Bobola previews work from the Schulte laboratory showing that the atypical homeodomain protein MEIS2 facilitates chromatin accessibility of transcriptionally inactive genes in neuronal differentiation.
Abstract
How transcription factors (TFs) control enhancer and promoter functions to effect changes in gene expression is an important question. In this issue, Hau et al. (2017. J. Cell Biol. https://doi.org/10.1083/jcb.201701154) show that the TALE TF MEIS recruits the histone modifier PARP1/ARTD1 at promoters to decompact chromatin and activate transcription.
Changes in gene expression underlie development, disease, and evolution, and they are largely controlled by transcription factors (TFs). Recently, next-generation sequencing technologies have greatly advanced our understanding of how TFs read the genome to identify their cognate enhancers and promoters. However, an important outstanding question is how the binding of TFs and cofactors regulates RNA polymerase II activity. The DNA-binding TFs myeloid ecotropic viral integration site (MEIS) and pre–B-cell leukemia homeobox (PBX) are members of the three–amino acid loop extension (TALE) class of homeodomain-containing proteins. They are essential in development, where they control limb, axial skeleton, face, eye, brain, and heart formation. In addition, they function as oncogenes in several forms of cancer. In line with their broad range of functions, MEIS and PBX occupy tens of thousands of sites in the genome (Penkov et al., 2013; Amin et al., 2015), and, consistent with their strong tendency to heterodimerize, their genomic sites are largely overlapping. PBX1 constitutively binds chromatin before lineage-specific TFs, suggesting that it acts as a pioneer TF (Berkes et al., 2004; Magnani et al., 2011), whereas MEIS has a positive effect on transcription (Huang et al., 2005; Choe et al., 2009). Nonetheless, dissecting the dynamics of TALE interactions and the exact contribution of the individual partners to the functional complex has been difficult.
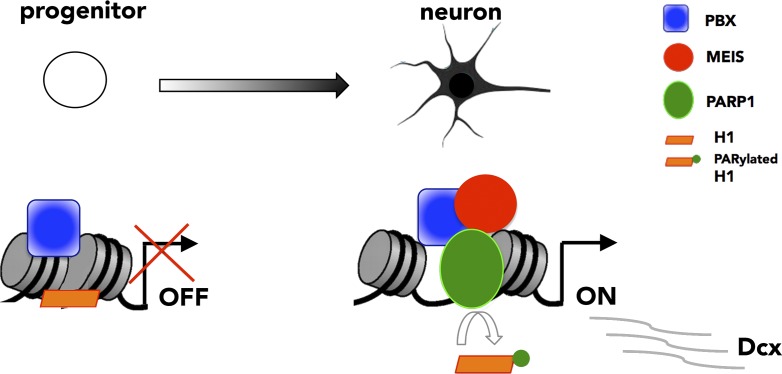
In this issue, Hau et al. successfully address these issues by following the activation of the TALE target Dcx while adult progenitor cells differentiate into neurons. Dcx is an early and global marker of neuronal differentiation, whose expression requires PBX1 and MEIS2 (Agoston et al., 2014; Grebbin et al., 2016). Progenitor cells in the adult brain subventricular zone (SVZ) do not express Dcx, which is then rapidly transcribed by all newly generated neurons. Taking advantage of an in vitro model system that recreates the first hours of neuronal differentiation, the study by Hau et al. (2017) neatly unravels the dynamics of Dcx transcriptional activation. The authors use chromatin immunoprecipitation to interrogate TALE binding to the Dcx promoter. They find that, in progenitor cells, where Dcx is off, PBX is the only TALE TF that binds the Dcx promoter. Differentiation cues cause MEIS accumulation in the nucleus, and, 5 h after the beginning of differentiation, Hau et al. (2017) show that MEIS is bound with PBX at Dcx regulatory regions, where it rapidly recruits the poly-ADP-ribose polymerase I (PARP1). The authors identify PARP1 in a protein complex with MEIS2 in neuroblastoma cells and at regions of ongoing neurogenesis. PARP1 transfers ADP-ribose moieties onto histone H1 and decreases its affinity for DNA (Tulin and Spradling, 2003; Kim et al., 2004; Krishnakumar et al., 2008), thus facilitating the assembly of the transcriptional machinery at promoters. Hau et al. (2017)’s model is that local, MEIS-dependent recruitment of PARP1 modifies and clears histone H1 from the promoter, promotes loading of RNA polymerase II, and allows Dcx transcription to proceed (Fig. 1).
Figure 1.
MEIS recruits PARP1 to activate Dcx transcription. In progenitor cells, Dcx is off. PBX (blue square) binds the Dcx promoter on closely spaced nucleosomes. After differentiation, MEIS (red circle) binds PBX and recruits PARP1 (green oval). Poly-ADP-ribosylation of linker histone H1 (orange rectangle) by PARP1 leads to H1 eviction and nucleosome decompaction, allowing Dcx transcription to begin.
Thus, once recruited to the Dcx promoter by PBX, MEIS orchestrates specific chromatin dynamics that culminate in the transcription of Dcx. This is not the first time MEIS has been linked to transcriptional activation. In fact, TALE TFs are known to be essential for the activation of Hox-regulated promoters (Choe et al., 2014). Their main function is to prepare or poise promoters for transcription, which is then effectively triggered by binding of the TF HOXB1. Turning on transcription by rapid and transient recruitment of PARP1 at the Dcx promoter is an entirely new modality of MEIS transcriptional regulation.
Hau et al. (2017) extend these findings to additional genes expressed at the onset of neural differentiation. By monitoring changes in global transcription of differentiating progenitor cells treated with an inhibitor of PARP1, the authors identify additional PARP1-dependent transcripts. Consistent with MEIS mediating PARP1 recruitment to activate transcription, a subset of these PARP1-dependent transcripts is found to be regulated by MEIS and to display MEIS–PARP1 complexes at their promoters.
Of note, previous work by the same group established a general requirement for MEIS and PBX in adult neurogenesis in the SVZ in mice (Agoston et al., 2014; Grebbin et al., 2016). MEIS and PBX (Agoston et al., 2014; Grebbin et al., 2016), as well as PARP1 (Hau et al., 2017), are also essential for in vitro differentiation of progenitor cells into neurons. Thus, the mechanism described by Hau et al. (2017) is causally linked to the physiological changes that accompany differentiation of progenitor cells into neurons and couples dynamics of TF binding with cell differentiation.
By monitoring TALE binding to a functional target gene at short temporal intervals within a context where TALE instructs transcriptional changes underlying cell fate, Hau et al. (2017)’s results yield additional surprises. First, the formation of MEIS-PBX dimers takes place in a temporal, sequential order: one partner, PBX1, is already bound to its target site in closed chromatin, which is consistent with PBX pioneering activity. The other partner, MEIS2, joins in later. Second, MEIS binding is dynamic, and MEIS transiently associates with chromatin, a feature never detected previously. Despite being a winning approach, the focus on a single gene inevitably raises the most compelling question: How widespread are the mechanisms identified?
Whereas the ubiquitous distribution of PARP1 lends itself to a potentially broad use of this mechanism in gene activation, local and specific recruitment of PARP1 relies on the transient accumulation and binding of MEIS, a feature not observed in other systems. On the contrary, genome-wide studies paint a constitutive binding picture of MEIS, which occupies many sites, largely shared across different tissues (Amin et al., 2015). It is possible—as argued by Hau et al. (2017)—that, by averaging different cell types, high-throughput experiments may fail to detect transient MEIS binding. Still, MEIS distribution does not appear to be restricted to narrow temporal windows during development. If PARP1 is ubiquitous and MEIS binds constitutively across many sites, which do not largely change across tissues, how could MEIS specifically recruit PARP1 to lineage-specific genes? MEIS binding at chromatin is enhanced by cooperation with HOXA2 (Amin et al., 2015). HOXA2/MEIS combinatorial binding increases MEIS binding levels at selected sites, leading to gene activation. These observations highlight an alternative mechanism to the transient nuclear accumulation of MEIS proteins, whereby MEIS binding levels can locally increase as a result of cooperative binding with HOX and possibly other tissue-specific TFs. Higher MEIS binding levels equate with longer permanence of MEIS on chromatin. In turn, a more stable association of MEIS with DNA may increase the likelihood of interacting with PARP1 and holding it in place for sufficient time to modify histone H1.
Unlike the instance of Dcx, where TALE TFs bind relatively close to the transcriptional start site (TSS), TALE binding is largely detected in intergenic regions and at long distances from genes’ TSSs (Penkov et al., 2013; Amin et al., 2015). It will therefore be important to test whether proximal TALE binding is a requirement for PARP1-mediated eviction of histone H1 at promoters or whether TALE bound at distant enhancers can equally recruit and position PARP1 at TSSs, perhaps via enhancer/promoter interaction.
If and how the mechanism used by TALE to activate transcription of neural genes can be exported to other systems controlled by TALE is an intriguing question. For this, it will be critical to investigate the contribution of PARP1 to TALE transcriptional activation in different contexts. In any case, the work of Hau et al. (2017) provides exciting new mechanistic insights into the ordered sequence of events that takes place at chromatin to drive neural differentiation. In combination with high-throughput studies to identify functional binding sites in vivo, studies at the resolution of specific targets, such as the current work of Hau et al. (2017), are crucial to address the dynamics of TF-mediated transcriptional activation and promise to unlock the mechanisms underlying TALE function in development and disease.
Acknowledgments
We apologize to those authors whose work on this topic could not be cited because of space limitations.
This work was supported by the Biotechnology and Biological Sciences Research Council (grant BB/N00907X/1) and Medical Research Council (grant MR/L009986/1) to N. Bobola.
The author declares no competing financial interests.
References
- Agoston Z., Heine P., Brill M.S., Grebbin B.M., Hau A.-C., Kallenborn-Gerhardt W., Schramm J., Götz M., and Schulte D.. 2014. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development. 141:28–38. 10.1242/dev.097295 [DOI] [PubMed] [Google Scholar]
- Amin S., Donaldson I.J., Zannino D.A., Hensman J., Rattray M., Losa M., Spitz F., Ladam F., Sagerström C., and Bobola N.. 2015. Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell. 32:265–277. 10.1016/j.devcel.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes C.A., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., and Tapscott S.J.. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 14:465–477. 10.1016/S1097-2765(04)00260-6 [DOI] [PubMed] [Google Scholar]
- Choe S.K., Lu P., Nakamura M., Lee J., and Sagerström C.G.. 2009. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev. Cell. 17:561–567. 10.1016/j.devcel.2009.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S.K., Ladam F., and Sagerström C.G.. 2014. TALE factors poise promoters for activation by Hox proteins. Dev. Cell. 28:203–211. 10.1016/j.devcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebbin B.M., Hau A.C., Groß A., Anders-Maurer M., Schramm J., Koss M., Wille C., Mittelbronn M., Selleri L., and Schulte D.. 2016. Pbx1 is required for adult subventricular zone neurogenesis. Development. 143:2281–2291. 10.1242/dev.128033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau A.-C., Grebbin B.M., Agoston Z., Anders-Maurer M., Müller T., Groß A., Kolb J., Langer J.D., Döring C., and Schulte D.. 2017. MEIS homeodomain proteins facilitate PARP1/ARTD1-mediated eviction of histone H1. J. Cell Biol. 10.1083/jcb.201701154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Rastegar M., Bodner C., Goh S.L., Rambaldi I., and Featherstone M.. 2005. MEIS C termini harbor transcriptional activation domains that respond to cell signaling. J. Biol. Chem. 280:10119–10127. 10.1074/jbc.M413963200 [DOI] [PubMed] [Google Scholar]
- Kim M.Y., Mauro S., Gévry N., Lis J.T., and Kraus W.L.. 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 119:803–814. 10.1016/j.cell.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R., Gamble M.J., Frizzell K.M., Berrocal J.G., Kininis M., and Kraus W.L.. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 319:819–821. 10.1126/science.1149250 [DOI] [PubMed] [Google Scholar]
- Magnani L., Ballantyne E.B., Zhang X., and Lupien M.. 2011. PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet. 7:e1002368 10.1371/journal.pgen.1002368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkov D., Mateos San Martín D., Fernandez-Díaz L.C., Rosselló C.A., Torroja C., Sánchez-Cabo F., Warnatz H.J., Sultan M., Yaspo M.L., Gabrieli A., et al. 2013. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Reports. 3:1321–1333. 10.1016/j.celrep.2013.03.029 [DOI] [PubMed] [Google Scholar]
- Tulin A., and Spradling A.. 2003. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 299:560–562. 10.1126/science.1078764 [DOI] [PubMed] [Google Scholar]