Murley et al. show that sterol transport proteins regulate target-of-rapamycin signaling at membrane contact sites in budding yeast. Ltc3/4 localize to a previously unknown region of the plasma membrane and, with other proteins, inhibit TORC2-Ypk1 signaling. At ER–vacuole contact sites, Ltc1 regulates the formation of sterol-enriched membrane domains that correlate with reduced TORC1 signaling.
Abstract
Membrane contact sites (MCSs) function to facilitate the formation of membrane domains composed of specialized lipids, proteins, and nucleic acids. In cells, membrane domains regulate membrane dynamics and biochemical and signaling pathways. We and others identified a highly conserved family of sterol transport proteins (Ltc/Lam) localized at diverse MCSs. In this study, we describe data indicating that the yeast family members Ltc1 and Ltc3/4 function at the vacuole and plasma membrane, respectively, to create membrane domains that partition upstream regulators of the TORC1 and TORC2 signaling pathways to coordinate cellular stress responses with sterol homeostasis.
Introduction
Compartmentalization is a key determinant of emergent biological functions. As such, multiple distinct mechanisms for cellular compartmentalization exist. Membrane-bound organelles and nonmembranous structures such as stress granules create compartments that serve to organize specialized functions in eukaryotic cells. In addition, specialized domains within membrane lipid bilayers exist such as dynamic plasma membrane (PM) lipid rafts and more stable structures such as caveoli and eisosomes, and these serve to regulate biochemical reactions underlying membrane-coupled cell signaling pathways and membrane dynamics (Harder and Simons, 1997; Anderson, 1998; Veatch and Keller, 2002; Douglas and Konopka, 2014).
As for other compartments, membrane domains can create regulation and/or drive cooperative reactions by increasing the effective concentration of proteins and lipids localized within them as, for example, in the clustering of dynein motors on sterol-enriched domains to facilitate active phagosome transport (Rai et al., 2016). Membrane domains can also function in a regulatory manner by excluding factors. This is the case for PM T cell receptor signaling domains, which form at immunological synapses between antigen-presenting cells and T cells and exclude the transmembrane phosphatase CD45 to permit phosphorylation of the T cell receptor and activation of downstream signaling events (Su et al., 2016).
Membrane domain biogenesis is driven by multivalent protein interactions and by lipid partitioning, which together create a phase transition within the membrane bilayer. Certain lipid species such as sterols and sphingolipids have a greater propensity to phase separate into liquid-ordered regions that can selectively recruit membrane-associated proteins, such as for example glycosylphosphotidylinositol-anchored and acylated proteins (Harder and Simons, 1997; Simons and Ikonen, 1997; Veatch and Keller, 2002). Close apposition of membrane bilayers, which occurs at regions termed membrane contact sites (MCSs), can also facilitate membrane domain compartment biogenesis. MCSs are created by actively tethering partner membranes together, which defines the geometry of the contact site. In vitro MCS model membrane systems have demonstrated that the physical dimension of a membrane-tethering protein is a critical determinant of protein sorting at these regions, independent of lipid composition—a phenomenon that likely contributes, for example, to the exclusion of CD45 from the immunological synapse (Schmid et al., 2016). In biological membranes, however, membrane domain formation involves the cooperative contributions of protein and lipids, which is underscored by the observation that MCS tethers are often membrane-associated proteins that also directly mediate lipid transport between membranes (AhYoung et al., 2015; Gatta et al., 2015; Murley et al., 2015).
We and others recently described a widely conserved family of ER membrane–associated Ltc/Lam proteins that within yeast cells localize to MCSs and function as tethers (Elbaz-Alon et al., 2015; Gatta et al., 2015; Murley et al., 2015). Ltc proteins also bind and selectively facilitate the intermembrane transfer of sterol lipids (Gatta et al., 2015; Murley et al., 2015). In yeast, the paralogs Ltc3/4 are localized to ER–PM contact sites, and cells lacking Ltc3/4 are sensitive to amphotericin B, an antifungal compound that targets the PM sterols and/or sphingolipids (Gatta et al., 2015). Ltc1 is localized to ER–mitochondria and ER–lysosome/vacuole contacts, where it forms via physical interactions with its partners Tom70/71 and Vac8, respectively (Elbaz-Alon et al., 2015; Murley et al., 2015). Although Ltc1 is a shared component of two distinct MCSs, its functions at each MCS are separable. At ER–mitochondria contacts, it functions with another tethering/lipid transport complex, the ER–mitochondria encounter structure, to maintain yeast cell viability, and at ER–vacuolar contacts, Ltc1 specifically facilitates the stress-dependent formation of sterol-enriched, liquid-ordered membrane domains on the vacuolar membrane (Murley et al., 2015).
In this study, we explored how Ltc1 and Ltc3/4 contribute to the formation of membrane domains at their respective MCSs. Our data indicate that Ltc3/4 localize to a previously undescribed domain of the yeast PM that we term the “membrane compartment of Ltc3/4” (MCL). Our data indicate that Ltc3/4 and additional MCL components contribute to PM homeostasis by regulating target of rapamycin complex 2 (TORC2) in a manner dependent on the sterol-binding properties of Ltc4 likely through partitioning of the key upstream TORC2 regulators Slm1/2. Analogously, our data indicate that Ltc1 actively creates sterol-enriched vacuole membrane domains that regulate TORC1 signaling by spatially segregating upstream TORC1 regulators, suggesting a role in the integration of nutrient sensing and stress responses with sterol homeostasis in the cell.
Results and discussion
Ltc3/4 define a new PM compartment at ER–PM contacts: The MCL
The yeast PM is actively organized by lipid and protein components into distinct membrane domains. The best-characterized PM domains include the membrane compartment of Pma1 (MCP), the membrane compartment of Can1/eisosome (MCC) domain, and the membrane compartment of TORC2 (MCT) compartment (Merzendorfer and Heinisch, 2013). Contact sites between the cortical ER and PM in yeast are known to occupy ∼40% of the surface area of the PM in yeast, but the role of ER–PM contact sites in the creation and maintenance of these PM domains has not been well characterized (Manford et al., 2012). To address this question, we examined whether the ER sterol transporters, Ltc3/4, localized to ER–PM contact sites, contribute to PM domain organization.
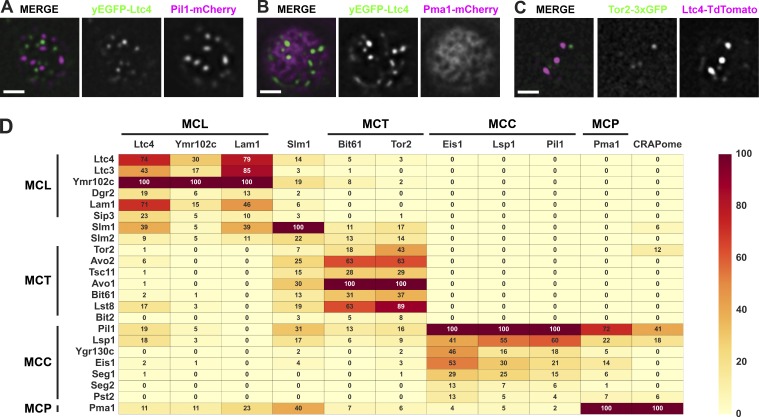
First, we assessed whether Ltc3/4 selectively localized to a specific PM domain by comparing the localization of Ltc4, which colocalizes with Ltc3, with known MCP, MCC, and MCT domain markers using fluorescence microscopy. Functional Ltc4-GFP or Ltc4-TdTomato localized to cortical foci at regions of ER–PM contact in cells as previously described (Fig. S1, A and B; Gatta et al., 2015), but did not significantly colocalize with markers for the MCC (Pil1-mCherry; Figs. 1 A and S1 C), MCP (Pma1-mCherry; Figs. 1 B and S1 D), or MCT (Tor2-3×GFP; Figs. 1 C and S1 E; and Bit61-GFP; Fig. S1 E). These observations suggest the possibility that Ltc3/4 localize to an uncharacterized region of the yeast PM.
Figure 1.
MCL is a novel PM domain. (A–C) Ltc4 does not colocalize with known PM domains. Cells expressing the indicated fluorescent protein fusions were grown to mid-log phase and imaged as described in the Microscopy and image analysis section of Materials and methods. A single focal plane from the top or bottom of cells is shown. Bars, 2 µm. (D) Proteomic analysis of Ltc4 identifies components of a novel PM domain, distinct from characterized MCL, MCC, and MCP domains. Endogenously tagged C-terminal GFP fusions to the indicated proteins—except for Tor2, which was tagged internally with 3×GFP—were immunopurified from whole-cell lysates using anti-GFP antibodies. Proteins in purified fractions were identified using MS/MS and filtered for nonspecific interactions as described in the Immunopurifications and proteomic analyses section of Materials and methods. The NSAF value for each identified protein is the mean of two experiments and is expressed as a percentage of the protein with the highest mean NSAF value from each immunopurification. NSAF values are measures of protein abundance that account for total spectra and proteins identified in the sample as well as the size and number of trypsin sites in the identified proteins. The NSAF value is color coded according the heat map as indicated. The CRAPome score is the percentage of experiments a protein is identified in the CRAPome database.
To test this idea, we assessed the molecular composition of the PM region marked by Ltc3/4 using proteomics. Specifically, to identify proteins associated with and/or in proximity to Ltc4, we immunopurified Ltc4-GFP using anti-GFP antibodies from cross-linked yeast whole-cell lysates and identified proteins from purifications by tandem mass spectrometry (MS; MS/MS). Using this approach, as shown in Fig. 1 D, the most abundant proteins in the Ltc4 purifications were three pairs of paralogous proteins: Ltc4 and its paralog Ltc3, the putative lipid transport paralogs Lam1/Sip3, and two poorly characterized WD-repeat paralogs, Ymr102c/Dgr2. To further test whether these components represent a protein–protein interaction (PPI) network, reciprocal purifications of Ymr102c-GFP or Lam1-GFP were performed using anti-GFP antibodies from yeast cross-linked extracts and analyzed by MS/MS. Consistently, proteomics identified the same three paralog pairs as the most abundant proteins in these purifications (Fig. 1 D). Also, consistent with cytological data (Fig. 1, A–C), known components of the MCT, MCC, and MCP in purified fractions of Ltc4-GFP, Lam1-GFP, and Ymr102c-GFP were significantly less abundant (Fig. 1 D). Indeed, the MCC components Pil1 and Lsp1 and the MCP component Pma1 were likely present as consequences of nonspecific interactions as suggested by their CRAPome scores (Fig. 1 D).
To more thoroughly define the boundaries of the Ltc4 PPI network, we characterized yeast MCT, MCC, and MCP PM domain PPI networks. We purified proteins localized to MCT (Tor2-3×GFP and Bit61-GFP), MCC (Pil1-GFP, Eis1-GFP, and Lsp1-GFP) and MCP (Pma1-GFP) from cross-linked extracts using anti-GFP antibodies and identified associated proteins by MS/MS. Our proteomic data indicate that for each domain marker, the most abundant associated proteins identified were reported to be localized within that domain, consistent with published work (Fig. 1 D). Components of the recently described membrane compartment of Wsc1 (MCW) PM compartment were not identified in any of our immunopurifications of components from MCL or other PM domains (Kock et al., 2016). This further supports the model that Ltc3/4, Lam1/Sip3, and the WD-repeat proteins Ymr102c/Dgr2 comprise a specific and solitary PPI network.
We directly examined whether Ltc4 colocalized with Lam1/Sip3 and Ymr102c/Dgr2 by fluorescence microscopy. GFP tagging of endogenous loci revealed that the fluorescent signal from Dgr2-GFP was too dim under our conditions for analysis. However, as shown in Figs. 2 (A and B) and S1 F, similar to Ltc4, Ymr102c-GFP, Lam1-GFP, and Sip3-GFP labeled cortically localized focal structures in cells. In addition, these structures significantly colocalized with focal structures labeled with Ltc4-TdTomato (Fig. 2, A and B; and Fig. S1 F). Time-lapse imaging of cortical Ltc4-GFP focal structures in live yeast cells indicated that they possess minimal dynamics (Fig. S1 G). Thus, together with proteomic data, cytological data support the existence of a novel distinct domain of the PM, which we term the MCL, comprised of the paralogous protein pairs of Ltc3/4, Lam1/Sip3, and Dgr2/Ymr102c.
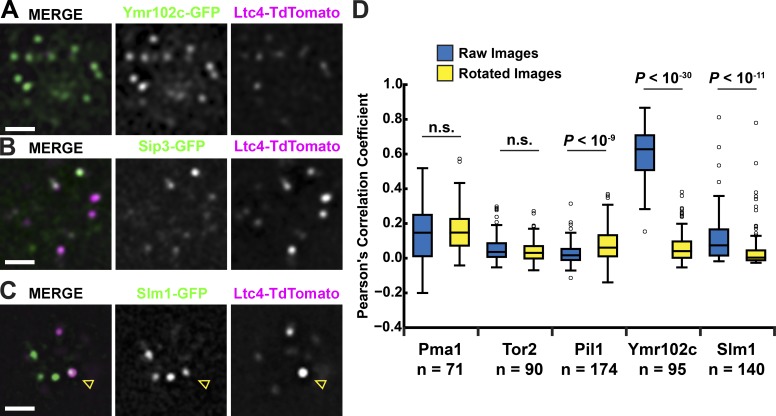
Figure 2.
Slm1/2 localize to the MCL. (A–C) Ltc1 colocalizes with MCL proteins and partially colocalizes with Slm1. Cells with the indicated fluorescent protein fusions were grown to mid-log phase and imaged as described in the Microscopy and image analysis section of Materials and methods. A single focal plane from the top or bottom of a cell is shown. Arrowheads in C indicate regions of colocalization between Ltc4 and Slm1. Bars, 2 µm. (D) Partial colocalization between Slm1 and Ltc4 is nonrandom. Pearson’s correlation coefficients between the fluorescent protein fusions to the indicated proteins and Ltc4 were measured in raw images of cells and images where the green channel was rotated 90°. P-values were calculated using the Mann–Whitney U test.
The TORC2 regulators Slm1/2 localize to the MCL
The paralogs Slm1/2 were also reproducibly identified as significant but less abundant proteins in purified fractions of all MCL components analyzed (Fig. 1 D). Slm1/2 are Pleckstrin homology and Bin, amphiphysin, and Rvs (BAR) domain–containing proteins that are thought to directly monitor PM phosphatidylinositol and sphingolipids and act with TORC2 and the downstream Akt kinase homologue Ypk1 as a PM quality surveillance system that regulates PM lipid homeostasis and the actin cytoskeleton. In response to PM mechanical stress or to the inhibition of sphingolipid biosynthesis by the serine palmitoyltransferase inhibitor myriocin, Slm1/2 dynamically repartition from the PM MCC domain to the MCT domain, where they recruit Ypk1, which is activated via TORC2 phosphorylation (Berchtold et al., 2012; Niles et al., 2012). In particular, TORC2-activated Ypk1 stimulates sphingolipid biosynthesis by phosphorylating the ceramide synthase components Lac1 and Lag1 (Muir et al., 2014). It also phosphorylates the ORM proteins, which de-represses sphingolipid biosynthesis (Roelants et al., 2011).
We assessed whether Slm1 was localized to the MCL by fluorescence microscopy. Slm1-GFP localized to cortical focal regions in cells as previously reported that were also, in a minority of instances, labeled by Ltc4-TdTomato (Fig. 2 C; Berchtold et al., 2012). To assess whether the Ltc4/Slm1 colocalization was significant, we compared the Pearson’s correlation coefficient obtained from pairs of respective raw images to the coefficient obtained from the cognate image pairs in which the green fluorescent signal image was rotated 90° (Fig. 2 D). A significant colocalization was expected if the Pearson’s coefficient from raw image pairs was greater relative to the rotated pairs. This analysis was performed on images from cells expressing Ltc4 and Slm1 and also, for validation, on images from Ltc4 and PM domain markers, Ymr102c (MCL), Tor2 (MCT), Pma1 (MCP), and Pil1 (MCC). As expected, for markers of the MCT, MCP, and MCC, the Pearson’s coefficient was either indistinguishable between raw and rotated images or significantly greater in the rotated image pairs as compared with the raw image pairs (Fig. 2 D). In contrast, the extent of colocalization between Ltc4 and the MCL component Ymr102c or between Ltc4 and Slm1 was greater in raw relative to rotated image pairs (Fig. 2 D). This analysis further substantiates the existence of a distinct MCL PM domain and indicates that Slm1 localizes to the MCL in addition to the MCT and MCC.
MCL function affects TORC2 signaling
The localization of Slm1/2 to the MCL suggests that this domain may contribute to the regulation of TORC2 signaling. To test this idea, we measured TORC2 activation by monitoring the phosphorylation of a variant of Ypk1, Ypk1(7A)-Myc, which only contains TORC2-specific phosphorylation sites (Muir et al., 2015). Phosphorylation of Ypk1(7A)-Myc was detected in yeast extracts by PhosTag SDS-PAGE, which selectively retards the migration of phosphorylated proteins, followed by Western analysis with anti-Myc antibodies. Using this methodology, as previously reported, we could detect both positive and negative regulation of TORC2 in cells via Ypk1(7A)-Myc hyper- and hypophosphorylation observed upon inhibition of sphingolipid biosynthesis with myriocin or hyperosmotic shock, respectively (Fig. S2 A; Muir et al., 2015).
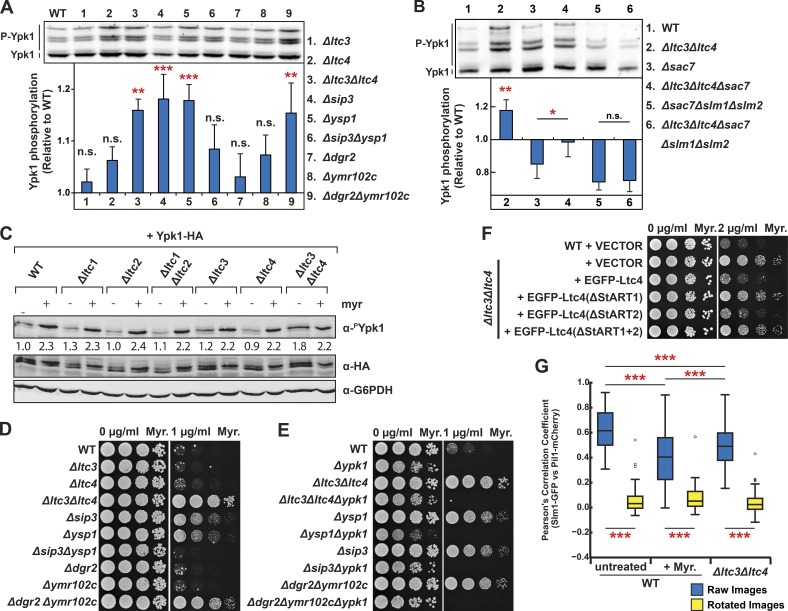
Using this assay, we examined TORC2 activity in cells harboring single and double paralogous deletions of MCL genes. In cells harboring double deletions of the LTC3/LTC4 paralogs or DGR2/YMR102c, we observed a significant increase in Ypk1(7A)-Myc phosphorylation relative to WT cells (Fig. 3 A). This is in contrast with cells harboring single deletions of either LTC3, LTC4, DGR2, or YMR102c, where no significant change in Ypk1(7A)-Myc phosphorylation was observed, suggesting that these paralogous gene pairs function redundantly. In addition, in cells harboring single deletions of either of LAM1 or SIP3 paralogs, but not in cells with the double LAM1/SIP3 deletions, we also observed significant activation of TORC2 demonstrated by increased phosphorylation of Ypk1(7A)-Myc relative to WT cells. Significant TORC2 activation was observed in the Δlam1 and Δsip3 single deletions but not in the double-deletion Δlam1Δsip3 cells. These phenotypes are indicative of a positive genetic interaction, suggesting that Lam1 and Sip3 physically interact within the MCL. These observations are consistent with proteomic and cytological data indicating a shared function of MCL components, and they indicate that disrupting the MCL compartment causes activation of TORC2, suggesting that the MCL may regulate TORC2 during cellular homeostasis.
Figure 3.
Defects in the MCL are sensed by TORC2-Ypk1 signaling through Slm1/2. (A and B) TORC2-mediated phosphorylation of Ypk1 was increased in MCL mutant cells in a Slm1/2-dependent manner. Proteins were extracted from cells of the indicated genotypes expressing the TORC2-specific Ypk1 variant Ypk1(7A)-Myc and separated by SDS-PAGE with PhosTag reagent to separate phosphorylated proteins as described in the Western blotting section of Materials and methods. P-values (*, P < 0.05; **, P < 0.01; ***, P ≤ 0.001) were calculated using a Fisher’s least significant difference post hoc test of a one-way ANOVA of repeated measurements from n = 6 (A) or 4 (B) independent experiments. Error bars represent SEM. (C) ER–PM Ltc proteins selectively regulate TORC2-specific Ypk1 phosphorylation. Cells of the indicated genotypes expressing plasmid-borne HA-tagged Ypk1 were grown to mid-log phase and treated with myriocin (Myr.) or vehicle only. Proteins were extracted and analyzed with Western blotting and quantitated as described in the Western blotting section of Materials and methods. To control for variation in Ypk1 expression, α-phospho–Ypk1(T662) signals were normalized by dividing by the signal derived from Ypk1-HA. Each value stated below the Western blots is relative to the normalized α-phospho–Ypk1(T662) signal in WT untreated samples. (D and E) Ypk1-dependent growth resistance to myriocin is observed in MCL mutant cells in which TORC2 is also activated. Cells of the indicated genotypes were grown to mid-log phase and spotted in 10-fold serial dilutions on YPD agar plates with the indicated concentrations of myriocin. (F) Ltc4 sterol binding activity is important to its function in the MCL domain. Cells harboring EGFP-Ltc4 or derivative mutations harboring StART domain deletions expressed from the native Ltc4 promoter on a low-copy plasmid were grown to mid-log phase and spotted in 10-fold serial dilutions on SD agar plates with the indicated concentrations of myriocin. (G) The localization of Slm1 to the MCC is altered in MCL mutant cells. WT cells (n = 80 cells), WT cells treated with myriocin (n = 53 cells), or Δltc3Δltc4 cells (n = 99 cells) expressing Slm1-GFP and Pil1-mCherry, an MCC marker, were imaged, and Pearson’s correlation coefficients were determined from single plane images taken at the tops and bottoms of cells. P-values (***, P ≤ 0.001) were calculated using Bonferroni’s post hoc test of a one-way ANOVA.
We tested whether Slm1 and Slm2 were required for activation of TORC2 in response to MCL perturbations by assessing Ypk1(7A)-Myc phosphorylation in cells harboring SLM1/SLM2 deletions. To maintain cell viability in the absence of Slm1 and Slm2, we deleted the SAC7 gene, which encodes a Rho1p GTPase switch that controls organization of the actin cytoskeleton in response to TORC2 activation (Audhya et al., 2004). We observed a decrease of Ypk1(7A)-Myc phosphorylation in Δsac7 relative to WT cells, however, consistent with a role for Ltc3/4 in regulating TORC2 Ypk1(7A)-Myc phosphorylation was significantly elevated in Δltc3Δltc4Δsac7 relative to Δsac7 cells, further suggesting that they may function in antagonistic pathways (Fig. 3 B). More importantly, however, Ypk1(7A)-Myc phosphorylation was indistinguishable between Δslm1Δslm2Δsac7 and Δltc3Δltc4Δslm1Δslm2Δsac7 cells (Fig. 3 B). These observations are consistent with activation of TORC2 in a Slm1/2-dependent manner in response to a loss of MCL function.
To substantiate observations made using the Ypk1(7A)-Myc reporter, we examined TORC2 signaling in Δltc3Δltc4 cells using an established phosphospecific antibody that specifically recognizes a Ypk1 TORC2–phosphorylated site (Niles et al., 2012, 2014). Consistently, using this assay, we observed a significant increase in TORC2-mediated phosphorylation of Ypk1 in Δltc3Δltc4 cells relative to WT cells (Fig. 3, A and C). In contrast, no significant changes in TORC2-Ypk1 signaling were observed in cells lacking either or both the Ltc1 and Ltc2 paralogs, which are homologous family members localized to contact sites between the ER and other organelles (Fig. 3 C; Murley et al., 2015). These observations suggest that sterol regulation of TORC2 signaling is specific to the MCL at ER–PM contact sites, consistent with a role of these contacts in the retrograde transport of sterols from the PM to the ER (Gatta et al., 2015; Murley et al., 2015).
As a consequence of its central role in the positive regulation of sphingolipid biosynthesis, yeast cells overexpressing Ypk1 are resistant to inhibition of growth caused by myriocin-dependent depletion of cellular sphingolipids. Thus, as a further test of a role of MCL function as an input to TORC2 signaling, we examined the growth of cells harboring MCL gene deletions on medium containing myriocin relative to WT. Consistent with our biochemical analysis, cells harboring deletions of MCL components that caused increased TORC2-dependent phosphorylation of Ypk1(7A)-Myc (Fig. 3, A and C) were also resistant to myriocin treatment relative to WT cells in a Ypk1-dependent manner (Fig. 3, D and E). This substantiates our conclusion that MCL function is tied to TORC2 regulation. We also used myriocin growth sensitivity to assess the role of sterol binding and transport activity of Ltc3/4 in Slm1/2-TORC2-Ypk1 regulation. Ltc4 possesses two StAR-related lipid transfer (StART)-like domains that have been previously shown to selectively bind and extract sterols from membranes (Gatta et al., 2015). We assessed the growth of Δltc3Δltc4 cells containing a plasmid-borne copy of Ltc4 lacking either or both StART1 and StART2 domains on media containing myriocin (Fig. 3 F). EGFP-Ltc4(ΔStART1), EGFP-Ltc4(ΔStART2), or EGFP-Ltc4(ΔStART1+2) localized to cortical foci similar to WT Ltc4, indicating that the Ltc4 StART variants are expressed and correctly targeted to ER–PM contacts (Fig. S2 B). In contrast to Ltc4, cells expressing either Ltc4(ΔStART1) or Ltc4(ΔStART1+2) failed to restore the myriocin growth sensitivity of Δltc3Δltc4 cells (Fig. 3 F). This observation suggests that MCL-mediated PM sterol homeostasis is a critical regulator of the TORC2–Ypk1 pathway.
Our data suggest the possibility that Ltc4-mediated PM sterol transport regulates TORC2-Ypk1 via altering the distribution Slm1/2 in a manner similar to that observed under conditions of sphingolipid depletion by myriocin (Berchtold et al., 2012; Niles et al., 2014). Consistent with this and similar to myriocin treatment, Slm1 localization to the MCC decreased in Δltc3Δltc4 cells relative to WT cells, as monitored by colocalization with Pil1 (Fig. 3 G).
At ER–vacuole contacts, Ltc1 regulates TORC1 signaling via formation of sterol enriched vacuolar membrane domains
Ltc1 is an ER transmembrane protein that is localized to ER–mitochondria and ER–vacuole contact sites. Similar to Ltc3/4, Ltc1 also functions in the formation membrane domains at MCSs. Specifically, under stress conditions, Ltc1 localized at ER–vacuolar contact sites facilitates the segregation of the vacuolar membrane into discrete micron scale sterol-enriched and –de-enriched domains. Interestingly, TORC1 has been proposed to function at the vacuole, similar to mTORC1 function at the lysosome in mammalian cells. In this context, our data indicating that the Ltc3/4 PM MCL domain regulates stress-dependent TORC2 signaling at ER–PM contact prompted us to ask whether Ltc1 might play an analogous role in regulating vacuolar-localized TORC1 signaling.
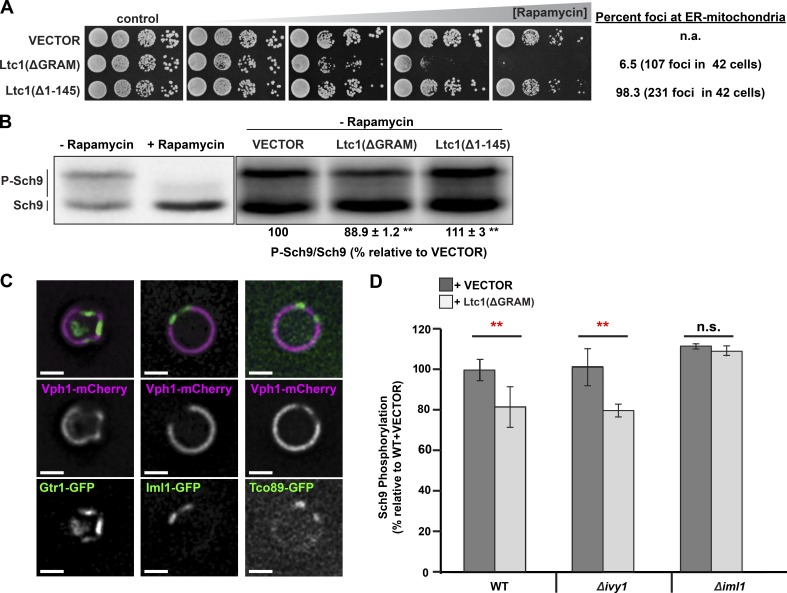
To specifically address whether Ltc1-dependent vacuole membrane domains function in TORC1 signaling, we targeted Ltc1 exclusively to ER–vacuole contacts in cells, a condition previously shown to induce vacuole domain formation in the absence of stress, and we assessed their growth sensitivity to the TORC1-specific inhibitor rapamycin (Murley et al., 2015). Our previous work demonstrated that Ltc1’s localization to ER–mitochondria contacts requires its glycosyltransferases, Rab-like GTPase activators, and myotubularins (GRAM) domain and the mitochondrial outer membrane import receptors Tom70/71 (Murley et al., 2015). Thus, to target Ltc1 exclusively to ER–vacuole contact sites, we expressed Ltc1(ΔGRAM) or deleted TOM70 and TOM71. WT cells expressing plasmid-borne Ltc1(ΔGRAM) were highly sensitive to low concentrations of rapamycin in comparison with cells harboring an empty vector or with cells expressing plasmid-borne Ltc1(Δ1–145), a variant that localized almost exclusively to ER–mitochondria contacts relative to Ltc1 and Ltc1(ΔGRAM) (Fig. 4 A). Consistently, deletion of the Ltc1 mitochondrial binding partner paralogs TOM70 and TOM71 also rendered cells hypersensitive to rapamycin in an Ltc1-dependent manner (Fig. S3 A). The correlation of Ltc1-dependent formation of vacuole domains with sensitivity to TORC1 inhibition suggests that cells with vacuolar domains have fewer active TORC1 complexes but maintain sufficient TORC1 activity to promote normal growth.
Figure 4.
Ltc1-dependent vacuole membrane domains inhibit TORC1. (A) Growth of cells with vacuole membrane domains in sensitive rapamycin. Cells harboring high-copy plasmids encoding Ltc1(ΔGRAM)-EGFP, Ltc1(Δ1–145)-EGFP, or an empty vector were spotted 10-fold in serial dilutions on agar plates containing 0, 1, 2, 3, or 4 nM rapamycin. Quantification of Ltc1 at ER–mitochondria contacts in cells was determined from cells expressing the Ltc1 variants and mitochondria matrix–targeted BFP. The percentage of Ltc1-EGFP foci associated with mitochondria in WT cells was 75% (n = 224 foci in 42 cells) under identical imaging conditions. (B) TORC1-dependent Sch9 phosphorylation is decreased in cells with vacuole membrane domains. Cells harboring an empty vector, Ltc1(ΔGRAM)-EGFP, or Ltc1(Δ1–145)-EGFP expressed from the endogenous Ltc1 promoter on a low-copy plasmid were grown to mid-log phase and treated with rapamycin (100 ng/µl) or vehicle-only control. Proteins were extracted and analyzed by SDS-PAGE as described in the Western blotting section of Materials and methods. (C) TORC1 and its regulators localize to sterol-enriched vacuole membrane domains. Cells with the indicated fluorescent protein fusions were grown to mid-log phase and then transferred to glucose-free media, incubated for 3 h, and imaged as described in the Microscopy and image analysis section of Materials and methods. A single focal plane is shown. Bars, 2 µm. (D) Iml1 is required for vacuole domain–mediated inhibition of TORC1 in cells. Cells of the indicated genotypes harboring an empty vector or Ltc1(ΔGRAM)-EGFP expressed from its own promoter were analyzed as in B. n = 6 samples per condition. Error bars represent the 95% confidence interval. P-values (**, P < 0.01) were calculated using Fisher’s least significant difference post hoc test on repeated measurements from a one-way ANOVA.
To directly assess TORC1 activity in cells, we monitored phosphorylation of the TORC1 target Sch9 in cell extracts. We used an established SDS-PAGE migration shift assay to interrogate the TORC1 phosphorylation of an Sch9 C-terminal 2-nitro-5-thiocyanatobenzoic acid cleavage fragment (CtermSch9; Urban et al., 2007). We found that that endogenous expression of Ltc1(ΔGRAM)-EGFP but not Ltc1(Δ1–145)-EGFP significantly reduced the amount of slower-migrating hyperphosphorylated species of CtermSch9 (Fig. 4 B). Cytological examination of cells indicated that ∼30% of cells (n = 42 cells) expressing Ltc1(ΔGRAM)-EGFP had vacuole domains, marked by the patterned localization of Vph1-mCherry. Similarly, ∼15% of Δtom70Δtom71 cells where Ltc1 is localized exclusively to ER–vacuole contacts have vacuole domains (Murley et al., 2015). Thus, vacuole domains in a minority of cells in a population (15–30%) correlate with an ∼10–15% decrease in Sch9 phosphorylation in the overall cell population (Fig. 4 B), suggesting that stress-induced vacuole domains significantly inhibit TORC1 signaling.
To address how vacuolar domains regulate TORC1 activity, we examined the vacuolar subdomain localization of TORC1 and its regulators during stress. Specifically, we examined the TORC1 subunit Tco89-GFP; the GTPase Gtr1, a subunit of the TORC1 upstream positive regulator exit from G0 (EGO) complex, which is the equivalent of the mammalian Ragulator complex subunits in the late endosomal/lysosomal adapter, MAPK, and MTOR activator (LAMTOR) complex; and the Gtr1 GTPase-activating protein (GAP) Iml1, a subunit of the upstream negative TORC1 regulator SEAC subcomblex inhibiting TORC1 signaling (SEACIT) complex that, together with Npr2/3, is equivalent to the mammalian GATOR1 complex (Bar-Peled et al., 2013; Panchaud et al., 2013). As previously described, we observed that Gtr1-GFP was localized to sterol-enriched domains and excluded from sterol–de-enriched domains marked by the vacuolar ATPase subunit Vph1-mCherry in cells (Fig. 4 C; Toulmay and Prinz, 2013). Similarly, both the Gtr1 GAP Iml1-GFP and the subunit of TORC1 Tco89-GFP also exclusively localized to sterol-enriched membrane domains (Figs. 4 C and S3 B).
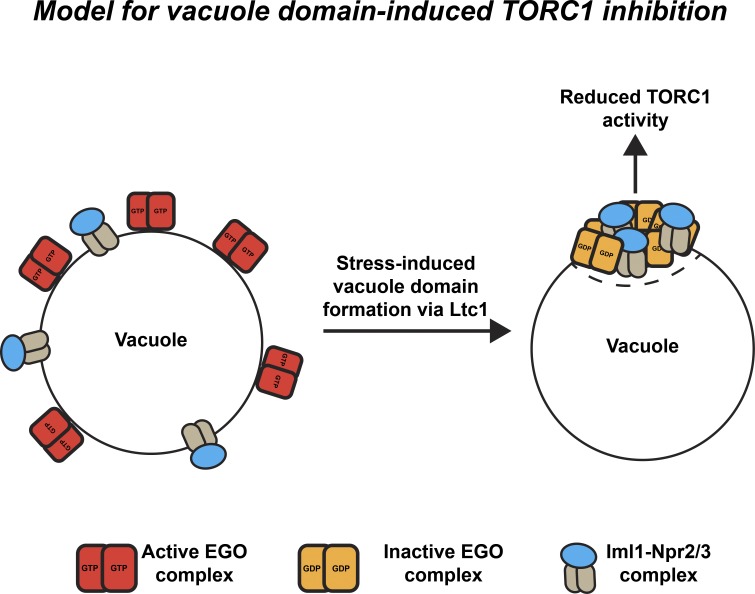
The localization of EGO and SEACIT complexes with TORC1 within sterol-enriched vacuolar domains raises the possibility that domain formation functions to inhibit TORC1 by increasing the concentration of the Gtr1 GAP Iml1 to promote the Gtr1 guanosine diphosphate (GDP)-bound state under stress. To test this, we induced vacuole domain formation by expressing Ltc1(ΔGRAM) under nonstressed conditions and assessed whether TORC1 activity was Iml1 dependent. As expected, deletion of IML1 slightly increased TORC1 activity as assessed by Sch9 phosphorylation (Fig. 4 D). In contrast with WT cells expressing Ltc1(ΔGRAM) (Fig. 4 D), however, we did not detect a significant difference in TORC1-dependent Sch9 phosphorylation in Δiml1 cells expressing Ltc1(ΔGRAM) relative to the vector-only control (Fig. 4 D). The lack of TORC1 inhibition observed in Δiml1 cells expressing Ltc1(ΔGRAM) expression was not a consequence of defective vacuolar domain formation, as assessed by fluorescence microscopy (Fig. S3 C). In addition, to date, the only protein complexes known to localize to sterol-enriched domains are TORC1, the EGO complex, SEACIT, and Ivy1, an inverted BAR domain protein that has also been linked to TORC1 regulation (Toulmay and Prinz, 2013; Numrich et al., 2015). Thus, we also examined the potential role of Ivy1 and observed no difference in TORC1 inhibition in Δivy1 cells expressing Ltc1(ΔGRAM) relative to WT cells expressing Ltc1(ΔGRAM) (Fig. 4 D). Therefore, although Ivy1 may play a role in TORC1 signaling related to vacuolar ATPase function as previously reported, it does not regulate TORC1 via vacuolar membrane domains, further indicating the specificity of the EGO–Iml1 signaling axis to vacuolar domains (Numrich et al., 2015). Although the exact mechanism is unclear, one possibility is that vacuole membrane domains inhibit TORC1 activity during stress by partitioning and concentrating the EGO complex with its inhibitory GAP Iml1 into sterol-enriched domains, thereby driving Gtr1 into its GDP-bound state to inactivate TORC1 (Fig. 5).
Figure 5.
Model for vacuole domain–induced inhibition of TORC1. In cells lacking vacuole domains, the EGO complex subunits Gtr1/2 on the vacuole surface bound in an active GTP-bound configuration because of the low effective concentration of the GAP Iml1. Vacuole domains induced by Ltc1 correlate with Iml1 coclustering and concentration with Gtr1/2 in sterol-enriched vacuole domains. The effective increase in Iml1 concentration may drive Gtr1/2 into a primarily GDP-bound configuration, resulting in inhibition of TORC1 activity.
Our findings highlight the important and general role that interorganelle tethering and lipid transport proteins play at MCSs in facilitating the formation of membrane domains that serve as regulatory hubs for cell signaling. Our data suggest that at ER–vacuole contact sites, Ltc1 facilitates the formation of sterol-enriched domains that copartition and possibly concentrate the TORC1 upstream positive Ragulator complex subunit Gtr1 in the EGO complex with its GAP, Iml1, to drive TORC1 inhibition (Fig. 5). Our data also raise the possibility that contact sites can also influence longer-distance changes in cellular compartments by altering the partitioning and subcellular localization of signaling proteins. Specifically, our results suggest that at ER–PM contact sites, Ltc3/4 may work within the newly defined PM domain MCL to alter the relative distribution of the upstream TORC2 regulators Slm1/2 between distinct compartments of the PM to alter TORC2 activity (Berchtold et al., 2012; Niles et al., 2012). Consistently, although Ltc1 and Ltc3/4 regulate TORC1 and TORC2 via different modes, a role of their sterol binding/transfer activity is likely to be important in both circumstances. Thus, our data suggest that sterols play a key general role in TORC1/2 regulation, a speculation consistent with the recent finding that cholesterol regulates the recruitment and activation of mTORC1 at the lysosomal surface in mammalian cells (Castellano et al., 2017). This finding raises the interesting question of whether sterol transport at MCSs is integrated with overall cellular sterol homeostasis and other nutrient signals via TORC1/2.
Our observations suggest that contact sites function in networks that both contribute and respond to alterations in cellular signaling pathways. In this context, given the proposed role of Ltc4 in retrograde transport of sterols from the PM to the ER, our data suggest that in addition to sphingolipids, PM sterols regulated by the MCL may function as upstream inputs for TORC2 regulation. In this context, published data indicate that the TORC2 target and Akt kinase homologue Ypk1 phosphorylates Ltc4 (Muir et al., 2014; Gatta et al., 2015). Thus, it is possible that within the MCL, Ltc4 also functions in a TORC2-regulatory feedback circuit to regulate PM sterol levels. These sensing and feedback pathways would provide a means of coordinating sterol and sphingolipid metabolism, which both play key roles in PM integrity (Berchtold et al., 2012; Niles et al., 2012, 2014). Our observations also suggest a mechanism for how the ER–vacuole contact site domain may function to sense and integrate nutrient responses to regulate TORC1 activity. Previous work has shown that glucose starvation but not amino acid starvation causes vacuole domain formation (Toulmay and Prinz, 2013). Our results suggest that vacuole membrane domain formation via Ltc1 under glucose-limiting conditions could mitigate TORC1 activation by otherwise sufficient levels of amino acids. Indeed, growing cells in media with poor carbon sources, such as raffinose or glycerol, reduces TORC1 activity, as measured by reduced Sch9 phosphorylation, despite the presence of sufficient amino acid levels (Urban et al., 2007). Thus, regulation of TORC1 by vacuolar domain formation could be used by the cell to divert ketogenic amino acids from protein synthesis toward scavenging pathways, such as autophagy, to provide acetyl-CoA for the TCA cycle when preferred carbon sources such as glucose are limiting.
Our observations regarding the regulation of TOR signaling in yeast are likely significant for other organisms given that LTC proteins are conserved throughout eukaryotes (e.g. VAD1 in Arabidopsis thaliana and GRAMD proteins in humans). The regulation of TORC1 by Ltc1-mediated vacuole membrane domains could have direct parallels to the regulation of mTORC1, as mTORC1 activation is also coupled to the mammalian lysosomal membrane and TORC1 regulators such as SEACIT (GATOR1 in mammals) and the EGO complex (Rag–Ragulator in mammals) are conserved. Indeed, LAMTOR1 in humans and Ego1 in yeast are N-terminally myristoylated and dipalmitoylated on a conserved Gly-Cys-Cys motif, which anchors Ragulator and EGO to lysosomal/vacuolar membranes, and mTORC1 recruitment has recently been shown to be influenced by lysosomal cholesterol (Ashrafi et al., 1998; Nada et al., 2009; Castellano et al., 2017). These posttranslational lipid modifications can bias proteins to sorting into liquid-ordered domains and thus may contribute to the sorting of LAMTOR and EGO complexes into sterol-induced liquid-ordered domains on mammalian lysosomes (Levental et al., 2010; Aicart-Ramos et al., 2011). Given the fundamental roles TOR complexes play in cell growth and proliferation and their roles in diseases such as cancer, it will be interesting to examine whether mammalian homologues of yeast Ltc proteins regulate TOR signaling via sterol homeostasis at MCSs.
Materials and methods
Yeast genetics and molecular biology
W303 (Rothstein, 1983), BY4741 (Brachmann et al., 1998), and SEY6210 (Robinson et al., 1988) strain backgrounds were used in this study. Yeast were transformed using the lithium acetate method.
To create EGFP-YSP2, the coding sequence of YSP2 was amplified from W303 yeast genomic DNA and inserted into p416MET25-yEGFP (Lackner et al., 2013) using the BamHI–SmaI sites. The ΔStART1, ΔStART2, and ΔStART1+2 domain mutants were made using isothermal assembly. The MET25 promoter from these plasmids was excised using SpeI and SacI and then replaced using the native promoter of YSP2 (bases 459 immediately 5′ of the start codon as in Gatta et al., 2015). pRS313::Ltc1(Δ145)-EGFP was created by isothermal assembly and included the 250 bp upstream of the start codon and an ADH1 terminator. All plasmids were verified by sequencing to ensure they had only desired mutations.
All yeast strains and other plasmids used in this study have been described previously and are listed in Table S1.
Microscopy and image analysis
Cells were grown to log phase (OD600 ∼0.6–1.0) at 30°C in the appropriate synthetic medium to select for plasmids, concentrated by centrifugation, deposited directly on a glass slide, and then sealed under a 1.5 coverslip using nail polish. Cells were imaged at 25°C.
Cells in Figs. 1 (A–C), 2 (A–C), 4 C, S1 (B–F), and S3 B were imaged on a DeltaVision real-time microscope (IX70; Olympus) using a 60× 1.40 NA objective lens (Olympus) and a 100-W mercury lamp (Applied Precision, Ltd.). Light microscopy images were collected using an integrated cooled charge-coupled device–based camera (CoolSNAP HQ; Photometrics) equipped with a Sony Interline Chip. Datasets were processed using the iterative constrained 3D deconvolution method in SoftWoRx software (Applied Precision, Ltd.) to remove out-of-focus light.
Cells depicted or analyzed in Fig. S2 B were imaged using the spinning-disk module of a Marianas SDC real-time 3D confocal-TIRF microscope (Intelligent Imaging Innovations) fit with a spinning-disk head (Yokogawa Electric Corporation), a 63× 1.40 NA (Fig. S2) or 100× 1.46 NA objective (Olympus; Figs. 3 B, 4 B, S1 B, D, and E; and Fig. S2), and an electron-multiplying charge-coupled device camera.
For assessing formation of vacuolar membrane domains, cells were grown for >12 generations to mid-log phase (to ensure no residual vacuole domains from cells in the colony) in synthetic complete (SC) media containing dextrose as a carbon source (synthetic dextrose; SD) and were then harvested by centrifugation. They were then resuspended in 50 µg/ml cycloheximide or SC media without glucose. For glucose starvation, cells were washed three times with water before resuspending them in an equal volume of SC glucose. All treatments lasted 1 h. Cells were imaged on the DeltaVision widefield deconvolution microscope described earlier in this section.
ImageJ (National Institutes of Health) and Elements software (Nikon) were used to assess colocalization of PM-localized proteins. ImageJ macros were used to crop images, split channels, and create images where the green channel was rotated 90° or unrotated. Background correction was done equally to all images of the same fluorescent proteins, and colocalization analysis was done using Elements.
Images were manipulated in Photoshop (Adobe) for linear adjustments to brightness and contrast.
Immunopurifications and proteomic analyses
C-terminally GFP-tagged proteins from the yeast GFP collection (except for Tor2-3×GFP) were immunopurified from whole-cell lysates as previously described (Lackner et al., 2013). Cells were grown overnight to log phase (OD600 = 1) in 500 ml YPD medium (2% glucose, 2% peptone, and 1% yeast extract supplemented with adenine and tryptophan), harvested by centrifugation, washed once with distilled water, and resuspended 1:1 with immunopurification lysis buffer (IPLB; 20 mM Hepes, pH 7.4, 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, and 0.6 M sorbitol) containing protease inhibitor cocktail Set 1 (PIC; EMD Millipore). Cell suspensions were flash-frozen dropwise in liquid nitrogen. Frozen cells were lysed in a freezer mill (Spex 6970 EFM) and using three lysing periods of 2 min at speed “7” with 2 min of chilling between each. Lysates were stored at −80°C. Frozen cell lysates were thawed in an RT water bath with additional PIC and then were clarified of unlysed cells and debris by centrifugation at 500 g for 5 min. Clarified lysates were treated with a reversible cross-linker dithiobis succinylmidyl propionate (Thermo Fisher Scientific) at a concentration of 1 mM for 30 min at 4°C, nutating at 50 rpm. The reaction was quenched by addition of tris, pH 7.5, to 100 mM and incubation on ice for 10 min. Membranes were solubilized by addition of digitonin to a final concentration of 1% and nutating at 4°C at 50 rpm. Lysates were clarified by centrifugation at 12,000 g for 10 min at 4°C. Clarified lysates were incubated with 50 µl of μMACs monoclonal mouse anti-GFP magnetic microbeads (Miltenyi Biotec) incubated at 4°C for 30 min. μMACS columns were equilibrated in IPLB + 1% digitonin + 1× PIC. Lysates and anti-GFP beads were flown over the column, and bound beads were washed three times with 800 µl IPLB + 0.1% digitonin + PIC and then twice more with 500 µl IPLB. On-bead digestion with trypsin was achieved by applying 25 µl of elution buffer 1 (2 M urea, 50 mM tris-HCl, pH 7.5, 1 mM DTT, and 5 µg/ml trypsin) to the column and incubating for 30 min. Digested proteins were eluted by adding 2× 50 µl of elution buffer 2 (2 M urea, 50 mM tris-HCl, pH 7.5, and 5 mM chloroacetamide). Eluted proteins were incubated overnight at 25°C and then quenched by addition of 1 µl of trifluoroacetic acid. Mock purifications from cells that did not express GFP were used as controls for nonspecific interactions.
Purified proteins were identified by MS/MS in the University of California, Davis, proteomics core. Downstream analysis was done using Scaffold (Proteomic Software Inc.) and a Python script, which is available as filterproteomics.txt. This script removes identified proteins that do not meet both of the following criteria: they are identified in duplicate experiments and their normalized spectral abundance factor (NSAF) value is 10-fold greater than from a weighted mean from mock purifications. Weighted averages from mock purifications using GFP antibodies are from eleven independent experiments. We normalized the NSAF values of proteins in this filtered list relative to the protein with the highest NSAF value for each purification. We rearranged these data in Excel (Microsoft) and applied a conditioned-color heat map into that shown in Fig. 1 D. A full list of proteins with raw NSAF and total spectral counts is included in Table S2.
Growth assays
Yeast strains were grown in liquid YPD or the appropriate synthetic medium to select for plasmids to mid-log phase. Cells were harvested by centrifugation and resuspended in sterile water to a concentration of 0.5 OD600 per ml for plating on YPD plates and 0.2 OD600 per ml for SD plates. 10-fold serial dilutions were made in water, and 4 µl (for YPD agar plates) or 10 µl (for minimal medium agar plates) of each of these dilutions were spotted onto plates containing the concentrations of myriocin, rapamycin, or AmB specified in the figure legends. Aqueous solutions spotted onto synthetic medium seemed to have a higher surface tension; therefore, we spotted a larger volume of a more diluted cell suspension to make larger spots that would allow easier visualization of growth differences. Plates containing AmB were poured in the afternoon and dried overnight at 30°C before spotting with yeast. AmB potency seemed to diminish over time. Control plates not containing drugs were made from the same batch of medium.
Western blotting
Monitoring of Ypk1 phosphorylation using Ypk1(7A)-Myc
Yeast strains harboring pFR252 (encoding Ypk1(7A)-Myc expressed from its own promoter) were grown overnight in SD leucine to log phase. Cultures were diluted to OD600 = 0.15 in 3 ml of YPD prewarmed to 30°C and grown for 3 h. Cells were collected by centrifugation at 1,000 g for 2.5 min and then resuspended in 1 ml of 0.255 M NaOH + 1% 2-mercaptoethanol. Cells were incubated on ice for 5 min before addition of 150 µl of 60% TCA and a further 15 min of incubation on ice. Precipitated proteins were collected by centrifugation at 16,000 g at 4°C for 2 min in microfuge tubes. The supernatant was decanted, and pellets were resuspended in the remaining buffer by pipetting before addition of 700 µl of ice-cold acetone. Precipitates were collected by centrifugation at 16,000 g for 2 min and then washed again with 700 µl of ice-cold acetone. It was important to invert the tubes so that the acetone rinsed out any residual TCA in the caps of the tubes. Acetone was carefully aspirated, and then the pellet was resuspended in 50 µl of MURB buffer (100 mM MES, pH 7; 3 M urea, 1% SDS, 0.1% bromophenol blue, and 5% 2-mercaptoethanol added fresh before each use). Samples were boiled in a heat block for 5 min and then clarified by centrifugation at 16,000 g for 1 min. 2 µl of the resulting supernatant was resolved by SDS-PAGE with gels containing 7% acrylamide (from a 29:1 acrylamide/bis-acrylamide solution), 20 µM PhosTag reagent (from a 1 mM stock), and 150 µM MnCl2. To ensure gel polymerization, freshly prepared ammonium persulfate solutions were used, and acrylamide gel solutions were degassed under vacuum in a 15-ml plastic conical tube until tapping the tube did not produce bubbles. Proteins were separated under a constant 110 V for 3.5 h. Gels were washed 3× for 10 min in transfer buffer containing 10 mM EDTA and once in transfer buffer before being transferred to nitrocellulose membranes (Bio-Rad Laboratories) at 110 V for 1.5 h. Membranes were blocked in TBST buffer + 5% nonfat milk for 1 h at RT before being probed with TBST + 5% milk with 1:500 anti-Myc antibody (9E10; Covance) overnight at 4°C. Membranes were washed 3× for 5 min with TBST, probed for 1 h at with fluorescently conjugated anti–mouse antibody at 1:10,000 dilution in TBST, and washed three more times with TBST for 5 min each. Membranes were imaged on an Odyssey scanner and analyzed using the freely available Image Studio Lite software (LI-COR Biosciences). Ypk1(7A)-Myc phosphorylation was determined by calculating the proportion of total Ypk1(7A)-Myc signal not found in the fastest-migrating (nonphosphorylated) band as a fraction of the total Ypk1(7A)-Myc signal, which includes all Ypk1(7A)-Myc species including the fastest migrating band.
Monitoring of Ypk1 phosphorylation using Ypk1(T662) antibodies
Cells expressing Ypk1-HA were grown overnight in SD-uracil media at 30°C to mid-log phase. Cells were treated with DMSO or 5 µg/ml myriocin for 1 h. Protein extracts were prepared using the NaOH cell lysis method described in the previous section. Proteins were resolved on 8% tris-glycine SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were probed with α-HA (12CA5; 1:5,000; Covance), α-G6PDH (1:100,000; Sigma-Aldrich), and α-phospho-Ypk (1:20,000; T662; described previously by Niles et al., 2012) primary antibodies, and visualized using the appropriate secondary antibodies conjugated to IRDye (1:5,000; LI-COR Biosciences) on the Odyssey Infrared Imaging system (LI-COR Biosciences). Images were quantified using ImageQuant software (GE Healthcare) and described as the difference relative to WT after normalizing to the α-HA signal.
Chemical fragmentation assays for monitoring Sch9 phosphorylation
Sch9 blots were performed similarly to experiments by Urban et al. (2007). Cells harboring Sch9-3×HA and either pRS313, pRS313::Ltc1(Δ145–360)-EGFP, or pRS313::Ltc1(Δ1–145)-EGFP in 3 ml of synthetic dextrose medium to mid-log phase (OD600 = 0.6). One tenth volume of 60% TCA was added to the cultures, which were incubated on ice for 10 min before collecting them in two steps of centrifugation at 16,000 g for 2 min in microfuge tubes. Supernatants were decanted, and the pellets were resuspended by pipetting in the remaining solution before addition of 700 µl of ice-cold acetone. Pellets were collected by centrifugation at 16,000 g for 2 min and then washed again with ice-cold acetone. Tubes were inverted so that residual TCA was removed from the caps. After carefully aspirating the acetone, the pellets were resuspended in 33 µl urea buffer (3 M urea, 50 mM tris, pH 7.6, 1% SDS, 50 mM NaF, 2.4 mM NaN3, 50 mM paranitrophenylphosphate, 50 mM β-glycerophosphate, 1× protease inhibitor cocktail I [EMD Millipore], 50 mM NaH2PO4, and 5 mM EDTA) and then heated to 65°C for 10 min. Samples were allowed to cool for 2 min before the addition of 10 µl of 0.5 M CHES buffer, pH 10.5, and 7 µl of 5 mM nitrothyocyanobenzoic acid. The samples turned yellow. Samples were incubated overnight at RT in the dark. 50 µl of a loading buffer were added to the samples, which were then boiled for 3 min. 20 µl of clarified lysate was separated on a 10% SDS-PAGE gel and blotted to nitrocellulose for 0.8 h at 100 V. The membrane was blocked for 1 h in TBST + 5% milk and probed overnight at 4°C with 1:1,000 anti-HA antibody in TBST + 5% milk. The blots were washed 3× for 5 min with TBST, probed for 1 h with 1:10,000 anti–mouse secondary antibody in TBST, and then washed 3× for 5 min before imaging on an Odyssey scanner and analysis using Image Studio Lite. Sch9 phosphorylation was determined by calculating the proportion of total Sch9Cterm signal not found in the fastest migrating band.
Online supplemental material
Fig. S1 contains additional images comparing Ltc4 localization to other PM domain markers. Fig. S2 contains blots showing increased Ypk1(7A)-Myc phosphorylation in response to myriocin treatment as well as images showing localization of Ltc4 mutants. Fig. S3 shows that Δtom70Δtom71 mutants are sensitive to rapamycin in an Ltc-1 dependent manner and that Iml1 is not required for vacuole domain formation. Table S1 shows all yeast strains and other plasmids used in this study. Table S2 shows a full list of proteins with raw NSAF and total spectral counts. A separate file is included containing the script “filterproteomics.txt.”
Acknowledgments
We would like to thank Dr. Michael Paddy in the University of California, Davis, Department of Molecular and Cell Biology Imaging Facility for advice with fluorescence microscopy. Also, we would like to thank members of the Nunnari laboratory for stimulating discussion.
J. Nunnari is supported by National Institutes of Health grants R37GM097432 and R01GM106019. W. Prinz is supported by intramural National Institutes of Health grants DK600004 and DK060105. J. Nunnari is on the Scientific Advisory Board of Mitobridge, Inc.
The authors declare no further competing financial interests.
Author contributions: A. Murley designed and performed experiments and wrote the manuscript. J. Nunnari designed experiments and wrote the manuscript. J. Yamada performed experiments. B.J. Niles performed experiments. A. Toulmay performed experiments. W.A. Prinz and T. Powers edited the manuscripts.
Footnotes
Abbreviations used:
- EGO
- exit from G0
- GAP
- GTPase-activating protein
- GDP
- guanosine diphosphate
- GRAM
- glycosyltransferases, Rab-like GTPase activators, and myotubularins
- IPLB
- immunopurification lysis buffer
- LAMTOR
- late endosomal/lysosomal adapter, MAPK, and MTO activator
- MCC
- membrane compartment of Can1/eisosome
- MCL
- membrane compartment of Ltc3/4
- MCP
- membrane compartment of Pma1
- MCS
- membrane contact site
- MCT
- membrane compartment of TORC2
- MS
- mass spectrometry
- NSAF
- normalized spectral abundance factor
- PM
- plasma membrane
- PPI
- protein–protein interaction
- SD
- synthetic dextrose
- SEACIT
- SEAC subcomplex inhibiting TORC1 signaling
- StART
- StAR-related lipid transfer
References
- AhYoung A.P., Jiang J., Zhang J., Khoi Dang X., Loo J.A., Zhou Z.H., and Egea P.F.. 2015. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc. Natl. Acad. Sci. USA. 112:E3179–E3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aicart-Ramos C., Valero R.A., and Rodriguez-Crespo I.. 2011. Protein palmitoylation and subcellular trafficking Biomembranes. 1808:2981–2994. 10.1016/j.bbamem.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Anderson R.G. 1998. The caveolae membrane system. Annu. Rev. Biochem. 67:199–225. 10.1146/annurev.biochem.67.1.199 [DOI] [PubMed] [Google Scholar]
- Ashrafi K., Farazi T.A., and Gordon J.I.. 1998. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J. Biol. Chem. 273:25864–25874. 10.1074/jbc.273.40.25864 [DOI] [PubMed] [Google Scholar]
- Audhya A., Loewith R., Parsons A.B., Gao L., Tabuchi M., Zhou H., Boone C., Hall M.N., and Emr S.D.. 2004. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 23:3747–3757. 10.1038/sj.emboj.7600384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., Spear E.D., Carter S.L., Meyerson M., and Sabatini D.M.. 2013. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 340:1100–1106. 10.1126/science.1232044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold D., Piccolis M., Chiaruttini N., Riezman I., Riezman H., Roux A., Walther T.C., and Loewith R.. 2012. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 14:542–547. 10.1038/ncb2480 [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., and Boeke J.D.. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Castellano B.M., Thelen A.M., Moldavski O., Feltes M., van der Welle R.E., Mydock-McGrane L., Jiang X., van Eijkeren R.J., Davis O.B., Louie S.M., et al. 2017. Lysosomal cholesterol activates mTORC1 via an SLC38A9–Niemann-Pick C1 signaling complex. Science. 355:1306–1311. 10.1126/science.aag1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas L.M., and Konopka J.B.. 2014. Fungal membrane organization: The eisosome concept. Annu. Rev. Microbiol. 68:377–393. 10.1146/annurev-micro-091313-103507 [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y., Eisenberg-Bord M., Shinder V., Stiller S.B., Shimoni E., Wiedemann N., Geiger T., and Schuldiner M.. 2015. Lam6 regulates the extent of contacts between organelles. Cell Reports. 12:7–14. 10.1016/j.celrep.2015.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta A.T., Wong L.H., Sere Y.Y., Calderón-Noreña D.M., Cockcroft S., Menon A.K., and Levine T.P.. 2015. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 4:e07253 10.7554/eLife.07253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T., and Simons K.. 1997. Caveolae, DIGs, and the dynamics of sphingolipid–cholesterol microdomains. Curr. Opin. Cell Biol. 9:534–542. 10.1016/S0955-0674(97)80030-0 [DOI] [PubMed] [Google Scholar]
- Kock C., Arlt H., Ungermann C., and Heinisch J.J.. 2016. Yeast cell wall integrity sensors form specific plasma membrane microdomains important for signalling. Cell. Microbiol. 18:1251–1267. 10.1111/cmi.12635 [DOI] [PubMed] [Google Scholar]
- Lackner L.L., Ping H., Graef M., Murley A., and Nunnari J.. 2013. Endoplasmic reticulum-associated mitochondria–cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA. 110:E458–E467. 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I., Grzybek M., and Simons K.. 2010. Greasing their way: Lipid modifications determine protein association with membrane rafts. Biochemistry. 49:6305–6316. 10.1021/bi100882y [DOI] [PubMed] [Google Scholar]
- Manford A.G., Stefan C.J., Yuan H.L., Macgurn J.A., and Emr S.D.. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23:1129–1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Merzendorfer H., and Heinisch J.J.. 2013. Microcompartments within the yeast plasma membrane. Biol. Chem. 394:189–202. 10.1515/hsz-2012-0241 [DOI] [PubMed] [Google Scholar]
- Muir A., Ramachandran S., Roelants F.M., Timmons G., and Thorner J.. 2014. TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. eLife. 3:e03779 10.7554/eLife.03779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A., Roelants F.M., Timmons G., Leskoske K.L., and Thorner J.. 2015. Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. eLife. 4:e09336 10.7554/eLife.09336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Sarsam R.D., Toulmay A., Yamada J., Prinz W.A., and Nunnari J.. 2015. Ltc1 is an ER-localized sterol transporter and a component of ER–mitochondria and ER–vacuole contacts. J. Cell Biol. 209:539–548. 10.1083/jcb.201502033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S., Hondo A., Kasai A., Koike M., Saito K., Uchiyama Y., and Okada M.. 2009. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK–ERK pathway to late endosomes. EMBO J. 28:477–489. 10.1038/emboj.2008.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles B.J., Mogri H., Hill A., Vlahakis A., and Powers T.. 2012. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. USA. 109:1536–1541. 10.1073/pnas.1117563109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles B.J., Joslin A.C., Fresques T., and Powers T.. 2014. TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Reports. 6:541–552. (published erratum appears in Cell Reports 2014. 6:592) 10.1016/j.celrep.2013.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numrich J., Péli-Gulli M.P., Arlt H., Sardu A., Griffith J., Levine T., Engelbrecht-Vandré S., Reggiori F., De Virgilio C., and Ungermann C.. 2015. The I-BAR protein Ivy1 is an effector of the Rab7 GTPase Ypt7 involved in vacuole membrane homeostasis. J. Cell Sci. 128:2278–2292. 10.1242/jcs.164905 [DOI] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.P., and De Virgilio C.. 2013. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6:ra42. doi: 10.1126/scisignal.2004112. [DOI] [PubMed] [Google Scholar]
- Rai A., Pathak D., Thakur S., Singh S., Dubey A.K., and Mallik R.. 2016. Dynein clusters into lipid microdomains on phagosomes to drive rapid transport toward lysosomes. Cell. 164:722–734. 10.1016/j.cell.2015.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.S., Klionsky D.J., Banta L.M., and Emr S.D.. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936–4948. 10.1128/MCB.8.11.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Breslow D.K., Muir A., Weissman J.S., and Thorner J.. 2011. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 108:19222–19227. 10.1073/pnas.1116948108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R.J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202–211. 10.1016/0076-6879(83)01015-0 [DOI] [PubMed] [Google Scholar]
- Schmid E.M., Bakalar M.H., Choudhuri K., Weichsel J., Ann H., Geissler P.L., Dustin M.L., and Fletcher D.A.. 2016. Size-dependent protein segregation at membrane interfaces. Nat. Phys. 12:704–711. 10.1038/nphys3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., and Ikonen E.. 1997. Functional rafts in cell membranes. Nature. 387:569–572. 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., and Vale R.D.. 2016. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 352:595–599. 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A., and Prinz W.A.. 2013. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 202:35–44. 10.1083/jcb.201301039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 26:663–674. 10.1016/j.molcel.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Veatch S.L., and Keller S.L.. 2002. Organization in lipid membranes containing cholesterol. Phys. Rev. Lett. 89:268101 10.1103/PhysRevLett.89.268101 [DOI] [PubMed] [Google Scholar]