Autosomal-recessive omodysplasia (OMOD1) is caused by mutations in glypican-6. Capurro et al. show that glypican-6 stimulates Hedgehog (Hh) signaling, and reduced Hh signaling may contribute to the pathogenesis of OMOD1.
Abstract
Autosomal-recessive omodysplasia (OMOD1) is a genetic condition characterized by short stature, shortened limbs, and facial dysmorphism. OMOD1 is caused by loss-of-function mutations of glypican 6 (GPC6). In this study, we show that GPC6-null embryos display most of the abnormalities found in OMOD1 patients and that Hedgehog (Hh) signaling is significantly reduced in the long bones of these embryos. The Hh-stimulatory activity of GPC6 was also observed in cultured cells, where this GPC increased the binding of Hh to Patched 1 (Ptc1). Consistent with this, GPC6 interacts with Hh through its core protein and with Ptc1 through its glycosaminoglycan chains. Hh signaling is triggered at the primary cilium. In the absence of Hh, we observed that GPC6 is localized outside of the cilium but moves into the cilium upon the addition of Hh. We conclude that GPC6 stimulates Hh signaling by binding to Hh and Ptc1 at the cilium and increasing the interaction of the receptor and ligand.
Introduction
Autosomal-recessive omodysplasia (OMOD1) is a skeletal dysplasia characterized by short stature, severely shortened limbs, and craniofacial dysmorphisms (Borochowitz et al., 1998; Elçioglu et al., 2004). Other variable abnormalities include cryptorchidism, hernias, congenital heart defects, and cognitive delay (Borochowitz et al., 1998; Elçioglu et al., 2004). OMOD1 was first described in 1989, and its incidence is still unknown. This genetic condition is caused by loss-of-function mutations of glypican 6 (GPC6; Campos-Xavier et al., 2009). Consistent with this finding, GPC6 is expressed in the proliferative zone of the mouse bone growth plate (Ahrens et al., 2009; Campos-Xavier et al., 2009).
GPCs are a family of proteoglycans that are linked to the plasma membrane through a glycosylphosphatidylinositol anchor. Six members of the GPC family have been identified in mammals (GPC1 to GPC6; Filmus et al., 2008; Filmus and Capurro, 2012). Structural features that are conserved across the family include the localization of 14 cysteine residues and of the domain in which the glycosaminoglycan (GAG) chains are inserted. This domain is located close to the C terminus, placing the GAG chains in proximity to the cell surface (Filmus et al., 2008). GPCs display several GAG attachment sites (from two in GPC3 to four in GPC5 and GPC6). The functional implications of the variation in the number of GAG chains are still unknown. Most GPCs have been shown to only carry heparan sulfate (HS) chains. Another remarkable feature of GPCs is that they do not display domains with obvious homology to characterized domains found in other proteins, suggesting that GPCs have unique functions.
GPCs regulate the signaling activity of various morphogens/growth factors, including Hedgehogs (Hhs; Desbordes and Sanson, 2003; Lum et al., 2003; Wilson and Stoeckli, 2013), FGFs (Hagihara et al., 2000; Yan and Lin, 2007), Wnts (Ohkawara et al., 2003; Song et al., 2005), and bone morphogenetic proteins (Jackson et al., 1997; Akiyama et al., 2008; Taneja-Bageshwar and Gumienny, 2013). In most cases, this regulatory activity is based on the ability of GPCs to either inhibit or stimulate the interaction of these growth factors with their signaling receptors. It is now well established that the structural features of GPCs combine with the set of growth factors and growth factor receptors present in a given cell type to determine GPC function.
Hhs are growth factors/morphogens that play a critical role in tissue patterning during embryonic development and in the preservation of homeostasis in the adult (Robbins et al., 2012; Briscoe and Thérond, 2013; Lee et al., 2016). Three Hhs have been identified in mammals: Sonic Hh (Shh), Indian Hh (Ihh), and Desert Hh. Whereas Shh is produced by many tissues, the expression of Ihh and Desert Hh is restricted to a few cell types. Ihh is the only Hh found in developing bone (St-Jacques et al., 1999). The main cell membrane receptor of Hh is Patched 1 (Ptc1), a 12-span transmembrane protein. In the absence of Hh, Ptc1 suppresses the activity of Smoothened (Smo), another transmembrane protein. Hh stimulation abrogates the inhibition of Smo by Ptc1. Then, activated Smo triggers a signaling cascade that induces target gene transcription through the Gli family of transcription factors (Ryan and Chiang, 2012; Briscoe and Thérond, 2013). Although the target genes that are regulated by Hh signaling are cell-type specific, in general this signaling pathway activates the expression of genes that play a critical role in the cell cycle (i.e., cyclin D1; Oliver et al., 2003) and in the regulation of cell survival (i.e., Bcl-2; Bar et al., 2007). In addition, the list of target genes includes several components of the Hh pathway, such as Gli1 and Ptc1, indicating the existence of feed-back loops. Consistent with the crucial role that Hhs play in tissue patterning during embryonic development, mutations of several components of the Hh signaling pathway cause various congenital diseases, including holoprosencephaly, brachydactyly, and several bone dysplasias (Zhang et al., 2006; Gao et al., 2009). In vertebrates, Hh binds to Ptc1 at the primary cilium (Rohatgi et al., 2007). Hh cannot activate signaling activity in cells without this appendage (Toftgård, 2009).
The long bones are formed during development by a process called endochondral ossification, which starts in the growth plates located at the ends of the bones. The zone of the growth plate furthest from the ossification front is populated by resting chondrocytes (Mackie et al., 2008). Next to the resting zone is the proliferative zone, which contains cells that are actively proliferating, display a flattened shape, and are packed in clusters. Proliferating chondrocytes then mature into quiescent and enlarged prehypertrophic chondrocytes. Moving toward the ossification front, these cells become hypertrophic chondrocytes, which produce and deposit an extracellular matrix that becomes invaded by blood vessels and precursors of osteoblasts (Mackie et al., 2008; Karsenty et al., 2009).
Ihh, which is produced by the prehypertrophic cells (St-Jacques et al., 1999; Long et al., 2001), stimulates cell-cycle progression in the proliferative cells of the growth plate, which express Ptc1 (Long et al., 2001). The key role of Ihh in bone growth is underscored by the striking phenotype of the Ihh-null mice, where all skeletal structures are substantially reduced in size (St-Jacques et al., 1999).
The ability of GPCs to regulate Hh signaling was first reported in Drosophila (Lum et al., 2003). In mammals, GPCs can act as stimulators or inhibitors of Hh signaling (Filmus and Capurro, 2014). GPC5 and GPC1 have been shown to act as positive regulators of Hh activity (Li et al., 2011; Wilson and Stoeckli, 2013; Witt et al., 2013). GPC3, however, acts as an Hh inhibitor (Capurro et al., 2008). We have shown that GPC5 promotes the binding of Hh to Ptc1, and we have proposed that this stimulatory activity is because of the fact that GPC5 can bind to both Hh and Ptc1 (Li et al., 2011). Conversely, GPC3 binds with high affinity to Hh, but it does not interact with Ptc1 (Capurro et al., 2008). The binding of Hh to GPC3 triggers the endocytosis and degradation of the GPC3/Hh complex (Capurro et al., 2008). Thus, GPC3 acts as a signaling inhibitor by competing with Ptc1 for Hh binding (Capurro et al., 2008). Loss-of-function mutations of GPC3 cause Simpson-Golabi-Behmel overgrowth syndrome (Pilia et al., 1996). Notably, with regards to long bone length, these patients display the opposite phenotype of the OMOD1 patients, but the role of GPC6 in Hh signaling remains unknown.
Based on the ability of several GPCs to regulate Hh activity and on the fact that the Hh signaling pathway plays an essential role in promoting the growth of developing bone by stimulating chondrocyte proliferation, we have hypothesized that GPC6 is required for optimal Hh activity in the developing bone and that OMOD1 is caused, at least in part, by reduced Hh activity caused by the lack of GPC6. Here, we present genetic and biochemical evidence to support this hypothesis. In addition, we describe critical features of the mechanism by which GPC6 stimulates Hh activity.
Results
Characterization of GPC6-null embryos
The generation of GPC6-null mice has been previously reported (Tang et al., 2010). The only information provided by the study was that the GPC6-null mice display embryonic lethality. By breeding GPC6 heterozygous mice, we have established that mice lacking GPC6 die at birth, as we found the expected Mendelian frequency of knockout (KO) embryos at E17.5–18.5 (Table 1), but we did not identify any live newborn KO mice.
Table 1. Generation of GPC6-null mice.
| Genotype | Number of mice | Expected frequency (%) | Observed frequency (%) |
|---|---|---|---|
| WT | 38 | 25 | 21.5 |
| Heterozygote | 91 | 50 | 51.4 |
| KO | 48 | 25 | 27.1 |
Breeding data (female Gpc6+/− × male Gpc6+/−) corresponding to 23 litters. A total of 177 embryos were collected at E17.5–E18.5.
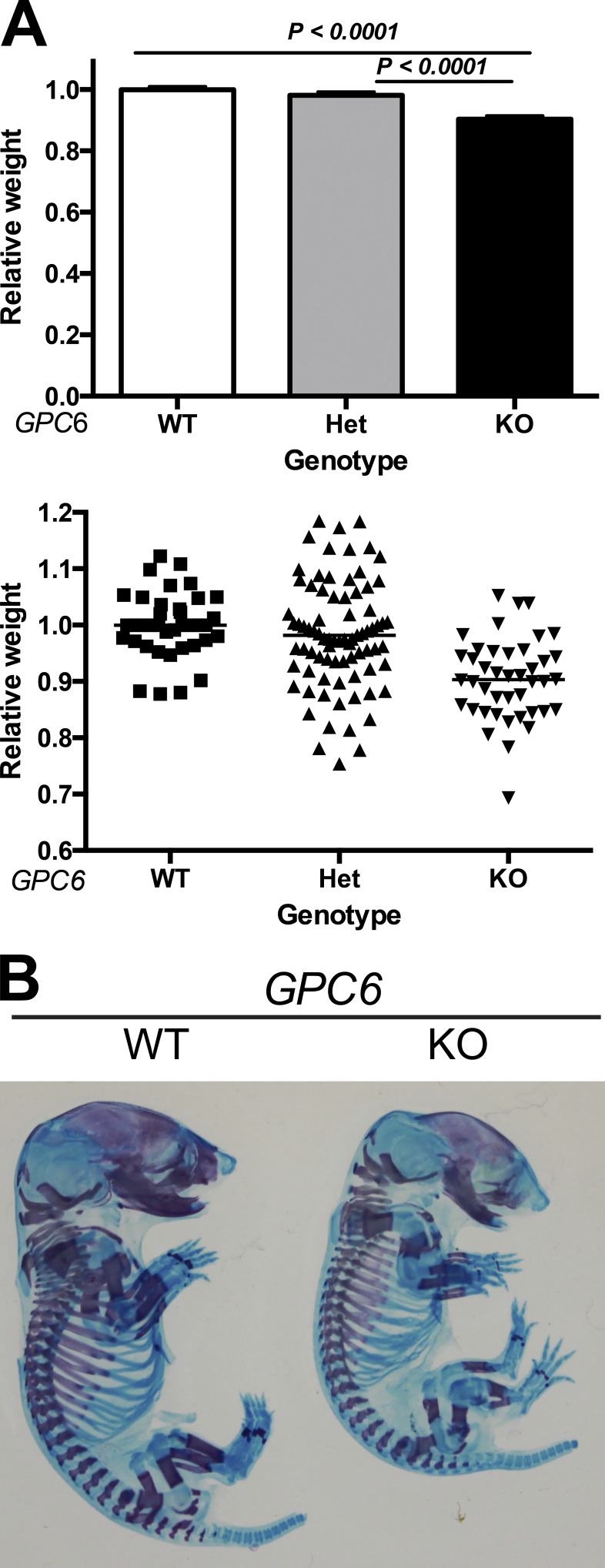
Compared with the WT littermates, E17.5–18.5 GPC6-null embryos are significantly lighter (Fig. 1 A). In contrast, GPC6 heterozygous and WT embryos have similar weight. Staining of E17.5 embryos with Alcian blue/Alizarin red revealed that the long bones of the GPC6-null embryos are shorter than those of the WT littermates (Figs. 1 B and 2 B). In addition, the presence of red staining indicates that, unlike in the Ihh-null mice (St-Jacques et al., 1999), the bones in the GPC6 KOs undergo differentiation/ossification. This is consistent with the observation that GPC6 is not expressed in the perichondrium, which plays a critical role in ossification, a process that is regulated by Ihh (Long et al., 2001). The cells from the perichondrium differentiate into osteoblasts, which produce the mineralized matrix required for bone formation. Notably, the long bone phenotype of the GPC6-null mice at the macroscopic level seems to be similar to that of the cartilage-specific Smo-KO mice. In these mice, the Hh pathway has been inactivated in the growth plate, but it is still active in the perichondrium (Long et al., 2001). The Alcian blue/Alizarin red staining of the embryos also revealed that the GPC6-null mice display a smaller skull and muzzle and a normal spine (Fig. 1 B).
Figure 1.
GPC6-null mice display reduced body size and bone abnormalities. (A, top) Relative body weights of E17.5–E18.5 embryos of the indicated genotype (WT, heterozygous [Het], and KO). The mean WT weight in each litter was arbitrarily assigned the value of 1. Bars represent mean relative weights + SEM. n = 38, 79, and 42 for GPC6 WT, heterozygous, and KO, respectively. Statistical analysis was performed by Student’s t test (unpaired two-tailed). (Bottom) Scatter plot representation of the data. (B) Alcian blue–Alizarin red staining of E17.5 embryos of the indicated genotypes.
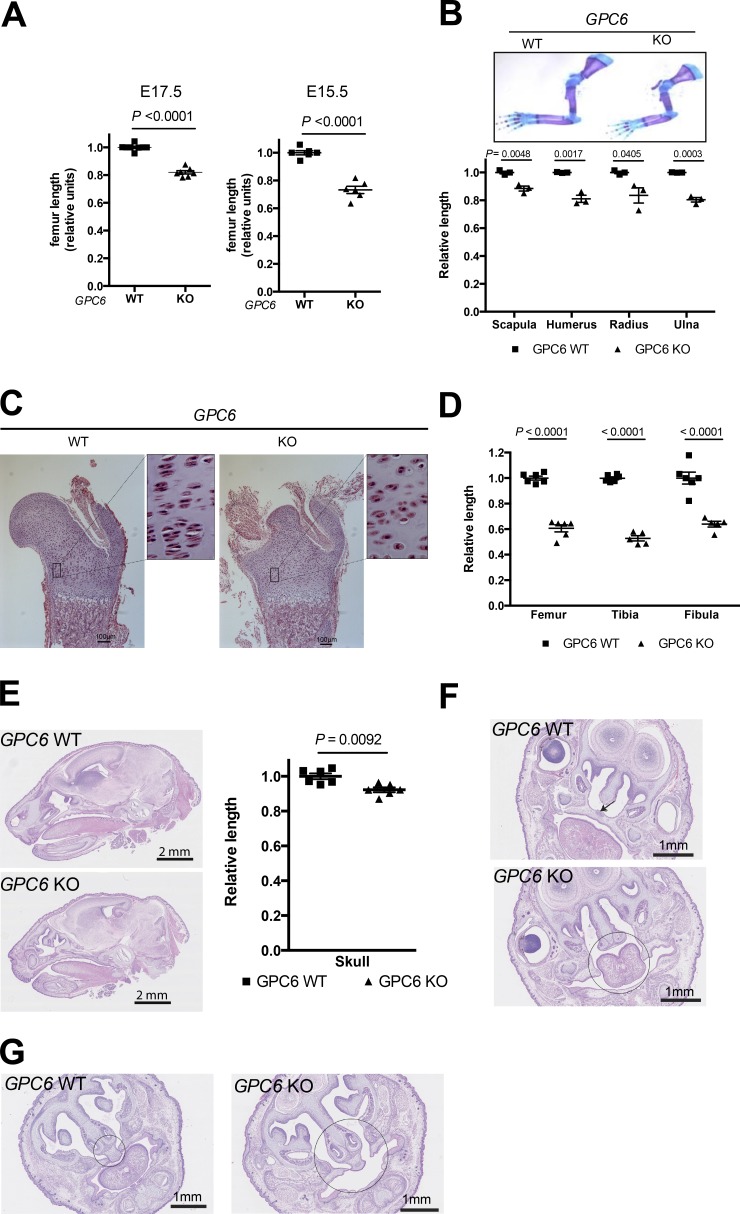
Figure 2.
GPC6-null mice display shorter long bones and face dysmorphisms. (A) Whole femurs were dissected from E17.5 or E15.5 embryos of the indicated genotypes. Images were acquired with a camera (DFC 295; Leica) using a stereoscope (M60; Leica), and femur length was measured using the Leica Application Suite (V3.8). Scatter plots with the mean ± SEM of eight WT and seven KO embryos are shown. Mean WT femur length in each litter was arbitrarily assigned the value of 1. (B, top) Upper limbs were dissected from GPC6 WT and GPC6 KO E18.5 embryos and stained with Alcian blue–Alizarin red. Images were acquired with a camera (DFC 295) using a stereoscope (M60). Representative bones are shown. (Bottom) Images from three WT and three KO femurs from different litters were measured with the Leica Application Suite (V3.8). The graph shows scatter plots with the mean ± SEM. (C) Proximal femoral metaphysis from E17.5 embryos of the indicated genotypes were stained with hematoxylin/eosin, and pictures were taken under the microscope. Insets represent an eight-times enlargement of the low-power picture. Bars, 100 µm. (D) The indicated bones were dissected from GPC6 WT (n = 6) and GPC6 KO (n = 6) E18.5 embryos and stained with Alcian blue–Alizarin red. Images were acquired, and the lengths of the diaphysis were measured as in B. The graph show scatter plots with the mean ± SEM. Mean WT bone length in each litter was arbitrarily assigned the value of 1. (E, left) Representative sagittal head sections corresponding to E18.5 GPC6 WT and GPC6 KO embryos. (Right) Skull length from the base of the skull to the end of nasal plenum was measured in GPC6 WT (n = 6) and KO (n = 6) embryos. Results are shown as a scatter plot with the mean ± SEM. Mean WT skull length in each litter was arbitrarily assigned the value of 1. (F and G) Representative cross-sections of the skulls of E18.5 embryos of the indicated genotypes illustrating the palatoschisis (arrow and circle; F) and nasal septal aplasia (circles; G) observed in the GPC6-null mice with a 100% penetrance (n = 5 of each genotype). Statistical analysis was performed by Student’s t tests (unpaired two-tailed).
Next, we measured the length of the femurs at E17.5 and E15.5. We found that these bones are significantly shorter in the GPC6-null embryos (Fig. 2 A). We also compared the length of the bones from the upper limbs. In all cases, the bones from the GPC6-null embryos were significantly shorter than those from WT embryos (Fig. 2 B). In fact, compared with the difference observed in body weight, the limbs of the GPC6-null mice were disproportionally smaller. These results are consistent with the phenotype of OMOD1 patients. Tissue sections of the femurs were prepared, stained with toluidine blue, and examined under a microscope. We found that the chondrocytes in the proliferative zone of the growth plate were disorganized, and they lacked the typical cell stacking in columns (Fig. 2 C). This phenotype had 100% penetrance. A similar disorganization of the growth plate has been reported in the Ihh-null mice (Koziel et al., 2005). We also compared the length of the diaphysis portion of E17.5 femurs, tibias, and fibulas. We found that the reduction in the length of the diaphysis of the GPC6-null embryos was even more pronounced than that of the whole bones (Fig. 2 D).
Another noticeable anatomical difference between the GPC6-null mice and the normal littermates was the size of the skull, which was significantly smaller in the KOs (Fig. 2 E). We also found that the GPC6-null mice display cleft palate (Fig. 2 F) and nasal septal aplasia (Fig. 2 G) with 100% penetrance. The spine and the extremities were normal, as in OMOD1 patients. It should be noted that GPC6 plays a role in the formation of excitatory synapses (Allen et al., 2012), but at this point in time, the cause for the premature death of the GPC6-null mice remains unknown.
Evaluation of Hh signaling and cell proliferation in femurs of GPC6-null embryos
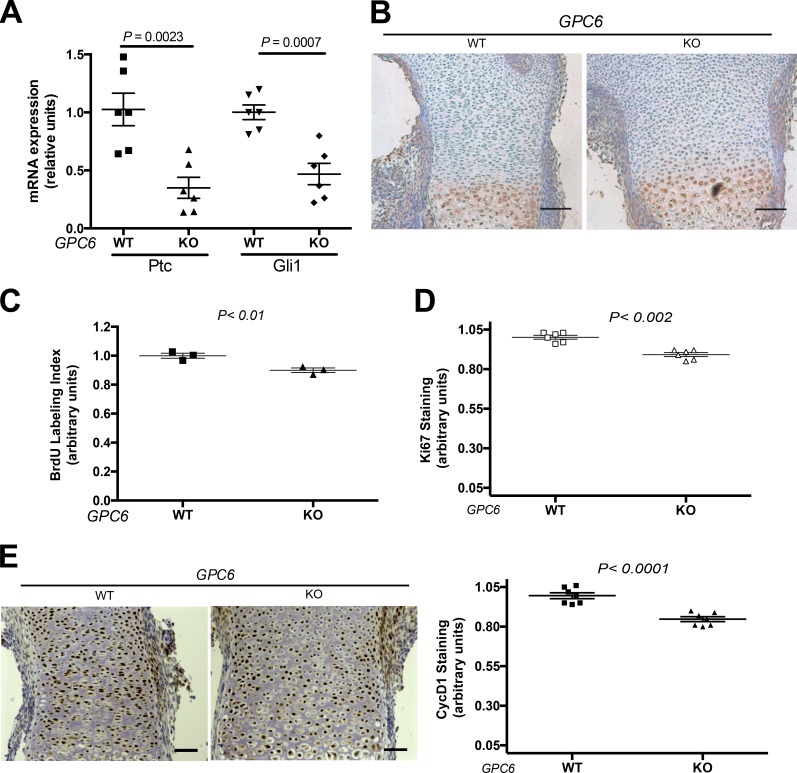
To compare the degree of activation of the Hh signaling pathway in the WT and KO mice, we assessed the levels of Ptc1 and Gli1 in the embryonic femurs. It is well established that Ptc1 and Gli1 are transcriptional targets of the Hh signaling pathway (Ingham et al., 2011). Femurs from 15.5-d embryos were obtained, and the levels of Ptc1 and Gli1 mRNAs were measured by real-time RT-PCR, as previously described (Capurro et al., 2008). We found that the levels of Ptc1 and Gli1 transcripts are significantly lower in the GPC6-null mice compared with normal littermates (Fig. 3 A). From these results, we conclude that GPC6 stimulates Hh signaling in the developing bone. It should be noted that, like the GPC6-null mice, the Ihh-null mice also display shorter long bones. However, the long bone phenotype of the Ihh-null mice is more severe than that of the GPC6 mutants. This is probably because of the fact that GPC6 stimulates Hh signaling, but it is not essential for Hh activity. In addition, unlike GPC6, the Ihh signaling receptor Ptc1 is expressed in the perichondrium (St-Jacques et al., 1999). Thus, in addition to displaying reduced chondrocyte proliferation, Ihh-null mice exhibit deficient osteoblast development (St-Jacques et al., 1999).
Figure 3.
GPC6-null femurs display reduced Hh signaling and chondrocyte proliferation. (A) Comparison of the levels of Ptc1 and Gli1 in E15.5 femurs. Ptc1 and Gli1 transcripts were measured by real-time RT-PCR using β-actin transcript levels as references. A total of six GPC6 WT and six GPC6-null (KO) femurs dissected from four litters of E15.5 embryos were analyzed. Mean WT value for each litter was arbitrarily considered as 1, and the relative levels of expression of the KOs were calculated. Scatter plots show the mean + SEM. (B) Immunohistochemical analysis of Ihh in E15.5 femur sections. Staining was performed in three littermate pairs of KO and WT embryos and one pair of KO and heterozygous embryos from three different litters. A representative result is shown. Bars, 100 µm. (C) BrdU labeling index of growth plates from femur sections dissected from E15.5 embryos. The results represent the mean + SEM of pairs of WT and KO mice from three different litters. The mean percentage of WT sections was defined as 1. All the nuclei in each femur section were counted (from 370 to 650 nuclei). Scatter plots show the mean ± SEM. (D) Immunohistochemical analysis of Ki67 in E15.5 femur growth plate sections. The percentage of positive cells was calculated. The results represent the mean ± SEM of pairs of WT and KO mice from three different litters. All the nuclei in each femur section were counted (from 370 to 650 nuclei). The mean percentage of WT sections was defined as 1. Scatter plots show the mean ± SEM. (E) Immunohistochemical analysis of cyclin D1 in E15.5 femur growth plate sections. Staining was performed in three pairs of KO and WT embryos from three different litters. (Left) A representative result is shown. Bars, 50 µm. (Right) Percentage of positive cells was calculated. The results represent the mean ± SEM of pairs of WT and KO mice from three different litters. All the nuclei in each femur section were counted (from 370 to 650 nuclei). The mean percentage of WT sections was defined as 1. Statistical analysis was performed by Student’s t test (unpaired two-tailed).
HS proteoglycans can play a role in the secretion of Hh (Hilton et al., 2005; Bandari et al., 2015). Thus, it could be hypothesized that the bone phenotype of the GPC6-null mice is caused by changes in the levels and/or extracellular distribution of Ihh. However, immunostaining and Western blot analysis of the femur growth plate showed that the expression levels and distribution of Ihh in the GPC6-null E17.5 embryos were similar to those of WT littermates (Fig. 3 B and Fig. S1). This is consistent with the fact that GPC6 is not expressed in the prehypertrophic zone, which is the area where Ihh is secreted (Campos-Xavier et al., 2009).
Ihh is a strong stimulator of chondrocyte proliferation during long bone development (Long et al., 2001). Based on this, we hypothesized that chondrocytes from the growth plates of GPC6-null mice should display reduced proliferation. To test this hypothesis, we compared the BrdU labeling index in growth plate sections of femurs from mutant mice and normal littermates. As shown in Fig. 3 C, we found that this index is significantly smaller in the GPC6-null femurs. We also assessed cell proliferation in the growth plate by staining tissue sections with an antibody against Ki67. Consistent with the results for the BrdU labeling index, we found that the growth plates of the GPC6-null femurs display a relative significantly lower percentage of Ki67-positive cells (Fig. 3 D).
In addition to showing a reduced BrdU index, the chondrocytes from the Ihh null also display lower levels of cyclin D1, an Hh signaling target gene whose expression is regulated by mitogens (Long et al., 2001). Therefore, we assessed the relative cyclin D1 levels in growth plate sections of femurs derived from GPC6-null embryos and normal littermates by immunostaining with an anti–cyclin D1 antibody. As expected, cyclin D1 levels were found to be significantly reduced in the mutant growth plates (Fig. 3 E).
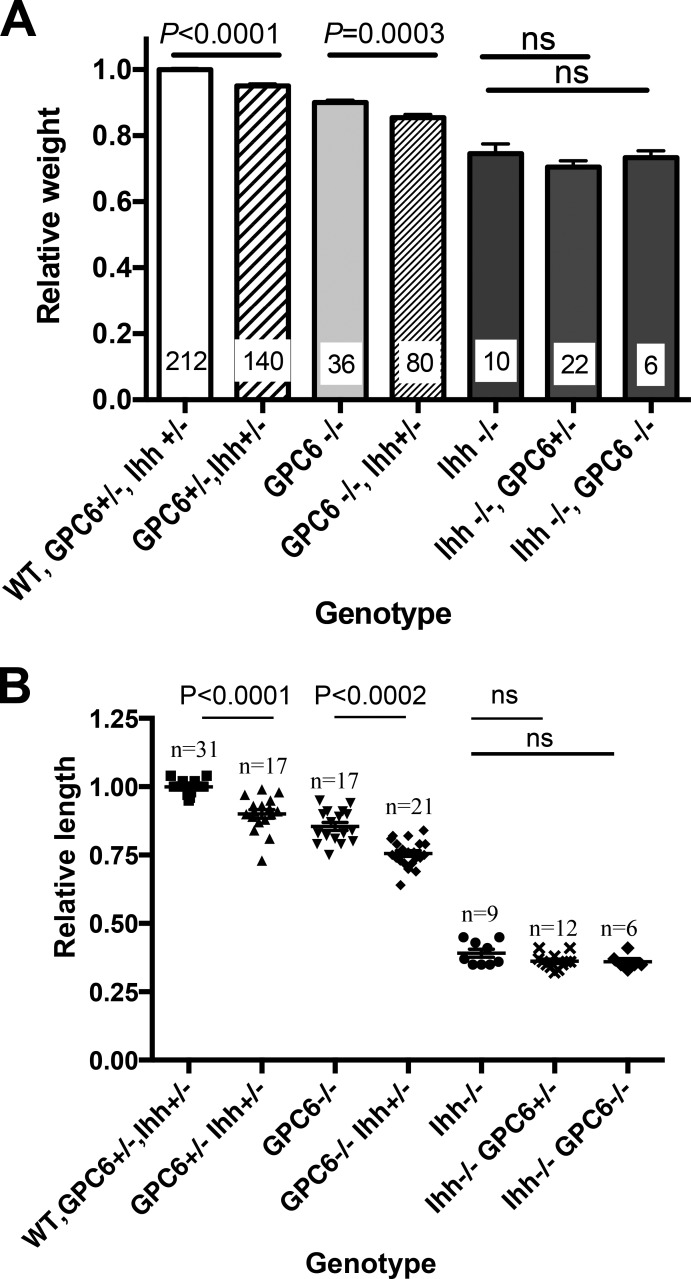
We have previously generated genetic evidence demonstrating that the Hh pathway mediates the growth-inhibitory effect of GPC3 by breeding GPC3 heterozygous and Ihh heterozygous mice (Capurro et al., 2009). Following a similar approach, we tested the hypothesis that the GPC6-induced stimulation of chondrocyte proliferation in the developing bone is mediated, at least in part, by the Hh signaling pathway. To this end, first, GPC6 and Ihh heterozygous mice were bred to generate double heterozygotes. It should be noted that GPC6 heterozygous (Fig. 1 A) and Ihh heterozygous mice (Long et al., 2006) display normal size and that Ihh KO mice are significantly smaller than normal mice (St-Jacques et al., 1999). Next, the double-heterozygous mice were bred, and every E17.5 embryo from each litter generated by the breeding was genotyped and weighted. In addition, we dissected the femurs of some of them, and bone length was measured. If, according to our hypothesis, GPC6 regulates long bone growth by stimulating Ihh pathway, it would be expected that GPC6/Ihh double heterozygotes would be smaller and display shorter long bones than GPC6 or Ihh single heterozygotes, that the GPC6 KO/Ihh heterozygotes would be smaller and display shorter long bones than the GPC6 KO/Ihh WT, and that the double-KO embryos would neither be smaller nor display shorter long bones than the Ihh KO/GPC6 WT embryos. However, if GPC6 regulates long bone growth independently of the Ihh pathway, it will be expected that the double-KO embryos will display a larger reduction in body weight and length of the long bones than that observed in each single KO.
The results shown in Fig. 4 are consistent with our hypothesis, as the double-heterozygous embryos are significantly smaller (Fig. 4 A) and display shorter femurs (Fig. 4 B) than the normal littermates, and the GPC6 KO/Ihh heterozygous embryos are significantly smaller and display shorter femurs than the GPC6 KO/Ihh WT. Furthermore, the double-KO embryos and the GPC6 heterozygous/Ihh KO embryos have similar weights and femur lengths as the GPC6 WT/Ihh KO embryos (Fig. 4, A and B). Thus, these results provide strong genetic support for the hypothesis that the Ihh pathway mediates the growth-stimulatory effect of GPC6 in the bones.
Figure 4.
Genetic interaction between GPC6 and the Hh signaling pathway. (A) GPC6/Ihh double-heterozygous mice were bred, and all E17.5 embryos from each litter were weighed and genotyped. To be able to compare embryos from different litters, the weight of embryos within a given litter was normalized by the mean weight of WT embryos, which was arbitrarily given a value of 1. Bars represent the mean weight + SEM for the indicated genotypes. The first bar at the right includes mice with the three different genotypes that display normal weight: GPC6+/+ Ihh+/+, GPC6+/− Ihh+/+, and GPC6+/+ Ihh+/−. Numbers on the bars represent the number of embryos that were weighted for each genotype. (B) Femurs from some of the litters generated by breeding the GPC6/Ihh double-heterozygous mice were dissected, and their length was measured as described in Fig. 2. Scatter plots with the mean ± SEM of the indicated number of embryos are shown. Mean WT femur length in each litter was arbitrarily assigned the value of 1. Statistical analysis was performed by Student’s t test (unpaired two-tailed).
Characterization of chondrocyte-specific GPC6-null mice
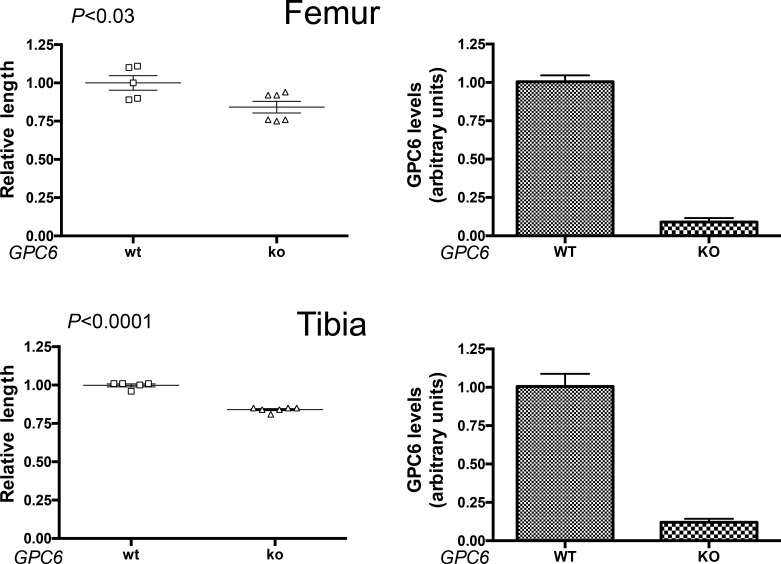
To completely rule out the possibility that the bone phenotype observed in the GPC6-null mice is caused, at least in part, by an indirect effect of the lack of GPC6 in other tissues/organs during development, chondrocyte-specific GPC6-null mice were generated. To this end, we crossed a mouse strain carrying a floxed allele of GPC6 with a strain carrying a Cre gene under the transcriptional control of the Col2α1 promoter. This approach has been previously used to generate chondrocyte-specific Ihh-null and Smo-null strains (Long et al., 2001; Razzaque et al., 2005). Mice carrying the chondrocyte-specific GPC6 deletion (Col2α1Cre; GPC6d/GPC6d) were born at the expected Mendelian frequency. Shortening of the long bones in newborn mutants was mild. However, by P15, a substantial difference in the length of the WT and mutant long bones was evident. Then, P21 mice were sacrificed, and femurs and tibia were dissected, and their lengths were measured. As shown in Fig. 5, these measurements confirmed that the Col2α1Cre;GPC6d/GPC6d mice display long bones that are significantly shorter than their normal littermates (Fig. 5). By performing real time RT-PCR, we verified that GPC6 expression was dramatically reduced in the mutant strain (Fig. 5). Tissue sections of the mutated growth plates were stained with toluidine blue and examined under the microscope. Similar to what was observed in the global GPC6-null strain, the proliferative zones in the growth plates of the tissue-specific mutants displayed a disorganized structure. Obvious facial dysmorphism was also observed (not depicted). Most likely, the delay in the manifestation of the bone phenotype in the chondrocyte-specific mutants compared with the global mutants is because of an incomplete and/or delayed deletion of the floxed GPC6.
Figure 5.
Chondrocyte-specific GPC6-null mice display shorter long bones. (Left) Femurs and tibia from WT and GPC6-null (KO) P21 mice were dissected, and their lengths were measured as described in Fig. 2. A total of five WT and six KO mice originated from four different litters were studied. Scatter plots with the mean ± SEM are shown. Mean WT femur length in each litter was arbitrarily assigned the value of 1. (Right) RNA was extracted from the measured bones, and the levels of GPC6 were assessed by real time RT-PCR. Bars represent the mean + SEM. Mean value for the WT in each litter was arbitrarily assigned the value of 1. Statistical analysis was performed by Student’s t test (unpaired two-tailed).
Effect of GPC6 on Hh signaling
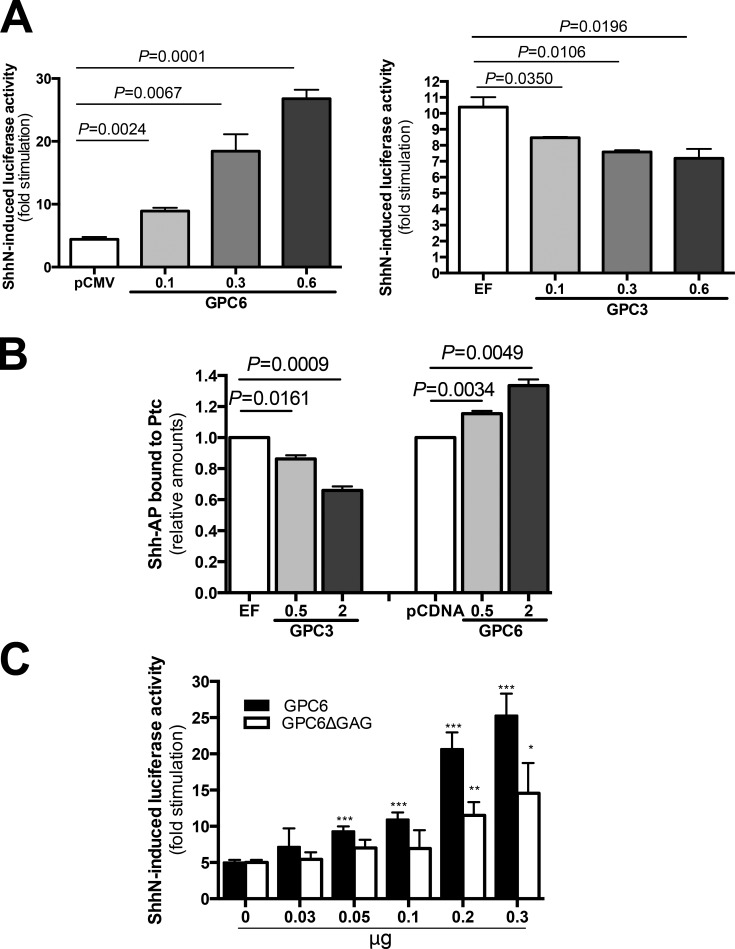
To investigate the mechanism by which GPC6 stimulates Hh signaling, we first tested the effect of GPC6 on Hh activity in NIH 3T3 cells by performing a luciferase reporter assay, in which luciferase expression was driven by an Hh-responsive promoter. As shown in Fig. 6 A and Fig. S2, this assay revealed that GPC6 strongly stimulates ShhN (Shh N-terminal domain) and fully mature Shh activity in a dose-dependent manner. As a control, we showed that, in the same assay, GPC3 displays an Hh-inhibitory activity (Fig. 6 A), as we and others have previously reported (Capurro et al., 2008; Williams et al., 2010). Therefore, this result provides additional evidence indicating that GPC6 stimulates Hh signaling.
Figure 6.
GPC6 stimulates the Hh signaling pathway. (A) Hh reporter assay. NIH3T3 cells were transfected with GPC6 (left) or GPC3 (right) or vector control (cytomegalovirus promoter [pCMV] and elongation factor promoter [EF]) along with a luciferase reporter driven by an Hh-responsive promoter and β-galactosidase. Transfected cells were stimulated during 48 h with ShhN- or control-conditioned medium. Then, luciferase and β-galactosidase assays were performed. Bars represent luciferase activity fold stimulation induced by Hh (mean + SEM of triplicates) normalized by β-galactosidase activity. One representative experiment of four is shown. (B) Increased binding of Shh to Ptc1 in the presence of GPC6. NIH3T3 cells expressing increasing amounts of GPC3 or GPC6 were incubated for 2.5 h at 4°C with Shh-AP or AP. Then, Ptc1 was immunoprecipitated, and the amount of Shh bound to the immunoprecipitated Ptc1 was determined by measuring AP activity. Bars represent the relative amount of Shh-AP bound to Ptc1 (mean + SEM of four independent experiments) after subtraction of the binding measured for AP alone. Shh-AP binding to the cells transfected with vector control was arbitrarily considered 1. (C) HS chains are required for maximum GPC6-induced stimulation of Hh signaling. NIH3T3 cells were transfected with increasing amounts of the indicated expression vectors along with a luciferase reporter driven by an Hh-responsive promoter and β-galactosidase. Then, luciferase activity in the presence and absence of Shh was performed as described in A. Bars represent luciferase activity fold stimulation induced by Hh (mean + SEM of triplicates) normalized by β-galactosidase activity. One representative experiment of two is shown. *, P < 0.05; **, P < 0.005; ***, P < 0.001. Statistical analysis was performed by Student’s t test (unpaired two-tailed).
Next, we sought to uncover the mechanism by which GPC6 stimulates Hh signaling. We have previously demonstrated that GPC5 increases Shh activity by promoting the binding of Shh to Ptc1 (Li et al., 2011). Therefore, we decided to investigate the effect of GPC6 on the interaction of these two proteins. To this end, we followed the same experimental design as the one we used to study the effect of GPC5 on the interaction of Shh and Ptc1 (Li et al., 2011). NIH 3T3 cells were transfected with different amounts of GPC6, and 2 d after transfection, the cells were incubated with Shh-AP fusion protein. Then, cells were lysed, Ptc1 was immunoprecipitated, and the levels of Shh-AP that coimmunoprecipitated with Ptc1 were measured. As shown in Fig. 6 B, we found that GPC6 significantly stimulates the binding of Shh to Ptc1 in a dose-dependent manner. It should be noted that, in this experimental protocol, the effect of GPC6 is most likely underestimated, as only a proportion (15–20%) of the cells uptake the transfected GPC6 cDNA, whereas Ptc1 is being immunoprecipitated from all the cells in the plate. As a control, we performed in parallel a similar experiment with GPC3. As previously reported, we observed that this GPC inhibits the interaction of Shh with Ptc1 (Capurro et al., 2008).
GAG chains are required for GPC5-induced stimulation of Hh signaling in NIH3T3 cells (Li et al., 2011). Therefore, we investigated whether the GAG chains are also required for GPC6-induced stimulation of Hh signaling. To this end, we generated a GPC6 mutant where the four attachment sites for the GAG chains have been mutated (GPC6ΔGAG) and tested the effect of this mutant on the Hh luciferase reporter assay in NIH 3T3 cells. We found that the nonglycanated GPC6 had a small stimulatory effect on Hh signaling that could only be seen when large amounts of this mutant were transfected (Fig. 6 C). This result suggests that the GAG chains play a key role in GPC6 activity. In addition, this result indicates that the mechanism by which GPC6 stimulates Hh signaling is somewhat different than that of GPC5, as the Hh-stimulatory activity of GPC5 was completely abrogated in the absence of the GAG chains (Li et al., 2011).
Analysis of the interaction of GPC6 with Shh, Ihh, and Ptc1
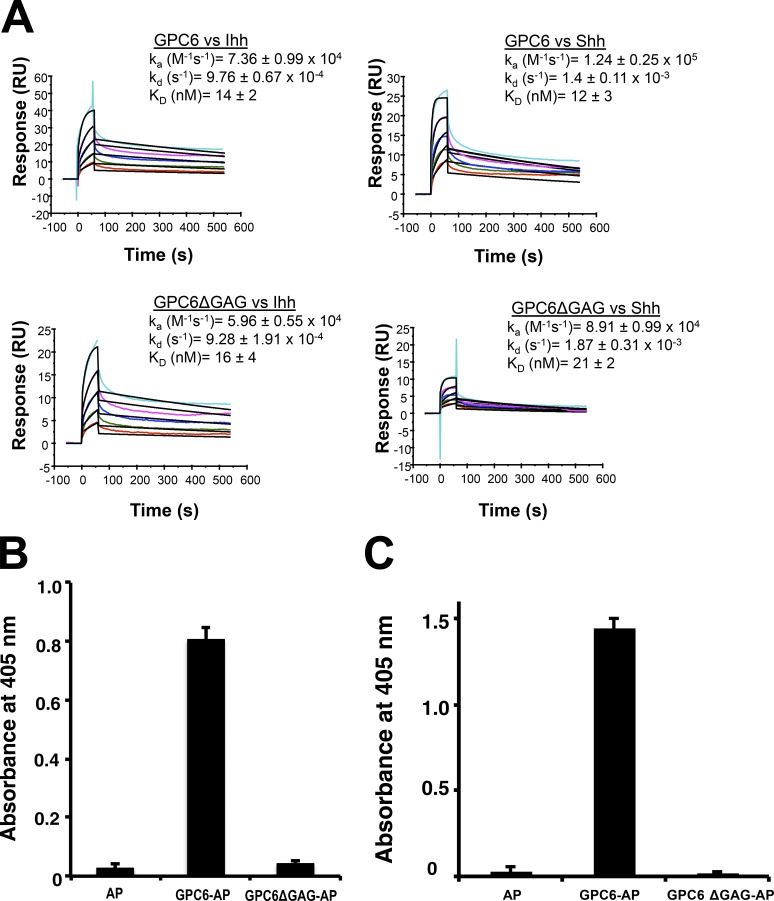
Next, we investigated the mechanism by which GPC6 stimulates the binding of Hh to Ptc1. We hypothesized that GPC6 can facilitate/stabilize the interaction of Hh with Ptc1. If this hypothesis were correct, it would be expected that GPC6 interacts with both Hh and Ptc1. First, we investigated whether GPC6 interacts with Shh by surface plasmon resonance (SPR) analysis. A GPC6-AP fusion protein (GPC6-AP) was purified from 293T cells and immobilized onto a sensor chip to measure its binding to recombinant Shh and Ihh. GPC6-AP was shown to bind to Shh and Ihh with comparable nanomolar dissociation constants (KD’s; 12 ± 3 nM and 14 ± 2 nM, respectively; Fig. 7 A). We also investigated the role of the core protein in the GPC6/Hh interaction. To this end, we performed SPR analysis with the nonglycanated GPC6ΔGAG mutant. We found that both Shh and Ihh bind to this mutant with relatively similar affinity to that observed for the glycanated GPC6 (KD of 21 ± 2 nM for Shh and 16 ± 4 nM for Ihh; Fig. 7 A), indicating that the GPC6 core protein is the main mediator of the GPC6/Hh interaction. However, these results do not exclude the possibility that the GAG chains of GPC6 also interact with Shh and Ihh.
Figure 7.
GPC6 interacts with Shh, Ihh, and Ptc1. (A) SPR analysis of the interaction of Shh and Ihh with GPC6-AP and GPC6ΔGAG-AP. Purified GPC6-AP and GPC6ΔGAG-AP were biotinylated and then immobilized on SA sensor chips (GPC6, 1,300 relative units [RU]; GPC6ΔGAG, 700 RU). Various concentrations of recombinant Shh or Ihh (from 62.5 to 1,000 nM) were injected to determine the binding profile. This experiment was repeated three times, and the results shown represent the mean ± SEM. (B) GPC6/Ptc1 pull-down assay. The interaction between Ptc1 and GPC6-AP or GPC6ΔGAG-AP was determined by a pull-down assay. Beads covered by Ptc1 were incubated with GPC6-AP–, GPC6ΔGAG-AP–, or vector control–conditioned media. Bars represent the mean of triplicates + SD. The experiment was repeated three times with similar results. A representative experiment is shown. (C) GPC6/Ptc1 cell-binding assay. 293T cells were transfected with a Ptc1 expression vector or vector control. Transfected cells were incubated with GPC6-AP–, GPC6ΔGAG-AP–, or AP-conditioned media containing equal AP concentrations for 2 h at 4°C. Cells were washed and lysed, and the AP activity of aliquots of cell lysates containing the same amount of protein was measured. Bars represent the mean of triplicates + SD. The experiment was repeated four times with similar results.
In light of the fact that Ptc1 could not be purified, we performed a pull-down assay to investigate the interaction of GPC6 and the GPC6ΔGAG mutant with Ptc1. We have previously used this approach to assess the interaction of GPC3 and GPC5 with Ptc1 (Capurro et al., 2008; Li et al., 2011). 293T cells were transiently transfected with an expression vector for hemagglutinin A (HA)–tagged Ptc1, and cell lysates were subsequently mixed with beads covered with an anti-HA antibody. Then, the beads were incubated with conditioned media containing equal concentrations of a GPC6-AP fusion protein or a GPC6ΔGAG-AP fusion protein. After washing, the amount of AP enzymatic activity that remained bound to the beads was measured. We found that, whereas there was a significant binding of WT GPC6, the GPC6ΔGAG mutant did not show any detectable binding (Fig. 7 B). This result indicates that GPC6 interacts directly or indirectly with Ptc1 through the GAG chains.
As an additional approach to assess the GPC6/Ptc1 interaction, we performed a cell-binding assay. To this end, 293T cells were transfected with Ptc1 or control vector and incubated with a GPC6-AP fusion protein or a GPC6ΔGAG-AP fusion protein for 2 h. After extensive washing, the cells were lysed, and AP activity that remained bound to the cell lysates was measured. Consistent with the pull-down assay results, we found significant binding of the WT GPC6 to the Ptc1-transfected cells. The binding was drastically reduced for the nonglycanated mutant (Fig. 7 C).
Localization of GPC6
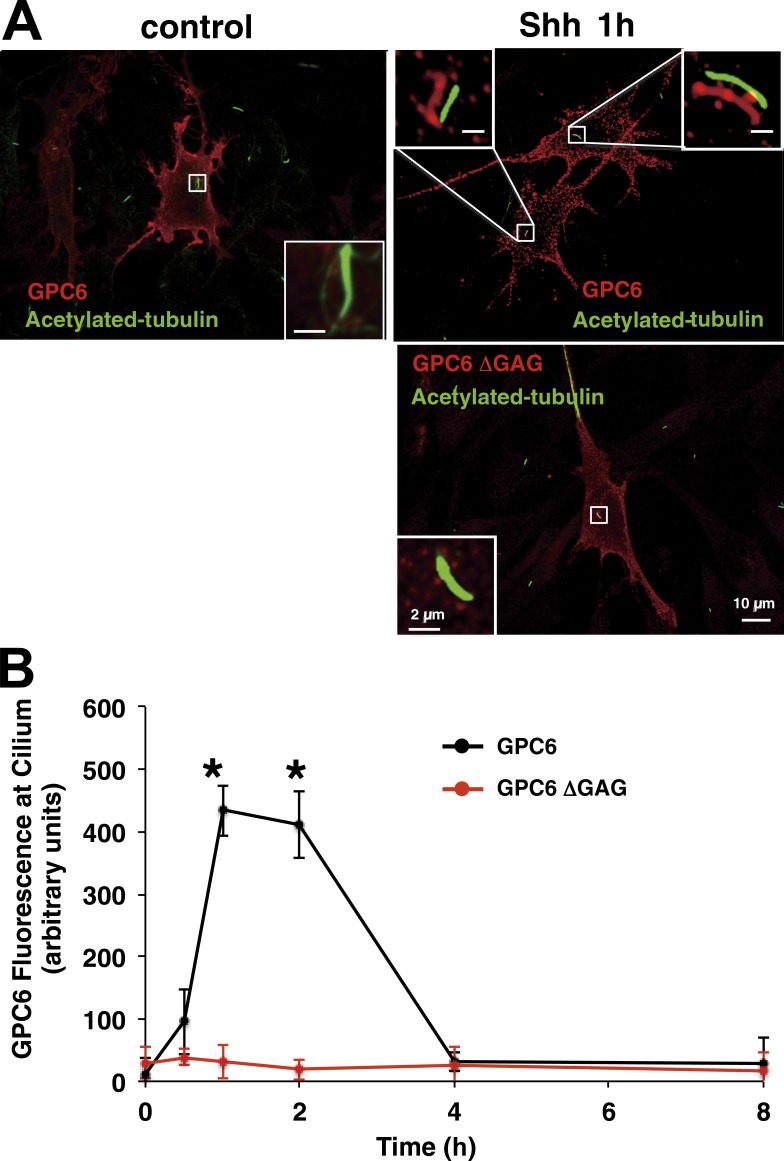
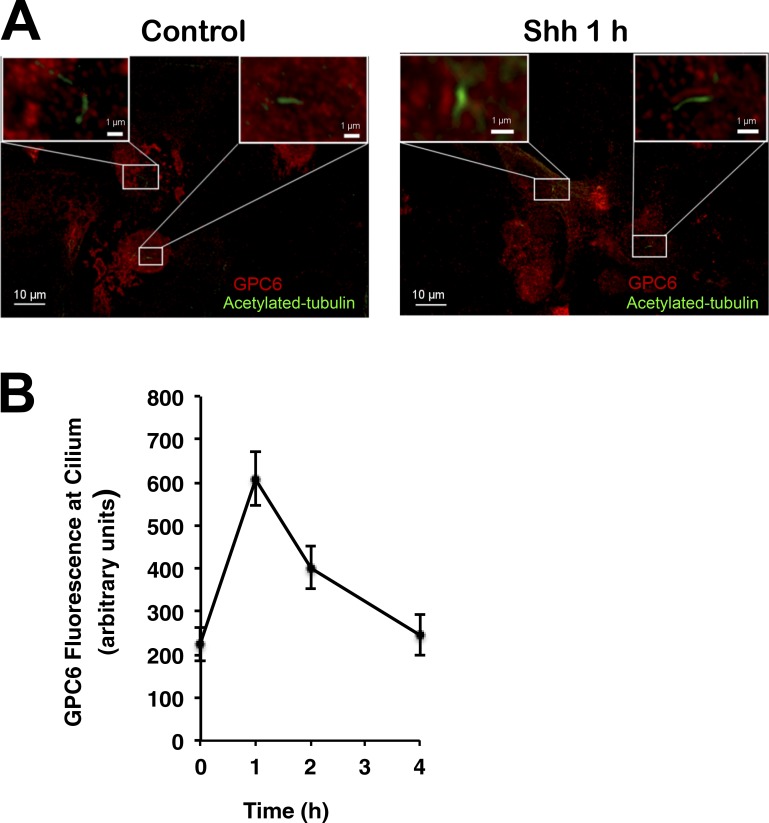
It is now well established that Hh signaling is triggered at the primary cilium and that the interaction between Hh and Ptc1 can occur at the ciliary membrane (Rohatgi et al., 2007). Therefore, we reasoned that, if GPC6 acts to facilitate and/or stabilize the Hh/Ptch1 interaction, this GPC should be found at the ciliary membrane. To determine the localization of GPC6, we transfected NIH 3T3 cells with GPC6 or GPC6ΔGAG and stained the transfected cells with antibodies against GPC6 and acetylated tubulin, a marker of primary cilium (Fig. 8 A). Surprisingly, we could not detect GPC6 in the cilium. However, when the transfected cells were incubated with Shh for 1 h at 37°C, most of the GPC6-expressing cells displayed GPC6 in their cilia (Fig. 8 A). This result strongly suggests that Shh induces the migration of GPC6 to the cilium. In contrast, the GPC6ΔGAG mutant did not move into the cilium after incubation with Shh (Fig. 8 A). In addition, we noticed that the GPC6 staining in the Shh-treated cells was dot like, whereas the staining pattern in the untreated cells or in the cells transfected with GPC6ΔGAG was more diffuse. Next, we quantified the fluorescence generated by ciliary GPC6 in cells incubated with Shh for different time periods. As shown in Fig. 8 B, we found that GPC6 can be detected at the cilia as soon as 30 min after Shh addition and that it migrates away from the ciliary membrane very quickly, reaching background levels after 4 h. However, GPC6ΔGAG was not found in the cilia at any of the investigated time points.
Figure 8.
Shh induces the migration of GPC6 to the cilia. (A) NIH 3T3 cells were transfected with GPC6 or GPC6ΔGAG and treated with Shh-conditioned medium or vector control–conditioned medium for 1 h. Then, cells were immunostained with antibodies for GPC6 (red) and acetylated tubulin (green). Boxes in the corners represent shifted overlays of enlargements of the indicated areas containing the cilia. (B) GPC6 or GPC6ΔGAG immunofluorescence at the cilium of transfected NIH 3T3 cells was measured before and after Shh treatment for the indicated time periods. The fluorescence intensities shown represent the mean of measurements performed in the following number of cells transfected with GPC6/GPC6ΔGAG: 30/25 (0 h), 22/11 (0.5 h), 30/25 (1 h), 39/10 (2 h), 10/26 (4 h), and 22/20 (8 h) ± SD. A two-tailed Student’s t test was applied for statistical analysis. *, P < 0.005 (significantly different).
It has been reported that, in the absence of Hh, Ptc1 is normally found in the ciliary membrane and that it moves out of the cilium upon Shh treatment (Rohatgi et al., 2007). Smo displays the opposite behavior (Rohatgi et al., 2007). First, we investigated the impact of GPC6 on the Shh-induced migration of Smo to the cilium. Transfected cells were stained at different time points after the addition of Shh with antibodies for anti–acetylated tubulin and Smo. We observed that GPC6 stimulated the accumulation of Smo in this appendage (Fig. 9 A). These changes were not observed when cells were transfected with GPC6ΔGAG. We also tried to investigate the effect of GPC6 on Ptc1 removal from the cilia. However, using commercially available antibodies, the detection of Ptc1 was not robust enough for accurate quantification.
Figure 9.
GPC6 stimulates the migration of Smo to the cilia after Shh treatment. (A) NIH 3T3 cells were transfected with GPC6, GPC6ΔGAG, or vector control. Transfected cells were treated with Shh for the indicated time periods, and cells were stained with antibodies against Smo and acetylated tubulin. The fluorescence intensities shown represent the mean of measurements performed in the following number of cells transfected with vector/GPC6/GPC6ΔGAG: 23/29/23 (0 h), 27/26/19 (1 h), 26/26/28 (2 h), and 26/37/22 (4 h) ± SD. *, P < 0.01 (significantly different). (B) NIH 3T3 cells were transfected with GPC6 or GPC6ΔGAG. Transfected cells were treated with SAG for the indicated time periods, and cells were stained with antibodies against GPC6 and acetylated tubulin. The fluorescence intensities shown represent the mean of measurements performed in the following number of cells transfected with GPC6/GPC6ΔGAG: 9/7 (0 h), 15/15 (0.5 h), 11/11 (2 h), and 10/10 (4 h) ± SD. Two-tailed Student’s t tests were applied for statistical analysis.
In principle, the Shh-induced migration of GPC6 to the cilium could be a direct consequence of the binding of Shh to GPC6, or it could represent an indirect consequence of the activation of the Hh signaling pathway. To distinguish between these two possibilities, we investigated the impact of treating the GPC6-transfected cells with Smo agonist (SAG), a small molecule that activates Hh signaling by binding to Smo. As shown in Fig. 9 B, we found that SAG treatment failed to induce GPC6 migration into the ciliary membrane, suggesting that the migration of this GPC to the cilium is a direct consequence of its interaction with Shh.
Finally, we examined whether Shh can induce the migration of endogenous GPC6 in primary chondrocytes isolated from E18.5 femurs. Fig. 10 shows that, like in the case of NIH 3T3 cells, treatment with Shh for 1 h induced a very clear migration of GPC6 into the cilium of chondrocytes and that its presence in this appendage is drastically reduced after 4 h of Shh treatment.
Figure 10.
Shh induces the migration of GPC6 in primary chondrocytes. (A) Chondrocytes isolated from E18.5 femurs were treated with Shh- or vector control–conditioned medium for 1 h. Then, cells were immunostained with antibodies for GPC6 (red) and acetylated tubulin (green). Boxes in the corners represent shifted overlays of enlargements of the indicated areas containing the cilium (bars, 1 µm). (B) GPC6 fluorescence at the cilia of chondrocytes was measured before and after Shh treatment for the indicated time points. The fluorescence intensities shown represent the mean of measurements performed in the following number of cells: 28 (0 h), 27 (1 h), 12 (2 h), and 12 (4 h) ± SD.
Discussion
In this study, we show that GPC6-null mice display significantly shorter long bones than normal littermates. In addition, the mutant mice present severe facial dysmorphism. Therefore, these observations indicate that GPC6-null mice represent a good model to study the molecular basis of OMOD1. We have also generated a mouse strain in which GPC6 has been specifically deleted in chondrocytes. Unlike the global KO, the chondrocyte-specific GPC6 mutants are viable, but they also display a significant shortening of the long bones. This confirms that GPC6 plays a direct role in bone development.
Notably, OMOD1 is the second human genetic condition characterized by abnormal bone development in which a member of the GPC family has been implicated. Simpson-Golabi-Behmel overgrowth syndrome is caused by loss-of function mutations in the GPC3 gene (Pilia et al., 1996). We have previously demonstrated that GPC3 acts as an inhibitor of Hh signaling in the embryonic bone, and we have proposed that the overgrowth observed in the Simpson-Golabi-Behmel patients is caused, at least in part, by increased Hh activity (Capurro et al., 2008, 2009). Interestingly, in this study, we have shown that, in the context of long bone development, GPC6 displays the opposite activity compared with GPC3: it acts to stimulate Hh signaling, strongly suggesting that OMOD1 is caused, at least in part, by reduced Hh activity in the developing long bones.
The present study provides extensive evidence that GPC6 stimulates Hh signaling. First, we showed that the expression of Gli1 and Ptc1, two well-established Hh pathway targets, is significantly reduced in the developing femurs of GPC6-null mice. Second, by breeding GPC6-heterozygous mice with Ihh-heterozygous mice, we provided genetic evidence indicating that the Ihh signaling pathway mediates the regulatory activity of GPC6 on embryonic bone growth. Third, we demonstrated that GPC6 can strongly stimulate Hh signaling in luciferase reporter assays. Fourth, we showed that GPC6 can bind with high affinity to Shh and Ihh. In addition, our finding that growth plate chondrocytes from GPC6-null developing bones display reduced proliferation compared with chondrocytes from normal littermates is also consistent with a role of GPC6 as a positive regulator of Hh signaling.
GPCs have been shown to regulate canonical (β-catenin dependent) and noncanonical (β-catenin independent) Wnt signaling. Wnt5a is a well-characterized stimulator of noncanonical Wnt signaling. Notably, Wnt5a-null mice display perinatal lethality, severe dwarfism, short limbs, and facial dysmorphisms (Yamaguchi et al., 1999). Thus, in principle, if GPC6 is a positive regulator of noncanonical Wnt signaling, it could be proposed that the phenotype of the GPC6-null embryos is caused, at least in part, by a reduction in noncanonical Wnt signaling. One of the well-established changes induced by Wnt5a in many cell types is the phosphorylation of Dvl3 and Dvl2, and Wnt5a-null embryos display a significant reduction in such phosphorylation (Ho et al., 2012). However, we did not detect any significant difference in the degree of Dvl3 and Dvl2 phosphorylation in the GPC6-null femurs compared with those of the WT littermates (Fig. S3). Thus, even though we cannot discard a role of the noncanonical Wnt pathway in the phenotype of the GPC6-null mice, it is highly unlikely that this pathway is an important mediator of the regulatory activity of GPC6 in bone development. In this regard, it should be noted that Wnt5a-null mice display a drastic reduction of Ihh expression in the developing long bones (Yang et al., 2003). Therefore, it is possible that the shortened bones in Wnt5a-null embryos are, at least in part, the indirect consequence of reduced Ihh signaling.
Our in vitro studies have shown that GPC6 stimulates Hh signaling by increasing the interaction of Shh with Ptc1 (Fig. 6 B). Based on this finding and on our results indicating that GPC6 interacts with both Shh and Ptc1 (Fig. 7), we propose that GPC6 increases Hh signaling by forming a complex with these two proteins that facilitates/stabilizes their interaction.
We demonstrated by SPR analysis that Shh and Ihh can directly bind with high affinity to the GPC6 core protein (Fig. 7). Previously, we had shown that the core protein of GPC3 can also bind with high affinity to Shh and Ihh (Capurro et al., 2008). Likewise, it has been reported that Drosophila Hh directly interacts with the core protein of the GPC Dlp (Yan et al., 2010). However, the interaction of GPC5 with Shh is mediated by the GAG chains (Li et al., 2011). The molecular basis of this differential interaction of the core proteins of various GPCs with Hhs remains to be uncovered.
We have also shown here that GPC6 can bind to Ptc1 and that this interaction is mediated by the GAG chains. Similar findings have been reported for GPC5 (Li et al., 2011). Therefore, we are proposing that GPC6 stimulates Hh signaling by binding to both Hh and Ptc1 and facilitating/stabilizing their interaction. As such, it would be expected that the nonglycanated GPC6ΔGAG would not be able to stimulate Hh signaling. Experimentally, we have found that high levels of expression of this mutant can exert a modest stimulatory effect on Hh activity (Fig. 6 C). This finding could be explained by the fact that Hh can bind with high affinity to the core protein of GPC6. This interaction would increase the concentration of Hh at the cell surface, therefore reducing the dimensionality of ligand movement and consequently increasing the frequency of encounters between Hh and Ptc1. This mechanism has been proposed to explain the stimulatory effects of many cell-surface proteoglycans on cell signaling (Schlessinger et al., 1995).
One of the most important findings of this study is that, whereas in the absence of Hh, GPC6 is located outside of the primary cilium, Hh addition results in the migration of GPC6 to this appendage, where Hh signaling is normally triggered (Fig. 8). Notably, ciliary accumulation can be detected as early as 30 min after Hh addition, but GPC6 does not stay very long in the cilium, becoming almost undetectable at this location after 4 h. Interestingly, we also observed that, in the presence of Hh, GPC6 stimulates the accumulation of Smo at this organelle (Fig. 9). The Hh-induced migration of GPC6 into the cilium was observed both in GPC6-transfected embryonic fibroblasts (Fig. 8) and in primary chondrocytes from the femur expressing endogenous GPC6. The localization of GPC6 at the cilium in the presence of Hh is consistent with its capacity to bind to Ptc1 and to stimulate Hh activity.
We have previously reported that GPC5 is located at the cilium even in the absence of exogenous Hh (Li et al., 2011). However, GPC3 is located outside of the cilium in the presence and absence of this growth factor (Li et al., 2011). Therefore, it seems that ciliary localization correlates with the capacity of a given GPC to stimulate Hh signaling.
Currently, the mechanism of Hh-induced migration to the cilium remains unknown. However, the fact that the nonglycanated mutant GPC6ΔGAG does not move to the cilium in the presence of Hh suggests that GPC6 might piggyback through the GAG chains with another protein that has the capacity to migrate to the cilium in the presence of Hh.
Materials and methods
Mouse strains
Gpc6-heterozygous mice (B6N.129S5-Gpc6tm1/Lex/Mmcd) were obtained from the Mutant Mouse Regional Resource Center at the University of San Diego. Ihh-heterozygous mice were purchased from the Jackson Laboratory. C57BL/6 WT mice were obtained from Charles River Canada Laboratories. All the procedures used for mouse experiments were approved by the Animal Care Committee of the Sunnybrook Research Institute.
To generate chondrocyte-specific GPC6-null mice, a mouse strain containing a floxed GPC6 allele was purchased from EUCOMM. The modified allele had two loxP sites flanking exon 3 of GPC6, as well as a neomycin gene and a LacZ reporter flanked by two flippase recognition target sites. These mice were crossed with C57BL/6N Flp-recombinase transgenic mice (under the control of the ActnB promoter) to remove the Neo marker and LacZ reporter gene in every cell. The resulting mice were crossed with B6;SJL-Tg(Col2α1-Cre)1Bhr/J transgenic mice (purchased from Jackson laboratories) expressing the Cre recombinase under the control of the Col2α1 gene promoter. As the Col2α1 gene is expressed in chondrocytes, this crossing leads to the excision of GPC6 exon 3 specifically in these cells and, therefore, to the generation of a cartilage-specific GPC6-null mouse. After backcross on a C57BL/6N background, mice heterozygous for the GPC6 KO allele in chondrocytes were mated between them to obtain cartilage-specific GPC6-null mice.
Breeding and processing of mice
GPC6 KO and WT littermates were obtained by mating GPC6 heterozygotes. Double GPC6-Ihh heterozygotes were first obtained by crossing GPC6 and Ihh heterozygotes and then mated to generate the GPC6/Ihh double-KO embryos. The body weights of E17.5–E18.5 embryos were obtained immediately after dissection. Then, tail tip samples were collected and subsequently genotyped by PCR. Whole embryos or dissected limbs were processed for Alcian blue–Alizarin staining. Tissue sections from dissected femurs were paraffin embedded for immunohistochemical analysis. Femoral RNA was isolated for real-time PCR assessment of Hh target levels. Whole-embryo histopathology was performed at the Centre for Modeling Human Disease, Toronto Centre for Phenogenomics.
Cell lines and plasmids
293T and NIH 3T3 cells (obtained from the ATCC) were cultured in DMEM supplemented with 10% FBS at 37°C in a humidified atmosphere with 5% CO2. Primary chondrocytes were generated from mouse embryos as previously described (Gosset et al., 2008) and cultured in DMEM supplemented with 10% FBS and 2 mM glutamine. Expression vectors for GPC6, GPC6ΔGAG, GPC3, GPC3ΔGAG, GPC6-AP, GPC6ΔGAG-AP, fully processed Shh, ShhN, AP, Shh-AP, and Ptc-HA were previously described (Capurro et al., 2008, 2014).
Analysis of BrdU incorporation
15.5-d pregnant mice were injected intraperitoneally 1 h before sacrifice with 400 µl of the BrdU labeling reagent (BrdU Labeling and Detection kit II; Roche). Embryonic femurs were dissected, fixed in 10% formalin overnight, and stored in 70% ethanol until processing. Paraffin-embedded sections were prepared, and BrdU staining was performed with an anti-BrdU mouse monoclonal antibody (clone BMG6H8; Roche), and a Mouse on Mouse (M.O.M.) Immunodetection kit (VECTOR Laboratories). Tissue sections were blocked in M.O.M. blocking reagent and then were incubated with an anti-BrdU antibody (1:10 in M.O.M. diluents) overnight at 4°C. After washing twice with PBS, a biotinylated anti–mouse IgG antibody was added for 10 min. Bound antibody was detected with Vectastain ABC and DAB. For quantification of BrdU labeling, all the BrdU-positive and -negative nuclei in each femur section were counted.
Quantification of Ptc1 and Gli1 transcripts
E15.5 femurs were dissected, and RNA was extracted using TRIzol reagent (Invitrogen). cDNA was generated by reverse transcription using oligo(dT)12–18 primers (Invitrogen). The relative levels of Ptc1 and Gli1 transcripts were measured by real-time RT-PCR, using β-actin transcript levels as standards. A sequence detection system (ABI Prism 7000; Applied Biosystems) and QuantiTect SYBR Green (Qiagen) were used. Sequence of the oligonucleotide primers used were: Gli1 forward (F), 5′-AGGGAGGAAAGCAGACTGAC-3′ and reverse (R), 5′-CCAGTCATTTCCACACCACT-3′; Ptc1 F, 5′-CAAGTGTCGTCCGGTTTGC-3′ and R, 5′-CTGTACTCCGAGTCGGAGGAA-3′; and β-actin F, 5′-TTCTACAATGAGCTGCGTGTG-3′ and R, 5′-GGGGTGTTGAAGGTCTCAAA-3′.
Immunohistochemistry
Embryonic femurs were dissected, fixed in 10% formalin overnight, and stored in 70% ethanol until processing. Paraffin-embedded femur growth plate sections were prepared from E15.5 embryos. Ihh, Ki67, and Cyclin D1 were immunostained with antibodies from Santa Cruz (sc-1196; 1:75), Cell Signaling (no. 12202; 1:400), and Abcam (ab134175; 1:200), respectively. DAB was used as a color-developing agent.
Hh reporter assay
NIH 3T3 cells were seeded in 6-well plates (220,000 cells/well) and transfected with an Hh-luciferase reporter, a β-galactosidase plasmid, and the indicated expression vectors. 1 d after transfection, cells were transferred to 24-well plates at 50% confluence, and in the next day, Shh-, ShhN-, or control-conditioned medium was added for 48 h. Then, cells were lysed, and luciferase activity was measured. This measurement was normalized by the corresponding β-galactosidase activity. The Shh-, ShhN-, and control-conditioned media containing 2% FBS were prepared by transfecting 293T cells with SHH or ShhN expression vectors or vector control, respectively. The media were collected 4 d after transfection.
Shh-Ptc1 binding assay
NIH 3T3 cells were transiently transfected with increasing amounts of GPC6, GPC3, or vector control. 2 d later, the transfected cells were incubated for 2.5 h at 4°C with conditioned medium containing a Shh-AP fusion protein or AP alone. After washing, cells were lysed, and the endogenous Ptc1 was immunoprecipitated. Then, the AP activity in the immunoprecipitate was measured with a p-Nitrophenyl phosphate tablet set (SIGMAFAST; Sigma-Aldrich). The AP activity of immunoprecipitates from cells incubated with AP alone were subtracted from each sample.
SPR analysis
This analysis was performed as previously described with a Biacore T200 system (GE Healthcare; Li et al., 2011). In brief, GPC6-AP and GPC6ΔGAG-AP fusion proteins were purified from conditioned medium generated by 293T cells transfected with the corresponding expression vectors. The purification consisted of an anion-exchange chromatography followed by an anti-AP affinity chromatography. Purified GPCs were biotinylated with EZ-link Sulfo-NHS-LC biotin (Thermo Fisher Scientific) and were then immobilized on streptavidin (SA) sensor chips (1,300 and 800 response units for GPC6-AP and GPC6ΔGAG-AP, respectively). Various concentrations (from 62.5 to 1,000 nM) of recombinant Shh or Ihh were injected to determine the binding profile. The flow rate was 30 µl/min. To control for nonspecific association of Shh or Ihh with the SA sensor chip, solutions of analyte were injected over the ligand-free surface and subtracted from binding data before analysis. All measurements were performed in triplicate. Sensorgrams were analyzed using the 1:1 ligand model provided in the BIAevaluation software (GE Healthcare) to calculate association and dissociation rate constants (ka and kd, respectively).
Pull-down assay
293T cells were transfected with an HA-tagged Ptc1 expression vector. 2 d after transfection, cells were lysed, and the lysates were incubated with an anti-HA monoclonal antibody (12CA5; Roche) that was attached to protein G–sepharose beads (Sigma-Aldrich). Then, the Ptc1-covered beads were blocked with 5% BSA in PBS containing 0.1% Triton X-100 for 90 min at room temperature, and aliquots containing equal amounts of beads were incubated for 1 h at room temperature with GPC6-AP, GPC6ΔGAG-AP, or AP control-conditioned media. After four washes with 20 mM Hepes, pH 7.4, 150 mM NaCl, and 0.25% Tween 20, the AP activity bound to the beads was measured with a p-Nitrophenyl phosphate tablet set (SIGMAFAST). The background binding was measured by incubating the beads with lysates of 293T cells transfected with an AP vector control, and it was subtracted from each sample measurement. The GPC-AP media were generated by transfecting 293T cells with GPC6-AP or GPC6ΔGAG-AP expression vectors.
GPC6-Ptc1 binding assay
293T cells were plated on 24-well plates and transfected with a Ptc1 expression vector or vector control (pcDNA). Transfected cells were incubated with GPC6-AP–, GPC6ΔGAG-AP–, or AP-conditioned media containing equal AP concentrations for 2 h at 4°C. Cells were extensively washed with PBS and lysed in 1% Triton and 10 mM Tris, pH 8.0, at room temperature for 5 min, and the AP activity of aliquots of cell lysates containing the same amount of protein was measured. GPC6-AP–, GPC6ΔGAG-AP–, or AP-conditioned media were generated by transfecting 293T cells with the corresponding expression vectors. 16 h after transfection, conditioned media were collected for 24 h in medium with 10% FCS.
Localization of GPC6, GPC6ΔGAG, and Smo
NIH 3T3 cells were transfected with the indicated expression vectors using Lipofectamine 3000 (InvitroGen). 1 d later, cells were split and plated on poly-l-lysine–treated coverslips. Then, the cells were starved in serum-free medium overnight, and Shh-conditioned media was added for the indicated time periods. Then, cells were incubated at 37°C in a humidified atmosphere with 5% CO2. After washing three times with PBS, cells were fixed with 4% paraformaldehyde in PBS for 10 min at 4°C. Fixed cells were blocked for 30 min with 2% BSA in PBS, and immunofluorescence analysis was performed. For GPC6 detection, cells were incubated with an anti-GPC6 antibody (AF2845; 1:500; R&D). After washing three times with PBS, Alexa Fluor–coupled secondary antibodies were added. Then, the cells were washed and permeabilized with 0.1% Triton X-100 in PBS for 10 min and blocked for 30 min in 2% BSA. Primary cilia were visualized with anti–acetylated tubulin mAb (T7451; 1:1,000; Sigma-Aldrich). Smo was visualized with anti-HA rabbit mAb (C29F4; Cell Signaling) or anti-Smo (ab38686; 1:1,000; Abcam). After washing three times with PBS, Alexa Fluor–coupled secondary antibodies were added in blocking solution at 1:1,000 dilution for 1 h at room temperature. Confocal images were generated using spinning-disk microscopy and a Zeiss LSM image browser. All images were taken with equivalent exposure time. Image analysis was performed using the Zen 2 program (blue edition). In brief, a mask, which was constructed by manually outlining cilia in the image of acetylated tubulin staining, was applied to GPC6 or Smo-stained images to measure fluorescence intensity at cilia. The background, which was subtracted from the fluorescence intensity in the cilia, was obtained by measuring several representative regions on the cell by moving the mask. For the localization of endogenous GPC6 in primary chondrocytes, only freshly derived cells were used (P1). Cells were starved in serum-free medium overnight before staining.
Online supplemental material
Fig. S1 shows Western Blot analysis of Ihh in E17.5 femurs. Fig. S2 shows an Hh reporter assay. Fig. S3 shows that Wnt signaling is not altered in GPC6-null femurs.
Acknowledgments
We thank Susan Newbigging from the Centre for Modeling Human Disease, Toronto Centre for Phenogenomics, for her help in the characterization of the GPC6-null mice and Ana Belinda Campos-Xavier and Laureane Mittaz-Crettol for their help in the generation of the tissue-specific mouse model.
This project was funded by the Canadian Institutes of Health Research (grant MOP142344 to J. Filmus) and by the Swiss National Research Foundation (grant 310030_132940 to L. Bonafe). Tomomi Izumikawa and Tomuyuki Kaneiwa were supported by Overseas Fellowships from the Japan Society for the Promotion of Science.
There authors declare no competing financial interests.
Author contributions: M. Capurro designed and performed the analysis of the phenotype of the global GPC6-null mice, the quantification of Ptc1 and Gli1, the analysis of the GPC6/Ihh double mutants, the Hh reporter assays, and the Shh-Ptc1 binding assay. T. Izumikawa designed and performed the Ptc1-GPC6 pull-down assay and the analysis of the localization of GPC6 and Smo in NIH 3T3 cells and primary chondrocytes. P. Suarez generated and analyzed the chondrocyte-specific GPC6-mutant mice. W. Shi performed the BrdU incorporation assay and the immunohistochemistry for the detection of cyclin D1, Ihh, and Ki67 and contributed to the performance of some Hh reporter assays and to the analysis of the localization of GPC6 and Smo in NIH 3T3 cells. M. Cydzik designed and performed the SPR analysis. T. Kaneiwa contributed to the performance of the GPC6 and Smo localization studies. J. Gariepy contributed to the writing of the manuscript. L. Bonaparte participated in the conception of the project, designed and analyzed the experiments with the chondrocyte-specific GPC6-mutant mice, and contributed to the writing of the manuscript. J. Filmus conceived the project, designed and analyzed the results of the experiments, and wrote the manuscript.
Footnotes
Abbreviations used:
- GAG
- glycosaminoglycan
- GPC
- glypican
- HA
- hemagglutinin A
- Hh
- hedgehog
- HS
- heparan sulfate
- Ihh
- Indian Hh
- KO
- knockout
- Ptc1
- Patched 1
- SA
- streptavidin
- SAG
- Smo agonist
- Shh
- Sonic Hh
- Smo
- Smoothened
- SPR
- surface plasmon resonance
References
- Ahrens M.J., Li Y., Jiang H., and Dudley A.T.. 2009. Convergent extension movements in growth plate chondrocytes require gpi-anchored cell surface proteins. Development. 136:3463–3474. 10.1242/dev.040592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T., Kamimura K., Firkus C., Takeo S., Shimmi O., and Nakato H.. 2008. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev. Biol. 313:408–419. 10.1016/j.ydbio.2007.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N.J., Bennett M.L., Foo L.C., Wang G.X., Chakraborty C., Smith S.J., and Barres B.A.. 2012. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 486:410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandari S., Exner S., Ortmann C., Bachvarova V., Vortkamp A., and Grobe K.. 2015. Sweet on Hedgehogs: regulatory roles of heparan sulfate proteoglycans in Hedgehog-dependent cell proliferation and differentiation. Curr. Protein Pept. Sci. 16:66–76. 10.2174/1389203716666150213162649 [DOI] [PubMed] [Google Scholar]
- Bar E.E., Chaudhry A., Farah M.H., and Eberhart C.G.. 2007. Hedgehog signaling promotes medulloblastoma survival via Bc/II. Am. J. Pathol. 170:347–355. 10.2353/ajpath.2007.060066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borochowitz Z., Sabo E., Misselevitch I., and Boss J.H.. 1998. Autosomal-recessive omodysplasia: prenatal diagnosis and histomorphometric assessment of the physeal plates of the long bones. Am. J. Med. Genet. 76:238–244. [DOI] [PubMed] [Google Scholar]
- Briscoe J., and Thérond P.P.. 2013. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14:416–429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Campos-Xavier A.B., Martinet D., Bateman J., Belluoccio D., Rowley L., Tan T.Y., Baxová A., Gustavson K.H., Borochowitz Z.U., Innes A.M., et al. 2009. Mutations in the heparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am. J. Hum. Genet. 84:760–770. 10.1016/j.ajhg.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro M.I., Xu P., Shi W., Li F., Jia A., and Filmus J.. 2008. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev. Cell. 14:700–711. 10.1016/j.devcel.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Capurro M.I., Li F., and Filmus J.. 2009. Overgrowth of a mouse model of Simpson-Golabi-Behmel syndrome is partly mediated by Indian hedgehog. EMBO Rep. 10:901–907. 10.1038/embor.2009.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro M., Martin T., Shi W., and Filmus J.. 2014. Glypican-3 binds to Frizzled and plays a direct role in the stimulation of canonical Wnt signaling. J. Cell Sci. 127:1565–1575. 10.1242/jcs.140871 [DOI] [PubMed] [Google Scholar]
- Desbordes S.C., and Sanson B.. 2003. The glypican Dally-like is required for Hedgehog signalling in the embryonic epidermis of Drosophila. Development. 130:6245–6255. 10.1242/dev.00874 [DOI] [PubMed] [Google Scholar]
- Elçioglu N.H., Gustavson K.H., Wilkie A.O., Yüksel-Apak M., and Spranger J.W.. 2004. Recessive omodysplasia: five new cases and review of the literature. Pediatr. Radiol. 34:75–82. 10.1007/s00247-003-1064-9 [DOI] [PubMed] [Google Scholar]
- Filmus J., and Capurro M.. 2012. The glypican family. In Extracellular Matrix: Pathobiology and Signaling. Karamanos N.K., editor. De Gruyter, Berlin/Boston: 209–220. 10.1515/9783110258776.209 [DOI] [Google Scholar]
- Filmus J., and Capurro M.. 2014. The role of glypicans in Hedgehog signaling. Matrix Biol. 35:248–252. 10.1016/j.matbio.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Filmus J., Capurro M., and Rast J.. 2008. Glypicans. Genome Biol. 9:224 10.1186/gb-2008-9-5-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Hu J., Stricker S., Cheung M., Ma G., Law K.F., Witte F., Briscoe J., Mundlos S., He L., et al. 2009. A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature. 458:1196–1200. 10.1038/nature07862 [DOI] [PubMed] [Google Scholar]
- Gosset M., Berenbaum F., Thirion S., and Jacques C.. 2008. Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3:1253–1260. 10.1038/nprot.2008.95 [DOI] [PubMed] [Google Scholar]
- Hagihara K., Watanabe K., Chun J., and Yamaguchi Y.. 2000. Glypican-4 is an FGF2-binding heparan sulfate proteoglycan expressed in neural precursor cells. Dev. Dyn. 219:353–367. [DOI] [PubMed] [Google Scholar]
- Hilton M.J., Gutiérrez L., Martinez D.A., and Wells D.E.. 2005. EXT1 regulates chondrocyte proliferation and differentiation during endochondral bone development. Bone. 36:379–386. 10.1016/j.bone.2004.09.025 [DOI] [PubMed] [Google Scholar]
- Ho H.Y.H., Susman M.W., Bikoff J.B., Ryu Y.K., Jonas A.M., Hu L., Kuruvilla R., and Greenberg M.E.. 2012. Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc. Natl. Acad. Sci. USA. 109:4044–4051. 10.1073/pnas.1200421109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P.W., Nakano Y., and Seger C.. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12:393–406. 10.1038/nrg2984 [DOI] [PubMed] [Google Scholar]
- Jackson S.M., Nakato H., Sugiura M., Jannuzi A., Oakes R., Kaluza V., Golden C., and Selleck S.B.. 1997. dally, a Drosophila glypican, controls cellular responses to the TGF-beta-related morphogen, Dpp. Development. 124:4113–4120. [DOI] [PubMed] [Google Scholar]
- Karsenty G., Kronenberg H.M., and Settembre C.. 2009. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 25:629–648. 10.1146/annurev.cellbio.042308.113308 [DOI] [PubMed] [Google Scholar]
- Koziel L., Wuelling M., Schneider S., and Vortkamp A.. 2005. Gli3 acts as a repressor downstream of Ihh in regulating two distinct steps of chondrocyte differentiation. Development. 132:5249–5260. 10.1242/dev.02097 [DOI] [PubMed] [Google Scholar]
- Lee R.T.H., Zhao Z., and Ingham P.W.. 2016. Hedgehog signalling. Development. 143:367–372. 10.1242/dev.120154 [DOI] [PubMed] [Google Scholar]
- Li F., Shi W., Capurro M., and Filmus J.. 2011. Glypican-5 stimulates rhabdomyosarcoma cell proliferation by activating Hedgehog signaling. J. Cell Biol. 192:691–704. 10.1083/jcb.201008087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F., Zhang X.M., Karp S., Yang Y., and McMahon A.P.. 2001. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 128:5099–5108. [DOI] [PubMed] [Google Scholar]
- Long F., Joeng K.S., Xuan S., Efstratiadis A., and McMahon A.P.. 2006. Independent regulation of skeletal growth by Ihh and IGF signaling. Dev. Biol. 298:327–333. 10.1016/j.ydbio.2006.06.042 [DOI] [PubMed] [Google Scholar]
- Lum L., Yao S., Mozer B., Rovescalli A., Von Kessler D., Nirenberg M., and Beachy P.A.. 2003. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 299:2039–2045. 10.1126/science.1081403 [DOI] [PubMed] [Google Scholar]
- Mackie E.J., Ahmed Y.A., Tatarczuch L., Chen K.S., and Mirams M.. 2008. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int. J. Biochem. Cell Biol. 40:46–62. 10.1016/j.biocel.2007.06.009 [DOI] [PubMed] [Google Scholar]
- Ohkawara B., Yamamoto T.S., Tada M., and Ueno N.. 2003. Role of glypican 4 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development. 130:2129–2138. 10.1242/dev.00435 [DOI] [PubMed] [Google Scholar]
- Oliver T.G., Grasfeder L.L., Carroll A.L., Kaiser C., Gillingham C.L., Lin S.M., Wickramasinghe R., Scott M.P., and Wechsler-Reya R.J.. 2003. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. USA. 100:7331–7336. 10.1073/pnas.0832317100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilia G., Hughes-Benzie R.M., MacKenzie A., Baybayan P., Chen E.Y., Huber R., Neri G., Cao A., Forabosco A., and Schlessinger D.. 1996. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat. Genet. 12:241–247. 10.1038/ng0396-241 [DOI] [PubMed] [Google Scholar]
- Razzaque M.S., Soegiarto D.W., Chang D., Long F., and Lanske B.. 2005. Conditional deletion of Indian hedgehog from collagen type 2α1-expressing cells results in abnormal endochondral bone formation. J. Pathol. 207:453–461. 10.1002/path.1870 [DOI] [PubMed] [Google Scholar]
- Robbins D.J., Fei D.L., and Riobo N.A.. 2012. The Hedgehog signal transduction network. Sci. Signal. 5:re6 10.1126/scisignal.2002906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., and Scott M.P.. 2007. Patched1 regulates hedgehog signaling at the primary cilium. Science. 317:372–376. 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Ryan K.E., and Chiang C.. 2012. Hedgehog secretion and signal transduction in vertebrates. J. Biol. Chem. 287:17905–17913. 10.1074/jbc.R112.356006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J., Lax I., and Lemmon M.. 1995. Regulation of growth factor activation by proteoglycans: what is the role of the low affinity receptors? Cell. 83:357–360. 10.1016/0092-8674(95)90112-4 [DOI] [PubMed] [Google Scholar]
- Song H.H., Shi W., Xiang Y.Y., and Filmus J.. 2005. The loss of glypican-3 induces alterations in Wnt signaling. J. Biol. Chem. 280:2116–2125. 10.1074/jbc.M410090200 [DOI] [PubMed] [Google Scholar]
- St-Jacques B., Hammerschmidt M., and McMahon A.P.. 1999. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13:2072–2086. 10.1101/gad.13.16.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja-Bageshwar S., and Gumienny T.L.. 2013. Regulation of TGFβ superfamily signaling by two separable domains of glypican LON-2 in C. elegans. Worm. 2:e23843 10.4161/worm.23843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Li L., Tang J., Li Y., Lin W.Y., Martin F., Grant D., Solloway M., Parker L., Ye W., et al. 2010. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 28:749–755. 10.1038/nbt.1644 [DOI] [PubMed] [Google Scholar]
- Toftgård R. 2009. Two sides to cilia in cancer. Nat. Med. 15:994–996. 10.1038/nm0909-994 [DOI] [PubMed] [Google Scholar]
- Williams E.H., Pappano W.N., Saunders A.M., Kim M.S., Leahy D.J., and Beachy P.A.. 2010. Dally-like core protein and its mammalian homologues mediate stimulatory and inhibitory effects on Hedgehog signal response. Proc. Natl. Acad. Sci. USA. 107:5869–5874. 10.1073/pnas.1001777107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.H., and Stoeckli E.T.. 2013. Sonic hedgehog regulates its own receptor on postcrossing commissural axons in a glypican1-dependent manner. Neuron. 79:478–491. 10.1016/j.neuron.2013.05.025 [DOI] [PubMed] [Google Scholar]
- Witt R.M., Hecht M.L., Pazyra-Murphy M.F., Cohen S.M., Noti C., van Kuppevelt T.H., Fuller M., Chan J.A., Hopwood J.J., Seeberger P.H., and Segal R.A.. 2013. Heparan sulfate proteoglycans containing a glypican 5 core and 2-O-sulfo-iduronic acid function as Sonic Hedgehog co-receptors to promote proliferation. J. Biol. Chem. 288:26275–26288. 10.1074/jbc.M112.438937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T.P., Bradley A., McMahon A.P., and Jones S.. 1999. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 126:1211–1223. [DOI] [PubMed] [Google Scholar]
- Yan D., and Lin X.. 2007. Drosophila glypican Dally-like acts in FGF-receiving cells to modulate FGF signaling during tracheal morphogenesis. Dev. Biol. 312:203–216. 10.1016/j.ydbio.2007.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Wu Y., Yang Y., Belenkaya T.Y., Tang X., and Lin X.. 2010. The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development. 137:2033–2044. 10.1242/dev.045740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Topol L., Lee H., and Wu J.. 2003. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development. 130:1003–1015. 10.1242/dev.00324 [DOI] [PubMed] [Google Scholar]
- Zhang W., Kang J.S., Cole F., Yi M.J., and Krauss R.S.. 2006. Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev. Cell. 10:657–665. 10.1016/j.devcel.2006.04.005 [DOI] [PubMed] [Google Scholar]