Figure 1. Identify the canonical residues supported CDR loop conformation and residues involved in contact with the antigen.
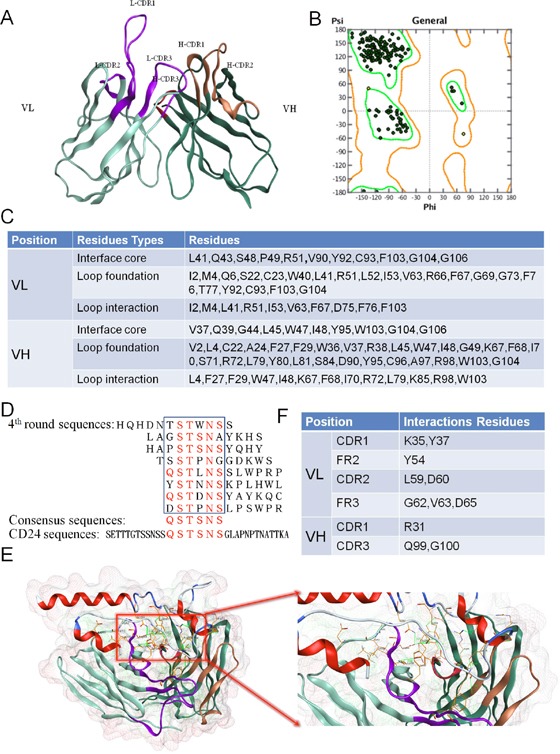
(A) 3-D structure of G7mAb Fv. MOE was be used to build the 3-D structure model of G7mAb Fv based on various antibody structures templates. (B) Fv structure evaluation with Ramachandran plot. Residues are all in the rendered regions. (C) Canonical residues in FRs. According to the 3-D structure, we identified three types of the canonical residues which were important for CDR conformation maintaining. (D) Epitope mapping of cG7. A dodecapeptide phage display library was screened against cG7. The consensus residues between the positive clones were QSTSNS, which were found in CD24 (residues 13-18). (E) Molecular docking of G7mAb Fv and CD24 based on epitope mapping. (F) Interactive residues in Fv. According to the complex structure, we identified the residues involved in contact with the antigen.
