Starvation-induced unconventional secretion of Acb1 requires ESCRT-I, -II, and -III and Grh1. Cruz-Garcia et al. report that SOD1 and its mutant form linked to amyotrophic lateral sclerosis are also secreted upon nutrient starvation in a Grh1- and ESCRT-I–, -II–, and -III–dependent process. The authors identify a conserved diacidic motif in Acb1 and SOD1 that is necessary for their export in yeast and human cells.
Abstract
The nutrient starvation-specific unconventional secretion of Acb1 in Saccharomyces cerevisiae requires ESCRT-I, -II, and -III and Grh1. In this study, we report that another signal sequence lacking cytoplasmic protein, superoxide dismutase 1 (SOD1), and its mutant form linked to amyotrophic lateral sclerosis (ALS), is also secreted by yeast upon nutrient starvation in a Grh1- and ESCRT-I–, -II–, and -III–dependent process. Our analyses reveal that a conserved diacidic motif (Asp-Glu) in these proteins is necessary for their export. Importantly, secretion of wild-type human SOD1 and the ALS-linked mutant in human cells also require the diacidic residues. Altogether, these findings reveal information encoded within the cytoplasmic proteins required for their unconventional secretion and provide a means to unravel the significance of the cytoplasmic versus the secreted form of mutant SOD1 in the pathology of ALS. We also propose how cells, based on a signal-induced change in cytoplasmic physiology, select a small pool of a subset of cytoplasmic proteins for unconventional secretion.
Introduction
A large number of proteins that lack a signal sequence for entering the conventional ER–Golgi complex pathway of secretion are exported by eukaryotic cells. These include but are not restricted to tissue transglutaminase, interleukin (IL)-1β and -1α, galectin-3, macrophage migration inhibitory factor, insulin-degrading enzymes, fibroblast growth factor 2, and diazepam-binding inhibitor (Kinseth et al., 2007; Rabouille et al., 2012; Malhotra, 2013). The challenge is to reveal the mechanism and physiological significance of this unconventional mode of secretion.
In search of a function for Golgi reassembling and stacking proteins (GRASPs), we discovered that deletion of the single GRASP gene in Dictyostelium discoideum blocked starvation-specific secretion of signal sequence lacking acyl-CoA–binding protein AcbA (Kinseth et al., 2007). This rekindled interest in this unconventional mode of secretion since its first description that signal sequence lacking IL-1β was secreted by activated macrophages (Auron et al., 1984; Rubartelli et al., 1990). Subsequent analysis revealed that the function of GRASP orthologues (Grh1 in budding yeast) in the secretion of AcbA orthologues (Acb1 in yeast) was conserved in eukaryotes (Duran et al., 2010; Manjithaya et al., 2010). GRASP proteins are also shown to have a role in the secretion of signal sequence lacking IL-1β (Dupont et al., 2011; Zhang et al., 2015) and insulin-degrading enzymes (Son et al., 2016). Altogether, these data lend confidence in the involvement of GRASP proteins in unconventional secretion. The Grh1-dependent export of Acb1 in yeast involves endosomal sorting complexes required for transport (ESCRT)-I, -II, and -III. Surprisingly, however, ESCRT-mediated secretion of Acb1 is independent of Vps4, which strongly suggests that its secretion is without incorporation into intraluminal vesicles at multivesicular bodies (Curwin et al., 2016). The secretion of Acb1 is therefore independent of exosome-mediated release of cytoplasmic contents. Autophagy-related proteins are also suggested to play a role in unconventional protein secretion, but it is unclear whether the dependence on Atg genes is direct or if they are required to keep cells in a response-competent status under nutrient starvation or stress conditions that result in the release of proteins such as Acb1 and IL-1β (Duran et al., 2010; Manjithaya et al., 2010; Dupont et al., 2011; Zhang et al., 2015).
Conditions that trigger secretion of Acb1 result in the biogenesis of a new Grh1-containing compartment called the compartment for unconventional protein secretion (CUPS) that appears in two sequential forms (Bruns et al., 2011; Curwin et al., 2016). Grh1 is first observed in a cluster of vesicles that are produced independently of COPII- and COPI-mediated export of membranes from the ER and the Golgi, respectively. Membrane export from late Golgi compartment, however, is necessary for production of these vesicular elements (Bruns et al., 2011; Cruz-Garcia et al., 2014). Clusters of Grh1-containing vesicles are then found surrounded by curved saccules (Curwin et al., 2016). The stability of this form of CUPS is dependent on the activity of the phosphatidylinositol 3-phosphate kinase Vps34 (Cruz-Garcia et al., 2014). For the sake of simplicity, we have decided to call the first stage of CUPS I-CUPS (for immature CUPS [cluster of Grh1-containing vesicles]), and we call the second stage mature CUPS (M-CUPS [Grh1-containing vesicles surrounded by curved saccules]). Acb1 is associated with M-CUPS, indicating their involvement in events leading to Acb1 secretion (Curwin et al., 2016). The pathway of unconventional secretory process is thus beginning to unravel.
The next obvious challenge in order to understand the mechanism of unconventional protein secretion is to identify whether there is a motif—a sequence—used by proteins to enter this pathway. An easy means to address this concern is to ask how many other proteins are secreted by the same pathway and then to test whether such proteins contain a common sequence that affects their secretion when mutated. We now present data that exactly address this issue. We report that signal sequence lacking cytoplasmic superoxide dismutase 1 (SOD1) is exported by the same pathway used for Acb1 secretion. In addition, we reveal that Acb1 and both yeast and human SOD1 contain a diacidic motif that is necessary for their release from cells. The description of our findings and their significance in understanding human pathology of mutations in SOD1 linked to amyotrophic lateral sclerosis (ALS) follows.
Results and discussion
SOD1 and Acb1 follow the same ER–Golgi independent secretory pathway
We have previously shown that Acb1 is secreted by yeast incubated in 2% potassium acetate for 2–3 h (Curwin et al., 2016). Extraction of the yeast cell wall was used to quantitate the secreted pool of Acb1. Lack of cofilin, a cytosolic protein of similar abundance, size, and charge as Acb1, in the extracted material served as a control for release of cytoplasmic content as a result of cell lysis during experimental manipulations (Curwin et al., 2016). In addition, presence of the cell wall protein Bgl2 was used as a control to monitor extraction efficiency in this assay (Curwin et al., 2016).
The cytosolic copper–zinc superoxide dismutase SOD1, which lacks a signal sequence, is released by several different mammalian cells (Mondola et al., 1996, 1998; Cimini et al., 2002). Although the mechanism of SOD1 export by cells is unclear, it has been reported that rat pituitary GH3 cells secrete SOD1 in response to depolarization through SNARE-dependent exocytosis, which allowed Santillo et al. (2007) to suggest the involvement of large dense-core vesicles containing SOD1. SOD1 has also been found associated with exosomal fraction obtained from tumor cell lines in high-throughput studies (ExoCarta database). Furthermore, immunochemical analysis has revealed that a fraction of extracellular human SOD1 is associated with exosomes secreted by mouse astrocytes and mouse motor neurons overexpressing wild-type human SOD1 (Gomes et al., 2007; Basso et al., 2013; Grad et al., 2014). Therefore, SOD1 is released by cells, and it might be associated with a membrane-bound compartment, but the conditions that trigger its release as well as the underlying mechanism are largely unknown.
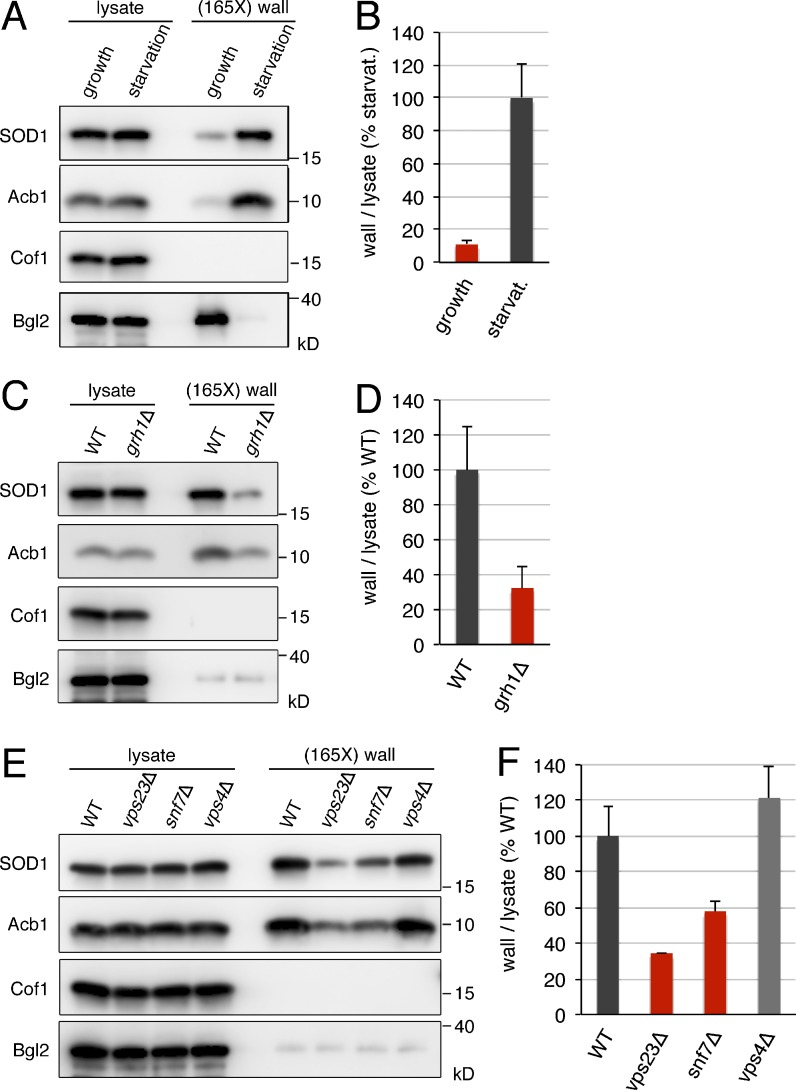
Budding yeast also express a signal sequence lacking SOD1 of 16 kD. Therefore, we tested whether SOD1 was secreted when cells are grown in nutrient-rich conditions or incubated in potassium acetate. Yeast were grown to mid-logarithmic phase in synthetic complete (SC) medium or cultured in 2% potassium acetate for 2.5 h. After collecting the cells, the cell wall was extracted as described previously and then Western blotted with specific antibodies to detect Acb1, SOD1, cofilin, and Bgl2 (Curwin et al., 2016). We detected a moderate amount of SOD1 in the cell wall of yeast grown in nutrient-rich medium (Fig. 1 A). However, cells incubated in potassium acetate revealed a ninefold increase in cell wall–extractable SOD1 compared with control cells (Fig. 1, A and B). This level of starvation-induced secretion of SOD1 was similar to that reported previously and shown in this study for Acb1 (Fig. 1 A; Curwin et al., 2016). Cofilin was not detected in the cell wall extracts obtained from starved cells or cells cultured in nutrient-rich medium (Fig. 1 A). These data indicate that, like Acb1, SOD1 secretion by yeast cells is stimulated by starvation in potassium acetate.
Figure 1.
SOD1 and Acb1 are secreted by the same pathway in response to nutrient starvation. (A and B) Secretion of SOD1 is induced by nutrient starvation. Wild-type cells were grown in nutrient-rich medium to mid-logarithmic phase (growth). An aliquot of this culture was washed twice and incubated in 2% potassium acetate for 2.5 h (starvation). Cell wall proteins were extracted from equal numbers of growing and starved cells followed by precipitation with TCA. Lysates and cell wall–extracted proteins (wall) were analyzed by Western blotting. SOD1 levels were quantified as the ratio of wall to lysate SOD1 and are represented as percentages of the starved wild-type culture. (C and D) Grh1 is required for SOD1 secretion. The ratio of wall to lysate SOD1 in wild-type and grh1Δ cells incubated in 2% potassium acetate for 2.5 h was determined as in A and B. (E and F) SOD1 secretion requires a subset of ESCRT proteins. The ratio of wall to lysate SOD1 in wild-type, vps23Δ, snf7Δ, and vps4Δ cells incubated in 2% potassium acetate for 2.5 h was determined as in A and B. (165X) wall indicates that the wall sample represents the material from 165-fold the number of cells analyzed in the lysate sample. Results in B, D, and F are shown as the means ± SEM of three experiments.
Secretion of Acb1 depends on Grh1 and a subset of ESCRT components (Curwin et al., 2016), and we tested their involvement in potassium acetate–induced SOD1 export. In GRH1-deleted cells, the level of SOD1 secreted to the cell wall after 2.5 h of starvation in potassium acetate decreased by 68% compared with the wild-type strain (Fig. 1, C and D). Yeast strains deleted of either VPS23 (ESCRT-I), VPS25 (ESCRT-II), or SNF7 (ESCRT-III) showed a similar reduction in the levels of cell wall–associated SOD1 as compared with the wild-type control (Fig. 1, E and F; and Fig. S1). However, as reported previously for the lack of requirement of ESCRT-0 and Vps4 in Acb1 secretion (Curwin et al., 2016), deletion of either VPS27 (ESCRT-0) or VPS4 did not have a significant effect on SOD1 export, and if anything, loss of Vps4 slightly increased the pool of secreted SOD1 (Fig. 1, E and F; and Fig. S1). Therefore, the quantitative effects of the deletions of GRH1 and the selected ESCRT genes on starvation-induced SOD1 secretion are similar to those previously reported for Acb1 release and reconfirmed in this study (Fig. 1, C and E; Curwin et al., 2016). These findings thus reveal that a new protein, SOD1, requires the same set of components as Acb1 for its export upon starvation.
Identification of a diacidic motif essential for starvation-induced SOD1 secretion
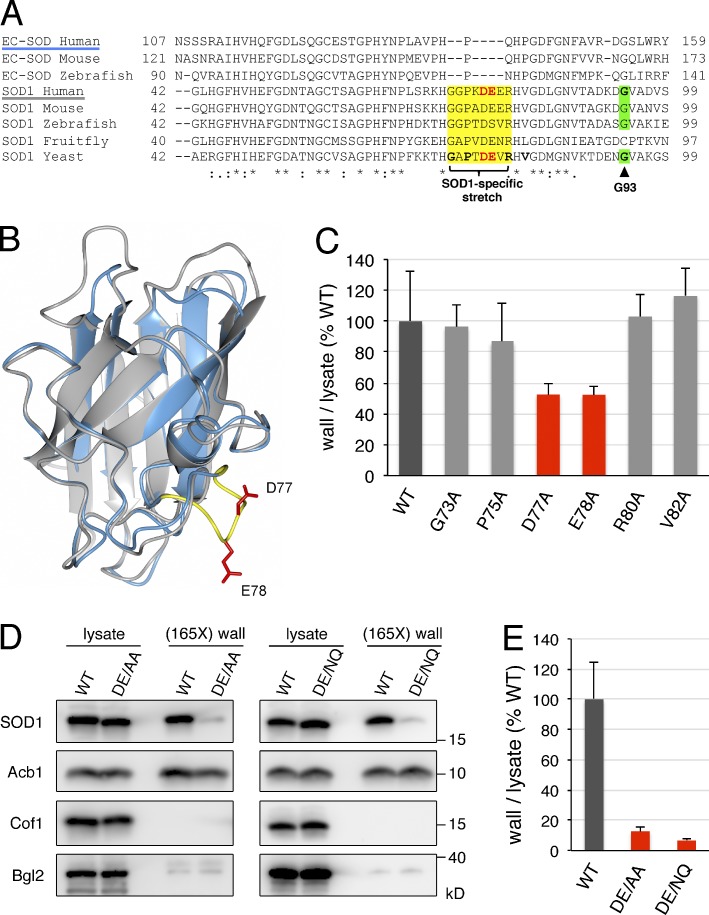
Eukaryotes are known to express two highly related classes of copper–zinc superoxide dismutase enzymes that play widespread roles in oxidative stress resistance and signaling. These two classes include the intracellular predominantly cytosolic SOD1 and the extracellular SOD (EC-SOD), which contains a signal sequence for entering the ER, thereby allowing its secretion via the conventional secretory pathway (Zelko et al., 2002). Sequence and structural analysis have revealed that EC-SOD and SOD1 diverged early in evolution (Zelko et al., 2002). In fact, the structural core of the ancestral SOD1 is highly conserved in EC-SOD and consists of a β-barrel motif with two large loops: the so-called electrostatic loop and the zinc-binding loop (Fig. 2, A and B; Zelko et al., 2002). We noticed that, within this structural core, EC-SOD and SOD1 share sequence homology with the exception of a short stretch of eight amino acids that are contained in SOD1 but not in EC-SOD (Fig. 2, A and B). This stretch (amino acids 73–GGPKDEER–80 in the sequence of human SOD1) is located in the zinc-binding loop and flanked by the conserved histidines H72 and H81, which coordinate the zinc cation. Taking into account that this short stretch is conserved in the SOD1 sequences from yeast to human and that it has been lost in the sequences of vertebrate EC-SOD, we hypothesized that it might be important for the unconventional secretion of SOD1.
Figure 2.
Identification of diacidic residues for SOD1 export. (A) Alignment of partial amino acid sequences of human, mouse, and zebrafish EC-SOD, and human, mouse, zebrafish, fruit fly, and budding yeast SOD1. The SOD1-specific stretch is shown in yellow, the residues in yeast SOD1 mutated to alanine in this study are in bold, the diacidic DE77/78 motif in human and yeast SOD1 are in bold red letters, and the G93 residue in human SOD1 mutated to alanine in ALS and its equivalent in yeast SOD1 are shown in bold and highlighted in green. Asterisks indicate positions that have a single fully conserved residue, colons indicate conservation between groups of strongly similar properties, and periods indicate conservation between groups of weakly similar properties. (B) Superimposition of the 3D structures of the human SOD1 monomer (PDB code: 2C9U) and human EC-SOD monomer (PDB code: 2JLP). SOD1 and EC-SOD are shown in gray and blue, respectively; the SOD1-specific amino acid stretch is shown in yellow, and side chains of D77 and E78 are in red. (C) Effect of single-alanine substitutions in the SOD1-specific stretch on yeast SOD1 secretion. Cells expressing either wild-type yeast SOD1 or single alanine substitutions in positions G73, P75, D77, E78, R80, or V82 of yeast SOD1 were grown to mid-logarithmic phase, washed twice, and incubated in 2% potassium acetate for 2.5 h. Cell wall proteins were extracted from equal numbers of cells followed by precipitation with TCA. Lysates and cell wall–extracted proteins were analyzed by Western blot. The ratio of wall to lysate SOD1 was determined and represented as percentages of the strain expressing wild-type SOD1. (D) The DE77/78 diacidic motif is essential for yeast SOD1 secretion. Representative cell wall extractions monitored by Western blotting of cells expressing either wild-type, DE77/78AA, or DE77/78NQ yeast SOD1 incubated for 2.5 h in 2% potassium acetate. (E) SOD1 levels were quantified, and the ratio of wall to lysate SOD1 was determined and represented as percentages of the starved wild-type culture. (165X) wall indicates that the wall sample represents the material from 165-fold the number of cells analyzed in the lysate sample. Results in C and E are shown as means ± SEM of at least three experiments.
We tested this hypothesis using nutrient starvation–induced SOD1 secretion by yeast cells. Budding yeast only express SOD1 and lack EC-SOD. We generated yeast strains where the five residues within this short stretch that are conserved between yeast and human SOD1 (G73, P75, D77, E78, and R80) were individually replaced with alanine in the genomic locus of SOD1 (Fig. 2 A). In addition, we substituted V82 to alanine, which is very close to this stretch of residues and is conserved in the sequences of SOD1 from yeast to human (Fig. 2 A). Substituting G73, P75, R80, or V82 with alanine did not affect starvation-induced secretion of SOD1 (Fig. 2 C). However, replacement of either D77 or E78 to alanine reduced the export of SOD1 in response to nutrient starvation by 48% with respect to the wild-type SOD1 strain (Fig. 2 C). The double-substitution DE77/78AA strongly decreased the secreted pool of SOD1 by 88% as compared with the wild-type cells (Fig. 2, D and E). We then tested whether the more conservative mutation DE77/78NQ affected SOD1 secretion. The results revealed that the DE77/78NQ substitution also strongly inhibits the export of SOD1 (Fig. 2, D and E). Importantly, the levels of Acb1 in the cell wall fraction were not affected by either the DE77/78AA or DE77/78NQ mutation in SOD1 (Fig. 2 D), indicating that the pathway of unconventional secretion is fully induced in response to nutrient starvation in the strains expressing these SOD1 mutants. Moreover, the intracellular levels of either the SOD1-DE77/78AA or -DE77/78NQ mutant were similar to the wild-type SOD1, suggesting that the stability of SOD1 was unaffected by these mutations (Fig. 2 D). These data therefore reveal, for the first time, a sequence in SOD1 that is required for its export from cells upon nutrient starvation.
Secretion of ALS-linked mutant SOD1 requires the diacidic motif
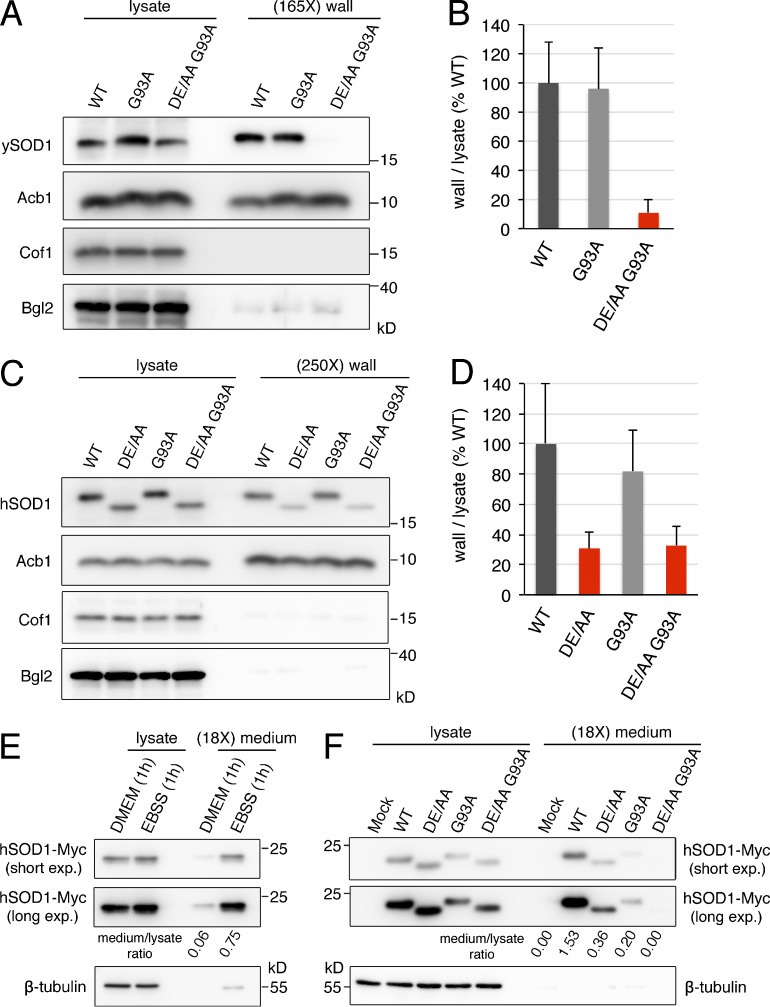
Mutations in human SOD1 are associated with familial ALS, a fatal neurodegenerative disease that results from selective dysfunction and death of upper and lower motor neurons in the spinal cord, brain stem, and cortex. Specifically, >150 mutations in SOD1 have been identified in 20% of familial ALS cases. Although the precise molecular mechanism or mechanisms by which mutations in SOD1 cause ALS are not completely understood, the collective evidence suggests that a toxic gain of function is conferred by these mutations (Bruijn et al., 2004; Robberecht and Philips, 2013). Active secretion of ALS mutants of SOD1 is suggested to contribute to the propagation of SOD1 toxicity along neuroanatomical pathways (Grad et al., 2014). In addition, extracellular ALS mutant SOD1 has been reported to activate microglia and subsequently induce noncell-autonomous neurotoxicity (Zhao et al., 2010). We therefore first asked whether the equivalent of an ALS-linked mutant in yeast SOD1 is secreted under nutrient starvation. To this end, the G93A mutation was introduced in the genomic copy of yeast SOD1 (Fig. 2 A), and the export of this form to the cell wall upon nutrient starvation was analyzed as described in Materials and methods. The levels of the secreted pool of this ALS-equivalent mutant were similar to those of the wild-type form of yeast SOD1 (Fig. 3, A and B). Next, we tested whether replacing the diacidic motif required for SOD1 secretion with alanine (DE77/78AA) affected the export of SOD1-G93A. Our analysis revealed that SOD1-DE77/78AA G93A is poorly secreted (Fig. 3, A and B).
Figure 3.
The diacidic residues are necessary for secretion of wild-type and the ALS mutant of human SOD1. (A and B) The yeast equivalent to the ALS-linked mutant G93A of human SOD1 is secreted in response to starvation in a diacidic sequence-dependent manner. Strains expressing the indicated versions of yeast SOD1 were grown to mid-logarithmic phase, washed twice, and incubated in 2% potassium acetate for 2.5 h. Cell wall proteins were extracted from equal numbers of cells followed by TCA precipitation. Lysates and cell wall fractions were analyzed by Western blotting. The ratios of wall to lysate SOD1 were determined and represented as percentages of the strain expressing wild-type SOD1. (165X) wall indicates that the wall sample represents the material from 165-fold the number of cells analyzed in the lysate sample. Results are shown as means ± SEM of four experiments. (C and D) The DE77/78 sequence is required for secretion of wild-type and G93A human SOD1 by yeast cells. The ratio of wall to lysate of human SOD1 in yeast strains expressing the indicated versions of human SOD1 incubated in 2% potassium acetate for 2.5 h was determined as in A and B. (250X) wall indicates that the wall sample represents the material from 250-fold the number of cells analyzed in the lysate sample. Results are shown as means ± SEM of three experiments. (E) Secretion of wild-type human SOD1 is induced by EBSS starvation in HeLa cells. HeLa cells were transfected with a vector encoding wild-type human SOD1-Myc. After 24 h, cells were washed and incubated in either serum-free DMEM or EBSS (starvation medium) for 1 h. Conditioned media were collected and precipitated with TCA. Lysates and medium fractions were analyzed by Western blotting with anti-Myc and anti–β-tubulin antibodies. (F) The DE77/78 motif determines secretion of wild-type and G93A human SOD1 by HeLa cells. HeLa cells transfected with either an empty Myc vector (Mock) or plasmids encoding the indicated versions of Myc-tagged human SOD1 were incubated in EBSS for 1 h, and the levels of SOD1-Myc and β-tubulin in the lysates and medium fractions were analyzed as in E. (18X) medium indicates that the medium sample represents the material from 18-fold the number of cells analyzed in the lysate sample. Two different exposures (short and long) of the anti-Myc blots are shown. The ratio of medium to lysate of SOD1-Myc for each condition was determined and is indicated under the corresponding lane of the anti-Myc blots. The results of the secretion assays in HeLa cells are representative of two experiments.
The DE77/78 sequence present in yeast SOD1 is conserved in human SOD1 (Fig. 2 A). Next, and considering that ALS-equivalent mutations in SOD1 from different eukaryotic model organisms do not cause the same pathogenic alterations as the corresponding ALS-linked mutations in human SOD1 (Bastow et al., 2016), we investigated whether the DE77/78 sequence is required for secretion of an ALS-linked mutant of human SOD1 expressed in yeast cells. Different forms of human SOD1 (wild-type, DE77/78AA, G93A, and DE77/78AA G93A) were integrated into the yeast genome, replacing the endogenous SOD1 ORF, and secretion of these human SOD1 versions was assessed using the cell wall extraction protocol followed by immunoblotting with an anti–human SOD1 antibody. Surprisingly, considering the moderate amino acid sequence identity between yeast and human SOD1 (56%), incubation in 2% potassium acetate for 2.5 h also induced the secretion of human SOD1 by yeast cells (Fig. S2). Moreover, and as observed for yeast SOD1, the G93A mutation did not affect the levels of cell wall–associated human SOD1, whereas the DE77/78AA substitution in both wild-type and G93A human SOD1 decreased the secreted pool of human SOD1 by 70% compared with wild-type human SOD1 (Fig. 3, C and D). These results indicate that secretion of an ALS-linked mutant of human SOD1 also depends on the DE77/78 sequence.
Finally, to test the relevance of the requirement of DE77/78 for SOD1 secretion in ALS pathology, the involvement of this sequence in the secretion of human SOD1 by human cells was analyzed. Export of yeast SOD1 and human SOD1 expressed in yeast was observed only upon nutrient starvation, so we tested whether a similar stress condition also triggered secretion of SOD1 by human cells. For this, transiently transfected HeLa cells expressing Myc-tagged wild-type human SOD1 were cultured for 1 h in either serum-free DMEM or Earle’s balanced salt solution (EBSS), a salt solution with lower glucose concentration than DMEM and lacking amino acids, which has been reported to stimulate unconventional secretion of IL-1β in a GRASP-dependent manner (Zhang et al., 2015). The conditioned media and the lysates were then collected and analyzed by Western blot with anti-Myc and anti–β-tubulin antibodies. We found low levels of SOD1-Myc in the conditioned medium from DMEM-cultured cells (Fig. 3 E). However, incubation in EBSS clearly increased the extracellular pool of SOD1 (Fig. 3 E). We then asked whether the DE77/78AA substitution affected the levels of secreted SOD1 after EBSS starvation. Similar to what we have observed for yeast SOD1, the secretion of the DE77/78AA form was severely inhibited with respect to wild-type SOD1 (Fig. 3 F). Finally, and as in the yeast system, we found that the DE77/78AA replacement strongly reduced the EBSS-induced extracellular levels of the ALS-linked mutant SOD1-G93A as compared with unmodified SOD1-G93A (Fig. 3 F). We also detected reduced levels of β-tubulin in the extracellular medium of transfected cells—even mock-transfected cells—incubated with EBSS, which did not correlate with the levels of SOD1-Myc and likely represent low nonspecific release of cytoplasmic content under the experimental conditions (Fig. 3, E and F).
Whereas incubation of cells in EBSS resulted in release of SOD1-G93A, we did not find SOD1-G93A in the medium of cells incubated in DMEM for 1 h (Figs. 3 F and S3). It has been reported that, in a neuronal cell line, SOD1, wild-type, and different ALS mutants including G93A are detected in the medium of cells cultured in DMEM for 3 d after transient transfection (Turner et al., 2005; Grad et al., 2014). But Grad et al. (2014) concluded thatEC-SOD1 detected under these experimental conditions of transient transfection is a result of cell death. Nevertheless, they report that SOD1 is contained in exosomes collected from cells stably expressing SOD1 and incubated overnight in DMEM. But what is not clear is whether the membrane-bound compartments called exosomes originate in the MVB and whether there is a secreted pool of SOD1 not contained in the exosomal fraction (Grad et al., 2014). It is important to note that we measured SOD1 secretion by HeLa cells after incubation in DMEM or in EBSS for only 1 h. Therefore, it is impossible to directly compare our findings in mammalian cells to those reported by others. Future studies on this issue should help resolve whether some cells secrete SOD1 under normal conditions, whether cells rely on more than one pathway to release SOD1, and what the physiologically relevant pool of secreted SOD1 is with respect to ALS.
Our data reveal that human cells secrete wild-type and ALS-linked mutant SOD1 upon nutrient starvation, and their secretion depends on the conserved diacidic residues.
Mutations in the diacidic motif do not affect intracellular localization or folding of SOD1
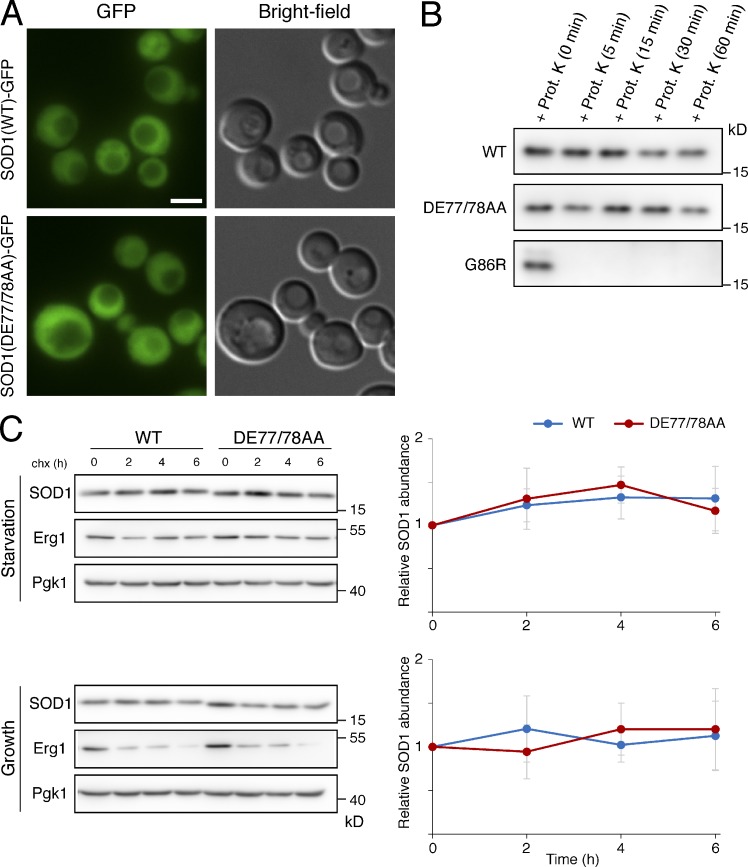
We investigated whether the DE77/78AA mutation alters intracellular distribution of SOD1. Examination of starved yeast cells by fluorescence microscopy revealed that SOD1-DE77/78AA tagged with GFP displays the same diffuse cytoplasmic distribution as the wild-type yeast SOD1 (Fig. 4 A), suggesting that the DE77/78AA substitution does not affect the intracellular location of SOD1.
Figure 4.
The DE77/78AA mutation does not alter the intracellular distribution or the folding status of yeast SOD1. (A) Cells expressing either C-terminally GFP-tagged wild-type SOD1 or the DE77/78AA mutant of SOD1-GFP were grown to mid-logarithmic phase, washed twice, incubated in 2% potassium acetate for 2 h, and examined by epifluorescence microscopy. Representative micrographs showing the cytoplasmic distribution of wild-type and DE77/78AA SOD1-GFP are presented. Brightfield images are shown as references. Bar, 2 µm. (B) Strains expressing the indicated forms of yeast SOD1 were starved in potassium acetate for 2 h. Total cell lysates were obtained, and equivalent amounts were incubated with 100 µg/ml proteinase K at 37°C for the indicated times. After terminating the protease reactions, samples were analyzed by Western blotting with an anti–yeast SOD1 antibody. (C) Exponentially growing cells carrying the indicated forms of yeast SOD1 were washed and suspended in 2% potassium acetate (Starvation) or maintained in nutrient-rich medium (Growth). Protein synthesis was inhibited with cycloheximide (chx), and samples were collected at the indicated time points for quantification of SOD1 by Western blotting with anti–yeast SOD1 antibody. Pgk1 was used as a loading control and was detected with anti-Pgk1 antibody. Erg1, a relatively short-lived protein in growth conditions, was used as a positive control and detected with anti-Erg1 antibody. Graphs show means of at least four independent experiments; error bars indicate SD.
We also evaluated whether the DE77/78AA mutation has an effect on SOD1 conformation. It is known that some of the ALS-linked mutations in human SOD1 induce protein misfolding, which strongly reduces the high resistance to protease degradation displayed by wild-type SOD1 (Ratovitski et al., 1999; Grad et al., 2011). We analyzed the folding status of the DE77/78AA mutant of yeast SOD1 by testing its susceptibility to proteinase K. A significant fraction of both wild-type and the DE/AA mutant of SOD1 was resistant to degradation after 1 h of incubation in the presence of proteinase K (Fig. 4 B). As a control, we used the equivalent of the ALS mutant G85R of SOD1, which was completely degraded after 5 min of incubation with the protease (Fig. 4 B). In vivo proteins failing to achieve correct folding are eliminated by the quality control mechanisms of the cell (Goldberg, 2003). To further probe for an effect of the DE77/78AA mutation on the folding status of SOD1, we performed cycloheximide shutoff experiments in yeast cultures. We found no difference between the stability of the wild-type and DE77/78AA SOD1 proteins, neither when the experiment was performed in starvation conditions nor when it was performed in cells growing in nutrient-rich medium (Fig. 4 C). These data suggest that the defect in the secretion of SOD1-DE77/78AA is not caused by global changes in its 3D structure that prevent recognition for capture into the unconventional secretory pathway.
Acb1 also requires a diacidic motif for its export
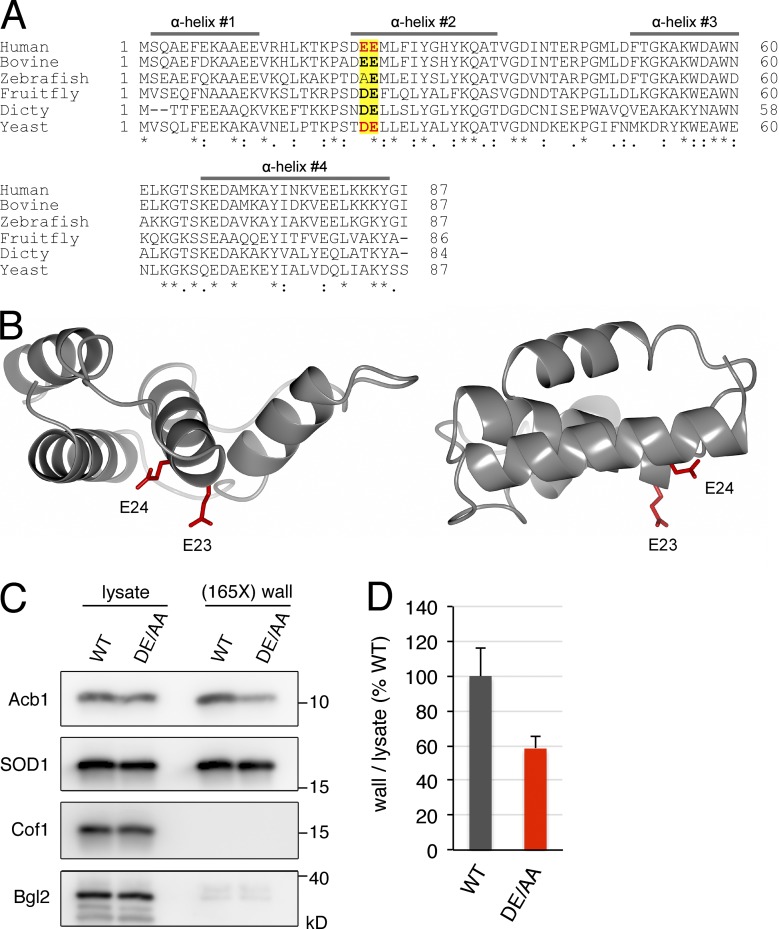
We next asked whether a similar diacidic motif was required for starvation-induced export of Acb1. Budding yeast Acb1 contains a single DE motif at positions 23 and 24, which is largely conserved from yeast to human (Fig. 5 A). This motif is located just at the beginning of the second α helix in the 3D structure of human Acb1 orthologue called acyl-CoA–binding protein (ACBP; Fig. 5, A and B). We generated a strain where DE23/24 residues were substituted by alanine in the genomic locus of ACB1. Using the cell wall extraction assay, we found that the levels of Acb1 secreted to the cell wall after 2.5 h of starvation were 42% lower in the Acb1-DE23/24AA strain compared with the control strain expressing wild-type Acb1 (Fig. 5, C and D). Therefore, a diacidic motif is also required for efficient secretion of Acb1 in response to nutrient starvation. The starvation-induced secretion of wild-type SOD1 was unaffected by the DE23/24AA mutation in Acb1 (Fig. 5 C).
Figure 5.
The diacidic residues DE23/24 are required for Acb1 secretion. (A) Amino acid sequence alignment of human, bovine, zebrafish, fruit fly, D. discoideum (Dicty), and budding yeast Acb1 orthologues. The four α helices of human ACBP are indicated by bars; the diacidic DE23/24 and EE23/24 motifs in yeast Acb1 and human ACBP, respectively, are in bold red letters; the position of this diacidic motif in Acb1 orthologues is indicated in yellow; and the conserved residues within the diacidic motif are in bold letters. Asterisks indicate positions that have a single fully conserved residue, colons indicate conservation between groups of strongly similar properties, and periods indicate conservation between groups of weakly similar properties. (B) Two views of the 3D structure of human ACBP (PDB code: 2FJ9). The side chains of E23 and E24 are in red. (C) The DE23/24 sequence is required for efficient Acb1 secretion. Representative cell wall extractions monitored by Western blotting of either wild-type or DE23/24AA Acb1–expressing cells incubated for 2.5 h in 2% potassium acetate. (D) Acb1 levels were quantified, and ratios of wall to lysate Acb1 were determined and represented as percentages of starved wild-type cultures. (165X) wall indicates that the wall sample represents the material from 165-fold the number of cells analyzed in the lysate sample. Results are shown as means ± SEM of four experiments.
Conclusion
The paucity in the number of proteins secreted under one experimental condition (for example, starvation) by a specific cell type is one reason for our inability to define whether the pathway of Acb1 secretion is a more general pathway and whether such unconventionally secreted proteins share a common theme for their capture and export from cells. Our findings reveal that cytoplasmic SOD1 is secreted under the same experimental condition (i.e., nutrient starvation) that triggers the release of Acb1. Moreover, like Acb1, secretion of SOD1 depends on the activity of Grh1 and ESCRT-I, -II, and -III proteins and is independent of Vps4. Therefore, based on our data, we conclude that SOD1 secretion is not by the conventional exosome-mediated export of cytoplasmic material. Identification of SOD1 as an unconventionally secreted protein of the same pathway as Acb1 has also helped us in discovering a common diacidic motif used by these two proteins for their export upon nutrient starvation. The discovery of a diacidic sequence for SOD1 secretion by human cells is particularly exciting because of the connection to ALS. The ability to block active secretion of wild-type and disease-associated mutant SOD1 provides a unique opportunity to test location-specific effects on neurotoxicity observed in the devastating pathology of ALS.
Interestingly, the negatively charged side chains of the residues of the diacidic motif face the solvent in the 3D structure of both human SOD1 and human ACBP (Figs. 2 B and 5 B). It is therefore likely that this motif binds specific proteins that promote their capture for export by the ER-independent pathway of secretion. It is important to note that the diacidic motif in Acb1 and SOD1 (D-E) is different from the diacidic motif (D-X-E) that captures and exports transmembrane proteins by Sec24 of COPII-coated vesicles at the ER (Barlowe, 2003). Moreover, COPII coats are not involved in the export of unconventionally secreted cargoes like Acb1 (Duran et al., 2010; Cruz-Garcia et al., 2014).
The finding of this diacidic motif for SOD1 and Acb1 secretion is clearly an important milestone, but it would be premature to conclude that this diacidic motif is the equivalent to the signal sequence, which is present at the N terminus of the secretory cargoes and used for their entry into the ER. The diacidic residues contained in Acb1 and SOD1 are more context dependent, and their position in the molecule is likely to act as a determinant for their export. But diacidic residues are present in many proteins, so how do cells exploit this relatively common motif to export a small pool of Acb1 and SOD1 upon nutrient starvation? We suggest that conditions that trigger secretion of Acb1 and SOD1—glucose and nitrogen deprivation—induce physicochemical changes in the cytoplasm that result in partial unfolding of a subset of proteins. In this partially unfolded state, proteins like SOD1 and Acb1 expose their diacidic residues and recruit chaperones. The chaperones prevent further collapse of their structure and subsequent degradation. A client present in limiting amounts binds the exposed diacidic motif, and this client-bound pool is processed for the next step in secretion that involves CUPS. The remaining chaperone-bound pool is excluded from export and degradation. This scheme provides a means to select, under a given condition, a small pool of a subset of cytoplasmic proteins for secretion without their entry into the ER. Identification of the cargo-specific binding partners and the specific chaperones will permit formal tests of this working hypothesis.
Materials and methods
Media, yeast strains, and plasmids
Yeast cells were grown in SC media (0.67% yeast nitrogen base without amino acids and 2% glucose supplemented with amino acid dropout mix; Sigma-Aldrich). All Saccharomyces cerevisiae strains used in this study are listed in Table S1. For deletion of GRH1, the ORF was replaced with a KanMX4 cassette using PCR-based targeted homologous recombination as previously described (Cruz-Garcia et al., 2014). To generate SOD1 mutant strains, the yeast SOD1 ORF was first PCR amplified from genomic DNA and cloned into the plasmid p416-ADH using the BamHI and XhoI sites. From this plasmid, the SOD1 ORF and CYC1 terminator were PCR amplified and introduced into the pFA6a-His3MX6 plasmid using the BamHI and XmaI sites to obtain the pFA6a-ySOD1-His3MX6 plasmid. Mutations in the SOD1 ORF were introduced using Gibson assembly (Gibson et al., 2009). The C-terminally GFP-tagged SOD1 wild-type or DE77/78AA mutant constructs were generated by replacing the SOD1 stop codon and CYC1 terminator with a fragment encompassing a linker-GFP-ADH1 terminator. For this, the SOD1 ORF was amplified from the respective construct described in this section, and the linker-GFP-ADH1 terminator fragment was PCR amplified from pYM44 (Janke et al., 2004). Both fragments were introduced in pFA6a-His3MX6 by Gibson assembly to obtain the GFP-tagged SOD1 wild-type or DE77/78AA constructs. To generate Acb1 mutant strains, we introduced the His3MX6 cassette into pRS416-PrACB1-Acb1-3×Flag (Duran et al., 2010) by Gibson assembly at the BglII site located in the ACB1 promoter region. Mutations in the ACB1 ORF were introduced by Gibson assembly. To generate strains expressing different forms of human SOD1 (hSOD1), the wild-type and G93A mutant hSOD1 ORFs were amplified from the pF146 pSOD1WTAcGFP1 and pF150 pSOD1G93AAcGFP1 plasmids, respectively (plasmids 26407 and 26411; Addgene; gifts from E. Fisher, University College London, London, England, UK) and used to replace the yeast SOD1 ORF in the pFA6a-ySOD1-His3MX6 construct using BamHI and XhoI sites. Gibson assembly was used to introduce the DE77/78AA substitution on the wild-type and G93A mutant hSOD1 plasmids. Strains expressing mutated yeast SOD1, SOD1-GFP, Acb1, or human SOD1 and the corresponding wild-type controls were then generated by PCR-based targeted homologous recombination using the respective constructs. Single colonies were selected and screened for the presence of the respective mutation by nucleotide sequence analysis. To generate plasmids for mammalian expression of C-terminally Myc-tagged human SOD1 forms, the ORFs of wild-type, DE77/78AA, G93A, and DE77/78AA G93A human SOD1 were amplified by PCR from the corresponding pFA6a-hSOD1-His3MX6 constructs and cloned into a pcDNA3.1-Myc vector. The pcDNA3.1-Myc vector was generated by inserting two aligned oligonucleotides coding for the Myc tag epitope followed by a stop codon into the XhoI and ApaI sites of the pcDNA3.1(+) plasmid. All constructs were confirmed by nucleotide sequence analysis covering the coding regions in the construct. SnapGene software (GSL Biotech) was used for molecular cloning design.
Assay for unconventional protein secretion in yeast cells
Yeast cells were inoculated at 0.004–0.008 OD600/ml in SC medium and grown overnight at 25°C. The following day, when cells had reached OD600/ml of 0.4–0.7, equal numbers of cells (15 OD600 units) were harvested, washed twice in sterile water, resuspended in 1.5 ml of 2% potassium acetate, and incubated for 2.5 h. Concomitant to this, growing cells were diluted in SC medium, continued growing in logarithmic phase, and then 15 OD600 units were harvested as before. The cell wall extraction buffer (100 mM Tris-HCl, pH 9.4, and 2% sorbitol) was always prepared fresh before use and kept on ice. To ensure no loss of cells and to avoid cell contamination in the extracted buffer, 2-ml tubes were siliconized with Sigmacote (Sigma-Aldrich) before collection. Cells were harvested by centrifugation at 800 g for 2.5 min at 4°C, then supernatant was removed, and 1.5 ml of cold extraction buffer was added. Cells were resuspended gently by inversion and incubated on ice for 10 min, after which they were centrifuged at 3,000 g for 2.5 min at 4°C, and 1.3 ml of extraction buffer was removed to ensure no cell contamination. The remaining buffer was removed, and the cells were resuspended in 0.75 ml of cold TE buffer (10 mM Tris-HCl, pH 7.5, and 1 mM EDTA) with protease inhibitors (aprotinin, pepstatin, and leupeptin; Sigma-Aldrich), and 10 µl was boiled directly in 90 µl of 2× sample buffer (lysate). To the cell wall–extracted protein fraction, 30 µg of BSA (Sigma-Aldrich) carrier protein and 0.2 ml of 100% TCA (Sigma-Aldrich) were added. Proteins were precipitated on ice for 1 h, centrifuged 16,000 g for 30 min, and boiled in 45 µl of 2× sample buffer (wall fraction). For immunoblotting, proteins (10 µl each of lysate or wall fraction) were separated in a 16.5% Tris-tricine peptide gel (Bio-Rad Laboratories) before transfer to nitrocellulose membrane. Rabbit anti–yeast SOD1 antibody was a gift from T. O’Halloran (Northwestern University, Chicago, IL). Rabbit anti-Cof1 antibody was a gift from J. Cooper (Washington University in St. Louis, St. Louis, MO). Rabbit anti-Bgl2 was a gift from R. Schekman (University of California, Berkeley, Berkeley, CA). Rabbit anti-Acb1 antibody has been described previously (Curwin et al., 2016). Goat anti–human SOD1 antibody was purchased from R&D Systems (AF3418).
Mammalian cell culture, transfection, and secretion assay
HeLa cells were grown in DMEM containing 10% FCS at 37°C in a 7% CO2 incubator. Plasmids were transfected in HeLa cells with X-tremeGENE 9 DNA Transfection Reagent (Roche) according to the manufacturer’s protocols. To measure the secretion of human SOD1-Myc, cells (2 × 105/well) were seeded in six-well plates and, after 24 h, were then transfected with 500 ng/well of plasmids encoding the indicated forms of human SOD1-Myc. 24 h after transfection, cells were washed three times with serum-free DMEM or EBSS (Invitrogen) and incubated with 1.5 ml/well of serum-free DMEM or EBSS for 1 h at 37°C. Then, medium was collected and centrifuged first at 1,000 g for 5 min and subsequently at 10,000 g for 30 min. The clarified conditioned medium was precipitated as described in the previous section after adding 30 µg of BSA and 0.2 ml of 100% TCA to each sample. The cells were lysed in 500 µl/well of TST buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, and protease inhibitors). A fraction of the lysate and of the concentrated medium was analyzed by Western blotting using rabbit anti-cMyc and mouse anti–β-tubulin antibodies (C3956 and T4026, respectively; Sigma-Aldrich).
Proteinase K sensitivity assay
After starvation in 2% potassium acetate for 2 h, cells were lysed by bead beating with glass beads in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, and protease inhibitors). Lysates were clarified by centrifugation at 16,000 g for 15 min, and supernatants were incubated with 100 µg/ml proteinase K (Merck) at 37°C. At the indicated time points, aliquots were removed and quenched by incubation with 10 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich). The samples were then analyzed by Western blotting with specific antibodies.
Cycloheximide shutoff
Cells grown overnight to mid-logarithmic phase (OD600/ml < 1) in SC medium were maintained in SC (growth) or washed and incubated in 2% potassium acetate (starvation) at 30°C. After 30 min, cycloheximide (Sigma-Aldrich) was added to 0.5 mg/ml. Culture aliquots were collected on ice at 0, 2, 4, and 6 h after cycloheximide addition and treated with 6 mM sodium azide. Whole-cell lysates were prepared as previously described (Kushnirov, 2000) and analyzed by Western blot. Rabbit anti-Erg1antibody was a gift from P. Carvalho (Sir William Dunn School of Pathology, Oxford, England, UK) and has been previously described (Foresti et al., 2014). Mouse anti-Pgk1antibody was purchased from Invitrogen (459250).
Epifluorescence microscopy
After incubation in 2% potassium acetate, ∼1.5 OD600 of cells was harvested by centrifugation at 3,000 g for 3 min, resuspended in a small volume of starvation medium, spotted on a microscopy slide, and subsequently live imaged at 25°C with a DMI6000 B epifluorescence microscope equipped with a DFC360 FX camera (Leica Microsystems) using an HCX Plan Apochromat 100× 1.4 NA objective lens. Images were acquired using LAS AF software (Leica Microsystems) with 0.2-s exposure times. Image processing was performed with ImageJ 1.45r software (National Institutes of Health).
Bioinformatic analysis
Amino acid sequences were downloaded from UniProt (UniProt Consortium, 2014) and aligned using Clustal Omega (Sievers et al., 2011). All structural figures were created using the CCP4MG program (McNicholas et al., 2011).
Online supplemental material
Fig. S1 shows the requirement of ESCRT-II, but not ESCRT-0, in SOD1 secretion by yeast cells. Fig. S2 shows that human SOD1 secretion by yeast cells is induced upon starvation in 2% potassium acetate. Fig. S3 shows that human SOD1-G93A is not detected in the conditioned medium of HeLa cells incubated in DMEM for 1 h. Table S1 lists the yeast strains used in this study.
Acknowledgments
We thank members of the Malhotra lab for valuable discussions. We thank Drs. Thomas O’Halloran, John Cooper, Randy Schekman, and Pedro Carvalho for sharing reagents.
We acknowledge support from the Spanish Ministry of Economy and Competitiveness through the program Centro de Excelencia Severo Ochoa 2013–2017 (SEV-2012-0208) and from the Centres de Recerca de Catalunya Program/Generalitat de Catalunya. Vivek Malhotra is an Institució Catalana de Recerca i Estudis Avançats professor at the Center for Genomic Regulation, and the work in his laboratory is funded by grants from the Spanish Ministry of Economy and Competitiveness’s Plan Nacional (BFU2013-44188-P) and Consolider (CSD2009-00016) as well as by the European Research Council (268692). The project has received research funding from the European Union. This paper reflects only the authors’ views. The European Union is not liable for any use that may be made of the information contained therein.
The authors declare no competing financial interests.
Author contributions: D. Cruz-Garcia and V. Malhotra conceptualized the experiments. D. Cruz-Garcia performed and analyzed most of the experiments, with contributions from N. Brouwers, J.M. Duran, G. Mora, and A.J. Curwin. D. Cruz-Garcia and V. Malhotra wrote the original and revised manuscripts with contributions from the other authors.
Footnotes
Abbreviations
- ACBP
- acyl-CoA–binding protein
- ALS
- amyotrophic lateral sclerosis
- CUPS
- compartment for unconventional protein secretion
- EBSS
- Earle’s balanced salt solution
- EC-SOD
- extracellular SOD
- ESCRT
- endosomal sorting complexes required for transport
- GRASP
- Golgi reassembling and stacking protein
- SC
- synthetic complete
- SOD
- superoxide dismutase
References
- Auron P.E., Webb A.C., Rosenwasser L.J., Mucci S.F., Rich A., Wolff S.M., and Dinarello C.A.. 1984. Nucleotide sequence of human monocyte interleukin 1 precursor cDNA. Proc. Natl. Acad. Sci. USA. 81:7907–7911. 10.1073/pnas.81.24.7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. 2003. Signals for COPII-dependent export from the ER: what’s the ticket out? Trends Cell Biol. 13:295–300. 10.1016/S0962-8924(03)00082-5 [DOI] [PubMed] [Google Scholar]
- Basso M., Pozzi S., Tortarolo M., Fiordaliso F., Bisighini C., Pasetto L., Spaltro G., Lidonnici D., Gensano F., Battaglia E., et al. 2013. Mutant copper-zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: Implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J. Biol. Chem. 288:15699–15711. 10.1074/jbc.M112.425066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow E.L., Peswani A.R., Tarrant D.S.J., Pentland D.R., Chen X., Morgan A., Staniforth G.L., Tullet J.M., Rowe M.L., Howard M.J., et al. 2016. New links between SOD1 and metabolic dysfunction from a yeast model of amyotrophic lateral sclerosis. J. Cell Sci. 129:4118–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruijn L.I., Miller T.M., and Cleveland D.W.. 2004. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 27:723–749. 10.1146/annurev.neuro.27.070203.144244 [DOI] [PubMed] [Google Scholar]
- Bruns C., McCaffery J.M., Curwin A.J., Duran J.M., and Malhotra V.. 2011. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J. Cell Biol. 195:979–992. 10.1083/jcb.201106098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini V., Ruggiero G., Buonomo T., Seru R., Sciorio S., Zanzi C., Santangelo F., and Mondola P.. 2002. CuZn-superoxide dismutase in human thymus: immunocytochemical localisation and secretion in thymus-derived epithelial and fibroblast cell lines. Histochem. Cell Biol. 118:163–169. [DOI] [PubMed] [Google Scholar]
- Cruz-Garcia D., Curwin A.J., Popoff J.-F., Bruns C., Duran J.M., and Malhotra V.. 2014. Remodeling of secretory compartments creates CUPS during nutrient starvation. J. Cell Biol. 207:695–703. 10.1083/jcb.201407119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwin A.J., Brouwers N., Alonso Y Adell M., Teis D., Turacchio G., Parashuraman S., Ronchi P., and Malhotra V.. 2016. ESCRT-III drives the final stages of CUPS maturation for unconventional protein secretion. eLife. 5:e16299 10.7554/eLife.16299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., and Deretic V.. 2011. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30:4701–4711. 10.1038/emboj.2011.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran J.M., Anjard C., Stefan C., Loomis W.F., and Malhotra V.. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188:527–536. 10.1083/jcb.200911154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O., Rodriguez-Vaello V., Funaya C., and Carvalho P.. 2014. Quality control of inner nuclear membrane proteins by the Asi complex. Science. 346:751–755. 10.1126/science.1255638 [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Goldberg A.L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature. 426:895–899. 10.1038/nature02263 [DOI] [PubMed] [Google Scholar]
- Gomes C., Keller S., Altevogt P., and Costa J.. 2007. Evidence for secretion of Cu,Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci. Lett. 428:43–46. 10.1016/j.neulet.2007.09.024 [DOI] [PubMed] [Google Scholar]
- Grad L.I., Guest W.C., Yanai A., Pokrishevsky E., O’Neill M.A., Gibbs E., Semenchenko V., Yousefi M., Wishart D.S., Plotkin S.S., and Cashman N.R.. 2011. Intermolecular transmission of superoxide dismutase 1 misfolding in living cells. Proc. Natl. Acad. Sci. USA. 108:16398–16403. 10.1073/pnas.1102645108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad L.I., Yerbury J.J., Turner B.J., Guest W.C., Pokrishevsky E., O’Neill M.A., Yanai A., Silverman J.M., Zeineddine R., Corcoran L., et al. 2014. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA. 111:3620–3625. 10.1073/pnas.1312245111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kinseth M.A., Anjard C., Fuller D., Guizzunti G., Loomis W.F., and Malhotra V.. 2007. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 130:524–534. 10.1016/j.cell.2007.06.029 [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. 2000. Rapid and reliable protein extraction from yeast. Yeast. 16:857–860. [DOI] [PubMed] [Google Scholar]
- Malhotra V. 2013. Unconventional protein secretion: an evolving mechanism. EMBO J. 32:1660–1664. 10.1038/emboj.2013.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R., Anjard C., Loomis W.F., and Subramani S.. 2010. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 188:537–546. 10.1083/jcb.200911149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicholas S., Potterton E., Wilson K.S., and Noble M.E.M.. 2011. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67:386–394. 10.1107/S0907444911007281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondola P., Annella T., Santillo M., and Santangelo F.. 1996. Evidence for secretion of cytosolic CuZn superoxide dismutase by Hep G2 cells and human fibroblasts. Int. J. Biochem. Cell Biol. 28:677–681. 10.1016/1357-2725(96)00004-0 [DOI] [PubMed] [Google Scholar]
- Mondola P., Annella T., Serù R., Santangelo F., Iossa S., Gioielli A., and Santillo M.. 1998. Secretion and increase of intracellular CuZn superoxide dismutase content in human neuroblastoma SK-N-BE cells subjected to oxidative stress. Brain Res. Bull. 45:517–520. 10.1016/S0361-9230(97)00438-3 [DOI] [PubMed] [Google Scholar]
- Rabouille C., Malhotra V., and Nickel W.. 2012. Diversity in unconventional protein secretion. J. Cell Sci. 125:5251–5255. 10.1242/jcs.103630 [DOI] [PubMed] [Google Scholar]
- Ratovitski T., Corson L.B., Strain J., Wong P., Cleveland D.W., Culotta V.C., and Borchelt D.R.. 1999. Variation in the biochemical/biophysical properties of mutant superoxide dismutase 1 enzymes and the rate of disease progression in familial amyotrophic lateral sclerosis kindreds. Hum. Mol. Genet. 8:1451–1460. 10.1093/hmg/8.8.1451 [DOI] [PubMed] [Google Scholar]
- Robberecht W., and Philips T.. 2013. The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 14:248–264. 10.1038/nrn3430 [DOI] [PubMed] [Google Scholar]
- Rubartelli A., Cozzolino F., Talio M., and Sitia R.. 1990. A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. EMBO J. 9:1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santillo M., Secondo A., Serù R., Damiano S., Garbi C., Taverna E., Rosa P., Giovedì S., Benfenati F., and Mondola P.. 2007. Evidence of calcium- and SNARE-dependent release of CuZn superoxide dismutase from rat pituitary GH3 cells and synaptosomes in response to depolarization. J. Neurochem. 102:679–685. 10.1111/j.1471-4159.2007.04538.x [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S.M., Cha M.-Y., Choi H., Kang S., Choi H., Lee M.-S., Park S.A., and Mook-Jung I.. 2016. Insulin-degrading enzyme secretion from astrocytes is mediated by an autophagy-based unconventional secretory pathway in Alzheimer disease. Autophagy. 12:784–800. 10.1080/15548627.2016.1159375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.J., Atkin J.D., Farg M.A., Zang D.W., Rembach A., Lopes E.C., Patch J.D., Hill A.F., and Cheema S.S.. 2005. Impaired extracellular secretion of mutant superoxide dismutase 1 associates with neurotoxicity in familial amyotrophic lateral sclerosis. J. Neurosci. 25:108–117. 10.1523/JNEUROSCI.4253-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium 2014. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42(D1):D191–D198. 10.1093/nar/gkt1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelko I.N., Mariani T.J., and Folz R.J.. 2002. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33:337–349. 10.1016/S0891-5849(02)00905-X [DOI] [PubMed] [Google Scholar]
- Zhang M., Kenny S.J., Ge L., Xu K., and Schekman R.. 2015. Translocation of interleukin-1β into a vesicle intermediate in autophagy-mediated secretion. eLife. 4:e11205 10.7554/eLife.11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Beers D.R., Henkel J.S., Zhang W., Urushitani M., Julien J.-P., and Appel S.H.. 2010. Extracellular mutant SOD1 induces microglial-mediated motoneuron injury. Glia. 58:231–243. 10.1002/glia.20919 [DOI] [PMC free article] [PubMed] [Google Scholar]