SUMMARY
The past 17 years have been marked by a revolution in our understanding of cellular multisubunit DNA-dependent RNA polymerases (MSDDRPs) at the structural level. A parallel development over the past 15 years has been the emerging story of the giant viruses, which encode MSDDRPs. Here we link the two in an attempt to understand the specialization of multisubunit RNA polymerases in the domain of life encompassing the large nucleocytoplasmic DNA viruses (NCLDV), a superclade that includes the giant viruses and the biochemically well-characterized poxvirus vaccinia virus. The first half of this review surveys the recently determined structural biology of cellular RNA polymerases for a microbiology readership. The second half discusses a reannotation of MSDDRP subunits from NCLDV families and the apparent specialization of these enzymes by virus family and by subunit with regard to subunit or domain loss, subunit dissociability, endogenous control of polymerase arrest, and the elimination/customization of regulatory interactions that would confer higher-order cellular control. Some themes are apparent in linking subunit function to structure in the viral world: as with cellular RNA polymerases I and III and unlike cellular RNA polymerase II, the viral enzymes seem to opt for speed and processivity and seem to have eliminated domains associated with higher-order regulation. The adoption/loss of viral RNA polymerase proofreading functions may have played a part in matching intrinsic mutability to genome size.
KEYWORDS: giant virus, mimivirus, NCLDV, phycodnavirus, poxvirus, RNA polymerase, vaccinia virus
INTRODUCTION
The multisubunit DNA-directed/dependent RNA polymerases (MSDDRPs) lie at the heart of the central dogma, are key enzymes of living systems, and are universally found in cellular organisms. While eubacteria and archaea possess a single such enzyme, eukaryotes possess at least three, namely, RNA polymerase I (pol I), pol II, and pol III, which specialize in the synthesis of rRNA, mRNA, and a collection of smaller RNAs such as tRNA and 5S rRNA, respectively. MSDDRPs are also encoded by DNA viruses that have a cytoplasmic transcriptional phase, such as the poxviruses. In this review, we address the viral MSDDRPs in the context of many recent advances in the structural and biochemical understanding of their cellular counterparts, along with the growing numbers of giant viruses encoding RNA polymerase subunits.
CELLULAR MSDDRPs
To paraphrase Dobzhansky, very little in transcription makes mechanistic sense except in the light of structural biology. Over the past 18 years, and ongoing, impressive efforts in X-ray crystallography, transitioning to cryo-electron microscopy (cryo-EM), have provided deep insights into the structural biology of the cellular MSDDRPs, including their architecture, evolution, structure-function relationships, and dynamics. Growing numbers of such studies have covered the enzymes from bacteria (1, 2), archaeal species (3–5), and eukaryotes (6–35), in various interaction states.
Functionally, all MSDDRPs share a set of common features, namely, the copying of a DNA template to newly synthesized RNA, stepwise enzyme translocation on the template during RNA synthesis, utilization of nucleoside triphosphate (NTP) substrates, pairing of the incoming nucleotide with the DNA template strand via Watson-Crick base pairing, and catalysis of nucleotide transfer via a metal-dependent mechanism. Architecturally, all MSDDRPs comprise two large subunits and a collection of smaller ones, in which the two large subunits have remained remarkably conserved across all domains of life, namely, the three cellular domains and the nucleocytoplasmic large DNA viruses (NCLDV) (36). Structural and biochemical studies have focused largely on two prototypical MSDDRPs, the bacterial enzyme and pol II from the budding yeast Saccharomyces cerevisiae (37, 38). While bacterial MSDDRP has just 4 distinct subunits, named α (two copies), β, β′, and ω, S. cerevisiae pol II has either 10 or 12. While the 10-subunit core enzyme comprising subunits Rpb1, -2, -3, -5, -6, and -8 to -12 is competent in transcription elongation, two additional, dissociable subunits, the Rpb4/7 heterodimer, are conditionally required for transcription initiation (39). Other eukaryotes always have the 12-subunit form. Of the two prototypical enzymes, the viral MSDDRPs more closely resemble the one from S. cerevisiae, so this will be described initially.
Structure-Based Models of Nonbacterial Transcription: Yeast pol II
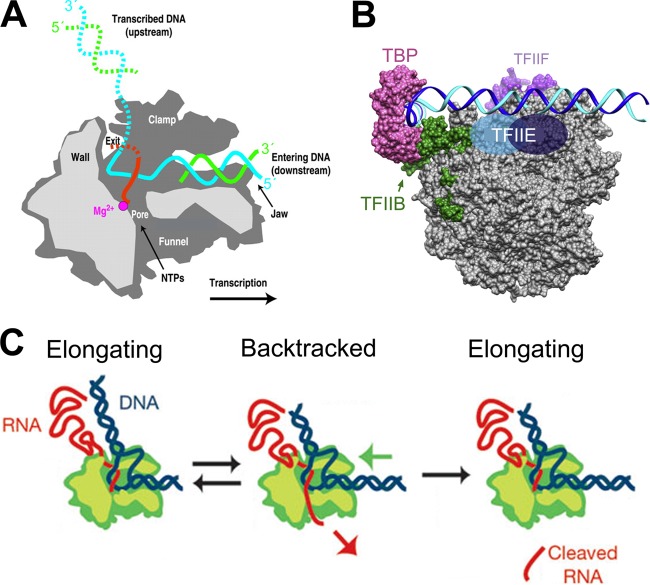
The earliest X-ray crystallographic structures of yeast pol II (6) showed the core (10-subunit) enzyme with an architecture comprising the two large subunits, RPB1 and -2, flanking a cleft that was deep and wide enough to accommodate a double-stranded DNA helix and which contained the catalytic center on its floor. An opening, or “pore,” in the floor immediately beneath the catalytic center exposed the DNA-RNA hybrid to an inverted funnel-shaped cavity on the outside of the enzyme, allowing incoming NTPs accesses to the active site (Fig. 1A). One end of the cleft opened at the downstream face of the polymerase (the “front” of the enzyme during its translocation along duplex DNA), while the other end was blocked by a “wall” structure positioned just beyond the catalytic center but prior to the upstream face of the polymerase (facing the promoter [Fig. 1A]). One side of the cleft formed a clamp which in the open state is wide enough for double-stranded DNA to enter but in the closed state is only wide enough for a single DNA strand (40) (Fig. 1A). Close to the downstream face of the enzyme, just within the cleft, structural features termed “jaws” were present, composed of yeast subunits Rpb5 (lower jaw) and Rpb1/9 (upper jaw). At the upstream face of the enzyme could be found a subassembly of four relatively small subunits (Rpb3, -10, -11, and -12), with the Rpb3/11 subunits corresponding, approximately, to subunits α/α′ in the bacterial MSDDRP. The above description accounts for 8 of the 10 core subunits. Additionally, Rpb6 formed a clamp across the cleft, while Rpb8 was located near to the Rpb3-10-11-12 subassembly (6). In the 12-subunit pol II holoenzyme, the Rpb4/7 heterodimer formed a “stalk” structure toward the enzyme's upstream face (9, 19).
FIG 1.
(A) Cutaway section schematic of a pol II transcribing complex (polymerase moving left to right). From the direction of view (“side” of pol II), the cutaway plane exposes nucleic acids and functional elements of the enzyme. Light gray, cut surfaces of the protein (front). Dark gray, receding surfaces. Right side, template and nontemplate strands (cyan and green, respectively) of the entering DNA duplex (the unwound portion of the nontemplate strand is not shown). Red, 3′ end of nascent RNA within the DNA-RNA hybrid minihelix. Magenta, catalytic metal. Of the two jaws, the cutaway reveals only the lower one. The far wall of the DNA-binding cleft forms the clamp structure (6, 18). For other details, see the text. (Adapted from reference 18 with permission [copyright 2002 National Academy of Sciences].) (B) Side view (as in panel A) of the surface-rendered (not cutaway) basal preinitiation complex showing pol II (gray), TBP (dark pink) and TFIIB (green) (131), TFIIF (purple), and the position of TFIIE (from cross-linking studies [132]) (blue). The closed-promoter DNA duplex encompassing the transcriptional start site is modeled, suspended above the pol II cleft via general transcription factors TBP, TFIIB, and TFIIE, in which TBP and TFIIB hold the upstream promoter DNA (100). (Adapted from reference 100 with permission from Elsevier.) (C) Backtracking. In the left and center panels, RNA polymerase can move forward (left) or backtrack with extrusion of the nascent transcript's 3′ end through the NTP entry pore leading to an arrested state (center). Transcript 3′ end cleavage (right) restores an elongation-competent complex. (Adapted from reference 133 by permission from Macmillan Publishers Ltd.)
As might be anticipated, pol I and pol III are closely related variants of pol II: five of the 10 subunits of yeast core pol II are identical across all three forms of eukaryotic MSDDRP, while another five are highly conserved between the three enzymes.
RNA Polymerase Function in the Context of Structure
Transcriptional elongation can be regarded as a continuous, dynamic “production line” in which the template enters the polymerase cleft at the downstream face and becomes unwound, with the template strand passing over the catalytic center where the 3′ end of a complementary nascent RNA transcript comprises the downstream terminus of a 7- to 8-base-pair DNA-RNA hybrid minihelix (Fig. 1A). This nascent transcript becomes extended with a complementary incoming ribonucleotide. At the upstream end of the minihelix, the nascent RNA is peeled away from the DNA template strand and channeled away from the polymerase (22). The template and nontemplate DNA strands are then free to reanneal and fully exit the enzyme assembly in an upstream direction.
A model for preinitiation complex formation has been developed in a minimal system comprising polymerase, DNA template, and the two general transcription factors (GTFs) TATA-binding protein (TBP) and TFIIB (13, 27–29). Here, promoter DNA is initially bound at the upstream face of the polymerase, with TBP binding both the promoter's minimal TATA element and factor TFIIB, which is in turn attached to the polymerase (Fig. 1B). The DNA duplex bends around the polymerase such that the region downstream of the promoter tracks above the enzyme's cleft. At this stage, however, DNA makes no direct contacts with the polymerase (Fig. 1B). As a result of breathing of the duplex, either via natural supercoiling or as induced by the factor TFIIF, an open state of the duplex is captured by TFIIB, and the flexible template strand subsequently descends into the cleft adjacent to the “wall” structure, where, upon reaching the catalytic center, RNA synthesis can commence in the presence of NTPs. Via its lowering into the cleft, the partially melted DNA has been reconfigured so that the downstream duplex can now enter the cleft from the downstream side with the nascent DNA-RNA hybrid helix climbing the wall behind the catalytic center and out of the cleft at the upstream side at an angle of approximately 90 to 105o to the incoming downstream duplex (8, 10) (Fig. 1A). During initiation, as downstream DNA enters the cleft and RNA synthesis proceeds to a 5-nucleotide (nt)-long transcript, a steric clash of the 5′ end of the nascent RNA with a finger domain of TFIIB that reaches into the cleft forces a decision between the abortion of transcription with minitranscript release or the destabilization of bound TFIIB (and of the whole initiation complex) followed by unhindered transcriptional elongation. At this point, transcripts of 7 nt and longer are able to interact with an unwinding site on the polymerase for the hybrid minihelix, leading to single-stranded RNA exit and DNA duplex rewinding at the upstream side (22). Elongation can now proceed.
The Intricate World of RNA Polymerase Backtracking
During mRNA synthesis, pol II moves forward along the DNA template as a “Brownian ratchet” (24, 41) (Fig. 1C). However, at certain DNA sequences the enzyme may pause, providing opportunities for “stop-go” transcriptional regulation at the level of elongation. In one notable example, partially elongated transcripts of the cellular heat shock gene pause for prolonged periods after synthesis of their 5′ ends, poised for rapid, factor-dependent reactivation later, in response to stress (42). Pausing also has roles in cotranscriptional RNA folding and processing, transcription termination, and genome stability. In addition, protein roadblocks such as nucleosomes or DNA-bound transcription factors can render transcriptional pausing unavoidable. Even on naked DNA under near-optimal conditions, pol II may persistently pause (43) at sequences where, for example, the DNA-RNA hybrid is weak (31).
During pausing as described above, or if a mismatched nucleotide has been misincorporated at the 3′ position of the transcript, the nascent RNA 3′ end may become “frayed,” i.e., disengaged from the DNA template strand at the polymerase catalytic center. In this case, the polymerase may move backwards on the template for a short distance (“backtracking”) (Fig. 1C). Though backtracking by just one or two residues may be reversible via one-dimensional forward diffusion of the enzyme (44), if backtracking continues for 8 or 9 nucleotides or more, the polymerase is likely to become arrested (incapable of spontaneously resuming forward elongation without the assistance of additional factors (reference 26 and references therein) (Fig. 1C). The arrested or backtracked enzyme is generally stable but inactive (41). In eukaryotes, it can be reactivated by transcription factor TFIIS (see below).
Structural correlate of backtracking.
As revealed from elegant structural studies, during pol II backtracking the “frayed” RNA 3′ end is extruded from the polymerase active site and through the NTP entry pore in the floor of the enzyme (Fig. 1A) and then into the “funnel” immediately below the pore on the outside surface of the polymerase (31). After backtracking for eight or more nucleotides, the extruded RNA is sufficiently long to bind a conserved “backtrack site” located along one side of the pore and into the funnel (31). Trapping of the RNA 3′ end at this site strongly inhibits further movement and is the basis for transcriptional arrest. The backtracked state is not equivalent to the forward (transcribing) state simply displaced backward along the nucleic acid scaffold but is instead a distinct and stable off-pathway state that involves structural changes leading to an inhibition of catalytic competency (41) and active retrograde movement (45). These changes are beyond the scope of this review. Where backtracking is less extensive, however, RNA interactions with the backtrack site may be partial and weak, and pol II may then spontaneously diffuse forward (31).
pol II reactivation by factor TFIIS.
Arrested pol II can be reactivated by the cellular transcription factor TFIIS via a mechanism involving cleavage of backtracked RNA at the catalytic center (46, 47) (Fig. 1C). TFIIS has three independently folding domains (references 47 and 48 and references therein). Domain 1 (amino acids [aa] 1 to 130, S. cerevisiae numbering) is not required for antiarrest functions (reference 48 and references therein) (see below). Domain II (aa 130 to 240) is tethered to domain III (aa 260 to 309) via a short linker, with domain II and the linker being responsible for pol II binding. Domain III is essential for the antiarrest activity of polymerase-bound TFIIS and for transcript cleavage (49).
TFIIS binds pol II near the rim of the funnel, extending domain III into the NTP entry pore so that a β-hairpin loop within a “Zn ribbon” region of domain III reaches the active site (11, 31). Transcript cleavage by TFIIS probably involves three charged residues within this hairpin (31) which complement the pol II active site and may help catalyze the necessary proton transfers (31). TFIIS also weakens pol II's grip on backtracked RNA at the backtrack site in the enzyme's funnel as follows. Via direct competition at the backtrack site, bound TFIIS displaces the backtracked RNA, which moves into a region of the pore that remains unblocked after TFIIS domain III insertion (31). The cleaved 3′ portion of the RNA, being already displaced from the polymerase, is released, leaving the enzyme poised for further NTP addition (41).
Intrinsic reactivation: pol II.
The pol II reactivation story does not quite end here: in addition to exploiting transcript cleavage factor TFIIS, pol II also has a very weak intrinsic cleavage activity arising from the nonessential (50) intrinsic subunit Rpb9 (44, 51, 52). Rpb9 has two Zn ribbons, one at each protein terminus. However, these are too distantly located and/or too tightly packed against the core enzyme to readily reach the NTP entry pore (51). It is unknown whether Rpb9's weak intrinsic cleavage activity arises from the vestigial activity of one or both of these suboptimally positioned ribbons directly (51) or through an ability of Rpb9 to allosterically reconfigure a key catalytic loop in the polymerase active site for transcript cleavage instead of polymerization (53). Whichever is the true mechanism, while TFIIS's in vivo role may be in the reversal of strong arrest (see above), Rpb9's role may be in the proofreading of nucleotide incorporation errors immediately after they occur, increasing pol II's transcriptional fidelity (52, 54, 55).
Intrinsic reactivation: pol I and pol III.
The jury is out on whether pol I and pol III also exploit TFIIS (56). Perhaps more importantly, however, pol I has a very effective intrinsic activity for the transcript 3′ end cleavage and the reactivation of backtracked complexes (57; see reference 44 and references therein), which is much more potent than the intrinsic activity of pol II (58). pol I's intrinsic cleavage activity arises from subunit A12.2 (Fig. 2), a homolog of Rpb9 (58). Like Rpb9, A12.2 possesses a Zn-binding β-ribbon at either protein terminus (being a much smaller subunit than Rpb9, A12.2 has just a flexible linker connecting the two ribbons). The C-terminal ribbon's hairpin includes counterparts of TFIIS's three catalytic residues for transcript cleavage (see above). In pol I “apo” structures (lacking nucleic acid), A12.2's N ribbon was positioned equivalently to that of Rpb9 in pol II (Fig. 2), but the C ribbon was positioned inside the NTP entry pore almost perfectly equivalent to the position of the TFIIS C ribbon in pol II structures (14, 15, 59). As in the TFIIS/pol II complex, the “catalytic” hairpin of A12.2 reached the polymerase active center (14), providing compelling structural evidence for involvement of A12.2's C ribbon in pol I's strong, intrinsic transcript 3′ cleavage activity through transient insertion into the NTP entry pore (Fig. 2). In the “transcribing” structure of pol I (16), i.e., in the presence of template DNA and partially elongated RNA, the A12.2 N ribbon remained unmoved, but the C ribbon was now displaced from the pore and invisible in the structure (Fig. 2). This underlined the apparently transitory nature of pore entry by A12.2's C ribbon.
FIG 2.
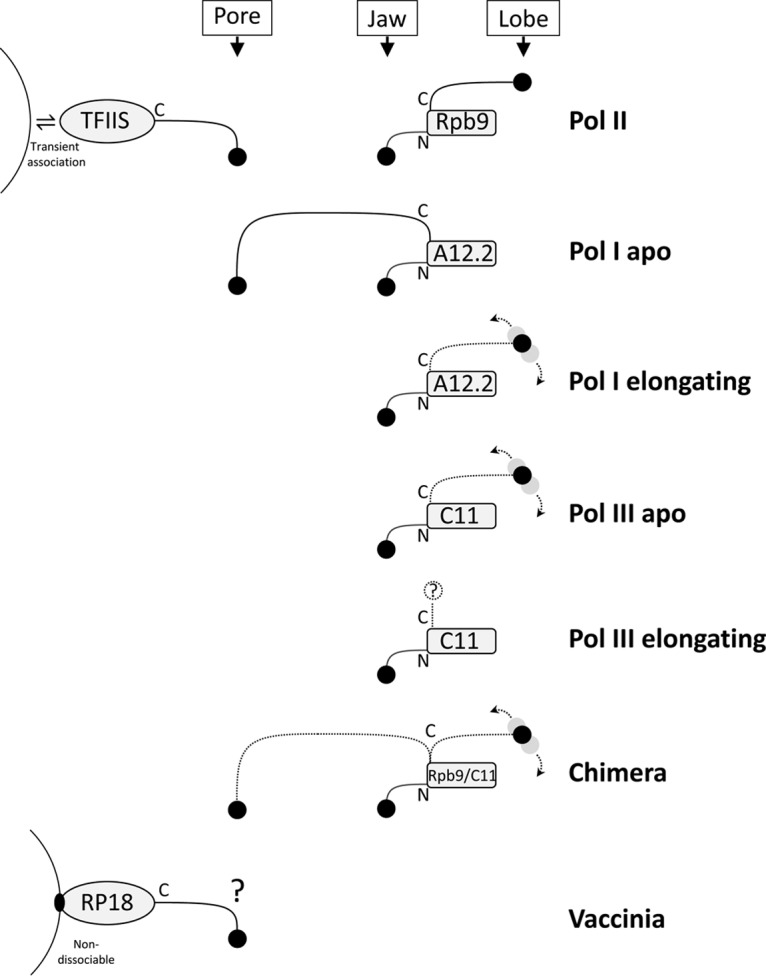
Schematic showing occupancy, by TFIIS and Rpb9 subunits and their equivalents, of three sites (NTP entry pore, jaw, and lobe, after reference 51) in coordination with transcript cleavage activities of pol I, pol II, pol III, Rpb9-C11 chimeric pol II (51), and vaccinia virus RNA polymerase. Subunit names are as given in the text. N- and C-terminal Zn ribbons of the subunits are shown as stalks. In the chimera, the C-terminal ribbon of pol II subunit Rpb9 is replaced with the equivalent ribbon pol III subunit C11 (51). Dotted arrows and gray circles denote mobility. For details, see the text.
Like pol I, pol III has a strong, intrinsic transcript cleavage activity (60). The pol III counterpart to A12.2 is subunit C11 which also has an N ribbon positioned in a similar way to that of Rpb9 (17) (Fig. 2). The C ribbon of C11 (61), however, in pol III apo structures (17) was far away from the position of A12.2's C ribbon in pol I. In the pol III elongating structure, the C ribbon was not visible at all (17) (Fig. 2). As with pol I, this suggested that the intrinsic C ribbon is mobile and is only temporarily recruited to the catalytic center (above, Fig. 2). In an elegant experiment, pol II's weak intrinsic transcript cleavage activity was rescued to strong pol III-type activity by substituting Rpb9's C ribbon with its counterpart from C11 (51). Structural analysis of the resulting chimera showed that the transplanted C ribbon was detached from the site occupied by Rpb9's native C ribbon on the surface of pol II and was mobile. Mutagenesis of “catalytic” residues in the transplanted C ribbon was consistent with its hairpin transiently inserting into the pol II NTP entry pore to complement residues at the active center (Fig. 2).
Thus, while evolution may have rendered the pol II system controllable by the dissociable factor TFIIS for regulated pausing, the endogenous cleavage activities of pol I and pol III would tend to favor the rapid, pause-free synthesis of their abundantly required transcripts (58, 62).
The TFIIS N-Terminal Domain Has Regulatory Roles in Transcriptional Initiation and Elongational “Stop-Go”
Among pol II transcription factors, some crossover between initiation and elongation activities is now recognized. For example TFIIF, long considered a transcription initiation factor, also has a role in transcriptional elongation (45, 63). In contrast, TFIIS, initially considered a pol II elongation factor, is now recognized to have a role in pol II initiation as indicated by yeast genetic analysis (64) and the finding of TFIIS in pol II preinitiation complexes (48). While TFIIS's domain III is entirely dispensable for initiation activity (48), TFIIS's domain I (see above) is centrally associated with initiation and also with the higher-order regulation of transcript cleavage for rescue from transcriptional arrest (reference 48 and references therein). Examples of such regulation would include the rescue activity of the multifunctional transcriptional regulator Ccr4-Not, which docks to TFIIS domain I (65). Other transcription elongation factors likely have comparable interactions with TFIIS (65–69). Consistent with its role in higher-order regulation, domain I is the most phylogenetically divergent portion of TFIIS and is also the most variable region of tissue-specific TFIIS isoforms and paralogs (70, 71).
ARCHAEAL MSDDRPs
Archaeal transcription systems appear to be a hybrid of the eukaryotic and bacterial systems (72), with the basal transcription apparatus being more eukaryote-like (73, 74) while the transcriptional regulatory factors are more bacterial (75, 76). Consistent with this, archaeal RNA polymerase subunit numbers and assignments are quite similar to those of yeast (77, 78). Three-dimensional similarities were borne out quite dramatically via X-ray crystallography of the archaeal enzymes from Sulfolobus solfataricus and Sulfolobus shibatae (two species of a thermoacidophile genus from the kingdom Crenarchaeota) in the presence and absence of DNA (3, 4, 79) and the enzyme from Thermococcus kodakarensis (from the kingdom Euryarchaeota [5]). Some differences between eukaryotic and archaeal enzymes include a missing domain in archaeal rpb5 forming the lower “jaw,” the distant structural relationship between archaeal Rpo8 and yeast Rpb8 (80), and the unique Rpo13 subunit in archaea (81). Indeed, Rpo8 and -13 are prominent in distinguishing MSDDRPs from different archaeal species and phyla (79). Recent studies have shown how virology and RNA polymerase structural biology in the archaea have the capacity to cross-inform (82).
MSDDRPs ACROSS ALL DOMAINS OF LIFE: THE NCLDV
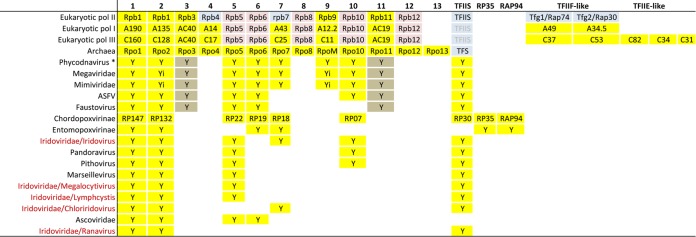
The elaboration of a broad clade of large DNA viruses termed the NCLDV is a recent development (83) arising from the paradigm-shifting discovery, in 2003 and since, of giant viruses (84) (also termed “megavirales” [85]; for reviews, see references 86 and 87). In addition to the giant viruses, the NCLDV “superclade” includes several of the established large-DNA virus families that feature a cytoplasmic stage, namely, the Poxviridae, Iridoviridae, Asfarviridae, Ascoviridae, and Phycodnaviridae (83) (Table 1). Among all of these viruses, the best studied is arguably vaccinia virus, a prototypical member of the Poxviridae. Having a cytoplasmic site of replication, the poxviruses encode their own transcription and RNA modification apparatus (88, 89), including a biochemically purified and characterized 8-subunit MSDDRP (90, 134). A 9th subunit, named RAP94, confers on the polymerase specificity for vaccinia virus early gene promoters via the heterodimeric vaccinia virus early gene transcription factor (91–93). At the protein sequence level, the two largest subunits of the vaccinia virus MSDDRP are unequivocally orthologous to the two large subunits of cellular enzymes (94–96), although the vaccinia virus largest subunit lacks a counterpart to the repeating C-terminal domain (CTD) found in eukaryotes. The smallest subunit of the vaccinia virus polymerase, RP07, shares sequence homology with the Rpb10 subunit of yeast pol II (97) (Table 2), and the vaccinia virus RP30 subunit has sequence homology to eukaryotic transcription elongation factor S-II (98), referred to here as TFIIS (see above).
TABLE 1.
| Family | Yr discovered | Host(s) | Replication site | Assembly site | Genome (kb) |
|---|---|---|---|---|---|
| Poxviridae | 1798? | Vertebrates, insects | Cytoplasm | Cytoplasm | Linear (130–380)c |
| Asfarviridae | 1921 | Pigs, warthogs, insects | Cytoplasm | Cytoplasm | Linear (170–190)c |
| Iridoviridae | 1966 | Fish, frogs, snakes, insects | Nucleus | Cytoplasm | Linear (102–212)d |
| Ascoviridae | 1983 | Insects, moths | Nucleus | Cytoplasm | Circular (157–186) |
| Phycodnaviridae | 1981 | Algae | Nucleus | Cytoplasm | Linear (100–560) |
| Mimiviridae | 2003 | Amoebae, zooplankton | Cytoplasm | Cytoplasm | Linear (∼1,200) |
| Marseilleviridae | 2009 | Amoebae | Cytoplasm | Cytoplasm | Circular (368) |
| Megaviridae | 2010 | Amoebae | Cytoplasm | Cytoplasm | Linear (1,208–1,259) |
| Pandoraviridae | 2013 | Amoebae | Cytoplasm | Cytoplasm | Linear (1,900–2,500) |
| Pithoviridae | 2014 | Amoebae | Cytoplasm | Cytoplasm | Linear (610)d |
| Faustovirus | 2015 | Vermamoeba vermiformisb | Cytoplasm | Cytoplasm | Circular (455–470)e |
Those families considered to be giant viruses, discovered starting in 2003, are shown in bold. Classification and tree topology are still developing, with, for example, the recently discovered Dinodinavirus, Faustovirus, Cedratvirus, Kaumoembavirus, and Mollivirus also being considered members of the NCLDV superclade.
A protist.
Has covalently cross-linked ends and inverted terminal repeats.
Circularly permuted and terminally redundant. The upper size limit is 303 kb if redundancy is included.
TABLE 2.
RNA polymerase subunit orthologs across all four domains of life, with emphasis on the NCLDVa
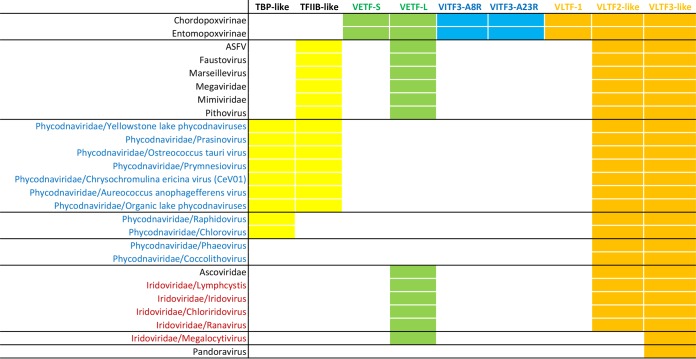
Y, ortholog found in a virus (among the NCLDV, only for vaccinia virus are orthologs named). Pink background, same polypeptide found in all three eukaryotic polymerases. Blue background, dissociable or not associated with the core polymerase. Khaki background, fused subunits. Yellow background, all other subunits. The Iridoviridae are shown by individual genera instead of family because of their divergence at the level of genus. Archaeal nomenclature is from reference 4. TFIIS is shown in gray font for eukaryotic pol I and pol III because it is unclear what role(s) this protein may play for these enzymes (56). All rows of the table are supported by complete proteomes. Table rows are in descending order of number of identified subunits. *, composite pattern over all phycodnaviruses; for individual viruses, see Table 3.
No three-dimensional structures are available for vaccinia virus or other NCLDV MSDDRPs or their subunits. However, the solved structures of yeast RNA polymerase II subunits Rpb5, -6, and -7 allowed them to be matched to the predicted secondary structures of vaccinia virus subunits RP22, RP19, and RP18, respectively (99) (Table 2). This left, as orphans, only the RP35 and RAP94 subunits from the vaccinia virus enzyme (99), and they remain so, without detectable orthologs outside the Poxviridae.
MSDDRP Subunit Assignments for the NCLDV
With the growing numbers of giant viruses being characterized, it seems apposite to revisit questions of RNA polymerase subunit assignments among the NCLDV. In our own assessment (Y. Mirzakhanyan and P. D. Gershon, unpublished data) (Tables 2 and 3), some NCLDV subunits seem to have been misannotated, while others may have been unrecognized in viral genomes. At the current state of reannotation (Mirzakhanyan and Gershon, unpublished data) (Tables 2 and 3), the enzymes from all NCLDV appear simpler than those of the eukaryotic cell, though some by not very much: among the 12 subunits of the yeast pol II/archaeal holoenzyme found in all eukaryotes and archaea, only Rpb4, -8, and -12 were universally missing among the viruses. In contrast, apart from the two large subunits (Rpb1 and -2), no subunit was universally conserved (though Rpb5 and TFIIS were almost so). Of the remaining subunits (Rpb3, -6, -7, -9, -10, and -11), although the Megaviridae, Mimiviridae, and some Phycodnaviridae encode representatives of each (Tables 2 and 3), they seem to be only sporadically present among other virus families. Extreme divergence cannot be ruled out as an explanation for the nondetection of viral homologs to these or of poxviral RP35 and RAP94 (see above). Biochemical isolation would be required to test the complete subunit composition of nonpoxviral MSDDRPs.
TABLE 3.
Breakout of phycodnavirus RNA polymerase and transcription apparatusa
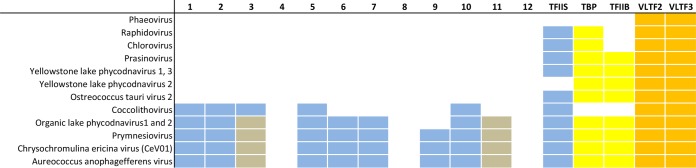
Numbers 1 to 12 refer to Rpb subunits. Background blue, gray, yellow, and orange refer to RNA polymerase (including TFIIS), RNA polymerase Rpb3/11 fusions, cellular transcription factor homologs, and vaccinia virus late transcription factor homologs, respectively. While one subgroup (represented by the prymnesioviruses, Chrysochromulina ericina virus [CeV01], and Aureococcus anophagefferens virus) seems to encode the most complete RNA polymerases of any NCLDV, another subgroup (Phaeovirus, Raphidovirus, Chlorovirus, Yellowstone lake phycodnavirus, and Ostreococcus tauri virus) seems to encode no RNA polymerase at all. All rows of the table are represented by complete proteomes.
The major compositional difference between the three major cellular RNA polymerases comprises the possession, by pol I and pol III, of additional subunits as fixed subcomplexes that are distant relatives of the pol II-dissociable GTFs TFIIF and TFIIE (100) (Table 2). Counterparts to these subcomplexes were universally absent from the NCLDV, rendering the viral polymerases more akin to pol II than to pol I or pol III in overall architecture. The roles of eukaryotic TFIIE and -F in transcription initiation include clamp opening (40), capture and stabilization of the open promoter complex, and “clearing” of protein obstructions in the cleft for loading of the template strand (101). This kind of functionality may not be critical for viral templates to achieve promoter opening and template strand loading, due to the A-T richness of viral transcriptional start sites, their genomes being noncircular in most cases (Table 1), and their encoding of topoisomerases.
Diversity of the Phycodnaviridae
Just one family within the NCLDV superclade, namely, the Phycodnaviridae, seemed to represent the full gamut of MSDDRP subunit diversity across all viruses, archaea, and eukaryotes: while one subgroup of phycodnaviruses encodes the most complete RNA polymerase noted in any NCLDV (Table 3), another subgroup within the same family seems to be unique among all NCLDV in encoding no RNA polymerase subunits at all (Table 3). The latter group presumably hijacks the host cell enzyme which, speculatively, transcribes the viral genome in combination with viral transcription factors. The above situation (factors encoded but no RNA polymerase) contrasts with the case for the Poxviridae, in which a functional MSDDRP is encoded, but there is a tantalizing partial reliance on cellular factors for intermediate- and late-stage transcription (88).
NCLDV with a Nuclear Phase: the Iridoviridae and Ascoviridae
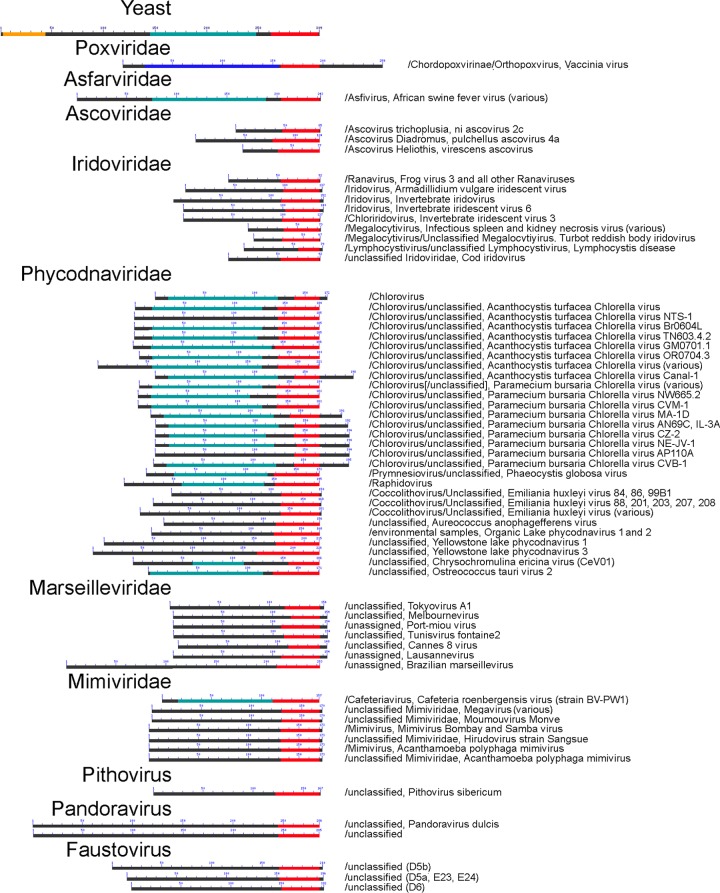
The Iridoviridae, which are diverse in terms of host range and gene content (102) were, in our annotation (Table 2), also diverse in MSDDRP subunit composition. Across Iridoviridae genera, numbers of recognizable subunits reflected, approximately, the complexity of the virus genome (Table 4). The best-characterized iridovirus at the molecular level is perhaps frog virus 3 (FV-3), a ranavirus whose genome contains a total of just 98 open reading frames (ORFs) (103). FV-3 early transcription is considered to be nuclear and mediated by host RNA polymerase II (104) with the aid of diffusible protein factors from the input virion (reference 105 and references therein). Late viral mRNAs are synthesized in the cytoplasm (106, 107). Although newly synthesized virus-encoded RNA polymerase is implicated in late transcription (108), only the two large RNA polymerase subunits were recognizable in Ranavirus genomes along with TFIIS (103) (Table 2). Assuming that all FV-3-encoded subunits have been accounted for, then either a eukaryote-like MSDDRP composed of just two or three subunits is sufficient for transcription, which would be unprecedented, or a host-virus chimera is used (which would also be novel). The Ascoviridae encode just four recognizable subunits (Table 2). Although the Ascoviridae and Iridoviridae are considered to be related, very little is known about ascoviral transcription (109). The Ascoviridae and Iridoviridae TFIIS homologs are notable in being unusually small (Fig. 3).
TABLE 4.
Iridoviridae by genus
| Genus | Type speciesa | No. of ORFs in type speciesb |
|
|---|---|---|---|
| Total | MSDDRP subunit | ||
| Iridovirus | IIV-6 | 468 | 6 |
| Lyphocystivirus | Lymphocystis disease virus, isolate China | 239 | 4 |
| Chloriridovirus | IIV-3 | 126 | 4 |
| Megalocytivirus | ISKNV | 125 | 4 |
| Ranavirus | FV-3 | 98 | 3 |
IIV, invertebrate iridescent virus; ISKNV, infectious spleen and kidney necrosis virus; FV, frog virus.
The type species is assumed to be representative. The MSDDRP subunits include TFIIS.
FIG 3.
Conserved domain search results for TFIIS across the NCLDV. Orange, turquoise, and red bars represent motif superfamilies for the N-terminal, central, and C-terminal domains (domains I, II, and III), respectively, of TFIIS. Blue bar, poxvirus RP30 superfamily. The region of highest conservation between TFIIS and RP30 is the Zn ribbon-containing domain III (98), which is involved in transcript cleavage. This region is universally found among NCLDV TFIIS/RP30 homologs. Proteins are shown aligned according to the C terminus of this domain. “Yeast” refers to S. cerevisiae.
Viral “Basal” Transcription Apparatus
In addition to RNA polymerase subunits, key, “basal” transcription factors for the NCLDV were also reannotated (Mirzakhanyan and Gershon, unpublished data) (Table 5). These efforts furthered the publicly available annotations substantially, though the task is not guaranteed to be complete. Factors annotated in Table 5 include orthologs of the vaccinia virus early transcription factor heterodimer (VETF-L and VETF-S), three of the four vaccinia virus late transcription factors (VLTF-1, VLTF-2, and VLTF-3), the obligate intermediate gene transcription factor heterodimer (VITF3-A8R and VITF3-A23R), and homologs of cellular factors TFIIB and TBP. Late factors VLTF-3 and VLTF-2 were found universally or almost universally, respectively, among the NCLDV (Mirzakhanyan and Gershon, unpublished data). The small subunit of VETF (VETF-S) was annotated conservatively in the NCLDV due to the presence of a functionally distinct paralog in the Poxviridae, namely, NPH I (helicase motifs are present in both). The VETF large subunit (VETF-L) was found in all NCLDV except the phycodnaviruses. Viruses lacking VETF-L tended to possess, instead, a TBP homolog (Table 5) suggesting complementarity between these two proteins, presumably in early promoter binding. Indeed, the core poxvirus early promoter region recognized by VETF-L (110) may be considered positionally comparable to the TATA box of core cellular promoters. The TFIIB homolog found in some NCLDV (Table 5) may represent a substitute for late factor VLTF-3 or intermediate factor VITF-3, or it may function in an additional (unknown) stage of viral transcription. A substantial overlap was apparent between the presence of TBP and TFIIB in the Phycodnaviridae (Table 5), consistent with them acting together in this family, in a “basal”-type transcription system. Somewhat counterintuitively, the Iridoviridae and Ascoviridae, whose early gene expression is considered to be dependent on the host transcription system (see above), encoded homologs of VETF-L, an early factor. Finally, despite the size and apparent sophistication of the pandoraviruses (with the largest genome yet found for any virus, nearly twice the size of its nearest rival [Table 1]), only a single homolog of a known viral or basal cellular transcription factor was detected among its genes. The transcription system of this virus may be highly divergent, or there may be a cryptic involvement of cellular proteins.
TABLE 5.
Transcription factors across the NCLDVa
Background green, blue, orange, and yellow refer to factors corresponding to poxvirus early, intermediate, and late transcriptional stages and unknown transcriptional stage, respectively. VETF-S was annotated conservatively for the NCLDV due to the presence of a functionally distinct paralog NPH-I in the Poxviridae. Font colors blue and red indicate Phycodnaviridae and Iridoviridae, respectively. Horizontal divisions denote patterns of presence/absence. For other details, see the text.
TFIIS
A TFIIS homolog was found to be nearly universally encoded across the NCLDV (Table 2), suggesting a fundamental role in transcription that transcends the deeply divergent evolutionary pathways of the different NCLDV families. Notably, a customized TFIIS is encoded even by those phycodnaviruses lacking an endogenous RNA polymerase (Table 3), suggesting that viral TFIIS is an equal-opportunity employer of viral or host cell polymerases and can displace the endogenous, host cell TFIIS from cellular pol II. The only two families/subfamilies lacking a TFIIS homolog were the ascoviruses and entomopoxviruses (Table 2). Tantalizingly, ascoviruses and entomopoxviruses are distinct among the NCLDV in infecting, primarily, lepidoptera (butterflies and moths), more specifically the family Noctuidae.
With regard to TFIIS function, the poxvirus homolog, RP30, is reported to have dual roles in transcription: as an intrinsic RNA polymerase subunit within the virion, where free RP30 does not exist (98), and as a free protein in the infected-cell cytoplasm, where it acts as an initiation factor for one of the three transcriptional classes in vaccinia virus, namely, the intermediate class (111). The intrinsic subunit has a presumed role in antiarrest (98), consistent with the nascent transcript cleavage activity demonstrated for purified ternary complexes of vaccinia virus early transcription complexes (135). The second function of RP30, as an initiation factor in vaccinia virus, has parallels with the finding of TFIIS in cellular pol II initiation complexes (see above). However, the absence of key intermediate transcription factors in NCLDV other than the Poxviridae (Table 5) suggests that this function is poxvirus specific. Interestingly, RP30's C-ribbon is flanked by a 62-aa C-terminal tail which, among the NCLDV, is also unique to the Poxviridae (Fig. 3). In the virion-packaged form of RP30, this Pro/Ser-rich tail was recently shown to be highly phosphorylated (112). It remains to be proven whether phosphorylation modulates a switch between RP30's two functions specifically in the Poxviridae.
Viral Subunit Function in the Context of Structure
With the wealth of structural information for cellular MSDDRPs alongside the revised NCLDV subunit annotations (see above), questions of structural and functional specialization in the viral enzymes can be addressed in relation to viral replication strategies. Some general themes are discussed below using, as a model, the best studied viral enzyme, namely, the one from vaccinia virus.
Dissociable Subunits in pol II Are Integral (Nondissociable) in the Vaccinia Virus Enzyme
For some subunits that are firmly attached to the vaccinia virus RNA polymerase, the cellular pol II equivalents are readily dissociable. Examples include the dissociable Rpb4/7 (“stalk”) complex of yeast pol II (see above), whose vaccinia virus equivalent, RP18, is nondissociable. Similarly, while transcription elongation factor TFIIS only transiently associates with pol II during transcription (see above), the vaccinia virus equivalent, RP30, is an integral subunit of the vaccinia virus enzyme, remaining stubbornly polymerase associated during gradient sedimentation and attempted column-based antibody affinity separation (98). The nondissociability of these subunits has parallels in the cellular realm: the pol I and pol III counterparts to pol II's stalk, namely, A14/A43 and C17/C25, respectively (Table 2) are, like the corresponding RP18 subunit of vaccinia virus, nondissociable (113). Moreover, the pol I and pol III functional equivalents of TFIIS/RP30 (subunits A12.2 and C11, respectively [see above]) are also nondissociable (14, 15, 17). Whether the giant virus homologs of TFIIS are integral to their core polymerases or dissociable is unknown.
Why might vaccinia virus RP30 be nondissociable? Just as pol I and pol III have refined their intrinsic antiarrest activity for the synthesis of relatively few, general-purpose RNAs in large quantities without “traffic jams” of paused or arrested polymerase (see above), so vaccinia virus may have opted for rapid waves of processive viral transcription, maximizing the accumulation of virus proteins and nascent virions before the host either dies or restricts the virus. Moreover, intrinsic antiarrest activity may have favored greater genomic sequence flexibility during the evolution and diversification of large viral genomes, especially if all transcriptional pausing or arrest sites could not be fully eliminated. Other possible explanations for a tight association of RP30 with vaccinia virus RNA polymerase, as suggested upon RP30's discovery and characterization (98), were to ensure RP30 packaging during virion assembly and its introduction into infected cells in stoichiometric amounts with the viral RNA polymerase.
Why is the vaccinia virus stalk subunit (RP18) nondissociable? During pol II initiation, the stalk has a role in transient opening of the clamp structure for entry of the template stand into the pol II cleft via either its own transient dissociation or its recruitment of initiation factor TFIIE that can actively open the clamp (114). Closure of the clamp around the template is associated with the presence of the stalk (19), specifically, the Rpb7 subunit (19). Within transcription elongation complexes, the clamp is always observed closed, even in the absence of Rpb4/7 (8). In pol I, pol III, and archaeal RNA polymerase (81), the stalk subunits (Table 2) are nondissociable, consistent with which, the pol I clamp appears to be permanently closed (15), or at least pol I has not yet been cocrystallized in the presence of putative factors that lead to transient clamp opening. In pol III (the apo enzyme), open and closed conformations of the clamp correspond to two distinct conformations of the nondissociable stalk (17). It seems that, as in pol I and pol III, vaccinia virus may have opted for a permanently closed clamp during elongation and the possibly greater processivity this may confer. Whatever mechanism of initiation is employed by pol I and pol III would presumably be reflected in the vaccinia virus enzyme.
pol II Proteins and Assemblies Are Reduced to Vestigial Stubs in Vaccinia Virus
While the RNA polymerase cleft and catalytic center are highly conserved in all domains of life, evolutionary plasticity seems to follow the smaller subunits. In the case of the NCLDV, this has included the formation of architectural “stubs,” as described below.
Stub 1: vaccinia virus subunit RP30.
Like vaccinia virus RP30, NCLDV TFIIS orthologs seem to mostly lack an N-terminal region sufficiently long to reflect the yeast N-terminal domain that mediates higher-order regulation of pausing (Fig. 3). The absence of this domain would suggest an unresponsiveness to higher-level transcriptional regulation (see above). This is consistent with the comparatively simple genomes and expression patterns typically found in viruses and with a presumptive need to get proteins made and virions assembled as rapidly as possible while escaping from cellular regulation. A possible exception, however, would be the large and sophisticated pandoraviruses, whose N-terminal domains approach the length of cellular TFIIS (Fig. 3). Consistent with this, the pandoraviruses currently having the largest genomes of any virus found, by a factor of 2 (115). Perhaps, as a viral genome approaches the complexity of the simplest cellular genome, there may be pressure to retain or invoke a more sophisticated regulatory mechanism for transcription elongation.
Stub 2: the “stalk.”
As discussed above, eukaryotic and archaeal MSDDRPs possess a “stalk” structure located toward the upstream face and which, in pol II, comprises the dissociable Rpb4/7 heterodimer (see above). Within this heterodimer, Rpb7 alone contacts the core polymerase (9) and is an essential subunit in yeast (9). Yeast Rpb4, which has regulatory roles in, for example, the stress response (117–119), is nonessential for viability (116). In pol I, “stalk” subunits (Table 2) provide a platform for the binding of initiation factors (58, 120–122). In all NCLDV, however, the polymerase stalk appears to be a “stub,” with a homolog of Rpb4 entirely missing from all viral MSDDRPs characterized thus far (Table 2).
Stub 3: the Rpb3-10-11-12 subassembly/subcomplex and the upstream face.
Biogenesis of the pol II core likely arises through three independent assembly subpathways, based around the two large subunits Rpb1 and Rpb2 and subunit Rpb3 (113). The Rpb3 subassembly comprises subunits Rpb3, -10, -11, and -12 (6). pol I, pol III, and archaeal RNA polymerase have equivalent subunits (Table 2). During biogenesis, the Rpb3-10-11-12 subcomplex is considered to nucleate the assembly of the holoenzyme, as the alpha subunit homodimer does in bacterial RNA polymerase (113). In mature pol II this subassembly is located on the upstream face (facing the promoter). Rpb12 is an essential subunit of pol I, pol II, and pol II in yeast and may have a role in maintaining the open promoter during initiation.
While all viruses lack a homolog to the essential eukaryotic rpb12, some (the Mimiviridae, the Megaviridae, Faustovirus, African swine fever virus, and some Phycodnaviridae) encode a fusion of subunits Rpb3 and Rpb11 (Table 2), and many encode, in addition, an Rpb10 homolog. The vaccinia virus enzyme, however, which is a good biochemical benchmark for the viral enzymes due to its extensive purification and characterization, is remarkable for the highly vestigial character of this subassembly: the only identifiable member is RP07, a homolog of the very small eukaryotic subunit Rpb10 (Table 2). This represents just 11% of the mass of the subassembly in pol II and is unprecedented among all cellular MSDDRPs (bacterial, eukaryotic, or archaeal).
Why is the Rpb3 subassembly minimal in many of the NCLDV? As with other “stubs” (see above), Rpb3 participates in regulatory interactions (reference 123 and references therein). Loss of subunits from the Rpb3 subassembly may therefore be associated with the elimination of higher-order regulatory interactions. Nonetheless, assuming that Rpb3 is central to polymerase assembly in all domains of life and with Rpb12 being an essential subunit apparently for promoter opening (124), it is unclear how these critical functions may be recapitulated in many or all of the NCLDV. It has been speculated that an “orphan” subunit in vaccinia virus of similar size, namely, RP35, may compensate for the absence of Rpb3 in the Poxviridae, although RP35 is clearly structurally unrelated (99). However, RP35 is not found outside the Poxviridae, with many NCLDV lacking both RP35 and Rpb3 (Table 2).
Stub 4: Rpb5, a “lower-jaw” subunit.
The Rpb5 subunit of yeast is a 215-aa two-domain protein (125). However, the corresponding archaeal subunit is only 84 aa in length due to an entirely missing N-terminal domain. Rpb5 is located at the lower “jaw” of pol II (see above). The presence/absence of the N domain is, in fact, not critical for polymerase function insofar as the archaeal and yeast subunits can cross-complement (125). However, in common with the above-described theme, Rpb5 seems to mediate regulatory interactions (126–128). In the Coccolithovirus genus of Phycodnaviridae as well as in two ascoviruses, the Rpb5 homolog is equivalent in size to the archaeal subunit. Although this may provide a means to escape regulatory interactions, the homologs in other NCLDV, including vaccinia virus, are close in size to the yeast protein. Whether the N domains of these homologs are sufficiently divergent to have adopted novel functions is unclear.
Subunit Rpb9: Modulation of Intrinsic Mutability?
The majority of the NCLDV examined, including vaccinia virus, lack an ortholog of pol II subunit Rpb9 (Table 2). In pol II, this subunit is implicated in transcriptional fidelity (via proofreading [see above]). In contrast to the delicately regulated and maintained eukaryotic cell, fidelity in virus transcripts may be unimportant, even to the extent that defective whole particles are typically well tolerated in virus biology. Thus, there may be few negative consequences of an occasional transcriptional error. Viruses, which are intrinsically mutagenic, do not have long-term health considerations at the level of the individual organism; only the population matters. Moreover, for mRNA in any living system, translational errors tend to swamp transcriptional ones by a substantial margin, providing an incentive for pol I and pol III to be error proof in the production of their highly recyclable and potentially mutagenic transcripts but not necessarily for pol II to be so in the production of mRNA (62). The loss of Rpb9 in the majority of NCLDV but its retention in others may have been a selectable property in maintaining a balance between fatal error and evolutionary velocity for genomes of various sizes experiencing various evolutionary pressures.
CONCLUSION
To summarize, while they retain sophistication, NCLDV RNA polymerases are to an extent stripped and honed with respect to their cellular counterparts during their adaptation and specialization for the purposes of speed, processivity, and escape from higher-order cellular control. From what we know so far of their structure-function relationships, these architectural changes seem rational, and they suggest that the many regions of viral polymerases that have universally survived the brutal journey of virus evolution are central to polymerase function, even if that function is not yet known for the cellular and viral enzymes. There is a lot we do not know about the functions of some of the smaller subunits and domains in polymerases in general, but there is every reason to believe that as structural biochemistry teaches us more, the bigger picture of subunit and domain changes and refinements found in each virus family and genus will begin to make sense also.
ACKNOWLEDGMENT
This work was supported by grant 1R21AI101577-01.
Note Added after Publication
This article was originally published on 12 July 2017. On 14 August 2017, the following changes were made.
In the originally published version, references 134 and 135 were omitted. These references were added to the References list and are now cited on page 8, line 6 (reference 134), and page 13, line 3 (reference 135).
In the originally published version, it was incorrectly stated that transcript cleavage activity has not, thus far, been demonstrated for the vaccinia virus polymerase. The sentence beginning on the first line of page 13, formerly reading “Although transcript cleavage activity has not, thus far, been demonstrated for the vaccinia virus polymerase the intrinsic subunit has a presumed role in antiarrest (98),” was changed to “The intrinsic subunit has a presumed role in antiarrest (98), consistent with the nascent transcript cleavage activity demonstrated for purified ternary complexes of vaccinia virus early transcription complexes (135).”
REFERENCES
- 1.Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA. 2015. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife doi: 10.7554/eLife.08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murakami KS. 2015. Structural biology of bacterial RNA polymerase. Biomolecules 5:848–864. doi: 10.3390/biom5020848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirata A, Klein BJ, Murakami KS. 2008. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451:851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korkhin Y, Unligil UM, Littlefield O, Nelson PJ, Stuart DI, Sigler PB, Bell SD, Abrescia NG. 2009. Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol 7:e1000102. doi: 10.1371/journal.pbio.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jun SH, Hirata A, Kanai T, Santangelo TJ, Imanaka T, Murakami KS. 2014. The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration. Nat Commun 5:5132. doi: 10.1038/ncomms6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 7.Cramer P, Bushnell DA, Kornberg RD. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 8.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 9.Armache KJ, Kettenberger H, Cramer P. 2003. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc Natl Acad Sci U S A 100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. 2016. Structure of transcribing mammalian RNA polymerase II. Nature 529:551–554. doi: 10.1038/nature16482. [DOI] [PubMed] [Google Scholar]
- 11.Kettenberger H, Armache KJ, Cramer P. 2004. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell 16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 12.Spahr H, Calero G, Bushnell DA, Kornberg RD. 2009. Schizosacharomyces pombe RNA polymerase II at 3.6-A resolution. Proc Natl Acad Sci U S A 106:9185–9190. doi: 10.1073/pnas.0903361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung AC, Cramer P. 2012. A movie of RNA polymerase II transcription. Cell 149:1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. 2013. RNA polymerase I structure and transcription regulation. Nature 502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Tornero C, Moreno-Morcillo M, Rashid UJ, Taylor NM, Ruiz FM, Gruene T, Legrand P, Steuerwald U, Muller CW. 2013. Crystal structure of the 14-subunit RNA polymerase I. Nature 502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 16.Neyer S, Kunz M, Geiss C, Hantsche M, Hodirnau VV, Seybert A, Engel C, Scheffer MP, Cramer P, Frangakis AS. 2016. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature 540:607–610. doi: 10.1038/nature20561. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJ, Sachse C, Muller CW. 2015. Molecular structures of unbound and transcribing RNA polymerase III. Nature 528:231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bushnell DA, Cramer P, Kornberg RD. 2002. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci U S A 99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bushnell DA, Kornberg RD. 2003. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc Natl Acad Sci U S A 100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bushnell DA, Westover KD, Davis RE, Kornberg RD. 2004. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science 303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 21.Westover KD, Bushnell DA, Kornberg RD. 2004. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Westover KD, Bushnell DA, Kornberg RD. 2004. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 23.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. 2006. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brueckner F, Cramer P. 2008. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol 15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan CD, Larsson KM, Kornberg RD. 2008. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell 30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. 2009. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science 324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. 2010. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. 2011. Initiation complex structure and promoter proofreading. Science 333:633–637. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami K, Elmlund H, Kalisman N, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Gibbons BJ, Levitt M, Kornberg RD. 2013. Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342:1238724. doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD. 2016. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell 166:1411–1422 e1416. doi: 10.1016/j.cell.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung AC, Cramer P. 2011. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 32.He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E. 2016. Near-atomic resolution visualization of human transcription promoter opening. Nature 533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sainsbury S, Niesser J, Cramer P. 2013. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493:437–440. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- 34.He Y, Fang J, Taatjes DJ, Nogales E. 2013. Structural visualization of key steps in human transcription initiation. Nature 495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung AC, Sainsbury S, Cramer P. 2011. Structural basis of initial RNA polymerase II transcription. EMBO J 30:4755–4763. doi: 10.1038/emboj.2011.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer M, Madoui MA, Gimenez G, La Scola B, Raoult D. 2010. Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4 domain of life including giant viruses. PLoS One 5:e15530. doi: 10.1371/journal.pone.0015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darst SA. 2001. Bacterial RNA polymerase. Curr Opin Struct Biol 11:155–162. doi: 10.1016/S0959-440X(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 38.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn CD, Lehmann E, Leike K, Sydow JF, Vannini A. 2008. Structure of eukaryotic RNA polymerases. Annu Rev Biophys 37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 39.Sampath V, Sadhale P. 2005. Rpb4 and Rpb7: a sub-complex integral to multi-subunit RNA polymerases performs a multitude of functions. IUBMB Life 57:93–102. doi: 10.1080/15216540500078905. [DOI] [PubMed] [Google Scholar]
- 40.Grunberg S, Hahn S. 2013. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci 38:603–611. doi: 10.1016/j.tibs.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nudler E. 2012. RNA polymerase backtracking in gene regulation and genome instability. Cell 149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. 2016. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell 62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svejstrup JQ. 2007. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci 32:165–171. doi: 10.1016/j.tibs.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Lisica A, Engel C, Jahnel M, Roldan E, Galburt EA, Cramer P, Grill SW. 2016. Mechanisms of backtrack recovery by RNA polymerases I and II. Proc Natl Acad Sci U S A 113:2946–2951. doi: 10.1073/pnas.1517011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweikhard V, Meng C, Murakami K, Kaplan CD, Kornberg RD, Block SM. 2014. Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc Natl Acad Sci U S A 111:6642–6647. doi: 10.1073/pnas.1405181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fish RN, Kane CM. 2002. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta 1577:287–307. doi: 10.1016/S0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 47.Wind M, Reines D. 2000. Transcription elongation factor SII. Bioessays 22:327–336. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA. 2007. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci U S A 104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Awrey DE, Shimasaki N, Koth C, Weilbaecher R, Olmsted V, Kazanis S, Shan X, Arellano J, Arrowsmith CH, Kane CM, Edwards AM. 1998. Yeast transcript elongation factor (TFIIS), structure and function. II. RNA polymerase binding, transcript cleavage, and read-through. J Biol Chem 273:22595–22605. [DOI] [PubMed] [Google Scholar]
- 50.Woychik NA, Lane WS, Young RA. 1991. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J Biol Chem 266:19053–19055. [PubMed] [Google Scholar]
- 51.Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P. 2011. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem 286:18701–18707. doi: 10.1074/jbc.M111.222273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walmacq C, Kireeva ML, Irvin J, Nedialkov Y, Lubkowska L, Malagon F, Strathern JN, Kashlev M. 2009. Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. J Biol Chem 284:19601–19612. doi: 10.1074/jbc.M109.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabart P, Jin H, Li L, Kaplan CD. 2014. Activation and reactivation of the RNA polymerase II trigger loop for intrinsic RNA cleavage and catalysis. Transcription 5:e28869. doi: 10.4161/trns.28869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nesser NK, Peterson DO, Hawley DK. 2006. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci U S A 103:3268–3273. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koyama H, Ueda T, Ito T, Sekimizu K. 2010. Novel RNA polymerase II mutation suppresses transcriptional fidelity and oxidative stress sensitivity in rpb9Delta yeast. Genes Cells 15:151–159. doi: 10.1111/j.1365-2443.2009.01372.x. [DOI] [PubMed] [Google Scholar]
- 56.Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. 2008. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev 22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tschochner H. 1996. A novel RNA polymerase I-dependent RNase activity that shortens nascent transcripts from the 3′ end. Proc Natl Acad Sci U S A 93:12914–12919. doi: 10.1073/pnas.93.23.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. 2007. Functional architecture of RNA polymerase I. Cell 131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 59.Kettenberger H, Armache KJ, Cramer P. 2003. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114:347–357. doi: 10.1016/S0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 60.Whitehall SK, Bardeleben C, Kassavetis GA. 1994. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J Biol Chem 269:2299–2306. [PubMed] [Google Scholar]
- 61.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. 1998. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev 12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alic N, Ayoub N, Landrieux E, Favry E, Baudouin-Cornu P, Riva M, Carles C. 2007. Selectivity and proofreading both contribute significantly to the fidelity of RNA polymerase III transcription. Proc Natl Acad Sci U S A 104:10400–10405. doi: 10.1073/pnas.0704116104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishibashi T, Dangkulwanich M, Coello Y, Lionberger TA, Lubkowska L, Ponticelli AS, Kashlev M, Bustamante C. 2014. Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc Natl Acad Sci U S A 111:3419–3424. doi: 10.1073/pnas.1401611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prather DM, Larschan E, Winston F. 2005. Evidence that the elongation factor TFIIS plays a role in transcription initiation at GAL1 in Saccharomyces cerevisiae. Mol Cell Biol 25:2650–2659. doi: 10.1128/MCB.25.7.2650-2659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dutta A, Babbarwal V, Fu J, Brunke-Reese D, Libert DM, Willis J, Reese JC. 2015. Ccr4-Not and TFIIS function cooperatively to rescue arrested RNA polymerase II. Mol Cell Biol 35:1915–1925. doi: 10.1128/MCB.00044-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J, Guermah M, Roeder RG. 2010. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palangat M, Renner DB, Price DH, Landick R. 2005. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc Natl Acad Sci U S A 102:15036–15041. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C, Yan H, Burton ZF. 2003. Combinatorial control of human RNA polymerase II (RNAP II) pausing and transcript cleavage by transcription factor IIF, hepatitis delta antigen, and stimulatory factor II. J Biol Chem 278:50101–50111. doi: 10.1074/jbc.M307590200. [DOI] [PubMed] [Google Scholar]
- 69.Elmendorf BJ, Shilatifard A, Yan Q, Conaway JW, Conaway RC. 2001. Transcription factors TFIIF, ELL, and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates. J Biol Chem 276:23109–23114. doi: 10.1074/jbc.M101445200. [DOI] [PubMed] [Google Scholar]
- 70.Labhart P, Morgan GT. 1998. Identification of novel genes encoding transcription elongation factor TFIIS (TCEA) in vertebrates: conservation of three distinct TFIIS isoforms in frog, mouse, and human. Genomics 52:278–288. doi: 10.1006/geno.1998.5449. [DOI] [PubMed] [Google Scholar]
- 71.Liao JM, Cao B, Deng J, Zhou X, Strong M, Zeng S, Xiong J, Flemington E, Lu H. 2016. TFIIS.h, a new target of p53, regulates transcription efficiency of pro-apoptotic bax gene. Sci Rep 6:23542. doi: 10.1038/srep23542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bell SD, Jackson SP. 1998. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol 6:222–228. doi: 10.1016/S0966-842X(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 73.Langer D, Hain J, Thuriaux P, Zillig W. 1995. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci U S A 92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zillig W, Stetter KO, Tobien M. 1978. DNA-dependent RNA polymerase from Halobacterium halobium. Eur J Biochem 91:193–199. doi: 10.1111/j.1432-1033.1978.tb20951.x. [DOI] [PubMed] [Google Scholar]
- 75.Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. 2003. The Lrp family of transcriptional regulators. Mol Microbiol 48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 76.Ouhammouch M. 2004. Transcriptional regulation in Archaea. Curr Opin Genet Dev 14:133–138. doi: 10.1016/j.gde.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 77.Huet J, Schnabel R, Sentenac A, Zillig W. 1983. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO J 2:1291–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Werner F. 2007. Structure and function of archaeal RNA polymerases. Mol Microbiol 65:1395–1404. doi: 10.1111/j.1365-2958.2007.05876.x. [DOI] [PubMed] [Google Scholar]
- 79.Wojtas MN, Mogni M, Millet O, Bell SD, Abrescia NG. 2012. Structural and functional analyses of the interaction of archaeal RNA polymerase with DNA. Nucleic Acids Res 40:9941–9952. doi: 10.1093/nar/gks692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kwapisz M, Beckouet F, Thuriaux P. 2008. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends Genet 24:211–215. doi: 10.1016/j.tig.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 81.Wojtas M, Peralta B, Ondiviela M, Mogni M, Bell SD, Abrescia NG. 2011. Archaeal RNA polymerase: the influence of the protruding stalk in crystal packing and preliminary biophysical analysis of the Rpo13 subunit. Biochem Soc Trans 39:25–30. doi: 10.1042/BST0390025. [DOI] [PubMed] [Google Scholar]
- 82.Sheppard C, Blombach F, Belsom A, Schulz S, Daviter T, Smollett K, Mahieu E, Erdmann S, Tinnefeld P, Garrett R, Grohmann D, Rappsilber J, Werner F. 2016. Repression of RNA polymerase by the archaeo-viral regulator ORF145/RIP. Nat Commun 7:13595. doi: 10.1038/ncomms13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yutin N, Koonin EV. 2012. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J 9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. 2003. A giant virus in amoebae. Science 299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 85.Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. 2013. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol 158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raoult D, Forterre P. 2008. Redefining viruses: lessons from Mimivirus. Nat Rev Microbiol 6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 87.Abergel C, Legendre M, Claverie JM. 2015. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol Rev 39:779–796. doi: 10.1093/femsre/fuv037. [DOI] [PubMed] [Google Scholar]
- 88.Moss B. 2013. Poxviridae, p 2129–2159. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 89.Moss B, Ahn B-Y, Amegadzie B, Gershon PD, Keck JG. 1991. Cytoplasmic transcription system encoded by vaccinia virus (minireview). J Biol Chem 266:1355–1358. [PubMed] [Google Scholar]
- 90.Baroudy BM, Moss B. 1980. Purification and characterization of a DNA-dependent RNA polymerase from vaccinia virions. J Biol Chem 255:4372–4380. [PubMed] [Google Scholar]
- 91.Ahn B-Y, Gershon PD, Moss B. 1994. RNA polymerase-associated protein Rap94 confers promoter specificity for initiating transcription of vaccinia virus early stage genes. J Biol Chem 269:7552–7557. [PubMed] [Google Scholar]
- 92.Ahn BY, Moss B. 1992. RNA polymerase-associated transcription specificity factor encoded by vaccinia virus. Proc Natl Acad Sci U S A 89:3536–3540. doi: 10.1073/pnas.89.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deng L, Shuman S. 1994. A role for the H4 subunit of vaccinia RNA polymerase in transcription initiation at a viral early promoter. J Biol Chem 269:14323–14328. [PubMed] [Google Scholar]
- 94.Broyles SS, Moss B. 1986. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: nucleotide sequence and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc Natl Acad Sci U S A 83:3141–3145. doi: 10.1073/pnas.83.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amegadzie BY, Holmes MH, Cole NB, Jones EV, Earl PL, Moss B. 1991. Identification, sequence, and expression of the gene encoding the second-largest subunit of the vaccinia virus DNA-dependent RNA polymerase. Virology 180:88–98. doi: 10.1016/0042-6822(91)90012-Z. [DOI] [PubMed] [Google Scholar]
- 96.Patel D, Pickup DJ. 1989. The second-largest subunit of the poxvirus RNA polymerase is similar to the corresponding subunits of procaryotic and eucaryotic RNA polymerases. J Virol 63:1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Amegadzie BY, Ahn BY, Moss B. 1992. Characterization of a 7-kilodalton subunit of vaccinia virus DNA-dependent RNA polymerase with structural similarities to the smallest subunit of eukaryotic RNA polymerase II. J Virol 66:3003–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahn B-Y, Gershon PD, Jones EV, Moss B. 1990. Identification of rpo30, a vaccinia virus RNA polymerase gene with structural similarity to a eucaryotic transcription elongation factor. Mol Cell Biol 10:5433–5441. doi: 10.1128/MCB.10.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Knutson BA, Broyles SS. 2008. Expansion of poxvirus RNA polymerase subunits sharing homology with corresponding subunits of RNA polymerase II. Virus Genes 36:307–311. doi: 10.1007/s11262-008-0207-3. [DOI] [PubMed] [Google Scholar]
- 100.Vannini A, Cramer P. 2012. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 101.Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P. 2016. Transcription initiation complex structures elucidate DNA opening. Nature 533:353–358. doi: 10.1038/nature17990. [DOI] [PubMed] [Google Scholar]
- 102.Eaton HE, Ring BA, Brunetti CR. 2010. The genomic diversity and phylogenetic relationship in the family iridoviridae. Viruses 2:1458–1475. doi: 10.3390/v2071458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tan WG, Barkman TJ, Gregory Chinchar V, Essani K. 2004. Comparative genomic analyses of frog virus 3, type species of the genus Ranavirus (family Iridoviridae). Virology 323:70–84. doi: 10.1016/j.virol.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 104.Goorha R. 1981. Frog virus 3 requires RNA polymerase II for its replication. J Virol 37:496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chinchar VG, Hyatt A, Miyazaki T, Williams T. 2009. Family Iridoviridae: poor viral relations no longer. Curr Top Microbiol Immunol 328:123–170. [DOI] [PubMed] [Google Scholar]
- 106.Willis DB, Granoff A. 1978. Macromolecular synthesis in cells infected by frog virus 3. IX. Two temporal classes of early viral RNA. Virology 86:443–453. [DOI] [PubMed] [Google Scholar]
- 107.Goorha R, Murti G, Granoff A, Tirey R. 1978. Macromolecular synthesis in cells infected by frog virus 3. VIII. The nucleus is a site of frog virus 3 DNA and RNA synthesis. Virology 84:32–50. [DOI] [PubMed] [Google Scholar]
- 108.Sample R, Bryan L, Long S, Majji S, Hoskins G, Sinning A, Olivier J, Chinchar VG. 2007. Inhibition of iridovirus protein synthesis and virus replication by antisense morpholino oligonucleotides targeted to the major capsid protein, the 18 kDa immediate-early protein, and a viral homolog of RNA polymerase II. Virology 358:311–320. doi: 10.1016/j.virol.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 109.Oliveira GP, Andrade AC, Rodrigues RA, Arantes TS, Boratto PV, Silva LK, Dornas FP, Trindade GS, Drumond BP, La Scola B, Kroon EG, Abrahao JS. 2017. Promoter motifs in NCLDVs: an evolutionary perspective. Viruses 9:16. doi: 10.3390/v9010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cassetti MA, Moss B. 1996. Interaction of the 82-kDa subunit of the vaccinia virus early transcription factor heterodimer with the promoter core sequence directs downstream DNA binding of the 70-kDa subunit. Proc Natl Acad Sci U S A 93:7540–7545. doi: 10.1073/pnas.93.15.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rosales R, Harris N, Ahn BY, Moss B. 1994. Purification and identification of a vaccinia virus-encoded intermediate stage promoter-specific transcription factor that has homology to eukaryotic transcription factor SII (TFIIS) and an additional role as a viral RNA polymerase subunit. J Biol Chem 269:14260–14267. [PubMed] [Google Scholar]
- 112.Ngo T, Mirzakhanyan Y, Moussatche N, Gershon PD. 2016. Protein primary structure of the vaccinia virion at increased resolution. J Virol 90:9905–9919. doi: 10.1128/JVI.01042-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wild T, Cramer P. 2012. Biogenesis of multisubunit RNA polymerases. Trends Biochem Sci 37:99–105. doi: 10.1016/j.tibs.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 114.Schulz S, Gietl A, Smollett K, Tinnefeld P, Werner F, Grohmann D. 2016. TFE and Spt4/5 open and close the RNA polymerase clamp during the transcription cycle. Proc Natl Acad Sci U S A 113:E1816–1825. doi: 10.1073/pnas.1515817113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 116.Woychik NA, Young RA. 1989. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol 9:2854–2859. doi: 10.1128/MCB.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bourbonnais Y, Faucher N, Pallotta D, Larouche C. 2001. Multiple cellular processes affected by the absence of the Rpb4 subunit of RNA polymerase II contribute to the deficiency in the stress response of the yeast rpb4(delta) mutant. Mol Gen Genet 264:763–772. doi: 10.1007/s004380000365. [DOI] [PubMed] [Google Scholar]
- 118.Kimura M, Suzuki H, Ishihama A. 2002. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol Cell Biol 22:1577–1588. doi: 10.1128/MCB.22.5.1577-1588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pillai B, Verma J, Abraham A, Francis P, Kumar Y, Tatu U, Brahmachari SK, Sadhale PP. 2003. Whole genome expression profiles of yeast RNA polymerase II core subunit, Rpb4, in stress and nonstress conditions. J Biol Chem 278:3339–3346. doi: 10.1074/jbc.M112180200. [DOI] [PubMed] [Google Scholar]
- 120.Engel C, Plitzko J, Cramer P. 2016. RNA polymerase I-Rrn3 complex at 4.8 A resolution. Nat Commun 7:12129. doi: 10.1038/ncomms12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. 2000. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J 19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Blattner C, Jennebach S, Herzog F, Mayer A, Cheung AC, Witte G, Lorenzen K, Hopfner KP, Heck AJ, Aebersold R, Cramer P. 2011. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev 25:2093–2105. doi: 10.1101/gad.17363311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.De Angelis R, Iezzi S, Bruno T, Corbi N, Di Padova M, Floridi A, Fanciulli M, Passananti C. 2003. Functional interaction of the subunit 3 of RNA polymerase II (RPB3) with transcription factor-4 (ATF4). FEBS Lett 547:15–19. doi: 10.1016/S0014-5793(03)00659-8. [DOI] [PubMed] [Google Scholar]
- 124.Reich C, Zeller M, Milkereit P, Hausner W, Cramer P, Tschochner H, Thomm M. 2009. The archaeal RNA polymerase subunit P and the eukaryotic polymerase subunit Rpb12 are interchangeable in vivo and in vitro. Mol Microbiol 71:989–1002. doi: 10.1111/j.1365-2958.2008.06577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sommer B, Waege I, Pollmann D, Seitz T, Thomm M, Sterner R, Hausner W. 2014. Activation of a chimeric Rpb5/RpoH subunit using library selection. PLoS One 9:e87485. doi: 10.1371/journal.pone.0087485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wei W, Dorjsuren D, Lin Y, Qin W, Nomura T, Hayashi N, Murakami S. 2001. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J Biol Chem 276:12266–12273. doi: 10.1074/jbc.M009634200. [DOI] [PubMed] [Google Scholar]
- 127.Lin Y, Nomura T, Cheong J, Dorjsuren D, Iida K, Murakami S. 1997. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J Biol Chem 272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 128.Cheong JH, Yi M, Lin Y, Murakami S. 1995. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J 14:143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Benamar S, Reteno DG, Bandaly V, Labas N, Raoult D, La Scola B. 2016. Faustoviruses: comparative genomics of new Megavirales family members. Front Microbiol 7:3. doi: 10.3389/fmicb.2016.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, Rossmann M, Colson P, Raoult D, La Scola B. 2015. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol 89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. 2009. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 132.Chen HT, Warfield L, Hahn S. 2007. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol 14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. 2003. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spencer E, Shuman S, Hurwitz J. 1980. Purification and properties of vaccinia virus DNA-dependent RNA polymerase. J Biol Chem 255:5388–5395. [PubMed] [Google Scholar]
- 135.Hagler J, Shuman S. 1993. Nascent RNA cleavage by purified ternary complexes of vaccinia RNA polymerase. J Biol Chem 268:2166–2173. [PubMed] [Google Scholar]