Abstract
Background
The cerebral subcortical deep gray matter nuclei (DGM) are a common, early, and clinically-relevant site of atrophy in multiple sclerosis (MS). Robust and reliable DGM segmentation could prove useful to evaluate putative neuroprotective MS therapies. The objective of the study was to compare the sensitivity and reliability of DGM volumes obtained from 1.5T vs. 3T MRI.
Methods
Fourteen patients with MS [age (mean, range) 50.2 (32.0–60.8) years, disease duration 18.4 (8.2–35.5) years, Expanded Disability Status Scale score 3.1 (0–6), median 3.0] and 15 normal controls (NC) underwent brain 3D T1-weighted paired scan-rescans at 1.5T and 3T. DGM (caudate, thalamus, globus pallidus, and putamen) segmentation was obtained by the fully automated FSL-FIRST pipeline. Both raw and normalized volumes were derived.
Results
DGM volumes were generally higher at 3T vs. 1.5T in both groups. For raw volumes, 3T showed slightly better sensitivity (thalamus: p = 0.02; caudate: p = 0.10; putamen: p = 0.02; globus pallidus: p = 0.0004; total DGM: p = 0.01) than 1.5T (thalamus: p = 0.05; caudate: p = 0.09; putamen: p = 0.03; globus pallidus: p = 0.0006; total DGM: p = 0.02) for detecting DGM atrophy in MS vs. NC. For normalized volumes, 3T but not 1.5T detected atrophy in the globus pallidus in the MS group. Across all subjects, scan-rescan reliability was generally very high for both platforms, showing slightly higher reliability for some DGM volumes at 3T. Raw volumes showed higher reliability than normalized volumes. Raw DGM volume showed higher reliability than the individual structures.
Conclusions
These results suggest somewhat higher sensitivity and reliability of DGM volumes obtained from 3T vs. 1.5T MRI. Further studies should assess the role of this 3T pipeline in tracking potential MS neurotherapeutic effects.
Keywords: Multiple sclerosis, Subcortical deep gray matter, Atrophy, 3T MRI, Brain segmentation
Background
The cerebral subcortical deep gray matter nuclei (DGM) are a common and clinically-relevant site of atrophy, beginning in the early stages of multiple sclerosis (MS) [1–4]. In addition, DGM atrophy is a feature of progressive forms of the disease and can be shown to progress in as little as 1 year [5, 6]. Given that few treatments are available for patients with progressive forms of MS [7], this represents a major unmet need, calling for the availability of new outcome measures to screen putative therapies. MRI-defined cerebral lesion activity and total burden, traditionally used in trials of patients with relapsing forms of MS [8–10], are less sensitive to change in patients with advanced disability and progressive forms of the disease [11, 12]. In one study, DGM atrophy assessment over 1 year was successful in demonstrating a treatment effect in MS [5]. Therefore, robust, sensitive, and reliable segmentation of DGM structures could prove useful in the evaluation of new MS therapies in all stages of the disease [6].
Field strength is known to bias the sensitivity and detectability of global cerebral MRI-based assessments of lesions and atrophy in MS [13–15]. The most commonly available MRI platforms employed for routine clinical care and research investigations are 1.5T and, less commonly, 3T. To date, it has not been clear whether longitudinal cerebral atrophy determinations would benefit from higher field strength acquisitions. The purpose of this study was to employ a fully automated freely available segmentation pipeline to compare the sensitivity and reliability of DGM volumetrics obtained from 1.5T vs. 3T MRI scans in normal controls (NCs) and patients with MS.
Methods
Subjects and neurologic examination
Fourteen patients with MS and 15 normal controls (NCs) were recruited to undergo brain MRI at both 1.5T and 3T. Table 1 shows the demographic and clinical characteristics of the subjects. The two groups differed on age (p = 0.0009) but not sex (p = 0.43). Patients met the International Panel criteria for either MS or a clinically isolated demyelinating syndrome (CIS) [16]. All patients underwent an examination by an MS specialist neurologist including evaluation of the Expanded Disability Status Scale (EDSS) [17] score and timed 25-ft walk (T25FW) [18]. At the time of MRI, 10 patients were on disease modifying therapy (DMT), while 4 patients were not. Among the DMTs used, four subjects were on dimethyl fumarate, three were on natalizumab, and one each were on fingolimod, glatiramer acetate, or cyclophosphamide. None of the DMTs were started in the 3 months before MRI. Our hospital’s human research ethics board (The Partners Human Research Committee) approved this study and written informed consent was obtained on all subjects. This work was presented in preliminary form at the 2015 annual meeting of the European Committee on Treatment and Research in Multiple Sclerosis (ECTRIMS), Barcelona, Spain; and at the 2016 annual meeting of the American Academy of Neurology, Vancouver, Canada.
Table 1.
Demographic and clinical characteristics
| MS (n = 14) | NC (n = 15) | |
|---|---|---|
| Sex ratio (women/men)a | 0.79 (11/3) | 0.60 (9/6) |
| Age, yearsa | 50.2 ± 8.2 (32–60) | 37.7 ± 9.6 (25–52) |
| MS disease category, n (%) | ||
| -Progressive relapsing MS | 1 (7%) | – |
| -Secondary progressive MS | 3 (21%) | – |
| -CIS or relapsing-remitting MS | 10 (71%) | – |
| Disease duration, yearsb | 18.4 ± 10.7 (8.2–35.5) | – |
| EDSS score | 3.1 ± 2.1 (0–6) (median 3.0) | – |
| Timed 25-ft walk, seconds | 6.2 ± 2.7 (3.5–13.0) | – |
Key: Data are mean ± standard deviation (range) unless otherwise indicated; MS multiple sclerosis, NC normal controls, CIS clinically isolated demyelinating syndrome, EDSS Expanded Disability Status Scale; aMS vs. NC were different on age (p = 0.0009) but not on sex (p = 0.43); bTime from first symptoms; n = number of subjects
MRI acquisition
All subjects underwent brain MRI at 1.5T (Signa; General Electric, Milwaukee, WI) and 3T (Skyra; Siemens, Erlangen, Germany). Scanner and acquisition details are show in Table 2. On both platforms, we obtained high-resolution 3D T1-weighted sequences covering the whole head. These were matched as closely as possible on voxel size and acquisition time, considering practical scan time limits for patient tolerability. Each sequence was optimized for signal-to-noise based on previous clinically-routine development. Each subject had a scan followed by a re-scan on the same day on each platform. Thus, at each field strength, two scans were acquired from each subject, where the subject was removed from the scanner between scans for a few minutes, and was repositioned and rescanned by the MRI technologist. For all subjects, except three, the 1.5T and 3T imaging was performed on the same day for a given subject. For the remaining three subjects, the interval between 1.5T and 3T acquistion was 6, 16, or 47 days. During the study period, there were no intervening scanner upgrades.
Table 2.
1.5T and 3T brain MRI acquisition protocols
| 1.5T | 3T | |
|---|---|---|
| Scanner manufacturer | GE Signa LX | Siemens Skyra |
| Operation system version | 11× | D13 |
| Coil | Quadrature head coil | 20-channel head and neck coil |
| Type of sequence | 3D SPGR | 3D MPRAGE |
| Acceleration factor for parallel imaging | N/A | 2 |
| Orientation | Sagittal | Sagittal |
| Field of view (cm) | 24 × 24 | 24 × 25.6 |
| Matrix size | 256 × 256 | 240 × 256 |
| Number of slices | 166 | 176 |
| Repetition time (msec) | 8.176 | 2300 |
| Echo time (msec) | 3.856 | 2.96 |
| Flip angle (degrees) | 20 | 9 |
| Voxel size (mm) | 0.938 × 0.938 × 1.2 | 1.0 × 1.0 × 1.0 |
| Scan time (minutes) | 6:24 | 5:12 |
| Number of signal averages | 1 | 1 |
Key: SPGR spoiled gradient recalled echo, MPRAGE magnetization-prepared rapid acquisition gradient echo
MRI analysis
All image pre-processing was performed using Jim software (v.7.0, Xinapse Systems Ltd., Northants, UK, http://www.xinapse.com/). For both the 1.5T and 3T images, the raw sagittal images did not yield adequate segmentation, particularly of the intracranial volume (ICV) cavity (“skull stripping”; data not shown). With optimization work, we determined necessary pre-processing steps, which were the same for the 1.5T and 3T images. First, all original DICOM images were converted to a Neuroimaging Informatics Technology Initiative (NIfTI) format, and their raw sagittal orientation was converted to axial. Then, 170 axial slices were extracted from each scan starting at the first slice showing the top of the head. This provided whole brain coverage in all patients extending to the foramen magnum. DGM (caudate, thalamus, globus pallidus, and putamen) volumes were obtained by a fully automated segmentation pipeline (FSL-FIRST, v. 5.0, The Analysis Group, Oxford, UK, http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FIRST) (Fig. 1). This pipeline was chosen for its free availability, full automation, and utility shown in detecting short term DGM atrophy and treatment effects in patients with MS [5, 6]. The ICV was the sum of gray matter, white matter and cerebrospinal fluid (CSF), which was obtained by applying these images to a fully automated algorithm (SIENAX, v. 5.0, The Analysis Group, Oxford, UK, http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/SIENA) [15, 19]. We assessed three volumetric measures of the DGM structures: 1) raw volumes, 2) those that were normalized by dividing by the subject’s ICV (“fractions” [1]); 3) those that were normalized by multiplying the raw volume by the whole brain SIENAX normalization factor (“normalized”).
Fig. 1.
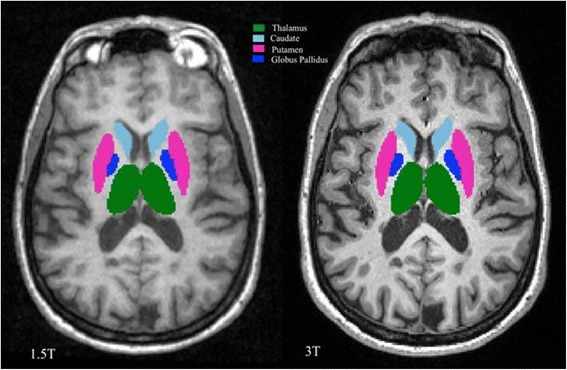
Representative anatomic slice showing segmentation of the cerebral subcortical deep gray matter (DGM) in one patient from 1.5T (left) and 3T (right) MRI scans. This is from a 51 year-old woman with multiple sclerosis and moderate physical disability. The total DGM volume was 28.4 ml at 1.5T and 29.3 ml at 3T. Component DGM structures are shown in different colors. The segmentation maps are overlaid to the original raw 3D T1-weighted images after re-sampling to the axial plane. Segmentation was performed by the fully automated FSL-FIRST pipeline. In the present study, we utilized the FSL-FIRST outputs to assess the volume of the thalamus, caudate, putamen, and globus pallidus (and their sum = total DGM)
Statistical Analysis
Statistical analyses included unpaired and paired t-tests, Fisher’s exact tests, and analysis of covariance with age as a covariate. Correlations were reported for MRI associations with age, EDSS score, and T25FW by Spearman coefficients. Sensitivity (differentiation between groups) was reported using mixed model analysis of covariance with age as covariate, comparing the methods for their ability to differentiate MS from NC by the interaction between method and group. Within-subject correlations were compared using the method of Meng et al. (1992) [20]. Reliability was reported using intraclass correlation coefficients (ICCs) [21] with 95% confidence intervals (CIs). The analysis was generated using SAS (v. 9.4, SAS Institute Inc., Cary, NC, http://www.sas.com/).
Results
Scan-rescan reliability: 1.5T vs. 3T
DGM scan-rescan reliability within groups, comparing field strengths, is shown in Table 3 for all three methods. Reliability was generally very high for both the 1.5T and 3T measurements. Regarding raw volumes, there was perhaps somewhat higher reliability at 3T and, across all comparisons, higher reliability for measuring total DGM than the individual structures. Regarding fractions and normalized volumes, there was a suggestion of higher reliability at 1.5T for the caudate. Comparing the three methods, there was slightly higher reliability for measuring raw volumes than fractions or normalized volumes. We determined that the latter were more reliable at 1.5T most likely due to increased accuracy vs. 3T in whole brain extraction, necessary for determination of the ICV and brain size (Fig. 2).
Table 3.
Deep gray matter data: scan-rescan reliability (within group and field strengths)
| MS (n = 14) | NC (n = 15) | |||
|---|---|---|---|---|
| 1.5T ICC (95% CI) | 3T ICC (95% CI) | 1.5T ICC (95% CI) | 3T ICC (95% CI) | |
| Volumes | ||||
| -Thalamus | 0.99 (0.96, 1.00) | 0.99 (0.98, 1.00) | 0.95 (0.87, 0.98) | 0.99 (0.97, 1.00) |
| -Caudate | 0.99 (0.98, 1.00) | 0.99 (0.98, 1.00) | 0.97 (0.93, 0.99) | 0.96 (0.88, 0.98) |
| -Putamen | 0.94 (0.85, 0.98) | 0.97 (0.90, 0.99) | 0.95 (0.87, 0.98) | 0.97 (0.93, 0.99) |
| -Globus pallidus | 0.94 (0.83, 0.98) | 0.98 (0.95, 0.99) | 0.95 (0.87, 0.98) | 0.98 (0.93, 0.99) |
| -Total DGM | 0.99 (0.97, 1.00) | 1.00 (0.99, 1.00) | 0.98 (0.94, 0.99) | 0.99 (0.98, 1.00) |
| Fractions | ||||
| -Thalamus | 0.98 (0.95, 0.99) | 0.95 (0.87, 0.98) | 0.88 (0.71, 0.96) | 0.93 (0.81, 0.97) |
| -Caudate | 0.98 (0.94, 0.99) | 0.94 (0.84, 0.98) | 0.96 (0.88, 0.98) | 0.88 (0.69, 0.95) |
| -Putamen | 0.92 (0.78, 0.97) | 0.92 (0.78, 0.97) | 0.93 (0.81, 0.97) | 0.95 (0.86, 0.98) |
| -Globus pallidus | 0.93 (0.80, 0.97) | 0.95 (0.85, 0.98) | 0.93 (0.81, 0.97) | 0.85 (0.63, 0.94) |
| -Total DGM | 0.98 (0.93, 0.99) | 0.93 (0.80, 0.97) | 0.94 (0.84, 0.98) | 0.89 (0.72, 0.96) |
| Normalized | ||||
| -Thalamus | 0.95 (0.87, 0.98) | 0.99 (0.96, 0.99) | 0.96 (0.89, 0.99) | 0.96 (0.88, 0.98) |
| -Caudate | 0.97 (0.90, 0.99) | 0.98 (0.95, 0.99) | 0.97 (0.93, 0.99) | 0.92 (0.78, 0.97) |
| -Putamen | 0.94 (0.83, 0.98) | 0.97 (0.92, 0.99) | 0.93 (0.81, 0.97) | 0.95 (0.86, 0.98) |
| -Globus pallidus | 0.92 (0.77, 0.97) | 0.97 (0.92, 0.99) | 0.97 (0.92, 0.99) | 0.93 (0.82, 0.98) |
| -Total DGM | 0.95 (0.87, 0.98) | 0.98 (0.95, 0.99) | 0.97 (0.93, 0.99) | 0.95 (0.85, 0.98) |
Key: MS multiple sclerosis, NC normal controls, DGM cerebral subcortical deep gray matter, CI confidence interval, ICC intraclass correlation coefficient; normalized = raw volume multiplied by SIENAX normalization factor
Fig. 2.

Examples of brain extraction masks obtained in the fully automated SIENAX pipeline, part of the process to determine intracranial volume. Both images are from a 51 year-old woman with multiple sclerosis and moderate physical disability. The brain parenchymal mask was more accurately obtained at 1.5T, whereas it is underestimated at 3T
DGM results: 1.5T vs. 3T
Using all three methods, DGM volumes were higher at 3T than 1.5T in both patients and controls (Tables 4 and 5). This was seen across all DGM structures examined, and significant for all comparisons (except thalamic fractions in the MS group). The effect sizes were generally larger for this field strength difference for raw volumes than fractions (Tables 4 and 5) or normalized volumes (Table 5). This effect was also reflected in total DGM data. For example, the total raw DGM volume was, on average, 3.9% higher in patients and 5.9% higher in controls (Table 5) at 3T vs. 1.5T (both p < 0.0001, Table 4). As shown in Figs. 3 and 4, we explored the possible causes for the increased DGM volume at 3T by performing expert, manual segmentation of one scan each from a healthy control and a patient with MS. The manual segmentations were then overlaid on the automated segmentation maps. The 3T images provided a more accurate (and larger) contour of DGM structures. This was most likely related to improved contrast at the ventricular CSF-tissue interfaces, and to a lesser extent, the gray-white edges.
Table 4.
Deep gray matter data: between group and between field strength comparisons
| MS group (n = 14) | NC group (n = 15) | pa, b MS vs NC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| DGM structure | 1.5T | 3T | p (MS) 1.5T vs. 3T | 1.5T | 3T | P (NC) 1.5T vs 3T | 1.5T | 3T | |
| Volumes (ml) | Thalamus | 14.4 (1.4) 11.7–16.3 |
14.7 (1.5) 12.6–16.7 |
0.003 | 15.2 (1.0) 12.9–16.3 |
16.0 (1.0) 13.6–17.1 |
<0.0001 | 0.05 | 0.02 |
| Caudate | 6.2 (0.9) 4.9–7.8 |
6.5 (0.9) 5.0–8.2 |
0.0001 | 6.9 (0.8) 5.2–8.6 |
7.3 (0.9) 5.6–9.4 |
<0.0001 | 0.09 | 0.10 | |
| Putamen | 8.7 (0.7) 7.4–9.7 |
9.0 (0.7) 7.6–10.1 |
<0.0001 | 9.6 (0.8) 8.6–11.7 |
10.1 (0.9) 9.1–12.6 |
<0.0001 | 0.03 | 0.02 | |
| Globus pallidus | 2.9 (0.3) 2.4–3.4 |
3.1 (0.4) 2.6–3.9 |
0.0004 | 3.3 (0.3) 2.9–3.7 |
3.6 (0.3) 3.2–4.0 |
<0.0001 | 0.0006 | 0.0004 | |
| Total DGM | 32.2 (2.8) 26.4–37.1 |
33.4 (3.0) 28.1–38.6 |
<0.0001 | 35.0 (2.6) 30.9–40.2 |
37.0 (2.6) 32.8–42.8 |
<0.0001 | 0.02 | 0.01 | |
| Fractions | Thalamus | 0.011 (0.0011) 0.0087–0.013 |
0.012 (0.00097) 0.0096–0.013 |
0.12 | 0.0116 (0.00058) 0.010–0.013 |
0.012 (0.00063) 0.011–0.013 |
0.0004 | 0.31 | 0.23 |
| Caudate | 0.0049 (0.00057) 0.0037–0.0056 |
0.0051 (0.00058) 0.0040–0.0059 |
0.007 | 0.0053 (0.00046) 0.0040–0.0060 |
0.0055 (0.00053) 0.0043–0.0066 |
0.0004 | 0.20 | 0.30 | |
| Putamen | 0.0068 (0.00055) 0.0055–0.0078 |
0.0071 (0.00057) 0.0057–0.0080 |
0.0001 | 0.0074 (0.00051) 0.0065–0.0081 |
0.0077 (0.00053) 0.0068–0.0088 |
<0.0001 | 0.20 | 0.20 | |
| Globus pallidus | 0.0023 (0.00021) 0.0018–0.0026 |
0.0024 (0.00024) 0.0020–0.0028 |
0.0002 | 0.0025 (0.00017) 0.0023–0.0028 |
0.0028 (0.00013) 0.0025–0.0030 |
<0.0001 | 0.009 | 0.003 | |
| Total DGM | 0.0252 (0.0020) 0.0197–0.0282 |
0.0261 (0.0019) 0.0213–0.0280 |
0.002 | 0.0268 (0.0013) 0.0236–0.0290 |
0.0282 (0.0013) 0.0256–0.0299 |
<0.0001 | 0.11 | 0.07 | |
Key: Data are mean (standard deviation) on first line; range on second line; Percent differences between 1.5T and 3T are shown in Table 5; DGM = cerebral subcortical deep gray matter; MS multiple sclerosis, NC normal controls; aage adjusted; bthe differentiation (sensitivity) between MS and NC was similar for 1.5 versus 3T (all p > 0.05)
Table 5.
Deep gray matter data: field strength comparisons: percent differences
| DGM structure | MS 1.5T vs. 3T | NC 1.5T vs. 3T | |
|---|---|---|---|
| Volumes | Thalamus | 2.7% | 5.7% |
| Caudate | 4.8% | 5.0% | |
| Putamen | 4.1% | 5.3% | |
| Globus pallidus | 7.9% | 10.4% | |
| Total DGM | 3.9% | 5.9% | |
| Fractions | Thalamus | 2.4% | 5.2% |
| Caudate | 4.5% | 4.5% | |
| Putamen | 3.7% | 4.8% | |
| Globus pallidus | 7.5% | 9.8% | |
| Total DGM | 3.6% | 5.4% | |
| Normalized | Thalamus | 3.2% | 4.6% |
| Caudate | 5.3% | 3.9% | |
| Putamen | 4.6% | 4.2% | |
| Globus pallidus | 8.4% | 9.2% | |
| Total DGM | 4.4% | 4.8% |
Key: For each subject, the percent difference for 3T data minus 1.5T data was calculated, using the 1.5T data as the denominator. The averages of those percentages are shown for each group. DGM = cerebral subcortical deep gray matter; MS multiple sclerosis, NC normal controls; normalized = raw volume x SIENAX normalization factor
Fig. 3.
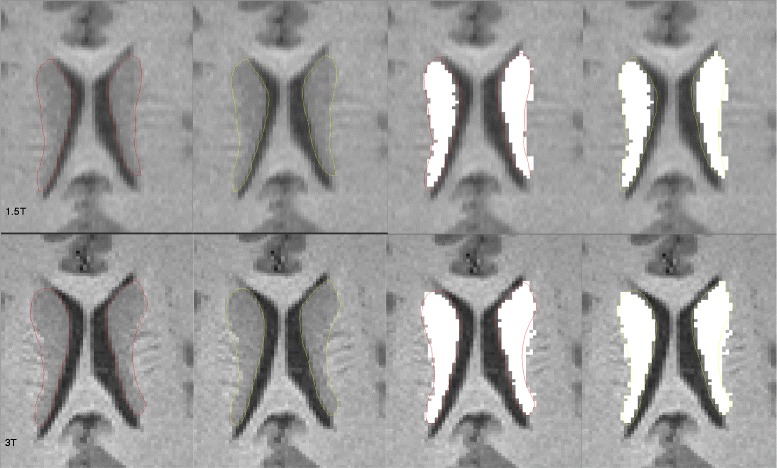
Manual (expert) segmentation overlaid with automated segmentation output (white areas) of the caudate nucleus in a healthy 33 year-old control subject from 1.5T (upper row) and 3T (lower row) 3D T1-weighted MRI scans. The total DGM volume was 36.6 ml at 1.5T and 38.9 ml at 3T. For illustrative purposes, the ground truth contours are shown in both red and yellow colors. Automated segmentation was performed by the fully automated FSL-FIRST pipeline. The 3T automated output typically provided a larger and more accurate contour than 1.5T. At 1.5T, the interface between the caudate and the medial aspects of the ventricular CSF is underestimated vs. 3T. The interface between the caudate and the adjacent (lateral) white matter appears to be captured similarly at both field strengths
Fig. 4.
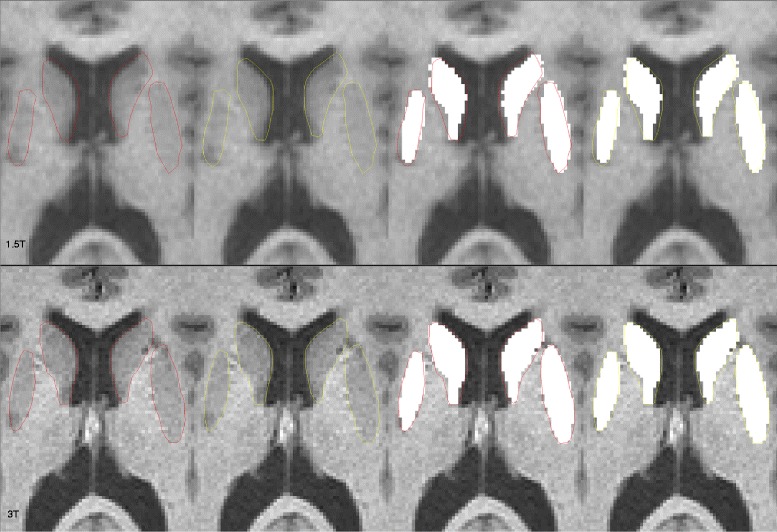
Manual (expert) segmentation overlaid with automated segmentation output (white areas) of the caudate nucleus and putamen from 1.5T (upper row) and 3T (lower row) 3D T1-weighted MRI scans. Images are from a 51 year-old woman with multiple sclerosis and moderate physical disability. The total DGM volume was 28.4 ml at1.5T and 29.3 ml at 3T. For illustrative purposes, the ground truth contours are shown in both red and yellow colors. Automated segmentation was performed by the fully automated FSL-FIRST pipeline. The 3T automated output typically provided a larger and more accurate contour than 1.5T. At 1.5T, the interface between the caudate and the anterior and medial aspects of the ventricular CSF is underestimated vs. 3T. The interface between the putamen and the adjacent (lateral) white matter appears to be larger and more accurate at 3T
Detection of DGM atrophy in MS vs. NC
A comparison of DGM volume and fraction in MS vs. NC is shown in Table 4. 3T showed slightly better differentiation of raw volumes (thalamus: p = 0.02; caudate: p = 0.10; putamen: p = 0.02; globus pallidus: p = 0.0004; total DGM: p = 0.01) than 1.5T (thalamus: p = 0.05; caudate: p = 0.09; putamen: p = 0.03; globus pallidus: p = 0.0006; total DGM: p = 0.02) for detecting DGM atrophy in MS vs. NC. Regarding the normalized volumes, atrophy was not detected in patients vs. NC in any structures at either field strength (p > 0.05) except for the globus pallidus at 3T (mean 4.4 vs. 5.0 ml, p < 0.05). Thus, thalamic and pallidal atrophy was detected slightly more definitively at 3T. Otherwise, the presence of atrophy in the other DGM structures was similarly detected at both 1.5T and 3T. Overall, these field strength differences in the ability to differentiate MS from NC were small (all p > 0.10). Comparing raw, fractional, and normalized volumes, there was slightly better sensitivity for detecting atrophy in the MS group for the raw volumes.
Correlation between DGM and age or disability in the MS group
With the exception of the normalized putamen volume at 1.5T (r = −0.56, p = 0.04), DGM raw or normalized volumes and fractions at 1.5T or 3T did not correlate with age; the correlations were also not different between field strengths (all p > 0.05, data not shown). Regarding the correlation between DGM and EDSS scores, significant negative relationships were found with normalized volumes at 1.5T in the thalamus (r = −0.61, p = 0.02), globus pallidus (r = −0.53, p = 0.0498), and total DGM (r = −0.56, p = 0.04) and at 3T in the putamen (r = −0.56, p = 0.04). No significant correlations were found at either field strength between DGM raw volumes or fractions and EDSS scores (all p > 0.05, data not shown). The correlations were not different between field strengths (all p > 0.05, data not shown). Regarding the correlation between DGM and T25FW, with the exception of total DGM normalized volume (r = −0.55, p = 0.04), none of the comparisons reached significance (data not shown).
Discussion
In this study, we explored the sensitivity and reliability of DGM volumes obtained from 1.5T vs. 3T and their clinical relevance. The first main finding was that volumes were generally higher at 3T. Secondly, 3T showed slightly better differentiation in the ability to detect atrophy in the thalamus and globus pallidus in MS vs. NC. Third, 3T showed slightly higher scan/re-scan reliability. We also noted that, regardless of field strength, volumes showed higher reliability than fractions, and total DGM volume was measured with higher reliability than the individual DGM nuclei. These data indicate that 1.5T and 3T are not interchangeable in measuring DGM volumes.
One clear finding was that raw volumes, fractions, and normalized volumes were higher at 3T vs. 1.5T across all DGM structures examined. In a previous study, volume biases were also detected based on field strength in comparing 1.5T to 3T using the FreeSurfer processing toolkit in 15 healthy elderly subjects; these occurred in either direction (not always higher at 3T) [22]. For example, the globus pallidus and thalamus showed significantly higher volumes at 1.5T but the amygdala was higher at 3T. Another study of whole brain volume measurements, comparing 1.5T and 3T MRI showed significant differences between platforms using the SIENAX toolkit [15]. Interestingly, this study found that the bias was in the opposite direction vs. the present study, with higher whole brain volumes measures from 1.5T. This was most likely related to an overestimation of brain volume at the sulcal-CSF interfaces at 1.5T due to partial volume averaging. In the present study, the DGM volumes from 3T may have been larger on the basis of more accurate detection of structure boundaries with CSF or adjacent white matter. Taken together, these results suggest that combining data across platforms and across field-strength introduces a bias that should be considered in the design of multi-site studies, such as clinical therapeutic trials.
In the detection of DGM atrophy in MS vs. NC in the present study, 3T showed slightly better differentiation. Similar findings were seen in a previous study in detecting hippocampal atrophy at 3T vs. 1.5T [23]. In that study, subjects who converted from mild cognitive impairment to Alzheimer disease within 3 years of baseline MRI showed significantly more atrophy in the cornu ammonis 1 region of the right hippocampus versus nonconverters at 3T but not at 1.5T. Another study, focusing on whole brain atrophy, showed a higher effect size for 3T in detecting brain atrophy in MS versus NC when compared to 1.5T [15]. These results parallel what has been shown regarding MS lesion detection at 3T and 7T, with an increase in the diagnostic yield in the detection of MS brain lesions compared to 1.5T [14, 24]; moreover, the brain lesion load at 3T showed a closer relationship to cognitive status than 1.5T [14]. These results underscore potential gains in sensitivity and validity in the MRI measurement of MS-related structural changes with ultra-high field strengths.
In the present study, we found atrophy of the DGM in patients with MS vs. NC in various nuclei. There are several mechanisms to consider in the pathogenesis of MS-related DGM atrophy. These include iron-deposition [25], oxidative stress [26], neurodegeneration [27], direct injury by the presence of DGM demyelinating lesions [28], and Wallerian degeneration due to damage of white matter tracts throughout the brain [29].
In a previous study, scan-rescan reliability for a variety of platforms at two field strengths was explored [30]. Their results showed that reliability of automatic brain morphometry obtained by FreeSurfer was generally higher with GE Signa Excite (1.5T) and Siemens Verio (3T) vs. Siemens Sonata (1.5T) and TrioTim (3T) acquisitions. The authors argued that, although TrioTim and Verio are both 3T MRI models from the same manufacture (Siemens), the results from the two machines differed significantly. Another group compared 3T Siemens scanners at seven sites to evaluate the difference between intra-scanner and inter-scanner reliability of lesion and atrophy-related MS volumetrics in a human phantom study using a variety of processing pipelines [31]; the authors showed that, despite protocol harmonization and the use of high-resolution sequences, a large degree of variability in the data was caused by inter-scanner effects. In the present study, we showed high scan-rescan reliabilities for both 1.5T and 3T in the assessment of DGM volumetry. In the measurement of DGM raw volumes, there was perhaps slightly higher reliability at 3T. However, for DGM fractions, there was perhaps somewhat higher reliability at 1.5T. Raw volumes showed slightly higher reliability than the other two normalized methods. Also, the most sensitivity in detecting DGM atrophy was observed with raw DGM volumes. This probably reflects the inaccurate estimation of ICV and the brain contour, necessary for normalization, due to higher susceptibility artifacts at 3T.
Several limitations of our study are worthy of comment. First, aside from the field strength difference, the two acquisitions differed on the scanner vendor, type of head coil, and use of parallel imaging (only at 3T). The voxel sizes also slightly differed. One should also consider the potential effect of DMTs on brain volume; first because of their partial but significant therapeutic effects on limiting the rate of atrophy in MS [3, 5, 6, 32]. Because most of our patients were receiving DMT, the generazibility of our results to untreated patients is not established by this study. Furthermore, there is the potential for individuals to show pseudoatrophy in the few months after their initiation of therapy (for some but not all DMTs) [33]. However, in the present study, none of patients had newly started their DMT in the previous 3 months, thus indicating that pseudoatrophy did not have a major effect on our results. In addition, the sample size was small and no longitudinal data were available to compare the rate of atrophy between the two acquisitions. We did not test other fully automated segmentation pipelines such as FreeSurfer and others that are available to measure DGM atrophy [31, 34–36]. Finally, our patient population was dominated by subjects with relapsing forms of MS, with only four people in our study having progressive forms of the disease. Thus, the generalizability of our results to the full MS spectrum would require further study. Thus, taken together, these caveats suggest other factors that could have influenced the differences we observed between the two MRI scan platforms.
Conclusion
We conclude that MRI scan acquisition field strength should be considered in the design of longitudinal studies and multicenter clinical trials. Such differences may introduce bias in the obtained data and results. If such consistency cannot be maintained, statistically corrective modelling may be considered [37, 38].
Acknowledgements
None.
Funding
There are no relevant external funding sources.
Availability of data and materials
The full datasets used and analyzed during the current study are available from the corresponding author by request.
Abbreviations
- CI
Confidence interval
- CIS
Clinically isolated demyelinating syndrome
- CSF
Cerebrospinal fluid
- DGM
Cerebral subcortical deep gray matter nuclei
- ICC
Intraclass correlation coefficient
- ICV
Intracranial volume
- MS
Multiple sclerosis
- NC
Normal controls
- NIfTI
Neuroimaging informatics technology initiative
- T25FW
Timed 25-ft walk
Authors’ contributions
RB conceived the study and designed the protocol. RC performed the image analysis. ST provided technical assistance and study supervision. SH provided statistical analysis. RB and RC drafted the manuscript. ST and SH edited the manuscript for critical content. RB provided overall study supervision. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Our hospital’s human research ethics board (The Partners Human Research Committee) approved this study and written informed consent was obtained on all subjects.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Renxin Chu, Email: renxin@bwh.harvard.edu.
Shelley Hurwitz, Email: hurwitz@hms.harvard.edu.
Shahamat Tauhid, Email: shahamat@bwh.harvard.edu.
Rohit Bakshi, Email: rbakshi@post.harvard.edu.
References
- 1.Houtchens MK, Benedict RH, Killiany R, Sharma J, Jaisani Z, Singh B, et al. Thalamic atrophy and cognition in multiple sclerosis. Neurology. 2007;69:1213–1223. doi: 10.1212/01.wnl.0000276992.17011.b5. [DOI] [PubMed] [Google Scholar]
- 2.Bergsland N, Horakova D, Dwyer MG, Dolezal O, Seidl ZK, Vaneckova M, et al. Subcortical and cortical gray matter atrophy in a large sample of patients with clinically isolated syndrome and early relapsing-remitting multiple sclerosis. AJNR Am J Neuroradiol. 2012;33:1573–1578. doi: 10.3174/ajnr.A3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakshi R, Dandamudi VSR, Neema M, De C, Bermel RA. Measurement of brain and spinal cord atrophy by magnetic resonance imaging as a tool to monitor multiple sclerosis. J Neuroimaging. 2005;15:30S–45S. doi: 10.1177/1051228405283901. [DOI] [PubMed] [Google Scholar]
- 4.Nourbakhsh B, Azevedo C, Maghzi AH, Spain R, Pelletier D, Waubant E. Subcortical grey matter volumes predict subsequent walking function in early multiple sclerosis. J Neurol Sci. 2016;366:229–233. doi: 10.1016/j.jns.2016.04.054. [DOI] [PubMed] [Google Scholar]
- 5.Dupuy SL, Tauhid S, Hurwitz S, Chu R, Yousuf F, Bakshi R. The effect of dimethyl fumarate on cerebral gray matter atrophy in multiple sclerosis. Neurol Ther. 2016;5:215–229. doi: 10.1007/s40120-016-0054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim G, Chu R, Yousuf F, Tauhid S, Stazzone L, Houtchens MK, et al. Sample size requirements for 1 year treatment effects using deep gray matter volume from 3T MRI in progressive forms of multiple sclerosis. Int J Neurosci. [Epub ahead of print]. doi:10.1080/00207454.2017.1283313. [DOI] [PubMed]
- 7.Ontaneda D, Fox RJ, Chataway J. Clinical trials in progressive multiple sclerosis: lessons learned and future perspectives. Lancet Neurol. 2015;14:208–223. doi: 10.1016/S1474-4422(14)70264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippi M, Rocca MA, Arnold DL, Bakshi R, Barkhof F, De Sefano N, et al. EFNS guideline on the use of neuroimaging in the management of multiple sclerosis. Eur J Neurol. 2006;13:313–325. doi: 10.1111/j.1468-1331.2006.01543.x. [DOI] [PubMed] [Google Scholar]
- 9.Filippi M, Wolinsky JS, Comi G. CORAL Study Group. Effects of oral glatiramer acetate on clinical and MRI-monitored disease activity in patients with relapsing multiple sclerosis: a multicentre, double-blind, randomised, placebo-controlled study. Lancet Neurol. 2006;5:213–220. doi: 10.1016/S1474-4422(06)70327-1. [DOI] [PubMed] [Google Scholar]
- 10.Zivadinov R, Bakshi R. Role of MRI in multiple sclerosis I: inflammation and lesions. Front Biosci. 2004;9:665–683. doi: 10.2741/1251. [DOI] [PubMed] [Google Scholar]
- 11.Li DK, Held U, Petkau J, Daumer M, Barkhof F, Fazekas F, et al. MRI T2 lesion burden in multiple sclerosis: a plateauing relationship with clinical disability. Neurology. 2006;66:1384–1389. doi: 10.1212/01.wnl.0000210506.00078.5c. [DOI] [PubMed] [Google Scholar]
- 12.Zurawski J, Lassmann H, Bakshi R. Use of magnetic resonance imaging to visualize leptomeningeal inflammation in patients with multiple sclerosis: A review. JAMA Neurol. 2017;74:100–109. doi: 10.1001/jamaneurol.2016.4237. [DOI] [PubMed] [Google Scholar]
- 13.Sicotte NL, Voskuhl RR, Bouvier S, Klutch R, Cohen MS, Mazziotta JC. Comparison of multiple sclerosis lesions at 1.5 and 3.0 Tesla. Investig Radiol. 2003;38:423–427. doi: 10.1097/01.RLI.0000065426.07178.f1. [DOI] [PubMed] [Google Scholar]
- 14.Stankiewicz JM, Glanz BI, Healy BC, Arora A, Neema M, Benedict RH, et al. Brain MRI lesion load at 1.5T and 3T versus clinical status in multiple sclerosis. J Neuroimaging. 2011;21:e50–e56. doi: 10.1111/j.1552-6569.2009.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu R, Tauhid S, Glanz BI, Healy BC, Kim G, Oommen VV, et al. whole brain volume measured from 1.5T versus 3T MRI in healthy subjects and patients with multiple sclerosis. J Neuroimaging. 2016;26:62–67. doi: 10.1111/jon.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58:840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 17.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/WNL.33.11.1444. [DOI] [PubMed] [Google Scholar]
- 18.Fischer JS, Rudick RA, Cutter GR, Reingold SC. The multiple sclerosis functional composite measure (MSFC): an integrated approach to MS clinical outcome assessment. National MS Society Clinical Outcomes Assessment Task Force. Mult Scler. 1999;5:244–250. doi: 10.1177/135245859900500409. [DOI] [PubMed] [Google Scholar]
- 19.Kalavathi P, Prasath VB. Methods on skull stripping of MRI head scan images–a review. J Digit Imaging. 2016;29:365–379. doi: 10.1007/s10278-015-9847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng X, Rosenthal R, Rubin DB. Comparing correlated correlation coefficients. Psychol Bull. 1992;111:172–175. doi: 10.1037/0033-2909.111.1.172. [DOI] [Google Scholar]
- 21.Winer BJ, Dr B, Michels KM. Statistical Principles in Experimental Design. 2. New York: McGraw-Hill; 1971. [Google Scholar]
- 22.Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, et al. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. NeuroImage. 2009;46:177–192. doi: 10.1016/j.neuroimage.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow N, Hwang KS, Hurtz S, Green AE, Somme JH, Thompson PM, et al. Comparing 3T and 1.5T MRI for mapping hippocampal atrophy in the Alzheimer's Disease Neuroimaging Initiative. AJNR Am J Neuroradiol. 2015;36:653–660. doi: 10.3174/ajnr.A4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kollia K, Maderwald S, Putzki N, Schlamann M, Theysohn JM, Kraff O, et al. First clinical study on ultra-high-field MR imaging in patients with multiple sclerosis: comparison of 1.5T and 7T. AJNR Am J Neuroradiol. 2009;30:699–702. doi: 10.3174/ajnr.A1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neema M, Arora A, Healy BC, Guss ZD, Brass SD, Duan Y, et al. Deep gray matter involvement on brain MRI scans is associated with clinical progression in multiple sclerosis. J Neuroimaging. 2009;19:3–8. doi: 10.1111/j.1552-6569.2008.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lassmann H, van Horssen J, Mahad D. Progressive multiple sclerosis: pathology and pathogenesis. Nat Rev Neurol. 2012;8:647–656. doi: 10.1038/nrneurol.2012.168. [DOI] [PubMed] [Google Scholar]
- 27.Frischer JM, Bramow S, Dal-Bianco A, Lucchinetti CF, Rauschka H, Schmidbauer M, et al. The relation between inflammation and neurodegeneration in multiple sclerosis. Brain. 2009;132:1175–1189. doi: 10.1093/brain/awp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison DM, Oh J, Roy S, Wood ET, Whetstone A, Seigo MA, et al. Thalamic lesions in multiple sclerosis by 7T MRI: Clinical implications and relationship to cortical pathology. Mult Scler. 2015;21:1139–1150. doi: 10.1177/1352458514558134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klawiter EC, Ceccarelli A, Arora A, Jackson JS, Bakshi S, Kim G, et al. Corpus callosum atrophy correlates with gray matter atrophy in patients with multiple sclerosis. J Neuroimaging. 2015;25:62–67. doi: 10.1111/jon.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C-Y, Liu HM, Chen SK, Chen YF, Lee CW, Yeh LR. Reproducibility of brain morphometry from short-term repeat clinical MRI examinations: a retrospective study. PLoS One. 2016;11:e0146913. doi: 10.1371/journal.pone.0146913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinohara RT, Oh J, Nair G, Calabresi PA, Davatzikos C, Doshi J, et al. Volumetric analysis from a harmonized multisite brain MRI study of a single subject with multiple sclerosis. AJNR Am J Neuroradiol. [Epub ahead of print]. 10.3174/ajnr.A5254. [DOI] [PMC free article] [PubMed]
- 32.Tsivgoulis G, Katsanos AH, Grigoriadis N, Hadjigeorgiou GM, Heliopoulos I, Kilidireas C, et al. The effect of disease modifying therapies on brain atrophy in patients with relapsing-remitting multiple sclerosis: a systematic review and meta-analysis. PLoS One. 2015;10:e0116511. doi: 10.1371/journal.pone.0116511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khoury SJ, Bakshi R. Cerebral pseudoatrophy or real atrophy after therapy in multiple sclerosis. Ann Neurol. 2010;68:778–779. doi: 10.1002/ana.22254. [DOI] [PubMed] [Google Scholar]
- 34.Ceccarelli A, Rocca MA, Pagani E, Colombo B, Martinelli V, Comi G, et al. Voxel-based morphometry study of grey matter loss in MS patients with different clinical phenotypes. NeuroImage. 2008;42:315–322. doi: 10.1016/j.neuroimage.2008.04.173. [DOI] [PubMed] [Google Scholar]
- 35.Benedict RH, Ramasamy D, Munschauer F, Weinstock-Guttman B, Zivadinov R. Memory impairment in multiple sclerosis: correlation with deep grey matter and mesial temporal atrophy. J Neurol Neurosurg Psychiatry. 2009;80:201–206. doi: 10.1136/jnnp.2008.148403. [DOI] [PubMed] [Google Scholar]
- 36.Popescu V, Schoonheim MM, Versteeg A, Chaturvedi N, Jonker M, Xavier de Menezes R, et al. Grey matter atrophy in multiple sclerosis: clinical interpretation depends on choice of analysis method. PLoS One. 2016;11:e0143942. doi: 10.1371/journal.pone.0143942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones BC, Nair G, Shea CD, Crainiceanu CM, Cortese IC, Reich DS. Quantification of multiple-sclerosis-related brain atrophy in two heterogeneous MRI datasets using mixed-effects modeling. Neuroimage Clin. 2013;3:171–179. doi: 10.1016/j.nicl.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chua AS, Egorova S, Anderson MC, Polgar-Turcsanyi M, Chitnis T, Weiner HL, et al. Handling changes in MRI acquisition parameters in modeling whole brain lesion volume and atrophy data in multiple sclerosis subjects: Comparison of linear mixed-effect models. Neuroimage: Clin. 2015;8:606–610. doi: 10.1016/j.nicl.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The full datasets used and analyzed during the current study are available from the corresponding author by request.


