Abstract
Voluntary movement is a fundamental way in which animals respond to, and interact with, their environment. In mammals, the main CNS pathway controlling voluntary movement is the corticospinal tract, which encompasses connections between the cerebral motor cortex and the spinal cord. Hereditary spastic paraplegias (HSPs) are a group of genetic disorders that lead to a length-dependent, distal axonopathy of fibres of the corticospinal tract, causing lower limb spasticity and weakness. Recent work aimed at elucidating the molecular cell biology underlying the HSPs has revealed the importance of basic cellular processes — especially membrane trafficking and organelle morphogenesis and distribution — in axonal maintenance and degeneration.
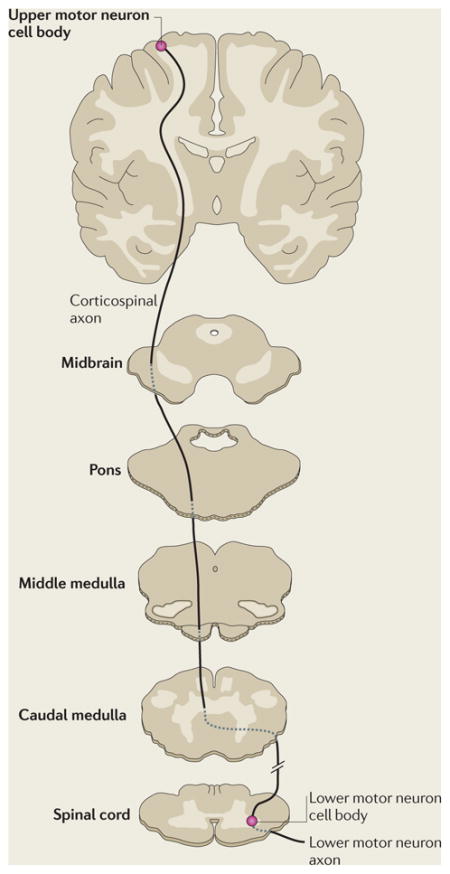
In humans, the pathways that comprise the voluntary motor system are arranged in two tiers (BOX 1). First, axons of the upper motor neurons, which originate in the cerebral motor cortex, pass through the medullary pyramids, where most axons decussate to form the lateral corticospinal tract in the spinal cord. These axons establish synapses directly or indirectly with the lower motor neurons in the spinal cord anterior horn. In the second tier, axons of the lower motor neurons synapse at neuromuscular junctions to mediate skeletal muscle contraction.
Box 1. The corticospinal tract and hereditary spastic paraplegias.
The main CNS motor pathway controlling voluntary movement is the corticospinal tract (see the figure). Its axons originate from upper motor neuron cell bodies in the cerebral motor cortex and pass to the medulla, where they form the medullary pyramids (hence the tract is sometimes termed the pyramidal tract). Most of the axons decussate at the caudal medulla, forming the lateral corticospinal tract. The fibres that do not decussate form the ventral corticospinal tract, and most of these axons decussate lower in the spinal cord. Axons of the lateral and ventral corticospinal tracts make direct or indirect synaptic connections (indirect connections are via interneurons) with lower motor neurons in the spinal grey matter. Lower motor neurons innervate skeletal muscles. Lesions of the corticospinal tracts cause spasticity.
The hereditary spastic paraplegias (HSPs) are in most cases caused by a length-dependent, distal degeneration of corticospinal tract axons. HSPs are estimated to have a prevalence of up to 18 per 100,000 of a population (5 per 100,000 is probably more accurate for North America and Northern Europe) and thus afflict several hundred thousand people worldwide95,96. HSPs can be divided into ‘pure’ forms, in which only spastic paraplegia occurs, and ‘complex’ or ‘complicated’ forms, characterized by additional clinical features2,5,97. Seminal studies recognized autosomal dominant, autosomal recessive and X-linked recessive inheritance patterns in both pure and complex HSPs. In Northern Europe and North America, autosomal dominant pure HSP is the most prevalent subtype, although in Mediterranean and North African countries autosomal recessive HSPs seem to be more common2,5,98.
More than 40 HSP loci have been mapped (named spastic gait or spastic paraplegia (SPG), from SPG1 to SPG48) and 20 HSP genes identified. Although the distinction between pure and complex forms of HSP remains clinically useful, this distinction is not always maintained at the molecular level. For example, although mutations in the gene encoding spastin (SPG4) usually cause pure HSP, they can also (although rarely) cause a complex phenotype99.
Distances traversed by the upper and lower motor neurons are extremely long, with axons reaching up to 1 m in length. This length is both an asset and a liability; it permits rapid relay of signals that mediate voluntary movement, but requires complex machineries for the proper intracellular sorting and distribution of proteins and organelles over long distances. The axoplasm can comprise over 99% of the total cell volume and is equipped with an elaborate cytoskeletal scaffold, mainly comprised of microtubules along which motor proteins mediate the selective transport of components. The interaction of intracellular cargos with a large number of diverse kinesin motor proteins — themselves associated with various adaptor proteins and other modulators of transport — permits tightly regulated, selective transport of organelles to growth cones during axonal development and to presynaptic terminals in mature neurons1.
Hereditary spastic paraplegias (HSPs) are a large and diverse group of genetic disorders characterized by progressive lower limb spasticity and weakness2 (BOX 1). These conditions highlight the clinical importance of understanding the mechanisms underlying axon maintenance and function, because in most HSP subtypes — including those on which this Review focuses — the spasticity is caused by a progressive distal axonopathy, mainly involving the longest corticospinal tract axons2,3. Interestingly, there is typically little neuronal cell death in HSPs. Studying HSPs therefore provides an important means to understand the specific molecular mechanisms of axonal maintenance and degeneration2. The insights gained may also be of relevance to more common neurological conditions in which axonopathy is a contributing feature, such as peripheral neuropathies, multiple sclerosis and motor neuron disease3,4.
The identification of genes that are implicated in HSPs has been fundamental to understanding the cellular biology and pathogenesis of this group of disorders. We now appreciate that most proteins encoded by ‘HSP genes’ fall into four main functional groups (TABLE 1). The largest group of HSP proteins are either known or thought to be involved in the intracellular trafficking, localization or shaping of membrane compartments (BOX 2), highlighting these processes as crucial to the maintenance of axonal health.
Table 1.
Known HSP genes, divided into functional groups
| Gene symbol | Protein name | Main phenotype | Cell biological function |
|---|---|---|---|
| Membrane traffic-related | |||
| SPG3A101 | Atlastin-1 | AD Pure | • ER morphogenesis • BMP signalling |
| SPG4 (also known as SPAST)11 | Spastin | AD Pure | • ER morphogenesis • Endosomal traffic • BMP signalling • Cytokinesis • Cytoskeletal regulation |
| SPG6 (also known as NIPA1)102 | NIPA1 | AD Pure | • Endosomal traffic • BMP signalling |
| SPG8 (also known as KIAA0196) 47 | Strumpellin | AD Pure | • Endosomal morphogenesis • Cytoskeletal regulation |
| SPG10 (also known as KIF5A) 82 | KIF5A | AD Complex | • Microtubule-based motor protein |
| SPG11 (REF. 103) | Spatacsin | AR Complex | • Membrane traffic? |
| SPG15 (also known as ZFYVE26)104,105 | Spastizin (also known as ZFYVE26 or FYVE-CENT) | AR Complex | • Membrane traffic? • Cytokinesis |
| SPG17 (also known as BSCL2)106 | Seipin | AD Complex | • ER membrane protein • Lipid droplet biogenesis |
| SPG20 (REF. 107) | Spartin | AR Complex | • Endosomal traffic • BMP signalling • Lipid droplet biogenesis • Mitochondrial? |
| SPG21 (REF. 108) | Maspardin | AR Complex | • Endosomal traffic |
| SPG31 (also known as REEP1)109 | REEP1 | AD Pure | • ER morphogenesis |
| Mitochondrial | |||
| SPG13 (also known as HSPD1)110 | HSP60 | AD Pure | • Mitochondrial chaperone |
| SPG7 (REF. 111) | Paraplegin | AR Complex | • Mitochondrial protease |
| Myelination | |||
| SPG2 (also known as PLP1)112 | PLP | XLR Complex | • Myelin protein |
| SPG35 (also known as FA2H)113,114 | Fatty acid 2-hydroxylase | AR Complex | • Hydroxylation of myelin lipids |
| Miscellaneous | |||
| SPG1 (also known as L1CAM)115 | L1CAM | XLR Complex | • Cell adhesion and signalling |
| SPG5 (also known as CYP7B1)116 | CYP7B1 | AR Pure | • Cholesterol metabolism |
| SPG39 (REF. 117) | Neuropathy target esterase | AR Complex | • Phospholipid homeostasis • Target of organophosphates |
| SPG42 (also known as SLC33A1)118 | SLC33A1 | AD Pure | • Acetyl-CoA transporter |
| SPG48 (also known as KIAA0415)119 | KIAA0415 | AR Complex | • DNA repair |
AD, autosomal dominant; AR, autosomal recessive; BMP, bone morphogenetic protein; CYP7B1, 25-hydroxycholesterol 7-alpha-hydroxylase; ER, endoplasmic reticulum; FA2H, fatty acid 2-hydroxylase; HSP60, heat shock protein 60; KIF5A, kinesin heavy chain isoform 5A; L1CAM, neural cell adhesion molecule L1; NIPA1, non imprinted in Prader-Willi/Angelman syndrome 1; PLP, myelin proteolipid protein; REEP1, receptor expression-enhancing protein 1; SPG, spastic paraplegia; SLC33A1, solute carrier family 33 (acetyl-CoA transporter), member 1; XLR, X-linked recessive.
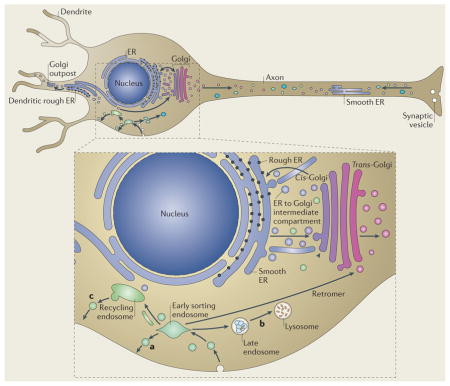
Box 2. Membrane traffic pathways in non-polarised cells and neurons.
The secretory and endocytic pathways are the main membrane traffic pathways in non-polarized cells (see the figure, blown-out part). In the secretory pathway (shown by a blue–purple gradient), cargo proteins are synthesised in the endoplasmic reticulum (ER). Cargoes are then trafficked to the Golgi through a tubulovesicular network, the ER to Golgi intermediate compartment. Post Golgi secretory vesicles and tubules are transported to several sites, including the plasma membrane and endosomes. In the endocytic pathway (shown in green), endocytosed material is delivered to early sorting endosomes. Cargo can then take three main routes: direct recycling back to the plasma membrane (a), delivery to the late endosomal and lysosomal degradative pathway (b), or a slow pathway back to the plasma membrane via perinuclear recycling endosomes (c). There are numerous cross-connections between the secretory and endocytic pathways — for example, the retromer complex sorts cargoes from early endosomes back to the trans-Golgi network. Long range movement of tubulovesicular cargoes occurs on cytoskeletal elements and uses motor proteins. For clarity, the recycling endosomal compartment and the Golgi are separated, but these organelles are pericentrosomal in most cells. In neurons the situation is more complex because the cells are polarized, with the somatodendritic membrane differing in composition to the axonal membrane. In addition to the core secretory pathway present in the cell soma, some dendrites also possess Golgi outposts. Cargo from ER within the dendrite may be transported to this, or may be transported to the somatic Golgi apparatus. The axon contains smooth ER continuous with the somatodendritic ER but is generally not regarded as containing rough ER or Golgi outposts. It also has endosomal components, including recycling endosomes, late endosomes and lysosomes, which are moved bidirectionally. At least some of these endosomes differ at the molecular level from somatodendritic endosomes — for example, the somatic early endosomal marker early endosome antigen 1 (EEA1) is not present in axons.
Recently, there have been substantial advances in our understanding of the function of this large group of HSP-associated proteins involved with membrane trafficking, and in this article we will review these advances, including the roles of HSP proteins in membrane modelling events, regulation of receptor-mediated signalling and membrane transport. Readers wishing to learn more about the clinical features, genetic epidemiology, pathology and molecular genetics of the HSPs are referred to other recent reviews2,5–10.
Membrane modelling and shaping
At least four HSP-associated proteins, including spastin (encoded by SPG4), atlastin-1 (encoded by SPG3A), receptor expression-enhancing protein 1 (REEP1; encoded by SPG31) and strumpellin (encoded by SPG8), are involved in membrane shaping and modelling events. This is an important subgroup, as mutations in the genes encoding these proteins cause up to 60% of HSP cases in North America and Northern Europe. This section will describe the role of these proteins and how their dysfunction might lead to HSPs.
Spastin: coupling membrane modelling to microtubule severing
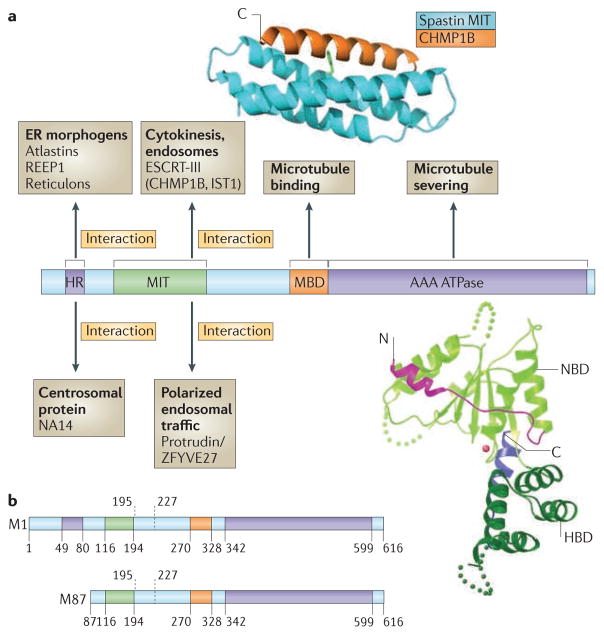
The gene encoding spastin was the first gene to be identified as a cause for autosomal dominant pure HSP11 (BOX 1). The gene encodes four main cellular isoforms of the spastin protein, which are generated by a combination of different promoter use, alternative initiation of translation from two different AUG start codons and differential exon splicing12,13 (FIG. 1). These four isoforms comprise a 616 amino-acid full-length protein (M1 spastin), a shorter isoform that lacks the first 86 amino acids of the full-length protein (M87 spastin) and splice variants of both of these, which lack a 32 amino-acid stretch encoded by exon 4 (REF. 12).
Figure 1. Spastin domain structure and interacting proteins.
Domains in the spastin protein. a | The hydrophobic region (HR) possibly forms an intramembrane hairpin loop. The microtubule interacting and trafficking domain (MIT) forms a three-helix bundle that interacts with a helix in the endosomal sorting complex required for transport III (ESCRT-III) proteins charged multivesicular body protein 1B (CHMP1B) and IST1. The microtubule binding domain (MBD) is necessary for spastin to bind to microtubules and is required for microtubule severing. The AAA (ATPases associated with diverse cellular activities) ATPase domain contains the enzymatic activity of the protein that is essential for microtubule breakage. The regions to which interaction sites with known binding partners have been narrowed are indicated. The structure of the interaction between the spastin MIT domain and CHMP1B is shown, as is the structure of the Drosophila AAA ATPase domain. The amino (N)- and carboxy (C)-terminal helices are shown in magenta and blue, respectively. NBD, nucleotide binding domain; HBD, helix bundle domain; REEP1, receptor expression-enhancing protein 1. The spastin structure and interaction with CHMP1B are reproduced, with permission, from REF. 30 © (2008) Macmillan Publishers Ltd. All rights reserved. The structure of the Drosophila AAA ATPase domain is reproduced, with permission, from REF. 18 © (2008) Macmillan Publishers Ltd. All right reserved. b | The domain structure of the M1 spastin isoform and the M87 spastin isoform, with amino-acid numbers indicated. The position of the alternatively spliced exon 4 is shown by the dashed lines.
There is overwhelming evidence from overexpression and knockdown studies in mammalian cultured cell lines, in vitro studies, structural studies and animal models that spastin can sever microtubules10,14. It is one of a small group of proteins that influence the cytoskeleton by causing internal breaks in microtubules, thereby regulating cellular functions that are dependent on these structures15. The carboxy-terminal half of spastin — from residue 270 onwards — contains the domains that are required for microtubule severing. This section of the protein contains a microtubule binding domain, situated between residues 270 and 328, and an AAA (ATPases that are associated with diverse cellular activities) ATPase domain, spanning residues 342–599 (REFS 11,16). AAA ATPase domains are found in a large family of proteins and typically use energy from ATP hydrolysis to catalyse conformational changes in target proteins17. Structural studies of recombinant spastin domains of Drosophila melanogaster and Caenorhabditis elegans have provided insights into its mode of action in microtubule severing (FIG. 1). Like most AAA domains, the AAA domain of spastin contains an α- and β-nucleotide-binding domain and a smaller four-helix bundle domain (HBD), which together comprise the enzymatic core of the protein. It contains an additional two helices, not found in AAA domains of other proteins, that interact with the nucleotide-binding domain. In the unbound state the nucleotide pocket is open and ATP binding is relatively inefficient because an extended loop that is likely to be involved in nucleotide contact and protomer–protomer interactions is positioned away from the nucleotide-binding pocket. In this non-ATP-bound state, spastin exists mainly as a monomer or a weak dimer. By contrast, ATP-bound spastin forms a hexameric ring with a prominent central pore. It has been suggested that polymerized spastin docks onto microtubules and destabilizes them by tugging the C terminus of tubulin through this central pore18.
The amino-terminal region of spastin contains sequences that mediate interaction with various adaptor proteins that recruit spastin to sites of action (FIG. 1). Most of these interactors are integral membrane or membrane-associated proteins, suggesting that spastin’s microtubule severing activity is targeted to specific membranes. In addition to showing partial colocalization with microtubules, endogenous spastin also localizes to the endoplasmic reticulum (ER) and can be recruited to endosomes19. Indeed, spastin shows an isoform-specific intracellular localization pattern, with the membrane-bound, larger M1 isoform predominantly found at structures of the early secretory pathway and to a much lesser extent on endosomes, whereas the shorter M87 isoform is present in a cytosolic pool that can be recruited to endosomes, but not to the early secretory pathway19.
The N-terminal region of spastin contains two well-characterized interaction domains that can explain its isoform-specific localization. First, a hydrophobic region that mediates interactions with at least three classes of early secretory pathway proteins — namely Atlastins, REEPs and Reticulons (see below) — lies within the beginning of spastin’s N-terminal region and is absent in the short M87 isoform20–22. Second, a microtubule interacting and trafficking (MIT) domain is present in all known spastin isoforms. It consists of a three-helix bundle formed by residues 116–194 of the full-length M1 protein (FIG. 1)23. MIT domains have been described in a number of proteins and typically mediate interactions with a group of proteins termed charged multivesicular body proteins (CHMPs)24. These CHMP proteins form a complex termed endosomal sorting complex required for transport III (ESCRT-III), and roles for this complex have been described in various membrane modelling processes, including viral budding, formation and sorting of cargoes (such as the epidermal growth factor (EGF) receptor) into the internal vesicles of late endosomal multivesicular bodies, and the final abscission stage of cytokinesis25,26. It has been suggested that ESCRT-III is a membrane scission machinery27. The MIT domain is necessary for the recruitment of spastin to endosomes, and spastin interacts strongly, through its MIT domain, with two ESCRT-III proteins, charged multivesicular body protein 1B (CHMP1B) and IST119,28,29. The crystal structure of the spastin MIT–CHMP1B complex reveals a non-canonical interaction site between the first and third helices of the spastin MIT domain and the C-terminal helical domain of the CHMP1B protein (FIG. 1). Interestingly, this C-terminal helix, which functions as an autoinhibitory domain that is exposed upon ESCRT-III oligomerization, provides an oligomerization-dependent switch for interaction with MIT-domain proteins, permitting selective recruitment30.
Spastin is required for completion of abscission at the end stage of cytokinesis, when the tubular midbody — which connects newly divided cells and is densely packed with an anti-parallel array of microtubules as well as a central collection of proteins known as the Flemming body — is resolved31,32. In cells lacking spastin, a microtubule disruption event that normally accompanies abscission does not occur, suggesting that it is caused by spastin-mediated microtubule severing19,30. The ESCRT proteins are also required for abscission, and there is evidence that these proteins participate in the resolution of the midbody membrane33,34. Endogenous spastin is localized at the periphery of the Flemming body within the midbody, and as its recruitment there depends on the MIT domain, it is thought to be mediated by one or more ESCRT-III proteins19,30. Thus, cytokinesis provides an example of how microtubule regulation can be linked to membrane modelling events through spastin. This link may be of relevance to the role of spastin in the nervous system, as loss of spastin reduces the rate of axonal branching in cultured primary neurons, and axonal branching also involves coordinated microtubule regulation and membrane modelling35,36.
Although spastin and the ESCRT-III proteins both localize to endosomes, a role for spastin in the known endosomal functions of the ESCRT complex has not been identified. For example, lack of spastin does not have a major effect on EGF receptor degradation19. Spastin also interacts with another endosomal protein, protrudin (also known as ZFYVE27), in HeLa cells37. Although this interaction has not yet been verified with the endogenous proteins in neurons, it is nonetheless interesting as lack of protrudin in cultured neurons inhibits neurite extension, whereas overexpression of protrudin promotes neurite extension38. Mechanistically, the effect of protrudin in neurite extension is mediated by Ras-related protein RAB11. Protrudin preferentially binds to the GDP-bound, inactive form of RAB11, which promotes directional membrane traffic38. The relevance of spastin to the action of protrudin on directional membrane traffic remains to be defined, but as neurite extension involves coordinated cytoskeletal remodelling and membrane trafficking, it is tempting to speculate that this might be another example of these processes being coupled through spastin.
Atlastin: a GTPase of the ER
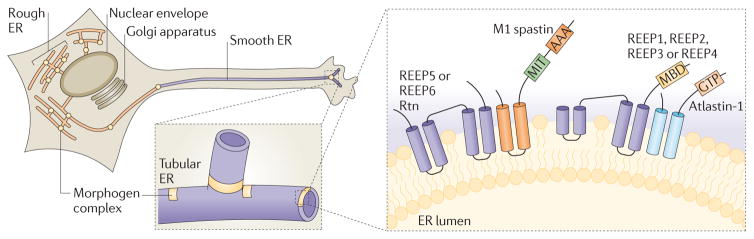
Mutations in SPG3A are the second most common cause of HSP, and the most common cause of early-onset disease. Atlastin-1, the protein encoded by SPG3A, is a member of a superfamily of dynamin-related GTPases, and recent studies have highlighted the crucial role of the Atlastin family in the formation of the ER network. Atlastin-1 is one of three mammalian Atlastin proteins that are thought to represent functional paralogues, and it is the only Atlastin that is highly expressed in the CNS; Drosophila spp. and C. elegans, for example, have only one Atlastin protein. Atlastin-related GTPases are present in all eukaryotic cells and are characterized by an N-terminal GTP-binding domain and two very closely spaced hydrophobic segments near the C terminus that probably form a hairpin transmembrane domain (see below)21,39. These multimeric, integral membrane GTPases localize predominantly to the tubular ER, but also to the ER to Golgi intermediate compartment (ERGIC) and cis-Golgi membranes. Atlastin GTPases are required for the formation of the three-way junctions in ER tubules — which give the characteristic, polygonal appearance of the ER in the cell periphery (FIG. 2) — by directly mediating homotypic fusion of ER tubules21,39,40. Consistent with their proposed role in the formation of the ER network, Atlastins localize to discrete sites along ER tubules, including at three-way junctions21,39.
Figure 2. The spastin–Atlastin–REEP–Reticulon complex at the ER.
Receptor expression-enhancing proteins (REEPs; Yop1p in yeast) and Reticulon proteins form large oligomers, referred to here as morphogen complexes, to shape the tubular endoplasmic reticulum (ER) network. Atlastin proteins (Sey1p in yeast) interact with REEPs and Reticulons and are enriched in puncta along the tubules (shown by yellow circles), including at three-way junctions. A blown-out image of the axon shows a tubular ER three-way junction. A nested blown-out image of a presumptive ER morphogen complex depicts the proposed membrane topologies for proteins involved in generating curvature of ER tubules, as well as mediating microtubule interactions and fusion of ER tubules. AAA, ATPases associated with diverse cellular activities (AAA) ATPase domain; GTP, Atlastin GTPase domain; MIT, microtubule-interacting and trafficking protein domain; MBD, microtubule-binding domain; REEP, receptor expression-enhancing protein; Rtn, reticulon.
Depletion of atlastin-1 in rat cortical neurons in primary culture inhibits axon elongation41, and the importance of ER morphology in the formation and maintenance of long processes is supported by studies of the Atlastin orthologue in Arabidopsis thaliana, RHD3. Mutant rhd3 plants have short, wavy root hairs and abnormal-appearing tubular ER bundles. Furthermore, the morphology of the ER changes noticeably during the elongation phase of root hair growth42. Thus, long cellular protrusions, such as plant root hairs and neuronal axons, are structures that are highly dependent on the dynamic morphology of the tubular ER.
Spastin, Atlastins and REEP1 are ER morphogens
The ER is a continuous membrane system that comprises the nuclear envelope, ribosome-studded peripheral sheets and a polygonal network of smooth tubules that extend throughout the cell. The mechanisms underlying the heterogeneous architecture of the ER have been clarified recently43. Several classes of proteins are important for the generation of tubular ER membranes, most notably the ER-shaping proteins of the REEP and Reticulon families. Although these families have little overall sequence homology to one another, they exhibit a common structural feature — elongated, hydrophobic segments that are predicted to form paired hairpin domains that partially span the membrane. With the majority of the protein domain localized to the outer leaflet of the phospholipid bilayer, ER-shaping proteins may generate curvature through hydrophobic wedging43,44.
In highly polarized cells such as neurons, distribution of ER domains is coordinated with cytoskeletal dynamics, mostly involving microtubules. Atlastins interact directly with spastin20,45, specifically through a predicted hydrophobic hairpin domain that is present only in the larger M1 isoform of spastin and possibly also through flanking hydrophilic sequences22. Expression of ATPase-defective M1 spastin causes a dramatic tubulation of the ER and redistribution of ER markers, including atlastin-1, onto abnormally thickened microtubule bundles20. In addition to interactions with tubule-shaping Reticulons, Atlastins and M1 spastin also seem to interact directly with REEPs in the tubular ER19,22,46 (FIG. 2). REEPs comprise six members in humans — REEP1 through to REEP6, with phylogenetic and structural distinctions between REEP1 through to REEP4 versus REEP5 and REEP6 (REF. 22). REEP1 through to REEP4 proteins harbour hydrophobic hairpins but also interact with microtubules through an extended C-terminal cytoplasmic domain that is enriched in basic amino acids22. These proteins may help to mediate the formation or stabilization of the tubular ER network, as deletion of the microtubule-interacting domain of REEP1 decreases the number of three-way junctions in the ER22. These interactions among REEPs, Atlastins, and spastin via hydrophobic hairpins provide a compelling mechanism for coupling ER membrane remodelling to cytoskeletal dynamics.
Strumpellin and endosomal tubulation
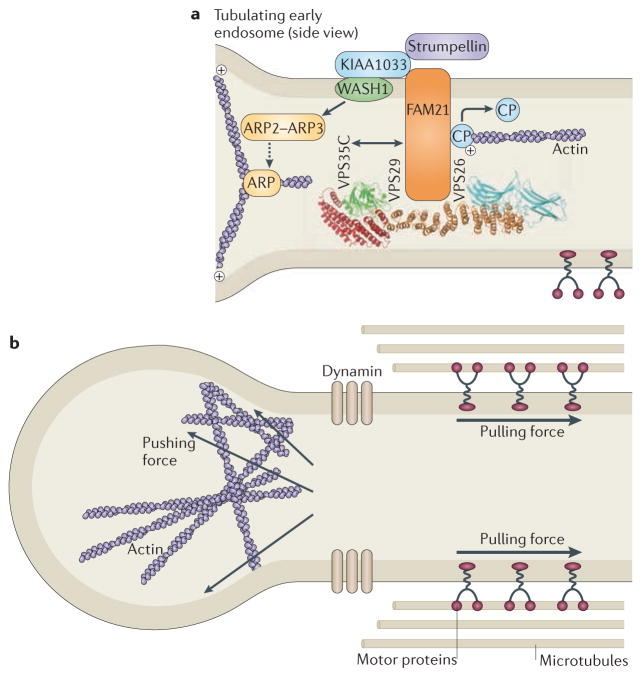
Mutations in KIAA1096 (also known as SPG8), the gene that encodes strumpellin, were identified as a cause of HSP in 2007 (REF. 47). Strumpellin has recently been identified as part of a large protein complex that associates with endosomes through an interaction with vacuolar protein sorting-associated protein 35 (VPS35). VSP35 is a component of retromer, an endosomal complex that is responsible for sorting certain cargoes from endosomes to the trans-Golgi network48–50 (BOX 2). Depletion of members of the strumpellin complex increases tubulation at early endosomes, resulting in impaired trafficking through early endosomal compartments, exemplified by impaired recycling of the transferrin receptor48–50. At least three members of the complex — Wiskott–Aldrich syndrome protein and SCAR homologue (WASH), family with sequence similarity 21 (FAM21) and actin capping protein — regulate actin dynamics. The complex generates an actin network on subdomains of early endosomes, for example by activating the actin-related protein 2 (ARP2)–ARP3 complex to initiate new actin filaments branching off existing filaments48,49,51 (FIG. 3). The increased tubulation associated with depletion of the strumpellin complex is thought to be due to a lack of the actin-driven force that is required for fission of tubular transport intermediates from the endosomal body48,49. The precise role of strumpellin in the complex remains unclear. However, the strumpellin–WASH complex does provide another example of an HSP protein being associated with coordinated membrane modelling and cytoskeletal organization. More recently, strumpellin has been shown to interact with valosin-containing protein (VCP; also known as p97), which is encoded by the gene that is mutated in frontotemporal dementia with Paget’s disease of bone and inclusion body myopathy52, although the relationship of this protein to the strumpellin–WASH complex has not yet been explored.
Figure 3. The strumpellin–WAsH complex.
a | The strumpellin–WASH (Wiskott–Aldrich syndrome protein and SCAR homologue) complex is recruited to endosomes by interaction between family with sequence similarity 21 (FAM21) and the vacuolar protein sorting-associated protein 35 (VPS35) component of the VPS26–VPS29–VPS35 cargo-selective retromer. Strumpellin probably interacts with the WASH complex via KIAA1033, but this remains to be proven. WASH interacts with, and activates, the actin-related protein 2 (ARP2)–ARP3 complex, which nucleates the formation of branched actin filaments. FAM21 has been proposed to interact with capping protein (CP) and promote its removal from the actin plus end, thus enhancing actin polymerization. b | In the absence of components of the WASH complex, early endosomal tubulation is enhanced, leading to the suggestion that the actin network generated by the complex is required to generate a pushing force on the tubule, which, combined with a pulling force generated by microtubule-based motors, promotes fission of the tubule by dynamin. The structure of the VPS26–VPS29–VPS35 complex is reproduced, with permission, from REF. 100 © (2007) Macmillan Publishers Ltd. All rights reserved.
Shaping defects and axonopathy
Spastin is a microtubule-severing protein that is recruited to membranes that are undergoing remodelling processes, atlastin-1 is a large integral-membrane GTPase of the ER that mediates homotypic fusion of ER tubules, REEP1 is an ER morphogen that links ER membranes to microtubules and strumpellin participates in a complex that is thought to link actin regulation to the fission of tubules from early endosomes. The common function of these HSP proteins in membrane shaping suggests that membrane modelling is mechanistically important in the pathophysiology of the disease. Defects in axonal growth or transport in animal models of HSP based on mutations in spastin and atlastin-1 also support a functional role for these proteins in axons41,53–55. How could membrane modelling events be linked to axonopathy? One possibility is that defects in membrane modelling events within the axon — events that are perhaps required for axonal functions such as synaptic plasticity or efficient axonal transport — are the primary cause of the disease. Although the link between these axonal functions and membrane shaping is currently unclear, it might be speculated that, for example, tubulation events at early endosomes could be part of a process required to distribute membranes from one site to another during synaptic plasticity. Correct morphogenesis of ER or other membrane compartments could be required to deliver membrane components with a particular composition to the distal axon, or could be important for bioenergetically efficient axonal transport, failure of which might lead to protein or lipid starvation of the distal axon. Alternatively, membrane modelling defects associated with mutated HSP proteins could affect the traffic of receptors that control specific signalling pathways that are important for axonal function. In fact, several proteins that, in mutated form, have been associated with HSPs regulate signalling pathways that are important for axonal function, as discussed in the following section.
Regulating receptor-mediated signalling
Bone morphogenetic protein signalling
Bone morphogenetic proteins (BMPs) are ligands of the transforming growth factor β (TGFβ) superfamily. BMP signalling has crucial roles in many developmental processes, including organogenesis, dorsoventral patterning, cellular differentiation and tissue remodelling. In Drosophila melanogaster and in mammals, the BMP signalling pathway is an important determinant of axonal growth and synaptic function56–60. Interestingly, impairment of BMP signalling in Drosophila melanogaster leads to axon transport defects61–63. In rodents, BMP signalling is upregulated after lesion of the corticospinal tract, and inhibition of this upregulation promotes axonal regrowth64.
HSP-associated mutations are found in at least four proteins — atlastin-1, non imprinted in Prader-Willi/Angelman syndrome 1 (NIPA1), spastin and spartin — that are inhibitors of BMP signalling65,66. The best characterized example is NIPA1, a polytopic integral membrane protein. Fly larvae lacking the Drosophila melanogaster homologue of NIPA1, spichthyin, show an increased number of synaptic boutons at neuromuscular junctions63. This axonal phenotype is associated with increased neuronal concentrations of phosphorylated mothers against decapentaplegic (MAD), a downstream messenger of BMP signalling, and it is suppressed by genetic manipulations that block BMP signalling63. NIPA1 and spichthyin are predominantly endosomal proteins that are also found at the plasma membrane, where they are subject to clathrin-mediated endocytosis65,67. They are thought to inhibit BMP signalling by binding to the type II BMP receptor and promoting its endocytic internalization and, at least in mammals, degradation in lysosomes63,65 (FIG. 4).
Figure 4. Model of NIPA1 action on bone morphogenetic protein receptor type-2 (BMPRII) traffic.
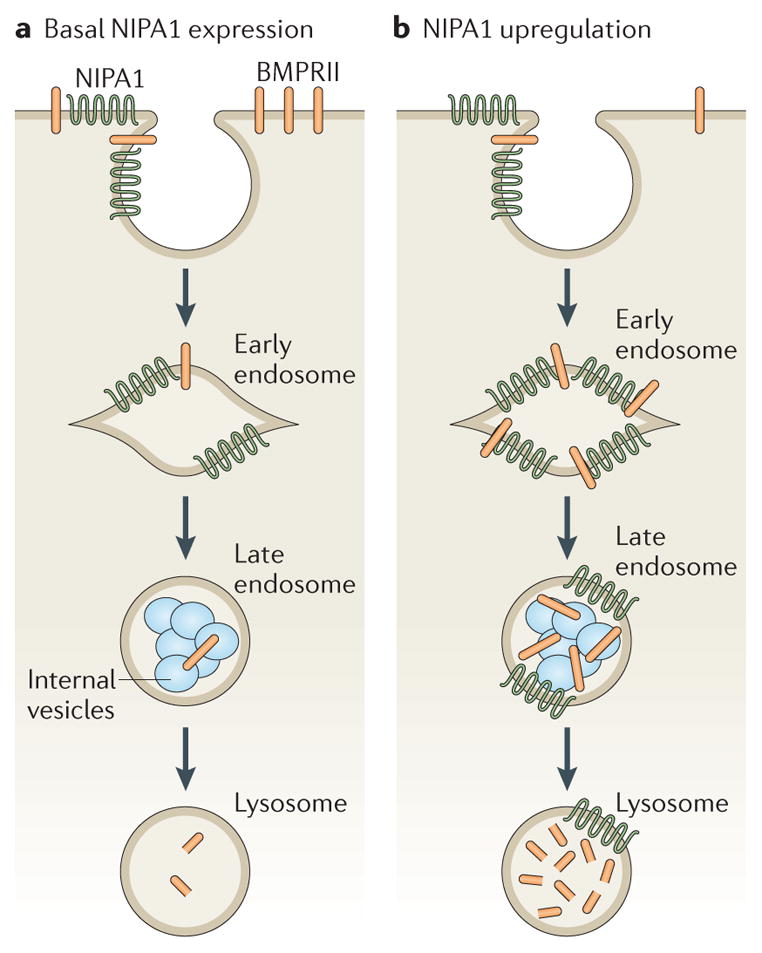
a | In cells that express normal levels of non imprinted in Prader-Willi/Angelman syndrome 1 (NIPA1), most bone morphogenetic protein receptor type-2 (BMPRII) is situated on the plasma membrane, with small amounts in early and recycling endosomal compartments. NIPA1 is found in endosomal compartments and at the plasma membrane. Lysosomal inhibition and cellular depletion of NIPA1 both increase cellular BMPRII levels, suggesting that NIPA1 promotes degradation of BMPRII in the lysosome. b | Consistent with this idea, overexpression of NIPA1 causes dramatic internalization of BMPRII into endosomal and lysosomal compartments, accompanied by increased lysosomal degradation of BMPRII. Certain features in these diagrams are conjectural — for example, it is not known whether the initial interaction between NIPA1 and BMPRII occurs at the plasma membrane or endosomes, whether NIPA1 is degraded in lysosomes or whether it can recycle back to the plasma membrane.
There has been some controversy over the pathological mechanism of NIPA1 in the causation of HSP. Studies examining the effect of NIPA1 disease-associated missense mutations showed that they cause the retention of NIPA1 in the ER and affect traffic of the protein through the secretory pathway67,68. This might cause disease through a gain-of-function induction of ER stress and the unfolded protein response (UPR) or by altering the trafficking of BMP receptors, with consequent effects on BMP signalling65,68.
The importance of dysregulated BMP signalling as a cause of axonal abnormalities in an in vivo vertebrate model of HSP has been confirmed by a recent study investigating atlastin-1 (REF. 66). Zebrafish morphants depleted of atlastin-1 had abnormal spinal motor axon morphology, particularly increased branching, as well as severely decreased larval mobility. The BMP signalling pathway was upregulated in these larvae, and inhibition of BMP signalling rescued the anatomical and behavioural phenotype of the atl1 knockdown zebrafish. Atlastin-1 partially colocalized with type I BMP receptors in neurite endosomes, suggesting that it may play a part in BMP receptor traffic66. Furthermore, depletion of spastin or spartin, which can both localize to endosomes, upregulates BMP signalling in mammalian cell lines through an as-yet-unknown mechanism65. Considered together, these results suggest that abnormal BMP signalling, probably caused by abnormal BMP receptor trafficking in many cases, could be a unifying mechanism in causing axonopathy in some classes of HSP. It will be important to determine whether the axonal phenotypes observed following depletion of several HSP proteins in primary neuronal cultures are caused by dysregulated BMP signalling. In particular, investigation of relevant HSP mouse models will now be crucial in determining whether abnormal BMP signalling has a pathological role in HSP and, if so, whether inhibition of BMP signalling by existing small-molecule inhibitors can rescue the disease phenotype69.
Other signalling pathways
In addition to BMP signalling, there is evidence that HSP-associated proteins regulate other signalling pathways. For example, the endosomal protein spartin is required for efficient EGF receptor degradation, and so probably regulates EGF signalling70. Spartin might also have a role in the regulation of signalling pathways by ubiquitin modification, as it interacts with a family of ubiquitin E3 ligases, including atrophin-1 interacting protein 4 (AIP4) and AIP5, which are known to regulate numerous pathways70–73. It is therefore possible that abnormality of spartin function might affect many axonal signalling pathways. Interestingly, spartin also regulates lipid droplet biogenesis by promoting AIP4-mediated ubiquitination of lipid droplet proteins71,73. In this regard it parallels the function of seipin, encoded by SPG17, which is an ER protein that functions in the formation of lipid droplets74. Although overexpression studies have suggested that the pathogenicity of seipin mutations is mediated by ER stress75,76, they could conceivably also affect lipid droplet biogenesis. Very little is known about the parts (if any) that lipid droplets play in axons. However, lipid droplets have been implicated in membrane biogenesis and cellular signalling, and could conceivably be involved in shaping organelles or regulating signalling pathways important for axonal function.
Mutations in the amyotrophic lateral sclerosis 2 (ALS2) protein alsin have been associated with spastic paraplegia and some patients with these mutations have a disease course that is more similar to the HSPs than to ALS. Mice that lack the gene encoding alsin show motor impairments and a distal axonopathy of the corticospinal tract that is characteristic of HSPs77,78. Alsin is a guanine nucleotide exchange factor for the small GTPases RAB5 and Ras-related C3 botulinum toxin substrate 1 (RAC1)79. Overexpression of alsin in neurons stimulated RAB5-dependent endosomal fusion, resulting in enlarged endosomes79, whereas RAB5-dependent endosomal fusion was impaired in neurons from alsin knockout mice80. Thus, abnormal endosomal function may be important in ALS2 pathogenesis, possibly through effects on AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) receptor trafficking, insulin-like growth factor 1 (IGF1) receptor trafficking or brain-derived neurotrophic factor (BDNF) receptor trafficking, which are disrupted in mice lacking alsin and which are components of a number of intracellular signalling cascades that control neuronal survival, plasticity and axonal morphology80,81.
Motor-based axonal transport
The identification of mutations in the gene kinesin family member 5A (KIF5A, also known as SPG10), which encodes the kinesin heavy chain isoform 5A (KIF5A), in families with pure and complex HSP indicates the importance of motor-based transport in the disease82,83. KIF5s are microtubule plus-end-directed, ATP-dependent motors that move cargo in the anterograde direction in axons84,85. Axonal transport fails in mutants of the Drosophila orthologue of KIF5A, resulting in axonal ‘log jams’ of immotile anterograde and retrograde cargo organelles86. In mammals, KIF5A has been characterized as a motor that is necessary for axonal transport of neurofilament subunits, although it has not been excluded from contributing to the transport of other anterograde cargoes, such as membrane vesicles87. KIF5 proteins also regulate transport of cargoes in dendrites and have roles in a number of membrane traffic pathways in the cell body, including in ER-to-Golgi and Golgi-to-ER traffic, Golgi to plasma membrane transport, and transport of recycling endosomes and of lysosomes84,88. It is not yet certain which of these diverse functions is impaired in patients with HSP who carry mutations of kinesin.
Nearly all KIF5A mutations are missense mutations and typically impair transport as they affect the kinesin motor domain82,83,89. The efficiency of cargo transport to the distal axon is thought to be affected either because the mutated KIF5A are slower motors or because they have reduced microtubule binding affinity and compete with other, wild-type motors for cargo binding sites90.
Axonal transport is also affected in mice that lack spastin, which have axonal swellings in which accumulated organelles are found54,55. Studies of axons from these mice have shown defects in both anterograde and retrograde transport, and these defects are associated with altered microtubule architecture within the axon. However, it is not clear whether these axonal transport defects are a direct result of dysregulation of axonal microtubules owing to lack of spastin, or an indirect effect of loss of spastin on membrane traffic pathways that control axonal signalling pathways54,55.
Membrane traffic and other axonopathies
The importance of organelle morphology and distribution for the maintenance of axons is also emphasized by genes that have been associated with other inherited, non-HSP axonopathies. These can include disorders involving peripheral nerves such as Charcot–Marie–Tooth (CMT) neuropathies and polyneuropathies with associated autonomic neuropathies (known as hereditary sensory and autonomic neuropathies (HSANs)). In a recent study91, mutations in the gene encoding protein FAM134B, a cis-Golgi apparatus-enriched member of the FAM134 family of proteins, were identified in patients with HSAN type II. FAM134B is enriched in dorsal root ganglion neurons, consistent with the clinical presentation of the disease91. The FAM134 proteins each have a pair of long hydrophobic segments, reminiscent of those in the Reticulons and REEP proteins. Furthermore, depletion of FAM134B causes prominent changes in Golgi morphology in neurons and in a tumour cell line derived from autonomic ganglion neurons, although effects on ER morphology have not been described91. More work will be needed to establish whether these proteins have a direct shaping function and how their dysfunction causes axonopathies. Even so, these disorders may highlight the implications of morphological defects in the ER and early secretory pathway in the pathogenesis of length-dependent axonopathies.
Studies into the pathogenesis of CMT neuropathies may be instructive, because these are also length-dependent axonopathies — although they primarily affect peripheral, rather than central, neurons. CMT1 is comprised of demyelination disorders and CMT2 of those that cause axonopathies. Axonal forms of CMT can be caused by mutations in a number of genes that encode proteins with functions in trafficking and organelle morphogenesis. In particular, CMT2A is caused by mutations in the gene encoding mitofusin 2 (MFN2), which regulates mitochondrial morphology by mediating mitochondrial fusion. Also, the CMT2B protein RAB7 is a small GTPase that regulates vesicle trafficking. Interestingly, RAB7 interacts with the SPG21 protein maspardin, another HSP-associated protein that localizes to endosomes92.
Lastly, ER-shaping mechanisms may have roles in related neurologic disorders such as familial ALS, in which both corticospinal and lower motor neurons are affected. In the superoxide dismutase 1 (SOD1) G93A transgenic mouse model for ALS, overexpression of the ER-shaping protein reticulon-4A selectively redistributed the ER chaperone protein disulphide isomerase and protected against neurodegeneration. Conversely, loss of reticulon-4A increased the severity of disease in SOD1 G93A mice93. Further supporting a role for ER morphogenesis in neurologic disorders, a mutant variant of vesicle-associated membrane protein-associated protein B (VAPB) that underlies another familial form of ALS (ALS8) is associated with the production of a novel form of organized, smooth ER94. Taken together, these studies support dysfunctional ER morphogenesis as a potential mechanism for multiple neurological diseases.
Conclusion
Over the past decade, many HSP-associated gene products have been identified and a small number of common themes for HSP pathogenesis are emerging. A large group of proteins associated with the majority of HSPs is involved in membrane trafficking processes; several of the proteins within this group, including those involved in the most common forms of HSP — spastin and atlastin-1 — shape membranes of the ER or endosomes. In addition, many proteins within this membrane traffic group are regulators of BMP signalling, a pathway that is probably regulated by endosomal or secretory pathway trafficking of BMP receptors.
However, many questions remain to be answered. Do other HSP-associated proteins that localize to the ER or endosomes also contribute to the shape of membranes? How might defects in organelle shaping cause axonopathy? There could be a direct and crucial requirement for membrane modelling events in the axon, or alternatively, abnormal membrane modelling could cause axonopathy by altering axonal BMP signalling. Is dysregulation of BMP signalling sufficient to cause axonopathy in mammals and, if so, by what mechanism? What mechanisms underlie phenotypic differences in HSPs? Perhaps the answer to this last question could be related to the specificity with which membrane modelling events are affected in different HSPs. Alternatively, uncomplicated HSPs could arise if BMP signalling is dysregulated in isolation, whereas complex HSPs might arise if additional signalling pathways are also involved. A variety of animal and cellular models are currently being developed to address these questions in the near future. This will raise the prospect of rationally designed therapies based on a thorough knowledge of the molecular and cellular pathology of distal axonopathies.
Acknowledgments
We are grateful to the members of our laboratories who have contributed to HSP-related work, and to the many HSP family members who have helped with our research. We thank T. Wahlig and H. Wahlig for their tireless work in promoting interactions among HSP researchers, clinicians and families. E.R. is a Wellcome Trust Senior Research Fellow in Clinical Science (grant 082381) and is also supported by the UK Medical Research Council, the Tom Wahlig Stiftung and the UK HSP Support Group. The work of C.J.O’K. on HSP is funded by Wellcome Trust (grant WT081386). C.B. is supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, US National Institutes of Health.
Glossary
- Upper motor neurons
Neurons whose fibres comprise descending pathways in the CNS and that are involved in voluntary control of skeletal muscle contraction. Corticospinal neurons are a type of upper motor neuron.
- Decussate
To cross the midline to reach the contralateral side of the nervous system.
- Paraplegia
Muscle weakness involving both legs.
- Spasticity
Increased muscle tone and deep tendon reflexes resulting from damage to the corticospinal tract.
- Protomer
A structural unit of an oligomeric protein.
- Early secretory pathway
A pathway through the endoplasmic reticulum (ER), ER-to-Golgi intermediate compartment and the cis-Golgi apparatus.
- Viral budding
The process by which an enveloped virus particle is released from the plasma membrane of a host cell.
- Abscission
The final stage of cytokinesis, when the midbody connecting two daughter cells is broken and sealed.
- Cytokinesis
The stage in cell division when the cytoplasm of a single cell is divided to form two daughter cells.
- Midbody
The tubular plasma membrane-bound structure that connects two daughter cells in the late stage of cytokinesis.
- Anti-parallel
Running side-by-side, but in opposite directions. A bundle of microtubules is anti-parallel if the microtubules of which it is comprised have plus ends facing both directions.
- Paralogues
Similar DNA and protein sequences (often distinct genes) within a species.
- Hydrophobic wedging
A mechanism for inducing membrane curvature by partitioning the bulk of a hydrophobic domain within the outer leaflet of the bilayer.
- Tubular transport intermediates
Membrane-bound, small, cigar-shaped organelles that are trafficked from one intracellular membrane compartment to another. They are distinguished by their shape from vesicular transport intermediates, which are spherical.
- Polytopic integral membrane protein
A protein that spans the membrane more than once because it has more than one transmembrane domain.
- Clathrin-mediated endocytosis
The major endocytic pathway, in which cells internalize extracellular or plasma membrane molecules into clathrin-coated vesicles. Once uncoated, the vesicles are capable of fusing with internal organelles, such as endosomes.
- Unfolded protein response
A cellular stress response that is triggered by excess of unfolded or misfolded proteins in the endoplasmic reticulum.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Craig Blackstone’s homepage: http://www.ninds.nih.gov/research/labs/410.htm
Cahir J. O’Kane’s homepage: http://www.gen.cam.ac.uk/research/okane.html
Evan Reid’s homepage: http://www.cimr.cam.ac.uk/investigators/reid/index.html
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Goldstein AY, Wang X, Schwarz TL. Axonal transport and the delivery of pre-synaptic components. Curr Opin Neurobiol. 2008;18:495–503. doi: 10.1016/j.conb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harding AE. The Hereditary Ataxias and Related Disorders. Churchill Livingston; Edinburgh: 1984. [Google Scholar]
- 3.DeLuca GC, Ebers GC, Esiri MM. Axonal loss in multiple sclerosis: a pathological survey of the corticospinal and sensory tracts. Brain. 2004;127:1009–1018. doi: 10.1093/brain/awh118. [DOI] [PubMed] [Google Scholar]
- 4.Fischer LR, et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Harding AE. Hereditary spastic paraplegias. Semin Neurol. 1993;13:333–336. doi: 10.1055/s-2008-1041143. [DOI] [PubMed] [Google Scholar]
- 6.Fink JK. Hereditary spastic paraplegia. Curr Neurol Neurosci Rep. 2006;6:65–76. doi: 10.1007/s11910-996-0011-1. [DOI] [PubMed] [Google Scholar]
- 7.McDermott CJ, Shaw PJ. Hereditary spastic paraplegia. Int Rev Neurobiol. 2002;53:191–204. doi: 10.1016/s0074-7742(02)53008-7. [DOI] [PubMed] [Google Scholar]
- 8.Reid E. The hereditary spastic paraplegias. J Neurol. 1999;246:995–1003. doi: 10.1007/s004150050503. [DOI] [PubMed] [Google Scholar]
- 9.Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 10.Reid E, Rugarli E. The Online Metabolic and Molecular Bases of Inherited Diseases. 2010 http://www.ommbid.com/OMMBID/the_online_metabolic_and_molecular_bases_of_inherited_disease/b/abstract/part28/ch228.1.
- 11.Hazan J, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nature Genet. 1999;23:296–303. doi: 10.1038/15472. This paper identified SPG4, which encodes the spastin protein, as the gene most commonly mutated in HSP. [DOI] [PubMed] [Google Scholar]
- 12.Claudiani P, Riano E, Errico A, Andolfi G, Rugarli EI. Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp Cell Res. 2005;309:358–369. doi: 10.1016/j.yexcr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Mancuso G, Rugarli EI. A cryptic promoter in the first exon of the SPG4 gene directs the synthesis of the 60-kDa spastin isoform. BMC Biol. 2008;6:31. doi: 10.1186/1741-7007-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salinas S, Carazo-Salas RE, Proukakis C, Schiavo G, Warner TT. Spastin and microtubules: Functions in health and disease. J Neurosci Res. 2007;85:2778–2782. doi: 10.1002/jnr.21238. [DOI] [PubMed] [Google Scholar]
- 15.Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol. 2007;176:995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- 18.Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. This paper presented the structure of the spastin AAA ATPase domain and shows how spastin assembles into a hexameric ring. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. This paper showed that spastin is recruited to membrane sites in an isoform-specific fashion. Together with reference 30, it showed that spastin is required for the completion of cytokinesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanderson CM, et al. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet. 2006;15:307–318. doi: 10.1093/hmg/ddi447. Together with reference 45, this paper presented the first evidence that two HSP proteins were binding partners. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. Together with reference 40, this paper showed that Atlastin GTPases have crucial functions in defining the morphology of the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. This paper showed that spastin, Atlastins and REEPs all act in concert to shape the ER reticular membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciccarelli FD, et al. The identification of a conserved domain in both spartin and spastin, mutated in hereditary spastic paraplegia. Genomics. 2003;81:437–441. doi: 10.1016/s0888-7543(03)00011-9. [DOI] [PubMed] [Google Scholar]
- 24.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 26.Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nature Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid E, et al. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- 29.Agromayor M, et al. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nature Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 32.Echard A, Hickson GR, Foley E, O’Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 35.Riano E, et al. Pleiotropic effects of spastin on neurite growth depending on expression levels. J Neurochem. 2009;108:1277–1288. doi: 10.1111/j.1471-4159.2009.05875.x. [DOI] [PubMed] [Google Scholar]
- 36.Yu W, et al. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol Biol Cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mannan AU, et al. ZFYVE27 (SPG33), a novel spastin-binding protein, is mutated in hereditary spastic paraplegia. Am J Hum Genet. 2006;79:351–357. doi: 10.1086/504927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirane M, Nakayama KI. Protrudin induces neurite formation by directional membrane trafficking. Science. 2006;314:818–821. doi: 10.1126/science.1134027. [DOI] [PubMed] [Google Scholar]
- 39.Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum Mol Genet. 2008;17:1591–1604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 41.Zhu PP, Soderblom C, Tao-Cheng JH, Stadler J, Blackstone C. SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum Mol Genet. 2006;15:1343–1353. doi: 10.1093/hmg/ddl054. [DOI] [PubMed] [Google Scholar]
- 42.Ridge RW, Uozumi Y, Plazinski J, Hurley UA, Williamson RE. Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol. 1999;40:1253–1261. doi: 10.1093/oxfordjournals.pcp.a029513. [DOI] [PubMed] [Google Scholar]
- 43.Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 44.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 45.Evans K, et al. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc Natl Acad Sci USA. 2006;103:10666–10671. doi: 10.1073/pnas.0510863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mannan AU, et al. Spastin, the most commonly mutated protein in hereditary spastic paraplegia interacts with Reticulon 1 an endoplasmic reticulum protein. Neurogenetics. 2006;7:93–103. doi: 10.1007/s10048-006-0034-4. [DOI] [PubMed] [Google Scholar]
- 47.Valdmanis PN, et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet. 2007;80:152–161. doi: 10.1086/510782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. Together with reference 49, this paper defined the role of the strumpellin–WASH complex in actin regulation and tubulation at endosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derivery E, et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 50.Harbour M, et al. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J Cell Sci. 2010;123:3703–3717. doi: 10.1242/jcs.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 52.Clemen CS, et al. Strumpellin is a novel valosin-containing protein binding partner linking hereditary spastic paraplegia to protein aggregation diseases. Brain. 2010;133:2920–2941. doi: 10.1093/brain/awq222. [DOI] [PubMed] [Google Scholar]
- 53.Wood JD, et al. The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum Mol Genet. 2006;15:2763–2771. doi: 10.1093/hmg/ddl212. [DOI] [PubMed] [Google Scholar]
- 54.Tarrade A, et al. A mutation of spastin is responsible for swellings and impairment of transport in a region of axon characterized by changes in microtubule composition. Hum Mol Genet. 2006;15:3544–3558. doi: 10.1093/hmg/ddl431. [DOI] [PubMed] [Google Scholar]
- 55.Kasher PR, et al. Direct evidence for axonal transport defects in a novel mouse model of mutant spastin-induced hereditary spastic paraplegia (HSP) and human HSP patients. J Neurochem. 2009;110:34–44. doi: 10.1111/j.1471-4159.2009.06104.x. [DOI] [PubMed] [Google Scholar]
- 56.Keshishian H, Kim YS. Orchestrating development and function: retrograde BMP signaling in the Drosophila nervous system. Trends Neurosci. 2004;27:143–147. doi: 10.1016/j.tins.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 57.O’Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Charron F, Tessier-Lavigne M. The Hedgehog, TGF-beta/BMP and Wnt families of morphogens in axon guidance. Adv Exp Med Biol. 2007;621:116–133. doi: 10.1007/978-0-387-76715-4_9. [DOI] [PubMed] [Google Scholar]
- 59.Wen Z, et al. BMP gradients steer nerve growth cones by a balancing act of LIM kinase and Slingshot phosphatase on ADF/cofilin. J Cell Biol. 2007;178:107–119. doi: 10.1083/jcb.200703055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lasorella A, et al. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 61.Aberle H, et al. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–558. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- 62.Ellis JE, Parker L, Cho J, Arora K. Activin signaling functions upstream of Gbb to regulate synaptic growth at the Drosophila neuromuscular junction. Dev Biol. 2010;342:121–133. doi: 10.1016/j.ydbio.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Shaw WR, Tsang HT, Reid E, O’Kane CJ. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nature Neurosci. 2007;10:177–185. doi: 10.1038/nn1841. Together with reference 65, this was the first paper to identify abnormal BMP signalling as a potential pathogenic mechanism in HSP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuura I, Taniguchi J, Hata K, Saeki N, Yamashita T. BMP inhibition enhances axonal growth and functional recovery after spinal cord injury. J Neurochem. 2008;105:1471–1479. doi: 10.1111/j.1471-4159.2008.05251.x. [DOI] [PubMed] [Google Scholar]
- 65.Tsang HT, et al. The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian BMP signalling. Hum Mol Genet. 2009;18:3805–3821. doi: 10.1093/hmg/ddp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fassier C, et al. Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nature Neurosci. 2010;13:1380–1387. doi: 10.1038/nn.2662. This paper suggests that abnormal BMP signalling is a cause of axonopathy in a vertebrate model of SPG3A. [DOI] [PubMed] [Google Scholar]
- 67.Goytain A, Hines RM, El-Husseini A, Quamme GA. NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. J Biol Chem. 2007;282:8060–8068. doi: 10.1074/jbc.M610314200. [DOI] [PubMed] [Google Scholar]
- 68.Zhao J, et al. Hereditary spastic paraplegia-associated mutations in the NIPA1 gene and its Caenorhabditis elegans homolog trigger neural degeneration in vitro and in vivo through a gain-of-function mechanism. J Neurosci. 2008;28:13938–13951. doi: 10.1523/JNEUROSCI.4668-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hao J, et al. In vivo structure-activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem Biol. 2010;5:245–253. doi: 10.1021/cb9002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bakowska JC, Jupille H, Fatheddin P, Puertollano R, Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol Biol Cell. 2007;18:1683–1692. doi: 10.1091/mbc.E06-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eastman SW, Yassaee M, Bieniasz PD. A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J Cell Biol. 2009;184:881–894. doi: 10.1083/jcb.200808041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edwards TL, et al. Endogenous spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases AIP4 and AIP5. Biochem J. 2009;423:31–39. doi: 10.1042/BJ20082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hooper C, Puttamadappa S, Loring Z, Shekhtman A, Bakowska J. Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biology. 2010;8:72. doi: 10.1186/1741-7007-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szymanski KM, et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daisuke I, Norihiro S. Molecular pathogenesis of seipin/BSCL2-related motor neuron diseases. Ann Neurol. 2007;61:237–250. doi: 10.1002/ana.21070. [DOI] [PubMed] [Google Scholar]
- 76.Farese RV, Jr, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C.-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamanaka K, Miller TM, McAlonis-Downes M, Chun SJ, Cleveland DW. Progressive spinal axonal degeneration and slowness in ALS2-deficient mice. Ann Neurol. 2006;60:95–104. doi: 10.1002/ana.20888. [DOI] [PubMed] [Google Scholar]
- 78.Deng HX, et al. Distal axonopathy in an alsin-deficient mouse model. Hum Mol Genet. 2007;16:2911–2920. doi: 10.1093/hmg/ddm251. [DOI] [PubMed] [Google Scholar]
- 79.Otomo A, et al. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet. 2003;12:1671–1687. doi: 10.1093/hmg/ddg184. [DOI] [PubMed] [Google Scholar]
- 80.Devon RS, et al. Als2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc Natl Acad Sci USA. 2006;103:9595–9600. doi: 10.1073/pnas.0510197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lai C, et al. Amyotrophic lateral sclerosis 2-deficiency leads to neuronal degeneration in amyotrophic lateral sclerosis through altered AMPA receptor trafficking. J Neurosci. 2006;26:11798–11806. doi: 10.1523/JNEUROSCI.2084-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reid E, et al. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) Am J Hum Genet. 2002;71:1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goizet C, et al. Complicated forms of autosomal dominant hereditary spastic paraplegia are frequent in SPG10. Hum Mutat. 2009;30:E376–385. doi: 10.1002/humu.20920. [DOI] [PubMed] [Google Scholar]
- 84.Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 85.Hirokawa N, Nitta R, Okada Y. The mechanisms of kinesin motor motility: lessons from the monomeric motor KIF1A. Nature Rev Mol Cell Biol. 2009;10:877–884. doi: 10.1038/nrm2807. [DOI] [PubMed] [Google Scholar]
- 86.Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xia CH, et al. Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J Cell Biol. 2003;161:55–66. doi: 10.1083/jcb.200301026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta V, Palmer KJ, Spence P, Hudson A, Stephens DJ. Kinesin-1 (uKHC/KIF5B) is required for bidirectional motility of ER exit sites and efficient ER-to-Golgi transport. Traffic. 2008;9:1850–1866. doi: 10.1111/j.1600-0854.2008.00811.x. [DOI] [PubMed] [Google Scholar]
- 89.Schüle R, et al. SPG10 is a rare cause of spastic paraplegia in European families. J Neurol Neurosurg Psychiatry. 2008;79:584–587. doi: 10.1136/jnnp.2007.137596. [DOI] [PubMed] [Google Scholar]
- 90.Ebbing B, et al. Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum Mol Genet. 2008;17:1245–1252. doi: 10.1093/hmg/ddn014. [DOI] [PubMed] [Google Scholar]
- 91.Kurth I, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nature Genet. 2009;41:1179–1181. doi: 10.1038/ng.464. [DOI] [PubMed] [Google Scholar]
- 92.McCray BA, Skordalakes E, Taylor JP. Disease mutations in Rab7 result in unregulated nucleotide exchange and inappropriate activation. Hum Mol Genet. 2010;19:1033–1047. doi: 10.1093/hmg/ddp567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang YS, Harel NY, Strittmatter SM. Reticulon-4A (Nogo-A) redistributes protein disulfide isomerase to protect mice from SOD1-dependent amyotrophic lateral sclerosis. J Neurosci. 2009;29:13850–13859. doi: 10.1523/JNEUROSCI.2312-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fasana E, et al. A VAPB mutant linked to amyotrophic lateral sclerosis generates a novel form of organized smooth endoplasmic reticulum. FASEB J. 2010;24:1419–1430. doi: 10.1096/fj.09-147850. [DOI] [PubMed] [Google Scholar]
- 95.Chen KM, Brody JA, Kurland LT. Patterns of neurologic diseases on Guam. Arch Neurol. 1968;19:573–578. doi: 10.1001/archneur.1968.00480060043005. [DOI] [PubMed] [Google Scholar]
- 96.Erichsen AK, Koht J, Stray-Pedersen A, Abdelnoor M, Tallaksen CM. Prevalence of hereditary ataxia and spastic paraplegia in southeast Norway: a population-based study. Brain. 2009;132:1577–1588. doi: 10.1093/brain/awp056. [DOI] [PubMed] [Google Scholar]
- 97.Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;1:1151–1155. doi: 10.1016/s0140-6736(83)92879-9. [DOI] [PubMed] [Google Scholar]
- 98.Silva MC, Coutinho P, Pinheiro CD, Neves JM, Serrano P. Hereditary ataxias and spastic paraplegias: methodological aspects of a prevalence study in Portugal. J Clin Epidemiol. 1997;50:1377–1384. doi: 10.1016/s0895-4356(97)00202-3. [DOI] [PubMed] [Google Scholar]
- 99.Nielsen JE, et al. Hereditary spastic paraplegia with cerebellar ataxia: a complex phenotype associated with a new SPG4 gene mutation. Eur J Neurol. 2004;11:817–824. doi: 10.1111/j.1468-1331.2004.00888.x. [DOI] [PubMed] [Google Scholar]
- 100.Hierro A, et al. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao X, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nature Genet. 2001;29:326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- 102.Rainier S, Chai JH, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am J Hum Genet. 2003;73:967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stevanin G, et al. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nature Genet. 2007;39:366–372. doi: 10.1038/ng1980. [DOI] [PubMed] [Google Scholar]
- 104.Hanein S, et al. Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am J Hum Genet. 2008;82:992–1002. doi: 10.1016/j.ajhg.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sagona AP, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nature Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 106.Windpassinger C, et al. Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nature Genet. 2004;36:271–276. doi: 10.1038/ng1313. [DOI] [PubMed] [Google Scholar]
- 107.Patel H, et al. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nature Genet. 2002;31:347–348. doi: 10.1038/ng937. [DOI] [PubMed] [Google Scholar]
- 108.Simpson MA, et al. Maspardin is mutated in mast syndrome, a complicated form of hereditary spastic paraplegia associated with dementia. Am J Hum Genet. 2003;73:1147–1156. doi: 10.1086/379522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Züchner S, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006;79:365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hansen JJ, et al. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet. 2002;70:1328–1332. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Casari G, et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 112.Saugier-Veber P, et al. X-linked spastic paraplegia and Pelizaeus–Merzbacher disease are allelic disorders at the proteolipid protein locus. Nature Genet. 1994;6:257–262. doi: 10.1038/ng0394-257. [DOI] [PubMed] [Google Scholar]
- 113.Dick KJ, et al. Mutation of FA2H underlies a complicated form of hereditary spastic paraplegia (SPG35) Hum Mutat. 2010;31:E1251–E1260. doi: 10.1002/humu.21205. [DOI] [PubMed] [Google Scholar]
- 114.Edvardson S, et al. Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am J Hum Genet. 2008;83:643–648. doi: 10.1016/j.ajhg.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jouet M, et al. X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nature Genet. 1994;7:402–407. doi: 10.1038/ng0794-402. [DOI] [PubMed] [Google Scholar]
- 116.Tsaousidou MK, et al. Sequence alterations within CYP7B1 implicate defective cholesterol homeostasis in motor-neuron degeneration. Am J Hum Genet. 2008;82:510–515. doi: 10.1016/j.ajhg.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Read DJ, Li Y, Chao MV, Cavanagh JB, Glynn P. Neuropathy target esterase is required for adult vertebrate axon maintenance. J Neurosci. 2009;29:11594–11600. doi: 10.1523/JNEUROSCI.3007-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin P, et al. A missense mutation in SLC33A1, which encodes the acetyl-CoA transporter, causes autosomal-dominant spastic paraplegia (SPG42) Am J Hum Genet. 2008;83:752–759. doi: 10.1016/j.ajhg.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Slabicki M, et al. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol. 2010;8:e1000408. doi: 10.1371/journal.pbio.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]