How nuclear pore complexes (NPCs) become segregated during mitosis is unclear. Suresh et al. reveal that Nup2 acts as a tether between NPCs and chromatin during mitosis. This effectively links DNA and NPC segregation and ensures accurate NPC inheritance to daughter nuclei.
Abstract
Transport through nuclear pore complexes (NPCs) during interphase is facilitated by the nucleoporin Nup2 via its importin α– and Ran-binding domains. However, Aspergillus nidulans and vertebrate Nup2 also locate to chromatin during mitosis, suggestive of mitotic functions. In this study, we report that Nup2 is required for mitotic NPC inheritance in A. nidulans. Interestingly, the role of Nup2 during mitotic NPC segregation is independent of its importin α– and Ran-binding domains but relies on a central targeting domain that is necessary for localization and viability. To test whether mitotic chromatin-associated Nup2 might function to bridge NPCs with chromatin during segregation, we provided an artificial link between NPCs and chromatin via Nup133 and histone H1. Using this approach, we bypassed the requirement of Nup2 for NPC segregation. This indicates that A. nidulans cells ensure accurate mitotic NPC segregation to daughter nuclei by linking mitotic DNA and NPC segregation via the mitotic specific chromatin association of Nup2.
Introduction
Nuclear pore complexes (NPCs) are macromolecular assemblies embedded in the nuclear envelope (NE) with an overall conserved structure from yeasts to vertebrates. NPCs are assembled from multiple copies of ∼30 different proteins called nucleoporins (“Nups”) and carry out regulated, bidirectional transport between the nucleus and cytoplasm (Strambio-De-Castillia et al., 2010). During mitotic onset, NPCs disassemble to varying degrees in different organisms, and this forms a basis behind classification of mitosis (De Souza and Osmani, 2007). The budding yeast Saccharomyces cerevisiae undergoes a “closed” mitosis during which the NPCs and NE remain intact (Iouk et al., 2002). In contrast, vertebrate cells undergo an “open” form of mitosis where NPC disassembly and NE breakdown (NEBD) occur such that nuclei are not present during mitosis (Güttinger et al., 2009). During open mitosis, soluble NPC proteins are released into the cell as stable subcomplexes (Belgareh et al., 2001), whereas the insoluble transmembrane NPC proteins are absorbed into the ER membrane (Yang et al., 1997; Daigle et al., 2001). Interestingly, some soluble NPC proteins locate to mitotic structures and play mitotic roles (Güttinger et al., 2009). Between the closed and open forms of mitosis, the model filamentous fungus Aspergillus nidulans undergoes a “semi-open” mitosis involving partial NPC disassembly from an intact NE, which results in nuclear–cytoplasmic mixing (De Souza et al., 2004). Partial NPC disassembly entails dispersal of 14 peripheral Nups by a process similar to the initial steps of NPC disassembly in vertebrates that involves mitotic NPC protein phosphorylation (De Souza et al., 2004; Osmani et al., 2006a; Laurell et al., 2011). The mitotic NIMA kinase, first identified in A. nidulans (Osmani et al., 1988) and later termed the Nek kinase family in human cells (Schultz et al., 1994), triggers NPC disassembly by phosphorylating Nup98 in A. nidulans and vertebrates (De Souza et al., 2004; Laurell et al., 2011). Although this is followed by dispersal of core scaffolding Nups in vertebrates, the 12 core Nups continue to remain at the NE during A. nidulans mitosis (Osmani et al., 2006a).
Another feature that distinguishes different modes of mitoses is the behavior of the nucleolus. During open mitoses, the nucleolus undergoes complete disassembly (Leung et al., 2004). In the closed mitosis of S. cerevisiae and Schizosaccharomyces pombe, the nucleolus does not disassemble and segregates with DNA (Granot and Snyder, 1991). However, in A. nidulans and the fission yeast Schizosaccharomyces japonicus, the nucleolus remains intact during early mitosis but disassembles during anaphase after the segregation of ribosomal DNA, which act as the nucleolar organizing regions (Ukil et al., 2009; Yam et al., 2011). During mitotic exit in A. nidulans, the NE undergoes fission events on either side of the nucleolus to generate a transient structure containing nucleolar proteins between the forming daughter nuclei (Ukil et al., 2009). Nucleolar disassembly then occurs in the cytoplasm, released nucleolar proteins are reimported into transport-competent G1 nuclei, and new nucleoli re-form (Ukil et al., 2009). In the fission yeast S. japonicus, NE tearing occurs during anaphase, though no NPC disassembly occurs (Aoki et al., 2011).
Postmitotic NPC reassembly occurs in a stepwise manner in vertebrates to reinstate nuclear transport (Dultz et al., 2008). This process is initiated on the chromatin surface in anaphase and is regulated by RanGTP that releases NPC proteins from the inhibitory action of importin β during mitotic exit to allow NPC reformation (Harel et al., 2003a; Walther et al., 2003b). Members of the Nup107-160 subcomplex associate with anaphase chromatin and form a “prepore” complex together with the nuclear basket components Nup153 and Nup50 to seed reassembly (Harel et al., 2003b; Walther et al., 2003a; Dultz et al., 2008). Interaction of the soluble Nups with the transmembrane NPC proteins results in the recruitment of the membrane-associated NPC components for completion of the reassembly process (Mansfeld et al., 2006; Rasala et al., 2008). NPC assembly analyses by ectopic chromatin targeting of proteins belonging to different NPC subcomplexes further supports a seeded, hierarchical view for reassembly, with Nup133 and Nup153 providing the initial “seeds” (Schwartz et al., 2015). In A. nidulans, the dispersed NPC components reassemble onto the existing prepore structure on the NE that is constituted by the Nup84-120 subcomplex analogous to the vertebrate Nup107-160 subcomplex (Osmani et al., 2006a). Therefore, A. nidulans and vertebrates share similarities in the initial steps of mitotic NPC disassembly and reassembly.
Although we have gained insight into the NPC reassembly process, how cells inherit equal NPC numbers after mitosis remains poorly understood in A. nidulans and vertebrates. In the budding yeast S. cerevisiae undergoing closed mitosis, NPCs have been shown to migrate from the mother into the bud cell (Khmelinskii et al., 2010). Studies have also revealed the involvement of the Nsp1p subcomplex in the delivery of NPCs from the mother to the daughter cell (Makio et al., 2013) and the role of a cytoplasmic pool of Nsp1p in NPC inheritance (Colombi et al., 2013). In the fission yeast S. japonicus, NPC segregation is facilitated by the chromatin-associated inner nuclear membrane (INM) protein Man1, leading to poleward NPC movement with chromatin during anaphase and telophase (Yam et al., 2013).
Nup2 is a NPC nuclear basket protein that assists in accelerating the rate of nuclear protein transport and recycling importin α to the cytoplasm in both yeast and human cells (Booth et al., 1999; Guan et al., 2000; Hood et al., 2000; Gilchrist et al., 2002; Lindsay et al., 2002; Matsuura et al., 2003; Matsuura and Stewart, 2005; Makise et al., 2012; Finn et al., 2013). Nup2 also interacts with active genes in the nucleoplasm and participates in gene regulation (Ishii et al., 2002; Dilworth et al., 2005; Schmid et al., 2006; Kalverda et al., 2010; Buchwalter et al., 2014). During semi-open mitosis in A. nidulans and open mitosis of vertebrates, Nup2 and its vertebrate orthologue Nup50 transition to chromatin during the initiation of mitosis (Osmani et al., 2006a; Dultz et al., 2008; Ohta et al., 2010; Markossian et al., 2015). However, the functional significance of their mitotic chromatin association is currently unknown. In this study, we investigated the significance of the chromatin association of Nup2 in A. nidulans and discovered the role of Nup2 in NPC segregation. Interestingly, the newly identified functions of Nup2 in NPC segregation seem to be independent of its well-studied nuclear transport roles but can be bypassed by providing a tether between NPCs and chromatin. We therefore present the first evidence of the involvement of the conserved NPC protein Nup2 in mitotic NPC segregation to daughter nuclei.
Results
Nup2 is required for the exclusive nuclear localization of NPC proteins
Similar to its vertebrate counterpart, Nup2 is essential for viability in A. nidulans, and we analyzed the phenotypes of the null mutant using the heterokaryon rescue technique (Osmani et al., 2006b). A. nidulans is a coenocytic filamentous fungus forming cells containing multiple nuclei in a common cytoplasm. The heterokaryon rescue technique uses the ability of A. nidulans to form balanced heterokaryons in which cells contain two genetically distinct types of nuclei in a shared cytoplasm. Such balanced heterokaryons form spontaneously after deletion of essential genes when the target gene is replaced by a nutritional marker gene. Under selection for the nutritional marker, nuclei that carry the deleted allele provide the nutritional marker function, whereas other, nondeleted nuclei provide the essential gene function. When asexual spores are formed from heterokaryons, the heterokaryotic state is broken because all spores contain a single nucleus. Growth of the uninucleated spores from heterokaryons on selective media restricts the growth of WT spores that lack the nutritional marker function but permits the growth of the null mutant to reach its terminal phenotype. Thus, the heterokaryon rescue technique enables analysis of null phenotypes of essential genes (Osmani et al., 2006b). Because the generation of asexual spores from the heterokaryon results in the generation of a mixture of WT and null mutant cells, single-cell but not population-based analyses must be used to study null phenotypes.
To test whether Nup2 is potentially inherited from heterokaryons into Nup2 null (Δnup2) spores, we deleted the nup2-GFP gene from a strain expressing GFP-tagged Nup2. This caused the generation of heterokaryons containing parental nup2-GFP nuclei and deleted Δnup2-GFP nuclei. If Nup2-GFP generated within the parent heterokaryon were inherited into the Δnup2-GFP spores, all spores generated from the heterokaryotic colony would contain Nup2-GFP. However, spores grown from the heterokaryon either contained Nup2-GFP (WT) or no GFP fluorescence (Δnup2-GFP; Fig. S1 A). This result demonstrates that although the nup2-GFP gene is expressed within the parent heterokaryon, very little of that protein gets into the spores containing Δnup2-GFP nuclei. In nonselective media, nup2-GFP spores germinated and grew normally, whereas the Δnup2-GFP spores failed to express the Nup2-GFP protein and revealed the expected null phenotypes (Fig. S1 B). Measurement of GFP intensity in spores from the heterokaryon revealed a signal reduction of more than 10-fold in Δnup2-GFP spores compared with nup2-GFP spores (Fig. S1 C). The data indicate minimal inheritance/expression of Nup2 in spores from the heterokaryon that carried the null allele, explaining the high penetrance of the phenotypes caused by nup2 deletion using this methodology.
Nup2 has been thoroughly studied regarding its interphase roles in facilitating nuclear transport in yeast and vertebrates (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000; Gilchrist et al., 2002; Lindsay et al., 2002; Matsuura et al., 2003; Matsuura and Stewart, 2005). However, A. nidulans Nup2 is dispensable for normal postmitotic import rates of NLS-dsRed, a nuclear transport marker (Toews et al., 2004; Markossian et al., 2015) that contains a monopartite variant of the classical, basically charged NLS near its C terminus. Although it is dispensable for NLS-dsRed import, Nup2 is required for the normal postmitotic nuclear import of the NPC basket–associated protein Mad1 (Markossian et al., 2015). We therefore investigated the behavior of core nucleoporins in the absence of Nup2. Surprisingly, core NPC proteins of different subcomplexes—namely, Nup84-GFP (Fig. 1 A), Nup170-GFP (Fig. 1 B), and the transmembrane Nup Ndc1-GFP (Fig. 1 C)—all exhibited marked defects in their normal exclusive NE location in cells without Nup2. In WT cells, the core Nups were present exclusively around the nuclear periphery (nuclei defined by accumulated NLS-dsRed) with little to no cytoplasmic signal. However, without Nup2, these core NPC proteins all exhibited prominent cytoplasmic accumulation of foci even though their nuclei still imported NLS-dsRed. The same was true for the peripheral NPC component Nup49-GFP (Fig. 1 D), an FG repeat Nup that occupies the central NPC channel. In many Nup2 null cells, the location of nuclei was difficult to determine by looking solely at the location of the tagged NPC proteins, even though the NLS-dsRed signal confirmed that transport-competent nuclei were present.
Figure 1.
Nup2 is required for the exclusive nuclear localization of NPC proteins. (A) Images of the core NPC protein Nup84-GFP in relation to NLS-dsRed in WT cells (SGS325) and Δnup2 cells (SGS330-H) during interphase. (B) Images of the core NPC protein Nup170-GFP in relation to NLS-dsRed in WT cells (SGS347) and Δnup2 cells (SGS351-H) during interphase. (C) Images of the transmembrane NPC protein Ndc1-GFP in relation to NLS-dsRed in WT cells (SGS284) and Δnup2 cells (SGS299-H) during interphase. (D) Images of the peripheral NPC protein Nup49-GFP in relation to NLS-dsRed in WT cells (SM112) and Δnup2 cells (SM118-H) during interphase. (E) Quantitation of the mean cytoplasmic intensity of Ndc1-GFP during G1 in WT versus Δnup2 cells (SGS317 and SGS320-H). ****, P < 0.0001. Error bars represent ± SD. DIC, differential interference contrast H, heterokaryotic strains. Bars, ∼5 µm.
Nup2 is required for NPC inheritance to the NE of daughter nuclei during mitosis
The mislocalization of NPC proteins to the cytoplasm without Nup2 could be a result of defects in the interphase de novo assembly pathway or defects in NPC segregation during mitosis. To address these possibilities, we followed the dynamics of NPC proteins during mitosis. The core NPC proteins Nup84 and Nup170 remain at the NE throughout mitosis, as shown by live-cell imaging with no signal apparent in the cytoplasm (Fig. S2, A and C). However, without Nup2 function, in addition to locating at nuclei, both Nup84 and Nup170 appeared within the cytoplasm in G1 immediately after completion of mitosis (Fig. S2, B and D). Similar defects in postmitotic distribution were observed for Ndc1, a core Nup containing five transmembrane domains (Fig. S2, E and F). By imaging Ndc1-GFP and Nup170-RFP in the same strain, we determined that both NPC proteins colocalized throughout cells lacking nup2 (Fig. S2 G). Measurement of the cytoplasmic signal of Ndc1-GFP in G1 revealed a 2.5-fold increase in cytoplasmic Ndc1-GFP in Nup2 null mutants compared with WT cells (Fig. 1 E). The analysis indicates that NPCs become mislocalized during mitosis but does not exclude the possibility of further de novo synthesis defects.
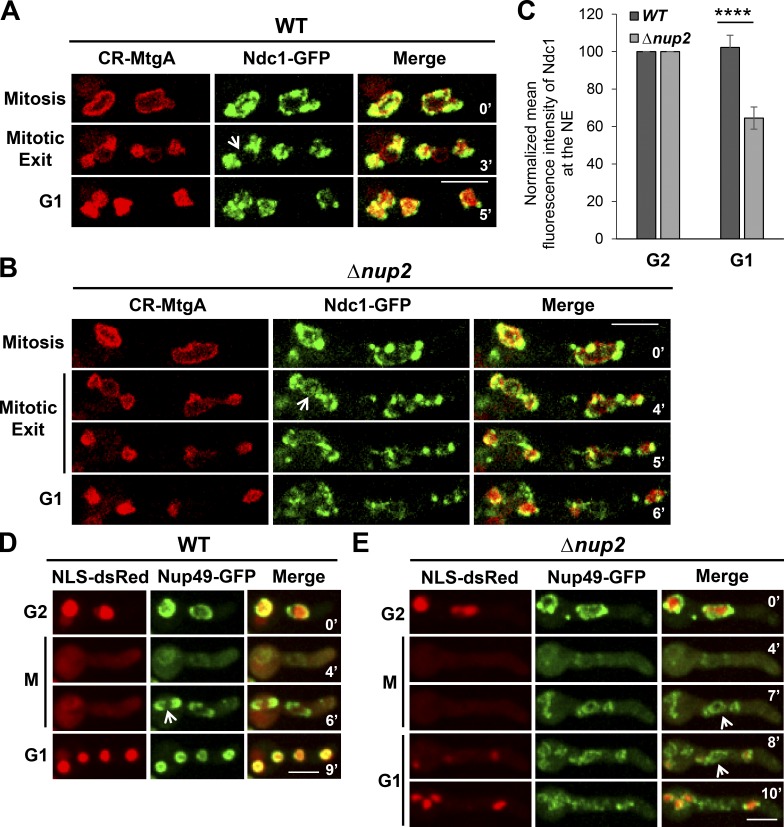
To determine when NPC proteins become uncoupled from the NE during mitosis, the behavior of Ndc1-GFP in relation to the INM marker CR-MtgA, an orthologue of S. pombe Bqt4 (Chemudupati et al., 2016), was tracked by live-cell imaging. During A. nidulans mitosis, the NE undergoes double restriction and fissions to generate two daughter nuclei. In the process, the nucleolus is separated into the cytoplasm surrounded by some NE membrane (Ukil et al., 2009; see Fig. 8). Importantly, NPCs are selectively segregated to the NE around chromatin in daughter nuclei and are excluded from the NE around the nucleolus (Fig. 2 A, arrow) via an unknown mechanism. Surprisingly, without Nup2, Ndc1-GFP occurred around all three NE compartments, failing to be excluded from the NE around the nucleolus (Fig. 2 B, arrow). Upon completion of mitosis, daughter nuclei carried the full complement of Ndc1-GFP in WT cells with no apparent Ndc1-GFP away from the nuclei (Fig. 2 A). However, in Nup2-deleted cells, the mislocated Ndc1-GFP was not incorporated into daughter nuclei but was instead misplaced into the cytoplasm (Fig. 2 B). In contrast, the INM protein CR-MtgA was partitioned exclusively around daughter nuclei with or without Nup2 (Fig. 2, A and B). Analysis of the NE-associated Ndc1-GFP signal during G2 and then G1 showed a ∼30% reduction in the signal corresponding to loss of Ndc1-GFP from the NE (Fig. 2 C). These results indicate that Nup2 is required for the normal mitotic segregation of NPC proteins to daughter nuclei via a mechanism involving attracting NPCs to the NE of daughter nuclei and away from the transient NE around the mitotic nucleolus.
Figure 8.
Model for the mitotic chromatin-associated functions of Nup2. Nup2 (red) coats chromatin (gray) during mitotic exit and serves as a chromatin-based attractant to help partition NPCs to daughter nuclei and exclude NPCs from around the nucleolar compartment (purple). Without Nup2, the chromatin-based cue for NPC partitioning is lost and NPCs remain at nuclear and nucleolar regions (arrow) during mitotic exit. After membrane fission to generate daughter nuclei, the nonnuclear NPCs are lost from the NE and locate to the cytoplasm in G1 (arrow) in the absence of Nup2 function.
Figure 2.
Nup2 mediates accurate mitotic segregation of NPC proteins to daughter nuclei. (A) Live-cell imaging of the INM protein CR-MtgA and the transmembrane NPC protein Ndc1-GFP in WT (SGS317) and (B) Δnup2 cells (SGS320-H) during mitosis, mitotic exit, and G1. Arrows indicate the predicted location of the nucleolus. The images have been overexposed to show the signal around the nucleolus. (C) Quantitation of the mean intensity of Ndc1-GFP at the NE defined by the INM protein CR-MtgA at G2 and then G1 from time-lapse imaging (SGS317 and SGS320-H). ****, P < 0.0001. Error bars represent ± SD. (D) Live-cell imaging of the peripheral NPC protein Nup49-GFP in relation to NLS-dsRed in WT cells (SM112) and Δnup2 cells (SM118-H; E) during G2-M-G1 transitions. Arrows indicate the predicted location of the nucleolus. Bars, ∼5 µm.
Because A. nidulans undergoes partial mitotic NPC disassembly accompanied by the release of nearly half of the NPC proteins from the mitotic NE, we next determined the fate of a peripheral NPC protein that disperses from the core NPC structure during prophase and returns to NPCs during mitotic exit (De Souza et al., 2004). This transition can be seen for Nup49-GFP in mitotic WT cells (Fig. 2 D). Although Nup49-GFP returned exclusively to daughter nuclei during mitotic exit in WT cells, it returned to both nuclear and nonnuclear locations in the absence of Nup2 (Fig. 2, D and E). The results indicate that core NPC proteins seed the reassembly of peripheral NPC proteins at daughter nuclei during mitotic exit in WT cells but also at ectopic nonnuclear locations without Nup2.
The functions of Nup2 in NPC segregation are independent of its conserved N- and C-terminal domains
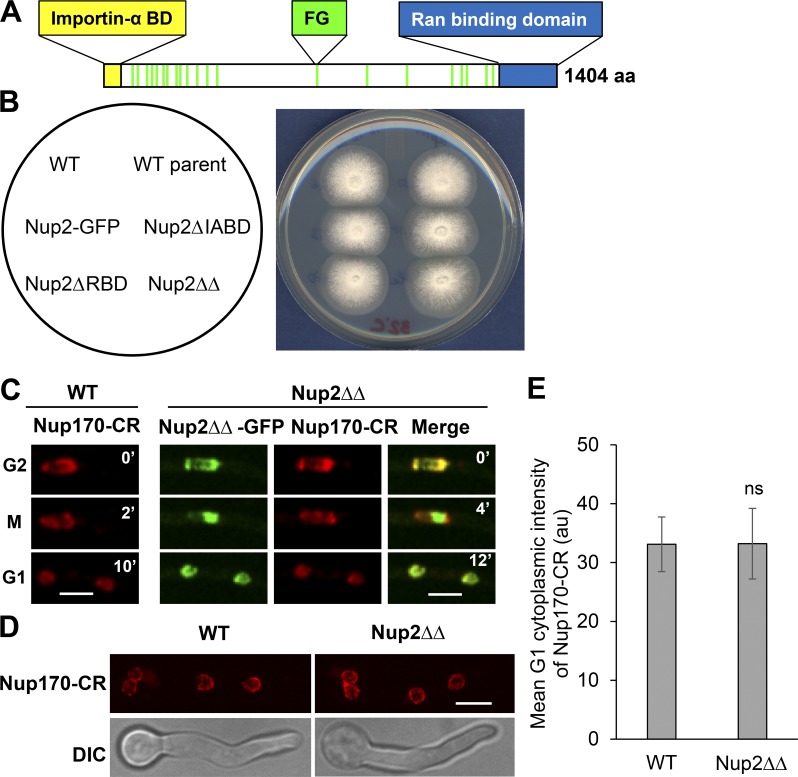
We then aimed to determine whether the roles of Nup2 in NPC segregation could be mediated by its well-known nuclear transport functions in interphase. Nup2 has conserved N- and C-terminal domains that interact with importin α and RanGTP (importin α–binding domain [IABD] and Ran-binding domain [RBD]; Fig. 3 A). These conserved domains help mediate the nuclear transport functions of Nup2 in yeast and vertebrates (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000; Lindsay et al., 2002; Matsuura et al., 2003; Matsuura and Stewart, 2005). Surprisingly, although they are known to facilitate nuclear transport (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000; Lindsay et al., 2002; Matsuura et al., 2003; Matsuura and Stewart, 2005), IABD and RBD were found to be dispensable for the essential functions of Nup2 in A. nidulans. The colonies of cells with single or double deletions (Nup2ΔΔ) of the domains grew like WT cells (Fig. 3 B), and the truncated versions exhibited unaltered interphase locations to the nuclear periphery and mitotic chromatin (Fig. S3, A–D). Because the lack of Nup2 functions leads to defects in mitotic partitioning of NPCs, the localization of Nup170-CR was investigated in Nup2ΔΔ-GFP cells during G2-M-G1 transitions (Fig. 3 C) and in asynchronous interphase cells (Fig. 3 D) and was found to be unaffected. Measurement of the mean cytoplasmic Nup170-CR signal in G1 revealed no difference between WT and Nup2ΔΔ cells (Fig. 3 E), indicating that NPC segregation proceeds normally in Nup2ΔΔ cells. Considering these factors, the analysis reveals that the essential functions of Nup2 and its roles in NPC segregation are independent of its conserved importin α and RanGTP interacting domains, which are known to facilitate interphase nuclear transport.
Figure 3.
The functions of Nup2 in NPC segregation are independent of its conserved N- and C-terminal domains. (A) Domain structure of Nup2 indicating IABD and RBD and the FG repeats. (B) Spot inoculum of cells in which endogenous Nup2 is replaced with versions of Nup2 lacking its IABD (SGS117) or RBD (SGS80) or both (ΔΔ, SGS147) displays comparable colony growth to WT cells (CDS746 and SO451) or Nup2-GFP cells (SO593) at 32°C. WT parent represents the strain in which the indicated modifications were performed. (C) Live-cell imaging of Nup170-CR in WT (CDS977) versus Nup2ΔΔ-GFP cells (SGS362) during G2-M-G1 transitions and (D) in interphase cells. (E) Quantitation of the mean cytoplasmic intensity of Nup170-CR during G1 in WT versus Nup2ΔΔ-GFP cells (CDS977 and SGS362). Error bars represent ± SD. Bars, ∼5 µm.
A central targeting domain within Nup2 is essential for localization, viability, and NPC segregation
We then identified domains of Nup2 required for its cell cycle–regulated locations and roles in NPC segregation. A study was completed in which regions of Nup2 tagged with GFP were expressed (Fig. S4 A) and their locations during the cell cycle followed (Fig. S4 B). This analysis revealed that a central region (domain 3, aa 401–1,200) of Nup2 tagged with GFP locates during interphase and mitosis like full-length Nup2-GFP. Comparing the locations of partially overlapping N- and C-terminal regions (domain 1, aa 1–1,000; and domain 2, aa 803–1,404) suggested that the domain responsible resides within a region between aa 401 and 802 (Fig. 4 A). To test this, the two halves of domain 3 were expressed (domain 4, aa 401–802; and domain 5, aa 803–1200) to reveal that domain 4 tagged with GFP locates during interphase and mitosis in the same manner as full-length Nup2-GFP (Fig. 4 B and Fig. S4 B). To test if domain 4 was being targeted via interactions with endogenous Nup2, the construct was expressed in cells in which Nup2 was deleted. In the absence of Nup2, domain 4–GFP locations during interphase and mitosis remained unchanged (Fig. 4 B). To determine if domain 4 is necessary for the targeting of Nup2, a construct lacking D4 (Nup2ΔD4-GFP) was used to replace endogenous Nup2. Nup2ΔD4-GFP did not locate like Nup2-GFP, was dispersed throughout cells, and failed to locate to mitotic chromatin during mitosis (Fig. 4 C and Fig. S4 C). This allele was also found to phenocopy a Nup2 null mutant, compromising further growth past the small germling stage (Fig. 4 D). We conclude that domain 4 of Nup2 is necessary and sufficient for interphase NPC and mitotic chromatin targeting.
Figure 4.
A central domain within Nup2 is required for Nup2 localization, cell growth, and mitotic NPC segregation. (A) Image depicting the domain 4 region of Nup2 that was GFP tagged and expressed under the alcA inducible promoter (Waring et al., 1989). (B) Domain 4 was imaged during mitosis in the presence (SM47) or absence of endogenous Nup2 (SM76-H). Bar, ∼5 μm. (C) Images of interphase cells expressing Nup2-GFP (SO593) or Nup2ΔD4-GFP (SM73-H). (D) Brightfield images of cells from spores of a heterokaryon containing either the WT Nup2 allele or the nup2ΔD4 allele grown for 3 d at 22°C on nonselective and selective media, respectively. Bar, ∼20 μm. (E) Images of Ndc1-GFP in relation to the INM protein CR-MtgA during interphase in WT (SGS317), nup2ΔD4 (SGS372-H), and ΔnupA cells (SGS322-H). Bar, ∼5 μm. (F) Quantitation of the mean cytoplasmic intensity of Ndc1-GFP during G1 in WT (SGS317), nup2ΔD4 (SGS372-H), and ΔnupA cells (SGS322-H). Error bars represent ± SD. H, heterokaryotic strains.
Because Nup2ΔD4-GFP is unable to localize normally during the cell cycle, we determined whether cells expressing Nup2ΔD4 as the only version of Nup2 would be impaired in NPC segregation. NPC segregation was found to be defective in Nup2ΔD4 cells, similar to Δnup2 cells (Fig. 4 E), and the mean cytoplasmic intensity of Ndc1-GFP in G1 was significantly higher than that of WT cells (Fig. 4 F). Together, the domain study indicates that a central targeting region is important for the NPC segregation functions of Nup2.
Previously, a protein named NupA was identified as being necessary for the localization of Nup2 to interphase NPCs and mitotic chromatin (Markossian et al., 2015). We therefore examined NPC segregation in NupA null mutants and found that it phenocopied nup2 deletion (Fig. 4, E and F).
Artificial bridging of NPCs to chromatin bypasses the need for Nup2 in mitotic NPC segregation and in the maintenance of normal nuclear structure
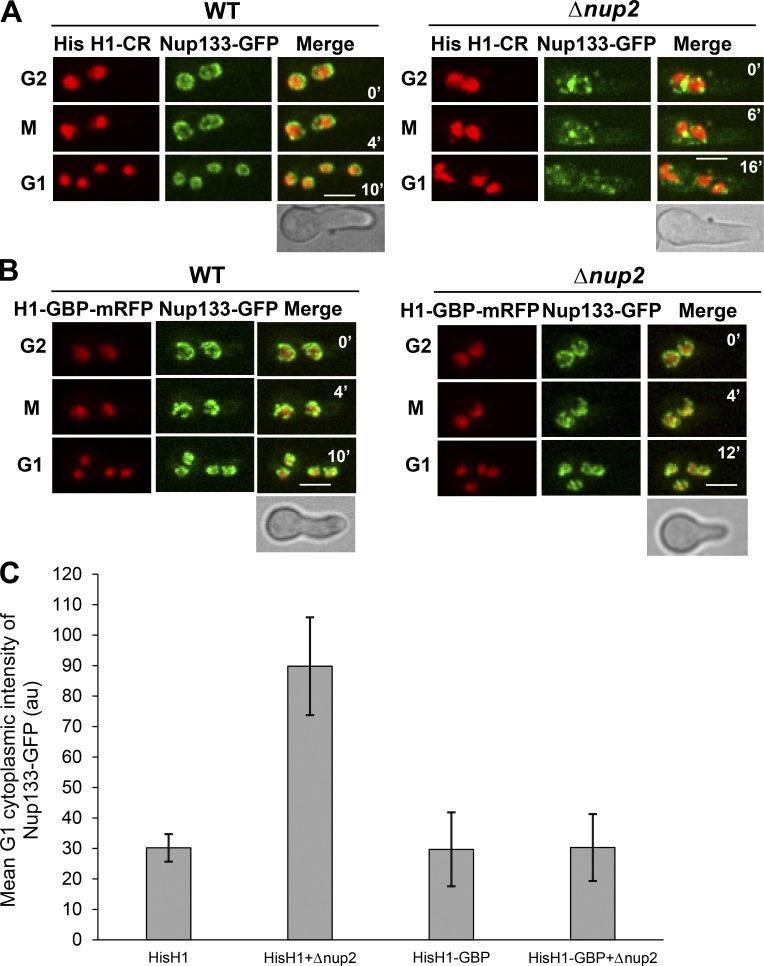
Because Nup2 moves from throughout chromatin to the chromatin periphery during anaphase (Markossian et al., 2015), we considered the possibility that it could mediate mitotic NPC segregation by acting as a linker between NPCs and chromatin. To test this, we provided an artificial link between NPCs and chromatin and examined whether it could bypass the need for Nup2 during NPC segregation. To provide a chromatin-based NPC linker, we used histone H1-mRFP tagged with the GFP-binding protein (GBP), a single-chain anti-GFP antibody that binds GFP with high affinity (Rothbauer et al., 2006, 2008). For the NPC GFP-tagged part of the linker, we chose the core NPC protein Nup133. Nup133 was chosen for this experiment because, unlike many other Nups, Nup133 can recruit other Nup subcomplexes when ectopically targeted to chromatin (Schwartz et al., 2015) and would be expected to be able to drive segregation of other NPC proteins along with it. Expression of Nup133-GFP with histone H1-GBP-mRFP from their native loci caused no adverse effects on cell growth (not depicted), and neither tagged protein displayed atypical locations (Fig. 5, compare WTs in A and B). Most notably, the artificial NPC–chromatin tether prevented the mislocalization of Nup133-GFP caused by the lack of Nup2. In Nup2 null cells lacking the NPC–chromatin tether, mislocalization of Nup133-GFP was apparent, with the protein locating not only around misshapen nuclei but also at foci within the cytoplasm (Fig. S5 A). In contrast, in the NPC–chromatin bridged cells, Nup133-GFP occurred only around the nuclear periphery, with no visible cytoplasmic signal (Fig. S5 B).
Figure 5.
Artificial bridging of Nup133 to histone H1 bypasses the requirement of Nup2 for its mitotic segregation. (A) Live-cell imaging of the core NPC protein Nup133-GFP in relation to histone H1-CR in WT cells (SGS392) and (B) Δnup2 cells (SGS393-H) during G2-M-G1 transitions. (B) Live-cell imaging of the core NPC protein Nup133-GFP in cells expressing histone H1-GBP-mRFP during G2-M-G1 transitions in WT (SGS386) and Δnup2 cells (SGS395-H). (C) Quantitation of the mean cytoplasmic intensity of Nup133-GFP during G1 in the backgrounds of A and B. Error bars represent ± SD. H, heterokaryotic strains. Bars, ∼5 µm.
The mislocalization of NPCs in the absence of Nup2 during mitosis was then tracked using live-cell imaging to determine if the NPC–chromatin tether remediated the mitotic NPC segregation defects (Fig. 5). Live-cell imaging of mitosis in Nup2 deleted cells confirmed that the NPC–chromatin tether corrected the mitotic NPC segregation defect caused by Δnup2 (Fig. 5, A and B), as further confirmed by quantitative analysis of the cytoplasmic levels of Nup133-GFP in G1 (Fig. 5 C).
To determine whether the location of other NPC proteins is corrected in the NPC–chromatin bridged cells without Nup2, we followed the peripheral NPC protein Nup49. During interphase, Nup49-CR located to cytoplasmic foci in the absence of Nup2 (Fig. 1 D, Fig. 2 E, and Fig. 6, A and B). This mislocalization of Nup49 was corrected in the NPC–chromatin bridged cells expressing histone H1-GBP-mRFP with no visible cytoplasmic red fluorescence signal without Nup2 (Fig. 6, A and B). After costaining with DAPI to visualize nuclei, quantitative analysis of cytoplasmic levels of Nup49-CR revealed a significantly higher cytoplasmic level of Nup49-CR when Nup2 was deleted in cells without the linker when compared with those expressing the linker (Fig. 6 C).
Figure 6.
Artificial bridging of NPCs to chromatin bypasses the need for Nup2 in NPC segregation. (A) Images of interphase Δnup2 cells expressing Nup133-GFP and Nup49-CR with (SO1726-1H) or without (SO1727-2H) the chromatin–GFP tether. Histone H1-GBP-mRFP in representative cells at the two- and four-nucleus cell stages (left and right panels, respectively). (B) Similar cells as in A but stained with DAPI to show the position of nuclei. (C) Quantitation of the mean cytoplasmic red fluorescence intensity representing Nup49-CR of cells with (SO1726-1H) or without (SO1727-2H) the NPC-H1 bridge provided by expression of histone H1-GBP-mRFP. Error bars represent ± SD. Asterisks indicate cells with dual red tags, Nup49-CR, and histone H1-GBP-mRFP. Bars, ∼5 µm.
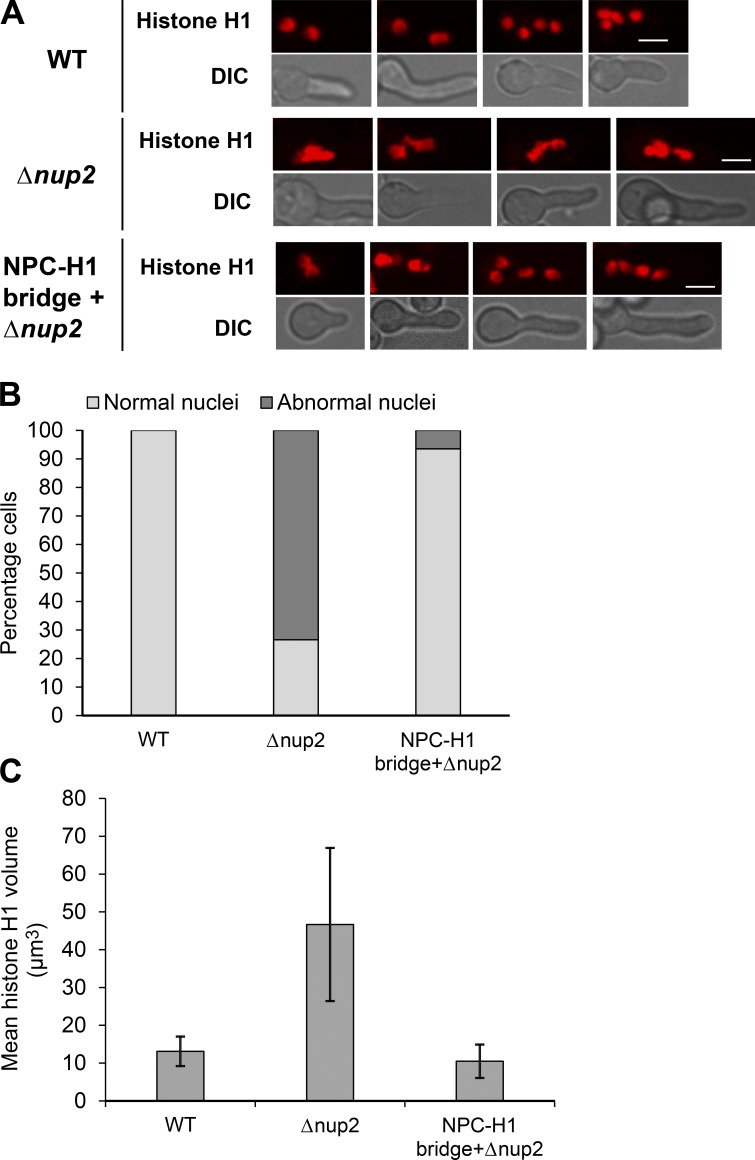
We then determined whether fixing NPC segregation corrected other defects caused by lack of Nup2. Deletion of Nup2 is known to compromise nuclear structure (Osmani et al., 2006a), and many cells without Nup2 function display fused nuclei with their nuclear chromatin appearing in unequal masses (see Δnup2 in Fig. 7 A and Δnup2 in Fig. S1 B, bottom). These nuclear configurations differ from those of WT cells, which have uniformly sized and more evenly distributed nuclei along their length (see WT in Fig. 7 A and WT in Fig. S1 B, bottom). The nuclear size, nuclear shape, and distribution defects caused by lack of Nup2 were largely rescued by the NPC–chromatin tether (Fig. 7 A). Quantitation of the number of cells with typical and atypical nuclear morphology (Fig. 7 B) and nuclear volumes (Fig. 7 C) revealed high penetrance of the rescue. The results indicate that supplying a tether between NPCs and chromatin rescues NPC missegregation and the abnormal nuclear structure of Nup2-deleted cells. The results are consistent with the proposal that one function of Nup2 is to act as a bridge between NPCs and chromatin to ensure mitotic NPC inheritance by daughter nuclei. However, although the NPC–H1 tether rescued the NPC segregation and nuclear structural defects caused by Δnup2, it was insufficient to rescue the lethal effects of Nup2 deletion (unpublished data). This indicates that Nup2 performs other functions, in addition to mitotic NPC segregation, that are essential for viability.
Figure 7.
Artificial bridging of NPCs to chromatin rescues abnormal nuclear morphology without Nup2. (A) Representative images of histone H1 in WT (SGS392), Δnup2 cells (SGS393-H), and Δnup2 cells expressing Nup133-GFP along with histone H1-GBP-mRFP (SGS395-H). (B) Quantitation of percentage cells with abnormal nuclei in WT (SGS392), Δnup2 cells (SGS393-H), and Δnup2 cells expressing Nup133-GFP along with histone H1-GBP-mRFP (SGS395-H). (C) Quantitation of the mean volume of histone H1 in WT (SGS392), Δnup2 cells (SGS393-H), and Δnup2 cells expressing Nup133-GFP along with histone H1-GBP-mRFP (SGS395-H). Error bars represent ± SD. Bars, ∼5 µm.
Discussion
Nup2 is essential in A. nidulans (Osmani et al., 2006a) and is required for mouse embryo development (Smitherman et al., 2000). Nup2 transitions onto mitotic chromatin in A. nidulans and vertebrate cells (Osmani et al., 2006a; Dultz et al., 2008; Markossian et al., 2015). In this work, we attempted to understand the potential mitotic functions of Nup2 and uncovered a mitotic exit function involving NPC partitioning to daughter nuclei. We propose that the mitotic location of Nup2 around chromatin during anaphase and telophase provides a bridge between segregating DNA and NPCs to ensure NPC inheritance during mitosis to the NE surrounding daughter nuclei. Suggestive of important nontransport roles for Nup2, its essential functions do not require its conserved N- and C-terminal domains that are involved with nuclear transport functions in yeast and vertebrate cells but do require its central domain. Interestingly, a recent study identified a 125-amino-acid-long, more structured region in budding yeast Nup2 sufficient for its NPC location (independent of its IABD, RBD, and FG repeat domains) that has transport-independent meiotic roles (Chu et al., 2017). Domain 4 of A. nidulans Nup2 also includes a more ordered region according to software predictions (http://iupred.enzim.hu/). It is therefore possible that a similar domain important for targeting Nup2 has also evolved in A. nidulans to perform additional functions not dependent on Nup2’s IABD, RBD, or FG repeat domains.
During A. nidulans mitosis, peripheral Nups disperse from the core NPC structures that remain embedded in the NE to provide open conduits through which proteins can pass, including tubulin, which forms the intranuclear mitotic spindle (Ovechkina et al., 2003). The core NPC structures are additionally thought to provide seeds to which dispersed Nups return during mitotic exit to reestablish regulated nuclear transport in daughter G1 nuclei. After anaphase, the A. nidulans NE forms three structures as the two daughter nuclei form and the nucleolus becomes separated from the nuclei (Fig. 2 A, mitotic exit; and Fig. 8 A, WT). During this process, the core NPC structures are preferentially segregated in the NE surrounding daughter nuclei, with none being left at the NE surrounding the nucleolus. How this preferential segregation of core NPCs is achieved was previously unknown. The current study provides support for a model in which the chromatin association of Nup2 provides a link between NPCs and chromatin during mitotic exit to ensure their mitotic segregation to daughter nuclei. In this model (Fig. 8, WT), chromatin-associated Nup2 acts as an attractant for the core NPC structure such that when the core structures encounter Nup2 at the surface of the chromatin of forming daughter nuclei, they remain there. This process effectively ensures that no NPCs are left behind in the transient NE surrounding the nucleolus and that all NPCs become located to the NE of daughter nuclei. Deletion analysis provides support for this model, as live-cell imaging shows that core NPC proteins fail to segregate exclusively to the NE of forming daughter nuclei in the absence of Nup2 function. Instead, a portion of the core structures remains within the NE around the nucleolus and then becomes misplaced outside the NE of daughter nuclei. These misplaced core structures retain the capacity to attract a dispersed peripheral Nup during mitotic exit and G1 such that a portion of the peripheral Nups also becomes misplaced to the cytoplasm. This results in the reduction of NPCs in daughter nuclei and aberrant cytoplasmic, NPC-like accumulation.
To determine if Nup2 might act as a tether between NPCs and chromatin to ensure their exclusive inheritance to daughter nuclei, we questioned whether providing cells with an alternative NPC–chromatin tether could bypass the requirement for Nup2 in NPC inheritance. We were able to successfully bypass the role of Nup2 in NPC inheritance by linking Nup133 to chromatin, and this artificial linkage additionally remediated several other nuclear architectural defects caused by lack of Nup2. However, although many mitotic nuclear defects caused by Nup2 deletion were corrected by an artificial NPC–chromatin tether, cells still failed to grow in the long term, indicating that Nup2 has additional essential functions beyond NPC inheritance that have not yet been determined.
In the fission yeast S. japonicus, which undergoes NE tearing during mitotic exit, the DNA-binding INM protein Man1 promotes equal partitioning of the nuclear membrane, nucleolus, and NPCs and may exert its function by linking NPCs to segregating chromatin (Yam et al., 2013). However, the Man1 orthologue in A. nidulans (Src1), although required for stable mitotic exit into G1, has no obvious direct impact on NPC segregation and does not cause accumulation of cytoplasmic NPC proteins when deleted (Liu et al., 2015). Interestingly, Man1 is not essential in S. japonicus, with its deletion causing a mild effect on doubling time (Yam et al., 2013). This might suggest that S. japonicus has an alternate additional pathway for NPC segregation that could potentially involve the Nup2-mediated mechanism.
The newly identified roles for A. nidulans Nup2 could be shared with the vertebrate orthologue Nup50. In vertebrates, NPC proteins distributed throughout the cytoplasm at mitosis are attracted to chromatin during mitotic exit (Walther et al., 2003b; Franz et al., 2007; Rasala et al., 2008). Two characteristics of Nup50 have been defined in vertebrate cells indicating that Nup50 could also act as an attractant at mitotic chromatin for NPC proteins. Nup50 locates on chromatin during mitosis (Dultz et al., 2008; Ohta et al., 2010), and it has additionally been shown to be able to recruit nucleoporins to ectopic sites (Schwartz et al., 2015). Mitotic chromatin-associated Nup50 might therefore define NPC reassembly sites on chromatin and provide seeds on segregating chromatin to achieve equal NPC numbers in daughter nuclei. The Nup2-mediated NPC seeding process might occur in parallel to the known ELYS-mediated assembly pathway (Rasala et al., 2008) or could act before this pathway because Nup50 is chromatin associated throughout mitosis whereas ELYS associates with late-anaphase chromatin (Franz et al., 2007; Gillespie et al., 2007). Because the A. nidulans orthologue of ELYS is far smaller, does not possess the AT hook motif that binds DNA, and is not required for NPC inheritance during mitosis (Liu et al., 2009), a Nup2-centric process on chromatin appears to be the critical pathway for NPC inheritance in A. nidulans.
Materials and methods
General techniques
Classical genetics techniques, media preparation, cultures, and transformation of A. nidulans were used as described previously (Pontecorvo et al., 1953; Osmani et al., 2006b; Markossian et al., 2015). The heterokaryon rescue technique was used to study the gene deletions of Nup2 and NupA as described previously (Osmani et al., 2006b). To assess phenotypes of Δnup2, ΔnupA, and nup2ΔD4 cells, spores from heterokaryons carrying the respective null alleles were grown on selective media to enable growth of the mutant but not the WT alleles. Strains used in the study are listed in Table S1. Strains CDS746 and CDS977 were obtained from C.P. De Souza, strain MC38 from M. Chemudupati, and strain HA424 from H.-L. Liu (all at The Ohio State University, Columbus, OH).
Imaging
Growth of cells for live-cell imaging was performed as described previously (Markossian et al., 2015). Images are all represented as maximum-intensity projections of multiple imaging planes, and no binning was used during image acquisition unless otherwise mentioned. Images were obtained using 60× or 100× objectives on two spinning disk confocal systems equipped with cameras with different resolutions. Spinning disk confocal system 1 (UltraVIEW Vox CSUX1 system; PerkinElmer) with a 60×/1.4 NA objective was used for most of the time-lapse imaging (Fig. 2, D and E; Fig. 3 C; Fig. 5, A and B; Fig. S2, A–F; Fig. S3; and Fig. S4 C) and for a few single still images (Fig. 7 A and Fig. S1, A and B). The microscope is equipped with 440-, 488-, 515-, and 561-nm solid-state lasers and a back-thinned electron multiplying charge-coupled device camera (Hamamatsu Photonics) on a Ti-E inverted microscope (Nikon). The distance between the imaging planes in a projected image is 0.8 μm. Using this setup, a resolution of 4.22 pixels/μm is obtained. Spinning disk confocal system 2 (UltraVIEW ERS; PerkinElmer) was used for all the other imaging. The system is equipped with a cooled, charge-coupled device camera (ORCA-AG; Hamamatsu Photonics) on an Eclipse (TE2000-U; Nikon). For most of the single still images (Fig. 1, Fig. 3 D, Fig. 4 E, Fig. 6 A, and Fig. S5), we used a 100×/1.4 NA objective. Using this setup, we obtained a resolution of 14.4 pixels/μm. The images in Fig. 6 B were captured with the same setup, but with 2 × 2 binning giving a resolution of 7.2 pixels/μm. For all of these images, the distance between the imaging planes is 0.4 μm. The images in Fig. 2 (A and B) were obtained using system 2 with a 60×/1.4 NA objective that provides a resolution of 8.7 pixels/μm. The images in Fig. 4 (B and C), Fig. S2 G, and Fig. S4 were obtained using system 2 with a 60×/1.4 NA objective, but with 2 × 2 binning giving a resolution of 4.4 pixels/µm.
Quantitation analysis
Image analysis was performed using the following software: (a) Volocity (PerkinElmer); (b) Ultraview; and (c) ImageJ (National Institutes of Health). Statistical analysis and graphs were created using Microsoft Excel. To test for statistical significance, a paired, two-tailed t test was used. Data points represent the mean ± SD.
For the analysis of maternal inheritance of Nup2, spores from a Nup2-GFP + Δnup2-GFP heterokaryon also expressing NLS-dsRed were cultured for 5 h in rich media along with GFP-negative cells expressing CR-MtgA (MC38) to account for the autofluorescence seen in spores. Whole-cell GFP intensity measurements were made by drawing regions of interest (ROIs) along the cell outline seen by brightfield imaging and measuring the mean values. The mean values measured in the GFP-negative cells were regarded as autofluorescence, subtracted from the mean values in Nup2-GFP and Δnup2-GFP cells, and plotted.
For measurements of the G1 cytoplasmic intensity of NPC proteins, images 6 min after nuclear division followed with nuclear markers were used to measure mean intensities. The nucleus was distinguished from the cytoplasm using nuclear markers CR-MtgA and/or NLS-dsRed. ROIs were drawn in the cytoplasm, and mean values were measured using Volocity software. Background correction was introduced by subtracting the mean values in the field adjacent to the cells of interest measured using the same ROIs.
For measurement of the Ndc1 signal at the NE (Fig. 2 C), the NE was defined using fixed thresholds for the INM protein CR-MtgA, and the mean GFP signal in the defined region was measured in G2 and G1. The G2 time point was chosen as the first time point before visible accumulation of Ndc1 to mitotic spindle poles, and the G1 time point was defined as 6 min after nuclear division. Background correction was introduced by measuring mean field intensities using ROIs drawn adjacent to the cells of interest and subtracting these from the mean Ndc1 intensities. The corrected values were further normalized to G2 values and expressed as a percentage.
For DAPI staining to image nuclei (Fig. 6 B), cells were fixed to preserve protein fluorescence using formaldehyde fixative for 10 min. The fixed cells were washed with water and mounted in 300 ng DAPI/ml in 80% Citifluor (Antifadent Mountant Solutions, Electron Microscopy Sciences) for microscopy. For the quantitation of the cytoplasmic Nup49-CR signal (Fig. 6 C), nuclei were distinguished from the cytoplasm by DAPI staining. ROIs were drawn in the cytoplasm, and mean fluorescence values were measured. Background correction was introduced by placing the same ROIs in the field adjacent to the cells of interest, obtaining a mean background value that was subtracted from the aforementioned fluorescence measurements and plotted.
For measurements of percentage cells with abnormal nuclei (Fig. 7), cells with more than two nuclei were used. The presence of clumped and large nuclei or the presence of nuclei of different sizes within a cell was considered abnormal. For measurement of histone H1 volumes, identical H1-RFP intensity thresholds were defined for the control and experimental samples using Volocity software, and volume measurements were obtained.
Generation of GBP clones
To generate plasmids with the GBP constructs, the coding sequence of GBP was amplified from the pc1336 plasmid obtained from H. Leonhardt (Ludwig-Maximilian University of Munich, Munich, Germany). The amplifying primer carried the sequence of the GA5 linker such that the resulting construct would carry either GA5-GBP or GA5-GBP-mRFP. The nutritional marker was also amplified and fused at the 3′ end of GBP or the GBP-mRFP cassette by fusion PCR. The PCR product was cloned into a pCR-Blunt II-TOPO vector (Invitrogen) and transformed into competent DH5α cells. Transformants were selected on Luria-Bertani plates with 50 μg/ml kanamycin. The clones were confirmed by restriction digestion followed by sequencing on both strands. GBP-mRFP was amplified from positive clones followed by fusion PCR for C-terminal tagging of histone H1.
Online supplemental material
Strains used in the study and their genotypes are given in Table S1. Fig. S1 shows that Nup2 is not inherited into spores from the heterokaryotic parent colony after nup2 deletion and heterokaryon rescue. Fig. S2 reveals that Nup2 is required for normal mitotic segregation of NPC proteins. Fig. S3 shows that Nup2 lacking its conserved IABD and RBD locates normally to the interphase nuclear periphery and mitotic chromatin. Fig. S4 represents a domain study of Nup2. Fig. S5 reveals that artificial bridging of NPCs to chromatin bypasses the need for Nup2 in NPC segregation.
Acknowledgments
We thank Colin P. De Souza, Hui-Lin Liu, and Mahesh Chemudupati for sharing strains. We are thankful to Dr. Heinrich Leonhardt for sharing the clone for GBP. We are grateful to the members of the Osmani laboratory for feedback and the undergraduate students Kimberlee Hunt and Leymaan Abdurehman for help with experiments.
This work was supported by a grant from the National Institutes of Health (GM042564) to S.A. Osmani.
The authors declare no competing financial interests.
Author contributions: S. Suresh, S. Markossian, and S.A. Osmani contributed to the experimental design and wrote the manuscript. S. Suresh, S. Markossian, A.H. Osmani, and S.A. Osmani performed experiments, analyzed data, and generated the figures.
Footnotes
Abbreviations used:
- GBP
- GFP-binding protein
- IABD
- importin α–binding domain
- INM
- inner nuclear membrane
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- RBD
- Ran-binding domain
- ROI
- region of interest
References
- Aoki K., Hayashi H., Furuya K., Sato M., Takagi T., Osumi M., Kimura A., and Niki H.. 2011. Breakage of the nuclear envelope by an extending mitotic nucleus occurs during anaphase in Schizosaccharomyces japonicus. Genes Cells. 16:911–926. 10.1111/j.1365-2443.2011.01540.x [DOI] [PubMed] [Google Scholar]
- Belgareh N., Rabut G., Baï S.W., van Overbeek M., Beaudouin J., Daigle N., Zatsepina O.V., Pasteau F., Labas V., Fromont-Racine M., et al. 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154:1147–1160. 10.1083/jcb.200101081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth J.W., Belanger K.D., Sannella M.I., and Davis L.I.. 1999. The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J. Biol. Chem. 274:32360–32367. 10.1074/jbc.274.45.32360 [DOI] [PubMed] [Google Scholar]
- Buchwalter A.L., Liang Y., and Hetzer M.W.. 2014. Nup50 is required for cell differentiation and exhibits transcription-dependent dynamics. Mol. Biol. Cell. 25:2472–2484. 10.1091/mbc.E14-04-0865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemudupati M., Osmani A.H., and Osmani S.A.. 2016. A mitotic nuclear envelope tether for Gle1 also affects nuclear and nucleolar architecture. Mol. Biol. Cell. 27:3757–3770. 10.1091/mbc.E16-07-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.B., Gromova T., Newman T.A.C., and Burgess S.M.. 2017. The nucleoporin Nup2 contains a meiotic-autonomous region that promotes the dynamic chromosome events of meiosis. Genetics. 206:1319–1337. 10.1534/genetics.116.194555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombi P., Webster B.M., Fröhlich F., and Lusk C.P.. 2013. The transmission of nuclear pore complexes to daughter cells requires a cytoplasmic pool of Nsp1. J. Cell Biol. 203:215–232. 10.1083/jcb.201305115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., and Ellenberg J.. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71–84. 10.1083/jcb.200101089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C.P., and Osmani S.A.. 2007. Mitosis, not just open or closed. Eukaryot. Cell. 6:1521–1527. 10.1128/EC.00178-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C.P., Osmani A.H., Hashmi S.B., and Osmani S.A.. 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14:1973–1984. 10.1016/j.cub.2004.10.050 [DOI] [PubMed] [Google Scholar]
- Dilworth D.J., Tackett A.J., Rogers R.S., Yi E.C., Christmas R.H., Smith J.J., Siegel A.F., Chait B.T., Wozniak R.W., and Aitchison J.D.. 2005. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell Biol. 171:955–965. 10.1083/jcb.200509061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., and Ellenberg J.. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180:857–865. 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn E.M., DeRoo E.P., Clement G.W., Rao S., Kruse S.E., Kokanovich K.M., and Belanger K.D.. 2013. A subset of FG-nucleoporins is necessary for efficient Msn5-mediated nuclear protein export. Biochim. Biophys. Acta. 1833:1096–1103. 10.1016/j.bbamcr.2012.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., and Antonin W.. 2007. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 8:165–172. 10.1038/sj.embor.7400889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist D., Mykytka B., and Rexach M.. 2002. Accelerating the rate of disassembly of karyopherin·cargo complexes. J. Biol. Chem. 277:18161–18172. 10.1074/jbc.M112306200 [DOI] [PubMed] [Google Scholar]
- Gillespie P.J., Khoudoli G.A., Stewart G., Swedlow J.R., and Blow J.J.. 2007. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr. Biol. 17:1657–1662. 10.1016/j.cub.2007.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot D., and Snyder M.. 1991. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil. Cytoskeleton. 20:47–54. 10.1002/cm.970200106 [DOI] [PubMed] [Google Scholar]
- Guan T., Kehlenbach R.H., Schirmer E.C., Kehlenbach A., Fan F., Clurman B.E., Arnheim N., and Gerace L.. 2000. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20:5619–5630. 10.1128/MCB.20.15.5619-5630.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttinger S., Laurell E., and Kutay U.. 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10:178–191. 10.1038/nrm2641 [DOI] [PubMed] [Google Scholar]
- Harel A., Chan R.C., Lachish-Zalait A., Zimmerman E., Elbaum M., and Forbes D.J.. 2003a Importin β negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell. 14:4387–4396. 10.1091/mbc.E03-05-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., and Forbes D.J.. 2003b Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 11:853–864. 10.1016/S1097-2765(03)00116-3 [DOI] [PubMed] [Google Scholar]
- Hood J.K., Casolari J.M., and Silver P.A.. 2000. Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-α export. J. Cell Sci. 113:1471–1480. [DOI] [PubMed] [Google Scholar]
- Iouk T., Kerscher O., Scott R.J., Basrai M.A., and Wozniak R.W.. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159:807–819. 10.1083/jcb.200205068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., and Laemmli U.K.. 2002. Chromatin boundaries in budding yeast: The nuclear pore connection. Cell. 109:551–562. 10.1016/S0092-8674(02)00756-0 [DOI] [PubMed] [Google Scholar]
- Kalverda B., Pickersgill H., Shloma V.V., and Fornerod M.. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 140:360–371. 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Keller P.J., Lorenz H., Schiebel E., and Knop M.. 2010. Segregation of yeast nuclear pores. Nature. 466:E1 10.1038/nature09255 [DOI] [PubMed] [Google Scholar]
- Laurell E., Beck K., Krupina K., Theerthagiri G., Bodenmiller B., Horvath P., Aebersold R., Antonin W., and Kutay U.. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 144:539–550. 10.1016/j.cell.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Leung A.K., Gerlich D., Miller G., Lyon C., Lam Y.W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., and Lamond A.I.. 2004. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166:787–800. 10.1083/jcb.200405013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay M.E., Plafker K., Smith A.E., Clurman B.E., and Macara I.G.. 2002. Npap60/Nup50 is a tri-stable switch that stimulates importin-α:β-mediated nuclear protein import. Cell. 110:349–360. 10.1016/S0092-8674(02)00836-X [DOI] [PubMed] [Google Scholar]
- Liu H.L., De Souza C.P., Osmani A.H., and Osmani S.A.. 2009. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol. Biol. Cell. 20:616–630. 10.1091/mbc.E08-06-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.L., Osmani A.H., and Osmani S.A.. 2015. The inner nuclear membrane protein Src1 is required for stable post-mitotic progression into G1 in Aspergillus nidulans. PLoS One. 10:e0132489 10.1371/journal.pone.0132489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio T., Lapeina D.L., and Wozniak R.W.. 2013. Inheritance of yeast nuclear pore complexes requires the Nsp1p subcomplex. J. Cell Biol. 203:187–196. 10.1083/jcb.201304047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makise M., Mackay D.R., Elgort S., Shankaran S.S., Adam S.A., and Ullman K.S.. 2012. The Nup153-Nup50 protein interface and its role in nuclear import. J. Biol. Chem. 287:38515–38522. 10.1074/jbc.M112.378893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J., Güttinger S., Hawryluk-Gara L.A., Panté N., Mall M., Galy V., Haselmann U., Mühlhäusser P., Wozniak R.W., Mattaj I.W., et al. 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 22:93–103. 10.1016/j.molcel.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Markossian S., Suresh S., Osmani A.H., and Osmani S.A.. 2015. Nup2 requires a highly divergent partner, NupA, to fulfill functions at nuclear pore complexes and the mitotic chromatin region. Mol. Biol. Cell. 26:605–621. 10.1091/mbc.E14-09-1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., and Stewart M.. 2005. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 24:3681–3689. 10.1038/sj.emboj.7600843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Lange A., Harreman M.T., Corbett A.H., and Stewart M.. 2003. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 22:5358–5369. 10.1093/emboj/cdg538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Bukowski-Wills J.C., Sanchez-Pulido L., Alves F.L., Wood L., Chen Z.A., Platani M., Fischer L., Hudson D.F., Ponting C.P., et al. 2010. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 142:810–821. 10.1016/j.cell.2010.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A.H., Davies J., Liu H.L., Nile A., and Osmani S.A.. 2006a Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell. 17:4946–4961. 10.1091/mbc.E06-07-0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A.H., Oakley B.R., and Osmani S.A.. 2006b Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat. Protoc. 1:2517–2526. 10.1038/nprot.2006.406 [DOI] [PubMed] [Google Scholar]
- Osmani S.A., Pu R.T., and Morris N.R.. 1988. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell. 53:237–244. 10.1016/0092-8674(88)90385-6 [DOI] [PubMed] [Google Scholar]
- Ovechkina Y., Maddox P., Oakley C.E., Xiang X., Osmani S.A., Salmon E.D., and Oakley B.R.. 2003. Spindle formation in Aspergillus is coupled to tubulin movement into the nucleus. Mol. Biol. Cell. 14:2192–2200. 10.1091/mbc.E02-10-0641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J.A., Hemmons L.M., MacDonald K.D., and Bufton A.W.. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141–238. [DOI] [PubMed] [Google Scholar]
- Rasala B.A., Ramos C., Harel A., and Forbes D.J.. 2008. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell. 19:3982–3996. 10.1091/mbc.E08-01-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., Gahl A., Backmann N., Conrath K., Muyldermans S., Cardoso M.C., and Leonhardt H.. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 3:887–889. 10.1038/nmeth953 [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., and Leonhardt H.. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289. 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., and Laemmli U.K.. 2006. Nup-PI: The nucleopore-promoter interaction of genes in yeast. Mol. Cell. 21:379–391. 10.1016/j.molcel.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Schultz S.J., Fry A.M., Sütterlin C., Ried T., and Nigg E.A.. 1994. Cell cycle-dependent expression of Nek2, a novel human protein kinase related to the NIMA mitotic regulator of Aspergillus nidulans. Cell Growth Differ. 5:625–635. [PubMed] [Google Scholar]
- Schwartz M., Travesa A., Martell S.W., and Forbes D.J.. 2015. Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin. Nucleus. 6:40–54. 10.1080/19491034.2015.1004260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smitherman M., Lee K., Swanger J., Kapur R., and Clurman B.E.. 2000. Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol. Cell. Biol. 20:5631–5642. 10.1128/MCB.20.15.5631-5642.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J., Maurer P., Vogel F., and Schlenstedt G.. 2000. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol. Cell. Biol. 20:8468–8479. 10.1128/MCB.20.22.8468-8479.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., and Rout M.P.. 2010. The nuclear pore complex: Bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501. 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Toews M.W., Warmbold J., Konzack S., Rischitor P., Veith D., Vienken K., Vinuesa C., Wei H., and Fischer R.. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45:383–389. 10.1007/s00294-004-0495-7 [DOI] [PubMed] [Google Scholar]
- Ukil L., De Souza C.P., Liu H.L., and Osmani S.A.. 2009. Nucleolar separation from chromosomes during Aspergillus nidulans mitosis can occur without spindle forces. Mol. Biol. Cell. 20:2132–2145. 10.1091/mbc.E08-10-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B.B., Köcher T., Wilm M., Allen T., et al. 2003a The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 113:195–206. 10.1016/S0092-8674(03)00235-6 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Askjaer P., Gentzel M., Habermann A., Griffiths G., Wilm M., Mattaj I.W., and Hetzer M.. 2003b RanGTP mediates nuclear pore complex assembly. Nature. 424:689–694. 10.1038/nature01898 [DOI] [PubMed] [Google Scholar]
- Waring R.B., May G.S., and Morris N.R.. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene. 79:119–130. 10.1016/0378-1119(89)90097-8 [DOI] [PubMed] [Google Scholar]
- Yam C., He Y., Zhang D., Chiam K.H., and Oliferenko S.. 2011. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr. Biol. 21:1314–1319. 10.1016/j.cub.2011.06.052 [DOI] [PubMed] [Google Scholar]
- Yam C., Gu Y., and Oliferenko S.. 2013. Partitioning and remodeling of the Schizosaccharomyces japonicus mitotic nucleus require chromosome tethers. Curr. Biol. 23:2303–2310. 10.1016/j.cub.2013.09.057 [DOI] [PubMed] [Google Scholar]
- Yang L., Guan T., and Gerace L.. 1997. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 137:1199–1210. 10.1083/jcb.137.6.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]