It is unclear how progranulin deficiency causes frontotemporal dementia, a neurodegenerative disease characterized by TDP-43 inclusions. Chang et al. show that loss of progranulin causes impairment of autophagy and autophagy signaling, which leads to accumulation of pathological TDP-43 in neurons.
Abstract
Loss-of-function mutations in GRN cause frontotemporal dementia (FTD) with transactive response DNA-binding protein of 43 kD (TDP-43)–positive inclusions and neuronal ceroid lipofuscinosis (NCL). There are no disease-modifying therapies for either FTD or NCL, in part because of a poor understanding of how mutations in genes such as GRN contribute to disease pathogenesis and neurodegeneration. By studying mice lacking progranulin (PGRN), the protein encoded by GRN, we discovered multiple lines of evidence that PGRN deficiency results in impairment of autophagy, a key cellular degradation pathway. PGRN-deficient mice are sensitive to Listeria monocytogenes because of deficits in xenophagy, a specialized form of autophagy that mediates clearance of intracellular pathogens. Cells lacking PGRN display reduced autophagic flux, and pathological forms of TDP-43 typically cleared by autophagy accumulate more rapidly in PGRN-deficient neurons. Our findings implicate autophagy as a novel therapeutic target for GRN-associated NCL and FTD and highlight the emerging theme of defective autophagy in the broader FTD/amyotrophic lateral sclerosis spectrum of neurodegenerative disease.
Introduction
Frontotemporal dementia (FTD) is the second most common dementia in people under the age of 65 years and is characterized by changes in personality, social behavior, and communication as a result of progressive neurodegeneration in the frontal and temporal lobes of the brain (Rademakers et al., 2012). Haploinsufficiency of progranulin (PGRN) caused by heterozygous loss-of-function mutations in the GRN gene is a leading genetic cause of FTD, with over 70 distinct causal GRN mutations identified to date (Cruts et al., 2012). Patients with GRN mutations typically present with behavioral variant FTD, and their brain tissues show hyperphosphorylated, ubiquitin-positive intracellular inclusions containing cleaved forms of TDP-43 (transactive response DNA-binding protein of 43 kD), a pathology commonly found in both amyotrophic lateral sclerosis (ALS) and FTD (Neumann et al., 2006).
PGRN is a secreted glycoprotein composed of seven and one-half tandem repeats of a cysteine-rich granulin motif (Plowman et al., 1992). In the periphery, PGRN is expressed in a wide variety of tissues and cell types, including macrophages, neutrophils, adipocytes, epithelial cells, and sperm, whereas in the central nervous system, it is present in microglia and neurons (Anakwe and Gerton, 1990; Bateman et al., 1990; Bhandari et al., 1993; Chantry et al., 1998; Petkau et al., 2010; Matsubara et al., 2012). As such, PGRN is proposed to have pleiotropic effects, and several mechanisms relating PGRN deficiency and neurodegeneration have been proposed. Treatment of primary cultured neurons with recombinant PGRN promotes neurite outgrowth (Van Damme et al., 2008; Gao et al., 2010; Gass et al., 2012), whereas knockdown or deletion of GRN leads to decreases in neurite arborization and length (Tapia et al., 2011; Gass et al., 2012), highlighting the growth factor–like properties of PGRN and suggesting that neurodegeneration caused by PGRN deficiency may result from insufficient trophic signaling. Alternatively, GRN−/− microglia/macrophages exhibit an amplified inflammatory response when stimulated (Yin et al., 2010b; Martens et al., 2012), and GRN−/− mice display augmented microgliosis as they age (Ahmed et al., 2010; Yin et al., 2010a; Filiano et al., 2013), suggesting that PGRN is an antiinflammatory molecule and might function to prevent neurotoxicity caused by neuroinflammation. These findings notwithstanding, the molecular and cell biological mechanisms by which PGRN deficiency leads to neurodegeneration, and the connection between PGRN deficiency and intraneuronal TDP-43 accumulation, are not well understood.
Recently, it was discovered that complete loss of PGRN in humans leads to a disease seemingly distinct from FTD called neuronal ceroid lipofuscinosis (NCL), a neurodegenerative lysosomal storage disorder that manifests as progressive visual failure, cerebellar ataxia, and seizures, along with accelerated lysosomal accumulation of lipofuscin (an autofluorescent lipid-containing material; Smith et al., 2012; Almeida et al., 2016). This null phenotype implicates PGRN in an undefined lysosome-related function.
Macroautophagy (herein referred to as autophagy) is a lysosome-associated process in which cytoplasmic cargos destined for degradation (such as intracellular pathogens, organelles, or protein aggregates) are first sequestered into double membrane–containing vesicles termed autophagosomes. Once formed, these autophagosomes fuse with lysosomes, forming an autolysosome, in which the cargo is degraded and the resulting products recycled. Intracellular inclusions containing misfolded, aggregated proteins are a common feature in neurodegenerative diseases (Menzies et al., 2015), and mice with disruptions in autophagy within the central nervous system develop neurodegeneration (Hara et al., 2006; Komatsu et al., 2006), implicating autophagy as a potential mechanism for neurodegenerative disease pathology and an interesting target for therapeutic intervention.
In this study, we provide multiple lines of evidence at the molecular, cellular, and organismal levels showing that impairment of autophagy is a key feature of neurons and immune cells lacking PGRN. GRN−/− macrophages fail to properly clear intracellular bacteria because of autophagy impairments, and mice lacking PGRN succumb to typically nonlethal doses of infection. PGRN-deficient neurons display hypoactivity of key autophagy regulators and impaired autophagic flux and are more prone to accumulate pathological TDP-43. These findings newly establish PGRN as a regulator of autophagy, which is a unifying theme in ALS/FTD pathogenesis, and provide a cell biological mechanism by which PGRN deficiency results in lysosomal dysfunction, TDP-43 aggregation, and neurodegeneration.
Results
Altered lysosomal gene expression in cortical microglia/macrophages from PGRN-deficient mice
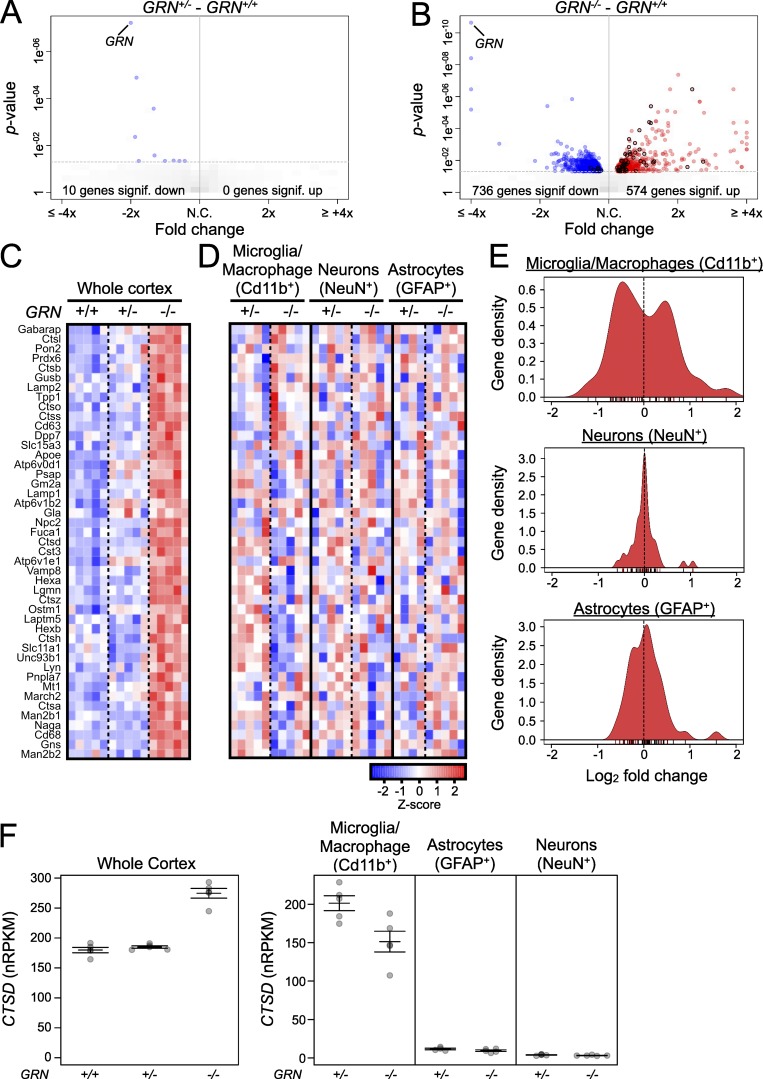
To obtain insight into the physiological function of PGRN, we used an unbiased transcriptomics (RNA-seq) approach to identify genes and pathways differentially regulated in the cerebral cortex of 12-mo-old GRN+/+ (WT), GRN+/− (heterozygous), and GRN−/− (KO) mice (Fig. 1). When we compared gene expression changes between GRN+/+ and GRN+/− mice, we found that despite a ∼50% reduction in GRN expression in heterozygote cortex, the transcriptome profiles of GRN+/+ and GRN+/− cortices were nearly identical, with only a handful of genes showing significant changes in gene expression with a p-value < 0.05 (Table S1), consistent with a previous study (Lui et al., 2016; Fig. 1 A). However, when we compared gene expression changes between GRN+/+ and GRN−/− cortex, we found significant up-regulation in 574 genes and down-regulation in 736 genes (Fig. 1 B). Gene ontology (GO) cellular component enrichment analysis of the 574 up-regulated genes in GRN−/− cortex revealed that the lysosome cellular component (GO: 0005764) accounted for ∼10% of the up-regulated gene set (Fig. 1, B and C), a greater than fivefold and highly significant overrepresentation (Table S2), suggesting altered lysosome or lysosome-related function in the brains of GRN−/− mice (Lui et al., 2016).
Figure 1.
Altered expression of lysosome-associated genes in brain and microglia/macrophages of GRN−/− mice. (A and B) Volcano plots (fold-change vs. p-value) of altered gene expression in whole cerebral cortex of heterozygous GRN+/− mice (A) or homozygous GRN−/− mice (B) relative to WT GRN+/+ mice (all 12 months old). Blue circles represent significantly down-regulated genes, and red circles represent significantly up-regulated genes. Circles with black outlines show genes with significant changes in expression from lysosome cellular component GO term (GO:0005764). n = 5 mice per group from one experiment. Significance cutoff at P < 0.05 using voom/limma. N.C., no change. (C) Heat map illustrating expression of a subset of lysosome-associated genes in whole cortex of GRN+/+, GRN+/−, and GRN−/− mice. The genes shown are those that were up in the GRN−/− cortex relative to GRN+/+ cortex and are members of lysosome cellular component GO term (GO:0005764; i.e., red circles with black outlines from B). n = 5 mice per group from one experiment. (D) Heat map showing expression of the same set of lysosome-associated genes shown in C in sorted microglia/macrophages, neurons, and astrocytes from GRN+/− and GRN−/− mice. Gene expression was normalized separately within each cell type. n = 5 mice from five experiments for all groups except GRN+/− astrocytes (n = 4 mice). (E) Gene density versus fold change plot of the data shown in D. n = 5 mice from five experiments for all groups except GRN+/− astrocytes (n = 4 mice). (F) Normalized reads per kilobase of transcript per million mapped reads (nRPKM) of cathepsin D (CTSD) in whole GRN+/+, GRN+/−, and GRN−/− cortex (left) and sorted microglia/macrophages, astrocytes, and neurons from GRN+/−, and GRN−/− cortex (right). n = 5 mice from five experiments for all groups except GRN+/− astrocytes (n = 4 mice). Error bars represent mean ± SEM.
The cerebral cortex contains multiple cell types, including neurons, astrocytes, and microglia, making overall gene expression changes in the bulk tissue difficult to interpret at the cellular level. To obtain a better understanding of the cell-type–specific changes in lysosomal gene expression within GRN−/− mice, we dissociated the cortices of aged GRN+/− and GRN−/− mice; used FACS to sort each sample into NeuN-positive neuron, GFAP-positive astrocyte, and Cd11b-positive microglia/macrophage populations; and performed comparative RNA-seq analysis on the sorted cells (Srinivasan et al., 2016). Analysis of several cell-specific markers confirmed the purity of our sorts (Fig. S1). Surprisingly, although the lysosome-related gene set was strongly up-regulated in GRN−/− whole cortical tissue (Fig. 1 C), the same lysosomal gene set showed no changes in neurons and astrocytes and bidirectional changes in microglia/macrophages, with approximately half of the lysosomal genes up-regulated and half down-regulated in microglia/macrophages (Fig. 1, D and E). It should be noted that microglia/macrophages express much higher levels of these lysosomal genes than either neurons or astrocytes (Fig. 1 F), and GRN−/− mice display accelerated gliosis as they age (Ahmed et al., 2010; Yin et al., 2010a; Wils et al., 2012). Thus, microglia/macrophages are the major contributor to lysosomal gene expression in whole cortex, and the increase in levels of lysosomal gene messenger RNA observed in GRN−/− cortex (Lui et al., 2016) is largely the result of a relative increase in microglia/macrophage numbers (i.e., microgliosis) as opposed to increased cellular expression (Fig. 1 F). However, results from our cell sorting experiment circumvent this caveat and reveal dysregulated lysosomal gene expression in GRN−/− myeloid cells, suggesting altered lysosome function.
Mice lacking PGRN have L. monocytogenes clearance defects
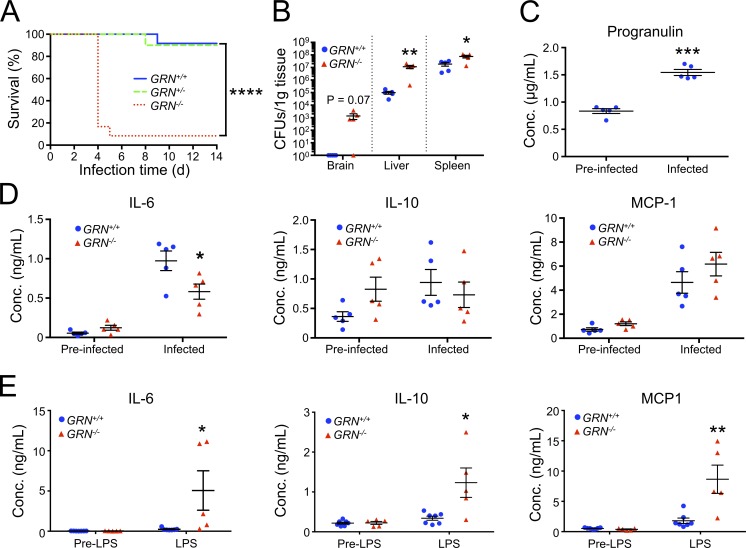
Lysosomes have a critical function in the autophagy pathway, fusing with autophagosomes to degrade unwanted cargo. Listeria monocytogenes is an intracellular bacterium that is cleared by macrophages, in part through xenophagy, a specialized form of autophagy that targets pathogens. Therefore, to test for lysosome-related impairments in GRN−/− macrophages in vivo, we infected GRN+/+, GRN+/−, and GRN−/− mice with a typically sublethal dose of L. monocytogenes and measured their ability to clear the bacteria and survive the infection. Although almost all GRN+/+ and GRN+/− mice survived infection through 2 wk, more than 90% of GRN−/− mice died within 5 d postinfection (Fig. 2 A). Consistent with a previous finding (Yin et al., 2010b), the increased susceptibility of GRN−/− mice to L. monocytogenes infection was accompanied by an increased bacterial burden in the brain, liver, and spleen (Fig. 2 B), demonstrating their inability to properly clear the pathogen.
Figure 2.
Impaired defense against Listeria monocytogenes in mice lacking PGRN. (A) 14-d survival curve of GRN+/+ (solid blue line), GRN+/− (dashed green line), and GRN−/− (dotted red line) mice after intravenous L. monocytogenes infection. n = 12 mice per group from one experiment. ****, P < 0.0001 GRN+/+ versus GRN−/− using a log-rank test. (B) L. monocytogenes burden in GRN+/+ (blue circles) and GRN−/− (red triangles) mice 2 d after infection. n = 5 mice per group from one experiment. *, P < 0.05 and **, P < 0.01 versus GRN+/+ using an unpaired t test with Welch’s correction. (C) Serum PGRN levels in response to L. monocytogenes infection. n = 5 mice per group from one experiment. ***, P < 0.001 versus pre-infected using a paired t test. (D) Serum IL-6, IL-10, and MCP-1 levels in GRN+/+ (circles) and GRN−/− (triangles) mice after intravenous L. moncytogenes infection. n = 5 mice per group from one experiment. *, P < 0.05 versus GRN+/+ using an unpaired t test. (E) Serum IL-6, IL-10, and MCP-1 levels in GRN+/+ (circles) and GRN−/− (triangles) mice after treatment with LPS. n = 5 mice per group from one experiment. *, P < 0.05 and **, P < 0.01 using an unpaired t test. Error bars represent mean ± SEM.
PGRN has previously been reported to have antiinflammatory properties (Zhu et al., 2002; Yin et al., 2010b), prompting us to ask whether this contributed to the L. monocytogenes clearance impairment. Twenty-four hours after infection with Listeria, we observed serum PGRN and cytokine levels rise in GRN+/+ mice (Fig. 2, C and D). However, we saw no evidence of heightened systemic inflammation in GRN−/− mice in response to L. monocytogenes infection when compared with GRN+/+ mice, despite the increased bacterial burden (Fig. 2 D). On the other hand, dosing GRN−/− mice with LPS stimulated a larger increase in the release of several proinflammatory cytokines than in GRN+/+ mice (Fig. 2 E), consistent with previous reports (Yu et al., 2016). Thus, we hypothesize that the inability for GRN−/− mice to effectively clear L. monocytogenes is caused by a defect in xenophagy.
PGRN is required for efficient macrophage clearance of L. monocytogenes via xenophagy
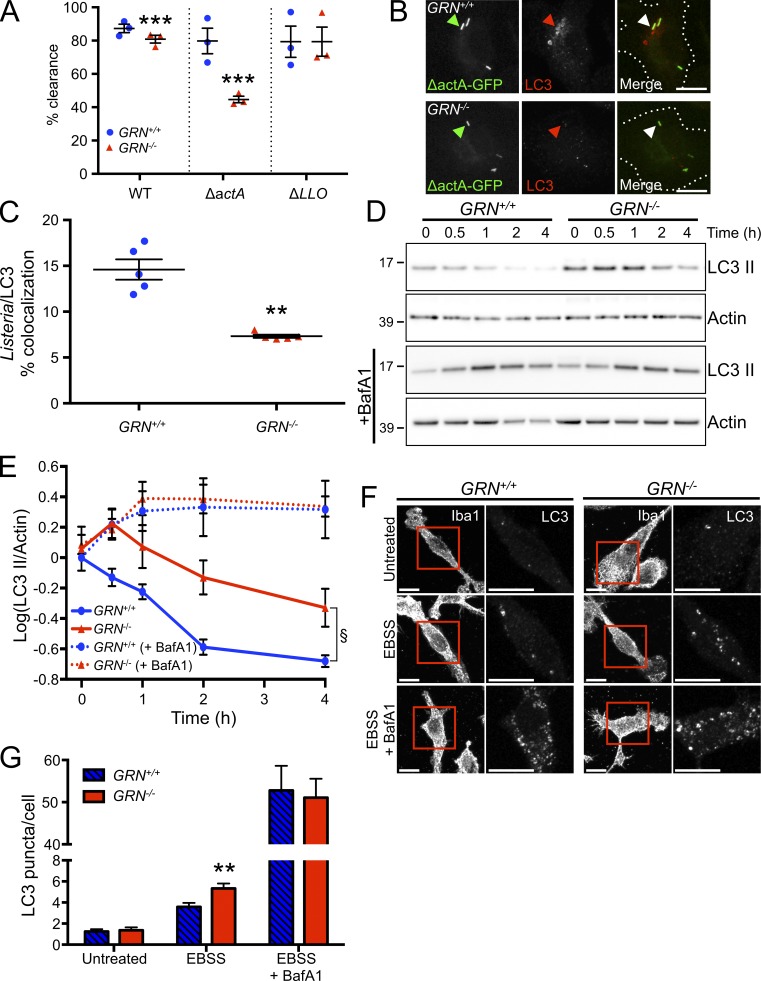
To assess the ability of GRN−/− macrophages to clear intracellular pathogens via xenophagy, we infected cultured GRN+/+ and GRN−/− BMDMs with L. monocytogenes and measured the percentage of bacteria cleared 6 h after infection. Consistent with our hypothesis and what we observed in vivo, infected GRN−/− BMDMs displayed an ∼10% impairment in clearance of WT L. monocytogenes at this time point when compared with GRN+/+ BMDMs (Fig. 3 A).
Figure 3.
Impaired xenophagy in macrophages from GRN−/− mice. (A) L. monocytogenes clearance assay of BMDMs from GRN+/+ (blue circles) and GRN−/− mice (red triangles) in response to the indicated strain of L. monocytogenes. n = 3 independent experiments, with each experiment performed in quadruplicate (mean value of each independent experiment plotted in graph). ***, P < 0.001 versus GRN+/+, using a linear least squares regression analysis. (B) Representative micrographs from n = 5 independent experiments showing BMDMs from GRN+/+ (top row) and GRN−/− (bottom) mice infected with GFP-labeled ΔactA L. monocytogenes (left) and stained for LC3 (middle). Example bacteria are marked with an arrowhead. Bars, 10 µm. (C) Quantification of L. monocytogenes/LC3 colocalization from experiments as shown in B. n = 5 independent experiments. **, P < 0.01 versus GRN+/+ using an unpaired t test. (D) Representative Western blot from n = 4 independent experiments of cell lysates from GRN+/+ and GRN−/− BMDMs starved for the indicated time in the absence (top row) and presence (middle row) of 100 nM bafilomycin A1 and probed for LC3 and actin. The actin (loading control) immunoblots shown in Figs. 3 D and S2 A are identical; this actin immunoblot, as well as LC3 (Fig. 3 D) and p62 (Fig. S2 A), were obtained by reprobing of the same Western blot. Molecular mass is indicated in kilodaltons. (E) Quantification of LC3 II levels from experiments as in D. n = 4 independent experiments. §, P = 0.0028 for GRN−/− versus GRN+/+ using two-way ANOVA and Dunnett’s multiple comparisons test (0.5 h = P < 0.01, 1 h = P < 0.05, 2 h = P < 0.0001, and 4 h = P < 0.01). (F) Representative images from n = 3 independent experiments of GRN+/+ (left) and GRN−/− (right) BMDMs treated with the indicated compounds before staining for LC3. Bars, 10 µm. (G) Quantification of number of LC3 puncta per cell from experiments as shown in F. n = 56, 47, 39, 60, 67, 65 cells from three independent experiments for the groups listed left to right. **, P < 0.01 versus GRN+/+ using a Mann–Whitney U test. Error bars represent mean ± SEM. EBSS, Earle’s balanced salt solution.
L. monocytogenes employs countermeasures to escape xenophagy and destruction in lysosomes. In particular, these pathogens can escape autophagic detection through recruitment of host cytoskeletal proteins to the bacterial surface via the bacterial protein Actin assembly-inducing protein (ActA; Yoshikawa et al., 2009). L. monocytogenes lacking ActA are more susceptible to xenophagy than WT bacteria (Yoshikawa et al., 2009). When we infected GRN+/+ and GRN−/− BMDMs with ΔactA mutants, the GRN−/− BMDMs showed a more pronounced (∼45%) deficit in bacterial clearance when compared with GRN+/+ BMDMs (Fig. 3 A). In addition, when we infected macrophages with ΔactA L. monocytogenes and measured the number of bacteria localized to autophagosomes 1 h postinfection, there was a significant reduction in the colocalization of the bacteria and the autophagosome marker LC3 in GRN−/− BMDMs (Fig. 3, B and C). Importantly, when we infected BMDMs with L. monocytogenes lacking the virulence factor listeriolysin O (a pore-forming toxin essential for phagosomal escape and entry into the cytosol), which renders these mutant Listeria insusceptible to autophagy, we observed no difference in bacterial clearance between the GRN+/+ and GRN−/− BMDMs (Fig. 3 A). Together, these results indicate a xenophagy impairment in GRN−/− BMDMs and provide a cell biological explanation for the inability of GRN−/− mice to clear L. monocytogenes and survive the infection.
PGRN is required for proper autophagy
Autophagy is induced in macrophages not only upon intracellular infection, but also by other stimuli such as nutrient starvation. To address whether other forms of autophagy are also impacted in the absence of PGRN, we used amino acid starvation to induce autophagy in BMDMs and measured autophagic flux by monitoring the change in levels of the autophagosome marker protein, microtubule-associated protein 1A/1B-light chain 3 (LC3 II; Klionsky et al., 2016). Basal LC3 II levels were slightly higher in GRN−/− BMDMs relative to GRN+/+ BMDMs, although this did not reach statistical significance (Fig. 3, D and E). Upon starvation, LC3 II levels increased in GRN−/− BMDMs and then declined more slowly when compared with GRN+/+ BMDMs, implying altered autophagy in the absence of PGRN (Fig. 3, D and E). An identical result was observed when we measured starvation-induced degradation of the autophagy adaptor, p62 (Fig. S2, A and B). Notably, when we added the V-ATPase inhibitor bafilomycin A1 to block the degradation of autophagosomes during autophagy by inhibiting autophagosome-lysosome fusion, the rate of LC3 II formation was identical between GRN+/+ and GRN−/− BMDMs (Fig. 3, D and E), suggesting that the autophagosome accumulation and slower decay of LC3 II in GRN−/− BMDMs is caused by the impaired clearance of autophagosomes (Klionsky et al., 2016).
As an alternative approach to monitoring autophagy, we counted the number of autophagosomes by LC3 immunocytochemistry. At basal state, both GRN+/+ and GRN−/− BMDMs had very few detectable LC3-positive puncta (Fig. 3, F and G). Upon amino acid starvation for 4 h, the number of autophagosomes in both GRN+/+ and GRN−/− BMDMs increased, but PGRN-deficient BMDMs contained ∼50% more autophagosomes than GRN+/+ BMDMs (Fig. 3, F and G). Similar to what was observed biochemically, starvation of the cells in the presence of bafilomycin A1 resulted in increased numbers of autophagosomes that were indistinguishable between GRN+/+ and GRN−/− BMDMs (Fig. 3, F and G), further supporting the notion of impaired autophagosome clearance in GRN−/− BMDMs. Together, these results demonstrate that PGRN is involved not only in xenophagy but also in starvation-induced autophagy in BMDMs.
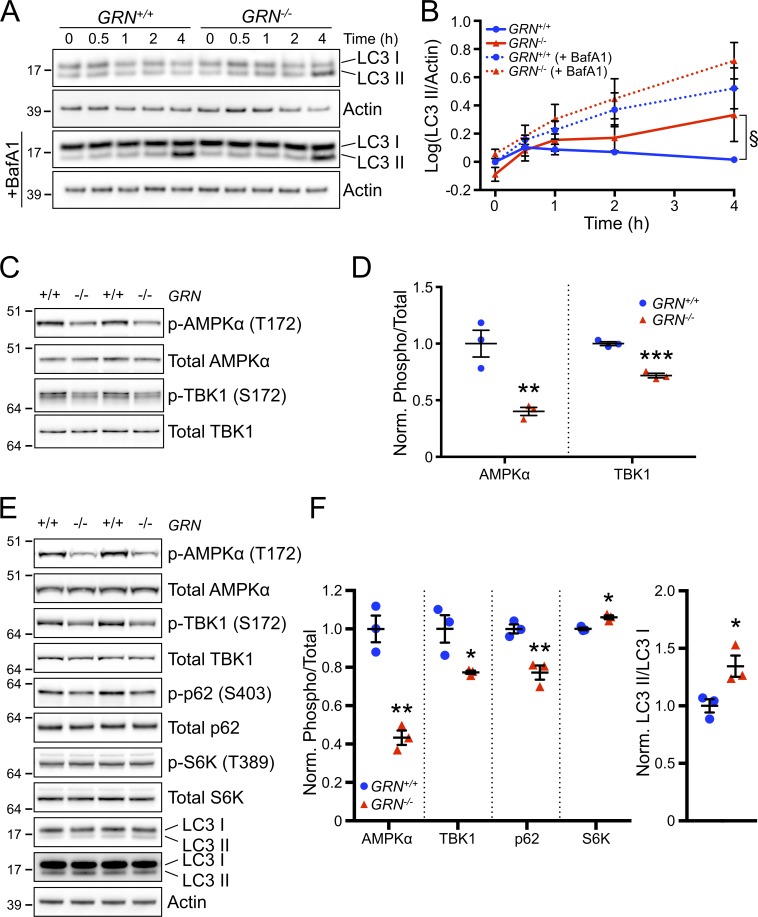
A pathological hallmark of GRN-associated frontotemporal lobar degeneration is the accumulation of ubiquitin- and TDP-43–positive inclusions specifically in the soma and dendrites of neurons (Baker et al., 2006; Cruts et al., 2006; Neumann et al., 2006). PGRN has been detected in both neurons and microglia in the brain, leading us to hypothesize that the autophagy defects observed in BMDMs may extend to neurons and may contribute to the accumulation and aggregation of TDP-43 in neurons. Indeed, clearance of pathological forms of TDP-43 through autophagy has been previously reported (Wang et al., 2012; Barmada et al., 2014). To test whether GRN−/− neurons are impaired in autophagy, we induced autophagy in cultured cortical neurons from GRN+/+ and GRN−/− mice by nutrient starvation and assessed turnover of LC3 II. Induction of autophagy in GRN+/+ neurons led to very little change in LC3 II levels over time, likely because of highly efficient autophagosome clearance in neurons (Boland et al., 2008). However, similar to what was observed in BMDMs, GRN−/− neurons had altered LC3 II kinetics, with an approximately threefold accumulation of LC3 II after 4 h of starvation (Fig. 4, A and B). When we blocked autophagosome degradation with bafilomycin A1, both GRN+/+ and GRN−/− neurons showed comparable, elevated levels of LC3 II (Fig. 4, A and B), suggesting that similar to BMDMs, the accumulation of autophagosomes in GRN−/− neurons is caused by an impairment of autophagosome clearance.
Figure 4.
Impaired autophagy and autophagy signaling in GRN−/− neurons. (A) Representative Western blot from n = 3 independent experiments of cell lysates from GRN+/+ and GRN−/− cortical neurons starved for the indicated times in the absence (top rows) or presence (bottom) of 100 nM bafilomycin A1 and probed for LC3 and actin. (B) Quantification of LC3 II levels from experiments as in A. n = 3 independent experiments. §, P = 0.038 for GRN−/− versus GRN+/+ using two-way ANOVA and Dunnett’s multiple comparisons test (0.5 h = not significant [n.s.], 1 h = P < 0.05, 2 h = P < 0.01, and 4 h = P < 0.0001). (C) Representative Western blot from n = 3 independent experiments of cell lysates from cultured GRN+/+ and GRN−/− cortical neurons probed with the indicated antibodies. (D) Quantification of phosphorylated/total signal for AMPK and TBK1 from experiments as shown in C. n = 3 independent experiments. **, P < 0.01 and ***, P < 0.001 versus GRN+/+ using unpaired t test. (E) Representative Western blot of whole cortex from ∼18-mo-old GRN+/+ and GRN−/− mice probed with the indicated antibodies. n = 3 mice per group from one experiment. (A, C, and E) Molecular mass is indicated in kilodaltons. (F) Quantification of phosphorylated/total signals for the indicated proteins (left) and LC3 II/LC3 I signal (right) from experiments as shown in E. n = 3 mice per group from one experiment. *, P < 0.05 and **, P < 0.01 using an unpaired t test. Error bars represent mean ± SEM.
To gain insight into the mechanism by which loss of PGRN leads to impaired autophagy, we probed GRN+/+ and GRN−/− neurons for alterations in the autophagy signaling pathway. 5′-Adenosine monophosphate–activated protein kinase (AMPK) is a central regulator of autophagy, stimulating autophagy through both the activation of the autophagy-initiating kinase ULK1 (Unc-51 like autophagy activating kinase 1) and the inhibition of mammalian target of rapamycin complex 1 (mTORC1). AMPK exists as a heterotrimeric complex comprising a catalytic α subunit and regulatory β and γ subunits. Phosphorylation of threonine-172 on the α subunit by one of several known upstream kinases is required for activation of AMPK and thus serves as a marker for monitoring AMPK activity (Klionsky et al., 2016). Western blot of cultured cortical neurons from GRN+/+ and GRN−/− mice revealed a ∼60% decrease in AMPK Thr-172 phosphorylation, indicating reduced activity of AMPK in the absence of PGRN (Fig. 4, C and D). Similarly, we found an ∼30% decrease in phosphorylation on serine-172 of TANK-binding kinase 1 (TBK1; also called NAK), a positive regulator of autophagy (Matsumoto et al., 2011; Wild et al., 2011; Pilli et al., 2012; Fig. 4, C and D). Phosphorylation on serine-172 via trans-autophosphorylation is required for TBK1 kinase activity. Thus, these results demonstrate that PGRN is necessary for the normal activation of autophagy regulators AMPK and TBK1 in cultured neurons.
Next, we assessed whether deficits in autophagy pathway signaling were also present in vivo in the brain. Cortical tissue from aged GRN−/− mice displayed reduced phosphorylation of AMPK (T172) and TBK1 (S172; Fig. 4, E and F), similar in degree to that seen in cultured neurons, implying hypoactivity of autophagy. Consistent with attenuated TBK1 activity, phosphorylation of p62 (also known as SQSTM1, a well-characterized autophagy receptor phosphorylated by TBK1 on serine-403 to enhance its activity; Matsumoto et al., 2011) was reduced by ∼25% in GRN−/− brain (Fig. 4, E and F). Additionally, mTORC1 activity, measured via phosphorylation of its substrate, p70 S6 kinase, at residue threonine-389, showed a modest but statistically significant increase. We also discovered an ∼35% increase in the levels of autophagosome marker LC3 II when normalized to cytosolic LC3 I, suggesting autophagosome accumulation in the cortex (Fig. 4, E and F). Although the signaling changes in whole cortex could potentially come from many different cell types within the brain, these results demonstrate that PGRN is required for normal activation of the key autophagy regulators AMPK and TBK1 and further support the idea that PGRN deficiency causes an autophagy defect in macrophages and neurons.
PGRN stimulates autophagy
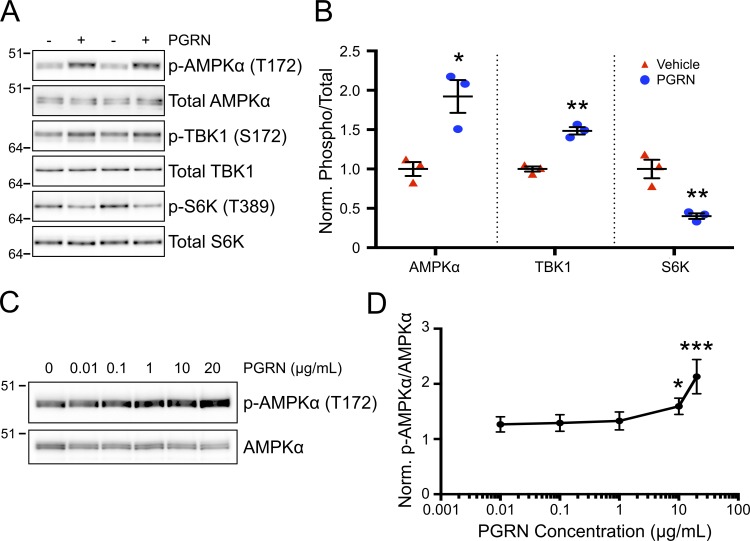
PGRN has been reported to stimulate several signal transduction pathways, including the PI3K/Akt and MAPK/ERK pathways, in a variety of different cell types including neurons (Bateman and Bennett, 2009; Xu et al., 2011). We tested whether PGRN was sufficient to stimulate autophagy signaling in neurons. GRN−/− neurons were treated with recombinant mouse PGRN for 4 h and autophagy pathway activity was measured. Upon treatment with PGRN, we observed ∼90% and ∼50% increases in phosphorylated AMPK (T172) and TBK1 (S172) levels, respectively, whereas phosphorylation of the mTORC1 substrate S6K (T389) declined ∼60% (Fig. 5, A and B). This stimulation of autophagy occurred in a dose-dependent manner, with an ∼60% increase in phosphorylated AMPK (T172) levels detected at 10 µg/ml PGRN (Fig. 5, C and D), a concentration that is comparable to that found in mouse serum after LPS administration (Fig. S3). Together, these results demonstrate not only that PGRN is necessary for proper autophagy pathway activation but also that purified recombinant PGRN is sufficient to stimulate autophagy signaling.
Figure 5.
Stimulation of autophagy signaling by PGRN. (A) Representative Western blot from n = 3 independent experiments of cell lysates from GRN−/− cortical neurons untreated or with 10 µg/ml recombinant mouse PGRN or vehicle and probed with the indicated antibodies. (B) Quantification of phosphorylated/total protein signals for the indicated proteins from experiments as shown in A. n = 3 independent experiments. *, P < 0.05 and **, P < 0.01 versus GRN+/+ using an unpaired t test. (C) Dose–response relationship for PGRN stimulation of AMPKα phosphorylation. Representative Western blot from n = 3 independent experiments of GRN−/− cortical cultures treated with the indicated concentrations of recombinant mPGRN and probed with the indicated antibodies. (A and C) Molecular mass is indicated in kilodaltons. (D) Quantification of phospho-AMPKα /total AMPKα signal from experiments as shown in C. n = 3 independent experiments. *, P < 0.05 and ***, P < 0.001 versus untreated using one-way ANOVA with Dunnett’s multiple comparisons test. Error bars represent mean ± SEM.
Pathogenic TDP-43 accumulates more rapidly in neurons lacking PGRN
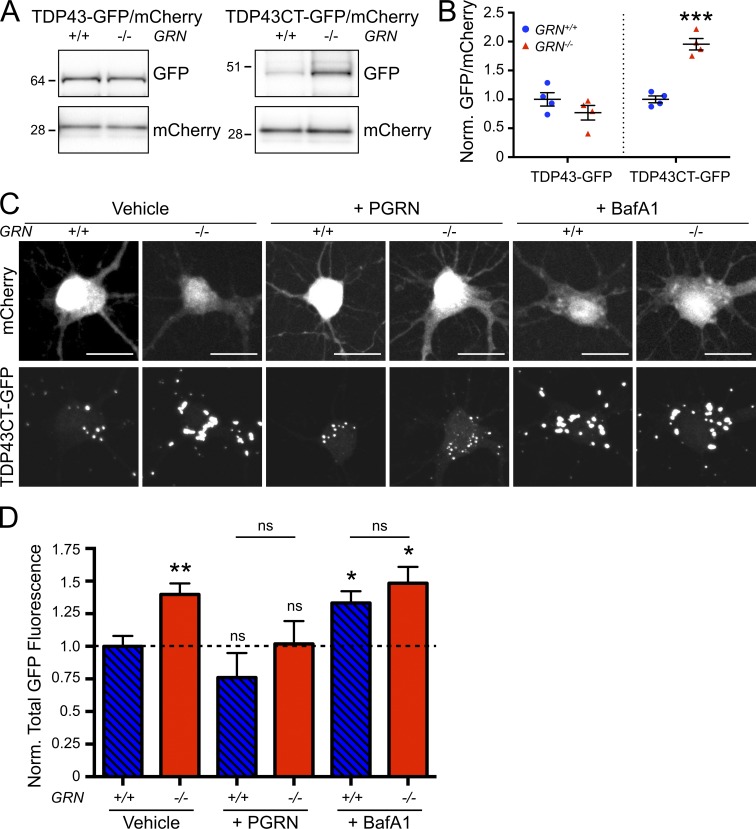
Accumulation of TDP-43–containing inclusions in neurons and glia is a pathological hallmark of GRN-associated frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U). Recent studies suggest that autophagy is an important pathway for clearance of pathological forms of TDP-43 (Caccamo et al., 2009; Barmada et al., 2014). We hypothesized that the autophagy impairment observed in GRN−/− neurons could result in reduced capacity to clear pathogenic TDP-43. We compared GRN+/+ and GRN−/− neurons after infection with an adeno-associated virus (AAV) expressing either full-length TDP-43 (TDP43-GFP) or a pathogenic C-terminal fragment of TDP-43 (TDP43CT-GFP). This fragment of TDP-43 is present in FTLD-U patient brains (Zhang et al., 2007), can be generated through the suppression of GRN expression (Zhang et al., 2007), and has previously been shown to be cleared by autophagy (Caccamo et al., 2009). Although full-length TDP43-GFP expression was similar between GRN−/− and GRN+/+ neurons (Fig. 6, A and B), levels of TDP43CT-GFP were elevated in GRN−/− neurons relative to GRN+/+ neurons after 10 d of expression (Fig. 6, A and B).
Figure 6.
Accelerated accumulation of pathogenic TDP-43 in GRN−/− neurons. (A) Representative Western blot from n = 4 independent experiments of GRN+/+ and GRN−/− cortical cultures infected with AAV expressing mCherry along with either full-length TDP43-GFP (left) or TDP43CT-GFP (right) and probed with the indicated antibodies. Molecular mass is indicated in kilodaltons. (B) Quantification of GFP/mCherry signal from experiments as shown in A. n = 4 independent experiments. ***, P < 0.001 versus GRN+/+ using an unpaired t test. (C) Representative images of GRN+/+ and GRN−/− hippocampal neurons cotransfected with mCherry (top row) and TDP43CT-GFP (bottom) and treated with vehicle (DMSO; left), 10 µM PGRN (middle), or 100 nM bafilomycin A1 (right). n = 158, 173, 37, 69, 147, and 96 cells per genotype and condition from eight, eight, three, three, eight, and eight independent experiments for the groups listed left to right. Bars, 25 µm. (D) Quantification of total GFP fluorescence per cell from experiments as shown in A. Data were normalized to the mean of GRN+/+ vehicle-treated cells. n = 158, 173, 37, 69, 147, and 96 cells per genotype and condition from eight, eight, three, three, eight, and eight independent experiments for the groups listed left to right. *, P < 0.05 and **, P < 0.01 versus vehicle-treated GRN+/+ using Kruskal–Wallis one-way ANOVA with Dunn’s multiple comparisons test. Error bars represent mean ± SEM. ns, not significant.
In a complementary approach, we transfected neurons with TDP43CT-GFP along with a cell marker, mCherry, and measured total GFP fluorescence per cell by microscopy. TDP43CT-GFP forms cytoplasmic clusters within transfected neurons (Fig. 6 C). Consistent with what was observed by Western blot using AAV to express TDP43CT-GFP (Fig. 6, A, B), GRN−/− neurons accumulated on average an ∼40% higher level of TDP43CT-GFP than GRN+/+ neurons (Fig. 6, C and D). Importantly, treatment of GRN−/− neurons with recombinant PGRN was sufficient to rescue this effect (Fig. 6, C and D), whereas treatment with bafilomycin A1 resulted in an increase in TDP43CT-GFP that was no different between GRN+/+ and GRN−/− neurons (Fig. 6, C and D). Together, these results support the idea that impaired autophagy contributes to the accumulation of TDP-43 in GRN−/− neurons and that PGRN can stimulate clearance of TDP-43 through autophagy.
Discussion
Patients with heterozygous and homozygous GRN loss-of-function mutations develop FTLD-U and NCL, respectively, but the mechanisms of pathogenesis are not well understood. However, recent findings suggest that FTD and NCL share common pathological features and thus may be caused by overlapping disease mechanisms (Götzl et al., 2014). Our study offers novel insight into how PGRN deficiency contributes to disease pathology by providing the first demonstration of a potential mechanism that links PGRN to lysosome-mediated protein degradation. Using biochemical, cell biological, and functional experiments, we demonstrate impaired autophagy and hypoactivity of key autophagy regulators in macrophages and neurons from mice lacking PGRN. Interestingly, GRN−/− BMDMs have impaired ΔactA L. monocytogenes clearance yet exhibit normal clearance of ΔLLO bacteria, arguing against a global impairment in lysosome activity in GRN−/− cells and instead suggesting that the deficit may be restricted to specific lysosome-associated pathways and functions, such as autophagy. Likely as a consequence of the autophagy deficit, GRN−/− neurons more readily accumulate pathological forms of TDP-43 previously shown to be cleared via autophagy (Caccamo et al., 2009). Based on our findings, GRN now joins a growing list of genes that regulate autophagy (including VCP, CHMP2B, SQSTM1, TBK1, and OPTN; Watts et al., 2004; Skibinski et al., 2005; Le Ber et al., 2013; Freischmidt et al., 2015; Pottier et al., 2015), which, when mutated, cause FTD and/or ALS in humans and result in the formation of TDP-43–positive inclusions in neurons. The positive regulation of the autophagy pathway by PGRN reinforces the emerging unifying theme that autophagy defects play a causal role along the FTD/ALS spectrum and that autophagy is a critical clearance mechanism for TDP-43.
In addition to the aforementioned association with autophagy, an overwhelming amount of evidence links PGRN to lysosome-associated function: (1) Immunohistochemical experiments demonstrate that PGRN accumulates within lysosomes of microglia and neurons (Hu et al., 2010; Tanaka et al., 2013; Zhou et al., 2015), where it can regulate the acidification of lysosomes (Tanaka et al., 2017). (2) The promoter region of GRN contains two coordinated lysosomal expression and regulation sequences (Tanaka et al., 2013), which make it a likely target of transcription factor EB, a master regulator of lysosome biogenesis (Sardiello et al., 2009). (3) Patients with homozygous GRN mutations develop NCL, a lysosomal storage disorder (Smith et al., 2012). (4) Lysosomal alkalizing reagents such as bafilomycin A1 and chloroquine, which also inhibit autophagy, stimulate PGRN production in multiple cell types, including neurons (Capell et al., 2011). (5) Results from our unbiased RNA-seq analysis revealed significant changes in lysosomal gene expression in GRN−/− microglia/macrophages. This body of evidence, combined with our findings, strongly suggest that PGRN plays a role in the later, degradative stages of autophagy. However, our study does not directly demonstrate a specific defect in autophagosome clearance in PGRN deficient cells, so follow-up studies examining this process in greater detail are warranted.
Despite an observed autophagy impairment in both neurons and macrophages, our RNA-seq data on sorted neurons failed to detect changes in lysosome-related gene expression as seen in microglia/macrophages. It is possible that neurons may respond to impaired autophagy through posttranscriptional mechanisms rather than transcriptional. Another possible explanation is that the much lower expression of lysosome-related genes in neurons (Fig. 1 F) may preclude us from detecting the modest changes in lysosomal gene expression observed in microglia.
Given that GRN haploinsufficiency is sufficient to predispose humans to FTLD-U, it is notable that heterozygous GRN+/− mice had no detectable impairment in L. monocytogenes clearance in vivo and minimal changes in gene expression when compared with GRN+/+ mice, consistent with the lack of phenotypes in the GRN+/− mice reported previously (Kayasuga et al., 2007; Ahmed et al., 2010; Lui et al., 2016). One possible explanation for this discrepancy is that normal human serum PGRN concentrations (∼200 ng/ml) are only one fifth the normal serum PGRN levels found in mouse (Finch et al., 2009; Fig. S3). Moreover, whereas GRN+/− mice express half the normal serum PGRN levels of GRN+/+ mice (Fig. S3), FTD patients with heterozygous GRN mutations express only ∼25% of normal human levels (Finch et al., 2009). Thus, GRN+/− mice may not present with any FTLD-U phenotype because they still express relatively high levels of PGRN compared with humans with heterozygous GRN mutations.
How does PGRN regulate autophagy? Several lines of evidence suggest that PGRN lies upstream of AMPK and/or TBK1, two critical regulators of autophagy. We find that GRN−/− mice and cells display reduced AMPK and TBK1 phosphorylation, whereas addition of recombinant PGRN is sufficient to stimulate the phosphorylation of these proteins in cells. TBK1 functions in part by phosphorylating p62, OPTN, and other autophagy receptors, enhancing their ability to recruit cargo to autophagosomes (Matsumoto et al., 2011; Wild et al., 2011; Pilli et al., 2012). Consistent with TBK1 hypoactivity, macrophages lacking PGRN show an impaired capacity to traffic intracellular bacteria into autophagosomes and clear them, a trait shared with cells harboring TBK1, p62, or OPTN mutations (Thurston et al., 2009; Zheng et al., 2009; Wild et al., 2011). TBK1 is also a regulator of autophagosome maturation through its association with the Ras-like GTPase Rab8b (Pilli et al., 2012), and we find impaired autophagosome clearance in PGRN-deficient cells. Since GRN−/− mice and cells show a significant autophagy phenotype only under conditions of acute stress (starvation, bacterial infection, or overexpression of aggregating proteins), we hypothesize that the ∼50% reduced pAMPK and pTBK1 levels observed in PGRN-deficient cells are sufficient for maintaining normal basal autophagy in unstressed cells but become limiting when the demands for autophagy are increased. Over long periods of time (such as aging), a modest deficiency in autophagy signaling can translate to increased risk of neurodegenerative disease.
Several putative PGRN receptors have been reported, including sortilin (Hu et al., 2010), TNF receptor (Tang et al., 2011), Toll-like receptor 9 (Park et al., 2011), prosaposin/mannose-6-phosphate receptor complex (Zhou et al., 2015), and EphA2 (Neill et al., 2016), none of which have a well-defined relationship to TBK1, AMPK, or autophagy. PGRN deficiency might also affect autophagy-independent pathways to induce TDP-43 pathology and neurodegeneration, such as impairment in the nuclear import of cytoplasmic TDP-43 (Ward et al., 2014). Thus, TDP-43 pathology in FTD patients may accumulate as a result of a combination of altered cell biological functions, not solely impaired autophagic clearance of cytoplasmic TDP-43.
In light of our findings, several important questions remain. PGRN can exist either in a precursor form or can be proteolytically processed into a set of granulins. Our studies with GRN−/− cells do not functionally distinguish between these species. Our pharmacological experiments imply that recombinant PGRN in its unprocessed form can stimulate autophagy; however, it is possible that proteolytic cleavage to granulins is occurring in the culture medium or within cells after internalization of PGRN. An additional consideration is that although previous immunohistochemical studies have detected PGRN protein in microglia and neurons, but not in astrocytes (Ryan et al., 2009; Petkau et al., 2010), analysis of our RNA-seq data reveals that astrocytes express higher levels of GRN mRNA than neurons (Fig. S4), a result confirmed by another study (Zhang et al., 2014). Thus, although our study demonstrates autophagy impairments in both macrophages and neurons, this effect may not be strictly limited to these cell types within the brain and additional studies are warranted to further elucidate the cellular and molecular process by which PGRN stimulates the autophagy pathway.
Materials and methods
Experimental design
The objective of this study was to identify the role of PGRN in FTD pathogenesis. Identification of disrupted lysosome expression in the brains of GRN−/− mice was used to justify our follow up studies examining autophagy/lysosome function in GRN−/− cells. Sample sizes and endpoints for experiments were predetermined based on previous experience. All available samples or animals were included for statistical analysis. All in vitro studies were performed with a minimum of three independent experiments, based on previous studies, and as indicated in the figure legends. Image acquisition and analysis were performed using automated software and/or by a blinded observer.
Materials
Recombinant mouse PGRN (2557-PG; R&D Systems) and bafilomycin A1 (B1793; Sigma) were used.
Cloning
To generate the TDP43CT-GFP construct, we used a QuikChange Lightning mutagenesis kit (Agilent) to delete amino acids 2–215 from full-length human TDP-43 fused at the C terminus to GFP in a pCAGGS vector.
Mice
GRN−/− mice were generated by M. Nishihara (University of Tokyo, Tokyo, Japan) as described previously (Kayasuga et al., 2007). GRN+/+, GRN+/−, and GRN−/− mice were of the genetic background C57BL/6J. For in vitro neuron experiments, GRN+/+ neurons were cultured from GRN+/+ mice on a C57BL/6J background, acquired from Charles River Laboratory. For all other experiments, GRN+/+ colony mates were used as controls. For all L. monocytogenes studies, only male mice were used; otherwise, mice used were of mixed sex. The use of vertebrate animals was regulated and approved by the Genentech Institutional Animal Care and Use Committee.
Antibodies
For immunocytochemical studies, the following primary antibodies were used: LC3 mouse monoclonal (M152-3/1:50; MBL), Iba1 rabbit polyclonal (019-19741/1:1,000; Wako), and Lamp1 rat polyclonal (1D4B/1:300; DHSB). Alexa Fluor secondary antibodies were from ThermoFisher (1:400). For studies involving Western blots, the following primary antibodies were used: LC3 mouse monoclonal (M186-3/1:1,000; MBL), actin (A2228/1:5,000; Sigma), p-AMPK (T172) rabbit polyclonal (2535/1:1,000; CST), AMPK rabbit polyclonal (5831/1:1,000; CST), p-TBK1 (S172) rabbit polyclonal (5483/1:1,000; CST), TBK1 rabbit polyclonal (ab40676/1:1,000; Abcam), p-p62 (S403) rat polyclonal (D343-3/1:1,000; MBL), p62 mouse monoclonal (ab56416/1:1,000; Abcam), p-S6K (T389) rabbit polyclonal (AT-7159/1:1,000; MBL), S6K rabbit polyclonal (2708/1:1,000; CST), GFP chicken polyclonal (ab13970/1:5,000; Abcam), and mCherry rabbit polyclonal (ab167453/1:1,000; Abcam). For FACS, the following antibodies were used: Alexa Flour 488–conjugated anti-NeuN (MAB377X/1:1,000; Millipore), PE-conjugated anti-GFAP (561483/1:50; BD Biosciences), and APC-conjugated anti-Cd11b (561690/1:250; BD Biosciences).
Cell culture
For hippocampal and cortical neuron cultures, the hippocampus and/or cortex was isolated from embryonic day 16.5 GRN+/+ or GRN−/− mice and treated with 0.7 mg ml−1 papain (Worthington) and 0.01% DNase (Sigma) in Hanks buffered salt solution (ThermoFisher) for 20 min at 37°C. After digestion, tissue was washed three times to remove enzymes and then triturated first using a 5-ml plastic pipette, followed by a fire-polished Pasteur pipette. Dissociated cells were filtered through a 70-µm filter and centrifuged at 300 g for 5 min at 4°C. Neurons were resuspended in NbActiv4 (Brainbits), counted, and plated on 18-mm coverslips (Carolina Biological) coated with 0.1 mg ml−1 30- to 70-kD mol wt poly-d-lysine in 0.1 M Trizma buffer, pH 8.5 (Sigma) and 2.5 µg ml−1 natural mouse laminin in 0.1 M Trizma buffer, pH 8.5 (ThermoFisher) at a density of 7 × 105 cells per 60-mm dish for immunocytochemistry. For biochemistry experiments, cortical neurons were plated 5 × 105 cells per well in a 12-well plate. 1 h after plating, the media was removed and replaced with fresh NbActiv4. Neurons were fed by replacing one third of the medium once weekly.
To harvest BMDMs, femurs were isolated from mice and placed into ice-cold PBS containing 0.5% BSA and 2 mM EDTA. The ends of each femur were cut off, and the femur was flushed with 10 ml ice-cold PBS containing 0.5% BSA and 2 mM EDTA. The bone marrow was centrifuged at 300 g for 5 min at 4°C, the media was removed, and the marrow was resuspended in 2 ml ACK lysing buffer (ThermoFisher) and incubated for 5 min at 25°C. After lysis, the remaining cells were passed through a 70-µm filter and centrifuged at 300 g for 5 min at 4°C. The media was removed and the cells were resuspended in DMEM (ThermoFisher) containing 10% FBS (Sigma), 1% GlutaMAX (ThermoFisher), 1% nonessential amino acids (ThermoFisher), 1% sodium pyruvate (ThermoFisher), and 50 ng ml−1 M-CSF. Cells were plated on nontissue culture–treated Petri dishes and allowed to differentiate for 6 or 7 d, after which the macrophages were carefully scraped off and replated on plasticware or coverslips coated with 1 mg ml−1 poly-d-lysine in 0.1 M Trizma buffer, pH 8.5, for use the next day. For biochemistry experiments, BMDMs were plated 4 × 105 cells per well in a 12-well plate. For immunocytochemistry, BMDMs were plated 1.25 × 105 cells per 18-mm coverslip.
RNA-seq
Whole-cortex RNA extraction
Mice were perfused with PBS, a single cortical hemisphere of ∼12-mo-old GRN+/− and GRN−/− mice was cut in half, and each section was added with a 5-mm stainless steel bead and 1 ml Qiazol Lysis Reagent (Qiagen) to an individual 2-ml centrifuge tube. Tissues were homogenized using a TissueLyser (2 × 20 Hz, 2 min at 4°C; Qiagen). After homogenization, samples were centrifuged for 1 min at 6,000 g, and the resulting supernatants were transferred to a new tube and incubated for 5 min at 25°C. Samples were then added to a 15-ml MaXtract High Density tube (Qiagen) along with 200 µl chloroform and shaken vigorously for 15 s before incubation for 2 min at 25°C and centrifugation at 1,500 g for 5 min at 4°C. The resulting upper aqueous phase was transferred into a new tube, and RNA was purified using an RNeasy Plus mini kit (Qiagen). The two samples originating from different pieces of the same cortex were combined after purification. The concentration of RNA samples was determined using a NanoDrop 8000 (ThermoFisher), and the integrity of RNA was determined by Fragment Analyzer (Advanced Analytical Technologies).
Dissociated cell preparation, immunostaining, FACS, and RNA isolation
Dissociation and isolation of neurons, microglia/macrophages, and astrocytes were performed as previously described (Srinivasan et al., 2016). In brief, animals were perfused with ice-cold PBS. Cortices together with hippocampi were dissociated rapidly in Hibernate media (ThermoFisher). The tissue samples were minced with a razor blade and incubated in Accutase (Sigma) at 4°C for 30 min. Tissue pieces were spun at 2,000 g for 1 min, resuspended in 1 ml Hibernate buffer, and triturated to get single cells. Samples were allowed to settle by gravity (1 min on ice), and the cloudy cell suspension on top was removed and filtered through a 70-μm cell strainer into a clean tube. This step was repeated five times. The filtered suspension was layered on a Percoll gradient and spun at low speed to get rid of myelin debris from the top layer and then at high speed to pellet cells. Cell pellet was resuspended in 1 ml Hibernate, and 1 ml ice-cold 100% ethanol was added dropwise to the cell suspension. Cells were then pipetted twice gently to mix the ethanol evenly and then incubated on ice for 15 min. After incubation, cells were washed twice in Hibernate to get rid of ethanol and resuspended in 1 ml Hibernate together with the following antibody cocktail: NeuN-488 at 1:1,000, GFAP-PE at 1:100, and Cd11b-APC at 1:200. Cells were then washed and resuspended in 4 ml Hibernate together with 2 μl DAPI (1 mg/ml) and FACS sorted to get neurons, astrocytes, and myeloid cells (microglia/macrophages). Animals were always processed in pairs (1 WT and 1 KO) and sorted simultaneously in two BD ARIA sorters.
RNA-seq library generation and sequencing
For whole-cortex RNA, 1 μg total RNA was used as an input material for library preparation using TruSeq RNA Sample Preparation kit v2 (Illumina). Size of the libraries was confirmed using Fragment Analyzer (Advanced Analytical Technologies), and their concentration was determined by qPCR-based method using a Library quantification kit (KAPA). The libraries were multiplexed and sequenced on Illumina HiSeq2500 (Illumina) to generate 30 M of single-end 50-bp reads.
For sorted cells, cDNA was generated using up to 20 ng total RNA with the Ovation RNA-Seq System V2 (NuGEN) as previously described (Srinivasan et al., 2016). Generated cDNA was sheared using LE220 focused ultrasonicator (Covaris) and quantified by Qubit dsDNA BR Assay (Life Technologies). 1 µg sheared cDNA was taken into library generation, starting at the end repair step, using the TruSeq RNA Sample Preparation kit v2 (Illumina). Size of the libraries was confirmed using Fragment Analyzer (Advanced Analytical Technologies), and their concentration was determined using a Library quantification kit. The libraries were multiplexed and sequenced on an Illumina HiSeq2500 to generate 30 M of single-end 50-bp reads.
RNA-seq alignment and feature counting
HTSeqGenie (Pau and Reeder, 2017) was used to perform filtering, alignment, and feature counting. HTSeqGenie uses GSNAP (Wu and Nacu, 2010) to align reads to the genome. We used version GRCm38 of the mouse genome and gene models from our internal database mostly based on RefSeq. Only reads with unique genomic alignments were analyzed. Such reads whose alignments overlapped any exon of the gene model (even by a single base) were counted toward that gene.
RNA-seq normalization
Normalized reads per kilobase of transcript per million mapped reads (nRPKM) values were used as a normalized measure of gene expression. This statistic is an attempt to combine the best of DESeq size Factor–normalized counts (Love et al., 2014) and traditional reads per kilobase of transcript per million mapped reads (RPKM). RPKM is defined as
where total means total uniquely aligned. Normalized RPKM is defined as
Note that the only difference is in the “M” term. The adjusted total reads is a statistic that is proportional to DESeq size Factor but rescaled to the level of total uniquely aligned reads:
Note that this is a single normalization based on size factors, with an overall adjustment to approximate total reads, but it is not a double normalization.
RNA-seq differential gene expression
Differential gene expression was performed with the limma Bioconductor package, after transforming the count data with the voom function (Ritchie et al., 2015). A prefilter was applied, and only genes with at least 10 counts in at least three samples (of either condition) were analyzed. p-values for other reads were simply set to 1 and log-fold-changes to 0 for visualization purposes, but such genes were not included in the multiple testing correction.
Heat maps
For heat maps, nRPKM values were normalized separately within each of four groups of samples: bulk tissue and the three cell types. Within each group, Z scores were calculated by subtracting the mean and dividing by the standard deviation of expression for each gene.
Density plots
Density plots were calculated using R’s density function, with default arguments.
RNA-seq GO analysis was performed using the PANTHER classification system (Mi et al., 2013).
AAV production
Sequences encoding either TDP43-GFP or TDP43CT-GFP were subcloned into pD2-T2A-mCherry vector (Vector BioLabs) to generate C-terminal mCherry fusion proteins linked via a 2A “self-cleavable” peptide linker (Szymczak et al., 2004), driven by a CBh promoter. The subsequent construct was packaged into the AAV-DJ serotype and transfected into HEK293 cells for expression, and virus was harvested and purified by CsCl centrifugation (Vector BioLabs).
TDP43-GFP expression
Hippocampal neurons cultured 7–10 d in vitro (DIV) were transfected using Lipofectamine LTX with PLUS Reagent (ThermoFisher) according to manufacturer. For each coverslip, 5 µl Lipofectamine LTX Reagent and 1 µl PLUS was added along with 0.2 µg TDP43CT-GFP and 0.8 µg mCherry plasmids in 1 ml media. After a 1-h incubation (the end of this incubation period was considered t = 0 h), media was changed, and neurons were incubated for an additional 9 h before fixation. For bafilomycin A1 experiments, cultures were treated 5 h posttransfection with 100 nM bafilomycin A1.
For AAV infections, DIV 7 hippocampal neurons were infected at an MOI of 3.2 × 105 for 10 d before protein quantification.
Immunocytochemistry
Neurons and BMDMs were fixed in 4% paraformaldehyde in PBS for 20 min at 25°C, followed by permeabilization with 100 µg ml−1 digitonin (Promega) for 10 min at 25°C. Nonspecific binding of antibodies was blocked with 10% goat serum (Jackson) in PBS for 1 h at 25°C. Primary antibodies were applied overnight at 4°C, and secondary antibodies were applied for 1 h at 25°C. Coverslips were washed with PBS in between all steps and were mounted with ProLong Gold Antifade Reagent (ThermoFisher).
Image analysis
Z-stacks of cells were imaged using an inverted Zeiss 710 confocal microscope set at software-defined optimal resolution and intervals based on objective used. Images were captured in 8-bit color without binning. Maximum intensity projections of each Z-stack were generated for quantification. All image analysis was performed using Fiji (Schindelin et al., 2012). For TDP43CT-GFP quantification, a set threshold was applied to the GFP signal of all images, and the total fluorescence intensity within the threshold area was measured and normalized to GRN+/+ neurons.
Biochemistry
For whole cortical lysates, mice were perfused with PBS, and the cortices of 12- to 18-mo-old GRN+/+ and GRN−/− mice were isolated into 1 ml ice-cold homogenization buffer (0.32 M sucrose, 5 mM Hepes, protease/phosphatase inhibitors) per cortical hemisphere. Each sample was disrupted using 12 strokes from a glass dounce homogenizer. Cortical lysates were centrifuged at 1,000 g for 15 min at 4°C, and the protein concentration of each supernatant was measured via BCA (ThermoFisher). Samples were diluted to 3.5 mg ml−1 in homogenization buffer before the addition of LDS sample buffer (ThermoFisher) and sample reducing agent (ThermoFisher).
For in vitro studies examining starvation-induced autophagy, BMDMs and neurons were washed twice with prewarmed Earle’s balanced salt solution (Sigma) before final incubation in Earle’s balanced salt solution with or without bafilomycin A1. Afterward, cells were washed once with ice-cold PBS and scraped into 2× gel loading buffer (2× LDS sample loading buffer and 2× sample reducing agent).
For Western blot, all samples were heated at 70°C for 10 min and 20 µg of protein from each sample was run. For all Western blots with the exception of LC3, samples were run on a NuPAGE Novex 4–12% Bis-Tris protein gel (ThermoFisher) using MOPS buffer (ThermoFisher) and transferred onto 0.2 µm nitrocellulose via Trans-Blot Turbo transfer system (Bio-Rad). For LC3 measurements, samples were run on a NuPAGE 12% Bis-Tris protein gel (ThermoFisher) using MES buffer (ThermoFisher) and transferred onto a methanol-activated 0.2-µm PVDF membrane (ThermoFisher) via NuPAGE Novex transfer system (ThermoFisher) in NuPAGE transfer buffer (ThermoFisher) containing 10% methanol.
In vivo bacterial infection
105 L. monocytogenes (ATCC strain 43251, passaged twice through C57/Bl6 mice before use) in sterile saline were injected into the tail vein of 2- to 3-mo-old mice. For survival studies, survival was monitored daily after infection for 2 wk. For CFU studies, the brain, spleen, and liver were harvested, weighed, and placed in 5 ml (brain and spleen) or 10 ml (liver) PBS containing 0.1% Triton X-100. Organs were homogenized using a disposable tissue grinder (ThermoFisher), and 1:1 serial dilutions were made in PBS containing 0.1% Triton-X 100. 5 µl of each dilution was plated in triplicate on brain heart infusion agar and incubated at 37°C overnight. Colonies were counted and CFUs per 1 g tissue were calculated.
For cytokine measurements, 2- to 3-mo-old mice were injected intraperitoneally with 3 mg kg−1 LPS (2 mg ml−1 in PBS) from Escherichia coli (strain O111:B4; Sigma).
In vitro bacterial infection
BMDMs were plated 1.25 × 105 per well of a 48-well plate. L. monocytogenes growing in brain heart infusion broth were counted (1 OD = 1.2 × 109 bacteria), resuspended in DMEM without antibiotics, and added to the macrophages at the indicated MOI. Plates were centrifuged at 200 g for 5 min at 25°C and then incubated for 30 min at 37°C to allow time for infection (the end of this incubation period was considered t = 0 h). After the 30-min incubation, cells were washed once with PBS and switched to DMEM containing 100 µg ml−1 gentamycin (ThermoFisher) to remove extracellular bacteria. For L. monocytogenes/p62 colocalization experiments, ΔactA-infected BMDMs were fixed 1 h after infection. To lyse BMDMs for L. monocytogenes clearance assay, cells were washed once with PBS and lysed in PBS containing 0.1% Triton X-100 0 hr and 6 h after infection. 1:1 serial dilutions of the lysates were made in PBS containing 0.1% Triton-X 100, and 5 µl of each dilution was plated in triplicate on brain heart infusion agar and incubated at 37°C overnight. Colonies were counted, and the percentage of colonies at 6 h versus 0 h was calculated.
Serum cytokine and PGRN measurements
150 µl blood was collected from the submandibular vein of 2- to 3-mo-old mice 7 d before and 24 h after infection using a 4-mm Goldenrod animal lancet (Braintree Scientific). Blood was collected in a Microtainer tube with serum separator (BD) and allowed to coagulate for 30 min at 25°C before centrifugation at 15,000 g for 90 s. Mouse serum IL-6, IL-10, and MCP-1 measurements were performed using a Bio-Plex Pro Mouse Cytokine 23-Plex Assay (Bio-Rad). Mouse serum PGRN levels were measured using a PGRN Mouse EIA kit (Cayman Chemical).
Accession numbers
RNA-seq data have been deposited to the Gene Expression Omnibus and are available under the accession numbers GSE78747 (sorted cells) and GSE78748 (bulk tissue).
Statistical analysis
Statistical analysis was performed using Prism 6 (GraphPad) and JMP (SAS). Differences between means were determined using a two-tailed unpaired Student’s t test, one-way or two-way ANOVA corrected for multiple comparisons, or linear regression as described in the figure legends, and statistical significance was set at P < 0.05. All available samples or animals were included for statistical analysis. Datasets with n ≥ 8 were tested for normality using the D’Agostino–Pearson omnibus normality test. If datasets did not meet normality, then a nonparametric test was chosen to determine statistical significance.
Online supplemental material
Fig. S1 shows expression of representative macrophage-specific (left), astrocyte-specific (middle), and neuron-specific (right) genes in sorted cells from GRN+/− and GRN−/− cortex. Fig. S2 illustrates reduced starvation-induced clearance of p62 in GRN+/+ and GRN−/− BMDMs. Fig. S3 shows serum PGRN concentrations in GRN+/+ and GRN−/− mice in response to LPS. Fig. S4 shows expression of GRN in sorted microglia/macrophages, astrocytes, and neurons from GRN+/+ cortex. Table S1 shows gene expression changes in GRN+/− cortex relative to GRN+/+ cortex. Table S2 shows GO cellular component enrichment in GRN−/− cortex relative to GRN+/+ cortex.
Supplementary Material
Acknowledgments
We thank Masugi Nishihara for providing GRN−/− mice; Laurie Appling, C.K. Poon, Jovencio Borneo, Terence Ho, and Jim Cupp for FACS assistance; Andres Paler Martinez for Luminex assistance; Mariela Del Rio for animal husbandry assistance; Jochen Brumm for statistical assistance; and Aditya Murthy, Rajkumar Noubade, and Paolo Manzanillo for helpful discussions and guidance.
M.C. Chang, K. Srinivasan, B.A. Friedman, E. Suto, Z. Modrusan, W.P. Lee, J.S. Kaminker, D.V. Hansen, and M. Sheng are full-time employees of Genentech, Inc., a member of the Roche group. The authors declare no additional competing financial interests.
Author contributions: M.C. Chang designed and performed experiments, analyzed the results, and wrote the bulk of the manuscript. K. Srinivasan, B.A. Friedman, and E. Suto performed experiments, analyzed the results, and helped with manuscript writing. Z. Modrusan, W.P. Lee, J.S. Kaminker, and D.V. Hansen helped with experimental design and manuscript revision. M. Sheng directed the study and helped with manuscript writing.
Footnotes
Abbreviations used:
- AAV
- adeno-associated virus
- ALS
- amyotrophic lateral sclerosis
- AMPK
- 5′-adenosine monophosphate–activated protein kinase
- DIV
- days in vitro
- FTD
- frontotemporal dementia
- FTLD-U
- frontotemporal lobar degeneration with ubiquitin-positive inclusions
- GO
- gene ontology
- NCL
- neuronal ceroid lipofuscinosis
- nRPKM
- normalized RPKM
- PGRN
- progranulin
- RPKM
- reads per kilobase of transcript per million mapped reads
References
- Ahmed Z., Sheng H., Xu Y.-F., Lin W.-L., Innes A.E., Gass J., Yu X., Wuertzer C.A., Hou H., Chiba S., et al. 2010. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am. J. Pathol. 177:311–324. 10.2353/ajpath.2010.090915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M.R., Macário M.C., Ramos L., Baldeiras I., Ribeiro M.H., and Santana I.. 2016. Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol. Aging. 41:200.e1–200.e5. 10.1016/j.neurobiolaging.2016.02.019 [DOI] [PubMed] [Google Scholar]
- Anakwe O.O., and Gerton G.L.. 1990. Acrosome biogenesis begins during meiosis: evidence from the synthesis and distribution of an acrosomal glycoprotein, acrogranin, during guinea pig spermatogenesis. Biol. Reprod. 42:317–328. 10.1095/biolreprod42.2.317 [DOI] [PubMed] [Google Scholar]
- Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. 2006. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 442:916–919. 10.1038/nature05016 [DOI] [PubMed] [Google Scholar]
- Barmada S.J., Serio A., Arjun A., Bilican B., Daub A., Ando D.M., Tsvetkov A., Pleiss M., Li X., Peisach D., et al. 2014. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat. Chem. Biol. 10:677–685. 10.1038/nchembio.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., and Bennett H.P.J.. 2009. The granulin gene family: From cancer to dementia. BioEssays. 31:1245–1254. 10.1002/bies.200900086 [DOI] [PubMed] [Google Scholar]
- Bateman A., Belcourt D., Bennett H., Lazure C., and Solomon S.. 1990. Granulins, a novel class of peptide from leukocytes. Biochem. Biophys. Res. Commun. 173:1161–1168. 10.1016/S0006-291X(05)80908-8 [DOI] [PubMed] [Google Scholar]
- Bhandari V., Giaid A., and Bateman A.. 1993. The complementary deoxyribonucleic acid sequence, tissue distribution, and cellular localization of the rat granulin precursor. Endocrinology. 133:2682–2689. 10.1210/endo.133.6.8243292 [DOI] [PubMed] [Google Scholar]
- Boland B., Kumar A., Lee S., Platt F.M., Wegiel J., Yu W.H., and Nixon R.A.. 2008. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J. Neurosci. 28:6926–6937. 10.1523/JNEUROSCI.0800-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A., Majumder S., Deng J.J., Bai Y., Thornton F.B., and Oddo S.. 2009. Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J. Biol. Chem. 284:27416–27424. 10.1074/jbc.M109.031278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell A., Liebscher S., Fellerer K., Brouwers N., Willem M., Lammich S., Gijselinck I., Bittner T., Carlson A.M., Sasse F., et al. 2011. Rescue of progranulin deficiency associated with frontotemporal lobar degeneration by alkalizing reagents and inhibition of vacuolar ATPase. J. Neurosci. 31:1885–1894. 10.1523/JNEUROSCI.5757-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantry D., DeMaggio A.J., Brammer H., Raport C.J., Wood C.L., Schweickart V.L., Epp A., Smith A., Stine J.T., Walton K., et al. 1998. Profile of human macrophage transcripts: Insights into macrophage biology and identification of novel chemokines. J. Leukoc. Biol. 64:49–54. [DOI] [PubMed] [Google Scholar]
- Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.-J., et al. 2006. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 442:920–924. 10.1038/nature05017 [DOI] [PubMed] [Google Scholar]
- Cruts M., Theuns J., and Van Broeckhoven C.. 2012. Locus-specific mutation databases for neurodegenerative brain diseases. Hum. Mutat. 33:1340–1344. 10.1002/humu.22117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiano A.J., Martens L.H., Young A.H., Warmus B.A., Zhou P., Diaz-Ramirez G., Jiao J., Zhang Z., Huang E.J., Gao F.-B., et al. 2013. Dissociation of frontotemporal dementia-related deficits and neuroinflammation in progranulin haploinsufficient mice. J. Neurosci. 33:5352–5361. 10.1523/JNEUROSCI.6103-11.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch N., Baker M., Crook R., Swanson K., Kuntz K., Surtees R., Bisceglio G., Rovelet-Lecrux A., Boeve B., Petersen R.C., et al. 2009. Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain. 132:583–591. 10.1093/brain/awn352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A., Wieland T., Richter B., Ruf W., Schaeffer V., Müller K., Marroquin N., Nordin F., Hübers A., Weydt P., et al. 2015. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 18:631–636. 10.1038/nn.4000 [DOI] [PubMed] [Google Scholar]
- Gao X., Joselin A.P., Wang L., Kar A., Ray P., Bateman A., Goate A.M., and Wu J.Y.. 2010. Progranulin promotes neurite outgrowth and neuronal differentiation by regulating GSK-3β. Protein Cell. 1:552–562. 10.1007/s13238-010-0067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass J., Lee W.C., Cook C., Finch N., Stetler C., Jansen-West K., Lewis J., Link C.D., Rademakers R., Nykjær A., and Petrucelli L.. 2012. Progranulin regulates neuronal outgrowth independent of sortilin. Mol. Neurodegener. 7:33 10.1186/1750-1326-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götzl J.K., Mori K., Damme M., Fellerer K., Tahirovic S., Kleinberger G., Janssens J., van der Zee J., Lang C.M., Kremmer E., et al. 2014. Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 127:845–860. [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N.. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hu F., Padukkavidana T., Vægter C.B., Brady O.A., Zheng Y., Mackenzie I.R., Feldman H.H., Nykjaer A., and Strittmatter S.M.. 2010. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 68:654–667. 10.1016/j.neuron.2010.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayasuga Y., Chiba S., Suzuki M., Kikusui T., Matsuwaki T., Yamanouchi K., Kotaki H., Horai R., Iwakura Y., and Nishihara M.. 2007. Alteration of behavioural phenotype in mice by targeted disruption of the progranulin gene. Behav. Brain Res. 185:110–118. 10.1016/j.bbr.2007.07.020 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo-Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 12:1–222. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., and Tanaka K.. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Le Ber I., Camuzat A., Guerreiro R., Bouya-Ahmed K., Bras J., Nicolas G., Gabelle A., Didic M., De Septenville A., Millecamps S.; French Clinical and Genetic Research Network on FTD/FTD-ALS, et al. 2013. SQSTM1 mutations in French patients with frontotemporal dementia or frontotemporal dementia with amyotrophic lateral sclerosis. JAMA Neurol. 70:1403–1410. 10.1001/jamaneurol.2013.3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H., Zhang J., Makinson S.R., Cahill M.K., Kelley K.W., Huang H.-Y., Shang Y., Oldham M.C., Martens L.H., Gao F., et al. 2016. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell. 165:921–935. 10.1016/j.cell.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L.H., Zhang J., Barmada S.J., Zhou P., Kamiya S., Sun B., Min S.-W., Gan L., Finkbeiner S., Huang E.J., and Farese R.V. Jr. 2012. Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J. Clin. Invest. 122:3955–3959. 10.1172/JCI63113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T., Mita A., Minami K., Hosooka T., Kitazawa S., Takahashi K., Tamori Y., Yokoi N., Watanabe M., Matsuo E., et al. 2012. PGRN is a key adipokine mediating high fat diet-induced insulin resistance and obesity through IL-6 in adipose tissue. Cell Metab. 15:38–50. 10.1016/j.cmet.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Matsumoto G., Wada K., Okuno M., Kurosawa M., and Nukina N.. 2011. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell. 44:279–289. 10.1016/j.molcel.2011.07.039 [DOI] [PubMed] [Google Scholar]
- Menzies F.M., Fleming A., and Rubinsztein D.C.. 2015. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 16:345–357. 10.1038/nrn3961 [DOI] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., and Thomas P.D.. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41:D377–D386. 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill T., Buraschi S., Goyal A., Sharpe C., Natkanski E., Schaefer L., Morrione A., and Iozzo R.V.. 2016. EphA2 is a functional receptor for the growth factor progranulin. J. Cell Biol. 215:687–703. 10.1083/jcb.201603079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 314:130–133. 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- Park B., Buti L., Lee S., Matsuwaki T., Spooner E., Brinkmann M.M., Nishihara M., and Ploegh H.L.. 2011. Granulin is a soluble cofactor for toll-like receptor 9 signaling. Immunity. 34:505–513. 10.1016/j.immuni.2011.01.018 [DOI] [PubMed] [Google Scholar]
- Pau G., and Reeder J.. 2017. HTSeqGenie: A NGS analysis pipeline. R package version 3.16.0.
- Petkau T.L., Neal S.J., Orban P.C., MacDonald J.L., Hill A.M., Lu G., Feldman H.H., Mackenzie I.R.A., and Leavitt B.R.. 2010. Progranulin expression in the developing and adult murine brain. J. Comp. Neurol. 518:3931–3947. 10.1002/cne.22430 [DOI] [PubMed] [Google Scholar]
- Pilli M., Arko-Mensah J., Ponpuak M., Roberts E., Master S., Mandell M.A., Dupont N., Ornatowski W., Jiang S., Bradfute S.B., et al. 2012. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 37:223–234. 10.1016/j.immuni.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman G.D., Green J.M., Neubauer M.G., Buckley S.D., McDonald V.L., Todaro G.J., and Shoyab M.. 1992. The epithelin precursor encodes two proteins with opposing activities on epithelial cell growth. J. Biol. Chem. 267:13073–13078. [PubMed] [Google Scholar]
- Pottier C., Bieniek K.F., Finch N., van de Vorst M., Baker M., Perkersen R., Brown P., Ravenscroft T., van Blitterswijk M., Nicholson A.M., et al. 2015. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 130:77–92. 10.1007/s00401-015-1436-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R., Neumann M., and Mackenzie I.R.. 2012. Advances in understanding the molecular basis of frontotemporal dementia. Nat. Rev. Neurol. 8:423–434. 10.1038/nrneurol.2012.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.L., Baranowski D.C., Chitramuthu B.P., Malik S., Li Z., Cao M., Minotti S., Durham H.D., Kay D.G., Shaw C.A., et al. 2009. Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci. 10:130 10.1186/1471-2202-10-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T., et al. 2005. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 37:806–808. 10.1038/ng1609 [DOI] [PubMed] [Google Scholar]
- Smith K.R., Damiano J., Franceschetti S., Carpenter S., Canafoglia L., Morbin M., Rossi G., Pareyson D., Mole S.E., Staropoli J.F., et al. 2012. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 90:1102–1107. 10.1016/j.ajhg.2012.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K., Friedman B.A., Larson J.L., Lauffer B.E., Goldstein L.D., Appling L.L., Borneo J., Poon C., Ho T., Cai F., et al. 2016. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat. Commun. 7:11295 10.1038/ncomms11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., and Vignali D.A.A.. 2004. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 22:589–594. 10.1038/nbt957 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Matsuwaki T., Yamanouchi K., and Nishihara M.. 2013. Increased lysosomal biogenesis in activated microglia and exacerbated neuronal damage after traumatic brain injury in progranulin-deficient mice. Neuroscience. 250:8–19. 10.1016/j.neuroscience.2013.06.049 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Suzuki G., Matsuwaki T., Hosokawa M., Serrano G., Beach T.G., Yamanouchi K., Hasegawa M., and Nishihara M.. 2017. Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum. Mol. Genet. 26:969–988. [DOI] [PubMed] [Google Scholar]
- Tang W., Lu Y., Tian Q.-Y., Zhang Y., Guo F.-J., Liu G.-Y., Syed N.M., Lai Y., Lin E.A., Kong L., et al. 2011. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 332:478–484. 10.1126/science.1199214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia L., Milnerwood A., Guo A., Mills F., Yoshida E., Vasuta C., Mackenzie I.R., Raymond L., Cynader M., Jia W., and Bamji S.X.. 2011. Progranulin deficiency decreases gross neural connectivity but enhances transmission at individual synapses. J. Neurosci. 31:11126–11132. 10.1523/JNEUROSCI.6244-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston T.L.M., Ryzhakov G., Bloor S., von Muhlinen N., and Randow F.. 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10:1215–1221. 10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- Van Damme P., Van Hoecke A., Lambrechts D., Vanacker P., Bogaert E., van Swieten J., Carmeliet P., Van Den Bosch L., and Robberecht W.. 2008. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J. Cell Biol. 181:37–41. 10.1083/jcb.200712039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.-F., Guo B.-S., Liu Y.-C., Wu C.-C., Yang C.-H., Tsai K.-J., and Shen C.-K.J.. 2012. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc. Natl. Acad. Sci. USA. 109:15024–15029. 10.1073/pnas.1206362109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M.E., Taubes A., Chen R., Miller B.L., Sephton C.F., Gelfand J.M., Minami S., Boscardin J., Martens L.H., Seeley W.W., et al. 2014. Early retinal neurodegeneration and impaired Ran-mediated nuclear import of TDP-43 in progranulin-deficient FTLD. J. Exp. Med. 211:1937–1945. 10.1084/jem.20140214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts G.D.J., Wymer J., Kovach M.J., Mehta S.G., Mumm S., Darvish D., Pestronk A., Whyte M.P., and Kimonis V.E.. 2004. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 36:377–381. 10.1038/ng1332 [DOI] [PubMed] [Google Scholar]
- Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R., Richter B., Korac J., Waidmann O., Choudhary C., et al. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 333:228–233. 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils H., Kleinberger G., Pereson S., Janssens J., Capell A., Van Dam D., Cuijt I., Joris G., De Deyn P.P., Haass C., et al. 2012. Cellular ageing, increased mortality and FTLD-TDP-associated neuropathology in progranulin knockout mice. J. Pathol. 228:67–76. 10.1002/path.4043 [DOI] [PubMed] [Google Scholar]
- Wu T.D., and Nacu S.. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 26:873–881. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xilouri M., Bruban J., Shioi J., Shao Z., Papazoglou I., Vekrellis K., and Robakis N.K.. 2011. Extracellular progranulin protects cortical neurons from toxic insults by activating survival signaling. Neurobiol. Aging. 32:2326.e5–2326.e16. 10.1016/j.neurobiolaging.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Banerjee R., Thomas B., Zhou P., Qian L., Jia T., Ma X., Ma Y., Iadecola C., Beal M.F., et al. 2010b Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med. 207:117–128. 10.1084/jem.20091568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Dumont M., Banerjee R., Ma Y., Li H., Lin M.T., Beal M.F., Nathan C., Thomas B., and Ding A.. 2010a Behavioral deficits and progressive neuropathology in progranulin-deficient mice: A mouse model of frontotemporal dementia. FASEB J. 24:4639–4647. 10.1096/fj.10-161471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y., Ogawa M., Hain T., Yoshida M., Fukumatsu M., Kim M., Mimuro H., Nakagawa I., Yanagawa T., Ishii T., et al. 2009. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 11:1233–1240. 10.1038/ncb1967 [DOI] [PubMed] [Google Scholar]
- Yu Y., Xu X., Liu L., Mao S., Feng T., Lu Y., Cheng Y., Wang H., Zhao W., and Tang W.. 2016. Progranulin deficiency leads to severe inflammation, lung injury and cell death in a mouse model of endotoxic shock. J. Cell. Mol. Med. 20:506–517. 10.1111/jcmm.12756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-J., Xu Y.-F., Dickey C.A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., and Petrucelli L.. 2007. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 27:10530–10534. 10.1523/JNEUROSCI.3421-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.T., Shahnazari S., Brech A., Lamark T., Johansen T., and Brumell J.H.. 2009. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183:5909–5916. 10.4049/jimmunol.0900441 [DOI] [PubMed] [Google Scholar]
- Zhou X., Sun L., Bastos de Oliveira F., Qi X., Brown W.J., Smolka M.B., Sun Y., and Hu F.. 2015. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J. Cell Biol. 210:991–1002. 10.1083/jcb.201502029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Nathan C., Jin W., Sim D., Ashcroft G.S., Wahl S.M., Lacomis L., Erdjument-Bromage H., Tempst P., Wright C.D., and Ding A.. 2002. Conversion of proepithelin to epithelins: Roles of SLPI and elastase in host defense and wound repair. Cell. 111:867–878. 10.1016/S0092-8674(02)01141-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.