PARP1/ARTD1 induces chromatin opening by posttranslational modification of the linker histone H1, but how PARP1 is targeted to physiologically correct gene loci is poorly understood. Hau et al. show that in differentiating neurons, PARP1 is rapidly and specifically recruited to a neuron-specific promoter by the atypical homeodomain protein MEIS2.
Abstract
Pre–B-cell leukemia homeobox (PBX) and myeloid ecotropic viral integration site (MEIS) proteins control cell fate decisions in many physiological and pathophysiological contexts, but how these proteins function mechanistically remains poorly defined. Focusing on the first hours of neuronal differentiation of adult subventricular zone–derived stem/progenitor cells, we describe a sequence of events by which PBX-MEIS facilitates chromatin accessibility of transcriptionally inactive genes: In undifferentiated cells, PBX1 is bound to the H1-compacted promoter/proximal enhancer of the neuron-specific gene doublecortin (Dcx). Once differentiation is induced, MEIS associates with chromatin-bound PBX1, recruits PARP1/ARTD1, and initiates PARP1-mediated eviction of H1 from the chromatin fiber. These results for the first time link MEIS proteins to PARP-regulated chromatin dynamics and provide a mechanistic basis to explain the profound cellular changes elicited by these proteins.
Introduction
All cellular differentiation and dedifferentiation events require the de novo activation of previously silent genes, which are typically embedded in “closed” chromatin and tightly packed with the linker histone H1. H1 stabilizes the chromatin fiber and actively promotes epigenetic silencing by recruiting DNA methyltransferases, interfering with ATP-dependent chromatin remodeling, and preventing the deposition of activating epigenetic marks (Saeki et al., 2005; Yang et al., 2013). Removal of H1 and the subsequent local chromatin decompaction are thus critical early steps in the transcriptional activation of dormant genes. ADP ribosylation, the transfer of ADP-ribose moieties from NAD+ onto H1 by poly-ADP-ribose (PAR) polymerase 1 (PARP1/ARTD1), decreases H1 affinity for DNA, and thereby facilitates a wide variety of chromatin-dependent processes, including transcription (Poirier et al., 1982; Tulin and Spradling, 2003; Kim et al., 2004). PARP1 modulates the chromatin structure in different ways: it can serve as a structural component of condensed chromatin and favor transcriptional repression in its enzymatically inactive form, but it also induces chromatin opening by poly-ADP-ribosylation (PARylation) of chromatin components including itself and H1 once activated (Tulin and Spradling, 2003; Kim et al., 2004). In addition, PARP1 prevents removal of activating H3K4me3 epigenetic marks, promotes loading of RNA pol II at the promoters of positively regulated target genes, and participates in transcription elongation (Krishnakumar and Kraus, 2010; Gibson et al., 2016). Antagonism between H1 and PARP1 therefore not only determines whether an open or closed chromatin structure prevails around the transcriptional start sites of target genes, but also shapes the epigenetic landscape and controls the assembly of the core transcription machinery at these promoters. Considering these multifaceted functions of PARP1 in transcription regulation, surprisingly little is known about how this enzyme is targeted to the physiologically correct gene loci. Structural analyses established that upon DNA damage, PARP1 recognition of DNA single- and double-strand breaks triggers a series of conformational changes that induce unfolding and activation of the catalytic domain (Langelier et al., 2012; Dawicki-McKenna et al., 2015; Eustermann et al., 2015). How PARP1 is recruited to and activated at promoters of specific target genes, in contrast, has remained poorly defined.
Myeloid ecotropic viral integration site (MEIS) and pre–B-cell leukemia homeobox (PBX) transcription factors are members of the three-amino acid loop extension (TALE) atypical class of homeodomain (HD)-containing proteins. They regulate a broad range of developmental processes, including the development of the limbs, axial skeleton, face, brain, and heart, are necessary for cell lineage commitment of embryonic stem cells and have been recognized as oncogenes in several forms of cancer (Mercader et al., 1999; Selleri et al., 2001; Eklund, 2011; Ferretti et al., 2011; Vitobello et al., 2011; Paige et al., 2012; Wamstad et al., 2012; Golonzhka et al., 2015). Consistent with their varying physiological roles, genome-wide binding studies in embryonic tissues revealed association of MEIS or PBX proteins with thousands of sites in the genome (Penkov et al., 2013; Amin et al., 2015). Prominent features of MEIS and PBX include their strong tendencies to heterodimerize and their ability to bind DNA cooperatively with other transcription factors (Chan et al., 1994; Chang et al., 1997). Previous work established a general requirement for MEIS and PBX in adult neurogenesis in the subventricular zone (SVZ) in mice and showed that the known neurogenic activity of PAX6 in this system depends on MEIS (Agoston et al., 2014; Grebbin et al., 2016). Interestingly, PBX1 can bind silent chromatin at times when the overall chromatin structure still prevents access of most other transcription factors, suggesting that it acts as a pioneer transcription factor (Berkes et al., 2004; Magnani et al., 2011; Choe et al., 2014; Grebbin et al., 2016). The exact sequence of events that follows PBX1 binding to transcriptionally inactive gene loci, however, is poorly defined.
We developed an in vitro assay to follow the opening and activation of the promoter/proximal enhancer of the neuron-specific gene doublecortin (Dcx) at the time when neural progenitor cells begin to differentiate into neurons. Dcx is expressed by all newly generated neurons and therefore is an early and global marker of neuronal differentiation (Gleeson et al., 1999). In the SVZ stem cell niche, Dcx expression requires PBX1 and MEIS2, although the precise mechanisms by which these proteins act on the Dcx gene have not been described (Agoston et al., 2014; Grebbin et al., 2016). By examining Dcx transcriptional activation in stem and progenitor cells of the SVZ within the first hours of differentiation in vitro, we now show that PBX and MEIS cooperate to induce chromatin opening through recruitment of PARP1 and eviction of H1 from the chromatin fiber.
Results
Activation of the Dcx gene is accompanied by dynamic transcription factor binding to a conserved PBX/MEIS binding motif in the Dcx promoter/proximal enhancer
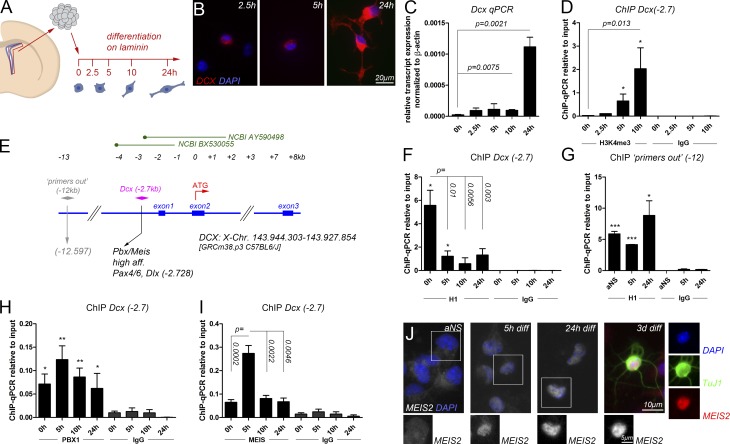
Freshly isolated neural stem and progenitor cells of the adult mouse SVZ can be cultured as adult neurospheres (aNSs) in the presence of EGF and fibroblast growth factor 2 (FGF2) but will undergo cellular differentiation in relative synchrony when plated on laminin in medium lacking EGF and FGF2 (Reynolds and Weiss, 1992; Costa et al., 2011). We took advantage of this system to follow the opening of the Dcx promoter/proximal enhancer within the first hours of differentiation in SVZ-derived stem and progenitor cells on population level by chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR; ChIP-qPCR). Progressive neuronal differentiation was evident in a gradual increase in neurite complexity, up-regulation of Dcx transcripts, and rising levels of H3K4me3 at the Dcx promoter within the first 24 h of differentiation (Fig. 1, A–D). We monitored the activation of the Dcx gene locus during these events, focusing on a PBX/MEIS recognition site, which is located within a previously characterized promoter/proximal enhancer of the Dcx gene (Karl et al., 2005; Agoston et al., 2014). This site (termed Dcx(−2.7) hereafter) is located 2,728 bp upstream of the Dcx translational start codon, carries PAX6 and distal-less homeobox (DLX) consensus motifs in its close proximity, and is bound and regulated by PBX1 and MEIS2 in neurons (Fig. 1 E; Agoston et al., 2014; Grebbin et al., 2016). Because dissociation of H1 from chromatin is an early step in the preparation of genes for transcriptional activation, we examined the compaction of the Dcx(−2.7) site with histone H1.4, a histone variant that has been shown to become dislodged from active promoters (Krishnakumar et al., 2008). Dcx(−2.7) was occupied by this histone H1 variant before differentiation was induced (0 h), but H1.4 was largely evicted 5 h after onset of differentiation (Fig. 1 F; the persisting H1.4 ChIP-qPCR signal after the 5-h time point likely results from undifferentiated cells still present in the culture or cells committed to glial differentiation). A site outside of the Dcx promoter/enhancer (12 kb upstream of the Dcx start codon, “primers out” (−12)) was also occupied by this H1 variant before differentiation and remained bound throughout the 24-h differentiation period (Fig. 1 G). We also examined Dcx(−2.7) for occupancy by the histone variant H1.0. H1.0 is widely expressed in the brain, and reduced expression of this H1 variant has been linked to disease progression in glioblastoma multiforme (GBM), rapidly proliferating and highly invasive brain tumors that are proposed to originate from germinal niches in the adult brain like the SVZ (García-Iglesias et al., 1993; Jackson and Alvarez-Buylla, 2008; Torres et al., 2016). In contrast with H1.4, however, H1.0 was not detected at Dcx(−2.7) in undifferentiated cells (Fig. S1 A). We therefore focused on H1.4 (abbreviated to H1 hereafter). Consistent with a role as priming factor, PBX1 occupied the Dcx(−2.7) site already at 0 h and over the following 24 h of differentiation, with a moderate increase in binding at 5 h (Fig. 1 H). Interestingly, ChIP-qPCR with an antibody that recognizes MEIS1 and MEIS2 (termed MEIS hereafter) revealed that the drop of H1 occupancy at the Dcx(−2.7) site at 5 h of differentiation coincided with a sharp and remarkably brief rise in MEIS binding to this site (Fig. 1 I). In the SVZ neurogenic niche in vivo, newly generated neurons can be recognized by their strong nuclear staining for MEIS2 (Agoston et al., 2014). To understand the strong increase of MEIS binding to the Dcx(−2.7) site at the 5-h time point, we monitored MEIS2 protein distribution during differentiation of SVZ-derived aNS by immunohistochemistry. MEIS2 immunoreactivity was very low in the aNS but accumulated in the cell nucleus after 5 h of differentiation and hence at the time when strong MEIS association with the Dcx(−2.7) site was detected by ChIP (Fig. 1 J). MEIS2 immunoreactivity in the cell nucleus increased even further at 24 h of differentiation to reach a level comparable to that seen in young neurons (differentiated from aNSs for 3 d and costaining for neuronal class III β-tubulin [βIII-tubulin, TUBB3, recognized by the TuJ1 antibody]), likely reflecting the involvement of MEIS2 in genetic programs in addition to Dcx transcriptional up-regulation (Fig. 1 J). MEIS2 immunoreactivity was not entirely absent from the nuclei of aNS, consistent with the fact that the MEIS2-antibody slightly enriched the Dcx(−2.7) site relative to the IgG control in these cells (Fig. 1, I and J). We therefore concluded that dismissal of histone H1 from a promoter-proximal site in the Dcx gene is a very early event of neuronal differentiation, which coincides with nuclear accumulation and strong but remarkably transient association of MEIS2 with the chromatin fiber at this position.
Figure 1.
Chromatin changes at the Dcx(−2.7) site during the first hours of neuronal differentiation. (A) Outline of the experiment. (B) Cellular morphology and DCX protein distribution at 2.5, 5, and 24 h of differentiation. (C) Dcx transcript expression at different times of differentiation. (D) ChIP-qPCR for H3K4me3 at Dcx(−2.7) at the times indicated, reflecting Dcx promoter activation beginning at 5 h of differentiation. (E) Relative position of the Dcx(−2.7) and “primers out” (−12) sites. (F–I) ChIP-qPCR at 0, 5, 10, and 24 h of differentiation: (F) H1 at Dcx(−2.7); (G) H1 at the promoter remote site “primers out” (−12); (H) PBX1 at Dcx(−2.7); (I) MEIS at Dcx(−2.7). (J) MEIS2 protein distribution in adult SVZ progenitor cells and after differentiation times of 5 h, 24 h, and 3 d. MEIS2 expression in the boxed cells is shown separately as single channel. The asterisks in F–I indicate statistically significant enrichment of ChIP with the antibodies indicated relative to ChIP with the IgG control antibodies for the same conditions, with *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical significance of ChIP results between experimental groups is given as p = numerical value. ChIP data are represented as means ± SEM. Samples sizes and the number of biological replicates are listed in Table S4.
PARP1/ARTD1 is recruited to the Dcx promoter/enhancer and induces poly-ADP modification of H1
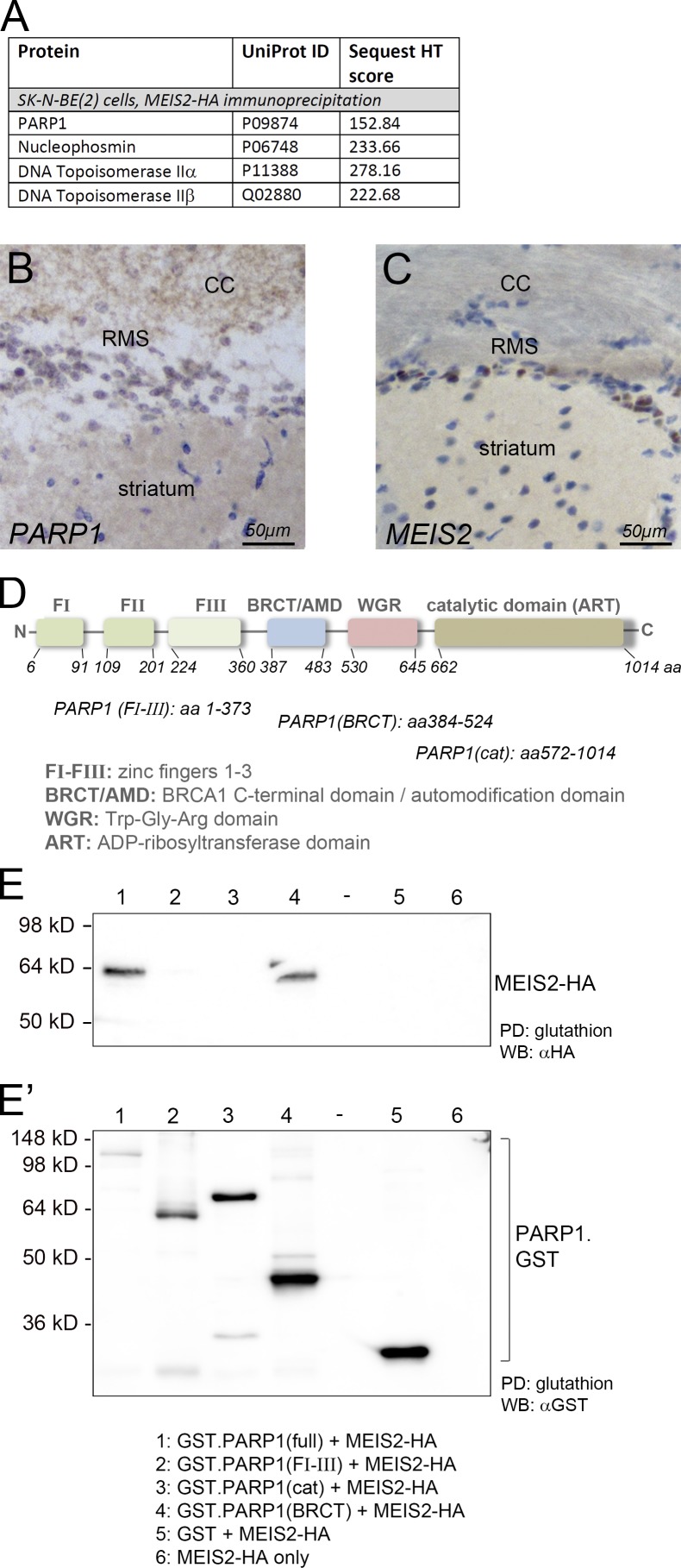
To gain insight into the underlying molecular mechanism, we isolated MEIS2-interacting proteins from extracts of Neuro2a (N2A) and SK-N-BE(2) neuroblastoma cells. Precipitation was performed in the presence of DNaseI to avoid unspecific enrichment of DNA-binding proteins by cellular DNA. Precipitates were separated by SDS-PAGE, and then prominent bands were isolated and analyzed by mass spectrometry. Interestingly, the predominant protein band isolated by MEIS2-GST pulldown from N2A cells corresponded with PARP1 (Fig. S1, B and C). PARP1 and several known subunits of the transcriptional PARP1 complex were also enriched by immunoprecipitation with HA-specific antibodies from SK-N-BE(2) cells stably expressing MEIS2-HA (Fig. 2 A; Ju et al., 2004). PARP1 is an abundant and ubiquitous nuclear protein, yet PARP1 immunohistochemical staining was particularly high in cells of the rostral migratory stream (RMS) compared with cells of the adjacent striatum, consistent with prominent PARP1 expression in adult-generated neuroblasts (Fig. 2, B and C). MEIS2/PARP1–containing protein complexes could also be isolated from E12.5 mouse embryonic forebrains and from midbrains and retinas of E2.5 chick embryos, three additional regions of ongoing neurogenesis in which MEIS2 is expressed (Fig. S1, D–F). MEIS2/PARP1-containing protein complexes hence exist in different vertebrate species in vivo. These observations raise the intriguing possibility that MEIS2 may modulate histone dynamics through recruitment of PARP1.
Figure 2.
MEIS and PARP1 interact. (A) Mass spectrometry scores of PARP1 and PARP1-interacting proteins copurifying with MEIS2 from nuclear extracts of SK-N-BE(2) neuroblastoma cells. (B and C) PARP1 (B) and MEIS2 (C) protein distribution in the RMS. MEIS2 and PARP1 protein staining is shown in brown, and nuclear counterstaining is in blue. CC, corpus callosum. (D) Domain structure of PARP1. (E) GST pulldown of HA-tagged MEIS2 with different PARP1–GST fusion proteins. The left panel shows a blot probed for HA detecting MEIS2-HA, and the right panel shows the same blot probed for GST detecting the different PARP1–GST fusion proteins. Because transfer of full-length PARP1–GST was incomplete because of its large size, higher-exposure images of the blot probed for HA and GST are shown in Fig. S2. The number of biological replicates performed for E are listed in Table S4. PD, pulldown; WB, Western blot.
As a starting point for testing this hypothesis, we performed pulldown experiments with extracts of HEK293T cells, which were transiently transfected with HA-tagged MEIS2 together with different domains of PARP1 fused to GST (Schreiber et al., 2002). HEK293T cells were chosen for their very low expression of endogenous MEIS2. PARP1 is a multidomain protein, which contacts specific DNA structures through N-terminally located zinc finger motifs, whereas the catalytic domain containing the ADP ribosyltransferase signature domain and responsible for the transfer of ADP-ribose moieties onto target proteins, resides in the C terminus of the protein. Enclosed between the DNA-binding and catalytic domains are a tryptophan-glycine-arginine–rich region and a BRCA1 C-terminal (BRCT) domain, which is known to serve as the protein–protein interaction domain (Fig. 2 D; Ji and Tulin, 2010). MEIS2 efficiently bound to full-length PARP1 as well as to the BRCT domain in these GST pulldown assays (Fig. 2, E and E′; and Fig. S2). In contrast, neither the zinc finger DNA-binding domains nor the catalytic domain successfully enriched MEIS2 (Figs. 2 E and S2). PBX1 was not present in the MEIS2/PARP1 precipitates, irrespective of whether the full-length protein or the BRCT domain was tested (Fig. S2). This argues that the mere binding of MEIS2 to PARP1 does not require PBX1.
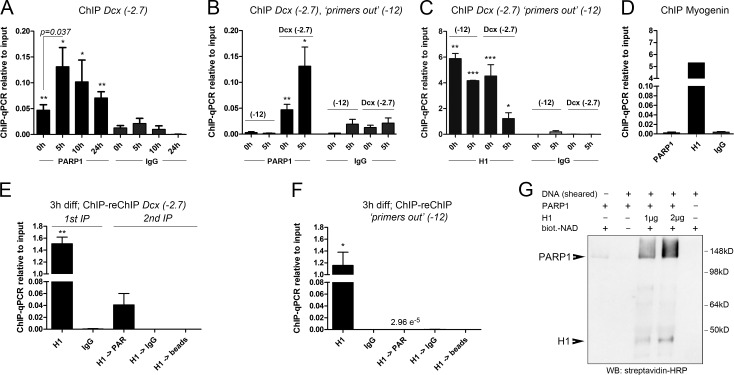
To relate these findings to the events that take place at the Dcx promoter/enhancer, we performed ChIP-qPCR experiments with SVZ-derived neural progenitor cells undergoing neuronal differentiation. We observed dynamic association of PARP1 with the promoter-proximal Dcx(−2.7) site, with low PARP1 binding at 0 h, strong binding at 5 h of differentiation, and declining levels thereafter (Fig. 3 A). PARP1 was not enriched at the promoter-distant site “primers out” (−12) at 0 or 5 h of differentiation (Fig. 3 B). In fact, association of PARP1 and H1 inversely correlated at both sites (Fig. 3, B and C). PARP1 was also not detected at a known PBX/MEIS binding site in the muscle-specific gene myogenin (Myog), demonstrating that PARP1 is not recruited to the promoter of a lineage-inappropriate gene (Fig. 3 D; Berkes et al., 2004).
Figure 3.
MEIS recruits PARP1 to the Dcx promoter/enhancer. (A) ChIP-qPCR for PARP1 at Dcx(−2.7) during neuronal differentiation. (B) Comparison of PARP1 binding to Dcx(−2.7) and “primers out” (−12) at 0 and 5 h; values for Dcx(−2.7) correspond with those shown in A. (C) ChIP-qPCR for H1 at Dcx(−2.7) and “primers out” at the times indicated. (D) ChIP-qPCR for PARP1 and H1 at the Myog promoter, showing reciprocal binding of PARP1 and H1. (E and F) ChIP-reChIP for H1 followed by PAR at Dcx(−2.7) (E) and “primers out” (F) at 3 h of differentiation. (G) In vitro PARylation assay of recombinant PARP1 and H1 in the presence of biotinylated (biot.) NAD+ and stimulated by the addition of low-molecular DNA fragments (sheared DNA), demonstrating efficient PARylation of H1 and autoPARylation of PARP1. The asterisks in A–F indicate statistically significant enrichment of ChIP with the antibodies indicated relative to ChIP with the IgG control antibodies for the same conditions, with *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical significance of ChIP results between experimental groups is given as p = numerical value. ChIP data are represented as means ± SEM. Samples sizes and the number of biological replicates are listed in Table S4. IP, immunoprecipitation; WB, Western blot.
Histone H1 is a major target of PARP1, and H1 PARylation leads to its depletion from polynucleosomes in vitro (Poirier et al., 1982). We therefore asked whether release of H1 from the Dcx(−2.7) site in vivo involved the attachment of PAR moieties to H1. Because eviction of H1 from Dcx(−2.7) was already maximal at 5 h of differentiation (Fig. 1 F), we examined chromatin from SVZ-derived neural progenitor cells at 3 h of differentiation, reasoning that enzymatic modification of H1 may be still ongoing at the Dcx promoter at this intermediate time. Indeed, ChIP-reChIP experiments with an antibody against H1 followed by ChIP with an antibody recognizing PAR revealed substantial PARylation of the H1 precipitate at the Dcx(−2.7) site but not at the promoter-distal site “primers out” (−12) (Fig. 3, E and F). PAR attachment thus reflects the preparation of a developmentally regulated promoter for transcriptional activation rather than a global response to DNA damage or cell death in early differentiating neurons. In support of this, only very few cells were immunoreactive for activated caspase 3 in our cultures at 5 h of differentiation (Fig. S3, A and B). These results demonstrate that programmed cell death is minimal during the first hours of neuronally directed differentiation in SVZ aNSs and therefore unlikely accounts for the local increase in PAR load at the Dcx promoter/enhancer seen during this time. Finally, and consistent with previously published results, recombinant PARP1 effectively PARylated recombinant H1 in an in vitro PARylation assay (Fig. 3 G; Poirier et al., 1982; Krishnakumar et al., 2008). MEIS2, in contrast, was not PARylated by PARP1 in vitro, suggesting that MEIS2 is not a PARP1 target but rather functions cooperatively with PARP1 (Fig. S3 C). In sum, PARP1 is rapidly and specifically recruited to the Dcx promoter/enhancer when neural progenitors undergo neuronal differentiation, and this recruitment is associated with PARylation and dismissal of histone H1 (and possibly additional proteins that coprecipitate with H1).
Recruitment of PARP1 to the Dcx promoter/enhancer and eviction of H1 from this site require MEIS2
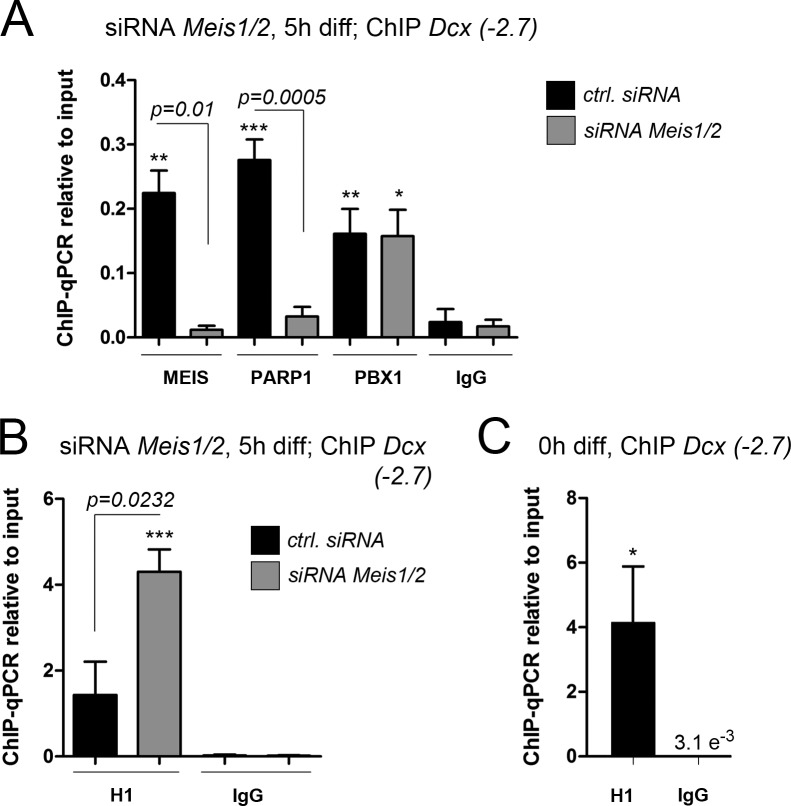
The striking temporal coincidence of H1 eviction from Dcx(−2.7) and transient binding of MEIS to this site motivated us to examine whether MEIS had a role in chromatin opening at the Dcx gene locus. SVZ-derived progenitor cells were transfected with a cocktail of validated siRNAs against Meis1 and Meis2 (to avoid up-regulation and compensation of Meis2 depletion by Meis1; Agoston et al., 2014), induced to differentiate 48 h later, and then were examined for protein association at the Dcx(−2.7) site after a 5-h differentiation period (Fig. 4). ChIP-qPCR verified efficient depletion of MEIS from this site, whereas PBX1 binding was unaffected, a finding that is consistent with our previous observation that PBX1 occupies the Dcx(−2.7) site before MEIS (Fig. 4 A; also see Fig. 1, H and I). Interestingly in Meis1/2-depleted cells, PARP1 binding to Dcx(−2.7) was virtually abolished, and H1 occupancy remained high at this site even though the cells were kept in the absence of EGF and FGF2 and allowed to attach to laminin, which under control conditions initiates rapid dismissal of H1 from the Dcx(−2.7) site (Fig. 4, A and B). In fact, after Meis1/2 knockdown, the H1 load at Dcx(−2.7) after 5 h of differentiation was comparable to the load seen in the aNS growing under nonadherent conditions and in EGF/FGF2-containing medium (Fig. 4, B and C). MEIS is thus a prerequisite for the loss of histone H1 from the Dcx promoter/proximal enhancer and the subsequent chromatin opening that occurs at the Dcx gene locus at the beginning of neuronal differentiation.
Figure 4.
MEIS recruits PARP1 to Dcx(−2.7). (A and B) ChIP-qPCR for the proteins indicated at 5 h of differentiation in cells transfected with Meis1/2-specific siRNAs (gray bars) or nontargeting control siRNAs (black bars): MEIS, PARP1, and PBX1 at Dcx(−2.7) (A), and H1 at Dcx(−2.7) (B). (C) ChIP-qPCR for H1 at Dcx(−2.7) in undifferentiated cell cultures. H1 is not released from the Dcx promoter/enhancer when cells are subjected to the cellular differentiation protocol under Meis-knockdown conditions. The asterisks indicate statistically significant enrichment of ChIP with the antibodies indicated relative to ChIP with the IgG control antibodies for the same conditions, with *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical significance of ChIP results between experimental groups is given as p = numerical value. ChIP data are represented as means ± SEM. Samples sizes and the number of biological replicates are listed in Table S4.
PARP1 is required for neuronal differentiation of SVZ-derived adult progenitor cells
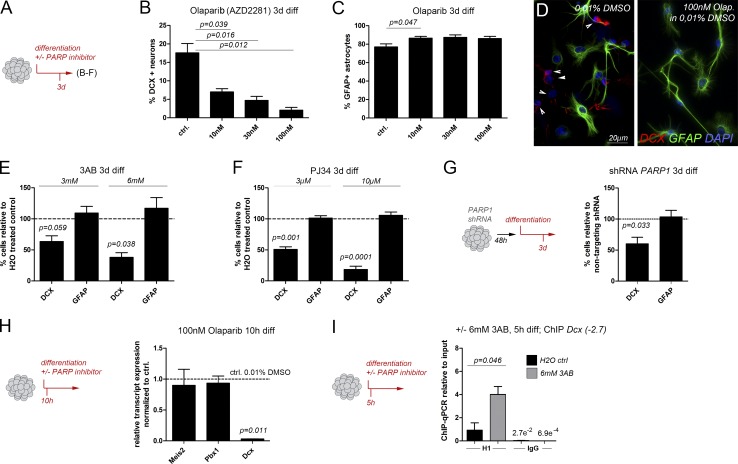
PARP1-deficient mice are viable and fertile, likely because of the compensatory effect of related enzymes like PARP2, but exhibit mildly defected SVZ neurogenesis at postnatal day 11 (P11; Wang et al., 1995; Plane et al., 2012). Because neurogenesis in the postnatal and adult brain differ considerably, we examined whether PAR synthesis also has a role in neurogenic differentiation of adult stem and progenitor cells. To exclude compensation between PARP1 and other PARylating enzymes, we blocked PARylating activity with a variety of pharmacological PARP inhibitors. Consistent with the important contribution of PARylating enzymes to cell survival, infusion of Olaparib, a selective inhibitor of PARP1 and PARP2 approved for the treatment of BRCA mutant ovarian cancer, into the SVZ in vivo had profound effects on tissue integrity (not depicted; Hassa, 2009). In addition, cell cycle exit and cellular differentiation of SVZ stem cells in vivo are profoundly influenced by signals from the stem cell niche, making it likely that broad application of a PARP inhibitor to the SVZ will cause confounding secondary effects, which may preclude the analysis of a direct influence on the stem and progenitor cell compartment of the niche (Bjornsson et al., 2015). We therefore turned to an in vitro approach. SVZ-derived progenitor cells were cultured as aNSs and differentiated in vitro in the presence or absence of pharmacological PARP inhibitors (Fig. 5). Addition of Olaparib to the culture medium during the differentiation regimen reduced the proportion of neurons (recognized by expression of Dcx or neuronal βIII-tubulin [TuJ1]) in a dose-dependent manner, as did two more general PARP-inhibitors, 3AB and PJ34 (Fig. 5, B–F). Reduced neurogenesis was accompanied by a mild but significant increase in the production of astrocytes expressing glial fibrillary acidic protein (GFAP), thus mimicking the neuronal-to-astroglial cell fate change that aNSs undergo in the absence of functional Meis1 and Meis2 (Agoston et al., 2014). ShRNA-mediated knockdown of PARP1 also reduced neurogenesis of SVZ-derived aNSs, albeit to a lesser extent than pharmacological PARP inhibition, presumably because other PARylating enzymes remained unaffected by the knockdown (Figs. 5 G and S3 D). PARP1 inhibition essentially abolished expression of Dcx but did not alter transcript levels of Pbx1 or Meis2 (Fig. 5 H). This observation argues that the observed defective neuronal differentiation is not caused by loss of expression of these two transcription factors but rather reflects a direct involvement of PARP1 in transcriptional up-regulation of Dcx. We therefore compared H1 occupancy at the Dcx promoter/enhancer by ChIP-qPCR in undifferentiated cells and after 5 h of differentiation in the presence or absence of 6-mM 3AB. Growth factor withdrawal and plating on laminin under control conditions caused release of H1 from the Dcx(−2.7) site (Fig. 5 I, black bar), whereas dismissal of H1 was blocked when neuronal differentiation was performed in the presence of 3AB (Fig. 5 I, gray bar). In fact, the ChIP-qPCR signals for H1 obtained from 3AB-treated differentiated cells were only slightly lower than those obtained from undifferentiated cells, suggesting that pharmacological inhibition of PARP-dependent PARylation “locks” the Dcx promoter/proximal enhancer in a chromatin state, which is similar to that present in progenitor cells (Fig. 5 I; compare with Fig. 1 F).
Figure 5.
PARP activity is required for neuronal differentiation and H1 eviction from the Dcx promoter/enhancer. (A) Schematic outline of the experiments shown in B–F. (B–F) Reduced neurogenesis and enhanced astrogliogenesis upon pharmacological PARP inhibition: (B and C) Proportions of neurons (B) and astrocytes (C) generated in the presence of increasing concentrations of Olaparib. (D) Representative images of cultures differentiated in the presence of Olaparib or 0.01% DMSO as control. Arrowheads indicate DCX-positive neuronal processes. (E and F) Neurons and astrocytes generated in the presence of 3AB (E) or PJ34 (F). (G, left) Outline of the experiment; (right) neuronal differentiation after shRNA-mediated knockdown of PARP1. (H, left) Outline of the experiment; (right) qPCR for Meis2, Pbx1, and Dcx transcripts in cells differentiated for 10 h by growth factor removal and plating on laminin in the presence of 100 nM Olaparib. Expression is shown relative to expression determined in cells treated with 0.01% DMSO (vehicle only). (I, left) Outline of the experiment; (right) ChIP-qPCR for H1 at Dcx(−2.7) in cells differentiated for 5 h in the presence of 6 mM 3AB (gray bars) or water as vehicle control (black bars). Statistical significance of ChIP results between experimental groups is given as p = numerical value. Data are represented as means ± SEM. Samples sizes and the number of biological replicates are listed in Table S4.
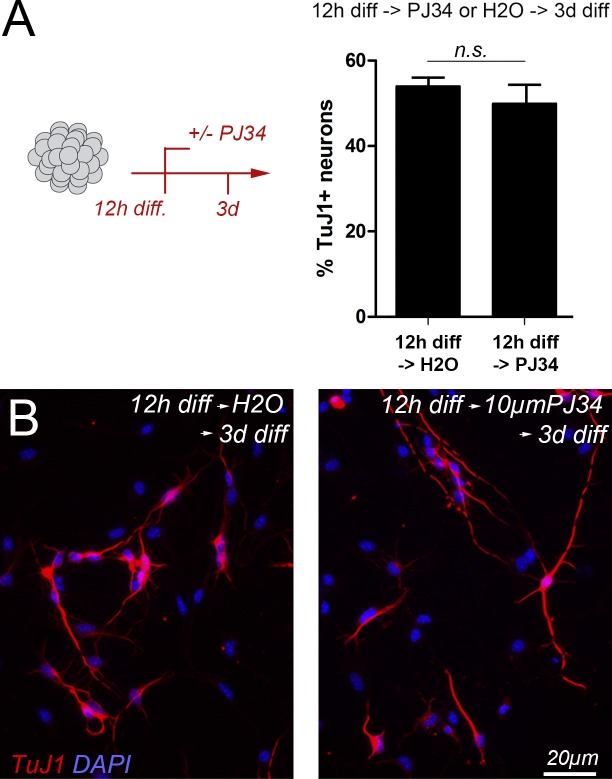
Importantly, neuron production was not compromised when neuronal differentiation was induced first and the PARP-inhibitor added 12 h later (Fig. 6). This observation clearly shows that progenitor cells need PARP1 activity to initiate neurogenic programs but not to execute later steps of neuronal maturation and differentiation, highlighting the importance of PARP1 for the initiation of MEIS-dependent developmental programs.
Figure 6.
Neuronal differentiation is not compromised when pharmacological inhibition of PARP follows the induction of cellular differentiation. (A, left) Outline of the experiment: differentiation was induced in primary SVZ progenitor cells by growth factor removal and plating on laminin 12 h before addition of PJ34 to the culture medium. Addition of water (diluent) served as control; (right) proportion of TuJ1+ neurons generated under both conditions; after 3 d of differentiation, 53.9 ± 3.6% (SD) of the cells differentiated into DCX+ neurons under standard conditions, and 49.9 ± 7.6% (SD) differentiated when PJ34 was added to the medium after the first 12 h of differentiation. (B) Representative micrographs of these experiments. Data are represented as means ± SEM, and the number of biological replicates is listed in Table S4.
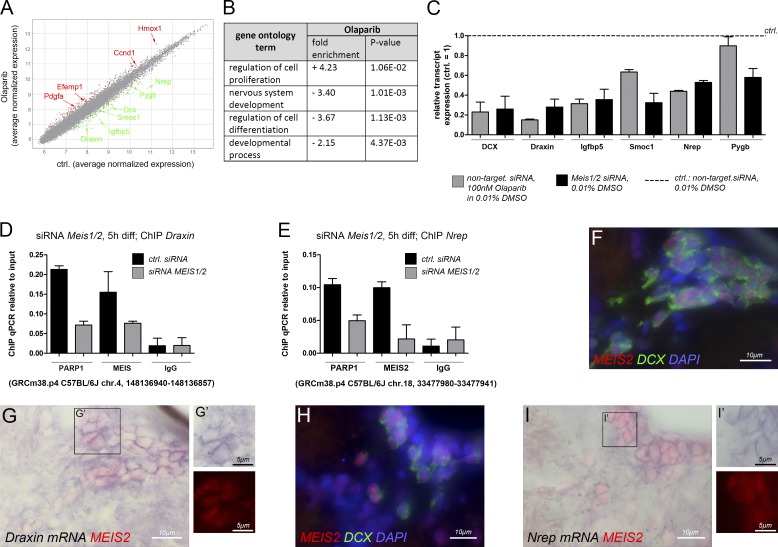
To investigate whether PARP1 is required for the activation of neurogenesis-associated genes other than Dcx, we performed genome-wide expression analysis with Affymetrix Mouse Gene 1.0 ST arrays. A short 10-h differentiation protocol was chosen to focus the analysis on direct target genes. Despite this brief differentiation regimen, expression of 55 genes was retained (significantly “up-regulated”), whereas expression of 94 genes failed to become induced (significantly “down-regulated”) after Olaparib treatment (relative to control; P ≤ 0.05; Fig. 7 A and Tables S1 and S5). The majority of up-regulated genes were related to the Gene Ontology (GO) term “regulation of cell proliferation,” an example being cyclin D1 (Ccnd1), indicating that the cells continued to proliferate even under differentiation-promoting conditions when PAR synthesis was blocked (Fig. 7, A and B). Interestingly, several of these genes were already linked to GBM. Example genes include heme oxygenase 1 (Hmox1) and platelet-derived growth factor α (Pdgfα), which are associated with disease progression or known to promote proliferation of GBM tumor-initiating cells, respectively (Ghosh et al., 2016; Sakakini et al., 2016). Most of the significantly down-regulated genes, in contrast, were related to GO terms consistent with neurogenesis, including “nervous system development” or “regulation of cell differentiation” and were significantly correlated with the UniGene Expression term “brain” (P = 1.703e-08, Fisher’s exact test; Fig. 7 B). Expression of many of them was particularly high in the SVZ, RMS, or olfactory bulb, three brain regions associated with adult neurogenesis. In fact, five of these genes, neuronal regeneration–related protein (Nrep; D0H4S114), insulin-like growth factor binding protein 5 (Igfbp5), SPARC-related modular calcium binding protein 1 (Smoc1), brain glycogen phosphorylase (Pygb), and Draxin (2610109H07Rik), emerged as novel markers for migrating neurons in the RMS (Fig. S4, A–F). Because PARP1 could not bind to the Dcx(−2.7) site when MEIS was depleted from the cells (Fig. 4 A), we examined whether expression of these five genes also depended on MEIS. Consistent with our ChIP-qPCR results, Dcx transcript levels were markedly reduced upon PARP inhibition and Meis knockdown (Fig. 7 C). In addition, Draxin, Igfbp5, Smoc, Nrep, and Pygb expression were also lower in Meis1/2 siRNA-treated cells than in cells differentiated under control conditions, arguing that these genes are coregulated by PARP1 and MEIS (Fig. 7 C). To test whether regulation may be direct, we searched for MEIS-binding motifs in the sequences encompassing 5–6 kb upstream of the start codons of these five genes using the MatInspector software package. Because MEIS2 forms heteromeric complexes with PAX and DLX proteins and taking into account the important roles of Pax6 and Dlx2 in adult SVZ neurogenesis, we concentrated on MEIS consensus motifs that were located in close proximity to putative PAX4/6 and DLX recognition sequences (Hack et al., 2005; Brill et al., 2008; Agoston et al., 2014). Sequence motifs fulfilling these criteria were identified upstream of the Draxin and Nrep start codons (Fig. S4 G). ChIP-qPCR with chromatin isolated from aNSs that had been differentiated for 5 h toward the neuronal lineage and antibodies specific for MEIS or PARP1 indeed enriched PARP1 and MEIS2 at positions 5.8 kb upstream of the Draxin and 3.3 kb upstream of the Nrep start codon, indicating that both genes may be direct MEIS/PARP1 targets (Fig. 7, D and E). Notably, PARP1 binding to these sites was reduced when cells were depleted for Meis1 and Meis2 before neuronal differentiation (Fig. 7, D and E). Consistent with direct regulation of Draxin and Nrep by MEIS2, MEIS2 protein colocalized with transcripts for both genes in neuroblasts of the adult SVZ in vivo (Fig. 7, F–I). In sum, with the caveat that the enhancer structure of Draxin and Nrep is not well defined at present, these results suggest that transcriptional activation of both genes is controlled by a MEIS/PARP1-dependent mechanism whereby MEIS mediates PARP1 recruitment to chromatin.
Figure 7.
Identification of PARP-regulated genes by Affymetrix Mouse Gene 1.0 ST arrays. (A) Scatter blot of differentially regulated genes in adult SVZ progenitor cells differentiated in the presence or absence of 100 nM Olaparib; representative significantly up-regulated genes are highlighted in red, and representative significantly down-regulated genes are in green. A list of all significantly differentially expressed genes is given in Table S5. (B) GO term enrichment analysis for genes differentially expressed after Olaparib treatment relative to the control. A more detailed collection of GO terms is shown in Table S1. (C) Transcript expression of six down-regulated candidate genes in SVZ-derived stem and progenitor cells differentiated for 10 h in vitro upon pharmacological inhibition of PARP1 and PARP2 or Meis1/2 knockdown, respectively, validated by qPCR. Gray bars, PARP-inhibition: transfection with nontargeting siRNAs and treatment with 100 nM Olaparib; black bars, Meis knockdown: transfection with Meis1/2-specific siRNAs and treatment with 0.01% DMSO. Transcript levels are expressed relative to those determined under control conditions (transfection with nontargeting siRNAs and differentiation in the presence of 0.01% DMSO). (D) ChIP-qPCR for MEIS2 and PARP1 at a consensus-binding motif for MEIS/PBX-type HD proteins upstream of the Draxin transcriptional start site in adult SVZ progenitor cells after 5 h of neuronally directed differentiation. Gray bars, cells transfected with Meis1/2-specific siRNAs; black bars, cells transfected with nontargeting control siRNAs. (E) ChIP-qPCR for MEIS2 and PARP1 under identical conditions as shown in D but binding to a consensus motif for MEIS/PBX-type HD proteins upstream of the Nrep transcriptional start site was assessed. (F–I) Transcript expression for Draxin (F and G) and Nrep (H and I) in the adult SVZ visualized by in situ hybridization in comparison with MEIS2 and DCX protein expression. (F and H) MEIS2 (red) and DCX (green) cell nuclei are counterstained with DAPI (blue). (G and H) Overlay of transcript expression (purple) and MEIS2 protein (red). The number of biological replicates is listed in Table S4.
Discussion
Cell fate acquisition and cellular differentiation require de novo transcriptional activation of previously dormant lineage-specific genes. One of the earliest steps in this process is the release of the linker histone H1 and the subsequent decompaction of the chromatin fiber, which is a prerequisite for the binding of other regulatory proteins. This decompaction has to occur in a highly selective manner: only cell lineage–specific genes may be activated, whereas lineage-inappropriate genes have to remain silent. Work over the last decades has shown that PARP1 induces chromatin accessibility by transferring ADP-ribose moieties onto H1 (Krishnakumar et al., 2008). PARP1 can be activated by various developmental signals and environmental cues, yet surprisingly little is known about how the enzyme is targeted to the physiologically correct gene loci. Focusing on the first hours of neuronal differentiation of primary SVZ-derived progenitor cells, we describe in this study a sequence of events by which PARP1 is rapidly and specifically recruited to the regulatory regions of neuron-specific genes by the TALE-HD protein MEIS2. These results establish a previously unrecognized role for MEIS proteins in the orchestration of chromatin dynamics.
Activation of the Dcx gene is accompanied by dynamic transcription factor binding to a conserved PBX/MEIS binding motif
With the help of an in vitro assay, where primary stem and progenitor cells are allowed to enter into a common differentiation program in relative synchrony, we show that MEIS association with a known PBX/MEIS recognition site in the Dcx promoter/proximal enhancer is highly dynamic, with a rapid, transient increase in binding occurring 5 h after experimental induction of differentiation. This brief association of MEIS with chromatin was unexpected, as previous studies had not hinted toward any particular temporal dynamic in the binding of MEIS proteins to DNA. In fact, previous genome-wide studies had identified MEIS–chromatin binding events primarily in intergenic regions (Penkov et al., 2013; Amin et al., 2015). Yet, in these studies, mixed cell populations were examined (whole-trunk tissue or branchial arches of mouse embryos, respectively), which may have precluded the identification of transient binding events that occur in a small portion of the cells. Dynamic association of MEIS within a defined promoter-proximal site may therefore have only become obvious in this study because we monitored MEIS binding to a single binding site at short temporal intervals and in primary cells that entered into the same differentiation program. The dynamic nature of MEIS chromatin binding reported in this study thus adds a new level of complexity for the interpretation of MEIS genome-wide chromatin association studies.
MEIS recruits PARP1/ARTD1 to induce H1 eviction from the Dcx promoter/enhancer
Previous work with mutant mice or using retroviral vectors has shown that MEIS and PBX protein activity are required for neuronal differentiation of adult SVZ stem and progenitor cells, but the underlying molecular mechanism remained incompletely defined (Agoston et al., 2014; Grebbin et al., 2016). We now show a physical interaction between MEIS2 and a PARP1-containing nuclear complex, linking MEIS to PARP1-dependent local decompaction of chromatin. Our comprehensive interaction studies revealed that MEIS2 specifically associates with the BRCT domain of PARP1 (Fig. 2). Interestingly, NMR studies of PARP1 protein bound to a synthetic construct that mimics a single-strand DNA break recently established that PARP1 activation at sites of DNA damage occurs as a series of conformational changes that allow for stepwise self-assembly and activation of an initially unstructured PARP1 polypeptide (Dawicki-McKenna et al., 2015; Eustermann et al., 2015). This sequence of events involves successive assembly of the three N-terminally located zinc finger domains over the DNA strand break followed by recruitment of the tryptophan-glycine-arginine–rich domain into the complex and association and unfolding of the C terminus to relieve of an autoinhibitory constraint within the catalytic domain (Dawicki-McKenna et al., 2015; Eustermann et al., 2015). The centrally located BRCT domain does not participate in this process and hence remains available for binding to additional proteins. These events were so far only observed on an oligonucleotide in vitro, and it therefore remains to be seen whether PARP1 undergoes similar conformational changes when it acts as transcriptional coactivator in the complex environment of the cellular genome in vivo. Nevertheless, through association with the BRCT domain, MEIS may recruit PARP1 to chromatin without interfering with PARP1 self-activation.
Taking into account published work, our results suggest a novel concept of how PBX and MEIS cooperate to initiate Dcx transcriptional activation: Before differentiation, PBX1 is already present at a defined position in the Dcx promoter/enhancer, essentially priming the gene for activation. As soon as neuronal differentiation is initiated, MEIS2, because of its strong affinity for PBX, recruits PARP1 to the PBX1-prebound site. Our concept thus centers around the known binding affinity of MEIS and PBX family proteins but extends current models of how PBX/MEIS dimers are formed as dimerization in our system occurs while one partner, PBX1, is already bound to its target site in closed chromatin, and the other partner, MEIS2, joins in later. PBX1 cannot bind to DNA on its own but may associate with other prebound proteins. A likely candidate is the paired-type transcription factor PAX6. PAX6 biases adult SVZ progenitor cells toward a neuronal cell fate, can perform pioneering function during adult SVZ neurogenesis, and physically interacts with both MEIS and PBX (Hack et al., 2005; Ninkovic et al., 2013; Agoston et al., 2014; Grebbin et al., 2016). Although additional work is needed to decipher the precise mode of PBX1 binding to closed chromatin, recruitment of PARP1 to a PBX1-bound site in the Dcx promoter/proximal enhancer by MEIS leads to PARP1-mediated eviction of H1 from this site and thereby facilitates Dcx gene expression. Notably, the known neurogenic activity of PAX6 in the SVZ stem cell niche is abrogated when MEIS is knocked down, establishing MEIS as essential cofactor of PAX6 in this system (Hack et al., 2005; Agoston et al., 2014). PAX6 in turn recruits a Brg1/BAF-containing chromatin remodeling complex to neuron-specific genes to facilitate nucleosome mobility during transcription (Ninkovic et al., 2013). Because H1 is known to prevent access of Brg1/BAF to chromatin, MEIS-mediated eviction of H1 has to precede PAX6/BAF-induced core nucleosome mobility, providing a molecular basis for MEIS-PAX6 cooperation in SVZ neurogenesis (Saeki et al., 2005).
MEIS/PARP-regulated chromatin dynamics may contribute to disease
Because TALE-HD proteins have been implicated not only in the regulation of embryonic development but also in different forms of cancer, the mechanism identified in this study also offers an explanation for the oncogenic potential of some TALE-HD proteins. PBX1 is a proposed pioneer factor whose binding was detected at thousands of genomic sites in embryonic tissues or cancer cells (Magnani et al., 2011; Penkov et al., 2013). Elevated MEIS expression, in turn, is seen in several forms of cancer. Based on the present results, we propose that rising MEIS levels may cause up-regulation of oncogenic pathways through the erroneous recruitment of PARP1 to transcriptionally inactive but PBX1-primed genes. Our findings thus on one hand suggest a novel mechanism for how selectivity can be achieved in targeting PARP1 to the promoters of downstream genes. On the other hand, they provide a new framework for understanding how MEIS proteins can act at the top of cell fate hierarchies in development, tissue homeostasis, and disease, as well as shed new light on their roles in cancer.
Material and methods
Cultivation, differentiation, and manipulation of SVZ aNSs
All procedures involving animals were approved by the local animal care committee as well as the government of Hessen and are in accordance with German and European Union regulations. Cells were isolated from 9–12-wk-old C57BL/6 mice of mixed gender and propagated under nonadherent conditions for no more than 5 d to obtain primary aNSs or after one passage and a further 3 d of in vitro culture to obtain secondary aNSs following previously described procedures (Brill et al., 2008; Agoston et al., 2014). Differentiation was induced by plating dissociated cells at a density of 1–2 × 105 cells per cm2 in medium without EGF/FGF2 on laminin-coated surfaces. For differentiation times exceeding 24 h, the medium was supplemented with 2 ng/ml bFGF and 20 ng/ml brain-derived neurotrophic factor (PeproTech). For neuronally directed differentiation, primary aNS cells were transduced with Pax6 as described previously by Hack et al. (2005) and differentiated 48 h later (Fig. S5). Pax6 transduction biased SVZ-derived progenitor cells toward the neuronal lineage but did not initiate neurogenic programs on its own as it neither induced signs of neuronal differentiation, increased its own binding to Dcx(−2.7), or altered the expression of neuron-specific genes when cells were kept in the presence of EGF/FGF2 and hence without exogenous stimulus to differentiation (Fig. S5). To inhibit PARP1 enzymatic activity during the differentiation process, the medium was supplemented with PJ34 hydrochloride (Tocris Bioscience), 3AB (Sigma-Aldrich), or Olaparib (AZD2281; Lynparza; Selleckchem) in the listed concentrations. Controls were treated with equal amounts of the corresponding diluents (0.001–0.01% DMSO or water, depending on the type and concentration of the inhibitor). Knockdown of Meis1 and Meis2 was performed as described previously (Agoston et al., 2014), and knockdown of PARP1 was achieved by transduction with PARP1-specific pGIPZ lentiviral vectors (clone ID numbers are shown in Fig. S3 D; GE Healthcare/Open Biosystems).
Immunohistochemistry
For immunofluorescence analysis, cells were fixed with 4% PFA in PBS, pH 7.5, for 10 min at RT and then washed in PBS at 4°C. Primary and secondary antibodies were diluted in 10% goat serum and 0.5% Triton X-100 in PBS. Primary antibodies were applied overnight at 4°C. Samples were washed three times with PBS for 5–10 min each. Secondary antibodies were applied for one hour at RT. The samples were washed with PBS, cell nuclei were stained with DAPI, and samples were mounted with Vectashield (Vector Laboratories). Chromogen staining was performed on cryosections with a DISCOVERY XT automated staining system, with antigen retrieval protocol Conditioner #1, Omni-Map HRP detection (Ventana Medical Systems, Inc.), and counterstaining with hematoxylin; mounting medium was Entellan (Merck). The images shown in Figs. 5 D and 6 B, and S3 A were taken at RT with an Eclipse 80i microscope, Plan Fluor 20× differential interference contrast M (0.50 NA) or Plan Fluor 40× differential interference contrast M (0.75 NA) objective lenses, and a DS-Qi1 MC-U3 camera (Nikon). The confocal images shown in Fig. 1 J were taken at RT with a TE2000-E confocal microscope, a Plan Fluor 40× oil immersion lens (1.30 NA), and a C1 camera with optical sections of 1–2-µm intervals (Nikon). Acquisition software was NIS elements 4.10 (Nikon). A minimum of 1,000 cells per condition and experimental repeats were photographed, and cells were counted blind. SD was calculated between independent replicate experiments with paired Student’s t tests (Prism 5.01; GraphPad Software). If necessary, brightness and contrast were moderately enhanced in Photoshop (CS4; Adobe) across the entire image, and no further image processing was performed. The antibodies used are listed in Table S2.
In situ hybridization followed by immunofluorescence detection
14-µm cryosections were postfixed in 4% PFA for 10 min at RT, washed in PBS, and incubated in 200 µl hybridization buffer (50% formamide, 5× Denhardt’s solution, 0.25 mg/ml baker’s yeast tRNA, 0.2 mg/ml salmon sperm DNA, and 5× saline-sodium citrate [SSC]) for 3 h at RT. Probes were nt 1,168–1,897 of NCBI accession number NM_027426 for Draxin and nt 489–1,140 of NCBI accession number NM_001109988 for Nrep. Hybridization was performed in 3 ng/µl digoxigenin (DIG)-labeled, Draxin- or Nrep-specific RNA probes in hybridization mix overnight at 66°C. Sections were washed once in 5× SSC for 5 min at 66°C, once in 2× SSC for 5 min at 66°C, once in 0.2× SSC/50% formamide for 20 min at 66°C, and once in 0.2× SSC for 10 min at RT. Sections were rinsed in 100 mM Tris, pH 7.5, and 150 mM NaCl, and then were blocked in 1% blocking reagent (Roche) in 100 mM Tris, pH 7.5, and 150 mM NaCl for 1 h at RT and incubated in anti–DIG–alkaline phosphatase Fab fragments (1:1,000; Roche) in 100 mM Tris, pH 7.5, and 150 mM NaCl for 1.5 h at RT. Color reaction was performed with 3.5 µl/ml 5-bromo-4-chloro-3-indolyl phosphate P-toluidine and 4.5 µl/ml nitrotetrazolium blue chloride in 100 mM Tris, pH 9.5, 100 mM NaCl, and 5 mM MgCl2 under visual inspection. Sections were rinsed in water and washed twice for 10 min in Tris-buffered saline, pH 7.5. Immunofluorescence detection of MEIS2 and DCX was performed following standard procedures involving antigene retrieval with citrate buffer for 40 min and with the antibodies listed in Table S2. Images were taken at RT with an Eclipse 80i microscope and a Plan Apochromat VC 60× 1.2 water immersion lens and a DS-Fi1-U3 camera (Nikon). Acquisition software was NIS elements (4.3). Brightness and contrast were moderately enhanced in Photoshop across the entire image, and no further image processing was performed.
Analysis of PARP1-containing protein complexes
Protein lysates were generated from SK-N-BE(2) or Neuro2a (N2A) cells, from manually dissected retinas or dorsal mesencephalic vesicles of E2.5 white leghorn chicks, or from isolated forebrain vesicles of E12.5 embryonic C57BL/6 mice. SK-N-BE(2) cells were a gift from M. Mittelbronn (University Hospital Frankfurt, Frankfurt, Germany) and authenticated by short tandem repeat profiling performed at the Leibniz Institute DSMZ. The N2A cell lines were purchased from ATCC (CCL-131). Both cell lines were regularly tested for mycoplasma contamination by PCR (PCR Mycoplasma Test kit; AppliChem). Subcellular fractionation, immunoprecipitation, and GST pulldown assays were performed as described previously, except precipitation was performed in the presence of DNaseI to avoid unspecific coprecipitation of DNA-binding proteins with DNA fragments present in the lysates (Agoston and Schulte, 2009). Antibodies for immunoprecipitation are listed in Table S2. For pulldown experiments with GST-PARP1 fusion proteins, HEK293T cells were transfected with 0.112 fmol pcDNA3 carrying hMEIS2D N-terminally fused to a single HA tag and 0.29 fmol of the PARP1-GST fusion constructs (a gift from V. Schreiber; Schreiber et al., 2002) in the mammalian expression vector pBC-GST (Chatton et al., 1995). To control for unspecific binding, cells were transfected with 0.112 fmol pcDNA3-MEIS2d-HA together with 0.29 fmol pBC-GST or with 0.112 fmol pcDNA3-MEIS2d-HA together with 0.29 fmol pBluescript. Cell transfection was performed by calcium phosphate transfection following standard procedures. 48 h after transfection, cell extracts were prepared by subcellular fractionation as described previously by Agoston et al. (2014). Nuclear and cytoplasmic extracts were combined and treated with 100 U RNase-free DNaseI (Roche) for 30 min at 4°C. Extracts were briefly centrifuged, and the supernatants were incubated for 2.5 h at 4°C under constant overhead shaking with 60 µl Glutathione Magnetic Beads (Thermo Fisher Scientific) prewashed in 125 mM Tris-HCl, pH 8, and 150 mM NaCl2 containing cOmplete protease inhibitor (Roche) and PhosSTOP phosphatase inhibitors (Sigma Aldrich). Beads were collected by magnetic separation, and bound proteins were eluted by addition of 50 µl 2× LDS loading buffer (Invitrogen) and heating for 10 min at 72°C. Western blot analysis was performed following standard procedures with the antibodies listed in Table S2 and visualized using an Odyssey Fc imager (LI-COR Biosciences).
ChIP
ChIP was performed as described previously (Kutejova et al., 2008; Agoston et al., 2014). In brief, aNS cells were transduced with Pax6 as described previously by Agoston et al. (2014) and in vitro differentiated by growth factor withdrawal and plating on laminin for the times indicated (Fig. S5). Cells were washed in PBS, and cross-linking was performed for 10 min at RT in 2% PFA made from freshly prepared 18.5% PFA. The reaction was quenched by the addition of glycine (final concentration, 100 mM) for 5 min at 4°C in an overhead rotator. Cells were pelleted by centrifugation for 5 min at 3,000 rpm and 4°C and then washed once in PBS containing cOmplete protease inhibitor. The pellet of 107 cells was resuspended in 1 ml buffer L1 (50 mM Tris, pH 8.0, 2 mM EDTA, 0.1% IGEPAL-CA630, 10% glycerol, and cOmplete protease inhibitor) and incubated on ice for 5 min. Cells were pelleted by centrifugation (3,000 rpm for 5 min at 8°C) and resuspended in 1.2 ml SDS lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% SDS, and cOmplete protease inhibitor). Cell lysis was verified by visual inspection. Chromatin was sheared to a mean length of 100–500 bp with a Bioruptor Plus (4°C; Diagenode) with cycle numbers optimized for each cell population and pelleted by centrifugation (13,000 rpm for 10 min at 8°C). Chromatin fragment length was verified by agarose gel electrophoresis. Chromatin was precleared by incubation for 2 h at 4°C, rotating, with 100 µl of 1:1 mixture of protein A/protein G DynaBeads (Thermo Fisher Scientific). DynaBeads were equilibrated in ChIP dilution buffer (50 mM Tris-HCl, pH 8.1, 200 mM NaCl, 5 mM EDTA, 0.5% IGEPAL-CA630, and cOmplete protease inhibitor). Immunoprecipitation was performed with chromatin corresponding with ∼1.6 × 106 cells per individual reaction with the antibodies listed in Table S2 in a volume of 1.8 ml in ChIP dilution buffer overnight at 4°C while rotating. Chromatin-immune complexes were collected by incubation with 50 µl of a 1:1 mixture of protein A/protein G DynaBeads (equilibrated in ChIP-dilution buffer) for 1 h at 4°C while rotating. Bead chromatin complexes were washed once in low-salt wash buffer (20 mM Tris-HCl, pH 8.1, 150 mM NaCl, 2 mM EDTA, 0.55% Triton X-100, 0.1% SDS, and cOmplete protease inhibitor) followed by three washes each with 800 µl of the following buffers: (a) high-salt wash buffer (20 mM Tris-HCl, pH 8.1, 500 mM NaCl, 2 mM EDTA, 1% IGEPAL-CA630, 0.1% SDS, and cOmplete protease inhibitor), (b) LiCl wash buffer (20 mM Tris-HCl, pH 8.1, 500 mM LiCl, 2 mM EDTA, 1% IGEPAL-CA630, 0.1% SDS, and cOmplete protease inhibitor), and (c) Tris-EDTA (TE) buffer with cOmplete protease inhibitor. Each wash step was performed for 10 min on ice without agitation. Beads were resuspended in 100 µl of 2% SDS in TE per immunoprecipitation, and the DNA was eluted first at 25°C for 15 min with vigorous shaking (1,400 rpm) followed by resuspension in another 100 µl of 2% SDS in TE and elution at 65°C for 15 min with vigorous shaking. Supernatants were combined, substituted with NaCl to a final concentration of 250 mM, and incubated for 5 h at 65°C at 1,400 rpm while shaking. DNA was purified with MinElute PCR purification columns (QIAGEN) according to the manufacturer’s instructions except that 800 µl buffer ERC per samples was used. DNA was eluted in 50 µl elution buffer per sample. ChIP precipitates were assessed by quantitative real-time PCR with the primers listed in Table S3 and absolute QPCR SYBR Green Fluorescein Mix (Thermo Fisher Scientific) on a CFX Touch or MyiQ Real-Time PCR detection system (Bio-Rad Laboratories). Enrichment of the precipitated DNA was determined relative to the input (1:100) as 100 × 2(Ct adjusted input − Ct immunoprecipitate).
For ChIP-reChIP, the first precipitation was performed with 2 µg α-H1 antibody or 2 µg control IgG (Merck; EMD Millipore). All reactions were set up in duplicates of which one sample served to validate the efficiency of the first precipitation and the other sample was used for the second precipitation. Immunoprecipitation was performed for 4–6 h at 4°C with gentle agitation. Chromatin-immune complexes were collected by adding 50 µl of a 1:1 mixture of Protein A/Protein G DynaBeads followed by incubation for 2 h at 4°C with gentle agitation. Bead-chromatin–immune complexes were washed as described earlier in this section, with the exception that each step was performed for 10 min at 4°C with gentle agitation. To validate the efficiency of the first ChIP, DNA was eluted for qPCR as described previously by Kutejova et al. (2008). Chromatin-immune complexes were isolated by incubating the beads twice in 100 µl elution buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 1% SDS, and 50 mM NaHCO3) for 30 min at 37°C. The eluates were combined, diluted in a ninefold excess of ChIP dilution buffer (0.5% IGEPAL CA-630, 5 mM EDTA, pH 8, 200 mM NaCl, and 50 mM Tris, pH 8.1) and incubated with 2 µg PAR-specific antibodies or IgGs overnight at 4°C with gentle agitation. Chromatin-immune complexes were collected by 50 µl protein G DynaBeads. DNA was purified with MinElute columns as described in the previous paragraph and analyzed by qPCR. Primers were designed around sequences encompassing TF-binding motifs as identified with the MatInspector software package (Genomatix). Standard error was calculated between biological replicates. Statistical significance was determined by unpaired Student’s t test, and comparison between three or more groups was performed by one-way ANOVA followed by Bonferroni’s Multiple Comparisons post hoc test. Statistical significance was assumed when *, P < 0.05; **, P < 0.01; ***, P < 0.001.
RNA isolation, cDNA transcription, and quantitative real-time PCR
RNA was isolated with the RNeasy Mini kit (QIAGEN) including on-column DNaseI digestion. mRNA was reverse transcribed with the RevertAid First strand cDNA synthesis kit (Thermo Fisher Scientific) followed by qPCR with the Absolute QPCR SYBR Green Fluorescein Mix on a CFX Touch Real-Time PCR detection system. Primer sequences are given in Table S3. Gene expression was normalized to β-actin by using the 2-ΔΔCT method. qPCR measurements of each sample were performed in triplicate. Results are plotted as SEM. Statistical significance was determined by unpaired Student’s t tests.
In vitro PARylation assay
GST-coupled MEIS2 was isolated from bacterial lysates transformed with pGEX4T1-Meis2 as described previously (Agoston and Schulte, 2009) and used at 1 µg per reaction. Calf thymus histone H1 was purchased from EMD Millipore and used at 1 and 2 µg per reaction as indicated. Human recombinant PARP1 enzyme (Enzo Life Sciences) was incubated with biotinylated NAD+ (Trevigen) in 50 mM Tris, pH 8, 25 mM MgCl2, and 1 mM DTT in the presence or absence of 2.5 µg sonicated linear double-stranded salmon sperm DNA (Sigma Aldrich) per reaction as indicated. To control for background PARylating activity, PARP1 or biotinylated NAD+ were omitted from the reaction, or 6 mM 3AB was added to block PARP1 enzymatic activity pharmacologically. Reactions were incubated for 30 min at 37°C, and Western blot analysis was performed with streptavidin-linked HRP (BioLegend).
Expression profiling
3 × 106 first-passage aNS cells per experiment were transfected with 50 pmol nontargeting siRNAs (Silencer Select; Thermo Fisher Scientific). 4 h later, cells were dissociated for 5 min at 37°C in 500 µl accutase, washed once in culture medium, and incubated for 5 h with medium containing pCLIG-Pax6 viral particles and either 100 nM Olaparib in 0.01% DMSO or 0.01% DMSO alone as control. Cells were collected by centrifugation, washed in culture medium, and cultured for 36 h under nonadherent conditions in the continuing presence of 100 nM Olaparib in 0.01% DMSO or 0.01% DMSO alone. Differentiation was performed by growth factor withdrawal and plating on laminin as substrate for 10 h. Cells were harvested by scraping off in culture medium, pelleted for 3 min at 1,000 rpm at RT, resuspended in 1 ml ice-cold PBS, and centrifuged again at 6,000 rpm and 4°C for 2 min. RNA isolation was performed with RNeasy Plus Micro kits (QIAGEN) according to the manufacturer’s instructions. RNA concentration and quality was assessed with an Experion RNA StdSens Analysis kit (Bio-Rad Laboratories). RNA was amplified using the Ovation Pico WTA System, samples were labeled with the Encore biotin module (NUGEN), and then samples were washed and stained using the hybridization, wash, and stain kit (Affymetrix). Mouse Gene 1.0 ST arrays (Affymetrix) were hybridized with 2.5 mg labeled cDNA each. Staining and scanning (Gene ChIP Scanner 3000 7G; Affymetrix) were done according to the Affymetrix expression protocol. Statistical analysis was done with the statistical computing environment R. Additional software packages (affy, geneplotter, multtest, and vsn) were taken from the Bioconductor project. For microarray preprocessing, probe level normalization was performed using the variance stabilization method (vsn; Huber et al., 2002). To reduce the dimension of the microarray data, data were filtered with an intensity filter (intensity of a gene should be >100 in ≥0.25% of the samples if the group size is equal) and a variance filter (interquartile range of log2 intensities should be ≥0.5 if the group size is equal). P-values were calculated with two-sample t tests (variance = equal) to identify genes that are differentially expressed between two groups. For multiple testing problems, we used a false discovery rate (FDR; Hochberg and Benjamini, 1990). Fold changes (FCs) between the two groups were calculated for each gene (FC expression level ≤1.5 and FDR <0.06). The lists of differentially expressed genes were filtered with FDR and FC criteria. The array data were submitted to NCBI Gene Expression Omnibus under the accession number GSE74314.
GO term analysis was performed with the PANTHER Overrepresentation Test (release 20150430; http://geneontology.org/) on the GO Ontology database released 2015-08-06 with Bonferroni’s correction for multiple testing.
To validate candidate gene expression by qPCR and to compare their transcript levels in Olaparib-treated cultures and after Meis1/2 knockdown, three treatment regimes were applied: (a) transfection with nontargeting siRNAs (4390846; 50 pmol transfected per 2 × 106 cells) and then incubation and differentiation in 0.01% DMSO (control); (b) transfection with Silencer Select siRNAs targeting Meis1 and Meis2 (25 pmol each transfected per 2 × 106 cells) followed by incubation and differentiation in 0.01% DMSO; and (c) transfection with nontargeting siRNAs followed by incubation and differentiation in 100 nM Olaparib in 0.01% DMSO. Each experiment was performed in duplicates. siRNA sequences were Meis1: 5′-UCAUGAUAUUUGUCGCGAC-3′ and Meis2 5′-CAGUGAAGAUGUAACAAGA-3′. Both siRNAs were mixed at equal molar ratios, and RNA duplexes were transfected with Metafectene Pro (Biontex). After siRNA transfection, cells were grown for 48 h as free-floating spheres before they were induced to differentiate for 10 h by removal of EGF/FGF2 from the medium and plating on laminin-coated tissue culture dishes. RNA isolation and assessment of concentration and quality were performed as described in the previous paragraph.
Protein identification using liquid chromatography–coupled tandem mass spectrometry
Protein affinity purification and digest
For isolation of proteins that copurify with MEIS2 in a GST pulldown assay, GST-coupled MEIS2 (MEIS2-GST) and GST alone were isolated from bacterial lysates transformed with pGEX4T1-MEIS2 and pGEX4T1 as described previously (Agoston and Schulte, 2009). Protein lysates were generated from ∼5 × 107 N2A cells as described in the Analysis of PARP1-containing protein complexes section and divided equally between GST pulldown experiments with MEIS2-GST and GST alone. Pulldown followed published protocols. Samples were separated by SDS-PAGE, stained with Colloidal Coomassie, and then prominent protein bands visible in the MEIS2-GST precipitate were excised from the gel. As controls, equal-sized gel bands at corresponding positions in the gel were collected from the GST pulldown. Generally, gel slices were reduced (5 mM DTT), alkylated (20 mM iodoacetymide), and subsequently digested using Trypsin (1:15; o/n). The supernatants containing the proteolytic peptides were stored at −80°C until injection into the liquid chromatography tandem mass spectrometry system.
For isolation of proteins that copurify with HA-tagged MEIS2 protein by immunoprecipitation, ∼5 × 107 SK-N-BE(2) cells stably expressing MESI2D-HA from a tetracycline-inducible promoter were used. Protein extracts were prepared 24 h after transgene expression was induced by doxycycline stimulation. HA-tagged MEIS2 proteins were isolated by immunoprecipitation with 2 µg antibodies against the HA probe (rb; Y-11; Santa Cruz Biotechnology, Inc.) as described in the Analysis of PARP1-containing protein complexes section. SK-N-BE(2)–MEIS2-HA cells not treated with doxycycline were also used for immunoprecipitation with HA-specific antibodies and served as controls. Proteolytic cleavage of immunocomplexes was performed on the Dynabead-bound proteins using Lys-C and trypsin as described previously (Fischer et al., 2015).
Liquid chromatography tandem mass spectrometry analyses
The proteolytic digests were loaded using a nano-HPLC (Dionex RSLCnano) on reverse-phase columns (trapping column: Acclaim PepMap c18, particle size 2 µm, L = 20 mm; analytical column: Acclaim PepMap c18, particle size 2 µm, L = 25 cm; Thermo Fisher Scientific) and eluted in organic phase gradients (Buffer A: 95% H2O, 5% DMSO, and 0.1% formic acid; Buffer B: 80% acetonitrile, 15% H2O, 5% DMSO, and 0.1% formic acid). Typically, gradients were ramped from 4 to 48% buffer B in 80 min at flowrates of 300 nl/min. Peptides eluting from the column were ionized online using a Nanospray Flex Ion source and analyzed in an Orbitrap Elite mass spectrometer. Mass spectra were acquired over the m/z range 350–1,600 at a resolution of 120,000, and sequence information was acquired by computer-controlled, data-dependent automated switching to tandem mass spectrometry mode using collision energies based on mass and charge state of the candidate ions (FTIT and TOP15).
Data processing
The datasets were processed using the Proteome Discoverer software package (version 2.1.0.81; Thermo Fisher Scientific). For the data obtained with the human cell line SK-N-BE(2) (shown in Fig. 2), proteins were identified by matching the derived mass lists against a customized Swissprot Homo sapiens database (TaxID, 9606; downloaded from Swissprot.org with common contaminants added) using Sequest (Thermo Fisher Scientific). For the results obtained with the murine Neuro2a cell line (shown in Fig. S1 C), the datasets were processed using a standard proteomics script with the software DataAnalysis 4.0 (Service Pack 1; Build 253; Bruker) and exported as MASCOT generic files. Proteins were identified by matching the derived mass lists against the NCBI RefSeq Non-Redundant Proteins database on a local MASCOT server (Matrix Science). In general, a mass tolerance of 10 ppm for parent ion spectra and 0.6 D for fragment ion spectra, two missed cleavages, oxidation of Met (dynamic modification), acetylation of the protein N terminus (dynamic modification), and carbamidomethyl cysteine (fixed modification) were selected as matching parameters in the search program. Results were evaluated using a percolator node (high-confidence q-value; FDR < 0.01) to exclude false-positive evaluations.
Accession nos.
The array data have been uploaded to NCBI Gene Expression Omnibus under the accession number GSE74314. The mass spectrometry proteomics data have been deposited to the PRIDE online repository under accession no. PXD007078 (Vizcaíno et al., 2016).
Online supplemental material
Fig. S1 contains supporting data showing that histone H1 variant 4 is a prominent linker histone present at the Dcx(−2.7) site in adult neural stem and progenitor cells and that MEIS2 and PARP1 physically interact in different tissues of ongoing neurogenesis in mouse and chick embryos. Fig. S2 presents higher-exposure images of the blots shown in Fig. 2 and shows that MEIS2 and PARP1 can form heterodimers in solution without PBX1. Fig. S3 contains supporting data showing that cells immunopositive for activated caspase 3 are rare during the first 24 h of in vitro differentiation of SVZ stem and progenitor cells, that MEIS2 is not PARylated by PARP1 in vitro, and that the shRNAs used for PARP1 knockdown effectively deplete PARP1 protein. Fig. S4 shows transcript expression of Igfbp5, Smoc1, Nrep, Pygb, and Draxin in the adult mouse forebrain and the relative position of consensus motifs for MEIS/PBX, PAX, and DLX transcription factors in the upstream sequences of the Dcx, Draxin, and Nrep genes. Fig. S5 contains supporting data showing that Pax6 transduction does not lead to neuronal gene expression before aNSs are induced to differentiate by growth factor withdrawal and plating on laminin. Table S1 contains the results of the GO term enrichment analysis of genes deregulated upon neuronally directed in vitro differentiation in the presence of Olaparib. Table S2 gives antibody specifications. Table S3 lists primer sequences. Table S4 lists the sample sizes and number of biological repeats of all experiments shown. Table S5 lists all significantly up- or down-regulated genes identified in the Affymetrix expression profiling experiments with FC values, FDRs, and statistical significance.
Acknowledgments
We thank A. Buchberg, M. Mittelbronn, M. Cleary (Stanford University, Stanford, CA), and V. Schreiber (Université de Strasbourg, Strasbourg, France) for reagents.
The work was supported by the Schram Foundation (T287/21795/2011) and the Deutsche Forschungsgemeinschaft grant SCHU1218/3-1 to D. Schulte.
The authors declare no competing financial interests.
Author contributions: A.-C. Hau, B.M. Grebbin, Z. Agoston, M. Anders-Maurer, T. Müller, A. Groß, and J. Kolb performed experiments; J.D. Langer contributed the mass spectrometry analyses; C. Döring contributed the Affymetrix expression data; and D. Schulte conceived the study, supervised the experiments, and wrote the manuscript.
Footnotes
Abbreviations used:
- aNS
- adult neurosphere
- BRCT
- BRCA1 C-terminal
- ChIP
- chromatin immunoprecipitation
- Dcx
- doublecortin
- DLX
- distal-less homeobox
- FC
- fold change
- FDR
- false discovery rate
- GBM
- glioblastoma multiforme
- GFAP
- glial fibrillary acidic protein
- GO
- Gene Ontology
- HD
- homeodomain
- MEIS
- myeloid ecotropic viral integration site
- PAR
- poly-ADP-ribose
- PBX
- pre–B-cell leukemia homeobox
- qPCR
- quantitative PCR
- RMS
- rostral migratory stream
- SVZ
- subventricular zone
- TALE
- three-amino acid loop extension
References
- Agoston Z., and Schulte D.. 2009. Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development. 136:3311–3322. 10.1242/dev.037770 [DOI] [PubMed] [Google Scholar]
- Agoston Z., Heine P., Brill M.S., Grebbin B.M., Hau A.-C., Kallenborn-Gerhardt W., Schramm J., Götz M., and Schulte D.. 2014. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development. 141:28–38. 10.1242/dev.097295 [DOI] [PubMed] [Google Scholar]
- Amin S., Donaldson I.J., Zannino D.A., Hensman J., Rattray M., Losa M., Spitz F., Ladam F., Sagerström C., and Bobola N.. 2015. Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell. 32:265–277. 10.1016/j.devcel.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes C.A., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., and Tapscott S.J.. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 14:465–477. 10.1016/S1097-2765(04)00260-6 [DOI] [PubMed] [Google Scholar]
- Bjornsson C.S., Apostolopoulou M., Tian Y., and Temple S.. 2015. It takes a village: Constructing the neurogenic niche. Dev. Cell. 32:435–446. 10.1016/j.devcel.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill M.S., Snapyan M., Wohlfrom H., Ninkovic J., Jawerka M., Mastick G.S., Ashery-Padan R., Saghatelyan A., Berninger B., and Götz M.. 2008. A dlx2- and pax6-dependent transcriptional code for periglomerular neuron specification in the adult olfactory bulb. J. Neurosci. 28:6439–6452. 10.1523/JNEUROSCI.0700-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.K., Jaffe L., Capovilla M., Botas J., and Mann R.S.. 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 78:603–615. [DOI] [PubMed] [Google Scholar]
- Chang C.P., Jacobs Y., Nakamura T., Jenkins N.A., Copeland N.G., and Cleary M.L.. 1997. Meis proteins are major in vivo binding partners for wild-type but not chimeric Pbx proteins. Mol. Cell. Biol. 17:5679–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton B., Bahr A., Acker J., and Kedinger C.. 1995. Eukaryotic GST fusion vector for the study of protein-protein associations in vivo: application to interaction of ATFa with Jun and Fos. Biotechniques. 18:142–145. [PubMed] [Google Scholar]
- Choe S.K., Ladam F., and Sagerström C.G.. 2014. TALE factors poise promoters for activation by Hox proteins. Dev. Cell. 28:203–211. 10.1016/j.devcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.R., Ortega F., Brill M.S., Beckervordersandforth R., Petrone C., Schroeder T., Gotz M., and Berninger B.. 2011. Continuous live imaging of adult neural stem cell division and lineage progression in vitro. Development. 138:1057–1068. 10.1242/dev.061663 [DOI] [PubMed] [Google Scholar]
- Dawicki-McKenna J.M., Langelier M.F., DeNizio J.E., Riccio A.A., Cao C.D., Karch K.R., McCauley M., Steffen J.D., Black B.E., and Pascal J.M.. 2015. PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol. Cell. 60:755–768. 10.1016/j.molcel.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund E. 2011. The role of Hox proteins in leukemogenesis: Insights into key regulatory events in hematopoiesis. Crit. Rev. Oncog. 16:65–76. 10.1615/CritRevOncog.v16.i1-2.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S., Wu W.F., Langelier M.F., Yang J.C., Easton L.E., Riccio A.A., Pascal J.M., and Neuhaus D.. 2015. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell. 60:742–754. 10.1016/j.molcel.2015.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., Li B., Zewdu R., Wells V., Hebert J.M., Karner C., Anderson M.J., Williams T., Dixon J., Dixon M.J., et al. 2011. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell. 21:627–641. 10.1016/j.devcel.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Langer J.D., and Osiewacz H.D.. 2015. Identification of potential mitochondrial CLPXP protease interactors and substrates suggests its central role in energy metabolism. Sci. Rep. 5:18375 10.1038/srep18375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Iglesias M.J., Ramirez A., Monzo M., Steuer B., Martínez J.M., Jorcano J.L., and Alonso A.. 1993. Specific expression in adult mice and post-implantation embryos of a transgene carrying the histone H10 regulatory region. Differentiation. 55:27–35. 10.1111/j.1432-0436.1993.tb00030.x [DOI] [PubMed] [Google Scholar]
- Ghosh D., Ulasov I.V., Chen L., Harkins L.E., Wallenborg K., Hothi P., Rostad S., Hood L., and Cobbs C.S.. 2016. TGFβ-responsive HMOX1 expression is associated with stemness and invasion in glioblastoma multiforme. Stem Cells. 34:2276–2289. 10.1002/stem.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B.A., Zhang Y., Jiang H., Hussey K.M., Shrimp J.H., Lin H., Schwede F., Yu Y., and Kraus W.L.. 2016. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 353:45–50. 10.1126/science.aaf7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson J.G., Lin P.T., Flanagan L.A., and Walsh C.A.. 1999. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 23:257–271. 10.1016/S0896-6273(00)80778-3 [DOI] [PubMed] [Google Scholar]
- Golonzhka O., Nord A., Tang P.L.F., Lindtner S., Ypsilanti A.R., Ferretti E., Visel A., Selleri L., and Rubenstein J.L.R.. 2015. Pbx regulates patterning of the cerebral cortex in progenitors and postmitotic neurons. Neuron. 88:1192–1207. 10.1016/j.neuron.2015.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebbin B.M., Hau A.-C., Groß A., Anders-Maurer M., Schramm J., Koss M., Wille C., Mittelbronn M., Selleri L., and Schulte D.. 2016. Pbx1 is required for adult SVZ neurogenesis. Development. 143:2281–2291. 10.1242/dev.128033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack M.A., Saghatelyan A., de Chevigny A., Pfeifer A., Ashery-Padan R., Lledo P.M., and Götz M.. 2005. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat. Neurosci. 8:865–872. 10.1038/nn1479 [DOI] [PubMed] [Google Scholar]
- Hassa P.O. 2009. The molecular “Jekyll and Hyde” duality of PARP1 in cell death and cell survival. Front. Biosci. 14:72–111. 10.2741/3232 [DOI] [PubMed] [Google Scholar]
- Hochberg Y., and Benjamini Y.. 1990. More powerful procedures for multiple significance testing. Stat. Med. 9:811–818. 10.1002/sim.4780090710 [DOI] [PubMed] [Google Scholar]
- Huber W., von Heydebreck A., Sültmann H., Poustka A., and Vingron M.. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 18(Suppl 1):S96–S104. 10.1093/bioinformatics/18.suppl_1.S96 [DOI] [PubMed] [Google Scholar]
- Jackson E.L., and Alvarez-Buylla A.. 2008. Characterization of adult neural stem cells and their relation to brain tumors. Cells Tissues Organs (Print). 188:212–224. 10.1159/000114541 [DOI] [PubMed] [Google Scholar]
- Ji Y., and Tulin A.V.. 2010. The roles of PARP1 in gene control and cell differentiation. Curr. Opin. Genet. Dev. 20:512–518. 10.1016/j.gde.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B.G., Solum D., Song E.J., Lee K.J., Rose D.W., Glass C.K., and Rosenfeld M.G.. 2004. Activating the PARP-1 sensor component of the Groucho/ TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell. 119:815–829. 10.1016/j.cell.2004.11.017 [DOI] [PubMed] [Google Scholar]
- Karl C., Couillard-Despres S., Prang P., Munding M., Kilb W., Brigadski T., Plötz S., Mages W., Luhmann H., Winkler J., et al. 2005. Neuronal precursor-specific activity of a human doublecortin regulatory sequence. J. Neurochem. 92:264–282. 10.1111/j.1471-4159.2004.02879.x [DOI] [PubMed] [Google Scholar]
- Kim M.Y., Mauro S., Gévry N., Lis J.T., and Kraus W.L.. 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 119:803–814. 10.1016/j.cell.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R., and Kraus W.L.. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell. 39:736–749. 10.1016/j.molcel.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R., Gamble M.J., Frizzell K.M., Berrocal J.G., Kininis M., and Kraus W.L.. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 319:819–821. 10.1126/science.1149250 [DOI] [PubMed] [Google Scholar]
- Kutejova E., Engist B., Self M., Oliver G., Kirilenko P., and Bobola N.. 2008. Six2 functions redundantly immediately downstream of Hoxa2. Development. 135:1463–1470. 10.1242/dev.017624 [DOI] [PubMed] [Google Scholar]
- Langelier M.-F., Planck J.L., Roy S., and Pascal J.M.. 2012. Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 336:728–732. 10.1126/science.1216338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L., Ballantyne E.B., Zhang X., and Lupien M.. 2011. PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet. 7:e1002368 10.1371/journal.pgen.1002368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N., Leonardo E., Azpiazu N., Serrano A., Morata G., Martínez C., and Torres M.. 1999. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 402:425–429. 10.1038/46580 [DOI] [PubMed] [Google Scholar]
- Ninkovic J., Steiner-Mezzadri A., Jawerka M., Akinci U., Masserdotti G., Petricca S., Fischer J., von Holst A., Beckers J., Lie C.D., et al. 2013. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 13:403–418. 10.1016/j.stem.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige S.L., Thomas S., Stoick-Cooper C.L., Wang H., Maves L., Sandstrom R., Pabon L., Reinecke H., Pratt G., Keller G., et al. 2012. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 151:221–232. 10.1016/j.cell.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkov D., Mateos San Martín D., Fernandez-Díaz L.C., Rosselló C.A., Torroja C., Sánchez-Cabo F., Warnatz H.J., Sultan M., Yaspo M.L., Gabrieli A., et al. 2013. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Reports. 3:1321–1333. 10.1016/j.celrep.2013.03.029 [DOI] [PubMed] [Google Scholar]
- Plane J.M., Grossenbacher S.K., and Deng W.. 2012. PARP-1 deletion promotes subventricular zone neural stem cells toward a glial fate. J. Neurosci. Res. 90:1489–1506. 10.1002/jnr.23040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier G.G., de Murcia G., Jongstra-Bilen J., Niedergang C., and Mandel P.. 1982. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. USA. 79:3423–3427. 10.1073/pnas.79.11.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds B.A., and Weiss S.. 1992. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 255:1707–1710. 10.1126/science.1553558 [DOI] [PubMed] [Google Scholar]
- Saeki H., Ohsumi K., Aihara H., Ito T., Hirose S., Ura K., and Kaneda Y.. 2005. Linker histone variants control chromatin dynamics during early embryogenesis. Proc. Natl. Acad. Sci. USA. 102:5697–5702. 10.1073/pnas.0409824102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakini N., Turchi L., Bergon A., Holota H., Rekima S., Lopez F., Paquis P., Almairac F., Fontaine D., Baeza-Kallee N., et al. 2016. A positive feed-forward loop associating EGR1 and PDGFA promotes proliferation and self-renewal in glioblastoma stem cells. J. Biol. Chem. 291:10684–10699. 10.1074/jbc.M116.720698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V., Amé J.C., Dollé P., Schultz I., Rinaldi B., Fraulob V., Ménissier-de Murcia J., and de Murcia G.. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277:23028–23036. 10.1074/jbc.M202390200 [DOI] [PubMed] [Google Scholar]
- Selleri L., Depew M.J., Jacobs Y., Chanda S.K., Tsang K.Y., Cheah K.S., Rubenstein J.L., O’Gorman S., and Cleary M.L.. 2001. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 128:3543–3557. [DOI] [PubMed] [Google Scholar]
- Torres C.M., Biran A., Burney M.J., Patel H., Henser-Brownhill T., Cohen A.S., Li Y., Ben-Hamo R., Nye E., Spencer-Dene B., et al. 2016. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science. 353:aaf1644 10.1126/science.aaf1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A., and Spradling A.. 2003. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 299:560–562. 10.1126/science.1078764 [DOI] [PubMed] [Google Scholar]
- Vitobello A., Ferretti E., Lampe X., Vilain N., Ducret S., Ori M., Spetz J.F., Selleri L., and Rijli F.M.. 2011. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev. Cell. 20:469–482. 10.1016/j.devcel.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44(D1):D447–D456. 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad J.A., Alexander J.M., Truty R.M., Shrikumar A., Li F., Eilertson K.E., Ding H., Wylie J.N., Pico A.R., Capra J.A., et al. 2012. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 151:206–220. 10.1016/j.cell.2012.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Q., Auer B., Stingl L., Berghammer H., Haidacher D., Schweiger M., and Wagner E.F.. 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9:509–520. 10.1101/gad.9.5.509 [DOI] [PubMed] [Google Scholar]
- Yang S.-M., Kim B.J., Norwood Toro L., and Skoultchi A.I.. 2013. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc. Natl. Acad. Sci. USA. 110:1708–1713. 10.1073/pnas.1213266110 [DOI] [PMC free article] [PubMed] [Google Scholar]