scale bars, 50 μm. (g) B10k.insHEL mice and NODk.insHEL mice were irradiated and reconstituted with NODk or B10k bone marrow, respectively, before aging for diabetes incidence (n = 7 and 6). *P < 0.05, **P < 0.001, ***P < 0.0001.
As T1D in humans and NOD mice is dependent on defects in immune tolerance, we tested the capacity of the insHEL transgene to act as a sensitizer for the development of immune-mediated diabetes by crossing diabetes-resistant B10k transgenic mice to mice on the Aire knockout background, which have defective thymic tolerance against pancreatic antigens16. B10k.Aire−/−.insHEL mice also developed diabetes, albeit at a lower rate than NODk.insHEL mice, in a process that was immune dependent (Fig. 1b). Before developing diabetes, NODk.insHEL mice but not B10k.insHEL mice demonstrated a reduction in pancreatic islet number (Fig. 1c). This reduction could be the consequence of an immune response directed against the HEL protein encoded on the H2k haplotype17 or it could represent an as yet undescribed beta cell defect. To discriminate between these two possibilities, we introgressed the insHEL transgene into the NODk.Prkdcscid and NODk.Rag1−/− lines. Despite the absence of T and B lymphocytes in both strains, insHEL transgenic NOD mice still developed diabetes (Fig. 1d,e). In immune-competent NOD.insHEL mice, we observed perivascular accumulation of lymphocytes around 4% of blood vessels in the pancreas (data not shown); however, no invasive insulitis was observed. Infiltrate was absent in NODk.Rag1−/−.insHEL mice (Fig. 1f), and chemokine expression was not induced (Supplementary Fig. 2). Finally, NOD mouse susceptibility to diabetes was extrinsic to the bone marrow, formally ruling out variation in the immune system as the primary driver of diabetes susceptibility in this model (Fig. 1g). Together, these results clearly demonstrate that NOD mice have a beta cell–intrinsic genetic defect, manifested by expression of the insHEL transgene.
Expression of HEL, commonly used as a model antigen, has not previously been reported to induce adverse functions, and the transgene did not interrupt gene expression (Supplementary Fig. 3). Further investigation found profound cellular stress induced by insHEL on both genetic backgrounds (Fig. 2a,b). Total insulin levels were increased in insHEL transgenic mice (Fig. 2c and Supplementary Fig. 4a–c); however, this masked a sharp decrease in the levels of mature, processed insulin (Supplementary Fig. 4d,e), with the insHEL transgene increasing proinsulin secretion in vivo (Fig. 2d–f) and in vitro (Supplementary Fig. 5a,b). In NODk.insHEL mice, this effect was associated with decreased abundance of prohormone convertase proteins (Supplementary Fig. 5c–i). The endoplasmic reticulum stress induced by the insHEL transgene therefore perturbs the correct processing of insulin, analogous to the Akita mouse18 and in line with the model of T2D disease progression.
Figure 2.
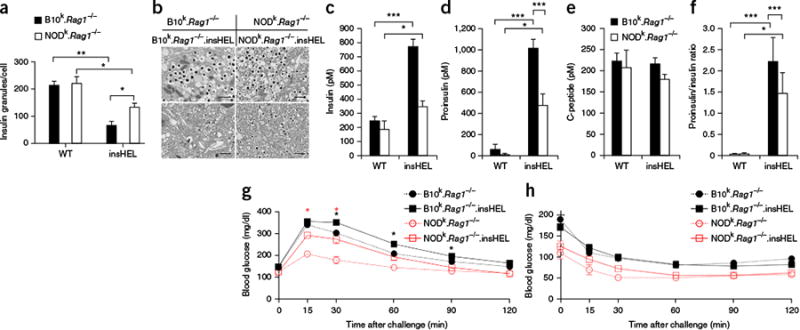
Transgene-induced beta cell stress results in disturbed insulin processing and glucose intolerance. (a)Electron microscopy of beta cells from wild-type and insHEL transgenic B10k.Rag1−/− and NODk.Rag1−/− mice at 12 weeks of age was used to assess the number of insulin granules per cellular cross-section (n = 3 mice/group). (b)Electron microscopy images of the cells described in a. Images are representative of three mice per group. Scale bars, 1 μm. (c–f) Fasting serum samples from B10k.Rag1−/−, B10k.Rag1−/−.insHEL, NODk.Rag1−/− and NODk.Rag1−/−. insHEL mice at 24 weeks of age were assessed by ELISA for insulin (n = 10, 33, 9 and 24 mice) (c), proinsulin (n = 31, 44, 9 and 26 mice) (d), C-peptide (n = 10, 33, 9 and 24 mice) (e) and proinsulin/insulin ratio (n = 10, 33, 5 and 24 mice) (f). (g,h) Blood glucose levels in 12-week-old B10k.Rag1−/−, B10k.Rag1−/−.insHEL, NODk.Rag1−/− and (non-diabetic) NODk.Rag1−/−.insHEL mice following a glucose tolerance test (n = 28, 47, 9 and 21 mice) (g) or an insulin tolerance test (n = 8, 17, 3 and 13 mice) (h). Data are shown as means ± s.e.m. *P < 0.05, **P < 0.001, ***P < 0.0001.
This similarity to T2D prompted an assessment of glucose tolerance. The insHEL transgene resulted in glucose intolerance in both strains, with elevated or delayed peak glucose levels (Fig. 2g and Supplementary Fig. 6a–e). The response to exogenous insulin injection, by contrast, remained normal (Fig. 2h), indicating that insHEL-mediated diabetes is a bona fide beta cell failure rather than a defect in peripheral insulin signaling. As both the B10 and NOD strains exhibited cellular stress and glucose intolerance, these data demonstrate that NOD mice possess an underlying genetic defect that manifests during beta cell stress, driving progression from subclinical glucose intolerance to clinical diabetes.
As diabetes was never observed in female mice (Supplementary Fig. 7a), we further sought to dissect the role of sex. Islets from insHEL transgenic NODk female mice exhibited intermediate signs of stress, with reduced size of insulin granules (Supplementary Fig. 7b) but no decrease in total number (Supplementary Fig. 7c). Likewise, beta cell granularity was only partially reduced in female mice (Supplementary Fig. 7d), with maintenance of insulin content (Supplementary Fig. 7e). As in male mice, the insHEL transgene resulted in increased insulin and proinsulin secretion (Supplementary Fig. 7f,g), whereas C-peptide levels were maintained (Supplementary Fig. 7h), despite the retention of PC1 and PC3 expression (Supplementary Fig. 7i,j). The net effect of this islet stress was mild glucose intolerance (Supplementary Fig. 7k); however, unlike male transgenic mice, females did not have a decrease in islet number (Supplementary Fig. 7l) and remained non-diabetic despite the addition of several stressors (Supplementary Fig. 7a). Finally, the unique susceptibility of the male mice was due to a sex hormone-dependent effect, as castration of males dramatically reduced diabetes incidence (Supplementary Fig. 7m).
Altered unfolded protein stress response in NOD islets
To determine the molecular mechanism of NOD mouse susceptibility to beta cell failure, we performed RNA sequencing (RNA-seq) on the islets of B10k.Rag1−/−, B10k.Rag1−/−.insHEL, NODk.Rag1−/−and (pre-diabetic) NODk.Rag1−/−.insHEL mice. This experimental design excluded any infiltrating lymphocytes or effects secondary to autoimmunity. At a global level, the greatest differences were observed between strains (Fig. 3a). Analysis of statistically significant changes in gene expression (Supplementary Data Set 1) identified the two non-transgenic parental strains (Fig. 3b) as having the greatest number of differences, largely at the lower end of the expression spectrum (Supplementary Fig. 8a) and with the affected genes not clustering in functional pathways (Fig. 3c and Supplementary Data Set 2). By contrast, the impact of insHEL, although globally smaller than the strain effect (Fig. 3a,b), was coordinated in functionally related gene sets (Fig. 3c and Supplementary Fig. 8b,c). The changes induced on the B10 and NOD backgrounds were highly related, with similar effects at both quantitative (Fig. 3a,b) and qualitative (Supplementary Fig. 9) levels. To build on the transcriptional analysis, we performed a proteomic analysis of B10k.Rag1−/−.insHEL and NODk.Rag1−/−.insHEL islets (Supplementary Data Set 3). Only 18 differences in protein expression were significant and reproducible across duplicate experiments, of which most reflected parental transcriptional changes (Supplementary Data Set 1). Together, these results indicate that there is no major quantitative defect in the UPR in NOD mice, suggesting that defects lie in the baseline strain variation, with pathogenic potential uncovered during cellular stress.
Figure 3.
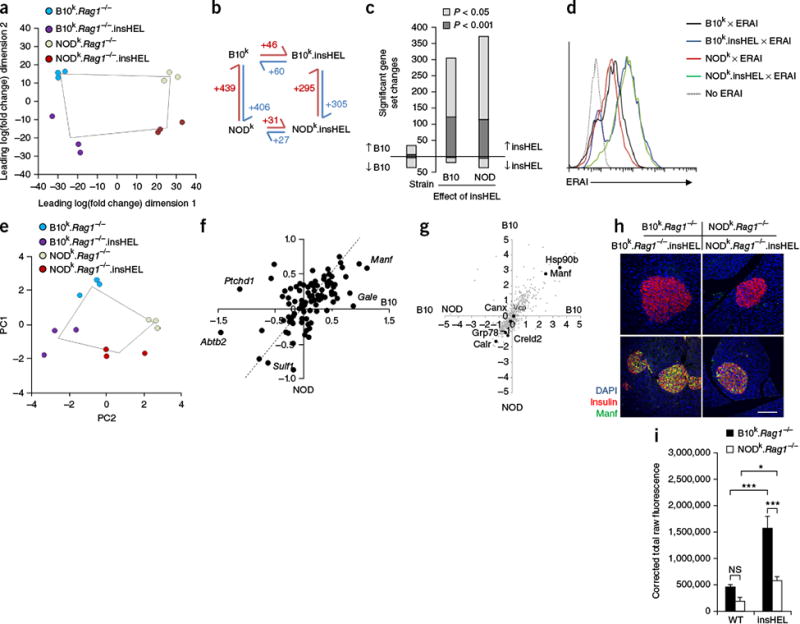
Qualitative rather than quantitative differences in the UPR on the B10 and NOD backgrounds. (a) Globa principal-components analysis (PCA) of RNA-seq data for islets (n = 3 mice/group). (b) Schematic of the number of significant (corrected P < 0.05) gene expression differences for pairs of sample conditions. (c) Number of significant gene set changes in gene set enrichment analysis (GSEA) using cutoffs of P < 0.05 and P < 0.001. (d) ERAI mice were crossed to B10k.Rag1−/−.insHEL and NODk.Rag1−/−.insHEL mice, and islets were analyzed by flow cytometry. Histograms show Xbp1s (Venus) expression in insulin-expressing beta cells. Results are representative of three experiments. (e) PCA of the Xbp1 response gene set in islets (n = 3 mice/group). (f) Scatterplot of individual Xbp1 response genes, showing the average log2-transformed fold change in expression induced by insHEL on the B10 background versus the NOD background. The dashed line indicates equivalent regulation; outlier genes are annotated. (g) Dot plot of mass spectrometry expression ratios, showing on each axis the log2-transformed ratio of expression in B10k.Rag1−/−.insHEL islets as compared to NODk.Rag1−/−.insHEL islets in duplicate experiments. All reproducibly detected proteins are displayed in gray, with black dots indicating annotated UPR-related proteins. (h,i) Immunofluorescence of wild-type and insHEL transgenic islets with a polyclonal antibody to insulin, antibody to Manf and DAPI. Staining is representative of three experiments (h), with Manf quantification (n = 10 mice/group) (i). Scale bar, 100 μm. Data are shown as means ± s.e.m. *P < 0.05, ***P < 0.0001. NS, not significant.
Notably, of the few proteins expressed at higher levels in B10k. insHEL islets than in NODk.insHEL islets, two (Hsp90b and Manf) are encoded by UPR-related genes. NOD mice have been suggested to have a defect in the UPR11, although the use of immunocompetent mice in published experiments means that the observation could be secondary. To determine whether NOD beta cells have a primary defect in activation of the UPR, we crossed the B10k.Rag1−/−.insHEL and NODk.Rag1−/−.insHEL strains with the endoplasmic reticulum stress–activated indicator (ERAI) transgenic mouse model, in which reporter activation is mediated by selective splicing of XBP1 mRNA during the UPR19. These crosses effectively replicate B10k.insHEL. ERAI (diabetes-resistant) mice and (B10k × NODk)F1.insHEL. ERAI (diabetes-susceptible) mice. Analysis of UPR reporter expression demonstrated equal induction of the reporter in both strains (Fig. 3d). As an independent approach to quantify the UPR, we analyzed the expression of Xbp1 target genes in the RNA-seq data set. Similar to the global results (Fig. 3a), analysis of the Xbp1 target gene set indicated large variation between the non-transgenic parental strains, but these strains showed similar induction with the insHEL transgene (Fig. 3e). At the individual gene level, most UPR-related genes demonstrated equivalent regulation by insHEL on the two genetic backgrounds at both the transcriptional (Fig. 3f) and protein (Fig. 3g) levels. Manf was the only UPR-related gene differentially expressed at both the mRNA and protein levels, with efficient induction by insHEL expression on the B10 background and poor induction on the NOD background (Fig. 3h,i). Together, these results demonstrate that insHEL induces the UPR in beta cells but, critically, does so to a similar extent in diabetes-resistant (B10) and diabetes-susceptible (NOD and (B10 × NOD)F1) hosts. Thus, rather than a global defect in inducing the UPR per se, NOD mice possess an intact UPR with differential transcriptional biases, resulting in a specific defect in upregulating Manf expression.
Genetic linkage of stress-induced diabetes in NOD mice
To determine the basis of NOD mouse susceptibility to insHEL-induced diabetes, we performed a genetic analysis. Initially, we used a candidate-based approach, testing the Idd loci associated most strongly with diabetes susceptibility in NOD mice20. NODg7 mice—with the diabetes resistance B10 alleles of Idd3, Idd5, Idd9, Idd10 and Idd18—were backcrossed onto the NODk.insHEL strain and monitored for diabetes development. Despite potent effects in NODg7 mice, none of these regions provided any protection against insHEL-mediated diabetes, even in combination (Fig. 4a). Next, we performed a linkage cross. As susceptibility of NOD mice to insHEL-induced diabetes was dominant (Fig. 1a), we generated a cohort of (NOD × B10)F2 mice. Of 331 (NOD × B10)F2.Rag1−/−.insHEL mice, 36% developed diabetes. Following genome-wide SNP genotyping, two distinct transgene-induced diabetes (Tid) loci were identified (Fig. 4b). Chromosome 13 linkage was observed with two peaks (Tid1 and Tid2) within a single Bayes interval (Fig. 4c), whereas chromosome 19 linkage was observed with a single distinct peak (Fig. 4d). The effect of each Tid locus was assessed by plotting the diabetes-free survival rate for (NOD × B10)F2.Rag1−/−.insHEL mice that were B10/B10, B10/NOD and NOD/NOD at each locus. For each locus, (NOD × B10)F2.Rag1−/−.insHEL.TidB10/B10 mice had significantly (P < 0.0001) lower diabetes rates than (NOD × B10)F2.Rag1−/−.insHEL.TidNOD/NOD mice (Fig. 4e–g). (NOD × B10)F2.Rag1−/−.insHEL.TidB10/B10 mice, with the B10 allele at each Tid locus, did not develop diabetes at all (Fig. 4h), demonstrating that the three Tid loci are independently sufficient and together necessary for NOD mouse susceptibility to insHEL-mediated diabetes.
Figure 4.
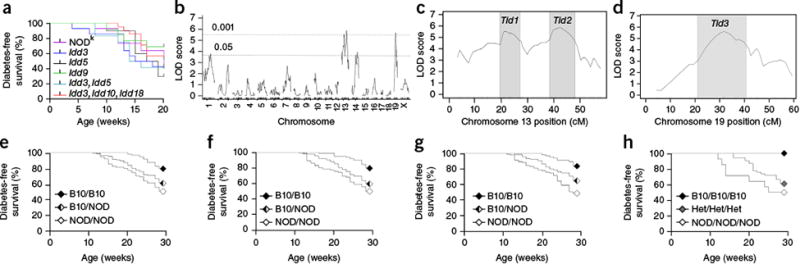
Genetic control of NOD mouse susceptibility to transgene-induced diabetes. (a) Diabetes incidence in insHEL transgenic male mice on the NODk background (n = 43) or on NODk congenic backgrounds homozygous for B6- or B10-derived chromosomal segments containing the diabetes resistance allele of Idd3 (n = 14), Idd5 (n = 10), Idd9 (n = 13), Idd3 and Idd5 (n = 12), and Idd3, Idd10 and Idd18 (n = 21). (b) A cohort of (NOD × B10)F2.Rag1-/-.insHEL male mice (n = 331) was assessed for diabetes incidence at 28 weeks of age and genotyped for 740 informative SNPs. Quantitative trait locus (QTL) association is shown across the genome. LOD, logarithm of odds. (c,d) LOD scores for chromosomes 13 (c) and 19 (d), indicating the LOD support intervals (LOD drop of 1) for the associated Tid1, Tid2 and Tid3 loci. (e–g) Diabetes development in the (NOD × B10)F2.Rag1-/-.insHEL cohort when stratified by genotype at the linkage SNP (rs13481783) in the Tid1 locus (n = 98, 157 and 75 mice) (e), at the linkage SNP (gnf13.088.732) in the Tid2 locus (n = 100, 163 and 67 mice) (f) and at the linkage SNP (rs6237466) in the Tid3 locus (n = 68, 166 and 96 mice) (g). (h) Diabetes development in the (NOD × B10)F2.Rag1-/-.insHEL cohort with the B10 homozygous, B10/NOD heterozygous (Het) and NOD homozygous genotypes at all Tid loci (n = 12, 54 and 14 mice).
To further dissect the genetic basis for NOD mouse susceptibility to stress-induced diabetes, we generated a list of potential candidate genes meeting one or more of four criteria: (i) homology with human genes associated with diabetes in GWAS; (ii) significant mRNA expression differences between strains (Supplementary Data Set 1); (iii) >2-fold difference in protein expression between strains (Supplementary Data Set 3); and (iv) islet expression with nonsynonymous polymorphisms. This latter class was generated from RNA-seq data by filtering for allelic variants (Supplementary Data Set 4). Having generated a list of potential candidate genes, we cross-referenced the list with the genetic linkage data to identify 25 candidate genes (Supplementary Table 1).
Of the candidate genes within the Tid1 and Tid2 loci, supporting evidence was found for Xrcc4. The Xrcc4 protein is a critical binding partner of DNA ligase IV (Lig4). Lig4 knockout mice develop spontaneous diabetes due to poor double-stranded DNA break (DSB) repair in beta cells, leading to senescence21,22. We found two polymorphisms in the NOD Xrcc4 allele (Fig. 5a–c). Molecular modeling of the Xrcc4-Lig4 complex predicted that the NOD polymorphisms reduce the stability of Xrcc4 by causing loss of the Asp125-Arg3 electrostatic interaction (Fig. 5d), affecting the Xrcc4-Lig4 complex (Supplementary Fig. 10). Investigation of embryonic fibroblasts showed reduced Xrcc4 expression in NODk-derived fibroblasts than in B10k-derived fibroblasts (Fig. 5e,f), accompanied by poorer DSB repair (Fig. 5g). To formally test the role of the NOD polymorphisms, we expressed the B10 and NOD alleles of Xrcc4 in Xrcc4-deficient CHO cells, finding defective DSB repair in Xrcc4NOD CHO cells as compared to Xrcc4B10 CHO cells (Fig. 5h). As these results suggested that NOD mice have suboptimal DSB repair, we investigated the in vivo context and found that insHEL drives accumulation of senescent beta cells with unrepaired damage on the NOD background (Fig. 5i,j). As the NOD Xrcc4 allele is shared with the T2D-prone DBA/2 strain, we crossed B10k.insHEL mice with DBA/2 mice and found that F1 transgenics became diabetic (Fig. 5k) and also developed an accumulation of unrepaired DSBs (Fig. 5l,m). Likewise, making B10k.insHEL mice heterozygous for the Lig4 hypomorph allele encoding p.Tyr288Cys (ref. 23) resulted in spontaneous diabetes (Supplementary Fig. 11). These results formally demonstrate that partial defects in the Lig4-Xrcc4 complex catalyze beta cell senescence and diabetes during stress. Further, they strongly implicate Xrcc4 polymorphism as the causative changes in Tid1 and Tid2.
Figure 5.
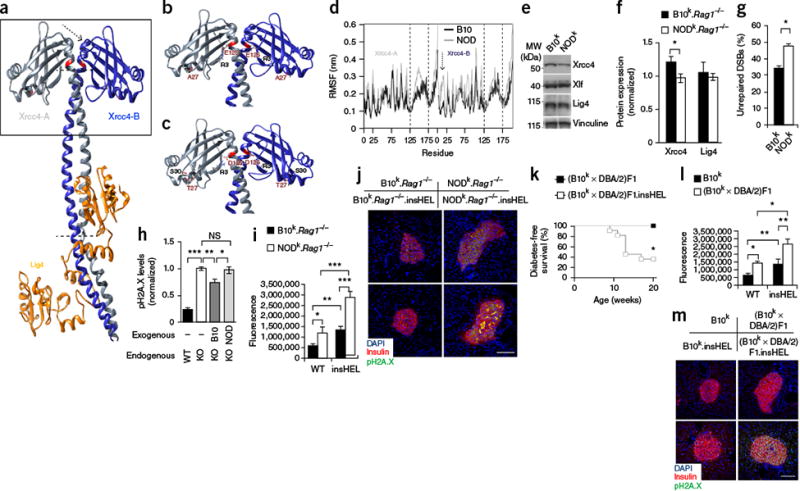
Xrcc4 mutation drives enhanced susceptibility to senescence. (a) Three-dimensional structure of the Lig4 complex, determined using the human structure as scaffolding. Modified residues are highlighted in red, and the arrow and dotted lines indicate regions of major instability. The box highlights the regions shown in detail in b and c. (b,c) Molecular interactions calculated along the represented trajectory for the B10 (b) and NOD (c) alleles of Xrcc4. (d) Per-residue root-mean-square fluctuation (RMSF) in Xrcc4 between 70–100 ns of simulation. Regions with a different fluctuation profile are highlighted by the dashed lines and one arrow, corresponding to the regions indicated in a. The results are representative of four simulations. (e,f) Representative immunoblotting of mouse embryonic fibroblasts (MEFs) for Xrcc4, Xlf and Lig4 (e), with quantification (n = 5 technical replicates/group) (f). (g) Proportion of MEFs that remained positive for H2A.X (Ser139) phosphorylation (pH2A.X; indicative of DSBs) after etoposide exposure (n = 6 technical replicates/group). (h) Wild-type CHO cells and Xrcc4-deficient (KO) CHO cells reconstituted with the B10 or NOD Xrcc4 allele were exposed to etoposide, and unrepaired DNA damage was quantified (n = 6–8 technical replicates/group). (i,j) Immunofluorescence with a polyclonal antibody to insulin, antibody to phosphorylated H2A.X and DAPI on pancreata from the B10k.Rag1−/− and NODk.Rag1−/− backgrounds. Quantification is shown for islet raw fluorescence in the channel for phosphorylated H2A.X (n = 19, 17, 20 and 20 sections) (i), with images of representative sections (j). Scale bar, 100 μm. (k) Diabetes incidence of (B10k × DBA/2)F1 and (B10k × DBA/2)F1.insHEL mice (n = 6 and 11 mice). (l,m) Immunofluorescence with a polyclonal antibody to insulin, antibody to phosphorylated H2A.X and DAPI on pancreata from the B10k and (B10k × DBA/2)F1 backgrounds. Quantification is shown of islet raw fluorescence in the channel for phosphorylated H2A.X (n = 19, 17, 16 and 20 sections) (l), with images of representative sections (m). Scale bar, 100 μm. Data are shown as means ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.0001; NS, not significant.
Of the candidate genes within Tid3, Glis3 was validated as the lead candidate. Glis3, identified by GWAS association, showed reduced islet expression in NODk mice (Fig. 6a). As Glis3 knockdown in INS-1 cells increases susceptibility to apoptosis24, we challenged B10k and NODk islets in vitro with high glucose concentrations (Fig. 6b) and induction of chemical stress (Fig. 6c), both of which caused significantly (P < 0.05) greater levels of apoptosis in NODk islets. In vivo assessment demonstrated that the insHEL transgene increases the apoptosis rate only on the NODk background (Fig. 6d,e), resulting in reduced beta cell mass (Fig. 6f). To formally test the ability of reduced Glis3 expression to drive diabetes in the insHEL model, we intercrossed Glis3 heterozygous mice with B10k.Rag1−/−.insHEL mice, with offspring showing glucose intolerance and increased levels of proinsulin secretion (Supplementary Fig. 12). Upon aging, Glis3 heterozygous mice (like NOD mice) developed diabetes (Fig. 6g). As Manf is a critical antiapoptotic factor for beta cells25 and is poorly upregulated in NODk.Rag1−/−.insHEL islets (Fig. 3f–h), we sought to determine whether Manf upregulation was dependent on Glis3 expression. Glis3 heterozygous mice failed to adequately induce Manf after insHEL expression (Fig. 6h,i). Together, these results provide a functional mechanism for the Tid3 linkage locus, whereby reduced Glis3 levels in NOD islets result in poor Manf induction during the UPR, allowing apoptosis of the stressed beta cells.
Figure 6.
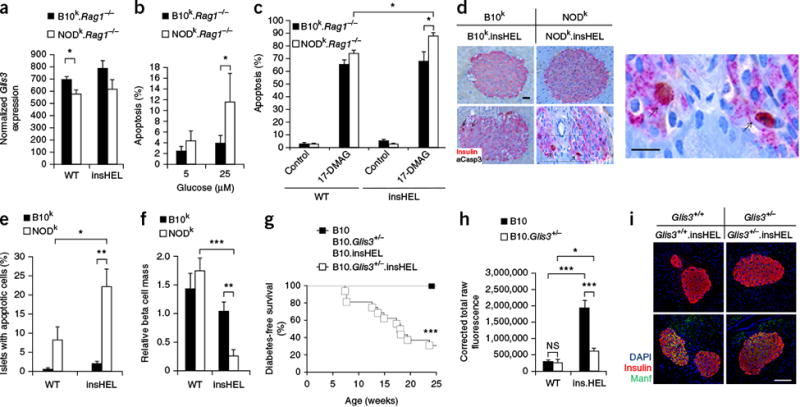
Reduced Glis3 expression results in enhanced susceptibility to apoptosis. (a) Islet expression of Glis3, based on RNA-seq analysis (n = 3 mice/group). (b) The percentage of apoptotic cells in the islets of wild-type and insHEL transgenic B10k.Rag1-/- and NODk.Rag1-/- mice after culturing in the presence of low (5 mM) and high (25 mM) concentrations of glucose (n = 5–6 replicates/group). (c) The percentage of apoptotic cells in the islets of B10k.Rag1-/- and NODk.Rag1-/- cells, with and without insHEL, after culturing in the presence of the geldanamycin analog 17-DMAG and a low (5 mM) glucose concentration (n = 6–18 replicates/group). (d–f) Representative sections (d), average observation of apoptosis in islets (e) and beta cell mass (f) after immunohistochemistry with a polyclonal antibody to insulin and antibody to activated caspase-3 (aCasp3) (n = 4–5 replicates/group). In d, the box highlights the magnified region. Arrows indicate example apoptotic cells. Scale bars, 100 μm. (g) Diabetes incidence in B10 (n = 6), B10.Glis3+/- (n = 11), B10.insHEL (n = 12) and B10.Glis3+/-.insHEL (n = 23) littermates. (h,i) Pancreas immunofluorescence with a polyclonal antibody to insulin, antibody to Manf and DAPI. Fluorescence in the channel for Manf was quantified in islets (n = 8, 8, 10 and 10 mice) (h), with representative staining shown (i). Scale bar, 100 μm. Data are shown as means ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.0001; NS, not significant.
Finally, as increased dietary fat has been demonstrated to reduce islet Glis3 expression26 and induce senescence in beta cells27, we sought to determine whether a 10% fat diet could induce diabetes in diabetes-resistant B10k.Rag1−/−.insHEL mice. We confirmed that elevated fat levels reduced Glis3 expression (Fig. 7a) and found a corresponding decrease in Manf levels (Fig. 7b). On the insHEL background, the diet with 10% fat eliminated the normal upregulation of Manf (Fig. 7c,d), recapitulating the NOD phenotype. Within weeks of the diet change, without obesity, the B10k.Rag1−/−.insHEL mice on a 10% fat diet developed diabetes (Fig. 7e). Together, these results demonstrate that a propensity to apoptosis and senescence, due to genetic background or diet, can convert subclinical cellular stress into insulin-dependent diabetes.
Figure 7.
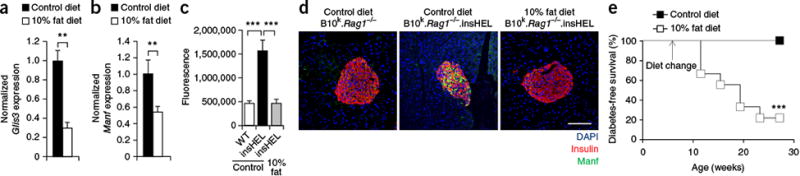
Dietary change recapitulates the effect of the NOD genetic background on a resistant mouse strain. B10k.Rag1−/−.insHEL mice were maintained on a control diet or converted to a 10% fat diet at 6 weeks of age. (a,b) Islet expression (quantitative PCR) of Glis3 (a) and Manf (b), normalized to Rpl37a expression (n = 6 mice/group). (c,d) Pancreas immunofluorescence with a polyclonal antibody to insulin, antibody to Manf and DAPI, with wild-type B10k.Rag1−/− mice and B10k.Rag1−/−.insHEL mice on a control diet and B10k.Rag1−/−.insHEL mice on a 10% fat diet. Quantification is shown of islet raw fluorescence in the channel for Manf (n = 8, 10 and 10) (c), with images of representative sections (d). Scale bar, 100 μm. (e) Diabetes incidence of B10k.Rag1−/−.insHEL mice fed with a control diet (n = 17) or a 10% fat diet (n = 9). Data are shown as means ± s.e.m. **P < 0.01, ***P < 0.0001.
Parallel transcriptional regulation in human islets
To determine whether the findings observed in mice were applicable to humans, we investigated whether the pathway identified in NOD mice also demonstrated genetic linkage to diabetes or glucose regulation traits in humans. GLIS3 polymorphisms have previously been associated with altered glucose regulation; we additionally identified nominally significant associations for MANF, XRCC4 and LIG4 polymorphisms (Supplementary Table 2). In an independent approach that takes into account environmental effects, we analyzed RNA-seq data from human pancreatic islets isolated from 119 donors, including 14 diagnosed with T2D28. To assess the validity of the Glis3-Manf relationship observed in mice, we investigated the relationship of these two genes in human islets. A trend toward reduced GLIS3 expression was observed in T2D islets, whereas MANF expression appeared unchanged (Supplementary Fig. 13). Critically, a significant positive relationship was observed between GLIS3 and MANF levels in human islets (Fig. 8a). Next, we investigated whether patients with T2D might exhibit reduced XRCC4 expression, analogous to the NOD polymorphisms. We found no change in XRCC4 expression in T2D islets (Fig. 8b); however, the levels of the obligate binding partner encoded by LIG4 were significantly reduced (Fig. 8c). In mice, Xrcc4 polymorphisms were associated with increased senescence; likewise, in patients with T2D, the levels of the senescence markers H2AFX (Fig. 8d) and CDKN1A (Fig. 8e) were increased. Finally, a direct relationship was observed between reduced LIG4 and increased H2AFX levels (Fig. 8f). Although the cause of co-regulation cannot be assessed in ex vivo human islets, the parallel with NOD mice strongly supports a conservation of diabetes susceptibility mechanisms across species.
Figure 8.
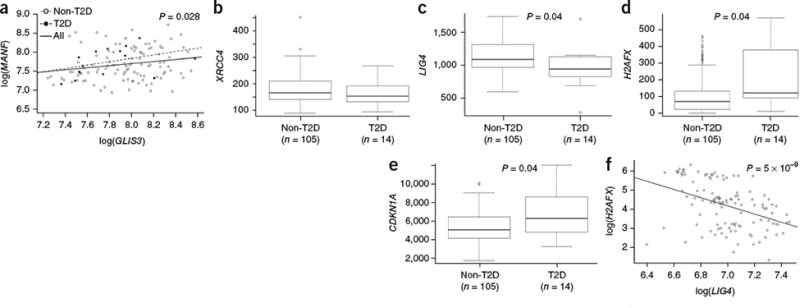
Molecular changes in the islets of patients with T2D mirror the processes altered in NOD mice. mRNA expression in human pancreatic islets from healthy individuals (n = 105) and those diagnosed with T2D (n = 14) was assessed through RNA-seq analysis. (a) Relationship between GLIS3 and MANF expression in healthy ndividuals (Spearman correlation P value = 0.043), individuals with T2D (Spearman correlation P value = 0.075) and all individuals (Spearman correlation P value = 0.028). (b-e) Expression of XRCC4 (b) LIG4 (c), H2AFX (d) and CDKN1A (e) in healthy islets as compared to i slets from patients withT2D (P values shown after multiple-testing correction). The median and interquartile range (IQR; box) are shown, with error bars indicating 1.5 times the IQR. Individual values are shown if beyond 1.5 times the IQR. (f) Relationship between H2AFX and LIG4 expression in human islets (Spearman correlation P value = 5 × 10-9).
Discussion
In this study, we have used the properties of the insHEL transgene as a sensitizer for beta cell failure. The association of insulin promoter–driven transgenes with diabetes induction has been observed before, such as with homozygosity of the insGFP allele29. It has typically been considered an epiphenomenon resulting from enhanced autoreactivity to a xenogenic peptide. However, we found here that the diabetes-inducing properties of one such transgene are not immunological in nature and rather result from unfolded protein stress, a physiologically relevant pathway. The usefulness of the insHEL transgene in this process lies in the subclinical nature of the stress it places upon the beta cell, which, being below the threshold for spontaneous failure, requires the addition of a second insult (genetic or environmental) for diabetes to develop.
At a molecular level, our findings demonstrate that the NOD mouse strain harbors a remarkable susceptibility to primary beta cell failure. No major differences were observed in induction of the UPR, despite previous studies suggesting a defect in NOD islets11. The primary difference between the strains was instead preexisting and emerged with the addition of stress. Thus, most UPR response proteins were equally expressed in both strains, including pro-apoptotic Chop30. Only a small subset of these proteins was expressed at higher levels in B10 islets; however, this subset included the antiapoptotic Manf protein31, the chaperone Hsp90b32, and Sel1 and Herp, rate-limiting components in unfolded protein degradation33,34. Together, these results reflect a differential outcome to cellular stress, with B10 islets capable of stressed survival whereas NOD islets respond with apoptosis and senescence.
At a genetic level, we identified two discrete Tid loci that are sufficient to account for the entire NOD susceptibility. It is notable that the Tid loci are distinct from the Idd regions mapped for autoimmune diabetes susceptibility; however, this result is expected as Idd mapping necessitated a backcross approach, which is unable to detect dominant Tid loci. Although we have focused on the beta cell–intrinsic aspects of NOD genetic susceptibility, in contrast to most previous studies, there are several reasons to expect that immunological and beta cell–intrinsic genetic factors synergize in the development of diabetes. First, the addition of autoimmune pressure in the form of Aire deficiency was enough to generate diabetes in a fraction of insHEL transgenic mice, even on a resistant background. Second, of the NOD.insHEL mice that develop diabetes, 42% produce antibodies against insulin (as compared to 0% of NODk mice; data not shown), indicating that islet death spurs an autoimmune reaction. Third, in our previous studies using HEL-reactive T cells to drive autoimmunity in insHEL transgenic mice, NOD.insHEL mice developed diabetes at a higher rate than Aire−/−.insHEL mice, despite having a milder immunological phenotype35,36. Together, these results point to a complex genetic origin of autoimmune diabetes in NOD mice, with interplay between defects in immunological tolerance and beta cell robustness. This interplay is reminiscent of investigations in Obese Strain (OS) chickens, which develop spontaneous autoimmune thyroiditis due to both excessive anti-thyroid immunity and susceptibility of the thyroid to apoptosis37.
Continued research into these phenomena may allow greater insight into the extent to which the target organ in human autoimmune disease is a passive victim or an active cause of autoimmune failure. In the case of T1D, environmental factors that produce beta cell–intrinsic effects may account for the growing T1D epidemic, in which T1D incidence is increasing at a rate of ∼3% per year38–40. Although genetic factors influencing beta cell defects will remain constant, our results demonstrate the capacity of dietary fat to substitute for genetic defects in the precipitation of diabetes. Saturated fatty acids drive the apoptosis and senescence of beta cells27,41, with increased oxidative stress42 and endoplasmic reticulum stress41. As increased body mass index is associated with earlier onset of T1D43, it is possible that dietary fat is acting as a sensitizer similar to insHEL, in effect lowering the threshold for autoimmune stress to precipitate clinical diabetes.
The male-specific susceptibility to diabetes in this model is in sharp contrast to the strong female bias in NOD mice. T1D, with a small male bias44, is distinct from many other autoimmune diseases, which manifest a strong female bias44. It is therefore reasonable to postulate that females have both a strong autoimmune predisposition and strong metabolic resistance (such as the ‘insulin advantage’; ref. 45), which roughly negate each other in T1D. In this regard, it is interesting to note that male susceptibility in our model was dependent on sex hormones and in humans the sex divergence in T1D rates occurs after puberty44. The insHEL transgene model therefore may be a useful tool to study the hidden metabolic susceptibility of males to T1D.
This study identifies beta cell failure as a mechanistic commonality between T1D and T2D. Although the initial decline in beta cell mass in T1D can be clearly attributed to autoimmunity, the later decline in mass may result from the remaining beta cells being forced into compensatory insulin overproduction. Under this scenario, the same processes may be triggered as in insHEL mice. Likewise, many competing hypotheses have been proposed for the decline of beta cell mass in T2D, including direct glucotoxicity, inflammation-induced cytokines46, stress caused by compensatory insulin overproduction12, dietary saturated fats41, obesity3 and autoimmunity47. The major difficulty in understanding this late-stage decline in beta cells lies in dissecting the primary cause. An advantage of the insHEL model is that beta cell death occurs in the absence of autoimmunity or insulin resistance. Effectively, the production of HEL mimics the compensatory upregulation of insulin, allowing the negative effects of to be studied in the absence of complicating factors. The molecular pathways to transgene-induced diabetes identified here were notably paralleled in human islets, making the insHEL mouse strain a promising model for the development of drugs to prevent decline in beta cell numbers.
URLs
Cytoscape, http://cytoscape.org/; Distiller software, http://www.matrixscience.com/Distiller.html; Enrichment Map, http://www.baderlab.org/software/enrichmentmap; MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp; Grace program, http://plasma-gate.weizmann.ac.il/Grace/; HTSeq, http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html; KNIME, http://www.knime.org/; Protein Data Bank, http://www. rcsb.org/pdb/home/home.do; ProteomeXchange Consortium, http://www.proteomexchange.org/; R, http://www.r-project.org/.
Online Methods
Mice
B10.BR-H2k/SgSnJ (B10k) mice are a C57BL/10 substrain congenic for the C57BR H2 locus, H2k, imported from Jackson Laboratories. NOD.BR-H2k/Wicker (NODk) mice are NOD mice congenic for the entire B10k H2 locus, H2k (ref. 48). The ILK-3 insHEL transgene encodes a membrane-bound (class I transmembrane region) HEL antigen whose expression is driven by the rat insulin (Ins2) promoter. It was introduced in C57BL/6J mice, which were backcrossed for >7 generations to the B10k (ref. 49) and NODk (ref. 36) backgrounds. B10k.Rag1−/−.insHEL transgenic mice were produced by crossing to the Rag1tm1Mom strain50, imported from Jackson Laboratories. B10k.Aire−/−. insHEL and B10k.Aire−/−.Rag1−/−.insHEL strains were created by intercrossing with the Aire knockout mouse51. (B10k × DBA/2)F1 and (B10k × DBA/2)F1.insHEL strains were created by intercrossing with DBA/2 mice from Jackson Laboratories. NODk.Rag1−/−.insHEL mice and NODk. scid.insHEL mice were produced by backcrossing to the NOD.Rag1tm1Mom strain (Jackson Laboratories) and NOD.CB17-Prkdcscid/Lt strain52 (ARC). NODk.insHEL mice were backcrossed to NOD.B10-Idd9/Wicker (Taconic strain 905)53, NOD.B10-Idd3/Idd10/Idd18 (Taconic strain 1538)54 and NOD.B10-Idd3/Idd5/Wicker (Taconic strain 1591)55 strains to create the NODk. insHEL.Idd9, NODk.insHEL.Idd3, NODk.insHEL.Idd10/Idd18, NODk. insHEL.Idd3/Idd10/Idd18, NODk.insHEL.Idd5 and NODk.insHEL.Idd3/Idd5 strains. B10k.Rag1−/−.insHEL and NODk.Rag1−/− mice were crossed to generate F1.Rag1−/−.insHEL mice, which were intercrossed to generate F2.Rag1−/−. insHEL mice. C57BL/6.ERAIGFP mice, previously generated by Miura et al.56, were crossed to B10k.Rag1−/−.insHEL and NODk.Rag1−/−.insHEL mice to generate B10.ERAIGFP.insHEL and (B6 × NOD)F1.ERAIGFP.insHEL mice, respectively. C57BL/6.Glis3+/− mice57 were crossed to B10k.Rag1−/−.insHEL mice to generate B10.Glis3+/−.insHEL mice. C57BL/6.Lig4+/Y288C mice23 were crossed to B10k.Rag1−/−.insHEL mice to generate B10.Lig4+/Y288C.insHEL mice. Radiation chimeras were constructed as described previously49. Mice were fed a standard chow diet, except in the 10% fat experiment, where mice were provided with a 10% high-fat diet (Sniff). Allocation to the treatment group was made randomly at weaning, at the cage level. All experiments were performed in accordance with the University of Leuven Animal Ethics Committee guidelines or the Australian National University Animal Ethics Committee. Sample sizes for mouse experiments were chosen in conjunction with the Animal Ethics Committee to allow for robust sensitivity without excessive use. Unless otherwise mentioned, male mice were exclusively used.
Diabetes incidence study
Mice were kept for 20–28 weeks and tested for diabetes monthly by blood glucose and weekly by urine assessment, with a positive indication being followed by twice-weekly blood testing. Mice were diagnosed as diabetic when the blood glucose concentration was over 260 mg/dl (14.4 mM) after 2–3 h of fasting for two sequential tests. Glucose and insulin tolerance tests were performed by injecting glucose (2 g/kg body weight) or insulin (1 U/kg body weight) intraperitoneally in mice fasted for 6–7 h. Tail vein blood was tested by a Contour glucometer. Assessments of plasma insulin, proinsulin and C-peptide levels were performed using commercial ELISA kits, according to the manufacturer's instructions (insulin, proinsulin and C-peptide mouse ELISA kits, R&D Systems Quantikine). Assays were performed with blinding, with mice coded by number until experimental end.
In vitro assessment of glucotoxicity
Islets were isolated from pancreata as previously described58. Pancreata were digested with 0.8 mg/ml collagenase NB 8 (Serva) at 37 °C, and islets were isolated through density-gradient centrifugation with Dextran T70 (Pharmacosmos) solution and handpicked under a dissection microscope. For the assessment of islet susceptibility to glucotoxicity, pancreatic islets of 10- to 12-week-old mice were incubated in 10% RPMI-1640 with 5 mM and 25 mM glucose using a previously described protocol59. 2.5 μM 17-DMAG was added to indicated cultures. The percentage of islet cell death was assessed after 3 d of culture. Islets were stained with propidium iodide (Life Technologies) and Hoechst 33342 (Life Technologies) and scored under a florescence microscope.
Imaging
Standard histology was performed using pancreata preserved in 4% formaldehyde. Hematoxylin and eosin staining was performed by Histology Consultation Services, and pathology reports were generated by BioGenetics (Greenbank).
For immunofluorescence staining of islets, frozen pancreas sections were stained with goat antibody to insulin A (sc-7839, Santa Cruz Biotechnology), rabbit antibody to glucagon (sc-7780, Santa Cruz Biotechnology), rabbit antibody to MANF (SAB3500384, Sigma-Aldrich), donkey anti-goat Alexa Fluor 546 (A-11056, Life Technologies), donkey anti-rabbit Alexa Fluor 488 (A21206, Life Technologies) and DAPI (D1306, Life Technologies). Paraffin pancreas sections were stained with goat antibody to insulin A (sc-7839, Santa Cruz Biotechnology), antibody to phosphorylatedH2A.X (ab11174, Abcam) and DAPI (D1306, Life Technologies) using pH 9.0 Tris antigen retrieval. Images were acquired with an LSM 510 Meta confocal microscope (Zeiss). Quantification was performed using ImageJ v1.48 to measure integrated density.
For immunohistochemistry staining of islets, pancreata were preserved in 10% neutral phosphate-buffered formalin, and slides were heat treated for 20 min in CC1 buffer (Tris-borate-EDTA buffer, pH 8.0–8.5) before staining with rabbit antibody to activated caspase-3 (Abcam, ab13847) and guinea pig antibody to insulin (A0564, Dako). Signal was developed using the ultraUview Universal DAB detection kit and Universal Alkaline Phosphatase Red Detection kit, respectively. Slides were then counterstained with hematoxylin and eosin. Cells were scored as apoptotic if they demonstrated both blebbing of the nucleus and extensive cytoplasmic staining of activated caspase-3.
Transmission electron microscopy was performed on freshly isolated pancreatic islets from 10- to 12-week-old mice, which were Epon-embedded according to a standard protocol. Ultrathin sections (50 nm) were prepared, counterstained with uranyl acetate and lead acetate, and imaged with a JEOL JEM1400 electron microscope. Five cells from each of 2–3 islets per mouse were imaged, and insulin granules were quantified using ImageJ.
Flow cytometry
Flow cytometry of pancreatic islet cells was performed as previously described60. Islet cells were fixed and permeabilized with the Foxp3 staining kit (eBioscience) and then stained with APC-conjugated antibody to insulin (clone 182410, R&D Systems), Alexa Fluor 488–conjugated antibody to GFP (Invitrogen) and antibody to glucagon (clone 199017, R&D Systems; PerCP-Cy5.5 labeled using the Lightning-Link labeling kit from Innova Biosciences).
Functional assays
MEFs were generated from B10k.Rag1−/− and NODk.Rag1−/−mice and were treated with 5 μM etoposide (Sigma-Aldrich) for 1 h. MEFs were allowed to recover for 15 h before fixation and staining with Alexa Fluor 647–conjugated antibody to phosphorylated H2A.X (Ser139; clone 2F3, Biolegend) for flow cytometry.
CHO cells and Xrcc4−/− CHO cells were a kind gift from P. Jeggo (University of Sussex). Xrcc4 expression vectors were constructed with the B10 or NOD allele of Xrcc4 (or GFP control), preceded by a chicken β-actin intron and followed by a sequence encoding GFP linked to the Xrcc4 C terminus via a T2A peptide, under the control of the chicken β-actin promoter and CMV enhancer. Xrcc4−/− CHO cells were transfected using Lipofectamine 3000 (Invitrogen), followed by treatment with 5 μM etoposide (Sigma-Aldrich) for 1 h, and cells were allowed to recover for 15 h. Cell sorting was performed for GFP+ CHO cells on a FACS Aria II (Becton Dickinson). GFP+ cells were stained with the Zombie Aqua Fixable Viability kit (Biolegend) before fixation and staining with Alexa Fluor 647–conjugated antibody to phosphorylated H2A.X (Ser139; clone 2F3, Biolegend) and propidium iodide (eBioscience) for flow cytometry analysis.
RNA sequencing analysis
Total RNA was prepared from isolated islets by RNeasy Mini kit (Qiagen). An RNA library was prepared with TruSeq stranded mRNA library prep (Illumina), and RNA-seq was performed using the HiSeq 2000 300-Gb flow cell. Sequence reads were mapped to the mouse reference genome (assembly GRCm38.73) using TopHat v2.0.8b61. For assessment of insHEL expression, reads were aligned to a custom genome build combining the Ensembl build of chicken chromosome 1, containing LYZ (ENSGALG00000009963) with its corresponding transcript ENSGALT00000016916, with the mouse genome (Ensembl builds Galgal4.75 and GRCm38.75, respectively) using TopHat v 2.0.11.
Only primary mapped reads and reads with mapping quality ≥20 were retained. Reads were counted at the gene level with the HTSeq package (htseqcount; version 0.5.4). Differentially expressed genes and PCA components were obtained with the DESeq package of Bioconductor (version 1.12.1; ref. 61). The resulting P values were corrected for multiple testing to control the false discovery rate (FDR). PCA plots were created using either the full genome, the Xbp1 gene set (defined by MSigDB; V$XBP1_01) or the ribosome gene set (KEGG_RIBOSOME). GSEA62 was performed on the GenePattern webserver using MSigDB annotated gene sets combining curated gene sets and motif gene sets. FDR cutoffs of <0.05 and <0.001 were used for significant enrichment. Using the <0.001 threshold, a network analysis was performed using Enrichment Map in Cytoscape.
Variant calling was performed after first removing duplicate reads from the TopHat mapped reads using SAMtools (rmdup; version 0.1.19). Base score recalibration and local realignment were carried out using the Genome Analysis Toolkit (GATK)63,64. SNPs were called simultaneously on 12 samples with the GATK UnifiedGenotyper procedure. SNPs were retained if their quality score was greater than 50 and if the coverage was above 10× for at least one sample. Secondary analysis removed variants that were not differentially present in the NOD and B10 base strains (requiring >75% variant reading in more than half the samples of one strain and <25% variant readings in more than half the samples of the reciprocal strain).
RNA-seq expression data on human pancreatic islets from n = 119 donors including n = 14 donors diagnosed with T2D were available from the HTL of the Lund University Diabetes Centre (LUDC) (see ref. 28 for details). Across-sample normalization for each was performed using the TMM normalization method65. Correlations between gene expression were calculated with the non-parametric Spearman method. Differential gene expression analysis for CDKN1A, H2AFX, CDKN1A, XRCC4 and LIG4 between diabetic individual and healthy controls was performed by non-parametric Mann-Whitney U test. All calculations were performed using R.
Immunoblotting
Samples were homogenized in lysis buffer and sonicated before immunoblot analysis. 10–20 μg of lysate was run on 4–12% Bis-Tris acrylamide gels (NuPAGE Precast Gel System, Life Technologies) before blotting on a PVDF membrane (GE Healthcare). Membranes were incubated with mouse antibody to Xrcc4 (1:500 dilution; Santa Cruz Biotechnology, sc-365055), rabbit antibody to Lig4 (1:250 dilution; Santa Cruz Biotechnology, sc-28232), rabbit antibody to XLF (1:2,000 dilution; Sigma-Aldrich, V9264), mouse antibody to vinculine (1:2,000 dilution; Sigma-Aldrich, V9264), rabbit antibody to PC1 and PC3 (1:1,000 dilution; Millipore, AB10553) or rabbit antibody to PC2 (1:200 dilution; Millipore, AB15610). Proteins were visualized using Western Lightning Plus-ECL (PerkinElmer) and the LAS-4000 imaging system (Fuji). Quantification was performed using AIDA software (Raytest, version 4.27).
Proteome analysis
Differential proteome analysis was performed on isolated islets from B10k.Rag1−/−.insHEL and NODk.Rag1−/−.insHEL mice, at 12 weeks of age, by post-metabolic peptide labeling using different N-hydroxysuccinimide (NHS) esters of propionic acid: [12C3]propionate or [13C3]propionate66. In brief, each sample was lysed by applying three freezethaw cycles and digested overnight at 37 °C by endoproteinase Lys-C (1:500 protease/substrate ratio). The generated peptides were labeled in 20 mM of the different NHS esters. Equal amounts of peptide material were mixed, and peptides were fractionated by reversed-phase HPLC66,67. Then, peptide fractions eluted 20 min apart were pooled to reduce the number of LC-MS/MS runs to 20 per sample. The obtained peptide mixtures were introduced into an LC-MS/MS system, the Ultimate 3000 RSLC nano (Thermo Fisher Scientific) in-line system connected to an LTQ Orbitrap Velos (Thermo Fisher Scientific), for analysis. Samples were first loaded on a trapping column (made in house; 100-μm internal diameter × 20 mm, 5-μm beads C18 Reprosil-HD, Dr. Maisch). After backflushing from the trapping column, the sample was loaded on a reversed-phase column (made in house; 75-μm internal diameter × 150 mm, 3-μm beads C18 Reprosil-HD, Dr. Maisch). Peptides were loaded with solvent A (0.1% trifluoroacetic acid and 2% acetonitrile) and separated with a linear gradient from 2% solvent A′ (0.1% formic acid) to 55% solvent B′ (0.1% formic acid and 80% acetonitrile) at a flow rate of 300 nl/min followed by a wash reaching 100% of solvent B′. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the ten most abundant peaks in a given MS spectrum. In the LTQ Orbitrap Velos, full-scan MS spectra were acquired in the Orbitrap at a target value of 1 × 106 with a resolution of 60,000. The ten ions with the highest intensity were then isolated for fragmentation in the linear ion trap, with a dynamic exclusion of 30 s. Peptides were fragmented after filling the ion trap at a target value of 1 × 104 ion counts. From the MS/MS data in each LC run, Mascot generic files were created using Distiller software (version 2.4.3.3; Matrix Science). While generating these peak lists, grouping of spectra was allowed in Distiller with a maximal intermediate retention time of 30 s, and a maximum intermediate scan count of 5 was used where possible. Grouping was performed with 0.005 Th precursor tolerance. A peak list was only generated when the MS/MS spectrum contained more than ten peaks. There was no deisotoping, and the relative signal-to-noise limit was set at 2. Peak lists were searched with the Mascot search engine (Matrix Science) using the Mascot Daemon interface (version 2.4; Matrix Science). Spectra were searched against the SwissProt database. Variable modifications were set to pyroglutamate formation of N-terminal glutamine and acetylation of the N terminus. Methionine oxidation was set as a fixed modification. Mass tolerance on peptide precursor ions was set to 10 ppm (with Mascot's 13C option set to 1), and mass tolerance on peptide fragment ions was set to 0.5 Da. The peptide charge was set to 1+, 2+, 3+, and the instrument setting was put on ESI-TRAP. The enzyme was set to endoproteinase Lys-C, allowing for one missed cleavage; cleavage was also allowed when lysine was followed by proline. Only peptides that were ranked first and scored above the corresponding Mascot threshold score, set at 99% confidence, were withheld. Identified peptides were quantified using Mascot Distiller Toolbox version 2.4.0 (MatrixScience) in the precursor mode. All data management was performed with ms_lims68. Further data analysis was performed using R embedded in Knime. The data were further filtered by removing all peptides smaller than 8 amino acids, and all protein identifications without two peptide identifications were removed.
Molecular modeling
The starting coordinates for the Xrcc4-Lig4 complex were taken from the crystal structure (3II6)69 deposited in the Protein Data Bank. Variants were directly added with the leap tool of Amber 14 upon topology creation. The two resulting systems have been immersed in a rectangular box and further solvated with TIP3P water molecules70 and neutralized by adding 16 Na+ counterions, for a total of 216,438 and 216,440 atoms for the Ala27.Glu125 and Thr27.Asp125 complexes, respectively. The topology of the systems was built using the Amber 10 all-atom force field, with the parmbsc0 force field corrections71, and the simulations were run with Amber 14. Relaxation of solvent molecules and Na+ ions was initially performed with subsequent steps of 200 ps at 50, 100, 150, 200 and 250 K up to a final temperature of 300 K in the NPT ensemble. The systems were simulated in the NVT ensemble for 100 ns each in periodic boundary conditions, using a cutoff of 10 Å for the evaluation of short-range non-bonded interactions and the Particle Mesh Ewald method for the long-range electrostatic interactions72. The temperature was kept constant at 300 K, using Langevin dynamics73, whereas pressure was fixed at 1 atm through the Langevin piston method74. The SHAKE75 and SETTLE76 algorithms were used to restrain bond lengths, for the solute and water molecules, respectively. The atomic positions were saved every 250 steps (0.5 ps) for the analysis that was performed with the Gromacs 4.6 package. Graphs and images were obtained with the Grace program and UCSF Chimera, respectively.
Genetic mapping
Mouse DNA was isolated with the DNeasy Blood and Tissue kit (Qiagen). SNP analysis was performed on the Illumina platform by employing the GoldenGate method and exported by GenomeStudio. 1,449 SNP loci were selected from the Wellcome-CTC mouse strain SNP set to genotype the (B10 × NOD)F2 mice. QTL analysis was performed on the basis of the presence or absence of a diagnosis of diabetes at 28 weeks of age. Standard interval mapping was performed using the R/qtl software package (version 1.23-16). LOD scores were calculated as a measurement for linkage at each marker. The statistical significance of the results was evaluated by testing 1,000 permutation replicates. Candidate intervals were defined using an LOD drop of 1.
SNP association
Human linkage between SNP variants and selected glycemic traits (T2D, HbA1C, fasting glucose, 2-h glucose, HOMA-B, fasting insulin, 2-h insulin, HOMA-IR, proinsulin levels) was reported from data hosted by the AMP T2D-GENES Program, Sigma (December 2015 data set).
Statistics
Statistics for transcriptomics, proteomics and genetic mapping are described in the relevant sections. Diabetes incidence curves were analyzed by log-rank test, with P < 0.05 used as the threshold for statistical significance. For other mouse experiments, statistical analysis was performed using Prism (GraphPad). A significance threshold of 5% in a Student's t test was maintained throughout the study, including hypothesis-based RNA-seq expression analysis. For human data set calculations, where data were not assumed to be normally distributed, analyses were performed using non-parametric Mann-Whitney U tests in R. No results were excluded from analysis.
Supplementary Material
Acknowledgments
The authors thank P. Jeggo, C. Mathieu, D. Gray and A. Goris for critical insights and D. Pombal, J. Sreenivasan, A. Bullman, M. Koina and T. Dagpo for technical assistance. We acknowledge the Human Tissue Laboratory (HTL) of the Lund University Diabetes Centre (LUDC) for providing high-quality data from human pancreatic islets. This work was supported by the VIB, European Research Council (ERC) and a Juvenile Diabetes Research Foundation (JDRF) Career Development Award (A.L.) and by the National Health and Medical Research Council of Australia (project grant 1028108; C.J.N.). N.O. and V. Lyssenko acknowledge support by a Strategic Research Grant from the Swedish Research Council (2009-1039).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Author Contributions: A.L., S.M.S., S.L., N.P., C.J.N., D.R.L. and C.C.G. designed the study. J.D., L.T., S.S., V.D.-A., J.E.G.-P., E.P., D.D.M., D.F., L.O., J.V., G.C.-R., J.D., L.S., S.M.S., J.E.D. and A.L. performed the experiments. E.J.C., N.O., V. Lyssenko, V. Lagou, J.A., K.G., D.L., M.A.L. and A.L. analyzed results. A.M.J. provided reagents. A.L. wrote the manuscript. All authors discussed results and read and approved the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Wicker LS, et al. Type 1 diabetes genes and pathways shared by humans and NOD mice. J Autoimmun. 2005;25(suppl.):29–33. doi: 10.1016/j.jaut.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Islam MS, Wilson RD. Experimentally induced rodent models of type 2 diabetes. Methods Mol Biol. 2012;933:161–174. doi: 10.1007/978-1-62703-068-7_10. [DOI] [PubMed] [Google Scholar]
- 3.Butler AE, et al. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 4.Bradfield JP, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifes multiple associated loci. PLoS Genet. 2011;7:e1002293. doi: 10.1371/journal.pgen.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium & Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium. Genome-wide transancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fortune MD, et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet. 2015;47:839–846. doi: 10.1038/ng.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senée V, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 8.Delépine M, et al. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 9.Janssens S, Pulendran B, Lambrecht BN. Emerging functions of the unfolded protein response in immunity. Nat Immunol. 2014;15:910–919. doi: 10.1038/ni.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tersey SA, et al. Islet beta-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes. 2012;61:818–827. doi: 10.2337/db11-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engin F, et al. Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5:211ra156. doi: 10.1126/scitranslmed.3006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Back SH, Kaufman RJ. Endoplasmic reticulum stress and type 2 diabetes. Annu Rev Biochem. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planas R, Pujol-Borrell R, Vives-Pi M. Global gene expression changes in type 1 diabetes: insights into autoimmune response in the target organ and in the periphery. Immunol Lett. 2010;133:55–61. doi: 10.1016/j.imlet.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Chaparro RJ, et al. Nonobese diabetic mice express aspects of both type 1 and type 2 diabetes. Proc Natl Acad Sci USA. 2006;103:12475–12480. doi: 10.1073/pnas.0604317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liston A, et al. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity. 2004;21:817–830. doi: 10.1016/j.immuni.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specifc T cells. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 17.Hill SW, Kipp DE, Melchers I, Frelinger JA, Sercarz EE. Multiple H-2 and non-H-2 genes controlling the antilysozyme response: alternative gene constellations can lead to responsiveness. Eur J Immunol. 1980;10:384–391. doi: 10.1002/eji.1830100513. [DOI] [PubMed] [Google Scholar]
- 18.Izumi T, et al. Dominant negative pathogenesis by mutant proinsulin in the Akita diabetic mouse. Diabetes. 2003;52:409–416. doi: 10.2337/diabetes.52.2.409. [DOI] [PubMed] [Google Scholar]
- 19.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 20.Wicker LS, Todd JA, Peterson LB. Genetic control of autoimmune diabetes in the NOD mouse. Annu Rev Immunol. 1995;13:179–200. doi: 10.1146/annurev.iy.13.040195.001143. [DOI] [PubMed] [Google Scholar]
- 21.Puebla-Osorio N, et al. A novel Ku70 function in colorectal homeostasis separate from nonhomologous end joining. Oncogene. 2014;33:2748–2757. doi: 10.1038/onc.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavana O, Puebla-Osorio N, Sang M, Zhu C. Absence of p53-dependent apoptosis combined with nonhomologous end-joining defciency leads to a severe diabetic phenotype in mice. Diabetes. 2010;59:135–142. doi: 10.2337/db09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 24.Nogueira TC, et al. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genet. 2013;9:e1003532. doi: 10.1371/journal.pgen.1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindahl M, et al. MANF is indispensable for the proliferation and survival of pancreatic beta cells. Cell Reports. 2014;7:366–375. doi: 10.1016/j.celrep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall E, et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC Med. 2014;12:103. doi: 10.1186/1741-7015-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sone H, Kagawa Y. Pancreatic beta cell senescence contributes to the pathogenesis of type 2 diabetes in high-fat diet–induced diabetic mice. Diabetologia. 2005;48:58–67. doi: 10.1007/s00125-004-1605-2. [DOI] [PubMed] [Google Scholar]
- 28.Fadista J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes infuencing glucose metabolism. Proc Natl Acad Sci USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leiter EH, et al. Unexpected functional consequences of xenogeneic transgene expression in beta-cells of NOD mice. Diabetes Obes Metab. 2007;9(suppl. 2):14–22. doi: 10.1111/j.1463-1326.2007.00770.x. [DOI] [PubMed] [Google Scholar]
- 30.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apostolou A, Shen Y, Liang Y, Luo J, Fang S. Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress–induced cell death. Exp Cell Res. 2008;314:2454–2467. doi: 10.1016/j.yexcr.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davenport EL, et al. Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood. 2007;110:2641–2649. doi: 10.1182/blood-2006-11-053728. [DOI] [PubMed] [Google Scholar]
- 33.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 34.Leitman J, et al. Herp coordinates compartmentalization and recruitment of HRD1 and misfolded proteins for ERAD. Mol Biol Cell. 2014;25:1050–1060. doi: 10.1091/mbc.E13-06-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liston A, et al. Gene dosage—limiting role of Aire in thymic expression, clonal deletion, and organ-specifc autoimmunity. J Exp Med. 2004;200:1015–1026. doi: 10.1084/jem.20040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesage S, et al. Failure to censor forbidden clones of CD4 T cells in autoimmune diabetes. J Exp Med. 2002;196:1175–1188. doi: 10.1084/jem.20020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wick G, et al. The role of genetically-determined primary alterations of the target organ in the development of spontaneous autoimmune thyroiditis in obese strain (OS) chickens. Immunol Rev. 1986;94:113–136. doi: 10.1111/j.1600-065x.1986.tb01167.x. [DOI] [PubMed] [Google Scholar]
- 38.Project Group DIAMOND. Incidence and trends of childhood type 1 diabetes worldwide 1990–1999. Diabet Med. 2006;23:857–866. doi: 10.1111/j.1464-5491.2006.01925.x. [DOI] [PubMed] [Google Scholar]
- 39.Patterson CC, Dahlquist GG, Gyürüs E, Green A, Soltész G. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: a multicentre prospective registration study. Lancet. 2009;373:2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- 40.Study Group TEDDY. The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Ann NY Acad Sci. 2008;1150:1–13. doi: 10.1196/annals.1447.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laybutt DR, et al. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia. 2007;50:752–763. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- 42.Oprescu AI, et al. Free fatty acid–induced reduction in glucose-stimulated insulin secretion: evidence for a role of oxidative stress in vitro and in vivo. Diabetes. 2007;56:2927–2937. doi: 10.2337/db07-0075. [DOI] [PubMed] [Google Scholar]
- 43.Betts P, Mulligan J, Ward P, Smith B, Wilkin T. Increasing body weight predicts the earlier onset of insulin-dependant diabetes in childhood: testing the ‘accelerator hypothesis’ (2) Diabet Med. 2005;22:144–151. doi: 10.1111/j.1464-5491.2004.01368.x. [DOI] [PubMed] [Google Scholar]
- 44.Gale EA, Gillespie KM. Diabetes and gender. Diabetologia. 2001;44:3–15. doi: 10.1007/s001250051573. [DOI] [PubMed] [Google Scholar]
- 45.Donahue RP, Bean JA, Donahue RA, Goldberg RB, Prineas RJ. Insulin response in a triethnic population: effects of sex, ethnic origin, and body fat. Miami Community Health Study Diabetes Care. 1997;20:1670–1676. doi: 10.2337/diacare.20.11.1670. [DOI] [PubMed] [Google Scholar]
- 46.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to infammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 47.Cervin C, et al. Genetic similarities between latent autoimmune diabetes in adults, type 1 diabetes, and type 2 diabetes. Diabetes. 2008;57:1433–1437. doi: 10.2337/db07-0299. [DOI] [PubMed] [Google Scholar]
- 48.Podolin PL, et al. I-E+ nonobese diabetic mice develop insulitis and diabetes. J Exp Med. 1993;178:793–803. doi: 10.1084/jem.178.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akkaraju S, et al. A range of CD4 T cell tolerance: partial inactivation to organspecific antigen allows nondestructive thyroiditis or insulitis. Immunity. 1997;7:255–271. doi: 10.1016/s1074-7613(00)80528-2. [DOI] [PubMed] [Google Scholar]
- 50.Mombaerts P, et al. RAG-1–deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 51.Ramsey C, et al. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet. 2002;11:397–409. doi: 10.1093/hmg/11.4.397. [DOI] [PubMed] [Google Scholar]
- 52.Prochazka M, Gaskins HR, Shultz LD, Leiter EH. The nonobese diabetic SCID mouse: model for spontaneous thymomagenesis associated with immunodeficiency. Proc Natl Acad Sci USA. 1992;89:3290–3294. doi: 10.1073/pnas.89.8.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyons PA, et al. The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137. Immunity. 2000;13:107–115. doi: 10.1016/s1074-7613(00)00012-1. [DOI] [PubMed] [Google Scholar]
- 54.Lyons PA, et al. Congenic mapping of the type 1 diabetes locus, Idd3, to a 780-kb region of mouse chromosome 3: identification of a candidate segment of ancestral DNA by haplotype mapping. Genome Res. 2000;10:446–453. doi: 10.1101/gr.10.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill NJ, et al. NOD Idd5 locus controls insulitis and diabetes and overlaps the orthologous CTLA4/IDDM12 and NRAMP1 loci in humans. Diabetes. 2000;49:1744–1747. doi: 10.2337/diabetes.49.10.1744. [DOI] [PubMed] [Google Scholar]
- 56.Mao C, Dong D, Little E, Luo S, Lee AS. Transgenic mouse model for monitoring endoplasmic reticulum stress in vivo. Nat Med. 2004;10:1013–1014. doi: 10.1038/nm1004-1013. author reply 1014. [DOI] [PubMed] [Google Scholar]
- 57.Kang HS, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol. 2009;29:6366–6379. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li DS, Yuan YH, Tu HJ, Liang QL, Dai LJ. A protocol for islet isolation from mouse pancreas. Nat Protoc. 2009;4:1649–1652. doi: 10.1038/nprot.2009.150. [DOI] [PubMed] [Google Scholar]
- 59.Gysemans CA, et al. Disruption of the γ-interferon signaling pathway at the level of signal transducer and activator of transcription-1 prevents immune destruction of beta-cells. Diabetes. 2005;54:2396–2403. doi: 10.2337/diabetes.54.8.2396. [DOI] [PubMed] [Google Scholar]
- 60.Kornete M, Beauchemin H, Polychronakos C, Piccirillo CA. Pancreatic islet cell phenotype and endocrine function throughout diabetes development in non-obese diabetic mice. Autoimmunity. 2013;46:259–268. doi: 10.3109/08916934.2012.752462. [DOI] [PubMed] [Google Scholar]
- 61.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghesquiere B, et al. Redox proteomics of protein-bound methionine oxidation. Mol Cell Proteomics. 2011;10006866:M110. doi: 10.1074/mcp.M110.006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thimiri Govinda Raj DB, et al. A novel strategy for the comprehensive analysis of the biomolecular composition of isolated plasma membranes. Mol Syst Biol. 2011;7:541. doi: 10.1038/msb.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helsens K, et al. ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics. 2010;10:1261–1264. doi: 10.1002/pmic.200900409. [DOI] [PubMed] [Google Scholar]
- 69.Sibanda BL, et al. Crystal structure of an Xrcc4–DNA ligase IV complex. Nat Struct Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- 70.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 71.Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 72.Cheatham TE, Miller JL, Fox T, Darden TA, Kollman PA. Molecular-dynamics simulations on solvated biomolecular systems—the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J Am Chem Soc. 1995;117:4193–4194. [Google Scholar]
- 73.Ceriotti M, Bussi G, Parrinello M. Langevin equation with colored noise for constant-temperature molecular dynamics simulations. Phys Rev Lett. 2009;102:020601. doi: 10.1103/PhysRevLett.102.020601. [DOI] [PubMed] [Google Scholar]
- 74.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-pressure molecular-dynamics simulation—the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 75.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-integration of Cartesian equations of motion of a system with constraints—molecular-dynamics of N-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 76.Miyamoto S, Kollman PA. Settle—an analytical version of the shake and rattle algorithm for rigid water models. J Comput Chem. 1992;13:952–962. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


