Serquiña and Ziegelbauer discuss Chiu et al.’s recent finding of a novel strategy for viral genome partitioning used by Kaposi’s Sarcoma–associated herpesvirus.
Abstract
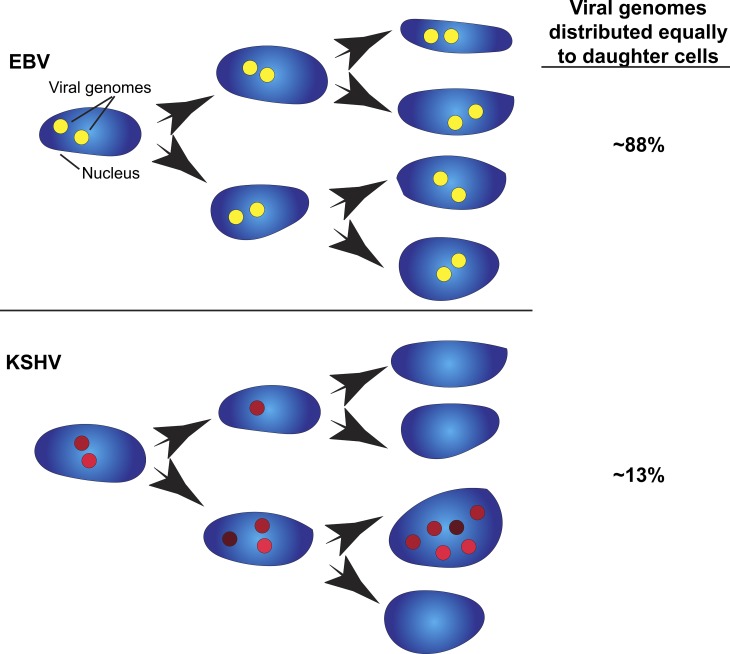
Herpesvirus genomes exist and replicate as episomes inside the host cell nucleus during latent infection. Chiu et al. (2017. J. Cell Biol. https://doi.org/10.1083/jcb.201702013) find that unlike Epstein–Barr virus, which partitions viral genomes faithfully during cell division, Kaposi’s Sarcoma–associated herpesvirus clusters viral genomes into loci that are distributed unequally to daughter cells.
After they first infect cells, many herpesviruses establish a latent lifecycle with the viral genome existing as circular genetic elements called episomes inside the host cell’s nucleus that are closely associated with, but not integrated into, the host DNA. During latent infection, episomes are replicated during S phase, but new viral particles are not produced and the infected cell survives. The maintenance of herpesvirus genomes as episomes contrasts significantly with retroviruses, which insert their genome within the host cell genome to ensure their replication and inheritance by daughter cells and establish long-term infection. For the herpesviruses, this lack of integration presents the problem of how to pass on their genome to host daughter cells after mitosis. In this issue, Chiu et al. have made some exciting, new discoveries about how Epstein–Barr virus (EBV) and Kaposi’s Sarcoma–associated herpesvirus (KSHV) tackle this challenge with different strategies.
EBV and KSHV are related herpesviruses: they can both infect B lymphocytes, have similar genomes, and cause B-lymphocyte malignancies. One interesting difference is that the viral protein EBV Nuclear Antigen 1 (EBNA1) attaches EBV episomes directly to human chromosomes by binding to AT-rich tracts of DNA (Lieberman, 2013). In contrast, KSHV uses the viral protein Latency-Associated Nuclear Antigen 1 (LANA1) to tether KSHV episomes to histones H2A and H2B (Barbera et al., 2006). In primary effusion lymphoma (PEL) cells from patients coinfected with EBV and KSHV, it had been observed that these tumor cells have more copies of KSHV than EBV genomes (Cesarman et al., 1995). In the laboratory, infection of primary cells can generate transformed B lymphocytes that faithfully retain the EBV genome (Lieberman, 2013). However, upon infection with KSHV, episomes are largely lost after time in cell culture systems of infection and the number of viral genomes per cell are varied (Adang et al., 2006; Lieberman, 2013). Together, these studies suggested that the processes for inheritance of EBV and KSHV genomes after their replication during cell division may be very different.
Chiu et al. (2017) investigated the strategies used for episome maintenance and distribution by these two viruses using three different approaches: FISH on patient-derived cell lines, live cell imaging, and computer modeling of episome distribution. Using quantitative FISH in fixed patient-derived PEL cells, the authors determined that the range of the number of episomes per cell and the FISH signal intensity (a measure of copy number) were greater with KSHV episomes than EBV. A computer model was generated using the FISH experimental data for viral genome distribution in fixed PEL cells using three parameters: cluster-formation after S-phase, cluster dissociation in the next G1-phase, and the size of the dissociated clusters. This model was fit with the FISH experimental data from assays with wild-type viral genomes and used to predict the cluster sizes and how they would be distributed in live cells during mitosis. To allow tracking in live cells, Chiu et al. (2017) constructed a 33-kb mini-episome that contained only the viral components necessary for latent genome maintenance in cells expressing the LANA1 protein (i.e., the KSHV terminal repeats [used for LANA1 binding to the episome]) and encoded copies of the lac operon (LacO) to enable fluorescence tracking in cells expressing a fusion of the Lac repressor with tdTomato. Three different assays were used to confirm that these miniKLacO plasmids behave like the normal KSHV episomes. In contrast to EBV genomes, the KSHV miniKLacO plasmids varied more in fluorescence intensity and were distributed unequally to daughter cells. The real-time tracking of the miniKLacO plasmids suggested that KSHV episomes may cluster into foci of multiple genome copies and, consequently, are not equally distributed upon host cell division as predicted by the computer model. This model was built with the FISH data, but was able to predict the cluster distributions observed in the live-cell imaging experiments.
In EBV and KSHV coinfected cells, Chiu et al. (2017) observed that EBV genomes segregated equally whereas KSHV genomes were clustered and remain aggregated when distributed into new cells, which suggests that there are some critical differences in the viral genomes or proteins expressed during latency responsible for these different inheritance strategies. The authors explored whether the LANA1 protein played a role in the mechanism for the KSHV clustering phenotype by replacing the histone-binding domains of LANA1 with the AT-hooks of EBNA1 (which EBV uses to attach its episome to AT-rich DNA sequences). The replacement of LANA1 with this LANA1/AT-hook fusion resulted in the loss of high copy number clusters of miniKLacO plasmids, confirming the hypothesis that the histone-binding function of LANA1 is required for the clustering of KSHV episomes. Previous structural studies of LANA1 have suggested oligomeric assembly of LANA1 and DNA (Domsic et al., 2013) and that binding of LANA1 dimers could facilitate compaction of the host genome into a 30-nm chromatin fiber (Chodaparambil et al., 2007). Chiu et al. (2017) suggest that LANA1 may similarly pack the KSHV genome if the DNA binding domain of one LANA1 dimer binds to the terminal repeat region of one KSHV episome while the other end of the dimer associates with the terminal repeats of another episome copy to promote condensation of viral DNA. The authors also note that the terminal repeats need to be in cis above eight copies for episome maintenance and clustering of the genomes. As different regions of LANA1 are known to interact with KSHV episomes and histones, the authors propose a model wherein LANA1 acts as a zipper: by binding to both the terminal repeats of the episomes and to the nucleosomes formed on the episomes with different regions of LANA1 dimers, LANA1 allows clustering of the episomes to form within nuclei during interphase. Another interesting observation from the LANA1/AT-hook experiments is that the segregation behavior of the LANA1/AT-hook was somewhere between the clustering and unequal distribution of KSHV episomes and the equal partitioning of EBV episomes. This suggests that neither the AT-hooks alone nor LANA1 alone determines the segregation behavior of their respective genomes; other cis-acting elements are proposed, but need to be elucidated.
Therefore, this work by Chiu et al. (2017) demonstrates that these two viruses use two distinct mechanisms for distributing their genomes during host cell division. EBV uses a mechanism to tether the EBV genome to host DNA, facilitating the equal distribution of episomes in daughter cells with a larger number of cells having at least one EBV genome. In contrast, KSHV uses a clustering strategy that promotes a higher number of episomes per cell, but at the expense of not passing on the episome to some daughter cells (Fig. 1). In general, the KSHV genome clusters double in intensity with each S-phase, but not always—some clusters do not continually increase in intensity during live-cell tracking across multiple divisions. During cell division, live imaging showed that some clusters disperse into smaller clusters, consistent with the model, which predicted a 31% chance of a cluster breaking up in G1. Currently, we do not know what factors determine whether KSHV genome clusters continue to increase in the number of copies per foci or disperse into smaller clusters. Further work is required to find out if there is a maximum size for a cluster or if there is a greater chance of a larger cluster breaking into smaller clusters.
Figure 1.
Clustering and unequal distribution of KSHV genomes. Nuclei from human cells are depicted with locations of viral episomes represented as filled circles (yellow and red for EBV and KSHV, respectively). The varied red intensities depict the different amounts of KSHV episome copies per foci. The intensity of EBV episome foci and the number of foci varied less in EBV-infected cells compared with KSHV-infected cells. EBV infection tends to equally distribute episomes to promote a higher number of infected cells. KSHV infection displays unequal distribution that promotes a higher number of episomes in a subset of nuclei.
Understanding the mechanisms of how viral episomes are distributed is important because these discoveries could lead to new ways of disrupting episome maintenance and ultimately eliminating herpesviruses in host cells. This approach may be more fruitful than CRISPR-based ways of targeting viral episomes because it may prove challenging for CRISPR to target so many episome copies inside infected cells. Additionally, specific methods to disrupt viral episome maintenance strategies would likely have little effect on uninfected cells. Identifying methods of increasing episome copy numbers may be useful to prevent episome loss and promote long term KSHV maintenance in difficult cell culture systems to study infection. The tools developed in this study could be applied in other systems to study clustering of other nonchromosomal genetic elements. Human cytomegalovirus, another herpesvirus that causes significant disease in immunocompromised patients and when transmitted in utero, has observable circularized viral genomes but the mechanisms for genome maintenance are still unclear (Tarrant-Elorza et al., 2014). Likewise, adeno-associated virus, currently a candidate as a gene therapy vector, persists as episomes (Penaud-Budloo et al., 2008) and it would be beneficial to determine how episomes carrying the gene of interest are distributed and maintained. The tools used by Chiu et al. (2017) could also be applied to nonviral genomic elements; for example, it remains to be determined if the 2-µm circle from yeast uses a clustering mechanism for distribution to daughter cells (Liu et al., 2016).
The clustering mechanism used by KSHV likely favors a more rapid increase in episome copy numbers but only within a subset of cells. Previous evidence indicates that there are more copies of KSHV episomes than EBV in infected cells, which raises the question of whether KSHV copy numbers need to be above a certain threshold to maintain latency. However, one significant cost of the KSHV clustering system is a decrease in the number of cells that receive an episome. Does this strategy of partitioning genomes only to a subpopulation cause the more indolent course of KSHV oncogenesis compared with EBV-associated tumors? Further structural studies and high resolution microscopy will likely complement this recent work by Chiu et al. (2017) and together answer many of the questions about the specific mechanisms of episome maintenance and its consequences for different latent viral infections.
Acknowledgments
We regret not being able to cite more articles because of space limitations.
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health (1ZIABC011176). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no competing financial interests.
References
- Adang L.A., Parsons C.H., and Kedes D.H.. 2006. Asynchronous progression through the lytic cascade and variations in intracellular viral loads revealed by high-throughput single-cell analysis of Kaposi’s sarcoma-associated herpesvirus infection. J. Virol. 80:10073–10082. 10.1128/JVI.01156-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera A.J., Chodaparambil J.V., Kelley-Clarke B., Joukov V., Walter J.C., Luger K., and Kaye K.M.. 2006. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 311:856–861. 10.1126/science.1120541 [DOI] [PubMed] [Google Scholar]
- Cesarman E., Moore P.S., Rao P.H., Inghirami G., Knowles D.M., and Chang Y.. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 86:2708–2714. [PubMed] [Google Scholar]
- Chiu Y.-F., Sugden A.U., Fox K., Hayes M., and Sugden B.. 2017. Kaposi’s sarcoma–associated herpesvirus stably clusters its genomes across generations to maintain itself extrachromosomally. J. Cell Biol.:jcb.201702013 10.1083/jcb.201702013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodaparambil J.V., Barbera A.J., Lu X., Kaye K.M., Hansen J.C., and Luger K.. 2007. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat. Struct. Mol. Biol. 14:1105–1107. 10.1038/nsmb1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsic J.F., Chen H.S., Lu F., Marmorstein R., and Lieberman P.M.. 2013. Molecular basis for oligomeric-DNA binding and episome maintenance by KSHV LANA. PLoS Pathog. 9:e1003672 10.1371/journal.ppat.1003672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman P.M. 2013. Keeping it quiet: Chromatin control of gammaherpesvirus latency. Nat. Rev. Microbiol. 11:863–875. 10.1038/nrmicro3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.T., Chang K.M., Ma C.H., and Jayaram M.. 2016. Replication-dependent and independent mechanisms for the chromosome-coupled persistence of a selfish genome. Nucleic Acids Res. 44:8302–8323. 10.1093/nar/gkw694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaud-Budloo M., Le Guiner C., Nowrouzi A., Toromanoff A., Chérel Y., Chenuaud P., Schmidt M., von Kalle C., Rolling F., Moullier P., and Snyder R.O.. 2008. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 82:7875–7885. 10.1128/JVI.00649-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant-Elorza M., Rossetto C.C., and Pari G.S.. 2014. Maintenance and replication of the human cytomegalovirus genome during latency. Cell Host Microbe. 16:43–54. 10.1016/j.chom.2014.06.006 [DOI] [PubMed] [Google Scholar]