TRIM37 gene mutations cause muscle–liver–brain–eye (mulibrey) nanism: a rare autosomal recessive, prenatal onset growth disorder. Wang et al. find that TRIM37 ubiquitylates and stabilizes PEX5, the receptor for import of peroxisomal matrix proteins, suggesting that mulibrey nanism is a new peroxisomal biogenesis disorder.
Abstract
Most proteins destined for the peroxisomal matrix depend on the peroxisomal targeting signals (PTSs), which require the PTS receptor PEX5, whose deficiency causes fatal human peroxisomal biogenesis disorders (PBDs). TRIM37 gene mutations cause muscle–liver–brain–eye (mulibrey) nanism. We found that TRIM37 localizes in peroxisomal membranes and ubiquitylates PEX5 at K464 by interacting with its C-terminal 51 amino acids (CT51), which is required for PTS protein import. PEX5 mutations (K464A or ΔCT51), or TRIM37 depletion or mutation, reduce PEX5 abundance by promoting its proteasomal degradation, thereby impairing its functions in cargo binding and PTS protein import in human cells. TRIM37 or PEX5 depletion induces apoptosis and enhances sensitivity to oxidative stress, underscoring the cellular requirement for functional peroxisomes. Therefore, TRIM37-mediated ubiquitylation stabilizes PEX5 and promotes peroxisomal matrix protein import, suggesting that mulibrey nanism is a new PBD.
Introduction
Peroxisomes are single membrane–bound organelles found in most eukaryotic cells. They house many metabolic pathways, most notably for β oxidation of fatty acids, as well as the production and degradation of hydrogen peroxide and other reactive oxygen species (Smith and Aitchison, 2013). Human diseases caused by peroxisomal disorders highlight the necessity of this organelle. Peroxisomal diseases fall into two categories: single-enzyme defects and peroxisomal biogenesis disorders (PBDs; Waterham et al., 2016). PBDs are more complex in etiology in that many peroxisomal enzymes are affected, generally via lack of peroxisomal protein import (Waterham et al., 2016). Earlier work in yeast, together with genetic phenotype complementation of peroxisome-deficient CHO mutant cells or human patient fibroblasts, identified PEX genes (encoding peroxins) necessary for peroxisomal biogenesis (Erdmann, 2016; Honsho et al., 2016). So far, 14 complementation groups (CGs) in PBDs have been identified based on the PEX gene mutation and clinical phenotypes (Ebberink et al., 2012; Waterham et al., 2016). Deficiency of PEX5 protein causes PBDs of CG2, manifesting peroxisomal matrix protein import defects (Dodt et al., 1995; Wiemer et al., 1995).
Peroxisomal biogenesis involves the assembly of peroxisomal membrane proteins (PMPs), followed by the import of matrix proteins (Ma et al., 2011). The latter depends on distinct peroxisomal targeting signals (PTSs): PTS1, comprising a noncleaved C-terminal tripeptide, SKL, or its conserved variants (Gould et al., 1989); or PTS2, consisting of the nonapeptide sequence (R/K)(L/V/I/Q)XX(L/V/I/H/Q)(L/S/G/A/K)X(H/Q)(L/A/F) localized near the N terminus of the cargo protein (Swinkels et al., 1991). In mammals, PEX5 and PEX7 isoforms are the receptors for PTS1 and PTS2 cargoes, respectively, but PEX5 isoforms are required for both PTS1 and PTS2 protein import because, whereas both isoforms (PEX5S and PEX5L) bind PTS1 cargo directly, only the long isoform (PEX5L) interacts with PTS2 cargo indirectly via its interaction with PEX7 (Braverman et al., 1998; Otera et al., 2000). PTS protein import occurs through the following steps: receptor–cargo binding in the cytoplasm; docking of the receptor–cargo complex at peroxisomal membranes and translocation to the matrix; and cargo release and recycling of receptors to the cytosol for the next round of import (Ma et al., 2011).
Ubiquitylation regulates PEX5-mediated PTS protein import and stability. Monoubiquitylation at a conserved, N-terminal cysteine in PEX5 is essential for receptor stability and for recycling from peroxisomes to the cytosol during the matrix protein import cycle in yeast and mammals (Carvalho et al., 2007; Platta et al., 2007; Williams et al., 2007; Okumoto et al., 2011). PEX4 and UbcH5a/b/c family members serve as the E2 for cysteine monoubiquitylation in PEX5 of yeast and mammals, respectively (Wiebel and Kunau, 1992; Grou et al., 2008). Components of the RING complex (PEX2/PEX10/PEX12) serve as the E3 ligase for PEX5 in both yeast and mammals (Krause et al., 2006; Okumoto et al., 2014). In contrast to monoubiquitylation, polyubiquitylation targets yeast Pex5 for proteasomal degradation via the RADAR (receptor accumulation and degradation in the absence of recycling) pathway, a quality-control system preventing the accumulation of nonfunctional Pex5 on peroxisomal membranes (Kiel et al., 2005). PEX5 stability decreases in several human PBD patient cells (Dodt and Gould, 1996; Yahraus et al., 1996), suggesting that a similar quality-control mechanism may operate in mammals. Although conservative modes of PEX5 ubiquitylation exist in different organisms, the precise mechanisms of PEX5 ubiquitylation and the quality-control system remain to be established in mammals.
Muscle–liver–brain–eye (mulibrey) nanism is a rare autosomal recessive growth disorder of prenatal onset caused by TRIM37 gene mutations (Avela et al., 2000). TRIM37 is a member of the tripartite motif (RING, B-Box, and coiled-coil domains) family and has E3 ubiquitin ligase activity toward itself and histone H2A protein (Kallijärvi et al., 2005; Bhatnagar et al., 2014). TRIM37 localizes to peroxisomes (Kallijärvi et al., 2002). Patients with mulibrey nanism or PBDs have severe growth and development problems early in life (Avela et al., 2000; Steinberg et al., 2006).
In this study, we unveil a novel function of TRIM37 in regulating PTS protein import. TRIM37 interacts with PEX5 and monoubiquitylates PEX5 at K464. Monoubiquitylation by TRIM37 stabilizes PEX5 protein and effectively increases its binding to cargoes, which promotes PTS protein import. Either TRIM37 depletion in human cancer cell lines or TRIM37 mutation in human patient cells impairs PTS protein import, confirming that mulibrey nanism is a new type of PBD.
Results
TRIM37 is a PMP
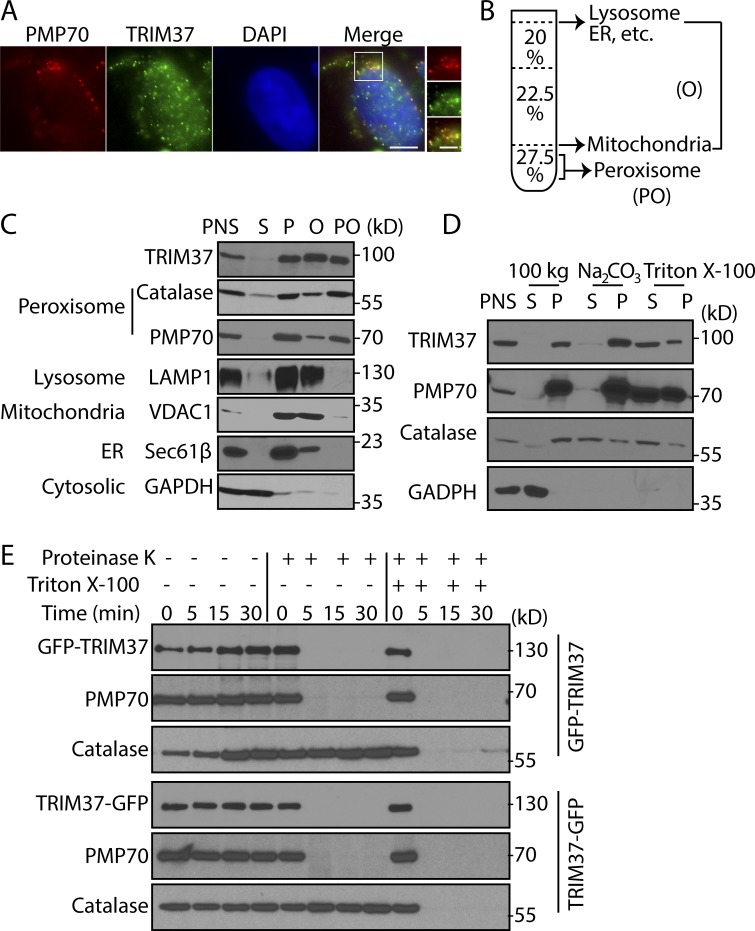
TRIM37 localizes to peroxisomes, but its exact location and functions were not elucidated (Kallijärvi et al., 2002). Immunostaining of endogenous TRIM37 confirmed its peroxisomal localization, showing colocalization with the PMP marker PMP70, although many dots containing TRIM37 were not peroxisomes (Fig. 1 A). Subcellular fractionation also established that TRIM37 localized in the organelle fractions (P), similar to the peroxisomal matrix protein, catalase; the PMP PMP70; and other organelle proteins: LAMP1 (lysosome), VDAC1 (mitochondria), and Sec61-β (ER). The cytosolic protein GAPDH was in the supernatant fraction (S; Fig. 1 C). To further demonstrate the subcellular location of TRIM37, peroxisomes were separated from other organelles using a density gradient that separates PMP70 and catalase from other organelle proteins, such as LAMP1, Sec61-β, and VDAC1 (Fig. 1, B and C). Consistent with the partial localization of TRIM37 to peroxisomes by immunostaining, ∼40% of TRIM37 was present in the peroxisome fraction (Fig. 1 C).
Figure 1.
Localization and topology of TRIM37 in peroxisomal membranes. (A) Subcellular localization of TRIM37. HepG2 cells were immunostained using antibodies against PMP70 and TRIM37. Bars: 5 µm; (insets) 2.5 µm. (B and C) Distribution of TRIM37 in peroxisome fractions. By using a density gradient method, other organelle proteins such as ER and lysosomes floated on the top, whereas mitochondrial proteins resided at the 22.5/27.5% interface. We pooled these two fractions as organelles (O) except peroxisomes. Peroxisomes (PO) floated slightly in the densest layer. (D) TRIM37 is an integral membrane protein. PNSs from HepG2 cells were fractionated differentially into cytosolic (S) and organelle (P) fractions by centrifugation at 100,000 g (100 kg). The P fractions were also extracted by either 0.1 M alkaline Na2CO3 or 0.5% Triton X-100, and the supernatants and pellets were collected. (E) Topology of TRIM37 fusion proteins. The organelle fractions from GFP-TRIM37– or TRIM37-GFP–expressing HEK 293T cells were either untreated or treated with proteinase K plus trypsin in the presence or absence of 1% Triton X-100 for the indicated times.
To address whether TRIM37 resides in the peroxisomal membrane or matrix, the organelle fractions were subjected to alkaline carbonate or Triton X-100 extraction, respectively. As expected for PMPs, PMP70 was resistant to carbonate but not Triton X-100 extraction, whereas catalase, a matrix enzyme, was mostly solubilized by both extractions (Fig. 1 D). Like PMP70, TRIM37 remained in the pellet after carbonate extraction but was solubilized by Triton X-100, showing that TRIM37 is a PMP.
To determine the topology of TRIM37, we fused GFP at the N (GFP-TRIM37) and C terminus of TRIM37 (TRIM37-GFP) and conducted protease protection assays. The region of PMPs (PMP70) facing the cytosol was degraded, whereas a matrix protein, catalase, was resistant to protease degradation (Fig. 1 E). In the presence of Triton X-100, however, both PMP70 and catalase were degraded. Because both GFP-TRIM37 and TRIM37-GFP proteins were degraded in the absence or presence of Triton X-100 (Fig. 1 E), TRIM37 is a PMP with both N and C termini facing the cytosol.
TRIM37 depletion or mutation results in defective peroxisomal matrix protein import
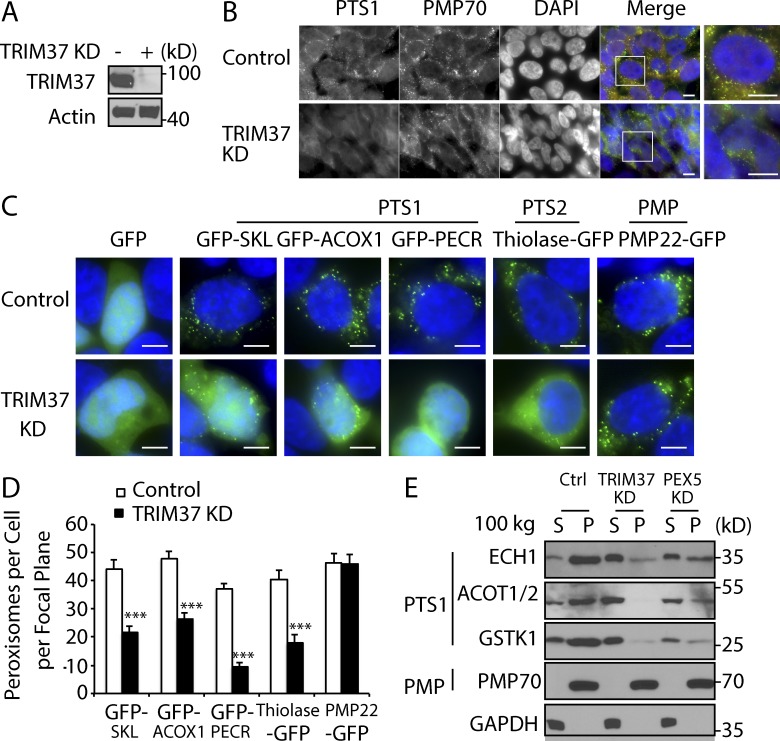
To study the role of TRIM37, we used a shRNA to knock down TRIM37 in HepG2 cells. TRIM37 was almost completely depleted in TRIM37 knockdown (KD) cells (Fig. 2 A). Fluorescence microscopy of endogenous PTS1-containing proteins (immunostained with the α-SKL antibody; Fig. 2 B), as well as GFP-PTS1 (GFP-SKL, GFP-ACOX1, and GFP-PECR) and PTS2 (thiolase-GFP) fusion proteins (Fig. 2, C and D), showed a severe mislocalization of matrix proteins from peroxisomes to the cytoplasm in TRIM37-depleted cells when compared with WT cells. The expression levels of proteins tested were unaffected by TRIM37 depletion (Fig. S1 A). However, no defect was observed in localization of the PMPs PMP70 (Fig. 2 B) and PMP22-GFP (Fig. 2, C and D) upon TRIM37 depletion, indicating a role of TRIM37 only in peroxisomal matrix protein import.
Figure 2.
Loss of TRIM37 impairs import of peroxisomal matrix proteins. (A) TRIM37 detection in HepG2 control and TRIM37 KD cells. (B) Immunostaining of peroxisomal proteins in HepG2 control and TRIM37 KD cells. Cells were costained with antibodies against PMP70 and PTS1 proteins. Bars: 10 µm. (C and D) Localization of peroxisomal matrix and membrane proteins in HepG2 control and TRIM37 KD cells. The tripeptide SKL was fused to the C terminus of GFP (GFP-SKL). Bars: 5 µm. The cDNAs encoding the PTS1-containing proteins (ACOX1, acyl-CoA oxidase 1; PECR, peroxisomal trans-2-enoyl-CoA reductase) were fused to the C terminus of GFP (GFP-ACOX1 and GFP-PECR). Thiolase and PMP22 were fused to the N terminus of GFP (thiolase-GFP and PMP22-GFP). These GFP constructs were transfected into HepG2 control and TRIM37 KD cells. Images were taken with the same exposure time for each construct (24 h after transfection). Nuclei were stained with DAPI. The expression levels of these constructs are equal in control and TRIM37 KD cells, as shown in Fig. S1 A. The number of punctate peroxisomes per cell was calculated and presented as mean ± SEM; ***, P < 0.001 (Student’s t test). The results are representative of three independent experiments. (E) Subcellular distribution of peroxisomal proteins in HepG2 control, TRIM37 KD, and PEX5 KD cells. Cytosolic (S) and organelle (P) fractions were separated by 100,000 g centrifugation of PNSs ECH1 (enoyl-CoA hydratase 1), ACOT1/2 (acyl-CoA thioesterase 1/2), GSTK1, GSTκ1, and GAPDH.
Similarly, subcellular fractionation assays of endogenous PTS1-containing proteins (ECH1, ACOT1/2, and GSTK1) in HepG2 cells showed their redistribution to the cytosolic fractions upon TRIM37 depletion, comparable to that seen in PEX5 KD cells (Fig. 2 E), but PMP70 distribution was unaffected. Fractionation results also showed that most GFP-SKL protein, but not PMP22-GFP or PEX13, was cytosolic upon TRIM37 depletion (Fig. S1, B and C). A peroxisomal matrix protein import defect was also observed in HEK 293T cells depleted for TRIM37 (Fig. S1, D–F), suggesting the TRIM37 function in humans is cell type independent. A similar peroxisomal matrix protein import defect was observed in another TRIM37 KD stable cell line (TRIM37 KD-2#) using a second shRNA (Fig. S1, G and H), excluding other off-target effects. Furthermore, GFP-ACOX1 relocalized to peroxisomes when TRIM37 protein expression was rescued in TRIM37 KD cells (Fig. S1, I and J).
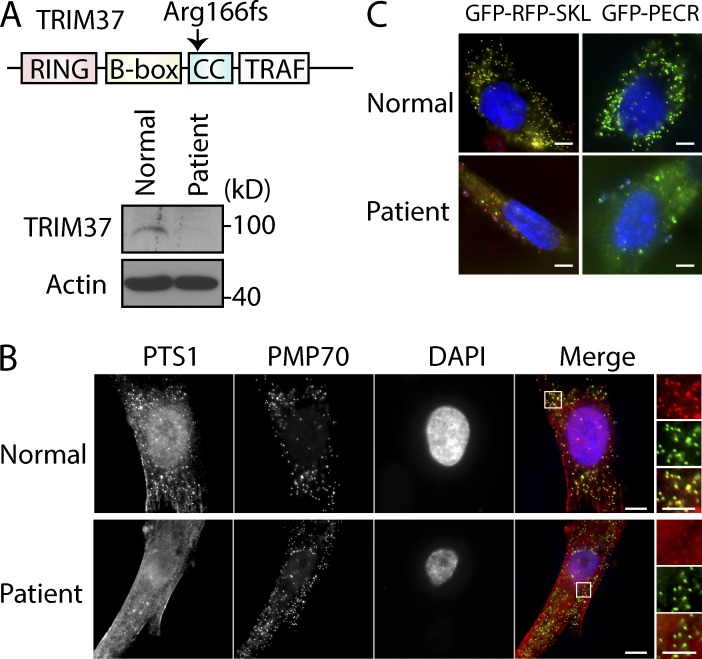
To test whether similar phenotypes were observed in patients with TRIM37 mutation, we examined PTS protein localization in fibroblasts from a mulibrey nanism patient with the most common mutation (Avela et al., 2000). This patient had an A to G transition in the 3′ splice acceptor sequence of intron 6 (c.493-2A>G), which is supposed to result in aberrant splicing and a frame shift (fs) mutation at Arg166, causing an early translation stop in the coiled-coil domain of TRIM37 (Fig. 3 A). As described previously (Avela et al., 2000), we confirmed that this TRIM37 gene mutation altered mRNA splicing and expressed no TRIM37 protein in the patient (Fig. S2, A and B; and Fig. 3 A). Immunostaining showed that most endogenous PTS1-containing proteins were cytoplasmic and only a small fraction colocalized with PMP70 in the patient fibroblasts (Fig. 3 B). Similarly, most PTS1 proteins (GFP-RFP-SKL and GFP-PECR) exogenously expressed in these patient cells were cytoplasmic, and only a few peroxisomal structures were seen in comparison with normal cells (Fig. 3 C). These data show that TRIM37 depletion or mutation causes an import defect for peroxisomal matrix proteins, but not for PMPs, manifesting a similar phenotype as PEX5 depletion or mutation.
Figure 3.
TRIM37 gene mutation causes a defect in the import of peroxisomal matrix proteins. (A) Diagram of TRIM37 structure and the fs mutation at Arg166. Loss of TRIM37 protein in human patient fibroblasts (AG02122) was verified by Western blot. (B) Normal (AG21802) or patient fibroblast cells (AG02122) were stained with antibodies against PTS1 proteins and PMP70. (C) GFP-RFP-SKL or GFP-PECR constructs were transfected into the fibroblasts. The images were taken 24 h after transfection. The expression of GFP-RFP-SKL and GFP-PECR constructs in patient cells was very weak, and hence relatively higher exposure times were used to capture the signal compared with those for normal cells. Bars: 10 µm; (insets) 5 µm.
The Arg166fs mutation should result in a translation fs at Arg166 and premature termination 10 amino acids downstream (Fig. S2 C). Overexpressing TRIM37 (Arg166fs)–GFP mutant protein in normal cells did not redistribute peroxisomal RFP-SKL (Fig. S2, D–F), excluding any dominant-negative effect of Arg166fs on peroxisomal matrix protein import.
TRIM37 is an E3 ligase for PEX5
In mammals, the import of PTS1 and PTS2 proteins requires PEX5, whose function depends on ubiquitylation (Dodt et al., 1995; Wiemer et al., 1995; Braverman et al., 1998; Thoms and Erdmann, 2006; Okumoto et al., 2014). The resemblance of phenotypes caused by TRIM37 and PEX5 deficiency prompted us to ask whether the E3 ligase TRIM37 ubiquitylates PEX5.
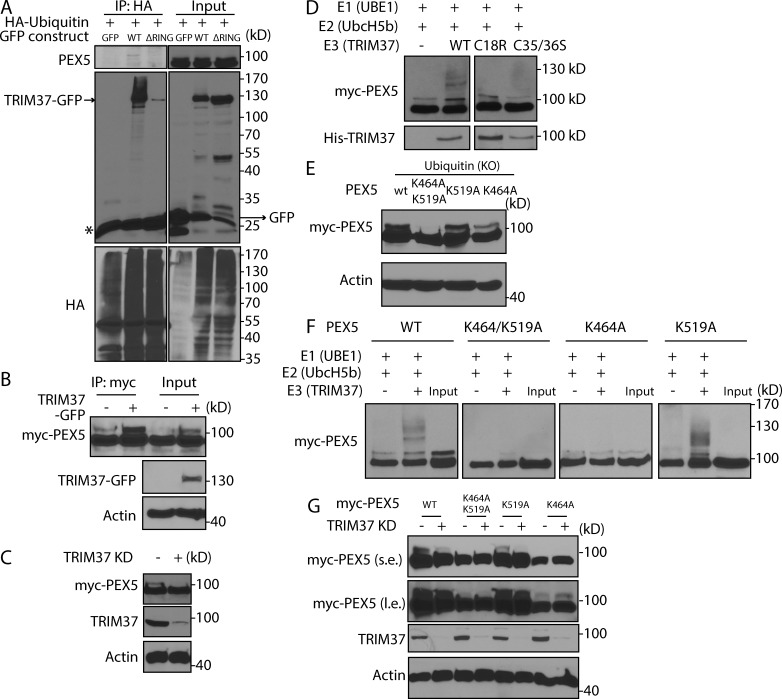
We tested whether TRIM37 expression level could modulate PEX5 ubiquitylation in vivo in HEK 293T cells. To follow the ubiquitylation status of endogenous PEX5, cells were transfected with HA-ubiquitin with or without TRIM37-GFP overexpression plasmids, and PEX5 ubiquitylation was monitored by detection of PEX5 in pull-down of HA-ubiquitin (Fig. 4 A, IP: HA). Ubiquitylated PEX5 was hardly detected in control (GFP) cells but was evident in TRIM37-GFP–overexpressing (WT) cells (Fig. 4 A, first and second lanes). The molecular weight of the ubiquitylated PEX5 suggested that the modification was monoubiquitylation (Fig. 4 A) and that deubiquitylase (USP2) treatment caused the disappearance of the slower migrating upper band of exogenously expressed myc-PEX5 protein (Fig. S3 A). This ubiquitylation was not observed in TRIM37 (ΔRING)–GFP-overexpressing cells (Fig. 4 A, third lane), suggesting its dependence on the RING domain, which is necessary for the E3 ligase activity of TRIM37. This requirement of TRIM37 for PEX5 monoubiquitylation in vivo was confirmed independently by showing that monoubiquitylation of exogenous PEX5 was also enhanced by TRIM37 overexpression and reduced by KD (Fig. 4, B and C).
Figure 4.
TRIM37 ubiquitylates PEX5 at K464. (A) HEK 293T cells were transfected with constructs overexpressing HA-ubiquitin (WT), plus GFP, TRIM37-GFP (WT), or TRIM37 (ΔRING)–GFP. The cell lysates (input) were used for HA-IP followed by detection with the indicated antibodies. The asterisk in the GFP blot indicates the IgG light chain. (B) HEK 293T cells were transfected with constructs expressing myc-PEX5 together with GFP(−) or TRIM37-GFP(+). Cell lysates were collected for the myc-IP. Myc-PEX5 and TRIM37-GFP proteins were detected by myc and GFP antibody, respectively. (C) HEK 293T control and TRIM37 KD cells were transfected with constructs expressing myc-PEX5. (D) Histidine-TRIM37 (WT) or the RING mutants (C18R and C35/36S) of TRIM37 were purified from bacterial lysates and used for in vitro ubiquitylation assays in the presence of E1 (UBE1), E2 (UbcH5b), and myc-PEX5 immunoprecipitated from HEK 293T cells overexpressing myc-PEX5. PEX5 ubiquitylation was detected by myc antibody. The amount of TRIM37 proteins was determined using histidine antibody before the ubiquitylation assay. (E) HEK 293T cells were transfected with the indicated PEX5 constructs together with constructs expressing HA-ubiquitin (KO). (F) In vitro ubiquitylation assay using myc-PEX5 (WT) or the indicated PEX5 mutants, UBE1, UbcH5b, and TRIM37 proteins. (G) HEK 293T control or TRIM37 KD cells were transfected with constructs expressing myc-PEX5 (WT) or the indicated PEX5 mutants, together with HA-ubiquitin (KO). s.e., short exposure; l.e., long exposure.
To examine whether TRIM37 ubiquitylates PEX5 directly in vitro, TRIM37 protein purified from bacteria and myc-PEX5 protein was immunoprecipitated from HEK 293T cells overexpressing myc-PEX5. Purified TRIM37 protein had E3 ligase activity in vitro as indicated by its self-ubiquitylation (Fig. S3 B). We used the E1 enzyme UBE1 and different E2 enzymes for in vitro ubiquitylation assays and found that UbcH5 family (UbcH5a/b/c) proteins catalyzed TRIM37-mediated PEX5 ubiquitylation (Fig. S3 C), analogous to the finding that the UbcH5 family is the E2 for PEX5 ubiquitylation on cysteine 11 (C11; Grou et al., 2008). The E3 ligase activity of TRIM37 depended on its RING domain, as mutations (C18R or C35/36S) that interfered with the RING structure (Kallijärvi et al., 2005; Bhatnagar et al., 2014) strongly reduced TRIM37-mediated PEX5 ubiquitylation (Fig. 4 D). In contrast to the results of the in vivo ubiquitylation experiments, the TRIM37 E3 ligase activity on PEX5 in vitro was manifested mainly by PEX5 poly- rather than monoubiquitylation. An explanation might be that the myc-PEX5 substrate purified from HEK 293T cells had already undergone monoubiquitylation in vivo, and excessive ubiquitylation components might cause sustained polyubiquitylation in vitro. To circumvent this problem, we used nonubiquitylated, purified PEX5 from Escherichia coli and found that both mono- and polyubiquitylation of PEX5 were catalyzed by TRIM37 (WT), but not its RING domain mutants (C18R or C35/36S; Fig. S3 D). Together, the in vitro and in vivo data show that TRIM37 directly monoubiquitylates PEX5.
TRIM37 ubiquitylates PEX5 at K464
Two sites were reported previously for PEX5 ubiquitylation. One is the nonconventional C11 ubiquitylation site, which is required for PEX5 recycling during PTS protein import (Okumoto et al., 2011); the other site is lysine 209 (K209), which is crucial for pexophagy induced by high levels of reactive oxygen species (Zhang et al., 2015). Surprisingly, TRIM37 still ubiquitylated PEX5 mutants lacking both these known sites (C11A/K209A; Fig. S3 E) in vitro, suggesting the existence of other ubiquitin acceptor sites for TRIM37-mediated PEX5 ubiquitylation. Most probably the sites were not cysteines because samples for Western blots were prepared in reducing conditions. By scanning lysines in PEX5, we found that K464 and K519 might be ubiquitin acceptor sites because their mutation caused an almost complete loss of PEX5 ubiquitylation in vivo (Fig. S3 F). To prove this, we made the double mutant PEX5 (K464/K519A) and expressed it with ubiquitin (KO), which has all its lysines mutated to arginines, thereby preventing polyubiquitylation and accumulating only monoubiquitylated species. As expected, the double mutant PEX5 (K464/K519A) lost all ubiquitylation; however, the single mutants, PEX5 (K519A) and PEX5 (K464A), were still monoubiquitylated in the presence of ubiquitin (KO) (Fig. 4 E). Therefore, K464 and K519 are the alternative sites for PEX5 monoubiquitylation.
To identify the functional TRIM37 site, we performed in vitro ubiquitylation assays using PEX5 mutants. TRIM37 could still (mono- and poly-) ubiquitylate PEX5 (K519A), but not K464/K519A or K464A (Fig. 4 F), providing direct evidence that K464 is the principal PEX5 ubiquitylation site targeted by TRIM37. Confirming this, TRIM37 failed to ubiquitylate the PEX5 (K464R) mutant in vitro (Fig. S3 G). Meanwhile, in vivo TRIM37 depletion reduced ubiquitylation of the K519A mutant (with K464 intact), comparable to that for PEX5 (WT) (Fig. 4 G). Additionally, TRIM37 depletion did not alter the pattern of ubiquitylation of PEX5 (K464A) (Fig. 4 G). These results show that TRIM37 monoubiquitylates PEX5 primarily at K464.
TRIM37 interacts with the C-terminal 51 amino acids (CT51) of PEX5
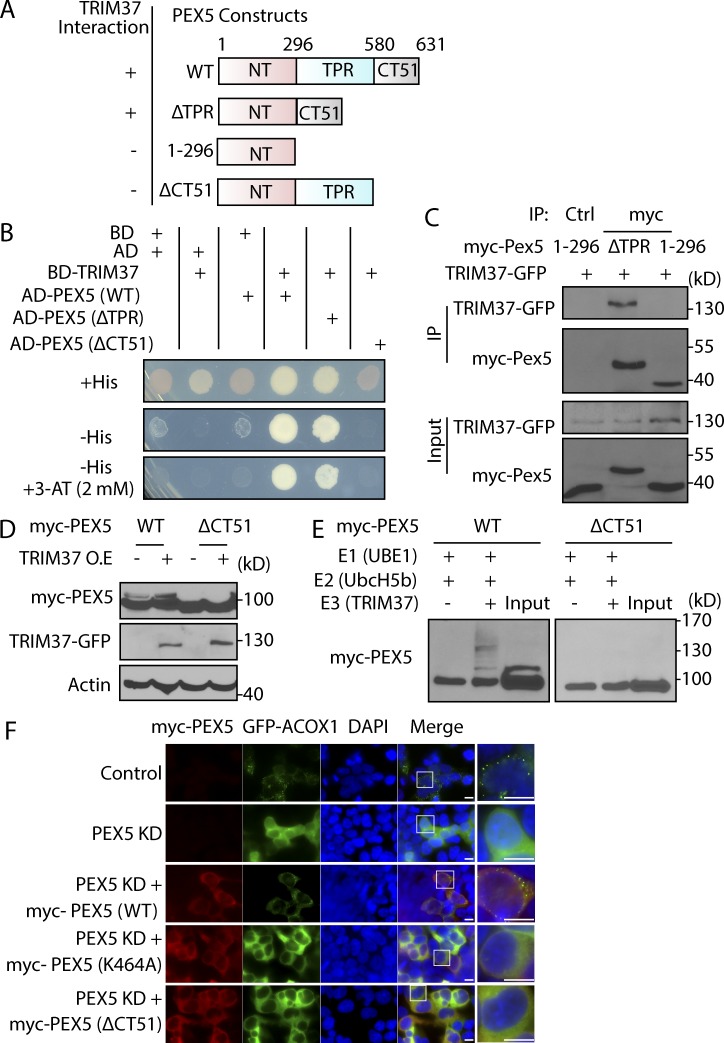
To explore how TRIM37 regulates PEX5 ubiquitylation, we mapped the TRIM37 interaction region on PEX5 using the yeast two-hybrid system and co-immunoprecipitation (IP) assays. PEX5 protein can be divided into three segments: the structurally disordered N-terminal region that binds to membrane peroxins (Saidowsky et al., 2001); a middle region comprising seven tetratricopeptide repeats (TPRs), which recognize the PTS1 cargoes (Gatto et al., 2000); and a CT51 region with no attributed function (Fig. 5 A). TRIM37 was fused to the GAL4-binding domain (BD) and several forms of PEX5 to the activation domain (AD), and their positive interactions were determined by growth in media without histidine (−His) and −His + 2 mM 3-amino-1,2,4-triazole (3-AT; more stringent condition). Cells transformed with BD-TRIM37 and AD-PEX5 (WT) grew on the −His and −His + 2 mM 3-AT selection plates (Fig. 5 B), indicative of their direct interaction. The AD-PEX5 (ΔTPR) mutant also interacted with BD-TRIM37; however, the CT51 deletion (ΔCT51) fully disrupted their interaction (Fig. 5, A and B), suggesting that CT51 contributes to the interaction of PEX5 with TRIM37.
Figure 5.
TRIM37 ubiquitylates PEX5 by interacting with CT51 of PEX5, which is required for peroxisomal matrix protein import. (A) Diagram of PEX5 constructs and summary of their interactions with TRIM37. (B) TRIM37 and PEX5 (WT or the indicated mutants) were fused to the GAL4-BD and -AD, respectively, and transformed into the yeast AH109 strain. (C) HEK 293T cells were transfected with TRIM37-GFP together with the indicated myc-PEX5 mutants. Cell lysates (input) were collected for myc-IP or control IgG-IP (Ctrl), followed by Western blot with the indicated antibodies. (D) HEK 293T cells were transfected with GFP(−) or TRIM37-GFP(+) together with constructs expressing myc-PEX5 WT or ΔCT51. Cells were collected for the indicated antibody analysis 24 h after transfection. (E) In vitro ubiquitylation assay using myc-PEX5 (WT) or (ΔCT51), UBE1, UbcH5B, and TRIM37. (F) pCMV-myc and GFP-ACOX1 constructs were transfected into HepG2 control and PEX5 KD cells. shRNA-resistant myc-PEX5 (WT), (K464A), or (ΔCT51) constructs, together with GFP-ACOX1, were transfected into HepG2 PEX5 KD cells. Myc antibody was used to stain overexpressed myc-PEX5. The localization of GFP-ACOX1 was observed by fluorescence microscopy. Bars: 10 µm.
The in vivo interactions between an E3 ligase and its substrate are very transient (Iconomou and Saunders, 2016), so not surprisingly, we were unable to detect the interaction between TRIM37 and PEX5 (WT) by co-IP assay (not depicted). Surprisingly, however, the interaction with TRIM37 was stabilized in the PEX5 (ΔTPR) mutant, as seen in our co-IP experiments (Fig. 5 C), and this interaction was abolished when CT51 was deleted (PEX5 1–296) (Fig. 5 C), further confirming that CT51 is required for PEX5 interaction with TRIM37.
Next, we asked whether the interacting region (CT51) was required for PEX5 ubiquitylation by TRIM37. As expected, deletion of CT51 (ΔCT51) abolished PEX5 ubiquitylation, and even TRIM37 overexpression, which enhanced ubiquitylation of PEX5 (WT), failed to restore PEX5 (ΔCT51) ubiquitylation in vivo (Fig. 5 D). Additionally, TRIM37 did not ubiquitylate PEX5 (ΔCT51) in vitro (Fig. 5 E), demonstrating that TRIM37 ubiquitylates PEX5 by interacting with the CT51 region.
To address whether the PEX5 (K464A) and PEX5 (ΔCT51) mutants retain their ability to import PTS proteins, we built shRNA-resistant PEX5 constructs (WT, K464A, and ΔCT51) and reexpressed them in PEX5 KD cells, respectively (Fig. 5 F). As expected, PEX5 KD resulted in cytoplasmic GFP-ACOX1 proteins, and reexpressing PEX5 (WT) rescued the import defect. However, neither PEX5 (K464A) nor PEX5 (ΔCT51) complemented the import defect (Fig. 5 F). These data strongly support our conclusion that TRIM37-mediated PEX5 ubiquitylation is essential for PTS protein import.
TRIM37 inhibits proteasomal degradation of PEX5
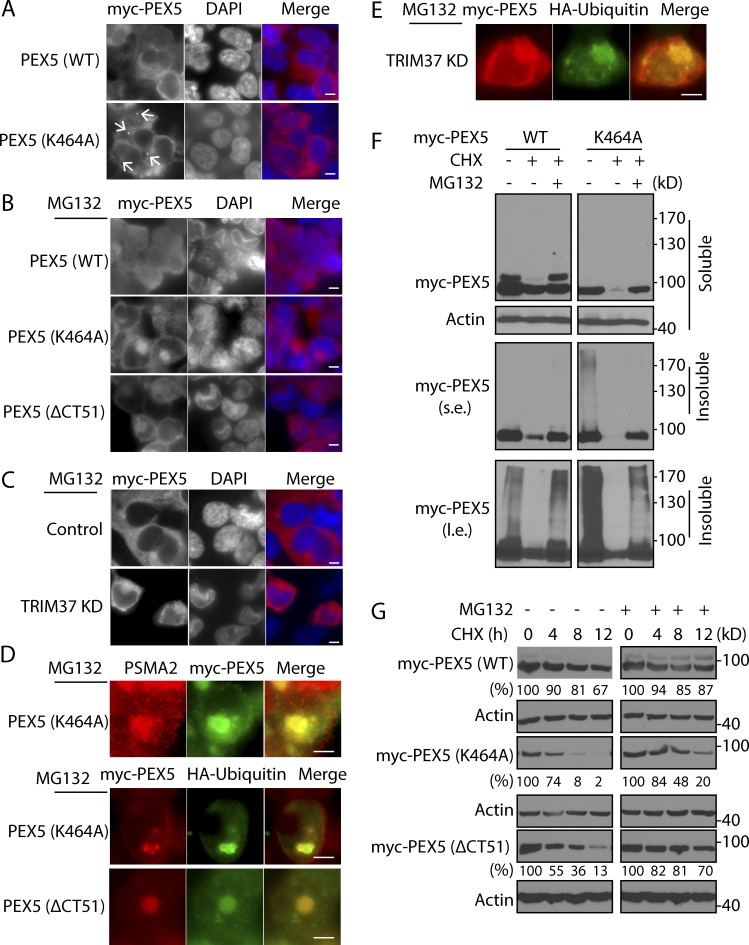
To understand the role of TRIM37-mediated ubiquitylation of PEX5 in the matrix protein import cycle, we studied the localization of PEX5 (K464A), which is not a substrate of TRIM37. As expected, PEX5 (WT) was cytosolic, but interestingly, the PEX5 (K464A) localized, in addition to the cytosol, in a single dot near the perinuclear region (Fig. 6 A, arrows), which resembles the cytoplasmic proteasome center (Wójcik and DeMartino, 2003). To further study the localization of the nonubiquitylated PEX5 pool, we localized PEX5 mutants, as well as the PEX5 (WT), in cells depleted of TRIM37 but in the presence of the proteasome inhibitor MG132, which blocks proteasomal degradation and induces the formation of a characteristic large aggresome near the perinuclear region (Wójcik and DeMartino, 2003). Confirming our hypothesis, in the presence of MG132, the PEX5 (K464A) and (ΔCT51) mutants that could not be ubiquitylated by TRIM37 localized mostly in a large aggregate near the nucleus, whereas these aggregates were far less abundant for PEX5 (WT) (Fig. 6 B). Notably, these PEX5 aggregates were observed in most TRIM37-depleted cells (Fig. 6 C). Finally, we confirmed that these large aggregates containing PEX5 (K464A or ΔCT51) or PEX5 (WT) in TRIM37 KD cells colocalized with ubiquitin aggregates and/or the proteasome marker PSMA2 (Fig. 6, D and E).
Figure 6.
TRIM37 inhibits polyubiquitylation-dependent proteasomal degradation of PEX5. (A) HEK 293T cells were transfected with myc-PEX5 (WT) or PEX5 (K464A) and immunostained with myc antibody. The arrows indicate the proteasome-like structure in myc-PEX5 (K464A)–expressing cells. (B and D) HEK 293T cells transfected with myc-PEX5 (WT), PEX5 (K464A), or PEX5 (ΔCT51) and HA-ubiquitin constructs were treated with 10 µM MG132 for 8 h. PEX5 was immunostained with myc antibody. Cells expressing PEX5 (K464A or ΔCT51) mutants were also costained with myc antibody and PSMA2 or HA antibody to localize the proteasomes and ubiquitin aggregates, respectively. (C and E) Myc-PEX5 and HA-ubiquitin were cotransfected into HepG2 control and TRIM37 KD cells. 24 h later, cells were treated with 10 µM MG132 for 8 h and stained with myc antibody. TRIM37 KD cells were also costained with myc and HA antibody to detect the colocalization of myc-PEX5 with ubiquitin aggregates. (F) HEK 293T cells transfected with myc-PEX5 (WT) or (K464A) were treated with 50 µg/ml CHX in the absence or presence of 10 µM MG132 for 6 h. Cell lysates were collected in 1% CHAPS buffer and centrifuged at 12,000 rpm for 5 min. The supernatants and pellets were collected as soluble and insoluble fractions, respectively. s.e., short exposure; l.e., long exposure. (G) Myc-PEX5 (WT), (K464A), or (ΔCT51) was transfected into HepG2 cells. 24 h later, cells were treated with 50 µg/ml CHX in the absence or presence of 10 µM MG132 for the indicated times. The quantifications of PEX5 protein (in soluble fractions) relative to actin are shown below myc-PEX5 blots. PEX5 protein amount at time 0 in each group was set to 100%. Bars: 5 µm.
We characterized in vivo PEX5 ubiquitylation by separating the cytosolic (soluble fraction) and proteasome-associated PEX5 (insoluble fraction; Fig. 6 F). The soluble fraction contained most, if not all, the monoubiquitylated PEX5 (WT), and the insoluble fraction contained some polyubiquitylated PEX5 species, as seen by the smeared, higher molecular weight bands (Fig. 6 F, left). The protein synthesis inhibitor cycloheximide (CHX) caused a small but significant decrease of PEX5 (WT) proteins and the disappearance of the smeared bands, but these effects were reversed by MG132. These results suggest that PEX5 is subject to a slow rate of proteasomal degradation in a polyubiquitylation-dependent manner. Interestingly, the K464A mutant appeared even more heavily polyubiquitylated (presumably at other ubiquitin acceptor sites in PEX5) relative to PEX5 (WT) in the basal state and therefore manifested a robust degradation by proteasomes because CHX treatment resulted in an almost complete loss of the mutant protein, which was reversed by MG132 (Fig. 6 F, right). These results suggested that monoubiquitylation of PEX5 at K464 by TRIM37 might have a stabilizing function because mutation of this site in PEX5 causes proteasomal degradation by polyubiquitylation at other sites, reminiscent of a mammalian equivalent of the yeast RADAR pathway that eliminates nonfunctional PEX5 from peroxisomes (Kiel et al., 2005; Léon et al., 2006).
To further validate this role of TRIM37-mediated monoubiquitylation of PEX5 as a stabilizing modification, we studied the half-life of PEX5 (WT) and the mutants. After inhibition of new protein synthesis by CHX, the half-lives of PEX5 (K464A) and PEX5 (ΔCT51) mutants were greatly reduced relative to that of WT PEX5 (Fig. 6 G). For PEX5 (WT) and both mutants, the half-lives were extended upon MG132 addition, showing that PEX5 is subject to proteasomal degradation (Fig. 6 G). Interestingly, we found that monoubiquitylated PEX5 in myc-PEX5 (WT) proteins and TRIM37 also decreased after CHX treatments, which were inhibited by MG132 treatment (Fig. 6 G, top; and Fig. S4 A). Considering the stabilizing role of TRIM37, this decrease of TRIM37 might cause the disappearance of the monoubiquitylated form of PEX5, which is inhibited when TRIM37 is stabilized by MG132.
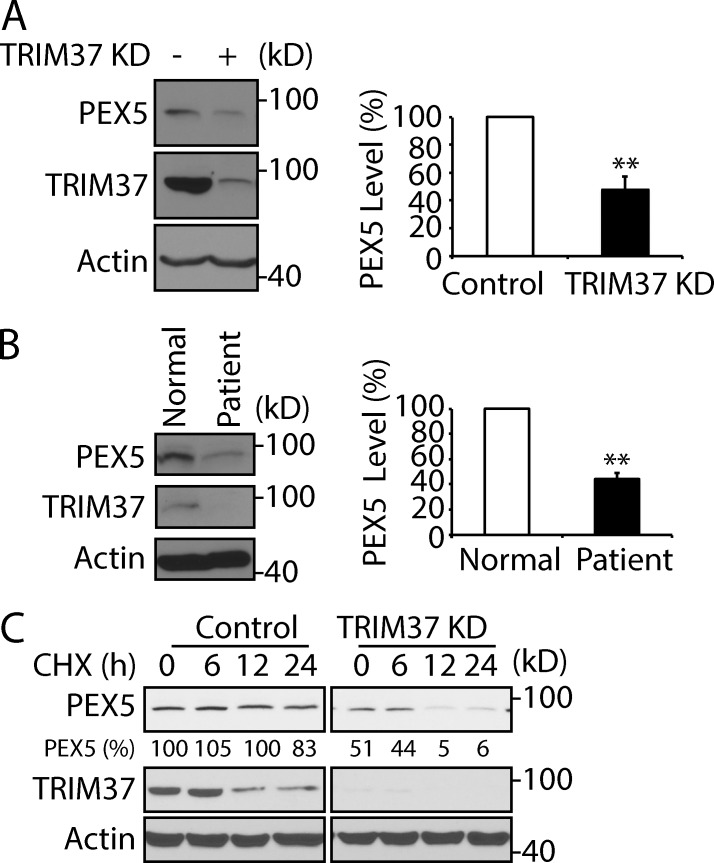
In agreement with the results obtained with these mutants, both TRIM37 depletion in HepG2 cells and TRIM37 mutation in patient cells resulted in a 40–50% reduction of endogenous PEX5 protein under normal growth conditions (Fig. 7, A and B). The half-lives of endogenous PEX5 proteins were strikingly reduced in TRIM37 KD cells compared with its slow degradation in control cells (Fig. 7 C). These multiple lines of evidence show clearly that K464 ubiquitylation in PEX5 by TRIM37 inhibits proteasome degradation of PEX5 and enhances its stability. Thus, TRIM37 is a positive regulator of PTS protein import and a negative regulator of proteasomal PEX5 turnover.
Figure 7.
Loss of TRIM37 reduces stability of PEX5 proteins. (A and B) PEX5 and TRIM37 protein levels in HepG2 control versus TRIM37 KD cells and in normal (AG21802) versus patient (AG02122) fibroblasts. Cell lysates were collected a day after cells were plated. Quantification was based on three independent experiments and presented as mean ± SEM (n = 3); **, P < 0.01 (Student’s t test). (C) PEX5 and TRIM37 stabilities in HepG2 control versus TRIM37 KD cells after 50 µg/ml CHX treatment for the indicated times. The amount of PEX5 proteins normalized to the loading control, actin, in control, and normal cells was set to 100%.
Binding of PEX5 to PTS1 cargoes does not require PEX5 ubiquitylation by TRIM37
The initial step in the peroxisomal import of PTS1-containing cargoes is the recognition and binding of the PTS in the cytoplasm by the TPR domain of PEX5 (aa 296–580; Gatto et al., 2000). TRIM37-mediated PEX5 ubiquitylation (K464) occurs in the TPR domain, so we asked whether this ubiquitylation might also affect PEX5 binding to its cargoes.
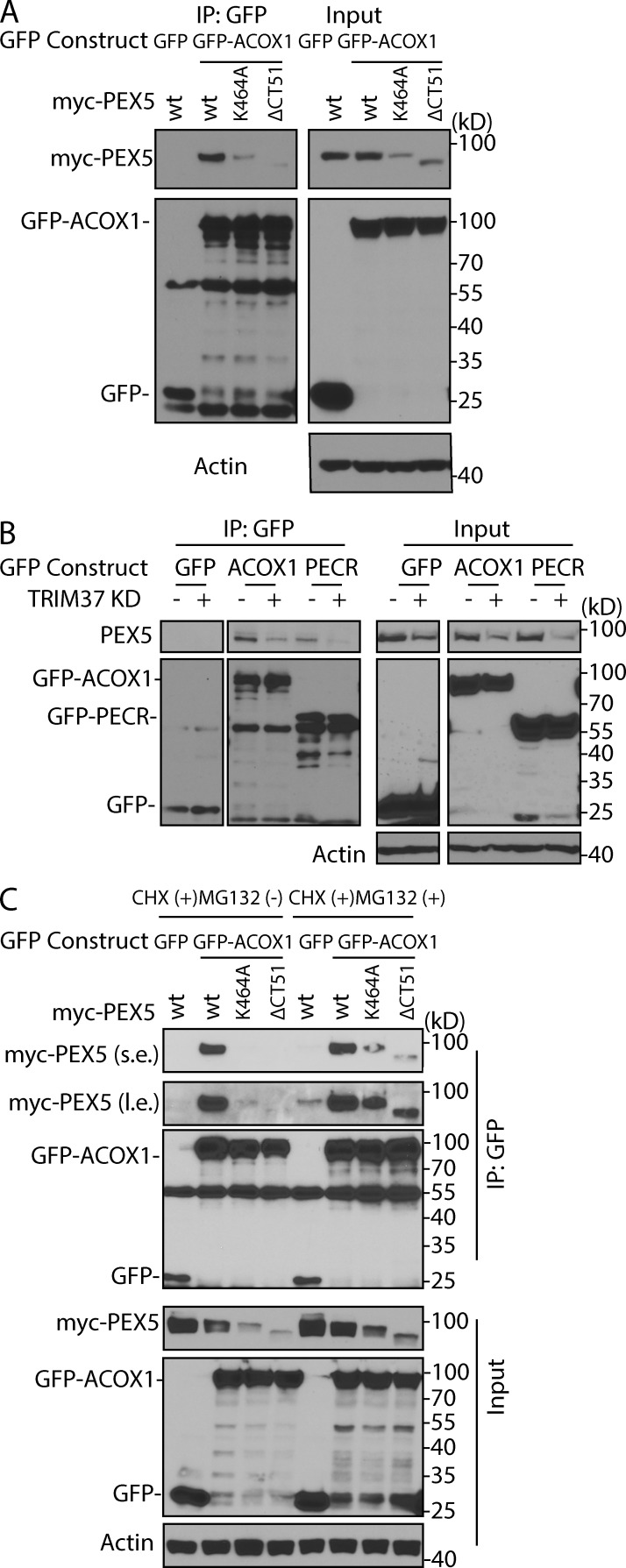
We characterized the affinity of PEX5 for different PTS1-containing cargoes (GFP-RFP-SKL, GFP-ACOX1, and GFP-PECR) and found that PEX5 bound strongly to ACOX1 (Fig. S4 B), making it the most suitable cargo for our studies. The binding of ACOX1 to PEX5 mutants (K464A and ΔCT51) was affected (Fig. 8 A, IP: GFP), but the relative differences in the binding were comparable with relative differences in the levels of PEX5 (WT), (K464A), and (ΔCT51) in the protein lysates (Fig. 8 A, input). Similarly, the interaction between PEX5 and PTS cargoes (GFP-ACOX1 and GFP-PECR) in TRIM37-depleted cells was reduced (Fig. 8 B, IP: GFP), but this reduction also reflected the PEX5 levels in the protein lysates (Fig. 8 B, input). To prove that reduced interactions of PEX5 mutants with PTS cargoes were caused by variations in PEX5 protein levels, MG132 was used to inhibit the CHX-induced degradation of PEX5 (K464A and ΔCT51) proteins (Fig. 8 C). The interactions of the two PEX5 mutants with ACOX1 were hardly detected (CHX+ and MG132−) but were rescued when their protein levels were increased after MG132 addition (Fig. 8 C, CHX+ and MG132+). Therefore, the cargo binding ability itself is not affected by PEX5 mutation or by TRIM37 depletion, and the differences seen in the co-IP are mainly a result of the relatively low levels of the PEX5 mutants or endogenous PEX5 proteins in TRIM37-depleted cells, which are caused by the proteasomal degradation.
Figure 8.
Binding of PEX5 to PTS1 cargoes does not require ubiquitylation by TRIM37. (A) HepG2 cells were transfected with GFP and myc-PEX5 (WT) (negative control) or GFP-ACOX1 together with myc-PEX5 (WT), (K464A), or (ΔCT51). Cell lysates were collected for GFP-IP and Western blot detection of their association with PEX5. (B) HepG2 control and TRIM37 KD cells were transfected with GFP, GFP-ACOX1, or GFP-PECR. GFP-IP was performed to detect their interactions with endogenous PEX5. (C) HepG2 cells transfected with the indicated constructs were treated with 50 µg/ml CHX in the absence or presence of 10 µM MG132 for 6 h. Cells were collected for GFP-IP. s.e., short exposure; l.e., long exposure.
TRIM37 or PEX5 depletion induces apoptosis and results in increased sensitivity to oxidative stress
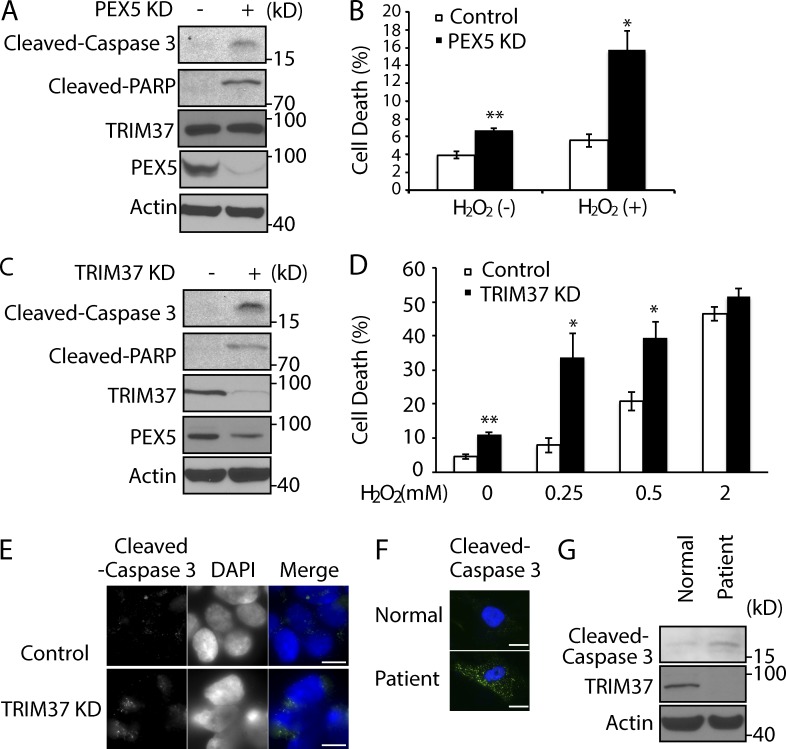
Supporting the essential cellular function of peroxisomes (Schrader and Fahimi, 2006), PEX5 depletion induced cell apoptosis, as indicated by the pronounced increase of the cleavage products of Caspase 3 and poly-ADP ribose polymerase and the percentage of dead cells detected by propidium iodide staining (Fig. 9, A and B). Not surprisingly, PEX5 depletion led to more cell death induced by oxidative stress (H2O2; Fig. 9 B). TRIM37 depletion also manifested a similar phenotype, with more cells undergoing apoptosis and enhanced sensitivity to H2O2 (Fig. 9, C and D). Immunostaining also showed that more cells underwent apoptosis after TRIM37 depletion, as indicated by the levels of cleaved Caspase 3 (Fig. 9 E). Consistently, basal apoptosis was also seen in fibroblasts of a patient with a TRIM37 gene mutation, as shown by immunostaining and Western blot detection for cleaved Caspase 3 proteins (Fig. 9, F and G). These results suggest that the PTS import defect caused by TRIM37 depletion might account for the apoptosis.
Figure 9.
TRIM37 or PEX5 depletion induces apoptosis and results in increased sensitivity to oxidative stress. (A and C) Lysates from HepG2 control versus PEX5 KD cells and control versus TRIM37 KD cells were immunoblotted for the indicated proteins. (B and D) HepG2 control versus PEX5 KD cells or control versus TRIM37 KD cells were either left untreated or were treated with 0.25 mM H2O2 (B) or the indicated H2O2 concentrations (C) for 3 h before propidium iodide staining for flow cytometry and cell death analysis. Data from triplicate samples in each group are presented as mean ± SEM and are representative of three independent experiments; *, P < 0.05; **, P < 0.01 (Student’s t test). (E and F) HepG2 control versus TRIM37 KD cells and normal versus patient fibroblast cells were immunostained with antibody that detects cleaved Caspase 3. (G) Cleaved Caspase 3 and TRIM37 levels in lysates from normal and patient cells (AG02122). Bars, 10 µm.
To address this directly, we overexpressed PEX5 protein in TRIM37 KD cells. As expected, the decrease of PEX5 protein by TRIM37 depletion was rescued in PEX5 overexpression in TRIM37 KD cells (Fig. S4 C). More importantly, the import of GFP-ACOX1 and GFP-PECR was also rescued in this cell line (Fig. S4 D). As a result, the cell apoptosis induced by TRIM37 KD was rescued by PEX5 overexpression, though partially, as indicated by a decrease of cleaved caspase 3 (Fig. S4 C). These results strongly indicate that the enhanced apoptosis caused by TRIM37 depletion depends, significantly but not completely, on PEX5-mediated peroxisomal protein import.
Discussion
We identified a novel E3 ligase, TRIM37, mediating PEX5 stability and consequently affecting PTS protein import into the peroxisomal matrix (Fig. 10). Several interesting results emerged from our studies. First, TRIM37 depletion in cancer cell lines or mutation in a mulibrey nanism patient impairs the import of peroxisomal matrix proteins, but not of PMPs, indicating that this disease is a new PBD. Second, TRIM37 ubiquitylates PEX5 at K464, which promotes efficient PTS import. Third, the deletion of the TRIM37-interacting region (CT51) results in loss of PEX5 ubiquitylation and a PTS protein import defect. Our mechanistic studies deciphered an unexpected role of TRIM37 in PEX5 ubiquitylation: it maintains PEX5 stability and abundance, and hence increases PTS cargo imports into the peroxisomal matrix. Thus, TRIM37 is a new positive regulator of PTS protein import.
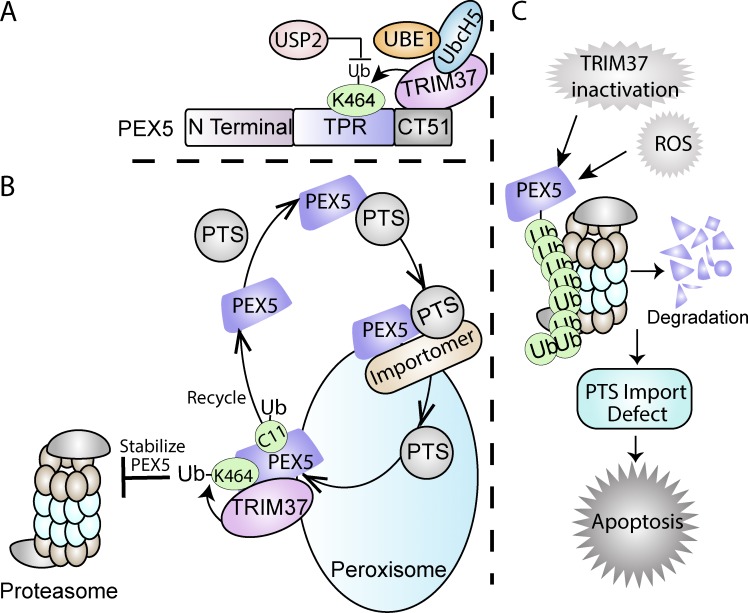
Figure 10.
Model for TRIM37-mediated PEX5 ubiquitylation. (A) TRIM37 ubiquitylates PEX5 at K464 by interacting with its CT51 in concert with the E1 enzyme, UBE1, and the E2 ubiquitin-conjugating protein UbcH5. The ubiquitylation at this site on PEX5 is reversed in vitro by USP2. (B) TRIM37-mediated monoubiquitylation of PEX5 at K464 stabilizes PEX5 by inhibiting its proteasome-mediated turnover via the RADAR pathway, thereby enhancing import of peroxisomal proteins. PEX5 plays a key role in the import of both PTS1- and PTS2-containing proteins into the peroxisomal matrix, and its ubiquitylation at C11 facilitates its recycling to the cytosol for multiple rounds of peroxisomal matrix protein import. (C) Multiple signals, such as the absence of TRIM37, PEX5 mutations that either prevent its interaction with (ΔCT51) or its ubiquitylation (K464A) by TRIM37, or high levels of reactive oxygen species relieve the negative regulation by TRIM37 of the mammalian RADAR pathway, making PEX5 susceptible to polyubiquitylation at other sites, followed by its proteasomal degradation. This depletion of PEX5 impairs peroxisomal matrix protein import and enhances cell apoptosis.
Polyubiquitylation occurs at one or two lysine residues near the N terminus of PEX5 in yeast, which serves as a quality-control system (also called the RADAR pathway; Léon et al., 2006) that prevents accumulation of the nonfunctional PEX5 at the peroxisomal membrane (Kiel et al., 2005). PEX5 stability is decreased in several human PBD patient cells (Dodt and Gould, 1996; Yahraus et al., 1996) and also in mulibrey nanism patients, suggesting that a similar quality-control mechanism may also exist in mammals. A very small amount of PEX5 (WT) is polyubiquitylated and undergoes a relatively slow proteasomal degradation under basal conditions, but this rate is dramatically enhanced when PEX5 ubiquitylation is inhibited by TRIM37 dysfunction, a property expected of a quality-control system. Additionally, we found that PEX5 is subject to enormous polyubiquitylation and degradation under oxidative stress (H2O2 treatment), a condition that disfavors peroxisomal matrix protein import (Fig. S5, A–C; Apanasets et al., 2014). These results suggest that there is also a proteasomal quality-control system in mammals to eliminate nonfunctional PEX5 proteins, which is negatively regulated by TRIM37 (Fig. 10 C). Interestingly, we found that TRIM37 protein was remarkably reduced in a CG4 (pex6 mutation) fibroblast cell line, in which PEX5 protein was also reduced (Fig. S5 D; Dodt and Gould, 1996; Yahraus et al., 1996), further supporting a role for TRIM37 in stabilizing PEX5 protein.
Exactly what the relative roles are for the proteasome versus pexophagy, which relies on PEX5 monoubiquitylation at K209 (Zhang et al., 2015), in PEX5 stability is uncertain at present, but the massive polyubiquitylation of PEX5 induced by H2O2 treatment and detected in the insoluble fraction (Fig. S5 A) is more reminiscent of the yeast RADAR pathway, in which nonfunctional PEX5 proteins are polyubiquitylated and subjected to proteasomal degradation and whose mammalian counterpart has not been described clearly.
Mulibrey nanism is an autosomal recessive, prenatal onset growth disorder with characteristic dysmorphic features, cardiopathy, and hepatomegaly caused by TRIM37 gene mutation (Avela et al., 2000; Karlberg et al., 2004). Despite the severe clinical symptoms, the physiological consequence of TRIM37 dysfunction remains elusive. Our results indicate that mulibrey nanism is a new type of PBD. TRIM37 is a PMP whose depletion or mutation causes a peroxisomal matrix protein import defect. The effect is direct because our mechanistic experiments demonstrate that TRIM37 functions through regulating PEX5 ubiquitylation and abundance. Furthermore, TRIM37 dysfunction also causes cell apoptosis in cancer cell lines and patient fibroblasts, which may result in the growth problems observed in mulibrey nanism patients (Karlberg et al., 2004), similar to that of PBDs caused by PEX5 mutations.
In contrast to our results, normal peroxisomes were reported in fibroblasts from a mulibrey nanism patient (Kallijärvi et al., 2002). The discrepancy likely lies in the complexities associated with the specific mutations observed in patients and the actual cellular phenotype resulting from these mutations (Jagiello et al., 2003). The cells from the patient we studied have the same TRIM37 genomic mutation as reported elsewhere (Kallijärvi et al., 2002), but because individuals with identical splice site mutations can show variable levels of aberrant splicing and TRIM37 protein expression (Jagiello et al., 2003), we took care to analyze only patient cell lines devoid of TRIM37 protein. We did also see normal PTS protein import in cells from a second patient with the same splice site mutation, which we attributed to the presence of normal TRIM37 mRNA and some TRIM37 protein expressed in the patient (Fig. S5, E–H). These observations further demonstrate that TRIM37 mutation contributes to abnormal cellular processes and human diseases by aberrant splicing. They also suggest that mutations at the splicing site are not the sole determining factor for TRIM37 deficiency, and they act together with other proteins, such as splicing factors acting in trans, for the pathogenesis of mulibrey nanism.
Interestingly, peroxisomes appear to be normal in TRIM37 knockout mice (Kettunen et al., 2016). To resolve this apparent discrepancy, we silenced TRIM37 in a mouse cell line, Raw264.7. Despite the almost complete depletion of TRIM37 in this cell line (Fig. S5 I), localization of both peroxisomal matrix and membrane proteins was normal in TRIM37 KD cells, as indicated by the distribution of GFP-PECR and PMP22-GFP, respectively (Fig. S5 J). However, in contrast to our results with human cells, the PEX5 protein level was unchanged in TRIM37 KD Raw264.7 cells compared with the control cells (Fig. S5 I). These results, together with the data from TRIM37 knockout mice, suggest that TRIM37 functions differently in humans and mice.
Besides its functions in peroxisomal biogenesis, TRIM37 is also an E3 ligase for histone 2A and promotes breast cancer development (Bhatnagar et al., 2014). It has recently been identified as a protein that regulates centrosome assembly (Balestra et al., 2013; Meitinger et al., 2016). These results may reflect the diverse functions of TRIM37. The links, if any, between these alternative functions of TRIM37 remain to be discovered.
Materials and methods
Reagents
The antibodies used were as follows: TRIM37 (sc-515044; Santa Cruz Biotechnologies), ECH1 (sc-515270; Santa Cruz Biotechnologies), ACOT1/2 (sc-373917; Santa Cruz Biotechnologies), GSTK1 (sc-515580; Santa Cruz Biotechnologies), ubiquitin (sc-8017; Santa Cruz Biotechnologies), PMP70 (sc-514728; Santa Cruz Biotechnologies), LAMP1 (sc-20011; Santa Cruz Biotechnologies), VDAC1 (sc-390996; Santa Cruz Biotechnologies), Sec61-β (sc-393633; Santa Cruz Biotechnologies), PSMA2 (2455; Cell Signaling Technology), cleaved Caspase 3 (9664; Cell Signaling Technology), cleaved poly-ADP ribose polymerase (5625; Cell Signaling Technology), GAPDH (2118; Cell Signaling Technology), actin (A1978; Sigma-Aldrich), myc (C3956; Sigma-Aldrich), PEX14 (ab109999; Abcam), HA (11867423001; Roche), GFP (632593; Clontech), and histidine (34660; QIAGEN). Antibodies against PEX5 and PTS1 were previously described by Wiemer et al. (1995). The inhibitors used were as follows: CHX (C104450; Sigma-Aldrich) and MG132 (C2211; Sigma-Aldrich). Components for in vitro ubiquitylation were purchased from Boston Biochem: UBE1 (E-305), E2 enzyme set (K-980B), and ubiquitin (U-100H). USP2 was provided by E. Bennett (University of California, San Diego).
Plasmids
The cDNA encoding TRIM37 was PCR amplified using a TRIM37 cDNA clone (TCH1003; Transomic) as a template and cloned into pEGFP-N1 (TRIM37-GFP) and pEGFP-C1 (GFP-TRIM37), respectively. pCMV-myc-PEX5 (WT) and (K209R) were gifts from C. Walker (Center for Translational Cancer Research, Texas A&M University, College Station, TX; Zhang et al., 2015). The PEX5 lysine mutants were constructed by PCR-directed mutagenesis using pCMV-myc-PEX5 (WT) as a template. PEX5 mutants encoding different domains were cloned into pCMV-myc plasmids. ACOX1 cDNA was cloned into pEGFP-C1 plasmid with the GFP tag at the N terminus. GFP-PECR, GFP-SKL, GFP-RFP-SKL, and thiolase-GFP were obtained as previously described (Amery et al., 2001; Mizuno et al., 2008; Nazarko et al., 2014). PMP22-GFP was provided by G. Luers (Institute of Anatomy, University of Marburg, Marburg, Germany). The cDNAs encoding TRIM37 or PEX5 were cloned into a pETDuet-1 vector and fused with a histidine and an S tag at the N and C termini, respectively. The shRNA-resistant PEX5 (WT), (K464A), or (ΔCT51) mutants were constructed by PCR-directed mutagenesis. HA-ubiquitin (WT) and HA-ubiquitin (KO) were purchased from Addgene (17608 and 17603). All constructs were sequenced.
Cell culture and DNA transfection
HEK 293T cells (ATCC), Raw264.7 (ATCC), and HepG2 (Zhang et al., 2015) were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin. Fibroblasts from healthy donors (AG21802) and two mulibrey nanism patients (AG02122 and AG02506) were obtained from the Coriell Institute and cultured in Eagle’s minimum essential medium supplemented with 15% FBS. All cells were incubated at 37°C in a humidified 5% (vol/vol) CO2 incubator. Cell transfection was performed by using X-tremeGENE HP DNA Transfection Reagent (6366546001; Sigma-Aldrich) according to the manufacturer’s instructions.
Subcellular fractionation
Fractionation assays were performed as described by Otera et al. (2000). In brief, ∼108 cells were collected in homogenization buffer (0.25 M sucrose and 5 mM Hepes-KOH, pH 7.4) with protease inhibitor cocktail (05892791001; Roche), followed by 30 strokes of an Elvehjem-Potter homogenizer. Postnuclear supernatant (PNS) fractions were obtained by centrifugation of homogenates at 750 g for 10 min. The PNSs were subjected to ultracentrifugation at 100,000 g for 40 min. The supernatants were collected as cytosolic fractions (S). The organelle pellet fractions (P; heavy and light mitochondrial and microsomal fractions) were suspended in urea buffer (120 mM Tris-HCl, pH 6.8, 4% SDS, 8 M urea, 0.1 M DTT, and 0.01% bromophenol blue). To dissect the subcellular localization of peroxisomal matrix and membrane proteins, the pellet fractions were extracted with either alkaline carbonate buffer (100 mM Na2CO3 and 10 mM Tris-HCl, pH 11.5) or detergent buffer (2% Triton X-100 and 1 M NaCl) and incubated for 30 min on ice, followed by ultracentrifugation at 100,000 g for 40 min. Supernatants and pellets were collected.
Protease degradation assay
TRIM37-GFP or GFP-TRIM37 constructs were transfected into HEK 293T cells. PNSs were separated into cytosolic and organelle fractions by centrifugation at 25,000 g. P fractions were treated with 150 µg/ml proteinase K plus 300 µg/ml trypsin in the presence or absence of 1% Triton X-100, or untreated in homogenization buffer on ice. The samples were collected at each time point by taking the samples into a TCA solution (13% final concentration) and stored at −80°C. Proteins were extracted by TCA precipitation (Wang and Subramani, 2017). In brief, the cell lysates in TCA solutions were centrifuged at 12,000 rpm for 5 min. The protein pellets were suspended and washed with 80% ice-cold acetone twice, followed by a Western blot analysis of the proteins.
Peroxisome isolation using density gradient
Peroxisomes were isolated by using a commercial kit (PEROX1; Sigma-Aldrich). PNSs from HepG2 cells were fractionated differentially into S and P fractions by centrifugation at 25,000 g. An Optiprep solution (22.5%) containing the P fraction was placed in the middle layer with 27.5 and 20% Optiprep solutions on the top and bottom, respectively, followed by ultracentrifugation at 100,000 g. Other organelle proteins such as ER and lysosomes floated on the top, and mitochondria proteins resided at the 22.5/27.5% interface, whereas peroxisomes slightly floated in the densest layer.
Immunofluorescence
Cells were grown on cover slides and fixed with 4% PFA for 10 min, followed by 5 min of 0.5% Triton X-100 permeabilization and 0.5 h of blocking with 1% BSA in PBS at room temperature. To detect the localization of TRIM37 in peroxisomes, cells were pretreated with 25 µg/ml digitonin for 5 min followed by the same procedures as described previously. The slides were then incubated with primary antibodies and the corresponding fluorescent dye–conjugated secondary antibodies for 2 h and 1 h, respectively. Nuclei were stained with DAPI (1 µg/ml). Images were captured using a plan apochromat 100× 1.40-NA oil immersion objective on a motorized fluorescence microscope (Axioskop 2 MOT plus; Carl Zeiss) coupled to a monochrome digital camera (AxioCam MRm; Carl Zeiss), and the pictures were processed using AxioVision software (Carl Zeiss).
In vitro ubiquitylation assay
The in vitro ubiquitylation assay was performed as described by Bhatnagar et al. (2014). PEX5 proteins were obtained either by myc-IP from myc-PEX5–overexpressing HEK 293T cells or by purification of bacterial histidine-PEX5 proteins. UBE1, different E2s, bacterially purified TRIM37 (E3), ubiquitin, myc-PEX5, histidine-PEX5 were added in a final 50 µl of buffer (50 mM Tris-Cl, pH 7.5, 5 mM MgCl2, 2 mM NaF, 10 mM DTT, and 2 mM ATP). The reactions were incubated at 30°C for 1.5 h and boiled with 2× SDS sample buffer for 5 min, followed by a Western blot detection of PEX5 ubiquitylation.
Yeast two-hybrid experiments
The GAL4-based Matchmaker yeast two-hybrid system (Clontech) was used as described previously by Farré et al. (2013). TRIM37 and PEX5 or PEX5 mutants were cloned into pGBKT7 (BD) and pGADT7 (AD) vector, respectively. The BD and AD plasmids were cotransformed into AH109 strains using synthetic dextrose (SD) lacking specified amino acids, i.e., SD (Leu−, Trp−) plates. Transformants were then grown on plates with His (SD [Leu−, Trp−]), −His (SD [Leu−, Trp−, His−]), and −His + 3-AT (SD [Leu−, Trp−, His−] with 2 mM 3-AT). The strains were incubated at 30°C until visible colonies appeared. Two transformants from each strain were tested. Transformants growing on −His and −His + 3-AT plates indicated the direct interaction of proteins expressed from the two constructs. 3-AT was used to reduce background growth.
Protein purification
The constructs pETDuet-1-TRIM37 or pETDuet-1-PEX5 were transformed into BL21 E. coli. Protein expression was induced by 0.2 mM IPTG at 22°C for 3 h. To solubilize PEX5, cell pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail). To solubilize TRIM37, cell pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.5% N-lauroylsarcosine, and protease inhibitor cocktail). The cell lysates were subjected to ultrasonication at 4°C, and cleared lysates were incubated with nickel–nitrilotriacetic acid agarose (30230; QIAGEN) at 4°C for 3 h. For purification of TRIM37, cleared lysates were diluted three times with buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2% Triton X-100) before nickel–nitrilotriacetic acid agarose binding. Beads were washed once with buffer (50 mM Tris-HCl, pH 7.5, and 20 mM imidazole) and eluted with buffer (50 mM Tris-HCl, pH 7.5, and 250 mM imidazole). Eluents were dialyzed against 20 mM Tris-HCl, pH 7.5.
Western blot and IP
Cells were collected in lysis buffer (40 mM Hepes, pH 7.4, 120 mM NaCl, 1 mM EDTA, 1.0% CHAPS, 10 mM pyrophosphate, 10 mM glycerophosphate, and protease inhibitor cocktails). The lysates were centrifuged, and supernatants were collected for Western blot analysis. To detect PEX5 polyubiquitylation in the insoluble fractions, the pellets were resuspended in urea buffer (one fifth of the volume of the 1.0% CHAPS lysis buffer). Equal volumes of the soluble and insoluble fractions were used for the immunoblots.
To study interactions between TRIM37 and PEX5 or the mutants, HEK 293T cells were transfected with TRIM37-GFP and myc-PEX5 (WT) or myc-PEX5 mutants (ΔTPR or 1–296). Cells were collected in lysis buffer (40 mM Hepes, pH 7.4, 120 mM NaCl, 1 mM EDTA, 0.3% CHAPS, 10 mM pyrophosphate, 10 mM glycerophosphate, and protease inhibitor cocktails). To study interactions between PEX5 and PTS cargoes, cells transfected with myc-PEX5 and various GFP-PTS constructs were collected in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2% Triton X-100, 10% glycerol, and protease inhibitors). Equal amounts of protein lysates (500 µg) were incubated with the corresponding IP antibody followed by addition of protein G beads. The IP beads were washed three times with lysis buffer and boiled for 10 min in 40 µl of 2× SDS sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 0.2 M DTT, and 0.02% bromophenol blue). The IP lysates were subjected to Western blot detection of the interacting proteins. To detect PEX5 ubiquitylation, cells transfected with HA-ubiquitin were collected in the denaturing lysis buffer (2% SDS, 150 mM NaCl, and 10 mM Tris-HCl, pH 8.0) as described previously by Choo and Zhang (2009), and the HA-IP was performed to detect the endogenous PEX5 by Western blot.
Generating stable KD cells
The shRNA constructs to KD TRIM37 or PEX5 were in pLKO.1 backbones purchased from Open Biosystems (RHS4533-EG4591 and RHS4533-EG5830). The two shRNA sequences targeting the TRIM37 gene were shRNA #1 (5′-TTCGAGAATATGATGCTGTGG-3′) and shRNA #2 (5′-TTCACTGGTAAAGTCTGGTGG-3′). The shRNA sequence targeting PEX5 was 5′-AATCCTGAGTTACATCCACAG-3′. The lentivirus system was used to establish KD stable cell lines as follows: the TRIM37 or PEX5 shRNA constructs were transfected together with the lentivirus packaging plasmids psPAX2 and pMD2.G. The virus supernatants were collected and filtered for target cell infection (HepG2 and HEK 293T cells). Surviving cell populations after puromycin selection (2 µg/ml) were subjected to KD analysis by Western blot. The TRIM37 shRNA #1 was also used for TRIM37 KD Raw264.7 cells because the mouse TRIM37 gene has the same targeting site.
Rescuing the TRIM37 KD cells
To confirm that the phenotype observed in the TRIM37 KD cells was caused by loss of the TRIM37 protein, HepG2 cells were infected with retroviruses expressing either the pBrit-empty vector or pBrit-TRIM37 cDNA (shRNA resistant) to select stable cells using 0.5 µg/ml puromycin. The pooled cells expressing the pBrit-empty vector were then infected with lentivirus expressing either a control shRNA or TRIM37 shRNA, and cells growing in 2 µg/ml puromycin were pooled as control and TRIM37 KD cells, respectively. Cells expressing shRNA-resistant TRIM37 cDNA were infected with lentivirus expressing TRIM37 shRNA, and stable cells growing in 2 µg/ml puromycin were pooled (short hairpin–resistant TRIM37 and TRIM37 KD). After 5 d, all three pooled stable cells were individually transfected with a plasmid transiently expressing GFP-ACOX1 cDNA and processed after 24 h for immunoblots of TRIM37 levels and for fluorescence localization of GFP-ACOX1.
DNA extraction and RT-PCR
Fibroblast cells were collected and the DNA was extracted by using the Wizard Genomic DNA Purification kit (A1125; Promega). The region surrounding the genomic mutation site (c.493-2A>G) was amplified by PCR with primers (forward, 5′-AGGAATCAAGAACCAAGT-3′; and reverse, 5′-ATGGCTATGAAAATTTTTCT-3′) and sent for sequencing. To analyze the TRIM37 mRNA expression, RNA was extracted from these fibroblast cells by TRIzol reagent (Invitrogen) and reverse transcribed according to the manufacturer’s instructions (4374966; Thermo Fisher). The cDNA was used as a template for amplification of the region surrounding the splicing mutation sites by PCR (forward, 5′-CCTTTAAACCTTTGGCAG-3′; and reverse, 5′-TGGTGCTCCACCTCCTGA-3′).
Flow cytometry
Cells were treated or not treated with different doses of H2O2 (H1009; Sigma-Aldrich) added directly into the culture media and digested to single cells using 0.25% trypsin. Cells were stained with 1 µg/ml propidium iodide in PBS. Channel FL3 (deep red fluorescence, excitation of 488 nm, and emission 670 long pass) was used to detect the propidium iodide signal using a FACSCalibur flow cytometer (Becton-Dickinson).
Statistics
P-values were calculated using a two-tailed unpaired Student’s t test. P-values of <0.05 were considered statistically significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Online supplemental material
Fig. S1 (related to Fig. 2) shows that the loss of TRIM37 impairs the import of peroxisomal matrix proteins. Fig. S2 (related to Fig. 3) shows that TRIM37 gene mutation results in the defective import of peroxisomal matrix proteins. Fig. S3 (related to Fig. 4) shows that TRIM37 ubiquitylates PEX5 at K464. Fig. S4 (related to Figs. 6, 8, and 9) and Fig. S5 present data for the Discussion section.
Acknowledgments
We thank all laboratory members for useful discussions.
This work was supported by National Institutes of Health grant 5RO1 DK41737 to S. Subramani.
The authors declare no competing financial interests.
Author contributions: W. Wang designed the experiments. W. Wang and Z.-J. Xia performed experiments and analyzed the results. J.-C. Farré provided critical advice and assistance for microscopy operation. S. Subramani guided and supervised the project. W. Wang, J.-C. Farré, and S. Subramani wrote the paper with input from all authors.
Footnotes
Abbreviations used:
- AD
- activation domain
- BD
- binding domain
- CG
- complementation group
- CHX
- cycloheximide
- fs
- frame shift
- IP
- immunoprecipitation
- KD
- knockdown
- mulibrey
- muscle–liver–brain–eye
- PBD
- peroxisomal biogenesis disorder
- PMP
- peroxisomal membrane protein
- PNS
- postnuclear supernatant
- PTS
- peroxisomal targeting signal
- RADAR
- receptor accumulation and degradation in the absence of recycling
- SD
- synthetic dextrose
- TPR
- tetratricopeptide repeat
References
- Amery L., Mannaerts G.P., Subramani S., Van Veldhoven P.P., and Fransen M.. 2001. Identification of a novel human peroxisomal 2,4-dienoyl-CoA reductase related protein using the M13 phage protein VI phage display technology. Comb. Chem. High Throughput Screen. 4:545–552. 10.2174/1386207013330832 [DOI] [PubMed] [Google Scholar]
- Apanasets O., Grou C.P., Van Veldhoven P.P., Brees C., Wang B., Nordgren M., Dodt G., Azevedo J.E., and Fransen M.. 2014. PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein. Traffic. 15:94–103. 10.1111/tra.12129 [DOI] [PubMed] [Google Scholar]
- Avela K., Lipsanen-Nyman M., Idänheimo N., Seemanová E., Rosengren S., Mäkelä T.P., Perheentupa J., Chapelle A.D., and Lehesjoki A.E.. 2000. Gene encoding a new RING-B-box-Coiled-coil protein is mutated in mulibrey nanism. Nat. Genet. 25:298–301. 10.1038/77053 [DOI] [PubMed] [Google Scholar]
- Balestra F.R., Strnad P., Flückiger I., and Gönczy P.. 2013. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell. 25:555–571. (published erratum appears in Dev. Cell. 2013. 26:220) 10.1016/j.devcel.2013.05.016 [DOI] [PubMed] [Google Scholar]
- Bhatnagar S., Gazin C., Chamberlain L., Ou J., Zhu X., Tushir J.S., Virbasius C.M., Lin L., Zhu L.J., Wajapeyee N., and Green M.R.. 2014. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 516:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman N., Dodt G., Gould S.J., and Valle D.. 1998. An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 7:1195–1205. 10.1093/hmg/7.8.1195 [DOI] [PubMed] [Google Scholar]
- Carvalho A.F., Pinto M.P., Grou C.P., Alencastre I.S., Fransen M., Sá-Miranda C., and Azevedo J.E.. 2007. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282:31267–31272. 10.1074/jbc.M706325200 [DOI] [PubMed] [Google Scholar]
- Choo Y.S., and Zhang Z.. 2009. Detection of protein ubiquitination. J. Vis. Exp. 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G., and Gould S.J.. 1996. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135:1763–1774. 10.1083/jcb.135.6.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G., Braverman N., Wong C., Moser A., Moser H.W., Watkins P., Valle D., and Gould S.J.. 1995. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 9:115–125. 10.1038/ng0295-115 [DOI] [PubMed] [Google Scholar]
- Ebberink M.S., Koster J., Visser G., Spronsen F., Stolte-Dijkstra I., Smit G.P., Fock J.M., Kemp S., Wanders R.J., and Waterham H.R.. 2012. A novel defect of peroxisome division due to a homozygous non-sense mutation in the PEX11β gene. J. Med. Genet. 49:307–313. 10.1136/jmedgenet-2012-100778 [DOI] [PubMed] [Google Scholar]
- Erdmann R. 2016. Assembly, maintenance and dynamics of peroxisomes. Biochim. Biophys. Acta. 1863:787–789. 10.1016/j.bbamcr.2016.01.020 [DOI] [PubMed] [Google Scholar]
- Farré J.C., Burkenroad A., Burnett S.F., and Subramani S.. 2013. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 14:441–449. 10.1038/embor.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto G.J., Jr., Geisbrecht B.V., Gould S.J., and Berg J.M.. 2000. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7:1091–1095. 10.1038/81930 [DOI] [PubMed] [Google Scholar]
- Gould S.J., Keller G.A., Hosken N., Wilkinson J., and Subramani S.. 1989. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108:1657–1664. 10.1083/jcb.108.5.1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grou C.P., Carvalho A.F., Pinto M.P., Wiese S., Piechura H., Meyer H.E., Warscheid B., Sá-Miranda C., and Azevedo J.E.. 2008. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J. Biol. Chem. 283:14190–14197. 10.1074/jbc.M800402200 [DOI] [PubMed] [Google Scholar]
- Honsho M., Yamashita S., and Fujiki Y.. 2016. Peroxisome homeostasis: Mechanisms of division and selective degradation of peroxisomes in mammals. Biochim. Biophys. Acta. 1863:984–991. 10.1016/j.bbamcr.2015.09.032 [DOI] [PubMed] [Google Scholar]
- Iconomou M., and Saunders D.N.. 2016. Systematic approaches to identify E3 ligase substrates. Biochem. J. 473:4083–4101. 10.1042/BCJ20160719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagiello P., Hammans C., Wieczorek S., Arning L., Stefanski A., Strehl H., Epplen J.T., and Gencik M.. 2003. A novel splice site mutation in the TRIM37 gene causes mulibrey nanism in a Turkish family with phenotypic heterogeneity. Hum. Mutat. 21:630–635. 10.1002/humu.10220 [DOI] [PubMed] [Google Scholar]
- Kallijärvi J., Avela K., Lipsanen-Nyman M., Ulmanen I., and Lehesjoki A.E.. 2002. The TRIM37 gene encodes a peroxisomal RING-B-box-coiled-coil protein: classification of mulibrey nanism as a new peroxisomal disorder. Am. J. Hum. Genet. 70:1215–1228. 10.1086/340256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallijärvi J., Lahtinen U., Hämäläinen R., Lipsanen-Nyman M., Palvimo J.J., and Lehesjoki A.E.. 2005. TRIM37 defective in mulibrey nanism is a novel RING finger ubiquitin E3 ligase. Exp. Cell Res. 308:146–155. 10.1016/j.yexcr.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Karlberg N., Jalanko H., Perheentupa J., and Lipsanen-Nyman M.. 2004. Mulibrey nanism: clinical features and diagnostic criteria. J. Med. Genet. 41:92–98. 10.1136/jmg.2003.014118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen K.M., Karikoski R., Hämäläinen R.H., Toivonen T.T., Antonenkov V.D., Kulesskaya N., Voikar V., Hölttä-Vuori M., Ikonen E., Sainio K., et al. . 2016. Trim37-deficient mice recapitulate several features of the multi-organ disorder Mulibrey nanism. Biol. Open. 5:584–595. 10.1242/bio.016246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel J.A., Emmrich K., Meyer H.E., and Kunau W.H.. 2005. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J. Biol. Chem. 280:1921–1930. 10.1074/jbc.M403632200 [DOI] [PubMed] [Google Scholar]
- Krause C., Rosewich H., Thanos M., and Gärtner J.. 2006. Identification of novel mutations in PEX2, PEX6, PEX10, PEX12, and PEX13 in Zellweger spectrum patients. Hum. Mutat. 27:1157 10.1002/humu.9462 [DOI] [PubMed] [Google Scholar]
- Léon S., Zhang L., McDonald W.H., Yates J. III, Cregg J.M., and Subramani S.. 2006. Dynamics of the peroxisomal import cycle of PpPex20p: Ubiquitin-dependent localization and regulation. J. Cell Biol. 172:67–78. 10.1083/jcb.200508096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Agrawal G., and Subramani S.. 2011. Peroxisome assembly: matrix and membrane protein biogenesis. J. Cell Biol. 193:7–16. 10.1083/jcb.201010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F., Anzola J.V., Kaulich M., Richardson A., Stender J.D., Benner C., Glass C.K., Dowdy S.F., Desai A., Shiau A.K., and Oegema K.. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 214:155–166. 10.1083/jcb.201604081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y., Kurochkin I.V., Herberth M., Okazaki Y., and Schönbach C.. 2008. Predicted mouse peroxisome-targeted proteins and their actual subcellular locations. BMC Bioinformatics. 9:S16 10.1186/1471-2105-9-S12-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko T.Y., Ozeki K., Till A., Ramakrishnan G., Lotfi P., Yan M., and Subramani S.. 2014. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J. Cell Biol. 204:541–557. 10.1083/jcb.201307050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K., Misono S., Miyata N., Matsumoto Y., Mukai S., and Fujiki Y.. 2011. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic. 12:1067–1083. [DOI] [PubMed] [Google Scholar]
- Okumoto K., Noda H., and Fujiki Y.. 2014. Distinct modes of ubiquitination of peroxisome-targeting signal type 1 (PTS1) receptor Pex5p regulate PTS1 protein import. J. Biol. Chem. 289:14089–14108. 10.1074/jbc.M113.527937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Harano T., Honsho M., Ghaedi K., Mukai S., Tanaka A., Kawai A., Shimizu N., and Fujiki Y.. 2000. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 275:21703–21714. 10.1074/jbc.M000720200 [DOI] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., and Erdmann R.. 2007. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177:197–204. 10.1083/jcb.200611012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidowsky J., Dodt G., Kirchberg K., Wegner A., Nastainczyk W., Kunau W.H., and Schliebs W.. 2001. The di-aromatic pentapeptide repeats of the human peroxisome import receptor PEX5 are separate high affinity binding sites for the peroxisomal membrane protein PEX14. J. Biol. Chem. 276:34524–34529. 10.1074/jbc.M104647200 [DOI] [PubMed] [Google Scholar]
- Schrader M., and Fahimi H.D.. 2006. Peroxisomes and oxidative stress. Biochim. Biophys. Acta. 1763:1755–1766. 10.1016/j.bbamcr.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Smith J.J., and Aitchison J.D.. 2013. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14:803–817. 10.1038/nrm3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S.J., Dodt G., Raymond G.V., Braverman N.E., Moser A.B., and Moser H.W.. 2006. Peroxisome biogenesis disorders. Biochim. Biophys. Acta. 1763:1733–1748. 10.1016/j.bbamcr.2006.09.010 [DOI] [PubMed] [Google Scholar]
- Swinkels B.W., Gould S.J., Bodnar A.G., Rachubinski R.A., and Subramani S.. 1991. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10:3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S., and Erdmann R.. 2006. Peroxisomal matrix protein receptor ubiquitination and recycling. Biochim. Biophys. Acta. 1763:1620–1628. 10.1016/j.bbamcr.2006.08.046 [DOI] [PubMed] [Google Scholar]
- Wang W., and Subramani S.. 2017. Assays to monitor pexophagy in yeast. Methods Enzymol. 588:413–427. 10.1016/bs.mie.2016.09.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham H.R., Ferdinandusse S., and Wanders R.J.. 2016. Human disorders of peroxisome metabolism and biogenesis. Biochim. Biophys. Acta. 1863:922–933. 10.1016/j.bbamcr.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Wiebel F.F., and Kunau W.H.. 1992. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature. 359:73–76. 10.1038/359073a0 [DOI] [PubMed] [Google Scholar]
- Wiemer E.A., Nuttley W.M., Bertolaet B.L., Li X., Francke U., Wheelock M.J., Anné U.K., Johnson K.R., and Subramani S.. 1995. Human peroxisomal targeting signal-1 receptor restores peroxisomal protein import in cells from patients with fatal peroxisomal disorders. J. Cell Biol. 130:51–65. 10.1083/jcb.130.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C., van den Berg M., Sprenger R.R., and Distel B.. 2007. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282:22534–22543. 10.1074/jbc.M702038200 [DOI] [PubMed] [Google Scholar]
- Wójcik C., and DeMartino G.N.. 2003. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35:579–589. 10.1016/S1357-2725(02)00380-1 [DOI] [PubMed] [Google Scholar]
- Yahraus T., Braverman N., Dodt G., Kalish J.E., Morrell J.C., Moser H.W., Valle D., and Gould S.J.. 1996. The peroxisome biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for stability of the PTS1 receptor. EMBO J. 15:2914–2923. [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T., Dere R., Tait-Mulder J., Lee J.H., Paull T.T., et al. . 2015. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 17:1259–1269. 10.1038/ncb3230 [DOI] [PMC free article] [PubMed] [Google Scholar]