Abstract
Background
Limited data are available on the prevalence of oncogenic driver mutations in Caucasian populations, and especially in Europeans.
Aim
To evaluate the targetable mutational spectra in unselected patients with lung adenocarcinoma in routine clinical practice from several French hospitals, using the same molecular platform.
Patients and Methods
Samples from 2,219 consecutive patients with histologically-proven advanced lung adenocarcinoma were centrally analysed at a referenced and certified diagnostic platform in order to test for activating and resistance mutations in EGFR, KRAS, BRAF, ERBB2 and PI3KCA. Demographic and clinical features were retrieved from the medical charts. Multivariate binary logistic regression was used to determine the independent predictive factors for the occurrence of specific mutations, in the whole study population or in selected subgroups.
Findings
The overall respective incidence of EGFR, KRAS, BRAF, ERBB2 and PI3KCA mutations was 10.5%, 0.9%, 25%, 1.5%, 2.1% and 1.4%, in our study sample including 87.4% white Caucasians, 10.8% Africans and 1.8% Asians; 60.6% men, 30.7% never smoker (median age: 68.3 years). Ethnicity was an independent predictor for EGFR, KRAS and ERBB2 gene abnormalities. In all cases, a significantly higher prevalence of targetable EGFR and ERBB2, and a lower prevalence of resistance KRAS mutations were observed in African women as compared to African men or Caucasians.
Conclusions
In real life conditions of routine genetic testing, we have identified subsets of patients with specific targetable activating somatic mutations according to ethnicity, who could preferentially benefit from anti-EGFR and anti-ERBB2 targeted therapies.
Keywords: NSCLC, genetic testing, genotype, EGFR, ERBB2
INTRODUCTION
The identification of EGFR mutations as activating mutations conferring sensitivity to anti-EGFR therapies like gefitinib or erlotinib was firstly reported in 2004 by 2 groups studying American and Asian patient populations [1–3]. They also observed that EGFR mutations in exons 18 to 21 were more prevalent in Japaneses (30%) as compared to Caucasians (only 7%). In 2009, the IRESSA Pan-Asia Study (IPASS), which compared the EGFR tyrosine kinase inhibitor (TKI), gefitinib, with carboplatin-paclitaxel cytotoxics, was conducted on patients of Asian descent with adenocarcinoma, as a mutation-enriching strategy in the study population. The trial demonstrated that gefitinib was associated with a 12-month progression-free survival rate of 24.9%, in comparison to 6.7% in the chemotherapy arm [4]. This key study has authorized the use of gefitinib as first-line therapy in patients whose metastatic adenocarcinoma carries EGFR mutations.
Women, and especially never smokers, also exhibit a higher EGFR mutational frequency, especially in Asian never smokers, with a rate ranging between 48% and 75.3% [5–8].
A number of institutions worldwide have integrated EGFR molecular profiling into routine lung cancer diagnosis to personalize treatment decisions, and especially the use of TKIs [9]. The vast amount of genomic information has shed light on high variations in EGFR mutational frequencies, ranging from 7% to 40.1% within the Asian population [10–17]. EGFR mutation frequency was lower in populations not of Asian origin, with a high genetic heterogeneity around the world [18]. Indeed, in Latin populations, either in Spain or in Latin America, the frequency of EGFR mutations in NSCLC has been reported to vary from 16.6% to 37% [19, 20]. In America where Latin-Americans or African-Americans with NSCLC are included in the screening for EGFR, the frequency of EGFR testing is approximately 25% [21–26]. In Europe, except for Spain, the frequency of EGFR mutations is the lowest reported, in approximately 10% of metastatic or advanced lung adenocarcinomas [27–32]. Nonetheless, only limited data on the prevalence of EGFR mutations are available in Caucasian populations, and especially in Europeans. Moreover, there are few data on the mutation rates of targetable oncogenic BRAF, ERBB2, KRAS or PI3KCA mutations across worldwide populations [33–35]. Corresponding information regarding the African population remains deficient except for African-Americans [21–26], although North Africans represent an important proportion of NSCLC patients daily treated in French hospitals.
The aim of our study was to analyze the mutational rate of EGFR, KRAS, BRAF, ERBB2 and PI3KCA in a large cohort of 2,219 unselected French patients presenting in routine clinical practice for first line treatment decision of metastatic or recurrent lung adenocarcinoma.
RESULTS
Characteristics of the French cohort of 2,219 patients with lung adenocarcinoma testing for EGFR mutations
All tumours were adenocarcinomas, and all patients had advanced stage disease. Eighty percent of the samples analysed were biopsies from lung tumors (through transbronchial fibroscopy or percutaneous scan-guided biopsy); the remaining 20% were surgical specimens or biopsies from metastatic lesions, mainly in supradiaphragmatic lymph nodes.
Patients were diagnosed at a median age of 68.3 years (range, 26.6 to 93.7 years; standard deviation, 11.1 years). There was a higher proportion of men (60.6% men vs 39.4% women) and of tobacco-exposed individuals (48.2% current smokers; 21.1% former smokers; 30.7% never smokers), with a median of 20 packs year for the smokers (Table 1).
Table 1. Clinical and demographic features of the whole study sample (N=2, 219).
| Feature | N | % | |
|---|---|---|---|
| Gender | Male | 1,345 | 60.6 |
| Female | 874 | 39.4 | |
| Smoking status | Never | 633 | 28.5 |
| Former | 434 | 19.6 | |
| Current | 993 | 44.7 | |
| Unknown | 159 | 7.2 | |
| Continent | Europe | 1,940 | 87.4 |
| Asia | 39 | 1.8 | |
| Africa | 240 | 10.8 | |
| Age | Median | 68.3 | |
| 1st-3rd quartile | 61.0-76.5 | ||
| Range | 26.6-93.7 | ||
Among the 2,219 consecutive patients that constitute our study population, most of them were Caucasian (1,940 patients; 87.4%); those of African ancestry (240 patients; 10.8%) were more common than those of Asian origin (39 patients; 1.8%).
Mutational spectrum of the study population
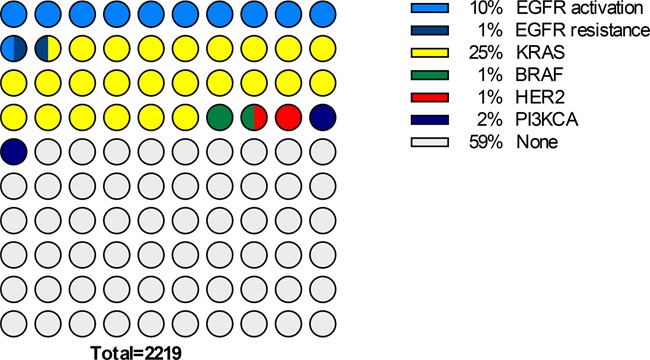
KRAS mutations were altogether the most common genetic abnormalities observed (Figure 1). Indeed, a fourth of the samples was mutated for KRAS exon 2 or 3 (95% Confidence Limits: 23.2%-26.8%), whereas 58.9% of the samples (95% CL: 56.9%-61.0%) did not show any genetic alteration within our panel. The observed prevalence of activating EGFR mutations (activating common and uncommon mutations in exon 18 to 21) was 10.5% (95% CL: 9.2%-11.8%); EGFR resistance mutations in exon 20 were present in only 0.9% of the samples (95% CL: 0.5% - 1.3%). Respective prevalence of BRAF, PI3KCA or ERBB2 alterations was 1.5% (1.0%-2.0%), 2.1% (1.5%-2.7%) and 1.4% (0.9%-1.9%).
Figure 1. Chart representing the proportions of patients with each mutation of the spectrum analysed, within the whole study sample (N=2, 219).
Frequency of EGFR mutations
Activating EGFR mutations have to date the most prominent therapeutic role in the clinical management of NSCLC. The independent predictors of activating EGFR mutations included gender, smoking status, age and ethnic origin (Table 2). Thus, activating EGFR mutations were almost thrice more common in women (17.7%) than in men (6.1%), and in non-smokers (74.5%) than in smokers (current: 26.2%; former: 25.7%). Older patients were also more likely to have a tumor with activating EGFR mutation, with a prevalence per age quartiles increasing from 7.7% in younger patients (< 61 years) to 15.6% in older patients (> 76 years). Patients of Asian descent had the highest prevalence of activating EGFR mutations (36.1%), in comparison to Caucasians (10.2%) or Africans (9.3%). However, the prevalence of mutated EGFR according to gender was more dissimilar in African patients (males: 5.2%; females: 29.3%) than in Caucasians (5.7% in men and 16.8% in women).
Table 2. Independent predictive factors for EGFR activating mutation, KRAS mutation and ERBB2 endoduplication in the whole study sample (multivariate analysis).
| Parameter | HR | 95% CL | p | |
|---|---|---|---|---|
| EGFR activating mutation | ||||
| Continent | <0.0001 | |||
| Europe | 1 | |||
| Asia | 5.65 | 2.67-12.0 | <0.0001 | |
| Africa | 1.27 | 0.77-2.10 | 0.35 | |
| Smoking status | <0.0001 | |||
| Never | 1 | |||
| Former | 0.12 | 0.04-0.32 | <0.0001 | |
| Current | 0.12 | 0.06-0.27 | <0.0001 | |
| Gender | <0.0001 | |||
| Male | 1 | |||
| Female | 3.50 | 2.59-4.71 | ||
| Age | 1.03 | 1.02-1.05 | <0.0001 | |
| KRAS mutation | ||||
| Continent | 0.01 | |||
| Europe | 1 | |||
| Asia | 0.24 | 0.07-0.79 | 0.019 | |
| Africa | 0.59 | 0.41-0.84 | 0.004 | |
| Age | 0.986 | 0.977-0.994 | 0.01 | |
| ERBB2 endoduplication | ||||
| Continent | <0.0001 | |||
| Europe | 1 | |||
| Asia | 9.96 | 2.77-35.8 | <0.0001 | |
| Africa | 4.32 | 1.80-10.4 | 0.001 | |
Subtypes of EGFR mutations
Subtypes of EGFR mutations were overall distributed as follows: L858R was observed in 63 patients (2.8%), del19 in 81 patients (3.7%), T790M and other rare ones in 13 (0.6%) patients each. However, the incidence of each mutation differed according to ethnicity. Thus, the ratio del19/L858R was 1.31 in Europeans, 2.00 in Asians and 0.86 in Africans, respectively. Additionally, the relative frequency of T790M among all EGFR mutations was 5.6% in Europeans, 14.3% in Asians, and 21.1% in Africans.
Frequency (prevalence) of KRAS mutations
KRAS mutations were the most commonly encountered genetic alterations in our study populations, and age and ethnic origin independently predicted for their occurrence (Table 2). Thus, their prevalence dropped from 28.1% in younger patients to 20.3% in older ones. Caucasians had the highest prevalence of KRAS mutations (26.3%), whilst Asians had the lowest (7.7%); Africans displayed an intermediate prevalence (17.2%).
The prevalence of mutated KRAS according to gender was most similar in patients with Caucasian background (males: 25.2% and females: 28.2%), intermediate in Africans (18.8% in men and 11.9% in women) and most dissimilar in Asians (males: 13.0%; females: 0%).
Frequency of BRAF, PI3KCA and ERBB2 mutations
Amidst the least frequent mutations, no independent predictive factor was identified for BRAF mutations, whereas increasing age had a positive predictive value for the occurrence of PI3KCA mutations (HR: 1.048; 95% CL: 1.018 to 1.078; p=0.002). Interestingly, ethnic origin was the only independent predictive factor for ERBB2 mutations (Table 2). Indeed, this genetic alteration occurred in 8.1% of Asian patients, in 3.3% of African ones, and in only 1.0% of those of Caucasian descent.
The prevalence of mutated ERBB2 according to gender was rather similar in patients with Asian (men: 8.7%; women: 7.7%) or Caucasian (0.9% and 1.3%) background, and fairly different in African patients (males: 2.1%; females: 9.3%).
Ethnic demographics
The observation that the mutational phenotype in patients from Africa appeared intermediate between that of Europeans and that of Asians prompted us to explore more in depth the effect of ethnic origin on the mutational phenotype of NSCLC patients treated in France. Hence, we focused on the subgroup of patients from Africa, which is often underrepresented and overlooked in Western studies in NSCLC, despite it accounts for a fairly substantial proportion of patients treated for NSCLC (in our study, 10.8%).
The clinical and demographic features of the 240 patients from Africa are summarized in Table 3. Thus, as compared to the other ethnic subgroups, African patients were more commonly males (81.7% versus 58.1%; p <0.0001) and smokers (never: 14.3% versus 35.1%; p=0.018), whereas no age difference with the rest of the patients was observed (median: 68.9 versus 68.2 years).
Table 3. Clinical and demographic features of the subset from Africa (N=240).
| Feature | N | % | |
|---|---|---|---|
| Gender | Male | 196 | 81.7 |
| Female | 44 | 18.3 | |
| Smoking status | Never | 34 | 14.3 |
| Former | 41 | 17.1 | |
| Current | 165 | 68.6 | |
| Age | Median | 68.9 | |
| 1st-3rd quartile | 59.6-75.1 | ||
| Range | 38.1-92.0 | ||
The mutational spectrum of African patients is shown in Figure 2. This ethnic group displayed a higher prevalence of EGFR resistance mutations (p=0.014), yet not of EGFR activating mutations (p=0.53), a higher prevalence of ERBB2 alterations (p=0.007); a lower prevalence of KRAS (p=0.004) and BRAF (p=0.045) mutations, and a not significant difference in PI3KCA (p=0.14).
Figure 2. Chart representing the mutational spectrum of the subset of patients of African descent (N=240).

However, subgroup analysis according to gender suggested that African women have a peculiar mutational spectrum, with an independently higher risk of presenting EGFR activating (HR: 2.05; 95% CL: 1.02-4.12; p=0.044) and EGFR resistance mutations (HR: 4.16; 95% CL: 1.17-14.77; p=0.028). Additionally, African women displayed a notably higher risk of having cancers harbouring ERBB2 mutations (HR: 8.01; 95% CL: 2.41-26.68; p=0.001), and a lower risk of KRAS mutations (HR: 0.34; 95% CL: 0.13-0.89; p=0.027).
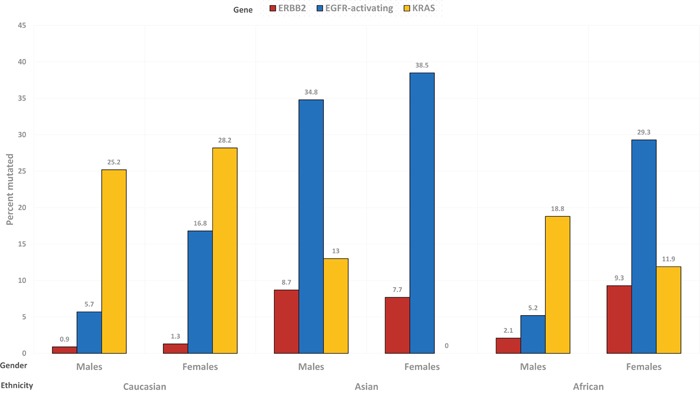
The observed prevalence of the main mutations, in man and women from the three ethnic groups is depicted in Figure 3.
Figure 3. Prevalence of ERBB2 (red), EGFR-activating (blue) and KRAS (yellow) mutations, in the subgroups determined by gender and ethnicity.
DISCUSSION
We examined 2,219 unselected patients treated in hospitals in Paris and suburbs (France) for metastatic or recurrent lung adenocarcinoma, who were systematically tested for EGFR and other targetable driver mutations in the daily clinical setting. We found, similarly to other groups, an overall rate of activating EGFR mutations of 10.5% [18, 32]. KRAS was the most prevalent mutation with 25.0% of positive cases, whereas BRAF, ERBB2 and PI3KCA mutations were relatively uncommon, with 1.5, 1.4, and 2.1% mutational rates, respectively. Gender, smoking habits, but also ethnicity, alone and combined, appeared to influence these figures. Indeed, in the subpopulation of migrants from Africa (1st or 2nd generations), the molecular phenotype appeared peculiar, with prominent gender-specific differences. For example, EGFR activating and resistance mutation rates were as high as 29.3% and 26.7% in African women, respectively, whilst respective rates being as low as 5.2% and 7.1% in African men. Similarly, and very interestingly, ERBB2 was mutated in 9.3% of African women, and in only 2.1% of men from Africa. Overall, rates of PI3KCA mutations were more frequent in females than in males from Africa, although not to a significant extent. These data could suggest that African women have a more genetically instable phenotype, even though KRAS was not more frequently mutated in women than in men from Africa. However, KRAS was altogether less frequently mutated in patients from Africa (17%) than in the global population (25%).
In Europe, the first reports of lung-cancer–specific EGFR mutations were described in Italy, and showed EGFR mutations in 10% of 375 lung adenocarcinomas [27], whereas in Spain it was described a rate of EGFR mutations of 16.6% [19]. Since the development of systematic EGFR mutational status testing in metastatic adenocarcinomas to identify patients who can benefit from first line TKIs, several reports have shown that the EGFR mutations prevalence is approximately 10% in Northern European populations [27–32]. In North America, several recent reports on the EGFR testing show a generally higher prevalence (approximately 25%), but most of them include a variety of heterogeneous racial groups and smoking habits [8, 26]. Similarly, it has been reported that the prevalence of EGFR mutations is 2 to 3-fold higher in Asians, with differences between Eastern and Western regions observed [10–17]. In the French population studied here, the subgroup from Asia was relatively underrepresented. However, in this ethnic subgroup counting 39 patients, we observed the expected higher EGFR mutational rate as compared to that in Caucasians, with a prevalence of 36.1%. Similarly, we observed, as already previously reported, that women and non-smokers exhibited more frequently EGFR activating mutations.
Thus, the analysis per ethnicity in our French unselected population showed similar EGFR mutational rates between Africans and Caucasians, despite an almost halved proportion of never smokers and a double proportion of males in our subgroup of patients of African descent.
Contrariwise, the KRAS mutational rate of Africans (17.2%) was intermediate between the highest one in Caucasians (26.3%) and the lowest one in Asians (7.7%). These results corroborate previous results showing that KRAS mutations are less common in Asians compared to Caucasians [6, 36, 37].
Similarly, the prevalence of ERBB2 mutation in patients of African origin (3.3%) was intermediate between that observed in Asian (8.1%) and Caucasian (1.0%) patients.
However, although the global prevalence of ERBB2 mutations was 1.4%, figure consistent with recent publications [38–41], the mutation rate of 3.3% observed here in the African population reaches an interesting threshold to be systematically screened for, in this selected subgroup of patients. This is even more clinically relevant since recent studies demonstrated a significant therapeutic benefit of trastuzumab (an anti-ERBB2 mAb) and of newer TKIs, such as afatinib and neratinib, which target both EGFR and ERBB2, in selected patients with lung adenocarcinoma [38, 42–46].
The prevalence of EGFR mutation has been recently reported in African populations. Errihani et al. [47] have studied a Moroccan population of 137 patients with an overall frequency of the EGFR mutations of 21%. However, they reported a high incidence of EGFR mutations with an unusual incidence of exon 21 L858R/exon 19 frameshift base pair deletions or exon 20 mutations. Nonetheless, they tested tumors on the French continent in a single laboratory, and did not mention to be from consecutive patients. We could therefore suspect a biased selection of patients presenting in Moroccan university hospitals who have then benefited abroad from oncogenetic testing. Other reports in American studies analysing African Americans subpopulations show a lower frequency of somatic driver mutations than in Caucasians [21]. We observed, nevertheless, that patients of African descent have a relatively higher incidence of T790M mutations (more than 4-fold), and a shift towards more frequent L858R rather than del19 (1.5-fold), in comparison with European patients.
Interestingly, some studies in Latin America populations report higher incidence of EGFR mutations than that found in Europe. A recent study shows an overall frequency of EGFR mutations of 26% in Latin America (from 14.4% in Argentina to 31% in Costa Rica) [48]. Interestingly, in Latin America, the frequency of KRAS mutations was as low as 14%. These results corroborate the importance of racial differences in the oncogenic driver gene mutations and could explain the higher incidence of EGFR mutations in Spain than in other European countries such as France or Germany. In a recent report, different EGFR and KRAS mutational ratio across South America was reported, and, allegedly, the migration of population through the Bering Strait could account for this observation [49].
Such migrations could explain the variations in the incidence of driver oncogene mutations across populations, and shed light on the importance of ethnic background to favour some genetic testing in subpopulations with the aim of increasing the number of patients who could benefit from targeted therapies.
Ethnic minorities in France, similarly to the rest of the European Community, tend to have immigrated mostly from Africa, rather than from Asia, contrarily to what observed in North America or Oceania. As a consequence, our unselected consecutive population included a higher proportion of Africans (10.8%) than of Asians (1.8%). Furthermore, smoking habits, especially in women, are probably different between ethnic groups, with 1st or 2nd generation North-Africans females being unlikely smokers. This social context could influence the mutational spectra of NSCLC diagnosed in French metropolitan areas in different ethnic subgroups and genders.
Our study, albeit large, has several limitations. Firstly, the assignment of ethnicity in this study was potentially prone to contain mistakes, yet it was based on the objective evaluation of birth places of the patient and of his/her parents. Indeed, French legislation forbids to record the ethnic origin of the patients. Hence, we were compelled to assume the ethnicity of each patient by considering the Country of birth. However, in the globalized world context, we fully acknowledge that the birthplace does not necessarily guarantee the ethnicity of the person. Nevertheless, by estimating the ethnic origin of each patient as that of the largest proportion of the population in the Country of birth, we can presume to have minimised the mismatches, albeit doubtlessly present, in our study. Moreover, our study population included a relatively low proportion of non-white Europeans, despite the multicultural milieu observed in the greater Paris region nowadays. In particular, given the relatively small number of patients of Asian descent in our study, the estimation of the frequency of rare mutations across subgroups could be rather imprecise. The rather low proportion of non-Europeans could be due to the age distribution, with several retired immigrants having decided to move back to their home Countries, while the younger ones remaining in France have not yet developed lung cancer. Furthermore, given the real-life approach of our study, we gathered data from several pathology centres, and some missing data could have occurred, especially concerning smoking habits. Nevertheless, our heterogeneous population encompasses all consecutive patients we could retrieve, without any selection except histology and exploitable tissue, thus reducing possible bias. Moreover, all our samples were analyzed by the same molecular platform within a relatively short time span, thus minimizing technical differences related to equipment or manpower.
In conclusion, we show interesting and clinically relevant variations of activating somatic mutations that are targetable in subgroups of patients according to the gene and patient's birthplace. In particular, African women exhibit molecular profiles in their tumor closer to those of Asian than Caucasian women, and they could dramatically benefit from anti-EGFR and anti-ERBB2 targeted therapies.
PATIENTS AND METHODS
This study was conducted in France during daily clinical practice for somatic EGFR mutations research and involved clinicians, pathologists and biologists. Clinical and biological dataavailable in patients’ medical records werecollected by the physician in charge of the patient. All the data were centralized in a database and made completely anonymous. The database has been declared to the CNIL (Commission nationale de l'informatique et des libertés; French dataprotection authority).
The study was conducted on a total of 2,240 consecutive patients with newly-diagnosed, histologically-proven metastatic or recurrent NSCLC and tested for EGFR mutational status in the routine clinical setting using the same somatic genetic testing strategy, between November 2011 and December 2013.
Mutational testing was performed at a single platform with expertise in both molecular and pathological diagnostics. The platform was ISO 15189 (for screening and identification of mutations in EGFR, KRAS and BRAF) and INCa (French National Institute of Cancer) certified. ALK or ROS1 translocations were not included in the analysis.
Age, gender and birthplace, collected by physicians at the first visit, were mentioned in the medical record of each patient. When birthplace was not specified in the medical record, it was found out from the personal identity number at the French Health care system (Sécurité sociale). For African or Asian descendants who were born in France, parents’ birthplace was considered when mentioned in the medical record. If this information was not available, patients were excluded from the study. Thus, full data were available for a total of 2,219 patients (out of 2,240), which were included in the analysis.
Patients were classified according tosSmoking status as never smokers, former smokers (≥ 5 years of quitting) or active smokers (including recent quitters, light or heavy smokers, with a cut off at 10 pack/years) and the number of pack/years of smoking (one pack/year is defined as 20 cigarettes per day during 1 year) was retrieved [50, 51].
Histology was assessed by specialist lung cancer pathologists using the WHO criteria, and adenocarcinoma described according to the International Association for the Study of Lung Cancer classification [52]. Histological type and grading was defined using haematoxylin and eosin (HE)-stained biopsies after serial sectioning. Immunohistochemistry used the recommended panel of antibodies for classification of NSCLC (TTF1, CK7, CK5/6 and/or p63). Clinico-pathological stage was assigned according to the 7th edition of the tumor-node-metastasis classification [53]. Squamous cell carcinoma samples were excluded from the study.
Mutational status analysis was performed using formalin-fixed and paraffin-embedded tumours containing more than 10% of cancer cells. DNA was extracted after overnight paraffin digestion at 56°C. All the known activation and resistance gene abnormalities in EGFR exons 18 to 21, KRAS exons 2 and 3, BRAF exons 11 and 15, ERBB2 exon 20 and PI3KCA exons 9 and 20 were screened. Genetic abnormalities were analysed using a combination of technologies. The screening of these gene abnormalities was performed for each exon using a High Resolution Melting (HRM) technology (Lightcycler 480, Roche), together with a concomitant search by allelic discrimination for the 7 most frequent gene abnormalities in KRAS exons 2 and 3; EGFR (L858R) and BRAF (V600E). Base pairs deletion in exon 19 of EGFR were analysed by fragment analysis (ABI 310, Applied). When gene abnormalities, especially uncommon mutations, were not observed by HRM, but only using the other techniques of identification, a Sanger sequencing was performed.
All sequence analyses were checked for plausibility. All techniques are broadly accepted for routine detection of EGFR mutational status. The laboratory of molecular pathology successfully completed the round robin test of the French quality initiative certified Societies [54]. It also got the ISO 15189 certification in 2013 for the screening and the identification of the EGFR, KRAS and BRAF mutations.
Statistical analysis
We used percentages for qualitative variables and mean and standard deviations for quantitative variables. Multivariate binary logistic regression was used to determine the independent predictive factors for the occurrence of specific mutations. In case of missing data (especially for smoking status), cases were excluded from the model. Differences in clinical characteristics according to ethnic origin were explored using Student's t or chi-squared tests. Significance was determined by a P-value < 0.05. All statistics were performed using SPSS software (SPSS version 17.0 for Windows, IBM Inc., Chicago, IL, USA).
Acknowledgments
This work was presented, in part, at the 15th World Conference on Lung Cancer, October 27th-30th 2013, Sydney, New South Wales, Australia (Oral Presentation: MO10.03: Decreased KRAS and increased HER2 OR PI3K mutations prevalence in the French emigrant population from African continent in lung adenocarcinoma metastatic cancer. Mohamed Bouchahda et al.).
Abbreviations
- NSCLC
non-small cell lung cancer
- EGFR
epidermal growth factor receptor
- ERBB2
human epidermal growth factor receptor 2 (avian erythroblastosis oncogene B2 homolog)
- KRAS
Kristen rat sarcoma oncogene homolog
- BRAF
murine sarcoma viral oncogene B homolog
- PI3KCA
phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha.
Author contributions
RS, NB and JH conducted the genotyping work, under the direction of AL, on tissues prepared by SM, SDD, AB, PV and CG's teams. AL assured also the funding of the study. The project was initially developed by MB and AL. JFM, PFI, JT, and MB were the main clinicians involved in the referral of patients for mutations assessment. The manuscript was mainly drafted by PFI and AL, and all co-authors approved its final version.
CONFLICTS OF INTEREST
All co-authors have no conflicts of interest to disclose.
FUNDING
There was no specific funding for this study; however, the molecular biology platform was supported in part by the French National Cancer Institute (INCa), by Paris South University (Université Paris Sud), and by Paris Public Hospital System (AP-HP).
REFERENCES
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Fang R, Sun Y, Han X, Li F, Gao B, Iafrate AJ, Liu XY, Pao W, Chen H, Ji H. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One. 2011;6:e28204. doi: 10.1371/journal.pone.0028204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HR, Shim HS, Chung JH, Lee YJ, Hong YK, Rha SY, Kim SH, Ha SJ, Kim SK, Chung KY, Soo R, Kim JH, Cho BC. Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer. 2012;118:729–739. doi: 10.1002/cncr.26311. [DOI] [PubMed] [Google Scholar]
- 7.Ren S, Kuang P, Zheng L, Su C, Li J, Li B, Chen X, Wang Y, KimCurran V, Liu L, Hu Q, Zhang J, Tang L, Zhou C. Analysis of driver mutations in female non-smoker Asian patients with pulmonary adenocarcinoma. Cell Biochem Biophys. 2012;64:155–160. doi: 10.1007/s12013-012-9384-8. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi N, Vanderlaan PA, Folch E, Boucher DH, Canepa HM, Kent MS, Gangadharan SP, Majid A, Kocher ON, Goldstein MA, Huberman MS, Costa DB. Smoking status and self-reported race affect the frequency of clinically relevant oncogenic alterations in non-small-cell lung cancers at a United States-based academic medical practice. Lung Cancer. 2013;82:31–37. doi: 10.1016/j.lungcan.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shtivelman E, Hensing T, Simon GR, Dennis PA, Otterson GA, Bueno R, Salgia R. Molecular pathways and therapeutic targets in lung cancer. Oncotarget. 2014;5:1392–1433. doi: 10.18632/oncotarget.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 11.Sonobe M, Manabe T, Wada H, Tanaka F. Mutations in the epidermal growth factor receptor gene are linked to smoking-independent, lung adenocarcinoma. Br J Cancer. 2005;93:355–363. doi: 10.1038/sj.bjc.6602707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, Ichimura K, Tsuda T, Yano M, Tsukuda K, Tabata M, Ueoka H, Tanimoto M, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- 13.Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, Chen YT, Lin YF, Chang WC, Kuo HP, Wu YC, Chen YR, Tsai SF. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res. 2004;10:8195–8203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- 14.Soung YH, Lee JW, Kim SY, Seo SH, Park WS, Nam SW, Song SY, Han JH, Park CK, Lee JY, Yoo NJ, Lee SH. Mutational analysis of EGFR and K-RAS genes in lung adenocarcinomas. Virchows Arch. 2005;446:483–488. doi: 10.1007/s00428-005-1254-y. [DOI] [PubMed] [Google Scholar]
- 15.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth JA, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 16.Sugio K, Uramoto H, Ono K, Oyama T, Hanagiri T, Sugaya M, Ichiki Y, So T, Nakata S, Morita M, Yasumoto K. Mutations within the tyrosine kinase domain of EGFR gene specifically occur in lung adenocarcinoma patients with a low exposure of tobacco smoking. Br J Cancer. 2006;94:896–903. doi: 10.1038/sj.bjc.6603040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Telbany A, Ma PC. Cancer genes in lung cancer: racial disparities: are there any? GenesCancer. 2012;3:467–480. doi: 10.1177/1947601912465177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang YL, Yuan JQ, Wang KF, Fu XH, Han XR, Threapleton D, Yang ZY, Mao C, Tang JL. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. 2016;7:78985–78993. doi: 10.18632/oncotarget.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 20.Arrieta O, Cardona AF, Federico Bramuglia G, Gallo A, Campos-Parra AD, Serrano S, Castro M, Aviles A, Amorin E, Kirchuk R, Cuello M, Borbolla J, Riemersma O, et al. Genotyping non-small cell lung cancer (NSCLC) in Latin America. J Thorac Oncol. 2011;6:1955–1959. doi: 10.1097/JTO.0b013e31822f655f. [DOI] [PubMed] [Google Scholar]
- 21.Cote ML, Haddad R, Edwards DJ, Atikukke G, Gadgeel S, Soubani AO, Lonardo F, Bepler G, Schwartz AG, Ethier SP. Frequency and type of epidermal growth factor receptor mutations in African Americans with non-small cell lung cancer. J Thorac Oncol. 2011;6:627–630. doi: 10.1097/JTO.0b013e31820a0ec0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leidner RS, Fu P, Clifford B, Hamdan A, Jin C, Eisenberg R, Boggon TJ, Skokan M, Franklin WA, Cappuzzo F, Hirsch FR, Varella-Garcia M, Halmos B. Genetic abnormalities of the EGFR pathway in African American Patients with non-small-cell lung cancer. J Clin Oncol. 2009;27:5620–5626. doi: 10.1200/JCO.2009.23.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinersman JM, Johnson ML, Riely GJ, Chitale DA, Nicastri AD, Soff GA, Schwartz AG, Sima CS, Ayalew G, Lau C, Zakowski MF, Rusch VW, Ladanyi M, Kris MG. Frequency of EGFR and KRAS mutations in lung adenocarcinomas in African Americans. J Thorac Oncol. 2011;6:28–31. doi: 10.1097/JTO.0b013e3181fb4fe2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang SH, Mechanic LE, Yang P, Landi MT, Bowman ED, Wampfler J, Meerzaman D, Hong KM, Mann F, Dracheva T, Fukuoka J, Travis W, Caporaso NE, et al. Mutations in the tyrosine kinase domain of the epidermal growth factor receptor in non-small cell lung cancer. Clin Cancer Res. 2005;11:2106–2110. doi: 10.1158/1078-0432.CCR-04-1853. [DOI] [PubMed] [Google Scholar]
- 25.Steuer CE, Behera M, Berry L, Kim S, Rossi M, Sica G, Owonikoko TK, Johnson BE, Kris MG, Bunn PA, Khuri FR, Garon EB, Ramalingam SS. Role of race in oncogenic driver prevalence and outcomes in lung adenocarcinoma: results from the Lung Cancer Mutation Consortium. Cancer. 2016;122:766–772. doi: 10.1002/cncr.29812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araujo LH, Lammers PE, Matthews-Smith V, Eisenberg R, Gonzalez A, Schwartz AG, Timmers C, Shilo K, Zhao W, Natarajan TG, Zhang J, Yilmaz AS, Liu T, et al. Somatic mutation spectrum of non-small-cell lung cancer in African Americans: a pooled analysis. J Thorac Oncol. 2015;10:1430–1436. doi: 10.1097/JTO.0000000000000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 28.Helland A, Skaug HM, Kleinberg L, Iversen ML, Rud AK, Fleischer T, Sagerup C, Solberg S, Jorgensen L, Ariansen S, Brustugun OT. EGFR gene alterations in a Norwegian cohort of lung cancer patients selected for surgery. J Thorac Oncol. 2011;6:947–950. doi: 10.1097/JTO.0b013e31820db209. [DOI] [PubMed] [Google Scholar]
- 29.Ludovini V, Bianconi F, Pistola L, Chiari R, Minotti V, Colella R, Giuffrida D, Tofanetti FR, Siggillino A, Flacco A, Baldelli E, Iacono D, Mameli MG, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2011;6:707–715. doi: 10.1097/JTO.0b013e31820a3a6b. [DOI] [PubMed] [Google Scholar]
- 30.Smits AJ, Kummer JA, Hinrichs JW, Herder GJ, Scheidel-Jacobse KC, Jiwa NM, Ruijter TE, Nooijen PT, Looijen-Salamon MG, Ligtenberg MJ, Thunnissen FB, Heideman DA, de Weger RA, Vink A. EGFR and KRAS mutations in lung carcinomas in the Dutch population: increased EGFR mutation frequency in malignant pleural effusion of lung adenocarcinoma. Cell Oncol (Dordr) 2012;35:189–196. doi: 10.1007/s13402-012-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gahr S, Stoehr R, Geissinger E, Ficker JH, Brueckl WM, Gschwendtner A, Gattenloehner S, Fuchs FS, Schulz C, Rieker RJ, Hartmann A, Ruemmele P, Dietmaier W. EGFR mutational status in a large series of Caucasian European NSCLC patients: data from daily practice. Br J Cancer. 2013;109:1821–1828. doi: 10.1038/bjc.2013.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Lena H, Ouafik L, Besse B, Rouquette I, Westeel V, Escande F, Monnet I, Lemoine A, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT) Lancet. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 33.Lim SM, Kim EY, Kim HR, Ali SM, Greenbowe JR, Shim HS, Chang H, Lim S, Paik S, Cho BC. Genomic profiling of lung adenocarcinoma patients reveals therapeutic targets and confers clinical benefit when standard molecular testing is negative. Oncotarget. 2016;7:24172–24178. doi: 10.18632/oncotarget.8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vigneswaran J, Tan YH, Murgu SD, Won BM, Patton KA, Villaflor VM, Hoffman PC, Hensing T, Hogarth DK, Malik R, MacMahon H, Mueller J, Simon CA, et al. Comprehensive genetic testing identifies targetable genomic alterations in most patients with non-small cell lung cancer, specifically adenocarcinoma, single institute investigation. Oncotarget. 2016;7:18876–18886. doi: 10.18632/oncotarget.7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, Zhang Y, Pan Y, Li Y, Hu H, Cai D, Li H, Ye T, Luo X, Zhang Y, Li B, Shen L, Sun Y, Chen H. Comprehensive investigation of oncogenic driver mutations in Chinese non-small cell lung cancer patients. Oncotarget. 2015;6:34300–34308. doi: 10.18632/oncotarget.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, Nafa K, Riedel ER, Hsu M, Pao W, Miller VA, Ladanyi M. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y, Ren Y, Fang Z, Li C, Fang R, Gao B, Han X, Tian W, Pao W, Chen H, Ji H. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol. 2010;28:4616–4620. doi: 10.1200/JCO.2010.29.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazieres J, Peters S, Lepage B, Cortot AB, Barlesi F, Beau-Faller M, Besse B, Blons H, Mansuet-Lupo A, Urban T, Moro-Sibilot D, Dansin E, Chouaid C, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31:1997–2003. doi: 10.1200/JCO.2012.45.6095. [DOI] [PubMed] [Google Scholar]
- 39.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 40.Reis H, Herold T, Ting S, Worm K, Huber U, Christoph DC, Eberhardt WE, Kostbade K, Kasper S, Stamatis G, Welter S, Darwiche K, Karpf-Wissel R, et al. HER2 expression and markers of phosphoinositide-3-kinase pathway activation define a favorable subgroup of metastatic pulmonary adenocarcinomas. Lung Cancer. 2015;88:34–41. doi: 10.1016/j.lungcan.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su PF, Shyr Y, Camidge DR, Sequist LV, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazieres J, Barlesi F, Filleron T, Besse B, Monnet I, Beau-Faller M, Peters S, Dansin E, Fruh M, Pless M, Rosell R, Wislez M, Fournel P, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: results from the European EUHER2 cohort. Ann Oncol. 2016;27:281–286. doi: 10.1093/annonc/mdv573. [DOI] [PubMed] [Google Scholar]
- 43.Mar N, Vredenburgh JJ, Wasser JS. Targeting HER2 in the treatment of non-small cell lung cancer. Lung Cancer. 2015;87:220–225. doi: 10.1016/j.lungcan.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 44.Song Z, Yu X, Shi Z, Zhao J, Zhang Y. HER2 mutations in Chinese patients with non-small cell lung cancer. Oncotarget. 2016;7:78152–78158. doi: 10.18632/oncotarget.11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricciardi GR, Russo A, Franchina T, Ferraro G, Zanghi M, Picone A, Scimone A, Adamo V. NSCLC and HER2: between lights and shadows. J Thorac Oncol. 2014;9:1750–1762. doi: 10.1097/JTO.0000000000000379. [DOI] [PubMed] [Google Scholar]
- 46.Riely GJ. Second-generation epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. J Thorac Oncol. 2008;3:S146–S149. doi: 10.1097/JTO.0b013e318174e96e. [DOI] [PubMed] [Google Scholar]
- 47.Errihani H, Inrhaoun H, Boukir A, Kettani F, Gamra L, Mestari A, Jabri L, Bensouda Y, Mrabti H, Elghissassi I. Frequency and type of epidermal growth factor receptor mutations in moroccan patients with lung adenocarcinoma. J Thorac Oncol. 2013;8:1212–1214. doi: 10.1097/JTO.0b013e31829f6b4a. [DOI] [PubMed] [Google Scholar]
- 48.Arrieta O, Ramirez-Tirado LA, Baez-Saldana R, Pena-Curiel O, Soca-Chafre G, Macedo-Perez EO. Different mutation profiles and clinical characteristics among Hispanic patients with non-small cell lung cancer could explain the “Hispanic paradox”. Lung Cancer. 2015;90:161–166. doi: 10.1016/j.lungcan.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Arrieta O, Cardona AF, Martin C, Mas-Lopez L, Corrales-Rodriguez L, Bramuglia G, Castillo-Fernandez O, Meyerson M, Amieva-Rivera E, Campos-Parra AD, Carranza H, Gomez de la Torre JC, Powazniak Y, et al. Updated frequency of EGFR and KRAS mutations in nonsmall-cell lung cancer in Latin America: the Latin-American Consortium for the investigation of lung cancer (CLICaP) J Thorac Oncol. 2015;10:838–843. doi: 10.1097/JTO.0000000000000481. [DOI] [PubMed] [Google Scholar]
- 50.Tammemagi MC, Lam S. Screening for lung cancer using low dose computed tomography. BMJ. 2014;348:g2253. doi: 10.1136/bmj.g2253. [DOI] [PubMed] [Google Scholar]
- 51.Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R. Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ. 2000;321:323–329. doi: 10.1136/bmj.321.7257.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, L; Sobin. International Association for the Study of Lung Cancer International Staging Committee; Participating Institutions. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 54.Dequeker EM, Keppens C, Egele C, Delen S, Lamy A, Lemoine A, Sabourin JC, Andrieu C, Ligtenberg M, Fetique D, Tops B, Descarpentries C, Blons H, et al. Three rounds of external quality assessment in France to evaluate the performance of 28 platforms for multiparametric molecular testing in metastatic colorectal and non-small cell lung cancer. J Mol Diagn. 2016;18:205–214. doi: 10.1016/j.jmoldx.2015.09.004. [DOI] [PubMed] [Google Scholar]


