Abstract
Thin filament length (TFL) is an important determinant of the force-sarcomere length (SL) relation of cardiac muscle. However, the various mechanisms that control TFL are not well understood. Here we tested the previously proposed hypothesis that the actin-binding protein nebulin contributes to TFL regulation in the heart by using a cardiac-specific nebulin cKO mouse model (αMHC Cre Neb cKO). Atrial myocytes were studied because nebulin expression has been reported to be most prominent in this cell type. TFL was measured in right and left atrial myocytes using deconvolution optical microscopy and staining for filamentous actin with phalloidin and for the thin filament pointed-end with an antibody to the capping protein Tropomodulin-1 (Tmod1). Results showed that TFLs in Neb cKO and littermate control mice were not different. Thus, deletion of nebulin in the heart does not alter TFL. However, TFL was found to be ~0.05 μm longer in the right than in the left atrium and Tmod1 expression was increased in the right atrium. We also tested the hypothesis that the length of titin’s spring region is a factor controlling TFL by studying the Rbm20ΔRRM mouse which expresses titins that are ~500 kDa (heterozygous mice) and ~1000 kDa (homozygous mice) longer than in control mice. Results revealed that TFL was not different in Rbm20ΔRRM mice. An unexpected finding in all genotypes studied was that TFL increased as sarcomeres were stretched (~0.1 μm per 0.35 μm of SL increase). This apparent increase in TFL reached a maximum at a SL of ~3.0 μm where TFL was ~1.05 μm. The SL dependence of TFL was independent of chemical fixation or the presence of cardiac myosin-binding protein C (cMyBP-C). In summary, we found that in cardiac myocytes TFL varies with SL in a manner that is independent of the size of titin or the presence of nebulin.
Keywords: Cardiac myocyte, Myofilament, Thin filament, Length regulation, Nebulin, Titin
1. Introduction
One of the hallmarks of the striated muscle sarcomere is its precise construction with centrally located thick filaments of well controlled length that overlap with thin filaments anchored to the Z-disks [1]. Thin filament length (TFL) varies in different muscle types and because TFL is an important determinant of force generation through its effect on the force-sarcomere length (SL) relation [2], mechanisms that control TFL need to be well understood. We studied two mechanisms that have been proposed to contribute to TFL control in the heart, one involving nebulin [3,4] and another titin [5]. Nebulin is a large protein (~800 kDa) that binds along the length of the actin filament of the sarcomere [6–8]. It is anchored by its COOH-terminal region in the Z-disk and its NH2-terminal region is located toward the pointed end of the thin filament [9]. Nebulin is likely to play multiple important roles in the structure and function of muscle [7,10–17], as revealed by mutations in nebulin that cause severe and debilitating nemaline myopathy [18,19].
Studies on a range of different skeletal muscles have shown that nebulin’s size and TFL are positively correlated, supporting that nebulin is important for TFL regulation [7]. In more recent work it was shown that the thin filament capping protein Tropomodulin-1(Tmod1) and the thin filament elongation factor Leimodin-2 (Lmod2) are also required for thin filaments to reach their mature length and that they regulate the length of a distal thin filament segment that is nebulin-free [5, 20–25]. Nebulin had long been assumed to be skeletal-muscle specific but studies on lower vertebrates have shown that nebulin can also be expressed at high levels in the heart [26]. Furthermore, studies on a conventional nebulin knockout (Neb KO) mouse provided evidence for expression of nebulin mRNA transcripts in the heart [27]. Although another study was unable to detect protein by Western blot [14], it is possible that small amounts of protein could have gone undetected. The notion that functionally relevant levels of nebulin exist in the heart is supported by RNA interference studies in cardiomyocytes that showed altered TFL when nebulin expression was suppressed [11]. It has been proposed that low levels of cardiac nebulin might regulate TFL by novel mechanisms in which a single nebulin molecule regulates the length of many thin filaments [4,28]. To critically test whether there is a role for nebulin in TFL control in cardiac muscle we made a cardiac-specific nebulin KO (αMHC Cre Neb cKO) and measured TFL as a function of sarcomere length (SL). We focused on left and right atrial tissues where nebulin expression has been shown to be most prominent [27]. We also examined Tmod1 and Lmod2 in these tissues. Overexpression of Tmod1 in the mouse heart leads to shorter thin filaments [29], consistent with increased pointed-end capping in these hearts. Recent studies showed that Lmod2 acts as a thin filament elongation factor that functions as an antagonist of Tmod1 [24,25].
The effect of titin on TFL was investigated as well. Titin molecules span from Z-disk to M-band regions of the sarcomere and their I-band spanning region functions as a molecular spring [30–32]. Alternative splicing results in size variants of titin’s spring region [33,34] and based on a TFL study in rabbit skeletal muscles that express various titin isoforms it was proposed that the length of titin’s spring region controls TFL [5]. To test this hypothesis we used Rbm20ΔRRM mice in which the RNA Recognition Motif of the splicing factor Rbm20 has been deleted [35]. As a result Rbm20ΔRRM mice express titins that are much longer than those in wild type mice. Hence by studying myocardium of Rbm20ΔRRM mice, we tested whether titin is a determinant of TFL in the heart.
Finally, we studied an unexpected phenomenon present in all tissue types and mouse models that we investigated, which is that TFL increases as sarcomeres were stretched. We examined whether this might have been caused by specimen preparation and studied unfixed specimen that were labeled in relaxing solution. Because the observed TFL increase with SL leveled off at the SL of ~3.0 μm, approximately where thin filament overlap with cMyBP-C ceases to exist, we tested the hypothesis that cMyBP-C causes an apparent TFL reduction in passive muscle, by opposing thin filament sliding into the A-band.
2. Methods
2.1. Animal models and tissue collection
All procedures were performed in compliance with institutional guidelines and had IACUC approval. A cardiac specific nebulin knockout mouse was produced by crossing the mouse in which loxP sites flank exon 3 of the nebulin gene (containing the start codon) [15] with a model that expresses Cre-recombinase under control of the cardiac specific αMHC promotor (tg(Myh6-cre)2182Mds/J from Jackson Laboratory). Details of the knockout strategy are provided in Figure S1A and B. Mice were studied that were heterozygous for αMHC-Cre and homozygous for the floxed nebulin allele and they are referred to as αMHC-Cre Neb cKO. In pilot experiments a MCK-Cre Neb cKO model was also studied which was obtained by crossing the floxed mouse with a mouse that expresses Cre under control of the muscle creatine kinase promotor (MCK), which causes deletion of nebulin in both skeletal muscle and the heart (tg(CKmmcre)5Khn/J from Jackson Laboratory). Rbm20ΔRRM mice were also used in which exons 6 and 7 are deleted from the Rbm20 gene, resulting in an in-frame deletion of the RNA recognition motif causing expression of aberrantly spliced large titin isoforms in the heart [35]. Mice (including littermate controls) were obtained by breeding parents that were heterozygous for the Rbm20ΔRRM allele. For details on the Rbm20ΔRRM model see [35]. Male mice homozygous for the cMyBP-C knockout mutation were also used. For details on this model see [36]. Mice were housed in a pathogen-free vivarium with 12 h light:12 h dark cycle and were studied at 2–3 mo of age, unless indicated otherwise. For tissue collection, mice were weighed, anesthetized with isoflurane and sacrificed by cervical dislocation. The hearts were rapidly excised and placed into a dish containing HEPES buffer ([in mmol/L] 133.5 NaCl, 5 KCl, 1.2 NaH2PO4, 1.2 MgSO4, 10 HEPES). All four chambers were removed, blotted and weighed separately.
2.2. Conscious echocardiography
Cardiac function was studied using echocardiography (Vevo2100, VisualSonics) on mice that were conscious (i.e., un-anesthetized). A standard short axis (M-mode) cine loop was recorded at the level of the papillary muscles to asses chamber dimensions posterior and anterior wall thickness, and cardiac function via fractional shortening and cardiac output. Data were analyzed in Vevo2100 software suite (VisualSonics).
2.3. Thin filament length measurement
The hearts were rapidly excised and placed in HEPES solution ([in mmol/L] 10 HEPES, 133.5 NaCl, 5 KCl, 1.2 NaH2PO4, 1.2 MgSO4) that contained protease inhibitors ([in mmol/L] 0.1 leupeptin, 0.1 E64, and 0.25 PMSF). For the studies on αMHC-Cre Neb cKO mice, the atrial tissue sections were permeabilized (skinned) in relaxing solution ([in mmol/L] 40 BES, 10 EGTA, 6.56 MgCl2, 5.88 Na-ATP, 1.0 DTT, 46.35 K-propionate, 15 creatine phosphate, pH 7.0) with protease inhibitors ([in mmol/L] 0.4 leupeptin, 0.1 E64, and 0.25 PMSF) containing 1% Triton X-100. Skinning was performed overnight on a 2D rocker at 4 °C and following washing in Triton-free relaxing solution the atria were cut open and pinned down flat in a dish while gently stretching the tissue in two dimensions. For the studies on the Rbm20ΔRRM model, LV papillary muscles was used in these studies because compared to atrial tissues, it is relatively easy to dissect muscle strips of uniform fiber direction. Muscles were dissected and placed in relaxing solution with 1% Triton X-100 and protease inhibitors (for solution composition, see above) for overnight on a 2D rocker at 4 °C. Skinned papillary muscles were rinsed and placed in a sylgard dish and both ends of the muscles were tied with sutures. Muscles were stretched by 7.5% slack length at a time for 3 times. For both tissue types (atria and papillary muscle) relaxing solution was replaced with 4% formaldehyde solution and muscles were fixed overnight. After fixation, muscles were washed with phosphate buffer saline (PBS) and embedded in Tissue-Tek O.C.T. compound (Ted Pella Inc.) and immediately frozen in 2-methylbutane-cooled in liquid N2 and then stored at −80 °C. The specimens were sectioned into 5 μm thick sections (Microm HM 550; Thermo Scientific) and placed on gelatin-coated coverglass. Fixed tissues were permeabilized again with 0.2% Triton X-100 in PBS for 20 min at room temperature on a light box, to bleach out the background fluorescence. Tissues were blocked with 2% bovine calf serum plus 1% normal donkey serum in PBS and incubated overnight with primary antibodies diluted in PBS. Tmod1 was detected using a polyclonal rabbit anti-Tmod1 antibody kindly provided by Dr. Carol Gregorio. F-actin was detected using Alexa Fluor 488-conjugated Phalloidin (A12379, Life Technologies). Alpha-actinin was detected using a mono-clonal mouse anti alpha-actinin antibody (A7811, Life Technologies). The tissues were washed with PBS for 30 min and incubated with secondary antibodies at room temperature in a dark box on 2D rocker for 2–4 h. The secondary antibodies were polyclonal Texas red-conjugated goat anti-rabbit IgG (T2767, Life Technologies) and polyclonal Alexafluor 350 Goat anti-mouse IgG (A21049, Life Technologies). Coverslips were mounted onto slides with Aqua Poly/Mount (Polysciences Inc.).
Some muscle preparations were also processed without formaldehyde fixation. Unfixed tissues were skinned overnight in relaxing solution as described above while others remained intact. Intact tissues were stretched in HEPES and skinned papillary muscles were stretched in relaxing solution. Tissues were then frozen in O.C.T as described above. To avoid protein degradation, extraction and activation of the un-fixed sections, a shortened immunofluorescence staining protocol was used and PBS was replaced with relaxing solution containing 30 mmol/L 2,3-Butanedione monoxime (BDM). Sections were permeabilized for 20 min, then blocked for 30 min. Primary incubation was for 3 h using the above described antibodies for both Tmod1 and alpha-actinin (in relaxing solution) followed by three 5 min washes, a 1 h secondary incubation, then three more 5 min washes. Coverslips were wet mounted on slides with relaxing solution and immediately imaged.
Images were captured using a Deltavision RT system (Applied Precision) with an inverted microscope (IX70; Olympus), a 100× (1.3 NA) objective, and a charge-coupled device camera (CoolSNAP HQ; Photo-metrics) using SoftWoRx 3.5.1 software (Applied Precision). The images were then deconvolved using SoftWoRx. An average of 10 fields were observed for each tissue section. Thin filament lengths and sarcomere lengths were obtained from deconvolved images of papillary muscles stained with fluorescently conjugated phalloidin and anti-Tmod antibodies. Deconvolved images were reopened in ImageJ (http://rsb.info.nih.gov/ij), then the 1D plot profile was determined along the myofibril direction. The plot profile was analyzed using fityk1.2.1 (http://fityk.nieto.pl). Well-defined peaks were fitted with Gaussian curves for Tmod and with a custom ‘rectangular Gaussian’ for phalloidin-stained images. In phalloidin images, a rectangle was fit to the phalloidin-stained region that was flanked by two half Gaussians (Fig. 1B). To account for actin overlap in the Z-disk which creates an increased intensity in the center of the rectangle, we developed a custom script designed for fityk that de-activates the center points within the rectangle fit. This improved the subsequent fit for the ‘rectangle +2 half Gaussian’ function. TFL was calculated as half the width of the rectangle plus half the width of the Gaussian fit at half height. SL was calculated from the distance between the centers of two adjacent rectangular Gaussian fits. In Tmod labeled images, TFL was calculated as half the distance between the two peak centers flanking either side of the Z-disk. SL was calculated as the sum of the distance between the two peak centers flanking either side of the Z-disk plus the average distance between both sets of adjacent peaks pairs centered on the M-line. Considering that it has been reported that only ~50% of the atrial cardiac cells contain nebulin transcript [27] we labeled atrial myocytes with anti-nebulin N-terminal antibodies, in an attempt to distinguish nebulin positive cells from nebulin negative cells. Mid-sarcomere labeling was extremely faint and hard to distinguish from background noise. (Parallel experiments on skeletal muscle fibers revealed very strong nebulin labeling, indicating that extreme weak labeling of atrial tissues was not due to technical problems.) TFLs were therefore analyzed in randomly selected cells and if TFL differences were to exist between cells they are expected to be represented in our data sets.
Fig. 1.
Thin filament length (TFL) measurement using deconvolution microscopy. A) Examples of αMHC-Cre Neb cKO myocardium triple labeled with tropomodulin1 (Tmod), phalloidin and α-actinin. B) Explanation of TFL measurement method using phalloidin-labeled sarcomeres. The densitometry profile was calculated along the length of the myofibril and a rectangle flanked on each side by a half Gaussian curve was fit to this profile (see Methods). In order to improve the goodness of fit of this rectangular-like equation, the increased density in the Z-disk region (due to thin filament overlap) was removed using an automated algorithm (see Methods). The width at half maximal height of the obtained best fit was taken as twice the TFL. C) The Tmod density peaks were fit with individual Gausian curves and TFL was defined as the distance between Gaussian peaks across the Z-disk divided by two. Sarcomere length (SL) was defined as indicated. (Note that the same SL was obtained as when using α-actinin to label Z-disks.) On this and other figures: cKO is αMHC-Cre Neb cKO.
2.4. Gel electrophoresis and Western blotting
Tissues were flash-frozen in liquid nitrogen and stored at −80 °C. Tissues were ground to a fine powder using glass pestles cooled in liquid nitrogen. After 20 min of at −20 °C, tissue was resuspended in a 1:1 mixture of an 8 M urea buffer [(in M) 8 urea, 2 thiourea, 0.05 Tris-HCl, 0.075 dithiothreitol, as well as 3% SDS and 0.03% bromophenol blue, pH 6.8] and 50% glycerol with protease inhibitors [(in mM) 0.04 E-64, 0.16 leupeptin, and 0.2 PMSF]. The solutions were mixed at 60 °C for four minutes, followed by ten minutes of incubation at the same temperature. Cell debris was removed via centrifugation at 13,000 rpm and the supernatant was divided into smaller aliquots and flash frozen for storage at −80 °C. Tmod1 (E-Tmod) and Lmod2 expression was quantified using Western blot. 10% acrylamide gels were run at 100 V for 1.5 h. Following this, they were transferred onto Immobilon-P PVDF membranes (Millipore, Billerica, MA, USA) using a semi-dry transfer unit (Bio-Rad, Hercules, CA, USA) for 2.5 h at 1.3 mA/cm2. Membranes were briefly stained with Ponceau S and scanned to quantify transferred protein levels. Following removal of the stain, membranes were incubated with primary antibody at 4 °C overnight. Primary antibodies were anti-Tmod1 (provided by Dr. Gregorio) and anti-Lmod2 (E-13, Santa Cruz Biotechnology)). Normalization was done through comparisons of ratios (Lmod2:Tmod1 and Right Atria:Left Atria) in left and right atrial samples from the same mouse. Myosin heavy chain (MHC) levels were determined from the Ponceau S stained membrane to account for loading differences. IR Western blots were analyzed using Odyssey Infrared Imaging System (Li-Cor Biosciences, NE, USA). Ponceau S images were analyzed with One-D scan EX (Scanalytics Inc., Rockville, MD, USA).
2.5. Statistics
Statistical analysis was performed in Graphpad Prism (GraphPad Software, Inc.), and included linear regression analysis and testing 1) whether slopes of linear fits are significantly different from zero, 2) whether slopes of different line fits are different from each other, and 3) if not, whether line offsets are different (Figs. 2, 3, 5, 6, S3, S4, and Table 1A). Results from 7 and 8 control muscles and 5 and 6 αMHC Cre Neb cKO muscle (one muscle per mouse) for right and left atrial studies (Figs. 2–4), respectively. Results obtained from 6 mice per genotype for the Rmb20 study (Fig. 5). Data from 8 control and 6 αMHC Cre Neb cKO mice in Table S1 and from 13 mice per genotype in Fig. S2. A one-way ANOVA with a post hoc analysis with a multiple testing correction (Tukey) was performed to assess differences in Fig. S1C and D, Table S1 and Table 1B; a two-way ANOVA was used in Fig 4. In Fig. S2 a paired t-test was used. Results are shown as mean ± SEM; p < 0.05 taken as significant. Symbols on figures: *p < 0.05, **p < 0.01, ***p < 0.001 versus results from control tissues.
Fig. 2.
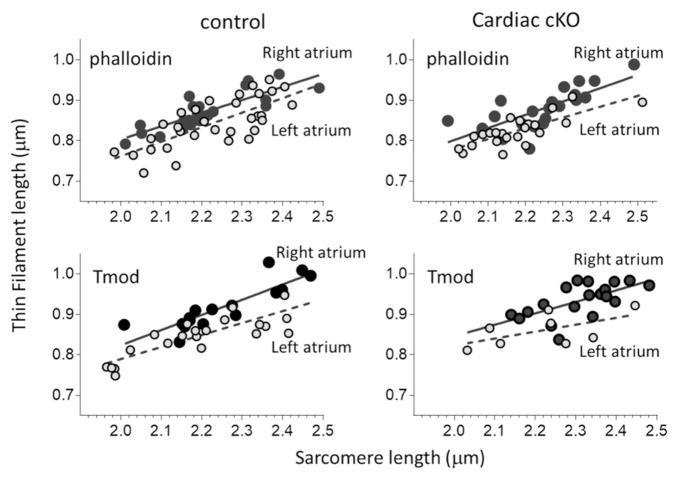
TFLs of αMHC Neb cKO (gray symbols) and control mice (black symbols) are indistinguishable. TFL in left atria (left panels) and right atria (right panels) obtained by phalloidin staining (top panels) or Tmod staining (bottom panels) were fit by linear regression lines. Regression lines of control and Neb cKO mice are indistinguishable. Also note that in all data sets TFL increases with SL.
Fig. 3.
TFL is longer in right (black symbols) than left (gray symbols) atria. TFL were measured by phalloidin (top panels) or Tmod (bottom panels) in control and αMHC Neb cKO atrial myocardium; TFL values were plotted versus SL. The slopes of all linear regression lines are significantly different from zero (p < 0.001) and do not differ between right and left atria. However, the y-axis offsets of the linear fits are significantly different between left and right atria. Note that this is the same data set as used in Fig. 2. (For details, see Table 1A.)
Fig. 5.
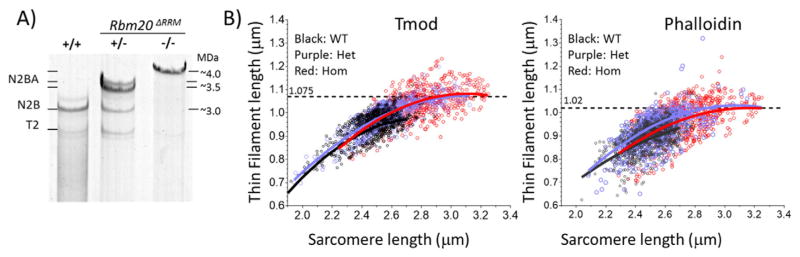
TFL in LV of WT, Het and Hom Rbm20ΔRRM mice. A) Compared to wildtype (+/+) mice, Rbm20ΔRRM mice express giant titin isoforms that are ~0.5 MDa larger in Het (±) and ~1.0 MDa larger in Hom (−/−) mice. B) LV muscle strips were stretched, labeled with Tmod and phalloidin, and TFL was measured in all 3 genotypes. Results were fit with second order polynomial equations. (Second order polynomial equations fit the data best, for a more limited SL range (as in Figs. 2, 3 and 6) linear regression lines were more suitable.) See text for details.
Fig. 6.
SL dependence of TFL in unfixed LV tissue and cMyBP-C KO mice. A) TFL measurements from intact unfixed LV papillary muscles are shown overlaid with those of fixed tissues (same data as Fig. 5B). B) Measurements in wildtype and cMyBPC KO mice. All linear regression fits have slopes that are significantly different from zero (p < 0.0001). In each panel linear regression fits are not different from each other (panel A: p = 0.11; B: 0.65). TFL values were obtained from Tmod labeling.
Table 1A.
Linear regression analysis of TFL (μm) vs SL (μm) relations.
| Phalloidin | p-Value for comparison between: | Tmod | p-Value for comparison between: | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| LA vs RA | Ctrl vs cKO | LA vs RA | Ctrl vs cKO | ||||
| CTRL: | Linear regress. fit | Slope/offset | Slope/offset | CTRL: | Linear regress. fit | Slope/offset | Slope/offset |
| LA | TFL = 0.32 SL + 0.15 | 0.7/b0.001 | NS/NS | LA | TFL = 0.30 SL + 0.20 | 0.3/b0.001 | NS/NS |
| RA | TFL = 0.36 SL + 0.42 | NS/NS | RA | TFL = 0.38 SL + 0.07 | NS/NS | ||
| αMHC Cre- Neb cKO: | αMHC Cre- Neb cKO: | ||||||
| LA | TFL = 0.322 SL + 0.14 | 0.5/b0.001 | N/S | LA | TFL = 0.17 SL + 0.48 | 0.4/b0.01 | NS/NS |
| RA | TFL = 0.27 SL + 0.25 | N/S | RA | TFL = 0.28 SL + 0.28 | NS/NS | ||
Fig. 4.
Comparison of TFLs in left versus right atria. TFL values (mean ± SEM) over the SL range 2.1–2.35 μm. Results obtained using phalloidin (left) or Tmod (right) labeling are shown. A two-way ANOVA (chamber type (left and right atria) and genotype (Ctrl and αMHC-Cre Neb cKO)) revealed significantly longer TFLs in the right atrium compared to left atrium irrespective of mouse model or method to measure TFL (phalloidin and Tmod). However, note that Tmod-based mean values exceed those of the phalloidin staining method (see Discussion for details). Note that this is the same data set as used in Fig. 2.
Table 1B.
TFL in atria of control (Ctrl) and αMHC cNeb KO mice.
| Phalloidin | Mean ± SEM values in SL range 2.1–2.35 μm: | Tmod | Mean ± SEM values in SL range 2.1–2.35 μm: | ||||
|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||
| Left atrium | SL (μm) | TFL (μm) | n | Left atrium | SL (μm) | TFL (μm) | n |
| Ctrl | 2.23 ± 0.02 | 0.84 ± 0.01 | 31 | Ctrl | 2.24 ± 0.04 | 0.86 ± 0.01 | 17 |
| cKO | 2.20 ± 0.02 (ns) | 0.83 ± 0.01 (ns) | 19 | CKO | 2.22 ± 0.04 (ns) | 0.86 ± 0.01 (ns) | 12 |
| Right atrium | Right atrium | ||||||
| Ctrl | 2.21 ± 0.03 | 0.87 ± 0.01 | 20 | Ctrl | 2.22 ± 0.03 | 0.91 ± 0.01 | 14 |
| cKO | 2.23 ± 0.02 (ns) | 0.87 ± 0.01 (ns) | 20 | CKO | 2.25 ± 0.02 (ns) | 0.92 ± 0.01 (ns) | 13 |
t-test; ns: non-significant vs Ctrl. No significant differences in mean SL of all 8 groups.
3. Results
3.1. αMHC Cre Neb cKO mouse model
To address the role of nebulin in determining cardiac muscle TFL, a mouse model was used in which exon 3 of the nebulin gene (containing the start codon) was floxed [15]. In pilot studies the floxed mouse was crossed with a mouse that expresses Cre recombinase under control of the muscle creatine kinase promoter (MCK Neb cKO), which causes deletion of nebulin in both skeletal and cardiac muscles (Figure S1 A and B). Whole heart weights of the MCK Neb cKO mice were significantly reduced relative to that of age-matched control mice (Fig. S1E, left). However, because the MCK Neb cKO model shows severe skeletal muscle myopathy [15] it was important to exclude systemic changes caused by the skeletal muscle myopathy. We therefore also crossed the floxed mouse with the cardiac-specific αMHC Cre mouse and studied offspring heterozygous for the αMHC Cre transgene and homozygous for the nebulin floxed allele (αMHC Neb cKO). These mice were born at the expected Mendelian ratio (in our breeding scheme 1 in 4) indicating that there is no embryonic lethality. Furthermore, αMHC Neb cKO mice reached adult age without an outwardly visible phenotype. Western blot analysis of atrial tissues revealed a low level of nebulin in control mice (<0.2% of levels in skeletal muscle) that was absent in αMHC Neb cKO mice (Fig. S1C and D). No nebulin could be detected in the ventricles of either Ctrl or αMHC Neb cKO mice (results not shown), consistent with previous studies [14]. The αMHC Neb cKO KO mice had a normal heart weight (Fig. S1E, right) and normal individual chamber weight (Fig. S1F). Echocardiography showed that chamber dimensions and systolic function were normal, e.g., the ejection fraction was 84.5 ± 2.5% in control and 83.5 ± 2.7% in αMHC Neb cKO mice (Table S1). Thus, by using a cardiac-specific nebulin KO, a skeletal muscle-induced cardiac phenotype was avoided and the effect of specifically deleting nebulin in the heart on TFL can be studied.
3.2. Thin filament length (TFL) measurement in αMHC Neb cKO mice
Deconvolution optical microscopy was used on sections of stretched atrial myocardium dissected from αMHC Neb cKO KO and control mice. Tissues were triple labeled using AlexaFluor488-conjugated phalloidin to detect F-actin and with anti-Tmod1 and anti-α-actinin antibodies. Left and right atria of Ctrl and αMHC Neb cKO mice showed clear labeling with the expected localizations for Tmod1 and α-actinin, see Fig. 1A. Using custom-written algorithms (Methods), we measured TFL and sarcomere length (SL) using phalloidin and Tmod labeled sarcomeres (Fig. 1B and C). We plotted TFL against SL for all 4 tissue types (left and right atria, Neb cKO and control) and superimposed Neb cKO and control data in Fig. 2 and left and right atrial data in Fig. 3. A striking observation is that in all data sets TFL increases with SL, an effect seen with both phalloidin and Tmod TFL measurement methods. All linear regression lines had slopes that were significantly different from zero. Furthermore, the slopes were indistinguishable when comparing Ctrl vs Neb cKO or left vs right atria (see Table 1A). Based on the slope values, increasing SL by 0.35 μm (which approximately occurs during diastole [37]) increases measured TFL by ~0.1 μm.
Linear regression lines were indistinguishable when comparing Neb cKO and control tissues (Fig. 2 and Table 1A) both as far as their slopes and offsets are concerned, showing that nebulin has no effect on TFL. The TFL vs SL regression lines had offsets that were significantly larger in the right vs left atria (Fig. 3 and Table 1A). This indicates that TFL is longer in right than in left atria. To explore this further, the mean SL and mean TFL in the SL range of 2.1–2.35 μm were determined. Within this range, the mean SL values of all studied groups were indistinguishable (Table 1B). TFL values were compared using a two-way ANOVA (independent variables: chamber type (left and right atria)) and genotype (Ctrl and Neb cKO)). The two-way ANOVA also showed that there was no genotype effect, confirming our conclusion from the linear regression analysis (above) that the cardiac-specific deletion of nebulin does not affect TFL. However, the analysis revealed significantly longer TFLs in the right atrium compared with the left atrium when staining for either F-actin or Tmod (Fig. 4), with TFL ~50 nm longer in the right atrium. This finding might be functionally relevant as a longer TFL will right-shift the force-SL relation [2] and allow the right chamber to accept larger blood volumes while still developing high force levels. Whether longer thin filaments are universal for all of the right side of the heart requires TFL measurements in the right ventricle, and is an interesting question for future study.
The expression levels of Tmod1 and Lmod2, two actin-binding proteins that have been proposed to function antagonistically in TFL control (see Introduction), were also studied. A simple explanation for the TFL difference between the two atrial chambers is that the Tmod1/Lmod2 ratio is reduced in the chamber with the longer TFL (right atrium). However, contrary to this expectation, Tmod1 expression was increased in the right atrium relative to the left atrium as was the Tmod1/Lmod2 expression ratio (Fig. S2). This suggests that TFL control in cardiac muscle is more complex than setting the expression ratio of capping (Tmod1) to elongation (Lmod2) factor.
3.3. Thin filament length (TFL) measurement in Rbm20ΔRRM mice
It has been hypothesized that titin plays a role in TFL regulation, based on a positive correlation observed between the size of titin and TFL in a range of skeletal muscles [5]. We tested this hypothesis by measuring TFL in LV myocardium of Rbm20ΔRRM mice that are deficient in the titin splicing factor Rbm20 [35]. Heterozygous Rbm20ΔRRM mice express ~3.5 MDa titin in the heart and homozygous Rbm20ΔRRM mice ~4.0 MDa titin, as opposed to ~3.0 MDa in wildtype mice (Fig. 5A). Thus Rbm20ΔRRM mice are ideal for testing the effect of titin size on TFL in the heart. Similar to in the atrial study, TFL increased with SL, in all 3 Rbm20ΔRRM genotypes, as assessed using either staining method (Fig. 5B). Despite stretching tissues to the same relative extent (Methods), SL in LV tissue from heterozygous and homozygous mice were longer than in WT tissues, which can be explained by their longer slack SL (SL at zero stretch) [35]. The longer SL range of Het and Hom mice revealed that TFL does not continue to rise but instead appears to level off. Based on a second order polynomial fit, a maximum is reached at a SL of ~3.0 μm with a TFL of 1.075 μm in Tmod labeled sarcomeres and 1.02 μm in phalloidin labeled sarcomeres (Fig. 5B).
We also performed a linear regression analysis using a SL range that was restricted to a maximal SL of 2.8 μm to ensure that all 3 Rbm20ΔRRM genotypes were well sampled. All linear regression lines had slopes that were significantly different from zero (p < 0.0001) and the slope values (in μm TFL per μm SL) were for Tmod 0.28, 0.32, 0.30 and for phalloidin 0.25, 0.28, 0.29 in WT, Het and Hom mice, respectively. This is consistent with results from atrial myocytes discussed above. By superimposing the atrial results (above) with the LV results it is clear that TFL in LV and LA is the same and that the RA has a longer TFL than LA or LV (Fig. S3).
The mean LV TFL in the SL range of 2.1–2.35 μm and in the 2.3–2.5 μm range are given in Table 2. Within these ranges no SL differences existed amongst all genotypes, making it possible to statistically compare the TFL results. Comparing TFL amongst the 3 studied genotypes revealed only a significant difference in the phalloidin-based TFL in the 2.3–2.5 μm SL range (Table 2). However, this difference was small (TFL was 0.014 μm longer in Het than in WT mice) and was not seen in the Het Tmod data set. Additionally no TFL difference was found when comparing Hom mice to WT mice or Hom to Het mice.
Table 2.
TFL in LV of Rbm20ΔRRM mouse.
| Mean ± SEM SL and TFL values in SL range 2.1–2.35 μm: | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Tmod | SL (μm) | TFL (μm) | n | Phalloidin | SL (μm) | TFL (μm) | n |
| WT | 2.237 ± 0.006 | 0.864 ± 0.005 | 82 | WT | 2.246 ± 0.006 | 0.825 ± 0.006 | 72 |
| Het | 2.248 ± 0.011(ns) | 0.867 ± 0.011 (ns) | 27 | Het | 2.249 ± 0.008 (ns) | 0.812 ± 0.013 (ns) | 26 |
| Hom | 2.262 ± 0.028 (ns) | 0.848 ± 0.0011 (ns) | 8 | Hom | ND | ND | |
| Mean ± SEM SL and TFL values in SL range 2.35–2.5 μm: | |||||||
|
| |||||||
| Tmod | SL (μm) | TFL (μm) | n | Phalloidin | SL (μm) | TFL (μm) | n |
|
| |||||||
| WT | 2.432 ± 0.002 | 0.946 ± 0.003 | 295 | WT | 2.430 ± 0.002 | 0.881 ± 0.003 | 339 |
| Het | 2.441 ± 0.003 (ns) | 0.947 ± 0.003 (ns) | 189 | Het | 2.424 ± 0.003 (ns) | 0.895 ± 0.003** | 309 |
| Hom | 2.442 ± 0.009 (ns) | 0.937 ± 0.0134 (ns) | 31 | Hom | 2.431 ± 0.006 (ns) | 0.883 ± 0.0121 (ns) | 62 |
p < 0.01 vs WT; ns: non-significant vs WT (ANOVA with Tukey’s multiple testing correction).
3.4. SL dependence of TFL
We studied the SL dependence of TFL in greater detail because the phenomenon is not expected by the sliding filament model of striated muscle that assumes constant thin and thick filament lengths [38]. Because fixation and specimen preparation is considered to cause myofilament shrinkage in electron microscopy studies [1] we tested whether this might also occur in optical microscopy. Experiments were performed on tissues that were not fixed and that were kept in relaxing solution during the full labeling protocol. It was found that measured TFL increased with SL in un-fixed tissues in a manner that was indistinguishable from that in fixed tissues (Fig. 6A), suggesting that the phenomenon is not an artifact of fixation or any other aspect of specimen preparation. Because the TFL vs SL relation leveled off at a SL of ~3.0 μm, where thin filament overlap with cMyBP-C ceases to exist (see Discussion), we next hypothesized that cMyBP-C exerts a force on the thin filament that opposes sliding into the C-zone of the sarcomere. Such force could result in a ‘waviness/crumpling’ of thin filaments giving rise to thin filaments that appear shorter when measured from Z-disk to pointed end. To test this hypothesis we studied the SL dependence of TFL in a cMyBP-C KO mouse model [36] where any opposing force due to cMyBP-C will be absent and thus TFL is predicted to be independent of SL. However, results show that in cMyBP-C KO mice TFL increased with SL in a manner indistinguishable from that of wildtype mice (Fig. 6B).
4. Discussion
The primary results from this study demonstrate for the first time that deleting nebulin specifically from the heart or altering the size of titin do not affect TFL in the cardiac sarcomere. Additional novel findings are that TFL is longer in the right atrium than in the left atrium and that TFL is not independent of SL but instead increases as sarcomeres are stretched. This phenomenon was evident in atrial and ventricular myocytes of all mouse models that were studied and was observed in both fixed and unfixed tissues.
TFL was measured using 2 distinct labeling methods, phalloidin to stain filamentous actin and anti-Tmod1 antibodies to label the pointed end of the thin filament. The two methods do not give TFL values that are exactly the same. In each of the 9 data sets that we collected TFL values based on Tmod localization exceeded that of values based on phalloidin staining (Tables 1B and 2) by on average 44 ± 5 nm. TFL values as measured by Tmod labeling were also reported as longer than TFL values determined by phalloidin-labeling in studies of cardiac myocytes by other investigators (e.g., 46 nm longer Tmod values in Littlefield and Fowler [39]). The phalloidin measurement is based on detecting the width at half maximal height of the filamentous actin stained region and reflects the mid-point of the TFL range that exists in the sarcomere (Fig. 1B). The fact that Tmod1 values exceed phalloidin values might mean that phalloidin does not bind the terminal actin molecules at the thin filament pointed ends. Alternatively Tmod may not interact with all thin filaments equally but preferentially cap those at the outer limit of the TFL range. Despite the fact that phalloidin and Tmod 1 results are not identical, the differences are small, and the conclusions of our studies are independent of the labeling method utilized.
Both labeling methods employed show a distinct SL dependence of TFL that has not been reported previously. TFL is ~0.8 μm at a SL of 2.0 μm and as sarcomeres are stretched, TFL increases initially linearly with SL and then levels off to reach a TFL maximum of ~1.05 μm (Fig. 5B). A non-constant TFL is also seen in cultured neonatal myocytes with a TFL vs SL slope similar to that of the adult mouse tissues (Fig. S4). Thus, the phenomenon is likely to be universal. The non-constant TFL complicates the comparison of TFL in different tissues and necessitates measurement of the TFL-SL relation and their statistical comparison (see also caption of Fig. S4).
The mechanism that underlies the SL dependence of TFL is unknown. It seems unlikely that it involves de novo assembly of F-actin since the phenomenon is observed in skinned muscle where G-actin is likely absent. The correlation between TFL and SL does not prove ‘cause and effect’, for example TFL might be constant but some sarcomeres always have long TFLs and these sarcomeres tend to have a long SL and other sarcomeres always have short TFLs and these sarcomere tend to have a short SL. In this interpretation when the muscle is stretched, the average TFL should not change. This is contrary to the TFL increase that we found in stretched muscle (until SL reaches ~3.0 μm). Additionally, when muscles are stretched to a long SL a subpopulation of long sarcomeres with short TFLs is not detected. Although we consider it more likely that in all sarcomeres TFL is similar, future experiments in which TFL is tracked in sarcomeres while the sarcomeres are stretched are needed to be conclusive. The increase in TFL in stretched sarcomeres is unlikely due to thin filament compliance that has been reported in X-ray diffraction studies [40], because the effect of compliance on TFL is relatively small. During maximal activation it causes thin filaments to stretch by ~0.25% (or ~3 nm per thin filament), almost two order of magnitude less than the TF length increase that we observed by passive stretching muscle (Fig. 5). Whether thin filament misalignment that gets worse with stretch results in an apparent increase in TFL was evaluated from the width of the Gaussian fits to the Tmod peaks. The half width at half maximal height (HWHM) showed only a minor increase with increase in SL for the cKO and a minor reduction for the Ctrl (Fig. S5). Thus, filament misalignment, as assessed from the Tmod peak width, is unlikely to explain our findings. Instead we propose that average TFL is constant in different sarcomeres (~ 1.05 μm, reached at SL 3.0 μm), and that the shorter TFL measured at SLs < 3.0 μm is due to thin filament ‘crumpling’ that arises because of resistance to thin filament sliding deep into the A-band region of the sarcomere. That such resistance exists is supported by earlier structural studies on passive skeletal muscle fibers that were forcibly shortened to below their slack length where thin filaments failed to slide into the bare zone of the A-band, causing them to become wavy and bend [41]. We noted that TFL is reduced only at SLs < ~3.0 μm, a length range at which thin filaments overlap with the C-zone, the cardiac myosin binding protein C (cMyBP-C) containing region of the sarcomere [42,43]. Because cMyBP-C has been reported to interact with actin [42] we tested whether cMyBP-C impedes sliding into the C-zone, resulting in a TFL that appears shorter. However, this mechanism is unlikely because TFL was also reduced at short SLs in the cMyBP-C KO mouse (Fig. 6B). Clearly, future structural research is required to gain insights into the mechanistic basis of the reduced TFL in short sarcomeres. Could the reduction of TFL at short SLs be functionally relevant during contraction? Assuming that the thin filament behavior that we report persists during calcium activation, a reduction in TFL is expected to left-shift the force-SL relation and result in higher forces on the ascending limb of this relation [2]. Considering that the heart functions largely on the ascending limb [44] of the force-SL relationship, a reduced TFL in short sarcomeres might thus have a beneficial effect on force development.
4.1. Increasing the size of titin does not alter TFL in the heart
The I-band spanning region of titin can vary greatly in size due to alternative splicing that involves the splicing factor Rbm20 [45]. Recently, a correlation between titin size and TFL was reported based on comparisons of multiple rabbit skeletal muscles [5], and it was suggested therefore that titin may play a role in TFL regulation. Titin could affect TFL directly (titin is known to interact with actin [46–49]) or indirectly, for example by altering the slack sarcomere length. We tested the proposal that titin affects TFL in the heart by using the Rbm20ΔRRM mouse model in which altered splicing results in titin molecules that are much larger than in wild type mice [35]. The results showed that TFL is independent of the size of titin (Fig. 5, Table 2). Since the physiological working range in the heart is shifted to longer SLs in the Rbm20ΔRRM mouse model [35], these findings also indicate that thin filament poly-merization is not affected by the proximity of its pointed end to the thick filament bare zone. Our findings are consistent with a recent study in a rat model with a spontaneous mutation in Rbm20 in which titin size and TFL were studied in skeletal muscle and no correlation between these two parameters was found [50]. Thus, increasing the size of titin has no effect on TFL in cardiac or skeletal muscle.
4.2. Nebulin does not play a role in TFL regulation in the heart
Nebulin is well suited to play an essential role in TFL control as it is a long filamentous protein that consists of ~175 actin-binding modules [6,7,51,52]. A role for nebulin in TFL regulation is supported by an earlier study on skeletal muscles in which TFL was found to correlate with the size of nebulin [7] and by recent studies that showed that deletion of nebulin from skeletal muscle results in TFLs that are on average shorter than in WT mice [7,10,11]. Whether nebulin plays a similar role in cardiac muscle has remained unclear. Although it is well recognized that expression of nebulin in mammalian cardiac muscle is low, there is evidence for higher levels of nebulin transcript in atria [27] and we found that indeed atria do express a small amount of nebulin protein (<0.2% of that in skeletal muscle). It has been suggested that small amounts of nebulin might be functionally relevant, for example if single nebulin molecules were to regulate the length of many thin filaments [4,28]. To address whether nebulin plays a role in TFL regulation in the heart, we made a cardiac-specific nebulin KO, the Neb cKO, in which nebulin deletion was directed by the αMHC promoter. These cardiac-specific nebulin knockout mice have normal cardiac function as measured by echocardiography and normal chamber weights (Table S1, Fig. S1F). TFL length measurements were similar in Neb cKO and control mice (Figs. 2 and 4, Table 1A and 1B) indicating that unlike in skeletal muscle, nebulin does not play a role in TFL regulation in the heart. Consistent with this notion is that mutations in the nebulin gene are known to cause a severe skeletal muscle dysfunction that underlies nemaline myopathy, with cardiac involvement in only rare cases [18,53].
This poses the question, why does nebulin play an essential role in skeletal muscle but not in cardiac muscle? To address this question it is helpful to evaluate the changes that occurred in the heart during evolution. High levels of a full-length nebulin are expressed in cardiac muscle of agnathans (a primitive class of jawless fish that is considered a mammalian precursor) and it is thought that during evolution the much smaller nebulette has evolved from this cardiac nebulin [26]. Nebulette is a cardiac-specific protein associated with the Z-disk that is comprised of a domain structure similar to the C-terminal (Z-disk) region of nebulin [54–56]. Thus, during vertebrate evolution the function of nebulin in the thin filament region of the cardiac sarcomere (i.e., outside the Z-disk) was present in agnathans but then was lost. It is interesting that agnathan sarcomeres have clear H-zones [26], suggesting that their TFL range is narrow, a feature that is also present in skeletal muscle but absent in mammalian cardiac muscle. For example, a recent electron tomography study revealed a 70 nm TFL range in skeletal muscle and 140 nm in mouse cardiac muscle [57]. A narrow TFL range is expected to result in a force-SL relation that has a well-defined plateau with sharp corners, whereas a broad TFL range will give rise to a less well defined force-SL relation with a plateau that is more ‘rounded’ and a longer optimal SL where maximal force is produced. Although this feature seems subtle, it might be beneficial considering that the heart functions largely on the ascending limb of the force-SL relation and extending this ascending limb to longer SLs might be beneficial to the Frank-Starling mechanism of the heart [44,58]. Additionally, it is also worth considering that nebulin not only impacts TFL but also the activation level of the thin filament during contraction. Studies in Neb KO and control mice have shown that during contraction of skeletal muscle the fraction of force generating crossbridges is much higher when nebulin is present, through an effect of nebulin on calcium sensitivity and crossbridge cycling kinetics [12,15,16]. This function of nebulin is expected to have been lost during evolution of the vertebrate heart. Although it might appear counterintuitive, this loss might actually have conferred an advantage to the heart. A low base level of activation opened up avenues for increasing thin filament activation levels acutely and as needed, such as through beta-adrenergic stimulation during the ‘fight or fiight’ response [59,60]. In contrast skeletal muscles can regulate force levels by varying the fraction of its fibers that are active [60]. It is less dependent therefore on myofilament-based force regulation and it maintained the high base level of thin filament activation derived from nebulin. Thus we propose that the loss of full-length nebulin during evolution of the mammalian heart is a case of ‘less is more’. This loss smoothed and shifted the peak of the force-SL curve to longer SL and made a wide range of rapidly tunable activation levels possible, so that when the needs of the circulatory system change, the output of the heart can be acutely matched.
Supplementary Material
Acknowledgments
We are grateful to Drs. Tom Doetschman and Teodora Georgieva in the GEMM core for gene targeting mouse services, to Dr. Sabine van Dijk for breeding cMyBP-C KO mice, to Drs. Danielle Buck and John E. Smith for breeding and genotyping of Neb cKO mice, Anne Wang for preparing neonatal cardiomyocyte cultures and Rachel Mayfield for TFL measurements of neonatal cardiomyocytes. We are also grateful to Dr. Carol Gregorio for antibody support and critical reviewing of the manuscript. This work was supported by National Institutes of Health [5R01AR053897 (HG), RO1 HL062881 (HG), R01 HL080367 (SPH), RO1HL123078 (CTP)], Muscular Dystrophy Association [295195], and a Foundation Building Strength.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.yjmcc.2016.04.013.
References
- 1.Sosa H, Popp D, Ouyang G, Huxley HE. Ultrastructure of skeletal muscle fibers studied by a plunge quick freezing method: myofilament lengths. Biophys J. 1994;67:283–292. doi: 10.1016/S0006-3495(94)80479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Granzier HL, Akster HA, Ter Keurs HE. Effect of thin filament length on the force-sarcomere length relation of skeletal muscle. Am J Phys. 1991;260:C1060–C1070. doi: 10.1152/ajpcell.1991.260.5.C1060. [DOI] [PubMed] [Google Scholar]
- 3.Labeit S, Ottenheijm CA, Granzier H. Nebulin, a major player in muscle health and disease. FASEB J. 2011;25:822–829. doi: 10.1096/fj.10-157412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horowits R. Nebulin regulation of actin filament lengths: new angles. Trends Cell Biol. 2006;16:121–124. doi: 10.1016/j.tcb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Castillo A, Nowak R, Littlefield KP, Fowler VM, Littlefield RS. A nebulin ruler does not dictate thin filament lengths. Biophys J. 2009;96:1856–1865. doi: 10.1016/j.bpj.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin JP, Wang K. Nebulin as a giant actin-binding template protein in skeletal muscle sarcomere. Interaction of actin and cloned human nebulin fragments. FEBS Lett. 1991;281:93–96. doi: 10.1016/0014-5793(91)80366-b. [DOI] [PubMed] [Google Scholar]
- 7.Kruger M, Wright J, Wang K. Nebulin as a length regulator of thin filaments of vertebrate skeletal muscles: correlation of thin filament length, nebulin size, and epitope profile. J Cell Biol. 1991;115:97–107. doi: 10.1083/jcb.115.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bang ML, Chen J. Roles of nebulin family members in the heart. Circ J. 2015;79:2081–2087. doi: 10.1253/circj.CJ-15-0854. [DOI] [PubMed] [Google Scholar]
- 9.McElhinny AS, Kazmierski ST, Labeit S, Gregorio CC. Nebulin: the nebulous, multifunctional giant of striated muscle. Trends Cardiovasc Med. 2003;13:195–201. doi: 10.1016/s1050-1738(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 10.Labeit S, et al. Evidence that nebulin is a protein-ruler in muscle thin filaments. FEBS Lett. 1991;282:313–316. doi: 10.1016/0014-5793(91)80503-u. [DOI] [PubMed] [Google Scholar]
- 11.McElhinny AS, Schwach C, Valichnac M, Mount-Patrick S, Gregorio CC. Nebulin regulates the assembly and lengths of the thin filaments in striated muscle. J Cell Biol. 2005;170:947–957. doi: 10.1083/jcb.200502158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bang ML, et al. Nebulin plays a direct role in promoting strong actin-myosin interactions. FASEB J. 2009;23:4117–4125. doi: 10.1096/fj.09-137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gokhin DS, Bang ML, Zhang J, Chen J, Lieber RL. Reduced thin filament length in nebulin-knockout skeletal muscle alters isometric contractile properties. Am J Phys Cell Phys. 2009;296:C1123–C1132. doi: 10.1152/ajpcell.00503.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witt CC, et al. Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J. 2006;25:3843–3855. doi: 10.1038/sj.emboj.7601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, et al. Nebulin deficiency in adult muscle causes sarcomere defects and muscle-type-dependent changes in trophicity: novel insights in nemaline myopathy. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandra M, et al. Nebulin alters cross-bridge cycling kinetics and increases thin filament activation: a novel mechanism for increasing tension and reducing tension cost. J Biol Chem. 2009;284:30889–30896. doi: 10.1074/jbc.M109.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Root DD, Wang K. High-affinity actin-binding nebulin fragments influence the actoS1 complex. Biochemistry. 2001;40:1171–1186. doi: 10.1021/bi0015010. [DOI] [PubMed] [Google Scholar]
- 18.Lehtokari VL, et al. Mutation update: the spectra of nebulin variants and associated myopathies. Hum Mutat. 2014;35:1418–1426. doi: 10.1002/humu.22693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottenheijm CA, et al. Thin filament length dysregulation contributes to muscle weakness in nemaline myopathy patients with nebulin deficiency. Hum Mol Genet. 2009;18:2359–2369. doi: 10.1093/hmg/ddp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gokhin DS, Fowler VM. A two-segment model for thin filament architecture in skeletal muscle. Nat Rev Mol Cell Biol. 2013;14:113–119. doi: 10.1038/nrm3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pappas CT, Krieg PA, Gregorio CC. Nebulin regulates actin filament lengths by a stabilization mechanism. J Cell Biol. 2010;189:859–870. doi: 10.1083/jcb.201001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gokhin DS, Fowler VM. Tropomodulin capping of actin filaments in striated muscle development and physiology. J Biomed Biotechnol. 2011;2011:103069. doi: 10.1155/2011/103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skwarek-Maruszewska A, et al. Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres. Mol Biol Cell. 2010;21:3352–3361. doi: 10.1091/mbc.E10-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukada T, et al. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J Cell Sci. 2010;123:3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pappas CT, et al. Knockout of Lmod2 results in shorter thin filaments followed by dilated cardiomyopathy and juvenile lethality. Proc Natl Acad Sci U S A. 2015;112:13573–13578. doi: 10.1073/pnas.1508273112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fock U, Hinssen H. Nebulin is a thin filament protein of the cardiac muscle of the agnathans. J Muscle Res Cell Motil. 2002;23:205–213. doi: 10.1023/a:1020909902462. [DOI] [PubMed] [Google Scholar]
- 27.Bang ML, et al. Nebulin-deficient mice exhibit shorter thin filament lengths and reduced contractile function in skeletal muscle. J Cell Biol. 2006;173:905–916. doi: 10.1083/jcb.200603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fowler VM, McKeown CR, Fischer RS. Nebulin: does it measure up as a ruler? Curr Biol. 2006;16:R18–R20. doi: 10.1016/j.cub.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Sussman MA, et al. Myofibril degeneration caused by tropomodulin overexpression leads to dilated cardiomyopathy in juvenile mice. J Clin Invest. 1998;101:51–61. doi: 10.1172/JCI1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, McClure J, Tu A. Titin: major myofibrillar components of striated muscle. Proc Natl Acad Sci U S A. 1979;76:3698–3702. doi: 10.1073/pnas.76.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furst DO, Osborn M, Nave R, Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J Cell Biol. 1988;106:1563–1572. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trombitas K, Jin JP, Granzier H. The mechanically active domain of titin in cardiac muscle. Circ Res. 1995;77:856–861. doi: 10.1161/01.res.77.4.856. [DOI] [PubMed] [Google Scholar]
- 33.Freiburg A, et al. Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ Res. 2000;86:1114–1121. doi: 10.1161/01.res.86.11.1114. [DOI] [PubMed] [Google Scholar]
- 34.Cazorla O, et al. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ Res. 2000;86:59–67. doi: 10.1161/01.res.86.1.59. [DOI] [PubMed] [Google Scholar]
- 35.Methawasin M, et al. Experimentally increasing titin compliance in a novel mouse model attenuates the Frank-Starling mechanism but has a beneficial effect on diastole. Circulation. 2014;129:1924–1936. doi: 10.1161/CIRCULATIONAHA.113.005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris SP, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 37.Chung CS, Granzier HL. Contribution of titin and extracellular matrix to passive pressure and measurement of sarcomere length in the mouse left ventricle. J Mol Cell Cardiol. 2011;50:731–739. doi: 10.1016/j.yjmcc.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 39.Littlefield R, Fowler VM. Measurement of thin filament lengths by distributed deconvolution analysis of fluorescence images. Biophys J. 2002;82:2548–2564. doi: 10.1016/S0006-3495(02)75598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huxley HE, Stewart A, Sosa H, Irving T. X-ray diffraction measurements of the extensibility of actin and myosin filaments in contracting muscle. Biophys J. 1994;67:2411–2421. doi: 10.1016/S0006-3495(94)80728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown LM, Gonzalez-Serratos H, Huxley AF. Structural studies of the waves in striated muscle flbres shortened passively below their slack length. J Muscle Res Cell Motil. 1984;5:273–292. doi: 10.1007/BF00713108. [DOI] [PubMed] [Google Scholar]
- 42.van Dijk SJ, Bezold KL, Harris SP. Earning stripes: myosin binding protein-C interactions with actin. Pflugers Arch. 2014;466:445–450. doi: 10.1007/s00424-013-1432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett P, Craig R, Starr R, Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J Muscle Res Cell Motil. 1986;7:550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 44.Kentish JC, Eter Keurs H, Ricciardi L, Bucx JJ, Noble MI. Comparison between the sarcomere length-force relations of intact and skinned trabeculae from rat right ventricle. Influence of calcium concentrations on these relations. Circ Res. 1986;58:755–768. doi: 10.1161/01.res.58.6.755. [DOI] [PubMed] [Google Scholar]
- 45.Guo W, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trombitas K, Granzier H. Actin removal from cardiac myocytes shows that near Z line titin attaches to actin while under tension. Am J Phys. 1997;273:C662–C670. doi: 10.1152/ajpcell.1997.273.2.C662. [DOI] [PubMed] [Google Scholar]
- 47.Yamasaki R, et al. Titin-actin interaction in mouse myocardium: passive tension modulation and its regulation by calcium/S100A1. Biophys J. 2001;81:2297–2313. doi: 10.1016/S0006-3495(01)75876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulke M, et al. Interaction between PEVK-titin and actin filaments: origin of a viscous force component in cardiac myofibrils. Circ Res. 2001;89:874–881. doi: 10.1161/hh2201.099453. [DOI] [PubMed] [Google Scholar]
- 49.Astier C, Raynaud F, Lebart MC, Roustan C, Benyamin Y. Binding of a native titin fragment to actin is regulated by PIP2. FEBS Lett. 1998;429:95–98. doi: 10.1016/s0014-5793(98)00572-9. [DOI] [PubMed] [Google Scholar]
- 50.Greaser ML, Pleitner JM. Titin isoform size is not correlated with thin filament length in rat skeletal muscle. Front Physiol. 2014;5:35. doi: 10.3389/fphys.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donner K, Sandbacka M, Lehtokari VL, Wallgren-Pettersson C, Pelin K. Complete genomic structure of the human nebulin gene and identification of alternatively spliced transcripts. Eur J Hum Genet. 2004;12:744–751. doi: 10.1038/sj.ejhg.5201242. [DOI] [PubMed] [Google Scholar]
- 52.Kazmierski ST, et al. The complete mouse nebulin gene sequence and the identifi-cation of cardiac nebulin. J Mol Biol. 2003;328:835–846. doi: 10.1016/s0022-2836(03)00348-6. [DOI] [PubMed] [Google Scholar]
- 53.Sarullo FM, et al. Nemaline myopathy and heart failure: role of ivabradine; a case report. BMC Cardiovasc Disord. 2015;15:5. doi: 10.1186/1471-2261-15-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moncman CL, Wang K. Nebulette: a 107 kD nebulin-like protein in cardiac muscle. Cell Motil Cytoskeleton. 1995;32:205–225. doi: 10.1002/cm.970320305. [DOI] [PubMed] [Google Scholar]
- 55.Millevoi S, et al. Characterization of nebulette and nebulin and emerging concepts of their roles for vertebrate Z-discs. J Mol Biol. 1998;282:111–123. doi: 10.1006/jmbi.1998.1999. [DOI] [PubMed] [Google Scholar]
- 56.Moncman CL, Wang K. Functional dissection of nebulette demonstrates actin binding of nebulin-like repeats and Z-line targeting of SH3 and linker domains. Cell Motil Cytoskeleton. 1999;44:1–22. doi: 10.1002/(SICI)1097-0169(199909)44:1<1::AID-CM1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 57.Burgoyne T, Muhamad F, Luther PK. Visualization of cardiac muscle thin filaments and measurement of their lengths by electron tomography. Cardiovasc Res. 2008;77:707–712. doi: 10.1093/cvr/cvm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Konhilas JP, Irving TC, de Tombe PP. Frank-Starling law of the heart and the cellular mechanisms of length-dependent activation. Pflugers Arch. 2002;445:305–310. doi: 10.1007/s00424-002-0902-1. [DOI] [PubMed] [Google Scholar]
- 59.Solaro RJ, Rarick HM. Troponin and tropomyosin: proteins that switch on and tune in the activity of cardiac myofilaments. Circ Res. 1998;83:471–480. doi: 10.1161/01.res.83.5.471. [DOI] [PubMed] [Google Scholar]
- 60.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.