Metastasis is the primary cause of cancer death. Weichand et al. describe a new mechanism explaining how tumor-associated macrophages contribute to metastatic spread, which involves promoting tumor lymphangiogenesis via S1P receptor 1 and the NLRP3 inflammasome.
Abstract
Metastasis is the primary cause of cancer death. The inflammatory tumor microenvironment contributes to metastasis, for instance, by recruiting blood and lymph vessels. Among tumor-infiltrating immune cells, tumor-associated macrophages (TAMs) take a center stage in promoting both tumor angiogenesis and metastatic spread. We found that genetic deletion of the S1P receptor 1 (S1pr1) alone in CD11bhi CD206+ TAMs infiltrating mouse breast tumors prevents pulmonary metastasis and tumor lymphangiogenesis. Reduced lymphangiogenesis was also observed in the nonrelated methylcholanthrene-induced fibrosarcoma model. Transcriptome analysis of isolated TAMs from both entities revealed reduced expression of the inflammasome component Nlrp3 in S1PR1-deficient TAMs. Macrophage-dependent lymphangiogenesis in vitro was triggered upon inflammasome activation and required both S1PR1 signaling and IL-1β production. Finally, NLRP3 expression in tumor-infiltrating macrophages correlated with survival, lymph node invasion, and metastasis of mammary carcinoma patients. Conceptually, our study indicates an unappreciated role of the NLRP3 inflammasome in promoting metastasis via the lymphatics downstream of S1PR1 signaling in macrophages.
Introduction
Tumor-associated inflammation contributes to all stages of tumor development (Hanahan and Weinberg, 2011). Among the tumor-infiltrating immune cells that propagate cancer, macrophages constitute a major population. Their density correlates with poor prognosis in a variety of solid tumors, including breast cancer, gastric cancer, lung cancer, and lymphoma (Noy and Pollard, 2014). It is currently unclear whether tumor-associated macrophage (TAM) subsets fulfill distinct tasks during tumor development. Differences in ontology as well as the microenvironmental niche, where TAMs reside, may generate cells with specific functional properties (Franklin et al., 2014; Noy and Pollard, 2014). Mechanisms of tumorigenic TAM activity in the primary tumor range from promoting tumor cell survival and therapeutic resistance and suppressing antitumor immunity, to fostering angiogenesis and invasiveness, which promotes metastatic spread (Noy and Pollard, 2014; Ruffell and Coussens, 2015). At secondary sites, macrophages are involved in tumor cell extravasation and promoting the survival of metastatic tumor cells (Kitamura et al., 2015). Because metastatic disease is the primary cause of patient death, there is an urgent need to understand molecular mechanisms by which the tumor microenvironment, including TAMs, promotes metastasis. Such an understanding may open potential therapeutic strategies to prevent or treat metastatic disease.
The sphingolipid sphingosine-1-phosphate (S1P) possesses pleiotropic biological functions and can regulate migration, proliferation, survival, and differentiation of cells (Kunkel et al., 2013). Because of these properties, S1P emerges as a mediator of tumor development. S1P is produced at cellular membranes by sphingosine kinases (SPHK1 and 2) via phosphorylation of sphingosine. S1P levels in normal tissues, with the exception of the circulation, are tightly restricted to picomolar concentrations by a specific S1P lyase or by S1P phosphatases. In tumors, the S1P rheostat is disturbed, leading to elevated S1P levels that promote tumor growth (Pyne and Pyne, 2010). The majority of S1P’s tumor-promoting effects are transmitted via one of its five distinct G protein–coupled receptors (S1PR1–5), but the individual contribution of distinct S1PRs is largely unknown. Recent studies connected S1PR1 signaling to persistent tumor-promoting STAT3 signaling in tumor and inflammatory cells in transplanted tumor models and experimental metastases (Lee et al., 2010; Deng et al., 2012). Moreover, targeting S1P/S1PR1 signaling attenuated tumor angiogenesis in xenografts (Visentin et al., 2006). In contrast, S1PR2 depletion promoted tumor growth and angiogenesis, confirming the sometimes antithetic actions of individual S1PRs (Weigert et al., 2011). These findings illustrate that targeting tumor-promoting S1PRs may be superior to targeting S1P or its enzymatic machinery itself. Our previous in vitro studies, using human macrophages, suggested that S1PR1 signaling induces a tumor-promoting, anti-inflammatory, and proangiogenic macrophage phenotype (Brecht et al., 2011; Weigert et al., 2011). This phenomenon might explain the anticancer effects observed in animal models upon blocking S1PR1. We therefore asked whether S1PR1 signaling in macrophages would be required for the protumor functions of TAMs in vivo.
Results
S1PR1 deficiency in macrophages prevents pulmonary metastasis
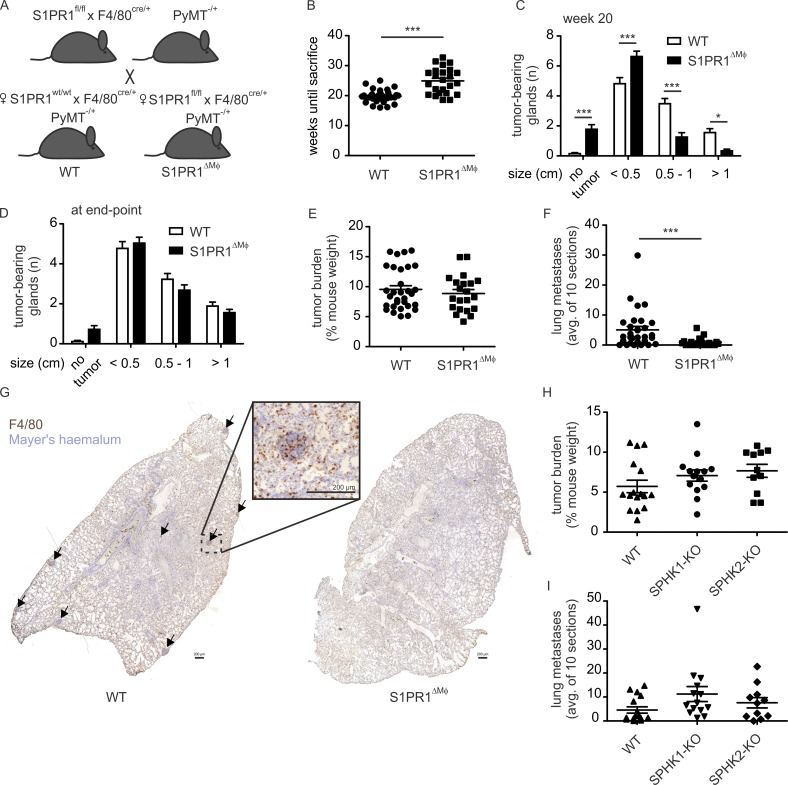
To explore S1PR1 signaling in TAMs, we crossed S1pr1fl/fl F4/80Cre/+ mice (Weichand et al., 2013) into a polyoma middle T (PyMT) background (Lin et al., 2003). These mice express the PyMT oncoprotein in the mammary epithelium, which results in the formation of autochthonous mammary tumors in each gland, starting at 8 wk of age and progressing to metastatic disease after 15 wks. The F4/80-Cre deleter strain was largely uncharacterized. By crossing F4/80Cre/+ mice into a reporter strain (mdTomato/meGFP mice), we observed that Cre recombinase was selectively active in subsets of F4/80hi macrophages but not in other cells expressing F4/80 such as monocytes and eosinophils (Fig. S1). Next, we compared primary tumor development between S1pr1wt/wt F4/80Cre/+ PyMT+/− (WT) and S1pr1fl/fl F4/80Cre/+ PyMT+/− (S1PR1ΔMΦ) animals (Fig. 1 A). Tumor size was scored, and animals were killed at the end point, when at least one tumor reached a size of >1 cm in diameter. We observed a moderate but significant delay in tumor development in S1PR1ΔMΦ compared with WT animals. This became apparent when comparing the time until death (Fig. 1 B) as well as tumor size distribution at 20 wk (Fig. 1 C). At the end point, tumor size distribution (Fig. 1 D) and tumor burden (Fig. 1 E) in S1PR1ΔMΦ animals were comparable to those of WT animals, indicating a minor delay rather than a major disturbance of tumor development. Despite an unchanged tumor burden and size distribution at the end point, the number of pulmonary metastases was strongly reduced (Fig. 1, F and G). Thus, S1PR1 deletion had a minor impact on primary tumor development but a major impact on distant metastasis. The enzymatic source of S1P in PyMT tumors appeared to be redundant, as tumor development in PyMT mice with a genetic deletion of either Sphk1 (SPHK1 KO) or Sphk2 (SPHK2 KO; Fig. 1, H and I) did not phenocopy that in S1PR1ΔMΦ mice. Tumor burden and pulmonary metastasis in these animals was unchanged compared with WT animals.
Figure 1.
S1PR1 deletion in macrophages prevents pulmonary metastasis in breast cancer. (A) Breeding strategy to generate S1pr1wt/wt F4/80Cre/+ PyMT+/− (WT) and S1pr1fl/fl F4/80Cre/+ PyMT+/− (S1PR1ΔMΦ) animals. (B) Weeks until death (until one tumor reached a diameter of >1 cm) in WT (n = 34) or S1PR1ΔMΦ (n = 24) mice. P-value was calculated using two-tailed Student’s t test; ***, P < 0.001. (C and D) The number of glands bearing tumors of a size indicated at 20 wks (C) or the day of death (D) in WT (n = 27 individual animals) or S1PR1ΔMΦ mice (n = 25). Data are means + SEM. P-values were calculated using two-way ANOVA with Bonferroni’s correction; *, P < 0.05; ***, P < 0.001. (E) Tumor burden at the day of death of WT (n = 31) or S1PR1ΔMΦ (n = 21) mice. P-value was calculated using two-tailed Student’s t test. Significant differences were not observed. (F) Number of metastatic nodules in lungs of WT (n = 30) or S1PR1ΔMΦ (n = 21) mice. P-value was calculated using two-tailed Student’s t test; ***, P < 0.001. (G) Representative sections of lung lobes stained with Mayer’s hemalum and an F4/80 antibody (brown). Arrows indicate metastatic lung nodules. Bars, 200 µm. (H and I) Tumor burden (H) and number of metastatic lung nodules (I) of C56BL/6 WT (16), SPHK1−/− (SPHK1-KO; 14), or SPHK2−/− (SPHK2-KO; 11) mice in the PyMT background. P-values were calculated using one-way ANOVA with Bonferroni’s correction. Significant differences were not observed.
S1PR1 deficiency affects neither tumor immune cell infiltrates nor the premetastatic niche
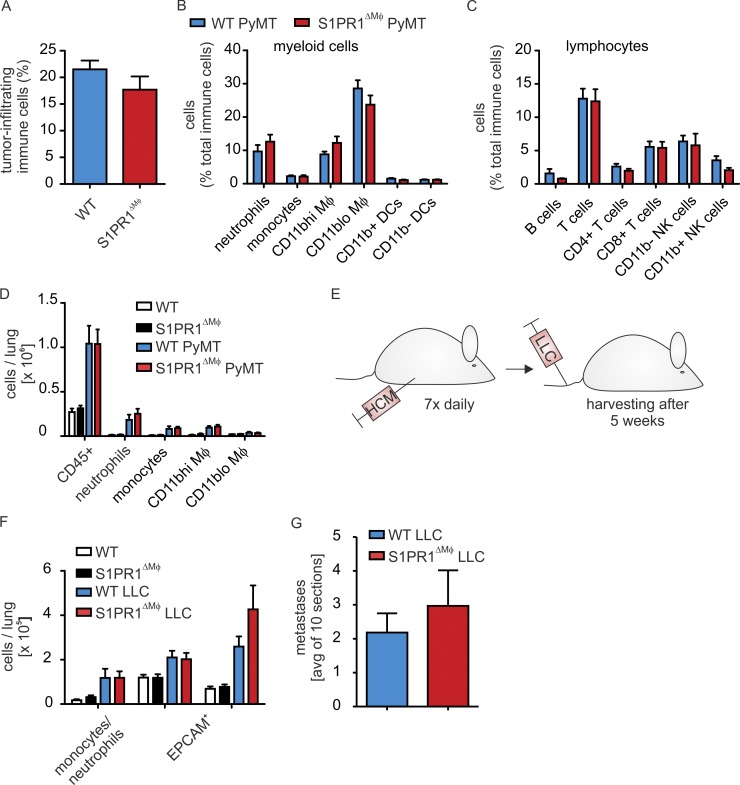
S1PR1 was previously connected to macrophage survival, migration, and/or proliferation (Weigert et al., 2011; Weichand et al., 2013), suggesting a putative change in the macrophage content at the primary tumor site to underlie reduced metastasis. Polychromatic flow cytometry (Fig. S2 A), however, revealed that immune cell infiltrates, including macrophages, were unaltered in S1PR1ΔMΦ versus WT animals (Fig. 2, A–C). Metastasis-associated macrophages (Qian et al., 2009), as well as immature CD11b+ myeloid cells (myeloid-derived suppressor cells; MDSCs; Deng et al., 2012) establish and maintain metastatic growth at distant sites. Their recruitment appears to be an essential component of the premetastatic niche (Sceneay et al., 2012). Despite reduced or absent metastasis, the lungs of S1PR1ΔMΦ animals did not show altered immune cell infiltrates, comprising myeloid cell subsets such as neutrophils and monocytes (that may include MDSCs) and resident alveolar or metastasis-associated macrophages (Fig. 2 D and Fig. S2 B). To further test the involvement of macrophage S1PR1 in establishing the premetastatic niche, we adopted an experimental metastasis assay. We primed mice with hypoxic tumor cell supernatants to create a prometastatic environment in the lung, followed by i.v. injection of cancer cells and monitoring of their pulmonary accumulation (Sceneay et al., 2012; Fig. 2 E). Priming with hypoxic tumor supernatants similarly increased lung myeloid cell infiltrates in both WT and S1PR1ΔMΦ animals (Fig. 2 F). Moreover, the number of epithelial cell adhesion molecule (EpCAM)–expressing tumor cells and the occurrence of macroscopic tumors was similar in both mouse strains (Fig. 2, F and G). Thus, loss of S1PR1 in macrophages did not affect immune cell accumulation at primary or secondary tumor sites and failed to support tumor cell growth in the lung.
Figure 2.
S1PR1 deletion in macrophages affects neither immune cell infiltration nor the premetastatic niche. (A–C) Immune cell populations in tumors of S1pr1wt/wt F4/80Cre/+ PyMT+/− (WT PyMT) and S1pr1fl/fl F4/80Cre/+ PyMT+/− (S1PR1ΔMΦ PyMT) mice were analyzed by polychromatic flow cytometry (n = 12 individual animals each). Relative amounts of total CD45+ immune cells (A), myeloid cell subsets (B), and lymphocyte subsets (C) are shown. Data are means + SEM. P-values were calculated using parametric two-tailed Student’s t test (A) or two-way ANOVA with Bonferroni’s correction (B and C). Significant differences were not observed. (D) Flow cytometric analysis of immune cell populations in lungs of S1pr1wt/wt F4/80Cre/+ (WT, n = 5), WT PyMT (n = 6), S1pr1fl/fl F4/80Cre/+ (S1PR1ΔMΦ; n = 5), and S1PR1ΔMΦ PyMT (n = 6) mice. Data are means + SEM. P-values were calculated using two-way ANOVA with Bonferroni’s correction. Significant differences were not observed. (E–G) Mice were injected with conditioned medium of hypoxic LLCs (HCM) daily for 7 d, followed by injection of LLCs into the tail vein. Two separate experiments using two to three animals of each group were performed. Lungs were harvested after an additional 5 wks (E). Schematic representation of experimental design. (F) Flow cytometric analysis of myeloid cells and EPCAM+ epithelial cells in lungs of untreated (n = 5 each) and HCM/LLC-treated (n = 6 each) WT and MΦ-S1PR1-KO mice. Data are means + SEM. P-values were calculated using two-way ANOVA with Bonferroni’s correction. Significant differences were not observed. (G) Number of metastatic nodules in lungs of HCM/LLC-treated WT and S1PR1ΔMΦ mice (n = 6 each). Data are means + SEM. P-values were calculated using nonparametric two-tailed Student’s t test. Significant differences were not observed.
S1PR1 deficiency prevents tumor lymphangiogenesis
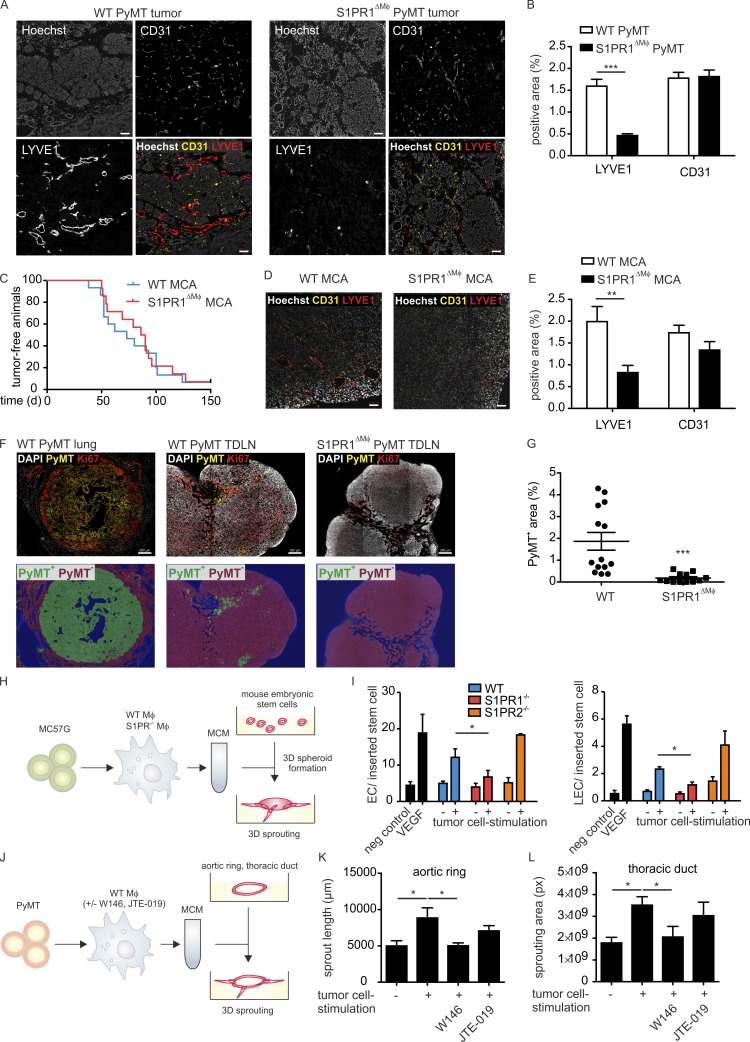
The circulatory system allows tumor cells to travel to distant sites. We asked whether the depletion of S1PR1 in macrophages would affect tumor angiogenesis. Analysis of mammary tumor sections from WT and S1PR1ΔMΦ animals revealed equivalent numbers of CD31hi endothelial cells. However, LYVE1-expressing lymphatic structures were almost absent in the PyMT tumors of S1PR1ΔMΦ mice, indicating dysfunctional lymphangiogenesis (Fig. 3, A and B). Lymphangiogenesis contributes to carcinoma metastasis in various ways. Lymph vessels may provide a route into the venous circulation, provide chemotactic factors for tumor cell mobilization, serve as a niche for cancer stem cells, or negatively modulate antitumor immunity (Alitalo, 2011; Karaman and Detmar, 2014). Macrophages appear to be crucial for tumor lymphangiogenesis by providing growth factors that regulate lymphatic endothelial cell (LEC) proliferation (Kubota et al., 2009; Gordon et al., 2010). This prompted the question whether dysfunctional tumor lymphangiogenesis caused by S1PR1 deletion in macrophages also occurred in other tumors. To evaluate the robustness of the phenotype, we used the methylcholanthrene (MCA)-induced fibrosarcoma model. MCA tumors are strictly inflammation driven and barely metastasize. Deletion of S1PR1 in macrophages did not impair the occurrence or development of MCA-induced fibrosarcomas (Fig. 3 C). Nevertheless, MCA-induced tumors of S1PR1ΔMΦ animals showed markedly disturbed lymphangiogenesis compared with WT mice, whereas angiogenesis, as seen in PyMT tumors, remained unaffected (Fig. 3, D and E). To analyze whether reduced lymphangiogenesis in PyMT tumors correlated with reduced migration of tumor cells to draining lymph nodes, we analyzed axillar lymph nodes of WT PyMT and S1PR1ΔMΦ PyMT mice for PyMT+ cell content and counterstained them with Ki67 to mark proliferating cells. Draining lymph nodes of WT PyMT mice contained areas of PyMT+ cells, but they were mostly absent in lymph nodes of S1PR1ΔMΦ PyMT mice (Fig. 3, F and G). The majority of these PyMT+ cells, however, did not coexpress Ki67: for instance, when compared with highly proliferating cells in the outer zones of PyMT lung metastases. This observation may suggest that PyMT tumor cells were in a transitioning state rather than forming solid metastases in draining lymph nodes. In conclusion, S1PR1ΔMΦ tumors showed markedly disrupted lymphangiogenesis, which correlated with reduced appearance of PyMT+ cells in draining lymph nodes and which may be causatively linked to reduced pulmonary metastasis by a number of the possible mechanisms, as indicated above.
Figure 3.
S1PR1 deletion in macrophages prevents tumor lymphangiogenesis. (A) Confocal microscopy images show LYVE1- and CD31-expressing cells in primary tumor sections of S1pr1wt/wt F4/80Cre/+ PyMT+/− (WT PyMT) and S1pr1fl/fl F4/80Cre/+ PyMT+/− (S1PR1ΔMΦ PyMT) animals. Nuclei were stained with DAPI. Bars, 50 µm. (B) Quantification of LYVE1- and CD31-positive area in confocal images of primary tumors of WT PyMT (n = 13 individual animals) and S1PR1ΔMΦ PyMT (n = 10) mice. Data are means + SEM. P-values were calculated using two-way ANOVA with Bonferroni’s correction. ***, P < 0.001. (C–E) S1pr1wt/wt F4/80Cre/+ (WT) and S1pr1fl/fl F4/80Cre/+ (S1PR1ΔMΦ) mice were injected with 100 µg MCA. (C) Tumor occurrence was monitored for up to 150 d (n = 15 per group). Data are means. P-values were calculated using log-rank test. Significant differences were not observed. (D) Confocal microscopy images show LYVE1- and CD31-expressing cells in primary tumor sections. Nuclei were stained with DAPI. Bars, 50 µm. (E) Quantification of LYVE1- and CD31-positive area in confocal images of primary tumors (n = 5 per group). Data are means + SEM. P-values were calculated using two-way ANOVA with Bonferroni’s correction. **, P < 0.01. (F) Microscopy images show PyMT- and Ki67-expressing cells in lung metastases and tumor-draining axillar lymph nodes of S1pr1wt/wt F4/80Cre/+ PyMT+/− (WT PyMT) and S1pr1fl/fl F4/80Cre/+ PyMT+/− (S1PR1ΔMΦ PyMT) animals. Nuclei were stained with DAPI. Bars, 200 µm. Bottom images indicate the PyMT+ area (green) analyzed with Inform. (G) Quantification of PyMT-positive area in images of draining axillar lymph nodes WT PyMT (n = 14) and S1PR1ΔMΦ PyMT (n = 12) mice. P-values were calculated using nonparametric two-tailed Student’s t test. ***, P < 0.001. (H–L) WT C57BL/6, S1pr1fl/fl F4/80Cre/+ and S1pr2−/− macrophages were stimulated with supernatants of MC57G fibrosarcoma (H) or PyMT mammary carcinoma (K and L) cells for 3 h. Medium was replaced with fresh macrophage medium, and macrophage supernatants were harvested after another 21 h. Macrophage supernatants were collected and added to 3D mouse embryoid bodies (EBs; H), aortic rings (K), or lymphatic rings (L) of WT mice. VEGF was used as a positive control in the EB assay. After another 5 d for EBs, 7 d for aortic rings, and 8–10 d for lymphatic rings, EC and LEC content in EBs was analyzed by flow cytometry, and aortic ring and thoracic duct sprouting were analyzed microscopically. (H) Graphical representation of the experimental setup of the EB assay. (I) Quantification of ECs (left) and LECs (right; n = 5 independent experiments). Data are means + SEM. P-values were calculated using nonparametric two-tailed Student’s t test. *, P < 0.05. (J) Graphical representation of the experimental setup of the aortic ring and lymphatic ring assays. (K) Quantification of aortic ring sprouting (n = 4 independent experiments). Data are means + SEM. P-values were calculated using nonparametric two-tailed Student’s t test. *, P < 0.05. (L) Quantification of lymphatic ring sprouting (n = 4 independent experiments). Data are means + SEM. P-values were calculated using nonparametric two-tailed Student’s t test. *, P < 0.05.
Next, we asked whether macrophage-derived soluble factors provoked lymphangiogenesis downstream of S1PR1. We used an in vitro model in which we stimulated bone marrow–derived macrophages (BMDMs) with tumor cell supernatants. The resulting macrophage supernatants were added to 3D mouse embryoid bodies to monitor their differentiation to vascular cells (Fig. 3 H). Supernatants of WT BMDMs stimulated with tumor cells generated significant amounts of vascular endothelial cells (VECs) and LECs (Fig. 3 I). The numbers of both VECs and LECs were reduced when using macrophages from S1pr1fl/fl F4/80Cre/+ but not S1pr2−/− mice (Fig. 3 I). These findings were confirmed in aortic ring and thoracic duct sprouting assays, in which sprouting angiogenesis of both lymph and blood vessels was increased upon stimulation with supernatants of WT BMDMs previously activated with tumor cells, but not when such supernatants were generated from macrophages with disrupted S1PR1 signaling (Fig. 3, J–L). Thus, S1PR1 signaling in macrophages in vitro promoted angiogenesis in addition to lymphangiogenesis via the production of soluble factors. We conclude that TAMs appear to be a redundant source of angiogenic, but a nonredundant source of lymphangiogenic, growth factors in vivo.
S1PR1 in lymph vessel–associated macrophages promotes NLRP3 expression and IL-1β production
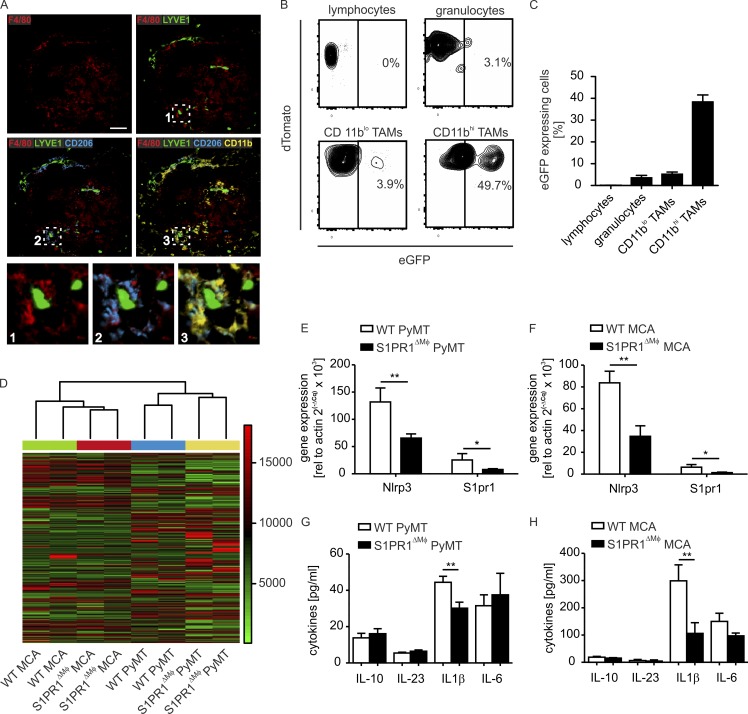
Using multi-epitope ligand cartography (Pierre et al., 2008), we detected colocalization of macrophages and lymph vessels in PyMT tumors. Lymph vessel–associated macrophages were characterized by high expression of CD11b and CD206 (Fig. 4 A). These cells constitute a minor subpopulation of TAMs in PyMT tumors that phenotypically resemble resident mammary gland macrophages (Franklin et al., 2014; Olesch et al., 2015). Importantly, CD11bhi TAMs showed a high level of Cre-dependent recombination in the F4/80Cre/+ background, whereas the major TAM subset in PyMT tumors, CD11blo CD11chi cells, did not (Fig. 4, B and C; and Fig. S1). These data point to the minor subset of CD11bhi macrophages as being responsible for facilitating S1PR1-dependent lymphangiogenesis. To identify molecular mechanisms of lymphangiogenesis downstream of S1PR1, we isolated CD11bhi TAMs from MCA and PyMT tumors of both WT and S1PR1ΔMΦ animals by FACS sorting and analyzed global gene expression. Unsupervised clustering of the resulting mRNA expression datasets revealed distinct gene expression profiles in WT versus S1PR1-KO TAMs from each model (Fig. 4 D). As expected by the heterogeneous nature of tissue macrophages, we observed major differences in gene expression profiles of PyMT TAMs versus MCA TAMs. However, we also identified a common cluster of genes whose expression differed between WT and S1PR1-KO macrophages of both models (Table 1). Focusing on genes down-regulated in S1PR1-KO TAMs, we noticed that a component of the NLRP3 inflammasome, Nlrp3 (or Cias1), was consistently and reproducibly (Fig. 4, E and F) altered. Activation of the NLRP3 inflammasome is a two-step process that requires initial expression of its components by stimuli such as TLR ligands. Second, NLRP3 activators (e.g., pathogen- or damage-associated molecular patterns or crystals) trigger the assembly of the inflammasome. This induces proteolytic activation of CASP1, which in turn cleaves pro–IL-1β to produce the mature cytokine (Kolb et al., 2014). The expression of other NLRP3 inflammasome components was not affected in S1PR1-KO TAMs. However, reduced NLRP3 expression in S1PR1-KO CD11bhi TAMs translated into significantly reduced IL-1β levels in PyMT- and MCA-induced tumor extracellular fluid of S1PR1ΔMΦ compared with WT animals (Fig. 4, G and H).
Figure 4.
S1PR1 deletion in macrophages reduces NLRP3 expression and IL-1β production. (A) Multiepitope ligand cartography analysis of macrophage subsets in a primary tumor section of a S1pr1wt/wt F4/80Cre/+ PyMT+/− (WT PyMT) mouse. Representative images of five independent experiments are shown. F4/80 (red) marks macrophages. Macrophages surrounding LYVE1+ (green) lymphatic vessels express CD206 (blue) and CD11b (yellow). Bar, 50 µm. (B and C) F4/80-driven Cre-dependent recombination in F4/80Cre/+ PyMT+/− mT/mG+/wt mice was determined by flow cytometry. (B) Representative contour plots show eGFP expression in tumor immune cell subpopulations. (C) Relative quantification of eGFP-expressing tumor immune cell subpopulations from tumors of four individual animals. Data are means + SEM. (D) Unsupervised hierarchical clustering of FACS-sorted WT PyMT, S1PR1ΔMΦ PyMT, WT MCA, and S1PR1ΔMΦ MCA CD11bhi TAM gene expression profiles. The dendrogram shows coclustering of PyMT- and MCA-derived TAM irrespective of genotype and coclustering of biological replicates each group. (E and F) Gene expression in FACS-sorted CD11bhi TAMs of WT PyMT and S1PR1ΔMΦ PyMT (n = 14 each; E) or WT MCA and S1PR1ΔMΦ MCA (n = 5 each; F) was determined by quantitative PCR. Data are means + SEM. P-values were calculated using two-way ANOVA with Bonferroni’s correction. *, P < 0.05; **, P < 0.01. (G and H) Cytokine concentrations in tumor extracellular fluid of WT PyMT and S1PR1ΔMΦ PyMT (n = 7 each; G) or WT MCA and S1PR1ΔMΦ MCA (n = 4 each; H) mice were measured using CBA. Data are means + SEM. P-values were calculated using two-way ANOVA with Bonferroni’s correction. **, P < 0.01.
Table 1. Gene expression in MΦ-S1PR1-KO versus WT CD11bhi TAMs.
| Fold change | P-value | Gene symbol |
|---|---|---|
| 2.0 | 0.0006 | Gdpd3 |
| 2.0 | 0.0191 | Xpr1 |
| 2.0 | 0.0064 | Mterfd2 |
| 2.0 | 0.0492 | Nrp2 |
| 1.6 | 0.0439 | Rnf121 |
| 1.6 | 0.0277 | Inpp4b |
| 1.6 | 0.0398 | Fusip1 |
| 1.6 | 0.0228 | Folr2 |
| 1.5 | 0.0325 | Aurka |
| 1.5 | 0.0336 | Pptc7 |
| 1.5 | 0.0461 | Lmna |
| 1.5 | 0.0364 | Birc5 |
| 1.5 | 0.0346 | Sept11 |
| 1.5 | 0.0188 | Nrp2 |
| 1.5 | 0.0362 | Baz2b |
| −1.5 | 0.0366 | Upf2 |
| −1.5 | 0.0345 | Chd1 |
| −1.5 | 0.0204 | Ppfia1 |
| −1.5 | 0.0102 | Wdr51b |
| −1.5 | 0.0142 | Arhgap22 |
| −1.5 | 0.0097 | Helb |
| −1.5 | 0.0146 | Cias1 |
| −1.5 | 0.0015 | Slc35c2 |
| −1.5 | 0.0386 | Mef2c |
| −1.6 | 0.0028 | Nlrp3 |
| −1.6 | 0.0473 | H3f3b |
| −1.6 | 0.0119 | Agl |
| −1.6 | 0.0004 | Abcd3 |
| −1.6 | 0.0030 | Slc35c2 |
| −1.7 | 0.0293 | Slc44a2 |
| −1.7 | 0.0348 | St3gal5 |
| −1.7 | 0.0027 | Anxa3 |
| −1.7 | 0.0083 | Olfml3 |
| −1.7 | 0.0033 | Tnfsf9 |
WT PyMT, S1PR1ΔMΦ PyMT, WT MCA, and S1PR1ΔMΦ MCA CD11bhi TAM (two replicates consisting of TAM from two to three individual animals for each group) were sorted by FACS and subjected to whole-genome mRNA array analysis. A common gene signature between WT and S1PR1ΔMΦ TAMs of the two tumor models is shown. P-values were calculated by two-sample t test after applying a variance filter to reduce microarray data complexity. The false discovery rate according to Benjamini and Hochberg was used to account for the multiple testing. Fold changes between the two groups of each supervised analysis were calculated for each gene.
S1PR1-dependent IL-1β release promotes lymphangiogenesis
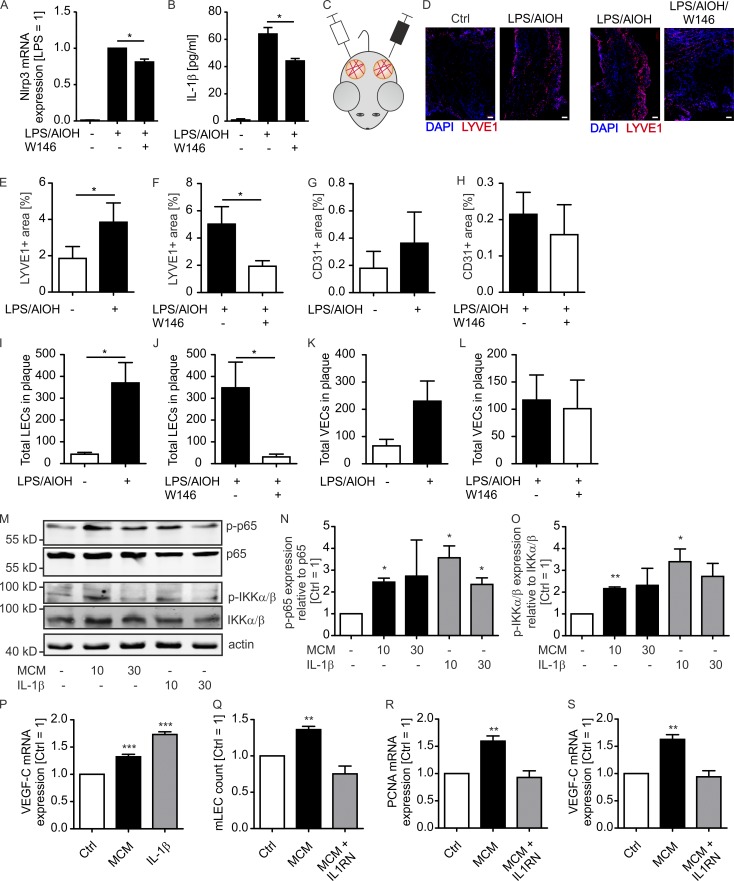
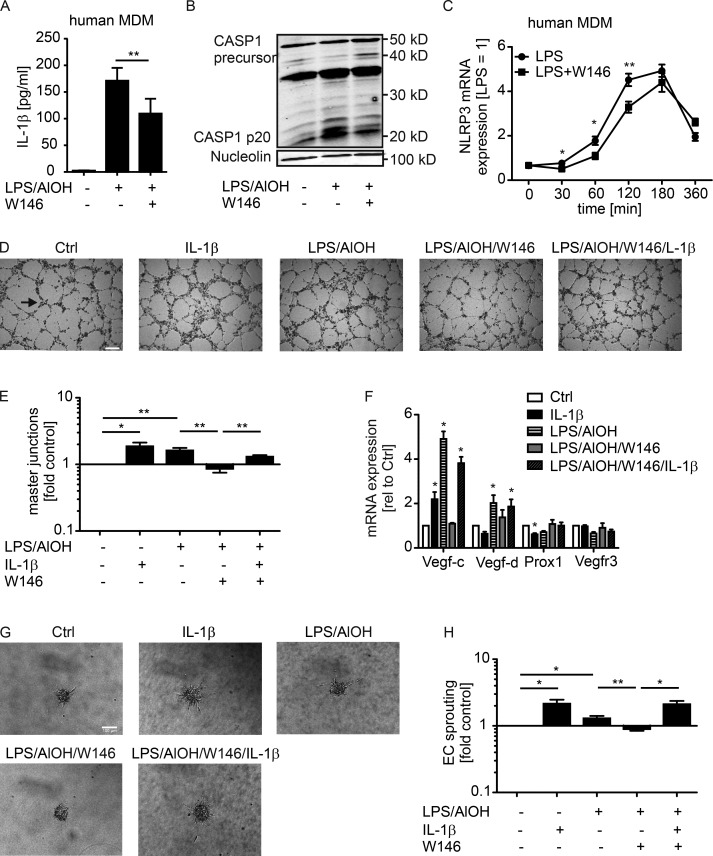
IL-1β levels and signaling are associated with breast cancer progression, invasiveness, and macrophage content (Jin et al., 1997; Kolb et al., 2014). We followed the idea that IL-1β, produced by macrophages downstream of S1PR1, directly affected lymphangiogenesis. To this end, we treated mouse BMDMs with LPS and AlOH to induce NLRP3 expression (Fig. 5 A), inflammasome activation, and IL-1β secretion (Fig. 5 B). Addition of the S1PR1 antagonist W146 reduced NLRP3 expression (Fig. 5 A) and IL-1β production (Fig. 5 B). To analyze whether IL-1β levels translate to prolymphangiogenic activity, we injected control macrophage versus LPS/AlOH-treated macrophage or LPS/AlOH-treated macrophage versus LPS/AlOH/W146-treated macrophage supernatants together with Matrigel into alternate flanks of WT mice (Fig. 5 C). LPS, AlOH, and W146 were removed 2 h after macrophage stimulation by washing. Matrigel plugs were maintained for 9 d and then analyzed by immunohistochemistry and flow cytometry for VEC and LEC content. LPS/AlOH-treated macrophage supernatants significantly increased LYVE1-expressing LEC content but not CD31-expressing VEC content in Matrigel, the latter being generally low compared with LYVE-1-expressing cells. The addition of W146 significantly reduced LEC but not VEC content (Fig. 5, D–H). Comparative results were obtained when analyzing total LEC versus VEC numbers by flow cytometry (Fig. 5, I–L). Thus, IL-1β production by macrophages correlated with lymphangiogenesis in vivo. To further analyze the impact of IL-1β on LECs, lung-derived mouse LECs were generated and cultured with IL-1β or supernatants of macrophages previously stimulated with tumor cell supernatants. Both treatments activated the NF-κB signaling pathway that operates downstream of IL-1R (Fig. 5, M–O) and induced vascular endothelial growth factor C (VEGF-C) expression (Fig. 5 P). Long-term (72-h) treatment of mouse LECs stimulated their proliferation, accompanied by enhanced proliferating cell nuclear antigen (PCNA) and VEGF-C expression. These responses were completely blocked when IL-1R antagonist was added together with macrophage supernatants (Fig. 5, Q–S). These data indicate that IL-1β in macrophage supernatants promotes LEC proliferation, which may occur via autocrine VEGF-C. We next used primary human LECs in an in vitro tube formation assay to investigate whether IL-1β directly acts on human LECs to promote lymphangiogenesis. Human monocyte-derived macrophages were stimulated with LPS/AlOH to activate the inflammasome. Also in human macrophages, cotreatment with W146 reduced IL-1β secretion, correlating with delayed NLRP3 expression and overall reduced inflammasome activity as determined by CASP1 processing (Fig. 6, A–C). The resulting macrophage supernatants, with or without addition of human recombinant IL-1β, were added to primary human LEC cultures on µ-slides to assay tube formation after 6 h. Adding IL-1β to control macrophage supernatants that did not contain IL-1β significantly promoted tube formation (Fig. 6, D and E). The same response was noticed with LPS/AlOH-treated macrophage supernatants. Attenuating IL-1β release with the S1PR1 antagonist W146 in LPS/AlOH-treated macrophages decreased tube formation, which was restored by supplementing with recombinant IL-1β. Conclusively, S1PR1-dependent generation of IL-1β directly acted on LECs to promote lymphangiogenesis (Fig. 6, D and E). Importantly, IL-1β as well as macrophage supernatants containing IL-1β induced VEGF-C expression in primary human LECs, whereas other factors involved in lymphangiogenesis such as VEGF-D, VEGFR3, and the LEC-specific transcription factor prospero homeobox protein 1 (PROX1) were not induced in this fashion (Fig. 6 F). These data support our findings in mouse LECs that autocrine VEGF-C production may be involved in IL-1β–induced lymphangiogenesis. As observed in the mouse system, S1PR1 signaling in macrophages in vitro induced not only LEC tube formation; sprouting of blood endothelial cells upon incubation with macrophage supernatants largely followed the pattern of the LEC tube formation assay (Fig. 6, G and H). Thus, IL-1β induced both blood and lymph angiogenesis in vitro. Nevertheless, at least in mice, IL-1β production by CD11bhi TAMs was not a major driver of angiogenesis, whereas it may be decisively involved in promoting lymphangiogenesis.
Figure 5.
S1PR1-dependent IL-1β formation by mouse macrophages promotes lymphangiogenesis in vivo. (A and B) Mouse BMDM were incubated with 100 ng/ml LPS, 100 µg/ml AlOH, and 1 µM S1PR1 antagonist W146 and washed after 2 h. (A) NLRP3 expression was determined by quantitative PCR (n = 3 independent experiments). Data are means + SEM. P-values were calculated using one-sample t test. *, P < 0.05. (B) IL-1β in supernatants was analyzed by CBA after 24 h (n = 3 independent experiments). Data are means + SEM. P-values were calculated using one-way ANOVA with Bonferroni’s correction. *, P < 0.05. (C–L) Supernatants of control versus LPS/AlOH-stimulated (D–F, I, and J) or LPS/AlOH-stimulated versus LPS/AlOH/W146-stimulated (D, G, H, K, and L) mouse BMDM mixed with Matrigel were implanted into alternate flanks of five WT mice each and maintained for 10 d. Two individual experiments using two or three animals were performed. (C) Schematic representation of experimental design. (D) Representative confocal microscopy images of LYVE1-expressing cells (nuclei stained with DAPI). Bars, 100 µm. (E–H) Quantification of LYVE1- and CD31-positive area. Data are means + SEM. P-values were calculated using nonparametric two-tailed Student’s t test. *, P < 0.05. (I–L) Matrigel was digested and VEC and LEC were quantified by flow cytometry. Data are means + SEM. P-values were calculated using nonparametric two-tailed Student’s t test. *, P < 0.05. (M–P) Mouse LECs isolated from lungs were stimulated with 10 ng/ml mouse recombinant IL-1β or supernatants of macrophages for 10 or 30 min (M–O) or 24 h (P). To generate macrophage supernatants, macrophages were stimulated with supernatants of PyMT mammary carcinoma cells for 3 h, followed by medium replacement and harvesting of macrophage supernatants after another 21 h. (M–O) Western blots show NF-κB-p65 and IKKα/β expression and phosphorylation. Representative Western blot images (M) and quantification of three independent experiments (N and O) are displayed. Data are means + SEM. P-values were calculated using one-sample t test *, P < 0.05. (P) VEGF-C expression was determined by quantitative PCR (n = 6 independent experiments). (Q–S) Mouse LECs were stimulated with macrophage supernatants for 72 h with or without the addition of 100 ng/ml mouse recombinant IL1RN (n = 4 independent experiments). (Q) Cell number was determined using a CASY cell counter, and PCNA (R) and VEGF-C (S) expression was determined by quantitative PCR. Data are means + SEM. P-values were calculated using one-sample t test. **, P < 0.01; ***, P < 0.001.
Figure 6.
S1PR1-dependent IL-1β formation by human macrophages promotes lymphangiogenesis in vitro. (A–C) Human monocyte-derived macrophages (MDMs) were incubated with LPS/AlOH and W146 and washed after 2 h. (A) IL-1β in supernatants was analyzed by CBA after 24 h (n = 9 independent experiments). Data are means + SEM. P-values were calculated using one-way ANOVA with Bonferroni’s correction. **, P < 0.01. (B) Representative Western blot of three independent experiments shows CASP1 proteolysis. (C) MDMs were incubated with LPS and W146 for the times indicated. NLRP3 expression was determined by quantitative PCR (n = 5 independent experiments). Data are means + SEM. P-values were calculated using nonparametric two-tailed Student’s t test. *, P < 0.05; **, P < 0.01. (D–H) Human primary LECs seeded on µ-slides (D, E, and H) or HUVECs grown as 3D spheroids (F and G) were stimulated with macrophage supernatant as described in A with or without 10 ng/ml recombinant IL-1β, or 10 ng/ml recombinant IL-1β was added alone. (D) Representative phase-contrast images show tube formation after 6-h culture (master junction indicated by arrow). Bar, 200 µm. (E) Quantification of master junctions relative to control (n = 6 independent experiments). Data are means + SEM. P-values were calculated using one-sample t test. *, P < 0.05; **, P < 0.01. (F) Vegfc, Vegfd, Vegfr3, and Prox1 expression were determined by quantitative PCR after 24 h (n = 4 independent experiments). Data are means + SEM. P-values were calculated using one-sample t test. *, P < 0.05; **, P < 0.01. (G) Representative phase-contrast images show human endothelial cell sprouting after 24-h culture. Bar, 100 µm. (H) Quantification of human endothelial cell sprouting relative to control (n = 4 independent experiments). Data are means + SEM. P-values were calculated using one-sample t test. *, P < 0.05; **, P < 0.01.
NLRP3 expression in macrophages correlates with survival and metastasis in human breast tumors
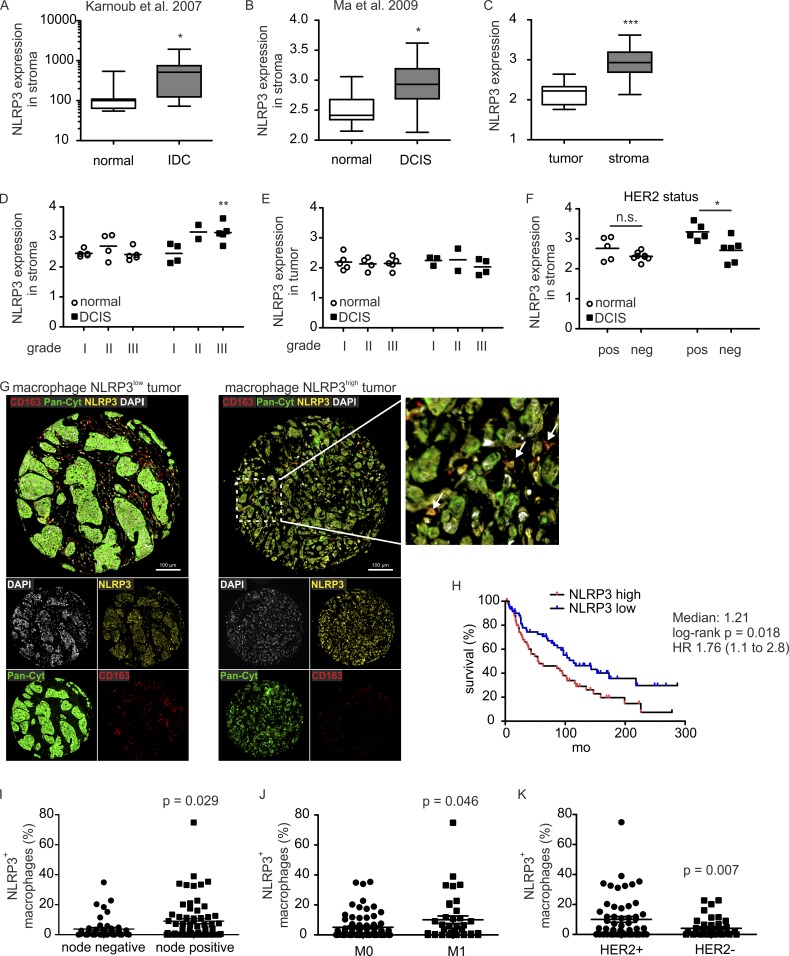
Our data link macrophage S1PR1 to NLRP3 expression, IL-1β production, and lymphangiogenesis. We reasoned that the NLRP3 inflammasome might also be relevant in human cancer. To investigate this hypothesis, we first explored publicly available human breast cancer datasets. Because macrophages do not express the highest levels of S1PR1 among stromal cell populations (e.g., http://www.biogps.org), and IL-1β expression itself does not coincide with the secretion of the active protein, we evaluated NLRP3 expression in stroma of breast cancer patients. Two gene expression datasets compared normal with breast tumor stroma (Karnoub et al., 2007; Ma et al., 2009). Analysis of these datasets revealed increased NLRP3 expression in stroma of inflammatory ductal carcinoma (Karnoub et al., 2007) and ductal carcinoma in situ (DCIS; Ma et al., 2009) compared with normal stroma (Fig. 7, A and B). The latter dataset further allowed the correlation of stromal NLRP3 expression with other tumor parameters. First, NLRP3 expression was higher in stroma compared with tumor cells (Fig. 7 C). Second, NLRP3 expression by DCIS stroma, but not normal stroma or tumor cells, increased with tumor grade, being significantly elevated in stroma of grade III tumors (Fig. 7, D and E). Finally, stromal cells of human epidermal growth factor receptor 2 (HER2)-positive tumors, a marker of aggressiveness and nodal involvement (Chikarmane et al., 2015), displayed higher levels of NLRP3 compared with stroma of HER2-negative tumors (Fig. 7 F). To be more specific in terms of stromal cell involvement, we analyzed tissue microarrays of human invasive breast cancer provided by the Cooperative Human Tissue Network and the Cancer Diagnosis Program for correlation of NLRP3 expression by macrophages with clinical parameters. Improved survival was noted for patients having low levels (below the median of NLRP3+ macrophages) of infiltrating NLRP3+ macrophages (Fig. 7, G and H), which indicates that high numbers of NLRP3-expressing macrophages confer a survival disadvantage for patients with invasive breast cancer. Moreover, lymph node invasion (Fig. 7 I) and distant metastasis (Fig. 7 J) were significantly associated with a higher NLRP3+ macrophage infiltrate. Correlation of HER2 positivity with higher NLRP3+ macrophage infiltrate was observed as well (Fig. 7 K). These data support the notion that enhanced NLRP3 expression in human breast tumor stroma, containing macrophages as the major cell type expressing a functional inflammasome (Kolb et al., 2014), correlates with disease aggressiveness and metastasis.
Figure 7.
NLRP3 expression in macrophages correlates with clinical parameters. Datasets of previous studies, Karnoub et al. (2007) (A) and Ma et al. (2009) (B–F), were analyzed regarding NLRP3 expression using GEO2R. (A and B) Expression of NLRP3 in normal tissue (A, n = 15; B, n = 14) compared with inflammatory ductal carcinoma (n = 7; A) or DCIS (n = 11; B) stroma. P-values were calculated using two-tailed Student’s t test. *, P < 0.05. (C) NLRP3 expression in tumor (n = 9) versus stromal (n = 11) cells. P-values were calculated using two-tailed Student’s t test. ***, P < 0.001. (D and E) NLRP3 expression correlated to tumor grade in unaffected (normal) compared with DCIS stroma (D) and epithelial (tumor; E) cells. P-values were calculated using two-way ANOVA with Bonferroni’s correction. **, P < 0.01. (F) NLRP3 expression relative to HER2 status in normal compared with DCIS stroma. P-values were calculated using two-way ANOVA with Bonferroni’s correction. n.s., not significant; *, P < 0.05. (G–K) Human CBCTR progression tissue microarrays were analyzed for correlation of macrophage NLRP3 expression with clinical parameters. 111 individual tissue cores of invasive human breast cancer were analyzed. (G) Representative microscopy images show cells expressing CD163, pan-cytokeratin (Pan-Cyt), and NLRP3 in mammary carcinoma cores. Nuclei were stained with DAPI. Bars, 100 µm. NLRP3-expressing macrophages (CD163+) are marked by arrows. (H) Survival rates of patients containing low or high numbers of NLRP3-expressing macrophages were compared. P-value was calculated using log-rank test. (I–K) Percentage of NLRP3+ macrophages relative to nodal status (I), occurrence of distant metastasis (M0, no metastasis; M1, metastasis; J), and HER2 status (K) are shown. P-values were calculated using two-tailed Student’s t test.
Discussion
Previous studies identified a prominent role of S1PR1 in tumor progression linked to persistent STAT3 activation in tumor and myeloid cells (Lee et al., 2010; Deng et al., 2012; Degagné et al., 2014). We previously noticed STAT3 signaling downstream of S1PR1 in human macrophages, which contributed to establishing an anti-inflammatory phenotype (Weis et al., 2009). However, in the present study, S1PR1 signaling in CD11bhi CD206+ TAMs did not affect typical STAT3 target genes in macrophages. Rather, a so-far-unexplored S1PR1 signaling circuit in macrophages promoted lymphangiogenesis via NLRP3-dependent IL-1β secretion.
Our global mRNA expression data in TAMs failed to identify previously described macrophage-derived prolymphangiogenic factors such as VEGF-C or VEGF-D as targets of S1PR1 signaling (Kerjaschki, 2005). Rather, NLRP3 expression and the concomitant IL-1β release promoted lymphangiogenesis. Although an involvement of VEGFs in the prolymphangiogenic properties of TAMs in breast tumors cannot be excluded, IL-1β appeared to be a nonredundant contributor, at least in vivo. An indirect contribution of IL-1β to inflammatory and tumor lymphangiogenesis, through the recruitment of macrophages, has been proposed before (Nakao et al., 2011; Watari et al., 2014). However, we suggest direct effects of IL-1β on LECs, which may evoke autocrine production of VEGF-C by lymphatic endothelial cells as indicated before (Ristimäki et al., 1998; Min et al., 2011). The NF-κB signaling pathway, which was activated by IL-1β in LECs in our system, was shown to increase VEGF-C (Zhu et al., 2016). Further studies are required to fully define molecular pathways underlying the prolymphangiogenic effect of IL-1β. There is evidence that IL-1β likely promotes pathological, tumor-associated lymphangiogenesis rather than physiological lymphangiogenesis. A number of germline mutations associated with lymphatic anomalies have been identified, albeit none of them in IL-1 signaling or production pathways (Brouillard et al., 2014). Moreover, macrophages, being the source of lymphangiogenic IL-1β in our study, contribute to pathological but not physiological lymphangiogenesis in adult mice (Kubota et al., 2009). Thus, the side effects of targeting IL-1β to block tumor lymphangiogenesis might be limited.
We propose the NLRP3 inflammasome as a target to prevent metastatic disease. Inflammasomes have been implicated in the development of tumors that are inflammation driven, such as gastric and colorectal cancer (Kolb et al., 2014). Also, chemical carcinogenesis in the inflammation-dependent MCA model demands IL-1β signaling (Krelin et al., 2007). However, the strong reduction of IL-1β in S1PR1 KO TAMs did not influence tumor development in the MCA model but prevented lymphangiogenesis. This different sensitivity to IL-1β may be explained by different sources of IL-1β during the course of tumor development or the close association of CD206+ TAMs with lymph vessels that allows high IL-1β levels in lymph vessel–associated niches in tumors. A similar mechanism is likely the basis for our finding that macrophage S1PR1-dependent IL-1β and potentially other soluble factors promote both angiogenesis and lymphangiogenesis in vitro, while affecting only lymphangiogenesis in two tumor models in vivo. A number of other myeloid cells in the tumor microenvironment are known to produce proangiogenic factors (Murdoch et al., 2008), which might override the reduced IL-1β levels in tumors containing S1PR1 KO TAMs. Interestingly, not just protumor properties of inflammasomes and their products have been described (Kolb et al., 2014). The NLRP3 inflammasome was protective in the azoxymethane/dextran sulfate sodium colitis-associated cancer model (Allen et al., 2010). Moreover, immunogenic chemotherapy relies on inflammasome activation in dendritic cells and subsequent IL-1β generation to mount protective immunity against tumors (Zitvogel et al., 2012). Therefore, it remains unclear whether interfering with S1PR1 may limit the success of immunogenic therapy.
It may be puzzling to assume an active NLRP3 inflammasome in low-grade inflammatory tumors such as PyMT tumors. Expression of inflammasome components is likely triggered by damage-associated molecular patterns derived from necrotic tumor cells that are found in areas of hypoxia and nutrient deprivation (Iyer et al., 2009; Kuraishy et al., 2011). Specific activators of the NLRP3 inflammasome such as ATP may be delivered from apoptotic tumor cells (Elliott et al., 2009), which also produce S1P (Weigert et al., 2010). Thus, cell death in tumors might drive lymphangiogenesis and concomitant metastasis by inducing the NLRP3 inflammasome in bystander myeloid cells.
Our data add to the emerging rationale to target S1P and its receptors in cancer. An S1P-neutralizing antibody (Visentin et al., 2006) and SPHK inhibitors are being tested in clinical trials (Kunkel et al., 2013). However, targeting specific S1PRs might be superior to targeting S1P or specific SPHKs, based on the sometimes antithetic function of S1PR signaling. For instance, S1PR2 signaling apparently possesses antitumor potential mainly by inhibiting angiogenesis (Du et al., 2010). Targeting S1PR1 would therefore not only block persistent STAT3 signaling in tumors (Lee et al., 2010; Deng et al., 2012; Degagné et al., 2014) as well as angiogenesis (Nagahashi et al., 2012), but might also redirect S1P toward S1PR2 to elicit its antitumor potential.
Preventing metastasis remains a major challenge in cancer therapy. An attractive approach might be targeting lymphangiogenesis in carcinomas. Although the connection of tumor lymphangiogenesis with lymph node invasion is well established, the contribution of tumor lymphangiogenesis to metastasis at distant sites is still controversially discussed. It has been argued whether or not lymph vessels can serve as a direct or indirect (through lymphovenous connections) route of metastatic spread (Ran et al., 2010; Stacker et al., 2014). Besides this controversy, experimental studies indicate that lymphatic vessels in tumors can suppress antitumor immunity and provide a niche for cancer stem cells, thereby promoting disease development and metastasis, without serving as a highway for cancer cells to distant sites (Alitalo, 2011; Card et al., 2014; Karaman and Detmar, 2014; Lund et al., 2016). Although the exact mechanism is not yet clear, our data suggest that targeting the S1PR1/NLRP3/IL-1β axis might be a way to limit metastatic disease, which appears to require the modulation of lymphangiogenesis.
Materials and methods
Animal experiments
Mouse care and experiments involving mice were approved by and followed the guidelines of the Hessian animal care and use committee. Female mice in the C57BL/6 background were used for all experiments. All treatments were initiated in animals at an age between 8 and 12 wks. SPHK1−/− and SPHK2−/− from Novartis were backcrossed for at least 10 generations into a C57BL/6 background. Mice with a deletion of S1pr1 in macrophages (S1pr1fl/fl F4/80Cre/+) were described before (Weichand et al., 2013). To validate the efficiency of F4/80-Cre–driven recombination, F4/80Cre/+ were crossed with mT/mG reporter mice (Muzumdar et al., 2007). All strains were crossed with mice expressing the PyMT oncoprotein under the mouse mammary tumor virus promoter (Lin et al., 2003), previously bred into a C57BL/6 background to induce mammary carcinoma. To induce fibrosarcoma, mice received a single s.c. injection of 100 µg MCA into their flanks. Experimental metastasis assays were performed essentially as described (Sceneay et al., 2012) using Lewis lung carcinoma (LLC) lung carcinoma cells (purchased from ATCC and regularly tested for mycoplasma contamination). Tumor growth was monitored using sliding calipers until tumors reached a size of >1 cm. Animals were perfused, and tumors, lungs, and draining lymph nodes were harvested for further analyses. For Matrigel plug assays, C57BL/6 mice received an s.c. injection of 0.5 ml Matrigel (BD Biosciences) mixture containing 0.0025 U/ml heparin and 25% (vol/vol) macrophage supernatants as indicated, along the dorsal midline on each site of the spine. After 10 d, mice were killed, and Matrigel plugs were harvested. Animals were randomly assigned to the different treatment groups. Matrigel assays were performed in a blinded manner. All other animal experiments were performed nonblinded.
Cell culture
LLC and E0771 breast cancer cells were cultured in RPMI 1640 supplemented with 10% FCS, 100 U/ml penicillin, and 100 µg/ml streptomycin. Primary human monocytes and macrophages were cultured in RPMI 1640 supplemented with 2.5% AB+ human serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. Mouse BMDMs were cultured in high-glucose DMEM with the addition of 10% FCS, 100 U/ml penicillin, and 100 µg/ml streptomycin. The mouse embryonic stem cell line CGR8 (provided by M. Wartenberg, University Hospital, Jena, Germany) was grown on gelatin-coated culture dishes with Glasgow minimum essential medium supplemented with 10% FCS, 2 mM l-glutamine, 50 µM β-mercaptoethanol, and 100 U/ml leukemia inhibitory factor (EMD Millipore) to prevent differentiation. All cells were cultured in a humidified atmosphere of 5% CO2 at 37°C.
Immunohistochemistry and immunofluorescence analyses
Investigators were blinded to group allocation during immunohistochemistry and immunofluorescence analyses. Primary tissue was Zn-fixed and paraffin-embedded or embedded in Tissue Tek (Sakura Finetek) for cryosections. For analyzing lung metastasis, lung sections were deparaffinized, stained with Mayer’s hemalum (Merck) and F4/80 antibodies (eBioscience), and examined under an Axioskop 40 microscope (Zeiss). Lung panorama pictures were produced using Autostitch v2.2 (CloudBurst Research). At least 10 independent sections of four different lung areas were analyzed. Immunofluorescence analysis of blood and lymph vessels from primary tumor and Matrigel plug sections was performed using LYVE-1 (R&D Systems) and CD31 (BD Biosciences) antibodies. For fluorescence detection, Alexa Fluor 488, 546, or 633 dye-conjugated secondary antibodies (Life Technologies) were used and counterstained with Hoechst H33342 (Merck) for nuclei. Fluorescent microscopy images were acquired using an automated confocal microscope (LSM 780; Zeiss). Vessel formation was analyzed by quantifying fluorescence positive area with Axiovision (Zeiss) in at least three independent sections. Multi-epitope ligand cartography was described before (Pierre et al., 2008). Samples were sequentially stained with PE-coupled LYVE-1 (BD Biosciences), F4/80 (eBioscience), CD11b, and CD206 (both from AbD Serotech) antibodies and separated by a bleaching step in an automated manner on an inverted, wide-field fluorescence microscope (Leica DM IRE2) with a cooled charge-coupled device camera (Apogee KX4; Apogee Instruments). Fluorescence images produced by each antibody were aligned pixel-wise. Images were corrected for illumination faults using flat-field correction (TIC Experiment Viewer; Meltec). For analyzing lymph node metastasis and tissue samples of human invasive breast cancer, provided by the Cooperative Human Tissue Network and the Cancer Diagnosis Program (other investigators may have received specimens from the same subjects), Opal Fluorescent IHC kits (PerkinElmer) were used according to the manufacturer’s instructions. Prepared paraffin slides were stained with primary antibodies targeting the PyMT protein and Ki67 (both from Abcam) for lymph nodes and pancytokeratin, CD163 (both from Abcam), and NLRP3 (Adipogen) for human tissue microarrays. Nuclei were counterstained with DAPI. Samples were acquired at 20× magnification using the Vectra3 automated quantitative pathology imaging system (PerkinElmer). InForm v2.1 (PerkinElmer) was used to quantify the PyMT-positive area in mouse lymph node sections and determine the percentage of NLRP3-expressing macrophages in human invasive breast cancer cores. Breast cancer cores were evaluated based on tissue integrity after staining and macrophage content; only cores containing macrophages were considered. 111 individual cores were suitable for analysis based on these criteria.
Flow cytometry
Tumor and lung single-cell suspensions were stained with fluorochrome-conjugated antibodies and analyzed on a LSRII/Fortessa flow cytometer or sorted using a FACSAria III cell sorter (both from BD Biosciences). Data were analyzed using FlowJo Vx (TreeStar). All antibodies and secondary reagents were titrated to determine optimal concentrations. CompBeads (BD) were used for single-color compensation to create multicolor compensation matrices. For gating, fluorescence minus one controls were used. The instrument calibration was controlled daily using Cytometer Setup and Tracking beads (BD Biosciences). Characterization of immune cell subsets in blood, spleen, liver, peritoneum, PyMT tumors, and lungs was performed essentially as described previously (Weigert et al., 2012; Olesch et al., 2015). Single-cell suspensions were created using the Miltenyi tumor dissociation kit and the GentleMACS isolator (Miltenyi Biotec) using standard protocols. The following antibodies were used: anti-CD3-PE-CF594, anti-CD4-V500, anti-CD11c-AlexaFluor700, anti-CD19-APC-H7, anti-CD326 (EpCAM)-BV711, anti-Ly6C-PerCP-Cy5.5 (BD Biosciences), anti-CD8-eFluor650, anti-CD11b-eFluor605NC (eBioscience), anti-CD45-VioBlue, anti-CD49b-PE, anti-MHC-II-APC (Miltenyi Biotec), anti-F4/80-PE-Cy7, and anti-Ly6G-APC-Cy7 (BioLegend). Single-cell suspensions of embryoid body sprouting assays were analyzed using anti-CD45-V500 (BD Biosciences), anti-CD31-PE-Cy7 (eBioscience), anti-podoplanin-Alexa Fluor 488 (BioLegend), and anti-LYVE1-PE (R&D Systems). Matrigel single-cell suspensions were analyzed using anti-CD45-VioBlue, anti-CD31-PE-Cy7, anti-CD146-Alexa Fluor 488, and anti-CD49f-PE-CF594 (BD Biosciences).
For FACS sorting of primary mouse TAMs, single-cell suspensions were stained with CD45, CD11b, CD11c, F4/80, Ly6C, and Ly6G antibodies and 7-aminoactinomycin D for dead cell exclusion. For isolation of mouse LECs from lung EC cultures, single-cell suspensions were stained with anti-CD31-PE-Cy7, anti-CD146-FITC (BD Biosciences), anti-CD204-PE (BioLegend), and anti-LYVE1-PE. Cell suspensions were filtered through a 30-µm cell strainer and diluted to the ideal concentrations for cell sorting. CD11bhi TAMs or mouse LECs were sorted into medium-prefilled FACS tubes at 4°C.
Whole-genome microarray analysis
WT and S1PR1 KO CD11bhi TAMs of PyMT and MCA tumors were sorted by FACS, and two to three samples each were pooled. RNA from pooled FACS-sorted CD11bhi TAMs was isolated using the RNeasy micro kit (Qiagen), quality controlled using the Agilent 2100 Bioanalyzer and pico RNA chips (Agilent), amplified using the µMACS SuperAmp Kit (Miltenyi) in-house, hybridized to mouse WG-6 v2 BeadChips in biological duplicates (TAMs from ≥ 4 individual mice), and scanned and quantile normalized at the microarray unit of the German Cancer Research Center (DKFZ) Genomics and Proteomics Core Facility. Statistical analysis was performed using the computing environment R. Additional software packages (geneplotter, genefilter, multtest) were taken from the Bioconductor project (Gentleman et al., 2004). Unsupervised hierarchical clustering was performed for the probe sets with SD >1 across all samples using the Manhattan distance and the mean linkage method. To reduce the dimension of the microarray data before conducting pairwise comparisons to analyze differential gene expression, a variance filter (the interquartile range of log2 intensities should be ≥0.5, if the group sizes were equal) was applied. The interquartile range of log2 intensities had to be at least 0.1 for unequal group sizes. After the global filtering, a two-sample t test was applied to identify genes that were differentially expressed between two groups. The false discovery rate according to Benjamini and Hochberg was used to account for the multiple testing. Fold changes between the two groups of each supervised analysis were calculated for each gene. The dataset is available at GEO: GSE71908.
Quantitative PCR
Quantitative PCR was performed as described previously (Weigert et al., 2012). For amplifying human Nlrp3, Vegfc, Vegfd, Vegfr3, Prox1, and actin and mouse Vegfc, QuantiTect Primer Assays (Qiagen) were used. Sequences of other mouse primers were as follows: ActB: forward, 5′-CAGCTTCTTTGCAGCTCCTT-3′; reverse, 5′-CACGATGGAGGGGAATACAG-3′; Casp1: forward, 5′-GCTTGAAAGACAAGCCCAAG-3′; reverse, 5′-GGCCTTCTTAATGCCATCAT-3′; IL-1β: forward, 5′-AGCTTCTCCACAGCCACAAT-3′; reverse, 5′-TGAAATGCCACCTTTTGACA-3′; IL-18: forward, 5′-GGCTGCCATGTCAGAAGACT-3′; reverse, 5′-GTGAAGTCGGCCAAAGTTGT-3′; Nlrp3: forward, 5′-ATTGCTGTGTGTGGGACTGA-3′; reverse, 5′-AACCAATGCGAGATCCTGAC-3′; Pcna: forward, 5′-AACTCCCAGAAAAGCAACAAGCA-3′; reverse, 5′-CGAGGAGGAATGAGAAGAAGACG-3′; and Pycard: forward, 5′-ACATGGGCTTACAGGAGCTG-3′; reverse, 5′-GCTGGTCCACAAAGTGTCCT-3′. Results were analyzed using Gene Expression Macro (Bio-Rad). Actin served as the internal control.
Cytokine quantification
Cytokines were isolated from tumor extracellular fluid as described (Eubank et al., 2009) and quantified in these samples and cell culture supernatants using cytometric bead array flex sets (mouse IL-6, IL-10, IL-23, and IL-1β and human IL-1β; BD Biosciences). Samples were acquired on a LSRII/Fortessa flow cytometer and evaluated with FCAP v3.0 (Soft Flow). Investigators were blinded to group allocation.
Macrophage generation
Human macrophages were differentiated from buffy coat–derived monocytes using 2.5% human AB+ serum as described previously (Weichand et al., 2013). The institutional ethics committee of the Goethe University Hospital, Frankfurt, Germany, waived the need for consent, because buffy coats were used anonymously for in vitro assays with no link to patient data. Mouse BMDMs were generated from 2E6 total bone marrow cells by culture in RPMI 1640 medium with 10% FCS and M-CSF (20 ng/ml) for 7 d.
Western analysis
Western analysis was performed as described (Weigert et al., 2010). Polyclonal antibodies against human pro-CASP1 and the p20 subunit, and against mouse phospho-IKKα/β (Ser176/180), IKKα/β, phospho-NF-κB p65 (Ser536), and NF-κB p65 (all from Cell Signaling Technology) were used. IRDye infrared secondary antibodies were visualized with the Odyssey infrared imaging system (LI-COR). Western blots were quantified using Odyssey v2.1.
Embryoid body assay
Sprouting assays using CGR8 cells were described before (Geis et al., 2015). 200 CGR8 cells/well were embedded in a collagen matrix containing 1.8 mg collagen type I/ml (BD Biosciences) in a 24-well plate and cultured for 5 d in the absence of leukemia inhibitory factor to allow embryoid body formation. Wells were covered with Iscove’s medium (negative control) or Iscove’s medium diluted 1:1 with supernatants of WT C57BL/6, S1pr1fl/fl F4/80Cre/+, and S1pr2−/− macrophages for an additional 5 d. Macrophages were stimulated with supernatants of MC57G fibrosarcoma cells for 3 h. Medium was replaced with fresh macrophage medium, and macrophage supernatants were harvested after another 21 h. Sprouting assays were analyzed by flow cytometry.
Aortic ring sprouting assay
Aortic ring sprouting assays were performed as described (Zippel et al., 2016). Aortas were removed, cleaned, and embedded in collagen (Corning) in a 48-well plate. After polymerization of the collagen gel, microvascular endothelial cell growth medium (Pelo Biotech) supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, and 2% mouse serum (made in-house from blood of C57/Bl6J mice: fresh blood was centrifuged at 800 g for 15 min, supernatant was collected, warmed to 60°C for 20 min, and sterile-filtered) were added into the wells. Tube-like structures were allowed to develop over 7 d upon stimulation with macrophage supernatant or medium containing IL-1β. The samples were fixed using formalin and Roti-Histofix 4% (Roth), endothelial cells were visualized with antibodies against CD31, and sprout length was quantified using ImageJ.
Lymphatic ring sprouting assay
Thoracic ducts were dissected from 2- to 3-mo-old C57Bl/6J mice and cut into 1-mm-long pieces. The explants were embedded in collagen type I (Corning) in a 24-well plate. After polymerization of the collagen gel, MCDB 131 supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, l-glutamine (10 mmol/l) and 10% FCS mixed with macrophage supernatant or media containing IL-1β as indicated at a 1:1 ratio was added. Rings were cultured at 5% oxygen for 8–10 d and washed two times with PBS; pictures were taken with a Canon EOS 700D. To quantify vessel outgrowth, binary images of photographs were taken under identical conditions of light, contrast, and magnification and analyzed using ImageJ.
Primary mouse LEC isolation and culture
Mouse pulmonary endothelial cells were isolated as described (Fleming et al., 2005). Lungs were removed from mice and kept in DMEM/F12 containing 10% FCS, 100 U/ml penicillin, 100 µg/ml streptomycin, and amphotericin B on ice. Lungs were chopped into small pieces and washed with HBSS (Ca2+/Mg2+ free). Lung pieces were passed through a 40-µm cell strainer and digested in HBSS containing antibiotics with 2.4 U/ml dispase II (Gibco) for 1 h at 37°C under orbital shaking. The cell suspension was filtered through a 40-µm cell strainer, and cells were pelleted by centrifugation at 1200 rpm for 3 min at 4°C. Cell pellets were resuspended in PBS with preincubated rat anti–mouse CD31 and goat anti–rat IgG microbeads (containing 0.5% BSA and EDTA) with gentle shaking. Microbead-coated endothelial cells were separated from nonendothelial cells using magnetic cell sorting with MACS MS columns (Miltenyi Biotec). Isolated endothelial cells were seeded on gelatin-coated culture dishes and expanded for 3 wks in DMEM/F12 (without phenol red) supplemented with 20% FCS, 0.4% endothelial cell growth supplement with heparin (ECGS/H; PromoCell), 0.1 ng/ml epidermal growth factor, 1 µg/ml hydrocortisone, 1 ng/ml basic fibroblast growth factor, antibiotics, and 2 mmol/l l-glutamine. LECs were purified by FACS sorting (defined as CD31+ CD146− CD204+, and LYVE1+ cells), expanded further, and used for downstream applications as indicated.
Primary human lymphatic endothelial cell culture
Primary human LECs were purchased from PromoCell and kept in endothelial cell growth medium MV2 including supplement mix (PromoCell). Cells were used up to passage 7.
Tube formation assay
For tube formation assays, 10,000 LECs per well were seeded on µ-slides (Ibidi) and analyzed according to the manufacturer’s instructions. Cells were treated with macrophage supernatants or human recombinant IL-1β (Peprotech) as indicated. Investigators were blinded to group allocation. Images were captured after 6 h using an inverted microscope (Axiovert 135; Zeiss) and camera (AxioCam MRc; Zeiss). The number of master junctions was quantified with the Angiogenesis Analyser using ImageJ.
Endothelial spheroid sprouting assay
Endothelial cell sprouting was investigated using a spheroid sprouting assay essentially as described before (Brecht et al., 2011). A Methocel solution composed of methylcellulose (1.2% [wt/vol], 4,000 centipoises; Sigma-Aldrich) was prepared in endothelial basal medium. HUVECs were suspended in HUVEC culture medium (endothelial basal media supplemented with 8% FCS, 1 ng/ml basic fibroblast growth factor, 0.1 ng/ml epidermal growth factor, 0.4% ECGS/H, 50 U/ml penicillin, 50 µg/ml streptomycin, and 10 mmol/l L-glutamine containing 20% Methocel). Drops of this suspension (500 cells per drop) were placed on culture plates and incubated upside down to form spheroids (Kelm et al., 2003). Spheroids were collected within 24 h and resuspended in a mixture of 80% Methocel, 10% FCS, and 10% MCDB131 medium. The spheroid suspension was mixed in equal amounts with collagen (1 µg/ml in Medium 199) and plated on a 48-well plate (75 spheroids per well). The resulting gels were covered with macrophage supernatant or medium containing IL-1β. After 24 h, pictures of endothelial cell sprouts were taken with a stereomicroscope (Axioscope; Zeiss). Each condition was tested in triplicate. The institutional ethics committee of the Goethe-University Hospital, Frankfurt, Germany, waived the need for consent because HUVECs were used anonymously for in vitro assays with no link to patient data.
Analysis of human breast cancer datasets
NLRP3 expression data from publicly available datasets of human breast cancer stroma (Karnoub et al., 2007; Ma et al., 2009) were retrieved using GEO2R and analyzed using GraphPad Prism v5.03.
Statistics
Data are presented as means ± SEM. Statistical comparisons between groups were performed with paired or unpaired two-tailed Student’s t test or one-sample t test for normalized data as indicated in the figure legends. One- or two-way analysis of variance (ANOVA) followed by Bonferroni’s posttest was used for multiple comparisons of data with normal distribution and equal variance (D’Agostino–Pearson omnibus normality test). Statistical analysis was performed with GraphPad Prism v5.03. Differences were considered significant at P < 0.05. No statistical test was used to predetermine sample size, and all samples were included in the analysis.
Online supplemental material
Fig. S1 shows the characterization of the F4/80-Cre mouse strain. Fig. S2 shows the gating strategy to identify immune cell subsets in mammary tumors and lungs.
Supplementary Material
Acknowledgments
We thank Praveen Mathoor and Margarete Mijatovic for excellent technical assistance and the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing the Illumina Whole-Genome Expression Bead chips and related services.
This work was supported by Deutsche Forschungsgemeinschaft (SFB 1039 TP A06, B04, and B06, Excellence Cluster Cardiopulmonary Systems), Deutsche Krebshilfe (109599 and 70112451), and Else Kröner-Fresenius-Stiftung (Else Kröner-Fresenius-Graduiertenkolleg and Translational Research Innovation Pharma). J. Mora was supported by Deutscher Akademischer Austauschdienst and the University of Costa Rica.
The authors declare no competing financial interests.
Author contributions: conceptualization: A. Weigert and B.B.; methodology: B. Weichand, R. Popp, J. Mora, E. Strack, K. Scholich, S.N. Syed, M. Jung, A. von Knethen, and A. Weigert; formal analysis: B. Weichand, C. Döring, and A. Weigert; investigation: B. Weichand, R. Popp, S. Dziumbla, J. Mora, E. Strack, E. Elwakeel, A.-C. Frank, K. Scholich, S. Pierre, S.N. Syed, C. Olesch, J. Ringleb, B. Ören, and A. Weigert; resources: R. Savai, B. Levkau, I. Fleming, and B. Brüne; writing of original draft: A. Weigert; manuscript review and editing: all authors; funding acquisition: I. Fleming, A. Weigert, and B. Brüne; visualization: A. Weigert and B.B.; supervision: I. Fleming, A. Weigert, and B. Brüne.
Footnotes
Abbreviations used:
- DCIS
- ductal carcinoma in situ
- EpCAM
- epithelial cell adhesion molecule
- LEC
- lymphatic endothelial cell
- MCA
- methylcholanthrene
- MDSC
- myeloid-derived suppressor cell
- PyMT
- polyoma middle T
- S1P
- sphingosine-1-phosphate
- S1PR
- S1P receptor
- SPHK
- sphingosine kinase
- TAM
- tumor-associated macrophage
- VEC
- vascular endothelial cell
- VEGF
- vascular endothelial growth factor
- VEGFR
- VEGF receptor
References
- Alitalo K. 2011. The lymphatic vasculature in disease. Nat. Med. 17:1371–1380. 10.1038/nm.2545 [DOI] [PubMed] [Google Scholar]
- Allen I.C., TeKippe E.M., Woodford R.M., Uronis J.M., Holl E.K., Rogers A.B., Herfarth H.H., Jobin C., and Ting J.P.. 2010. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207:1045–1056. 10.1084/jem.20100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht K., Weigert A., Hu J., Popp R., Fisslthaler B., Korff T., Fleming I., Geisslinger G., and Brüne B.. 2011. Macrophages programmed by apoptotic cells promote angiogenesis via prostaglandin E2. FASEB J. 25:2408–2417. 10.1096/fj.10-179473 [DOI] [PubMed] [Google Scholar]
- Brouillard P., Boon L., and Vikkula M.. 2014. Genetics of lymphatic anomalies. J. Clin. Invest. 124:898–904. 10.1172/JCI71614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card C.M., Yu S.S., and Swartz M.A.. 2014. Emerging roles of lymphatic endothelium in regulating adaptive immunity. J. Clin. Invest. 124:943–952. 10.1172/JCI73316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikarmane S.A., Tirumani S.H., Howard S.A., Jagannathan J.P., and DiPiro P.J.. 2015. Metastatic patterns of breast cancer subtypes: What radiologists should know in the era of personalized cancer medicine. Clin. Radiol. 70:1–10. 10.1016/j.crad.2014.08.015 [DOI] [PubMed] [Google Scholar]
- Degagné E., Pandurangan A., Bandhuvula P., Kumar A., Eltanawy A., Zhang M., Yoshinaga Y., Nefedov M., de Jong P.J., Fong L.G., et al. 2014. Sphingosine-1-phosphate lyase downregulation promotes colon carcinogenesis through STAT3-activated microRNAs. J. Clin. Invest. 124:5368–5384. 10.1172/JCI74188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Liu Y., Lee H., Herrmann A., Zhang W., Zhang C., Shen S., Priceman S.J., Kujawski M., Pal S.K., et al. 2012. S1PR1-STAT3 signaling is crucial for myeloid cell colonization at future metastatic sites. Cancer Cell. 21:642–654. 10.1016/j.ccr.2012.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Takuwa N., Yoshioka K., Okamoto Y., Gonda K., Sugihara K., Fukamizu A., Asano M., and Takuwa Y.. 2010. S1P(2), the G protein-coupled receptor for sphingosine-1-phosphate, negatively regulates tumor angiogenesis and tumor growth in vivo in mice. Cancer Res. 70:772–781. 10.1158/0008-5472.CAN-09-2722 [DOI] [PubMed] [Google Scholar]
- Elliott M.R., Chekeni F.B., Trampont P.C., Lazarowski E.R., Kadl A., Walk S.F., Park D., Woodson R.I., Ostankovich M., Sharma P., et al. 2009. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature. 461:282–286. 10.1038/nature08296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubank T.D., Roberts R.D., Khan M., Curry J.M., Nuovo G.J., Kuppusamy P., and Marsh C.B.. 2009. Granulocyte macrophage colony-stimulating factor inhibits breast cancer growth and metastasis by invoking an anti-angiogenic program in tumor-educated macrophages. Cancer Res. 69:2133–2140. 10.1158/0008-5472.CAN-08-1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming I., Fisslthaler B., Dixit M., and Busse R.. 2005. Role of PECAM-1 in the shear-stress-induced activation of Akt and the endothelial nitric oxide synthase (eNOS) in endothelial cells. J. Cell Sci. 118:4103–4111. 10.1242/jcs.02541 [DOI] [PubMed] [Google Scholar]
- Franklin R.A., Liao W., Sarkar A., Kim M.V., Bivona M.R., Liu K., Pamer E.G., and Li M.O.. 2014. The cellular and molecular origin of tumor-associated macrophages. Science. 344:921–925. 10.1126/science.1252510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis T., Döring C., Popp R., Grossmann N., Fleming I., Hansmann M.L., Dehne N., and Brüne B.. 2015. HIF-2α-dependent PAI-1 induction contributes to angiogenesis in hepatocellular carcinoma. Exp. Cell Res. 331:46–57. 10.1016/j.yexcr.2014.11.018 [DOI] [PubMed] [Google Scholar]
- Gordon E.J., Rao S., Pollard J.W., Nutt S.L., Lang R.A., and Harvey N.L.. 2010. Macrophages define dermal lymphatic vessel calibre during development by regulating lymphatic endothelial cell proliferation. Development. 137:3899–3910. 10.1242/dev.050021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: The next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Iyer S.S., Pulskens W.P., Sadler J.J., Butter L.M., Teske G.J., Ulland T.K., Eisenbarth S.C., Florquin S., Flavell R.A., Leemans J.C., and Sutterwala F.S.. 2009. Necrotic cells trigger a sterile inflammatory response through the Nlrp3 inflammasome. Proc. Natl. Acad. Sci. USA. 106:20388–20393. 10.1073/pnas.0908698106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Yuan R.Q., Fuchs A., Yao Y., Joseph A., Schwall R., Schnitt S.J., Guida A., Hastings H.M., Andres J., et al. 1997. Expression of interleukin-1β in human breast carcinoma. Cancer. 80:421–434. [DOI] [PubMed] [Google Scholar]
- Karaman S., and Detmar M.. 2014. Mechanisms of lymphatic metastasis. J. Clin. Invest. 124:922–928. 10.1172/JCI71606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub A.E., Dash A.B., Vo A.P., Sullivan A., Brooks M.W., Bell G.W., Richardson A.L., Polyak K., Tubo R., and Weinberg R.A.. 2007. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 449:557–563. 10.1038/nature06188 [DOI] [PubMed] [Google Scholar]
- Kelm J.M., Timmins N.E., Brown C.J., Fussenegger M., and Nielsen L.K.. 2003. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 83:173–180. 10.1002/bit.10655 [DOI] [PubMed] [Google Scholar]
- Kerjaschki D. 2005. The crucial role of macrophages in lymphangiogenesis. J. Clin. Invest. 115:2316–2319. 10.1172/JCI26354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., Qian B.Z., and Pollard J.W.. 2015. Immune cell promotion of metastasis. Nat. Rev. Immunol. 15:73–86. 10.1038/nri3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb R., Liu G.-H., Janowski A.M., Sutterwala F.S., and Zhang W.. 2014. Inflammasomes in cancer: A double-edged sword. Protein Cell. 5:12–20. 10.1007/s13238-013-0001-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krelin Y., Voronov E., Dotan S., Elkabets M., Reich E., Fogel M., Huszar M., Iwakura Y., Segal S., Dinarello C.A., and Apte R.N.. 2007. Interleukin-1β-driven inflammation promotes the development and invasiveness of chemical carcinogen-induced tumors. Cancer Res. 67:1062–1071. 10.1158/0008-5472.CAN-06-2956 [DOI] [PubMed] [Google Scholar]
- Kubota Y., Takubo K., Shimizu T., Ohno H., Kishi K., Shibuya M., Saya H., and Suda T.. 2009. M-CSF inhibition selectively targets pathological angiogenesis and lymphangiogenesis. J. Exp. Med. 206:1089–1102. 10.1084/jem.20081605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G.T., Maceyka M., Milstien S., and Spiegel S.. 2013. Targeting the sphingosine-1-phosphate axis in cancer, inflammation and beyond. Nat. Rev. Drug Discov. 12:688–702. 10.1038/nrd4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraishy A., Karin M., and Grivennikov S.I.. 2011. Tumor promotion via injury- and death-induced inflammation. Immunity. 35:467–477. 10.1016/j.immuni.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Deng J., Kujawski M., Yang C., Liu Y., Herrmann A., Kortylewski M., Horne D., Somlo G., Forman S., et al. 2010. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat. Med. 16:1421–1428. 10.1038/nm.2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E.Y., Jones J.G., Li P., Zhu L., Whitney K.D., Muller W.J., and Pollard J.W.. 2003. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163:2113–2126. 10.1016/S0002-9440(10)63568-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund A.W., Medler T.R., Leachman S.A., and Coussens L.M.. 2016. Lymphatic vessels, inflammation, and immunity in skin cancer. Cancer Discov. 6:22–35. 10.1158/2159-8290.CD-15-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.J., Dahiya S., Richardson E., Erlander M., and Sgroi D.C.. 2009. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 11:R7 10.1186/bcr2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y., Ghose S., Boelte K., Li J., Yang L., and Lin P.C.. 2011. C/EBP-δ regulates VEGF-C autocrine signaling in lymphangiogenesis and metastasis of lung cancer through HIF-1α. Oncogene. 30:4901–4909. 10.1038/onc.2011.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch C., Muthana M., Coffelt S.B., and Lewis C.E.. 2008. The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer. 8:618–631. 10.1038/nrc2444 [DOI] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., and Luo L.. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45:593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nagahashi M., Ramachandran S., Kim E.Y., Allegood J.C., Rashid O.M., Yamada A., Zhao R., Milstien S., Zhou H., Spiegel S., and Takabe K.. 2012. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 72:726–735. 10.1158/0008-5472.CAN-11-2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao S., Noda K., Zandi S., Sun D., Taher M., Schering A., Xie F., Mashima Y., and Hafezi-Moghadam A.. 2011. VAP-1-mediated M2 macrophage infiltration underlies IL-1β- but not VEGF-A-induced lymph- and angiogenesis. Am. J. Pathol. 178:1913–1921. 10.1016/j.ajpath.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R., and Pollard J.W.. 2014. Tumor-associated macrophages: From mechanisms to therapy. Immunity. 41:49–61. 10.1016/j.immuni.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesch C., Sha W., Angioni C., Sha L.K., Açaf E., Patrignani P., Jakobsson P.J., Radeke H.H., Grösch S., Geisslinger G., et al. 2015. MPGES-1-derived PGE2 suppresses CD80 expression on tumor-associated phagocytes to inhibit anti-tumor immune responses in breast cancer. Oncotarget. 6:10284–10296. 10.18632/oncotarget.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre S., Maeurer C., Coste O., Becker W., Schmidtko A., Holland S., Wittpoth C., Geisslinger G., and Scholich K.. 2008. Toponomics analysis of functional interactions of the ubiquitin ligase PAM (protein associated with myc) during spinal nociceptive processing. Mol. Cell. Proteomics. 7:2475–2485. 10.1074/mcp.M800201-MCP200 [DOI] [PubMed] [Google Scholar]
- Pyne N.J., and Pyne S.. 2010. Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer. 10:489–503. 10.1038/nrc2875 [DOI] [PubMed] [Google Scholar]
- Qian B., Deng Y., Im J.H., Muschel R.J., Zou Y., Li J., Lang R.A., and Pollard J.W.. 2009. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One. 4:e6562 10.1371/journal.pone.0006562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran S., Volk L., Hall K., and Flister M.J.. 2010. Lymphangiogenesis and lymphatic metastasis in breast cancer. Pathophysiology. 17:229–251. 10.1016/j.pathophys.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristimäki A., Narko K., Enholm B., Joukov V., and Alitalo K.. 1998. Proinflammatory cytokines regulate expression of the lymphatic endothelial mitogen vascular endothelial growth factor-C. J. Biol. Chem. 273:8413–8418. 10.1074/jbc.273.14.8413 [DOI] [PubMed] [Google Scholar]
- Ruffell B., and Coussens L.M.. 2015. Macrophages and therapeutic resistance in cancer. Cancer Cell. 27:462–472. 10.1016/j.ccell.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sceneay J., Chow M.T., Chen A., Halse H.M., Wong C.S., Andrews D.M., Sloan E.K., Parker B.S., Bowtell D.D., Smyth M.J., and Möller A.. 2012. Primary tumor hypoxia recruits CD11b+/Ly6Cmed/Ly6G+ immune suppressor cells and compromises NK cell cytotoxicity in the premetastatic niche. Cancer Res. 72:3906–3911. 10.1158/0008-5472.CAN-11-3873 [DOI] [PubMed] [Google Scholar]
- Stacker S.A., Williams S.P., Karnezis T., Shayan R., Fox S.B., and Achen M.G.. 2014. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer. 14:159–172. 10.1038/nrc3677 [DOI] [PubMed] [Google Scholar]
- Visentin B., Vekich J.A., Sibbald B.J., Cavalli A.L., Moreno K.M., Matteo R.G., Garland W.A., Lu Y., Yu S., Hall H.S., et al. 2006. Validation of an anti-sphingosine-1-phosphate antibody as a potential therapeutic in reducing growth, invasion, and angiogenesis in multiple tumor lineages. Cancer Cell. 9:225–238. 10.1016/j.ccr.2006.02.023 [DOI] [PubMed] [Google Scholar]
- Watari K., Shibata T., Kawahara A., Sata K., Nabeshima H., Shinoda A., Abe H., Azuma K., Murakami Y., Izumi H., et al. 2014. Tumor-derived interleukin-1 promotes lymphangiogenesis and lymph node metastasis through M2-type macrophages. PLoS One. 9:e99568 10.1371/journal.pone.0099568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichand B., Weis N., Weigert A., Grossmann N., Levkau B., and Brüne B.. 2013. Apoptotic cells enhance sphingosine-1-phosphate receptor 1 dependent macrophage migration. Eur. J. Immunol. 43:3306–3313. 10.1002/eji.201343441 [DOI] [PubMed] [Google Scholar]
- Weigert A., Cremer S., Schmidt M.V., von Knethen A., Angioni C., Geisslinger G., and Brüne B.. 2010. Cleavage of sphingosine kinase 2 by caspase-1 provokes its release from apoptotic cells. Blood. 115:3531–3540. 10.1182/blood-2009-10-243444 [DOI] [PubMed] [Google Scholar]
- Weigert A., Weichand B., and Brune B.. 2011. S1P regulation of macrophage functions in the context of cancer. Anticancer. Agents Med. Chem. 11:818–829. 10.2174/187152011797655096 [DOI] [PubMed] [Google Scholar]
- Weigert A., Weichand B., Sekar D., Sha W., Hahn C., Mora J., Ley S., Essler S., Dehne N., and Brüne B.. 2012. HIF-1α is a negative regulator of plasmacytoid DC development in vitro and in vivo. Blood. 120:3001–3006. 10.1182/blood-2012-03-417022 [DOI] [PubMed] [Google Scholar]
- Weis N., Weigert A., von Knethen A., and Brüne B.. 2009. Heme oxygenase-1 contributes to an alternative macrophage activation profile induced by apoptotic cell supernatants. Mol. Biol. Cell. 20:1280–1288. 10.1091/mbc.E08-10-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Huang Q., Huang Y., Zheng W., Hua J., Yang S., Zhuang J., Wang J., and Ye J.. 2016. Lipopolysaccharide increases the release of VEGF-C that enhances cell motility and promotes lymphangiogenesis and lymphatic metastasis through the TLR4- NF-κB/JNK pathways in colorectal cancer. Oncotarget. 7:73711–73724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippel N., Ding Y., and Fleming I.. 2016. A modified aortic ring assay to assess angiogenic potential in vitro. Methods Mol. Biol. 1430:205–219. 10.1007/978-1-4939-3628-1_14 [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Kepp O., Galluzzi L., and Kroemer G.. 2012. Inflammasomes in carcinogenesis and anticancer immune responses. Nat. Immunol. 13:343–351. 10.1038/ni.2224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.