Abstract
A class of curved DNA appears universally in eukaryotic genomic DNA at an average distance of ∼680 bp and shows nucleosome positioning activity by having high affinity for histone core particles in an orientation- and position-dependent manner. Here, we report that the enhancer activity at DNase I hypersensitive site 2 (HS2) of the human β-globin locus control region (β-LCR) can be modulated by the curved DNA located at a distance of two nucleosomes from HS2 and that the nucleosome at the curved DNA regulates nearby nucleosome phases as a key nucleosome. Erythroid-specific nucleosome phases which caused deviation of the NF-E2 (p18–p45 dimer) binding site from the nucleosome dyad axis were over-represented when the distance between the key nucleosome and HS2 exceeded 80 bp longer than the original length. At this state, enhancer activity was ∼50% of that in the original construct, presumably due to reduced binding of transcription factors.
INTRODUCTION
The nucleosome is the most basic unit of chromatin architecture and determines the large-scale chromatin structure as a building block and influences on transcription, recombination and other biological reactions (1). Although the importance of their localization has been reported (2,3), there is no consensus as to how this building block is arranged on the genomic DNA. Translational nucleosome positioning depends on local variations in DNA curvature, helical periodicity and/or boundary effects. When present in gene regulatory regions, nucleosomes can act as a barrier against transcriptional initiation (4) in a gene-specific manner (5). This repressive effect of chromatin is modulated at specific loci by rapid remodeling of the chromatin structure during gene activation. In order to adapt to the rapid changes in the environmental or cellular conditions, activation of genes should occur immediately and in a timely manner. Therefore, the configuration of chromatin, in particular around cis-acting elements, is crucial for gene activation.
One of the well-characterized examples of transcriptional activation and chromatin remodeling is the human β-globin locus. The human β-globin locus spans 70 kb containing five developmentally-regulated genes in the order of 5′-ɛ,Gγ,Aγ,δ,β-3′. Expression of these genes is restricted to erythroid tissues and regulated in a developmental stage-specific manner (6). The β-globin locus control region (β-LCR), a region upstream of the β-globin cluster containing a series of erythroid cell-specific and developmentally stable DNase I-hypersensitive sites (HSs) (7), is required for activation of all the genes. Locus control regions (LCR) maintain chromatin in an open configuration and override inhibitory influences on gene expression (8). The dominant role of the human β-LCR is to confer high-level transcription (9), but developmental regulation of the globin gene expression and the control of replication in this locus are independent of β-LCR (10). The human β-LCR is marked by erythroid-specific HSs (HS1–4) that lie 6–18 kb upstream of the embryonic β-globin gene (11,12). The functional activity of the β-LCR was further subdivided into discrete core regions 200–300 bp long, each coincident with the HSs in native chromatin (13,14). Most of the β-LCR activity resides within the HS2 and HS3 core elements. HS2 appears to be necessary and sufficient for full β-LCR activity in transgenic mice (15,16). This region contains multiple protein binding sites (14) and can act as an enhancer in transient expression experiments (17,18). Some of the trans-acting factors in this region are GATA-1, CACCC-binding proteins and NF-E2. GATA-1, an erythroid-specific transcription factor, can self-associate as well as interact with EKLF, an erythroid-specific factor that recognizes CACCC motifs, providing a potential basis for physical interactions among the regulatory regions (19,20). NF-E2, a bZIP transcription factor, is a heterodimer of 45- and 18-kDa subunits (21–23). Expression of NF-E2 p45 is primarily restricted to erythroid cells, while NF-E2 p18 appears to be ubiquitously expressed. There appears to be considerable functional redundancy among polypeptides that recognize NF-E2 sites, and while the p18-p45 NF-E2 dimer itself may be required to activate globin gene expression, other species may be able to participate in formation of the HSs (24). The overall stimulatory activity of the β-LCR (at least of HS2) in the chromatin environment of transgenic mice or in stably integrated constructs appears to depend on NF-E2 motifs (13,14,25–28).
Previously, we reported that a type of curved DNA appeared nearly periodically in eukaryotic genomes (29) including the human β-globin locus (30). These curved DNA sites were determined by the circular permutation assay (31), and their ‘bendability’ was dependent on their own nucleotide sequence. The curved DNA appeared to be ∼680 bp on average, which is almost the same length of four nucleosomes. We found that the length between the two neighboring sites of curved DNA at HS2 of the human β-LCR, ɛB-16 and ɛB-17, showed an irregularity, indicating that this region can accommodate five nucleosomes (32). NF-E2 binding sites and other cis-acting elements were present at the center between these curved DNA sites, and the center region could adapt to multiple nucleosome phases, although other regions around this accommodated almost only one phase. Thirty base pair sequences containing the bend centers cloned into a vector alone showed identical nucleosome phases to those observed with the in vitro and in vivo experiments, and removing the bend sites caused disruption of the phases at the bend sites as well as those in their direct vicinity (32). From these results, we hypothesized that the curved DNA has the ability to determine the nucleosome positions and act as an initiator of nucleosome phasing. Through such activities, they play important roles in various biological functions by affecting chromatin structure. In this study, we studied the relationship between the curved DNA at HS2 and enhancer activity to obtain evidence for this hypothesis.
MATERIALS AND METHODS
Chemicals
Restriction and modifying enzymes were purchased from New England BioLabs (Beverly, MA). K562 cells were supplied by the Human Science Research Resources Bank (Osaka, Japan). Cell culture materials were obtained from Gibco Laboratories (Grand Island, NY). Other chemicals used were of the highest quality commercially available and were purchased from Sigma Chemical Co. (St Louis, MO).
Cell lines and culture conditions
K562 erythroleukemia cells and its derivative transformants were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum in a humidified incubator with a 5% CO2 atmosphere.
Reconstitution of the nucleosomes in vitro
All procedures were carried out at 4°C. Nucleosomal core particles were prepared from chicken erythrocytes according to the method of Lutter (33). Briefly, erythrocytes were collected from 50 ml chicken blood and homogenized in 10 mM Tris–HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40 to prepare nuclei. The nuclei were suspended in 0.34 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM 2-mercaptoethanol, 0.5 mM spermidine, 0.15 mM spermine, 15 mM Tris–HCl pH 7.5, 1 mM CaCl2 and treated with 40 U/ml micrococcal nuclease for 2 min at 37°C. Histone core particles were recovered by centrifugation at 8000 g for 2 min, and after suspension in 0.2 mM EDTA pH 8.0, they were depleted of histone H1 with 0.6 M NaCl, 0.2 mM 2-mercaptoethanol, 5 mM Tris–HCl pH 7.5, and purified by gel filtration through Sepharose CL-6B (Pharmacia, Uppsala, Sweden) in the same buffer. Then, the core particles were dialyzed against 20 mM ammonium acetate, 0.2 mM EDTA, 2 mM 2-mercaptoethanol, 5 mM Tris–HCl pH 7.5 and concentrated to 0.2 mg/ml. Aliquots of 10 µg purified core particles were mixed with 20 µg XmnI-digested plasmid DNA containing the human β-globin HS2 region or its derivatives in 100 µl TEP (10 mM Tris–HCl pH 7.5, 0.1 mM EDTA, 0.1 mM PMSF) supplemented with 2 M NaCl. The mixtures were dialyzed against 1 l TEP with 0.4 M NaCl for 3 h and then against 1 l TEP with 16 mM NaCl overnight.
Southern blot analysis of the nucleosome phases
The nucleosomes were reconstituted in vitro and digested with micrococcal nuclease (MNase). MNase digestion was performed at the final concentration of 0.025 U/ml at 25°C for 1, 2 (where applicable), 5 and 10 min. DNA was purified and electrophoresed on 2% agarose gels, then transferred onto the Hybond plus membranes (Amersham, UK). DNA labeling, hybridization and detection were performed using the Alkphos Direct Labeling and Detection System (Amersham) according to the manufacturer’s instructions. The positions of the probes were: –11 627 to –11 499 (Probe A), –11 328 to –11 119 (Probe B), –11 118 to –10 949 (Probe C), –11 118 to –10 979 (Probe C′), –10 989 to –10 769 (Probe D), –10 768 to –10 569 (Probe E) and –10 688 to –10 609 (Probe E′) relative to the cap site of the ɛ-globin gene.
Reporter gene assay
DNA fragments containing HS2 of β-LCR and its derivatives were connected to the human Aγ-globin promoter region containing from –200 to +40 bp to the cap site, and cloned into the pGL3-Basic vector (Promega, Madison, WI). The constructs were co-transfected with pRL-CMV (Promega) as an internal control into K562 cells by electroporation. A luciferase assay was performed using a Dual Luciferase Reporter Assay System (Promega) and a Lumat LB 9507 luminometer (EG&G Berthold, Bad Wildbad, Germany). The transcriptional activities were normalized relative to the Renilla luciferase activities.
Establishment and characterization of stable transformants
K562 cells (1 × 107) were transfected with 10 µg luciferase reporter gene constructs containing HS2 of human β-LCR or its derivatives and the Aγ-globin promoter region (–200/+40), and 1 µg pTracer-CMV (Invitrogen Co., San Diego, CA). Cells were selected in Zeocin (100 µg/ml) for 2 weeks, and 10–20 stable clones were established for each construct. The luciferase activity was normalized relative to the protein concentration of each clone, which was determined by Bradford’s method using BSA as a standard (34). Genomic DNA of these clones was analyzed by Southern blotting. Aliquots of 15 µg DNA were digested to completion with XbaI, resolved on 0.8% agarose gels, and transferred onto Hybond plus membranes. A 32P-labeled luciferase cDNA fragment was used as a probe. The relative copy number of the integrated constructs was determined by analysis of the blot with a Phosphorimager (Molecular Dynamics, Sunnyvale, CA). The lowest copy number was arbitrarily designated as one copy.
In vivo footprinting
Nuclei were prepared from K562 or HeLa cells transfected with plasmid DNA and suspended in 0.34 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM 2-mercaptoethanol, 0.5 mM spermidine, 0.15 mM spermine, 15 mM Tris–HCl pH 7.5, 1 mM CaCl2. DNase I was added to nuclear suspensions to a final concentration of 35–350 U/ml, and reactions were carried out at 4°C for 10 min. Then, DNA was purified and used for primer extension as a template. Linear amplification of the digested fragment was carried out by PCR primer extension with a 32P end-labeled primer (5′-GGTCAGTGCCCCACCCCCGCCTT-3′; –10 918 to –10 896 relative to the cap site of the ɛ-globin gene) and the amplified fragments were resolved on 8% denaturing polyacrylamide gels. The plasmids containing NF-E2 subunits, p18 and p45, were kindly gifted by Dr M. Yamamoto (35). After preparation of the cording region fragments using these plasmids by PCR, the amplified fragments were inserted into pTracerCMV vector (Invitrogen) for expression of NF-E2 in HeLa cells.
RT–PCR
The first strand cDNA was synthesized from 5 µg RNA using Molony murine leukemia virus reverse transcriptase (TOYOBO, Osaka, Japan) and oligo(dT)-primers. The PCR primers were designed from the DDBJ database as follows: p18, 5′-ATGGATCCATGACGACTAATCCCAAGCCCAACAAGGCA-3′ and 5′-ATAAAGCTTCTAGGAGGCGGCTGAGAAGGGTACAGAGG-3′; p45, 5′-ATGGATCCATGCCCCCGTGTCCTCCTCAGCAGAACAGG-3′ and 5′-AGACCAGCTCAATCTGTAGCCTCC-3′; glyceraldehyde 3-phosphate dehydrogenase (G3PDH), 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′. The conditions for PCR were 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 2 min, followed by a single 10 min extension. The PCR fragments were analyzed by electrophoresis on 2% agarose gels and visualized by ethidium bromide staining.
RESULTS
Organization of nucleosomal phases by curved DNA
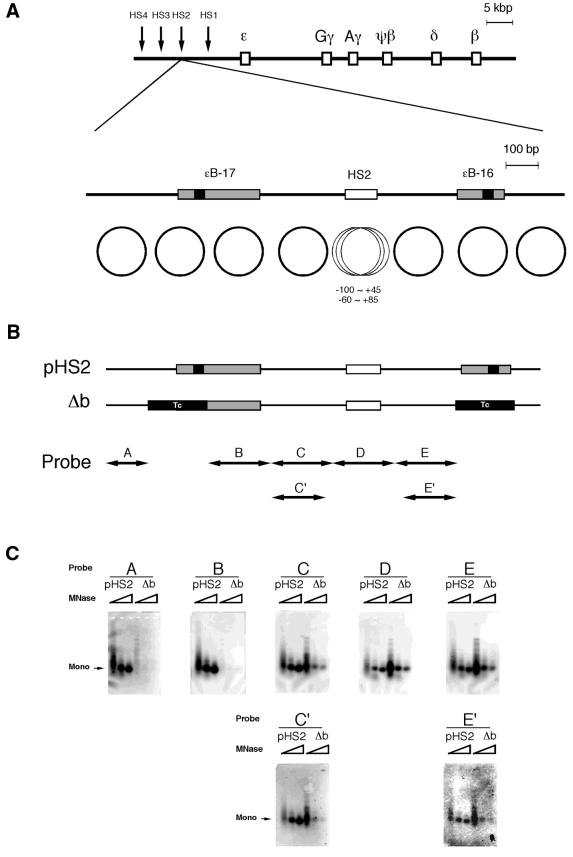
Two of the sites of curved DNA appearing regularly in β-LCR, ɛB-16 and ɛB-17, were mapped upstream and downstream of HS2, respectively (Fig. 1A). These curved DNA sites showed unique nucleosome phases at these positions, and there was a space for four nucleosomes between these nucleosomes (32). This space was actually filled with four nucleosomes with a unique phase when the locus was not open, in HeLa cells for example, whereas multiple phases were adopted at HS2 in the erythroid cell line K562. In HeLa cells, only the –60∼+85 phase was observed, while other phases including the –100∼+45 phase were observed in K562 cells. A deletion construct in which both ɛB-16 and ɛB-17 were replaced with non-curved DNA (Δb) was examined for nucleosome formation (Fig. 1B). While mononucleosome fragments resistant to micrococcal nuclease were observed with control pHS2 for all probes (probes A–E), they were not observed with Δb but only for probes A or B. Because the HS2 region indicated as a probe D in Figure 1 B showed multiple nucleosomal positions and some of these positions contained a part of the regions C or E, as we reported before (32), we applied Southern blotting using probes C′ and E′, in which the regions overlapping all nucleosome phases at HS2 were removed from probes C and E. Then reduced intensities of the fragments were observed. Only probe D showed identical intensities between Δb and pHS2. These observations indicated that when the curved DNA was removed, activity of nucleosome formation at the neighboring phases was lost. This suggests that nucleosomes at these curved DNA sites were used as key nucleosomes, determining the locations of the other neighboring phases. The effects of the key nucleosomes reached at least the second nucleosomes (see the results of probes C and C′). On the other hand, the nucleosome at HS2 was positioned independently, and its location was not uniquely determined but showed multiple phases.
Figure 1.
Nucleosomal phases in the presence or absence of curved DNA. (A) Maps of the human β-like globin gene cluster and DNase I HS2. The shaded and solid boxes are the sites of curved DNA determined by circular permutation assay and the DNA bend centers, respectively (32). The positions of the bend sites relative to the cap site of the ɛ-globin gene are: ɛB-17, –11 420 (SacI) to –11 166 (AflIII); ɛB-16, –10 558 (MunI) to –10 408 (EarI). The circles indicate the nucleosomal phases reported previously (32). (B) The mutant (Δb) was constructed by replacing the nucleosome regions containing the bend centers with fragments of identical length derived from pBR322 (positions 247–404 or positions 417–574 for ɛB-17 or ɛB-16, respectively) indicated as ‘Tc’ to remove bendability and nucleosome forming activity. The positions of the probes for Southern blot analysis correspond to the nucleosome positions. (C) Southern blot analysis of the in vitro reconstituted nucleosomes using plasmids, pHS2 and Δb. The plasmid DNA was digested with increasing amounts of micrococcal nuclease and the purified DNA was subjected to Southern blot analysis. The membrane was stripped and rehybridized with the probes shown as the horizontal arrows in (B). ‘Mono’ indicates the position of the mononucleosomes.
Positional effect of key nucleosome on enhancer activity
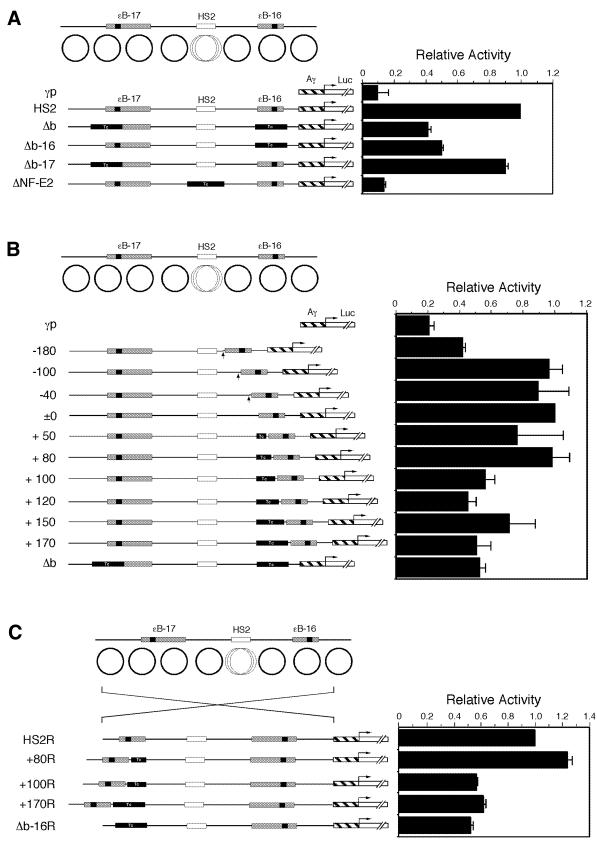
The effects of the key nucleosome on enhancer activity were examined by transient expression assay using reporter genes (Fig. 2). Constructs with the original and modified HS2 regions connected to the Aγ-globin gene promoter and the luciferase gene were introduced into K562 cells, and the luciferase activities relative to those of the HS2 constructs were examined. Reduced activities were observed with Δb and Δb-16, but not with Δb-17, suggesting that replacement of only ɛB-16 affected the activity (Fig. 2A). Here, we can speculate on two possibilities. One is that ɛB-16 region contains the important sequence(s) for globin gene expression, but it has been reported that deleting this region does not affect the gene expression (17). Another is that the length between the bend site and HS2 is important. To confirm this possibility, we further examined the effects of ɛB-16 by changing the distance between the curved DNA and HS2 (Fig. 2B). The reduction of the distance decreased the activity when the deletion starting from the linker region next to the key nucleosome at ɛB-16 reached –180 bp, probably due to the deletion of a functional GATA-1 motif (13) and other motifs for transcription factors (36). When the distance was increased by filling the nucleosomal linker next to ɛB-16 with non-curved DNA, there was a clear change in the activity between the +80 and +100 constructs. Constructs containing added distances longer than +80 bp showed a reduction of the activity to ∼50% of that of the original HS2 construct. This was also true when the space was filled with other non-curved DNA (data not shown). Reduction of the activity was also observed when the entire HS2 region was reversed (+100R, +170R and Δb-16R compared with HS2R and +80R; Fig. 2C), confirming that the effect of non-curved DNA was not due to the insulator activity (37) nor due to the change of the distances between the promoter and HS2.
Figure 2.
Effects of configuration around the HS2 region on the enhancer activity in transient expression assay. (A) Effects of replacement of curved DNA with non-curved DNA on the enhancer activity. Either or both the nucleosome regions of DNA bend sites, ɛB-16 and ɛB-17, or HS2 site were replaced with fragments of identical length derived from pBR322 (see Fig. 1 legend). (B) Effects of distance between ɛB-16 and NF-E2 sites on the enhancer activity. The distance between ɛB-16 and NF-E2 sites was changed by deleting or inserting fragments of various lengths into the nucleosome linker region next to ɛB-16. (C) Effects of the orientation of the original and modified HS2 regions on the enhancer activity. The constructs HS2R, +80R, +100R, +170R and Δb-16R contained the same fragments as in HS2, +80, +100, +170 and Δb-16 in the reverse orientation, respectively. Solid Tc boxes and hatched boxes indicate the non-curved DNA fragment and Aγ-globin promoter, respectively. The luciferase activities shown are the mean values of triplicate assays, and the horizontal bars represent standard deviations.
This reduced activity was observed again with the stable transformants where constructs containing a deletion (Δb) or insertions (+170 and +80) showed reduced luciferase activity (Fig. 3). The activities of transformants containing at least 10 clones for each construct showed the same tendency of reduced activities for Δb and +170 compared with HS2 and +80 (Fig. 3A). This was also reproduced with copy number-adjusted luciferase activities for five transformants from each construct (Fig. 3B), where reduction of the average activities of +170 and Δb constructs (2.29 and 2.58, respectively) was observed compared with HS2 and +80 constructs (4.81 and 4.85, respectively). Although Figure 3B shows some scatter, presumably due to the randomness of stable integration and the lack of flanking DNA insulators, the trends are real.
Figure 3.
Effects of configuration around the HS2 region on the enhancer activity in stable transformants. (A) Luciferase assay using stable transformants. Mixtures of stable transformants containing 10–20 different clones of each construct were collectively assayed for luciferase activities and the relative light units (RLUs) adjusted with total protein amounts were shown in the graph. (B) The relative luciferase activities of stable transformants normalized relative to the copy number of the integrated plasmid DNA. Five clones from each construct were subjected to Southern blot analysis to determine the relative copy number of the integrated plasmid DNA, which was used for correcting luciferase activities. A 32P-labeled luciferase cDNA fragment was used as a probe. The averages and the standard deviations are shown above. P values against HS2 constructs were assessed by student’s t-test.
Effect of elongation of distances between curved DNA and NF-E2 site on chromatin
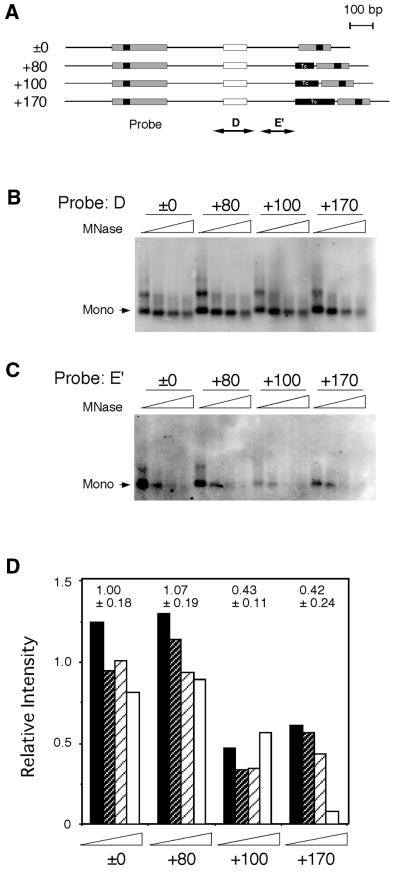
We next examined whether replacement of curved DNA at ɛB-16 and elongation of the distance affected the chromatin structure at and around HS2 (Fig. 4). Southern blot analysis of the constructs with various distances between the curved DNA and HS2 was performed with probes D and E′, which detected the nucleosomes at or next to HS2, respectively (Fig. 4A). The same membrane was used for both probes and the relative intensity of the probe E′ to that of probe D was calculated (Fig. 4B–D). The results clearly indicated that the relative intensity was lower for the +100 and +170 bp constructs while both the ±0 and +80 bp constructs retained similar degrees of resistance to micrococcal nuclease. This suggests that the nucleosomes located between the curved DNA and HS2 were influenced by elongation of the distance.
Figure 4.
In vitro nucleosome phases at HS2. (A) Constructs with various distances between ɛB-16 and NF-E2 sites were reconstituted with core particles in vitro. Reconstituted nucleosomes were digested with micrococcal nuclease, and DNA fragments were purified and subjected to Southern blot analysis. The probes corresponded to the nucleosome phase of the NF-E2 region (probe D) and the next phase toward ɛB-16 (probe E′). The same membrane was stripped and used for rehybridization. (B) Southern blot analysis of nucleosomes at NF-E2 site. (C) Southern blot analysis of nucleosomes in the region E′. (D) Relative degrees of nucleosome formation at region E′. The intensity of the bands was quantified by ImageQuant (Molecular Dynamics), and the degrees of nucleosome formation at region E′ relative to those of NF-E2 (determined with probe D) were calculated. The averages and the standard deviations, which were normalized with the average value for ±0 construct, are shown above.
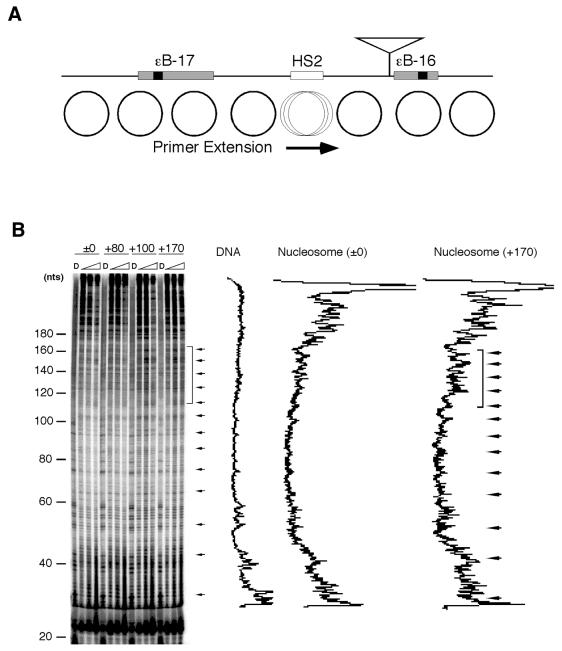
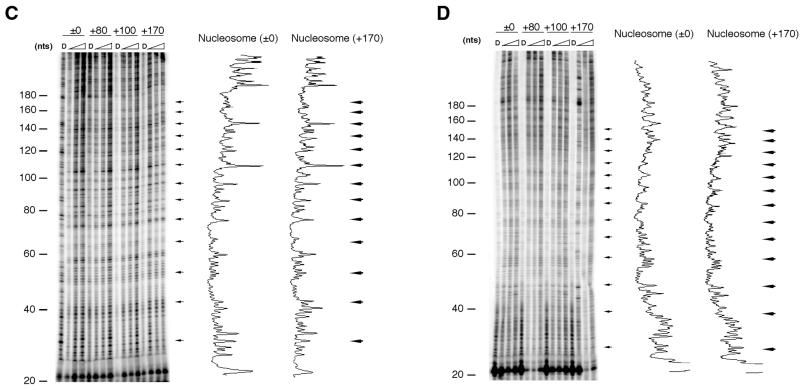
More precise nucleosome phases were determined by DNase I footprinting assay followed by primer extension (Fig. 5). From the presence of periodic 10-bp cleavage sites (arrowed in Fig. 5B), which corresponded to the rotational phases of the nucleosome at HS2, this assay detected an intact chromatin state. The result of the assay indicated that the region between 120 and 170 bp from the primer became more sensitive at distances longer than +100 bp (Fig. 5B, left panel). This region, which showed an increased DNase I sensitivity (Fig. 5B, marked by a square bracket), included the motifs for the erythroid-specific transcription factors HS2NF5 and GATA-1. The differences of the DNase I sensitivity between ±0/+80 and +100/+170 constructs were not observed in HeLa cells or in vitro reconstituted chromatin (Fig. 5C and D), suggesting that the erythroid-specific enhancer activity is involved in the chromatin state.
Figure 5.
In vivo and in vitro nucleosome phases at the HS2 region in constructs with various distances between ɛB-16 and NF-E2 sites. Locations of primer extension and insertion (shown by an inverted triangle) (A), and nucleosome phases in K562 cells (B), in HeLa cells (C) or in in vitro reconstituted chromatin (D) are shown. K562 and HeLa cells were harvested 24 h after transfection of the constructs ±0, +80, +100 or +170, and subjected to in vivo footprinting assay. The densitometrical patterns of the samples from naked DNA (labeled as D) and reconstituted nucleosomes in the ±0 and +170 constructs were visualized by ImageQuant. Horizontal arrows and a bracket indicate the periodic 10-bp cleavage sites of the nucleosome indicating rotational phases and a region with higher sensitivity found in the +100 and +170 constructs, respectively.
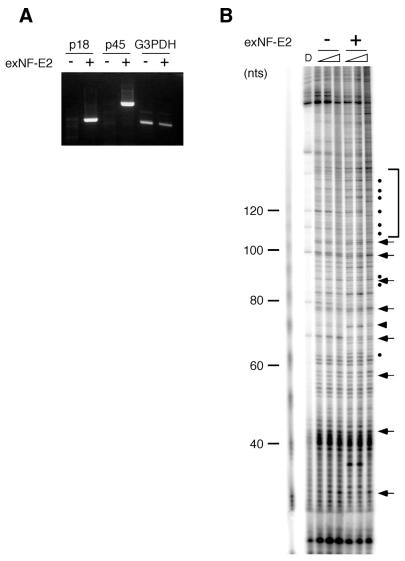
To elucidate the relationship to the erythroid-specific transcription factor NF-E2 on the differences of the DNase I sensitivity between ±0/+80 and +100/+170 constructs, DNase I footprinting assay was performed after plasmids expressing both p18 and p45 subunits of NF-E2 were introduced into HeLa cells (Fig. 6). Neither NF-E2 subunit p18 nor p45 was detected in HeLa cells by RT–PCR, while exogenous NF-E2 was detected after transfection (Fig. 6A). An increased DNase I sensitivity (shown by dots in Fig. 6B) of the region between 120 and 170 bp from the primer, which corresponded exactly to the region that was included in the –60∼+85 phase but was excluded in the –100∼+45 phase, was observed only in the presence of NF-E2 (Fig. 6B, marked by a square bracket). Note that an increased DNase I sensitivity was also observed next to the NF-E2 binding site (shown by arrowhead), which is characteristic to NF-E2 interaction to the motif (Y.Onishi and R.Kiyma, unpublished result). Other DNase I sensitive sites, around 90 nucleotides for example, were also observed. This result suggests that changes in the chromatin structure, as indicated by an increased DNase I sensitivity, was brought by NF-E2 binding.
Figure 6.
Effect of NF-E2 on nucleosome phases at the HS2 region. The plasmid ±0 construct was introduced into HeLa cells with or without the plasmids (exNF-E2) expressing NF-E2 subunits, p18 and p45. HeLa cells were harvested 24 h after transfection and subjected to RT–PCR (A) and in vivo DNase I footprinting assay (B). G3PDH expression was examined as a control in RT–PCR. Horizontal arrows and a square bracket indicate the periodic 10-bp cleavage sites of the nucleosome indicating rotational phases and a region with higher sensitivity found in the cells with exNF-E2, respectively. The DNA fragments showing increased sensitivities were marked by a closed triangle (at the NF-E2 binding site) or dots (at the other sites)..
DISCUSSION
Nucleosomes and modulation of transcription activity
Nucleosome positioning affects transcription efficiency. Wittig and Wittig (38) reported that, using the chicken tRNA gene, the functional state of a gene can be programmed by nucleosome position, and transcription of the gene is dependent on nucleosome position. In the mouse mammary tumor virus (MMTV) promoter, which is strongly induced by glucocorticoid hormone and NF1, transcriptional activation by glucocorticoid hormone induces nucleosome positioning in vivo (39). Moreover, it was demonstrated that rotational phasing and translational positioning affect the binding of NF1 to the MMTV promoter in vitro (40). We previously showed that a type of curved DNA appears regularly, nearly periodically in many locations, in eukaryotic genomic DNA (41,42). The biological importance of these curved DNA sites was elucidated for the globin genes. These sites were evolutionarily conserved among the human β-like globin genes as well as the β-like globin genes from other species. However, their nucleotide sequences were not conserved, and insertion of Alu and other elements and genome rearrangement did not change their relative locations. Transcriptional modulation by these sites was highlighted as these sites in the globin genes and other genes overlapped with silencer or repressor activity (43). The results shown here again strengthen the biological significance of these curved DNA sites and answer one of the basic questions: the significance of the average interval of 680 bp corresponding to roughly four nucleosomes. If the effect of a key nucleosome reaches the second nucleosome, a four-nucleosome interval would be convenient because all three nucleosomes located between two key nucleosomes could then be controlled. When there is a longer interval, such as that seen in HS2 where five nucleosomes can be accommodated, the nucleosome position and its effect on enhancer activity is influenced or regulated only by one of the key nucleosomes.
The effects of nucleosome positioning by the nucleosome located at ɛB-16, designated as a key nucleosome, were clearly shown by Southern blots (Figs 1C and 4). Insertion of non-curved DNA between the key nucleosome and the nucleosome located next to it changed the nucleosomal states not only at the position of insertion (data not shown) but also at the neighboring nucleosomes, as revealed by probe E′ (Fig. 4, the nucleosome next to the key nucleosome) and by in vivo DNase I footprinting (Fig. 5, the nucleosome at HS2). When the length of insertion reached at least 100 bp, nucleosome formation at the nucleosome next to the key nucleosome was reduced to ∼40% (Fig. 4). This change again affected the nucleosome phase at HS2 (Fig. 5), and was likely to change the enhancer function eventually (Figs 2B and 3).
Active and inactive states of enhancer and nucleosome phases
The –60∼+85 phase has a nucleosome dyad axis at its center where the NF-E2 binding site is located, and includes the binding sites of the erythroid-specific transcription factors HS2NF5 and GATA-1 (44). This phase was previously shown to be the only phase in HeLa cells (32), and thus, this is the original state where NF-E2 first interacts with the cognate motif. Since there is no NF-E2 in HeLa cells (Fig. 6A), chromatin there remains closed and the enhancer is inactive (Fig. 6B). In contrast, the –100∼+45 phase and several other minor phases were present in K562 cells which express NF-E2 (32). NF-E2 has been shown to undergo chromatin remodeling in the presence of nuclear factors (45). When the nucleosome is positioned at –60∼+85, the NF-E2 binding site is located at the dyad axis, and NF-E2 and other nuclear factors may have increased accessibility. If once NF-E2 and other factors bind to this region, the –100∼+45 phase and several other minor phases can be seen as a result of the chromatin remodeling (32). Therefore, in NF-E2-expressing HeLa cells, shifting of the –60∼+85 phase to the –100∼+45 phase was observed as an increased DNase I sensitivity (Fig. 6B, marked by a square bracket). Then the erythroid-specific transcription factors HS2NF5 and GATA-1 could interact with their binding sites, which was partially or completely covered with proteins resulting in reduced accessibility of DNase I as shown in K562 cells (Fig. 5B, region of ±0/+80 constructs marked by a square bracket).
When other phases were adopted by insertion of more than 80 bp between the key nucleosome and HS2, nucleosomes would have an inhibitory effect probably due to reduced accessibility of NF-E2, remodeling factors and/or nuclear factors. In cases of +100/+170 constructs, the region containing HS2NF5 and GATA-1 binding showed higher sensitivity to DNase I (Fig. 5B, marked by a square bracket). In fact, we observed reduced enhancer activity when this region is sensitive to DNase I (Figs 2B, 3 and 5). This was probably due to less factor binding caused by improper localization of the nucleosome, or its dyad axis, at this position. Localization of nucleosomes at HS2 was mostly influenced by the state of the nucleosome next to it as a result of cooperativity between nucleosomes, and the periodic DNA bend sites may provide the information of the nucleosomal configuration and organize in part the chromatin structure.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Y. Wada-Kiyama for helpful comments about this work. This work was supported by grants from the Ministry of Economy, Trade and Industry (formerly the Ministry of International Trade and Industry) and from the New Energy and Industrial Technology Development Organization (NEDO).
References
- 1.Kornberg R.D. and Lorch,Y. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell, 98, 285–294. [DOI] [PubMed] [Google Scholar]
- 2.Luger K., Mäder,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 3.Simpson R.T. (1991) Nucleosome positioning: occurence mechanisms and functional consequences. Prog. Nucleic Acid Res. Mol. Biol., 40, 143–184. [DOI] [PubMed] [Google Scholar]
- 4.Han M., Kim,U.-J., Kayne,P. and Grunstein,M. (1988) Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J., 7, 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyrick J.J., Holstege,F.C., Jennings,E.G., Causton,H.C., Shore,D., Grunstein,M., Lander,E.S. and Young,R.A. (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature, 402, 418–421. [DOI] [PubMed] [Google Scholar]
- 6.Karlsson S. and Nienhuis,A.W. (1985) Developmental regulation of human globin genes. Annu. Rev. Biochem., 54, 1071–1108. [DOI] [PubMed] [Google Scholar]
- 7.Forrester W.C., Thompson,C., Elder,J.T. and Groudine,M. (1986) A developmentally stable chromatin structure in the human β-globin gene cluster. Proc. Natl Acad. Sci. USA, 83, 1359–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenfeld G. (1992) Chromatin as an essential part of the transcriptional mechanism. Nature, 355, 219–224. [DOI] [PubMed] [Google Scholar]
- 9.Bender M.A., Bulger,M. and Groudine,M. (2000) β-globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol. Cell, 5, 387–393. [DOI] [PubMed] [Google Scholar]
- 10.Cimbora D.M., Schubeler,D., Reik,A., Hamilton,J., Francastel,C., Epner,E.M. and Groudine,M. (2000) Long-distance control of origin choice and replication timing in the human β-globin locus are independent of the locus control region. Mol. Cell. Biol., 20, 5581–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrester W.C., Takegawa,S., Papayannopoulou,T., Stamatoyannopoulos,G. and Groudine,M. (1987) Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res., 15, 10159–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuan D., Solomon,W., Li,Q. and London,I.M. (1985) The ‘β-like-globin’ gene domain in human erythroid cells. Proc. Natl Acad. Sci. USA, 82, 6384–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talbot D. and Grosveld,F. (1991) The 5′-HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J., 10, 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talbot D., Philips,P. and Grosveld,F. (1990) Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J., 9, 2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd J.A., Krakowsky,J.M., Crable,S.C. and Lingrel,J.B. (1992) Human γ- to β-globin gene switching using a mini construct in transgenic mice. Mol. Cell. Biol., 12, 1561–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morley B.J., Abbott,C.A., Sharpe,J.A., Lida,J., Chan-Thomas,P.S. and Wood,W.G. (1992) A single β-globin locus control region element (5′ hypersensitive site 2) is sufficient for developmental regulation of human globin genes in transgenic mice. Mol. Cell. Biol., 12, 2057–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ney P.A., Sorrentino,B.P., McDonagh,K.T. and Nienhuis,A.W. (1990) Tandem AP-1-binding sites within the human β-globin dominant control region function. Genes Dev., 4, 993–1006. [DOI] [PubMed] [Google Scholar]
- 18.Tuan D.Y., Solomon,W.B., London,I.M. and Lee,D.P. (1989) An erythroid-specific, developmental-stage-independent enhancer far upstream of the human ‘β-like globin’ genes. Proc. Natl Acad. Sci. USA, 86, 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crossley M., Merika,M. and Orkin,S.H. (1995) Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol., 15, 2448–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong Q. and Dean,A. (1993) Enhancer-dependent transcription of the ɛ-globin promoter requires promoter-bound GATA-1 and enhancer-bound AP-1/NF-E2. Mol. Cell. Biol., 13, 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews N.C., Erdjument-Bromage,H., Davidson,M.B., Tempst,P. and Orkin,S.H. (1993) Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature, 362, 722–728. [DOI] [PubMed] [Google Scholar]
- 22.Andrews N.C., Kotkow,K.J., Ney,P.A., Erdjument-Bromage,H., Tempst,P. and Orkin,S.H. (1993) The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc. Natl Acad. Sci. USA, 90, 11488–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ney P.A., Andrews,N.C., Jane,S.M., Safer,B., Purucker,M.E., Weremowicz,S., Morton,C.C., Goff,S.C., Orkin,S.H. and Nienhuis,A.W. (1993) Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol. Cell. Biol., 13, 5604–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotkow K.J. and Orkin,S.H. (1995) Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol., 15, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caterina J.J., Ryan,T.M., Pawlik,K.M., Palmiter,R.D., Brinster,R.L., Behringer,R.R. and Townes,T.M. (1991) Human β-globin locus control region: analysis of the 5′ DNase I hypersensitive site HS 2 in transgenic mice. Proc. Natl Acad. Sci. USA, 88, 1626–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caterina J.J., Ciavatta,D.J., Donze,D., Behringer,R.R. and Townes,T.M. (1994) Multiple elements in human β-globin locus control region 5′ HS 2 are involved in enhancer activity and position-independent, transgene expression. Nucleic Acids Res., 22, 1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis J., Talbot,D., Dillon,N. and Grosveld,F. (1993) Synthetic human β-globin 5′-HS2 constructs function as locus control regions only in multicopy transgene concatamers. EMBO J., 12, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D., Chang,J.C., Moi,P., Liu,W., Kan,Y.W. and Curtin,P.T. (1992) Dissection of the enhancer activity of β-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc. Natl Acad. Sci. USA, 89, 3899–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohki R., Hirota,M., Oishi,M. and Kiyama,R. (1998) Conservation and continuity of periodic bent DNA in genomic rearrangements between the c-myc and immunoglobulin heavy chain µ loci. Nucleic Acids Res., 26, 3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada-Kiyama, Y., Suzuki,S. and Kiyama,R. (1999) DNA bend sites in the human β-globin locus: Evidence for a basic and universal structural component of genomic DNA. Mol. Biol. Evol., 16, 922–930. [DOI] [PubMed] [Google Scholar]
- 31.Wu H.-M. and Crothers,D.M. (1984) The locus of sequence-directed and protein-induced DNA bending. Nature, 308, 509–513. [DOI] [PubMed] [Google Scholar]
- 32.Onishi Y., Wada-Kiyama,Y. and Kiyama,R. (1998) Expression-dependent perturbation of nucleosomal phases at HS2 of the human β-LCR: Possible correlation with periodic bent DNA. J. Mol. Biol., 284, 989–1004. [DOI] [PubMed] [Google Scholar]
- 33.Lutter L.C. (1978) Kinetic analysis of deoxyribonuclease I cleavages in the nucleosomal core: evidence for a DNA superhelix. J. Mol. Biol., 124, 391–420. [DOI] [PubMed] [Google Scholar]
- 34.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 35.Igarashi K., Itoh,K., Motohashi,H., Hayashi,N., Matsuzaki,Y., Nakauchi,H., Nishizawa,M. and Yamamoto,M. (1995) Activity and expression of murine small Maf family protein MafK. J. Biol. Chem., 270, 7615–7624. [DOI] [PubMed] [Google Scholar]
- 36.Elnitski L., Miller,W. and Hardison,R. (1997) Conserved E boxes function as part of the enhancer in hypersensitive sites 2 of the β-globin locus control region. J. Biol. Chem., 272, 369–378. [DOI] [PubMed] [Google Scholar]
- 37.Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–515. [DOI] [PubMed] [Google Scholar]
- 38.Wittig S. and Wittig,B. (1982) Function of a tRNA gene promoter depends on nucleosome position. Nature, 297, 31–38. [DOI] [PubMed] [Google Scholar]
- 39.Belikov S., Gelius,B., Almouzni,,G. and Wrange,Ö. (2000) Hormone activation induces nucleosome positioning in vivo. EMBO J., 19, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisfeld K., Candau,R., Truss,M. and Beato,M. (1997) Binding of NF1 to the MMTV promoter in nucleosomes: influence of rotational phasing, translational positioning and histone H1. Nucleic Acids Res, 25, 3733–3742. [DOI] [PMC free article] [PubMed]
- 41.Wada-Kiyama Y. and Kiyama,R. (1994) Periodicity of DNA bend sites in human ɛ-globin gene region: possibility of sequence-directed nucleosome phasing. J. Biol. Chem., 269, 22238–22244. [PubMed] [Google Scholar]
- 42.Wada-Kiyama Y. and Kiyama,R. (1995) Conservation and periodicity of DNA bend sites in the human β-globin gene locus. J. Biol. Chem., 270, 12439–12445. [DOI] [PubMed] [Google Scholar]
- 43.Kiyama R., Onishi,Y., Wanapirak,C. and Wada-Kiyama,Y. (1999) Regulation of transcription by bent DNA through chromatin structure. Gene Ther. Mol. Biol., 4, 327–332. [Google Scholar]
- 44.Lam L.T. and Bresnick,E.H. (1996) A novel DNA-binding protein, HS2NF5, interacts with a functionally important sequence of the human β-globin locus control region. J. Biol. Chem., 271, 32421–32429. [DOI] [PubMed] [Google Scholar]
- 45.Armstrong J.A. and Emerson,B.M. (1996) NF-E2 disrupts chromatin structure at human β-globin locus control region hypersensitive site 2 in vitro. Mol. Cell. Biol., 16, 5634–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]