Abstract
The antitumor antibiotic bleomycin causes two major lesions in the deoxyribose backbone of DNA: formation of 4′-keto abasic sites and formation of strand breaks with 3′-phosphoglycolate and 5′-phosphate ends. As a model for the 4′-keto abasic site, we have characterized an abasic site (X) in d(CCAAAGXACTGGG)·d(CCCAGTACTTTGG) by two-dimensional NMR spectroscopy. A total of 475 NOEs and 101 dihedral angles provided the restraints for molecular modeling. Four unusual NOEs were observed between each anomer of the abasic site and the neighboring bases. In addition, four coupling constants for adjacent protons of the deoxyribose of both the α and β anomers of the abasic site were observed. The modeling suggests that for both anomers the abasic site is extrahelical, without significant distortion of the backbone opposite the lesion. The coupling constants further allowed assignment of an unusual sugar pucker for each anomer. The unique position of the abasic site in our structural model for each anomer is discussed in terms of repair of such lesions in vivo.
INTRODUCTION
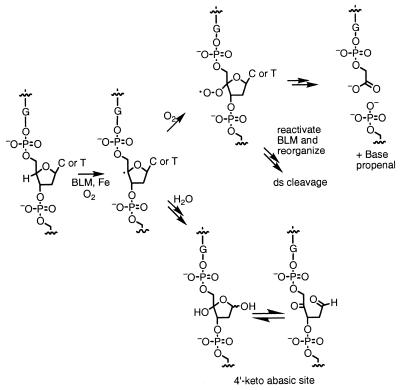
Bleomycins (BLMs) are antitumor antibiotics used clinically in the treatment of head and neck cancer, Hodgkin’s disease and testicular cancer (1,2). These natural products in the presence of their required cofactors, iron and oxygen, cause both single-strand (ss) and double-strand (ds) DNA damage (3–5). Our laboratory has long been interested in the mechanism of BLM-mediated ss and ds cleavage of DNA. BLM generates two types of sugar damage: 4′-keto abasic sites 3′ to guanines in an intact strand and 3′-phosphoglycolates, base propenals and 5′-phosphates in a cleaved strand (Fig. 1; 3–5). As a model for the 4′-keto abasic site, we have synthesized an abasic site (denoted by X) in d(CCAAAGXACTGGG)·d(CCCAGTACTTTGG) (1), where the intact duplex GTAC (T is the site of cleavage) is a hot-spot for BLM-induced ds cleavage (6). In this paper we present a structural model for each anomer of the abasic site in 1 using 2D NMR methods and molecular modeling. The long-range goal of these and other structural studies in our laboratory is to understand the structure of ds lesions created by a single BLM molecule. This information is a prerequisite for understanding BLM-mediated ds cleavage and the mechanism of the ds repair process.
Figure 1.
Proposed scheme for the formation of the two products resulting from BLM-induced H4′ abstraction. If oxygen adds into the radical, a 3′-phosphoglycolate lesion next to a 5′-phosphate end is generated. Under anaerobic conditions a 4′-keto abasic site results.
Cell viability and genome stability are continually threatened by DNA damage (7). Repair of this damage is essential for cell survival. The cytotoxicity of BLM is thought to be related to its ability to mediate ss or ds damage to the deoxyribose backbone of DNA (Fig. 1). Repair of this ss damage requires the base excision repair (BER) pathway (8–10). This pathway consists of a minimum of four enzymes in humans: a DNA glycosylase, a general damage apurinic/apyrimidinic (AP) endonuclease (APE1), DNA polymerase β and DNA ligase (11–13). The basis of recognition of different types of DNA damage remains a major focus of research. DNA base damage and repair have been studied most extensively. These studies reveal that the damaged base becomes extrahelical and binds to a specific pocket on the repair enzyme, where it is excised (14–18).
Removal of the base is only the first step in the repair process. An abasic site remains, which can also have detrimental effects on the cell. Approximately 10 000 abasic sites are generated per human cell per day, both spontaneously and enzymatically by the action of glycosylases (7). Since these sites are premutagenic (19), they must be repaired efficiently. APE1 (also known as HAP1, Ref-1 or Apex) is responsible for their recognition and repair (20,21). APE1 can also repair several other types of deoxyribose damage, such as 4′-keto abasic sites and 3′-phosphoglycolate lesions caused by BLM (8–10,22). The basis of recognition by APE1 of modified deoxyriboses is still unresolved.
The compound 3-hydroxy-2-(hydroxymethyl)tetrahydrofuran (THF) has often been used as a model for enzymatically generated abasic sites that exist as mixtures of α and β hemiacetals (23–26). Two structures of AP endonucleases (Escherichia coli endonuclease IV and APE1) bound to THF-containing oligonucleotides have recently been solved. Both reveal the THF flipped out of the helix into a specific binding pocket with the DNA bent away from the lesion (35° for APE1, 90° for endonuclease IV) (27,28). The base opposite the lesion is either stacked (27) or flipped out of the helix (28), depending on the enzyme.
Solution structures of abasic sites in naked duplex DNA using 2D NMR methods give a different picture. None of their structural models reveal an extrahelical deoxyribose or an extrahelical base opposite the lesion (29–33). Most THF analogs behave similarly (34–38). However, there is one report that a THF lesion opposite a pyrimidine is extrahelical (34), although the extrahelicity is sequence dependent (35). Since there have been no direct structural comparisons between abasic site- and THF-containing oligonucleotides in the same sequence context under the same conditions (29–38), it is unclear whether the THF analog is a good structural model for the abasic site. The limited number of structural models with abasic sites has prevented any general conclusions about recognition by repair endonucleases.
In this paper we describe the synthesis of 1, containing an abasic site (denoted by X), the assignment of the protons of the duplex DNA and NMR experiments to determine its structure. Using NMR-derived restraints, we have obtained two unique structural models: one for the α anomer and one for the β anomer. With both anomers the deoxyribose moiety is extrahelical, without significant distortion of the DNA backbone adjacent to and opposite the lesion. The conformations of the abasic site deoxyribose in this sequence context are compared to those observed for the various crystal structures of abasic sites in complex with repair enzymes and contrasted with abasic site structures in solution previously reported.
MATERIALS AND METHODS
Preparation of the abasic site
Phosphoramidites of dA, dC, dG and T, controlled pore glass columns (10 µmol, 500 Å pore size) and all other DNA synthesis reagents were purchased from Perkin Elmer-Applied Biosystems and the phosphoramidite of dU was purchased from Glen Research. Uracil DNA glycosylase was purchase from New England Biolabs (lot 1A).
Oligonucleotides d(CCAAAGUACTGGG) (2) and d(CCCAGTACTTTGG) (3) were synthesized on the 10 µmol scale in an automatic ABI 391 DNA synthesizer using standard coupling procedures provided by the manufacturer. The oligonucleotides were cleaved from the resin using ammonia, according to the standard procedures provided by the manufacturer, with the 5′-dimethoxytrityl protecting group left on.
Each oligonucleotide was purified by reverse phase high performance liquid chromatography (HPLC) (Alltech column, Econosil C18, 250 × 10 mm) using a 30 min linear gradient of 10–40% acetonitrile in 0.1 M triethylammonium acetate (TEAA) buffer, pH 7, at a flow rate of 3 ml/min. The major products eluted at 19 min for 2 and at 13 min for 3. The fractions containing the oligonucleotides were pooled and lyophilized to dryness. The remaining salt was removed by microdialysis against 2 l of water for 2 days. During dialysis the trityl group was cleaved from the oligonucleotide and was removed by filtration. Each oligonucleotide was then dried on a Speedvac and dissolved in 490 µl of water to give 1.8 µmol, 18% yield of 2 and ∼4.0 µmol, 40% yield of 3.
Compound 2 (450 µl, 3.7 mM) in water was added to 50 U uracil-DNA glycosylase in 110 µl of uracil-DNA glycosylase buffer (20 mM Tris–HCl, 1 mM EDTA, 1 mM DTT, 0.04 mg/ml BSA, 23 mM NaCl, pH 8). The reaction was incubated at 37°C for 14 h. Progress of the reaction was followed by monitoring uracil release. Reverse phase HPLC (Econosil C18, 250 × 10 mm column; Alltech) was then used to purify the oligonucleotide containing the abasic site (4), using a linear gradient of 5–30% acetonitrile in 0.1 M TEAA buffer, pH 7, at a flow rate of 3 ml/min. The major peak eluted at 16 min. The pooled fractions were concentrated to 5 ml in a lyophilizer. Compound 4 was isolated using a Dyonex Nucleopac PA 100 semiprep anion exchange column (250 × 9 mm) and a linear gradient from 25–55% buffer B (buffer A + 1 M NaCl) against buffer A (25 mM NaOAc, pH 6, 10% acetonitrile in water) over 30 min with a flow rate of 3 ml/min (compound 4, retention time 16 min, yield 90%; compound 2, retention time 23 min, yield 10%). Acetonitrile was removed from the sample. Since the abasic site decomposes in high salt conditions, 4 was dialyzed against 2 l of water for 2 days with one buffer change. Compound 4 was then lyophilized to dryness, dissolved in 1.5 ml of H2O and again dialyzed against 2 l of water.
A 1:1 ratio of 4 (1.5 µmol) (ɛ260 = 1.38 × 105 M–1) and 3 (1.5 µmol) (ɛ260 = 1.28 × 105 M–1) were mixed in 600 µl of 10 mM sodium phosphate buffer, pH 6.8, and the solution was heated to 70°C for 1 min and allowed to cool slowly to room temperature to give 1 (2.5 mM). A single peak eluting at 25 min (both ss oligonucleotides elute at 15 min under these conditions) observed by HPLC using the Dyonex anion exchange column confirmed generation of 1. This duplex was then lyophilized to dryness and exchanged twice in 600 µl of 99.999% D2O to give a 2.5 mM solution of 1 in 10 mM sodium phosphate buffer, pH 6.8. A second sample was prepared (3 mM) using the same protocol.
Samples were analyzed by electrospray mass spectrometry (ESI-MS) in the Vouros Laboratory at Northeastern University on a Finnigan LCQ Ion Trap Mass Spectrometer (San Jose, CA) with a standard electrospray source. The results are summarized in Table S1 in Supplementary Material.
Stability of the abasic site
Compound 1 was kept at 37°C for 1 week. An aliquot of 1 was then analyzed by reverse phase HPLC (Econosil C18, 250 × 4.6 mm column; Alltech) using a linear gradient of 5–30% acetonitrile in 0.1 M TEAA buffer, pH 7. Two major peaks of roughly equal volume eluted at 18.5 and 20 min. Compound 1 was treated with 10% piperidine in water and heated at 95°C for 90 min. Subsequent to piperidine treatment, an aliquot of 1 yielded three major peaks by reverse phase HPLC, eluting at 16 (∼25% of product), 17 (∼25% of product) and 19 min (∼50% of product).
NMR experiments
All NMR experiments were performed on custom-built 600 or 750 MHz NMR spectrometers at the Francis Bitter Magnet Laboratory. The acquired data were transferred to a Silicon Graphics workstation and processed using Felix software v.95 (Molecular Simulations Inc.). 1H chemical shifts were referenced to an internal standard, sodium 3-(trimethylsilyl)-1-propanesulfonate (TSP) at 0.00 p.p.m.
PECOSY, PCOSY, DQCOSY, TOCSY (30 or 60 ms mixing times) and NOESY (50, 100, 200 and 400 ms mixing times) experiments were recorded at 750 MHz at 20°C in D2O or at 5°C in 90% H2O/10% D2O. Datasets of 4096 × 512 complex points were acquired with spectral widths of 8000 Hz in both dimensions and 16 scans per t1 increment. During the relaxation delay period a presaturation pulse of 2.5 s was used for solvent suppression. For the NOESY experiments in 90% H2O/10% D2O a WATERGATE gradient pulse sequence (39) was used for water suppression and datasets with 4096 × 512 complex points were acquired with spectral widths of 15 000 Hz in both dimensions. For all these experiments spectra were zero filled to 4096 points in the t1 dimension. The data were processed with a combination of exponential and Gaussian weighting functions. Baselines were corrected with a polynomial or an automatic baseline correction routine in t2 when necessary.
The following experiments were run on the 600 MHz NMR spectrometer at 25°C in D2O. A 31P-decoupled PECOSY experiment was run with a spectral width of 6000 Hz in both dimensions and 4096 × 512 complex points to determine H2′/H2″ to H3′ coupling constants for the abasic site. A 31P-decoupled J-scaled 3′-4′ DQCOSY experiment with 4096 × 128 complex points (6000 × 2000 Hz spectral width) (40) was run to determine the H3′ to H4′ coupling constants for the abasic site. Coupling constants for the backbone were determined using an H3′ selective PHCOSY experiment (4096 × 64 complex points, 2000 × 500 Hz spectral width) with 256 t1 increments using a band-selective 180° pulse of EBURP shape to select the H3′ region (41,42). The following Karplus equation was used to derive backbone angles from these coupling constants: 3J(HCOP) = 15.3 cos2φ – 6.1 cosφ + 1.6 (43). Gradient-enhanced 31P-HSQC (spectral width 2000 × 500 Hz, 2048 × 128 complex points), 31P-HSQC (spectral width 6000 × 2000 Hz, 4096 × 128 complex points) and 31P-HCOSY (spectral width 6000 × 2000 Hz, 4096 × 128 complex points) experiments were also carried out (44,45). All 31P spectra were referenced indirectly through the gyromagnetic ratio to trimethyl phosphate by external calibration on TSP (46).
Molecular modeling: distance constraints
Distance constraints were derived from a 200 ms NOESY experiment at 750 MHz (no additional NOEs involving the abasic site were detected at shorter or longer mixing times). Peak volumes were assigned by visual inspection using H2′–H2″ NOEs and cytosine H5–H6 NOEs as a guide for strong NOEs. NOEs were classified as strong, medium and weak with distances of 1.8–3.5, 2.5–4.5 and 1.8–5.5 Å, respectively. One additional angstrom was added for distance constraints involving methyl groups. NOE volumes were also calculated with the peak pick protocol in Felix 95 and the measured volumes agreed well with those derived from visual inspection. Hydrogen bonds between base pairs were only included as restraints for those base pairs for which exchangeable imino protons could be assigned in the 200 ms NOESY spectrum in 90% H2O/10% D2O. For this reason no constraints were added for the terminal base pairs. All 475 experimentally determined NOEs were included in the molecular dynamics runs. Since the same number and types of NOEs were seen for both anomers, including the abasic site region, the same classes of NOE constraints were used in the molecular modeling protocol for both anomers.
Dihedral angle constraints
With the exception of the abasic site, the H1′–H2′ and H1′–H2″ coupling constants were consistent with a C2′ endo conformation for the deoxyribose. Therefore, the dihedral angle for H1′–C1′–C2′–H2″ was set at 30 ± 25° and that for H1′–C1′–C2′–H2′ was set at 150 ± 25°. For the abasic site, in addition to the H1′–H2′ and H1′–H2″ coupling constants, the H2′–H3′, H2″–H3′ and H3′–H4′ coupling constants allowed us to estimate the pseudorotation angle and pucker amplitude (47) and, consequently, a single solution was determined for the dihedral angles of the sugar ring using Karplus equations (48–50).
All four possible δ torsion angles were calculated for each abasic site anomer using standard methods (43) and separate calculations were performed for each of these solutions. Only the solution that consistently gave the lowest energy conformation was used in the final molecular modeling runs. A total of 101 dihedral constraints (46 backbone constraints and 55 deoxyribose constraints) were included in the molecular dynamics calculations.
Building of initial coordinate files
The abasic site-containing oligonucleotide was initially constructed in InsightII v.95 (Molecular Simulations Inc.) as double-stranded DNA (B-form) by removal of the base at the abasic site position and addition of OH in either the α or the β conformation. The parameter and topology files of the CHARMm force field (51) were modified separately for each abasic site anomer. In these two files the base was removed from a nucleotide unit and an OH was added at the 1′ carbon in the α or β position with the appropriate bonds, angles and electrostatic charges based on a hydroxyl bound to an sp3 carbon available in the force field. Each set of molecular coordinates for 1 built in InsightII was then read into XPLOR 3.851 on a Silicon Graphics (SGI) workstation to build the molecular structure file (psf) and molecular coordinate file (pdb) necessary for the molecular dynamics calculations, using the CHARMm force field. This rewriting of the initial pdb file built in InsightII is necessary to reconcile the InsightII naming conventions for protons with those of the force field used in the XPLOR calculations.
Molecular dynamics calculations
All molecular dynamics calculations were performed on the abasic site in vacuo. Initial coordinates for the abasic site were energy minimized at 300 K with 200 steps of conjugate gradient minimization. All experimental constraints were included in initial minimizations, molecular dynamics trajectories and final minimizations. Molecular dynamics trajectories were then run for 100 ps at 300 K. Throughout the energy minimizations and molecular dynamics trajectories, planarity constraints were included for the purine bases at 50 kcal mol–1 Å–2. The NOE force constant was 50 kcal mol–1 Å–2, while the dihedral angle force constant was 200 kcal mol–1 rad–2 for both anomers. These force constants were chosen to minimize NOE distance constraint and dihedral angle constraint violations. For the α anomer, lowering the force constant for the dihedral angle constraint to 40 kcal mol–1 rad–2 had no effect on the final structure. For the β anomer, lowering the force constant to 40 kcal mol–1 rad–2 caused a few dihedral angle constraint violations at the strand opposite the abasic site. Non-bonded interactions had a cut-off of 11.5 Å. The non-bonded interaction was switched from on to off between 9.5 and 10.5 Å. During the molecular dynamics calculations, atomic coordinates were written to a trajectory file every 0.2 ps throughout the 100 ps molecular dynamics trajectory. The structure was averaged over the last 20 ps of the run and the averaged structure was again subjected to 200 steps of conjugate gradient minimization. Ten structures were calculated for the α anomer and 20 structures were calculated for the β anomer. Coordinates for the final structures have been deposited in the PDB (α anomer, 1G5D; β anomer, 1G5E), in addition to the coordinates for an average minimized structure (α anomer, 1GIZ; β anomer, 1GJO).
Back-calculations
Back-calculations were performed in InsightII using the iterative relaxation matrix (IRMA) approach (52,53). The entire 200 ms NOESY spectrum at 750 MHz was back-calculated for the average of 10 abasic site structures for the α anomer. For the β anomer, the average of 10 structures with the abasic site in a flipped-out conformation were chosen for back-calculation. The spectra were calculated using the full relaxation matrix. Parameters for the IRMA calculations were set as follows: the rotational correlation time was 3 ns and the default T1 leakage was 10 s–1.
RESULTS
Preparation of the abasic site
Oligonucleotides d(CCAAAGUACTGGG) (2) and d(CCCAGTACTTTGG) (3) were synthesized and purified using standard procedures. The abasic site was generated in 2 by removing the uracil using uracil-DNA glycosylase. The resulting compound 4 was purified by reverse phase and anion exchange HPLC and appeared to be homogeneous (Supplementary Material, Fig. S1A). A 1:1 ratio of 4 and 3 was mixed, heated to 70°C and annealed to give duplex DNA (1). Anion exchange HPLC analysis of the resulting mixture under non-denaturing conditions showed a single species with a retention time of 25 min (Supplementary Material, Fig. S1B). The species had an ESI-mass spectrum consistent with the predicted structure. Furthermore, the 1D NMR spectrum of the duplex showed sharp peaks as expected for a stable duplex (Supplementary Material, Fig. S2). Linewidths in D2O are similar for the abasic site protons and the remaining protons of the oligonucleotide. The stability of 1 under the conditions used for NMR data acquisition was confirmed (as described in Materials and Methods).
Two samples of 1 were prepared. One (2.5 mM) was used to assign all the proton chemical shifts. The second (3 mM) was used to characterize the sugar pucker of the α and β anomers of the abasic site and to measure the coupling constants required to determine the backbone dihedral angles.
Assignment of the proton chemical shifts
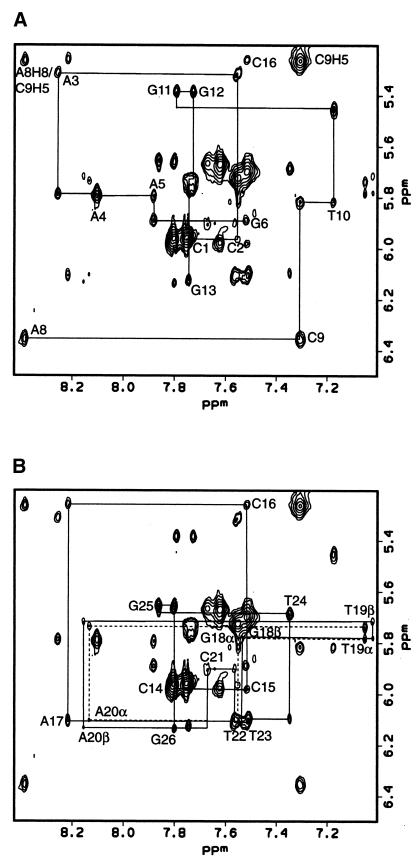
The proton chemical shifts for 1 were assigned by NOESY and TOCSY experiments in D2O using standard procedures (54). The base proton to base proton connectivity was established using a 400 ms NOESY experiment. Deoxyribose protons (H1′–H4′) were assigned for all residues using 30 and 60 ms TOCSY experiments, as were cytosine H5–H6 and thymine H6–methyl connectivities. The base proton to deoxyribose connectivity was established using mainly a 200 ms mixing time NOESY experiment, although 50, 100 and 400 ms mixing time NOESY experiments were also used in crowded regions. NOE walks were carried out for the base–H1′ (Fig. 2), base–H2′, base–H2″ and base–H3′ protons for all bases in the complementary strand and for C1–G6 and A8–G13 in the abasic site-containing strand. NOESY experiments (100, 200 and 400 ms) in 90% H2O/10% D2O were used to assign the adenine H2 and exchangeable imino and amino protons. The chemical shift assignments for the GXAC region are presented in Table 1 and the full chemical shift assignments for the 2.5 mM sample are summarized in Supplementary Material, Table S2. Two sets of chemical shifts were assigned for the deoxyribose protons of the abasic site (Table 1). Two distinct chemical shifts were also assigned to the G6 base and its H3′ protons, adjacent to the abasic site, and for many of the protons of G18, T19, A20 and C21. Complete NOE walks were accomplished from A17 to T22 for each set of NOEs. Thus two distinct conformations were observed for the backbone of the complementary strand in the GXAC region. With the exception of the chemical shifts for the abasic site, the chemical shift differences between the anomers in this region were small (0.02 p.p.m.) and the relative NOE intensities for the two sets of peaks were similar. Thus the differences in conformation of the strand complementary to the α and β anomer of the abasic site are expected to be small. A third conformation of the abasic site (∼5% of the total sample) is also apparent in some regions of the spectrum. It has not been investigated further due to its low abundance.
Figure 2.
NOE walks from H1′ to base protons for a 200 ms NOESY experiment at 750 MHz. (A) C1–G6 and A8–G13 are shown. (B) C14–G26 is shown. Where the α anomer differs from the β anomer (G18–C21), its NOE walk is shown as a dashed line. Connectivity cannot be seen between A20 and C21 for this region, but it is clearly seen in the H2′/H2″–base and H3′–base regions.
Table 1. Chemical shift assignments in p.p.m. for the GTAC region of the 2.5 mM sample of 1.
| Base |
H1′ |
H2′ |
H2″ |
H3′ |
H4′ |
H8/H6 |
H2/H5/Me |
NH imino |
NH2 |
| G6 α | 5.67 | 2.41 | 2.42 | 4.89 | 4.29 | 7.52 | 12.73 | ||
| G6 β | 5.67 | 2.41 | 2.42 | 4.92 | 4.29 | 7.55 | |||
| Abasic α | 5.26 | 1.96 | 1.83 | 4.49 | 4.38 | ||||
| Abasic β | 5.33 | 1.87 | 2.06 | 4.42 | 4.05 | ||||
| A8 | 6.35 | 2.81 | 2.96 | 5.00 | 4.48 | 8.39 | 7.70 | ||
| C9 | 5.81 | 1.90 | 2.45 | 4.72 | 4.22 | 7.31 | 5.26 | 6.73 | |
| 8.07 | |||||||||
| G18 α | 5.79 | 2.41 | 2.57 | 4.87 | 4.36 | 7.55 | 12.66 | ||
| G18 β | 5.79 | 2.40 | 2.56 | 4.87 | 4.36 | 7.53 | 12.66 | ||
| T19 α | 5.74 | 1.95 | 2.29 | 4.72 | 4.21 | 7.05 | 1.23 | 14.16 | |
| T19 β | 5.73 | 1.94 | 2.26 | 4.70 | 4.21 | 7.02 | 1.18 | 14.16 | |
| A20 α | 6.10 | 2.52 | 2.68 | 4.92 | 4.35 | 8.14 | 7.37 | ||
| A20 β | 6.11 | 2.51 | 2.68 | 4.92 | 4.35 | 8.16 | 7.37 | ||
| C21 α | 5.91 | 2.23 | 2.51 | 4.76 | 4.28 | 7.67 | 5.67 | ||
| C21 β | 5.91 | 2.22 | 2.51 | 4.76 | 4.26 | 7.67 | 5.67 |
Table 2. Chemical shifts in p.p.m. for the abasic site protons in the 3 mM sample.
| |
H1′ |
H2′ |
H2″ |
H3′ |
H4′ |
| α | 5.32 | 2.04 | 1.88 | 4.54 | 4.38 |
| β | 5.37 | 1.91 | 2.11 | 4.49 | 4.05 |
As noted above, two different samples of abasic site duplex have been utilized in acquisition of the data. Small chemical shift changes were observed between these samples (compare Tables 1 and 2). Chemical shifts for the 3 mM sample were generally shifted downfield by 0.02–0.05 p.p.m. from those assigned for the 2.5 mM sample. The changes in chemical shifts are probably due to minor changes in ionic strength between the two samples. Despite these chemical shift changes, the relative NOE intensities were similar. Thus no appreciable structural differences are expected for the two samples.
Assignment of the α and β anomers
Unambiguous assignment of the chemical shifts for the different anomers of the abasic site was straightforward based on the differences in the intensities of the NOEs between H1′ and H2′ versus H2″. For the β anomer, the H1′ proton is closer to the H2″ proton than to the H2′ proton, resulting in a larger NOE to the former. The reverse is true for the α anomer (Supplementary Material, Fig. S3). The unambiguous assignments of the H2′ and H2″ protons were based on the relative intensities of their NOEs to H3′. The H2′ chemical shift is downfield from the H2″ chemical shift in the α anomer. The reverse is true for all other deoxyriboses in the sample. The anomer ratio α:β is ∼60:40. A similar ratio is observed in the furanose form of 2-deoxy-d-ribose at pH 7 (55).
Establishing sugar puckers
Coupling constants for all deoxyriboses are consistent with the C2′ endo conformation expected for B-form DNA (Supplementary Material, Table S3), except for the abasic sites. Table 3 summarizes the assigned coupling constants for each abasic site deoxyribose. The H1′–H2′ coupling constant for the β anomer could not be determined from the H1′–H2″ multiplet in the PECOSY spectrum because of spectral overlap with the H1′–H2′ multiplet of C2. Similarly, spectral overlap prevented measurement of the H2′–H3′ coupling of the α anomer. Comparison of the experimentally determined coupling constants with the range of coupling constants expected for the possible pseudorotation angles showed that the α anomer is most consistent with an O1′ endo conformation and that the β anomer most closely corresponds to a C1′ exo conformation (47).
Table 3. JHH coupling constants in Hz for the two abasic site anomers.
| |
H1′–H2′ |
H1′–H2″ |
H2′–H3′ |
H2″–H3′ |
H3′–H4′ |
| α | 4.4 ± 0.5 | 1.3 ± 0.5 | Nd | 2.5 ± 0.5 | 8.0 ± 0.5 |
| β | Nd | 4.3 ± 0.5 | 7.3 ± 0.5 | <4.8 ± 1 | <6.8 ± 1 |
Nd, not determined due to spectral overlap.
Establishing the phosphodiester backbone conformation
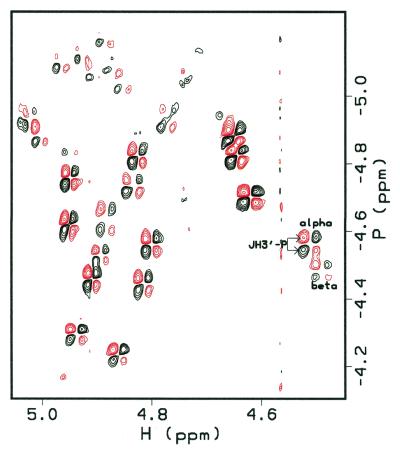
All H3′–31P coupling constants were determined using an H3′ selective {31P}-H heteronuclear correlation experiment (41,42), except for that at G18 due to spectral overlap (Supplementary Material, Table S4). Good dispersion of chemical shifts in the H3′ region made these assignments possible, including the area around the abasic site (Fig. 3). Since 31P chemical shifts are sensitive to O–P–O angles as well as backbone torsion angles (54), unusual 31P chemical shifts could be indicative of deviation from B-form or A-form DNA. However, all 31P chemical shifts, including those of the abasic site, were within the normal 31P envelope (approximately –4.7 ± 0.6 p.p.m., as referenced; 46), which confirmed that there are no significant distortions in the helix. All coupling constants are consistent with a B-form DNA conformation for 1, including the area around the abasic site.
Figure 3.
H3′-selective 31P HCOSY experiment at 600 MHz (41,42). All backbone coupling constants can be measured from these data, except that for G18 due to overlap. The abasic site anomers can be distinguished. Their chemical shifts are similar to the other 31P chemical shifts.
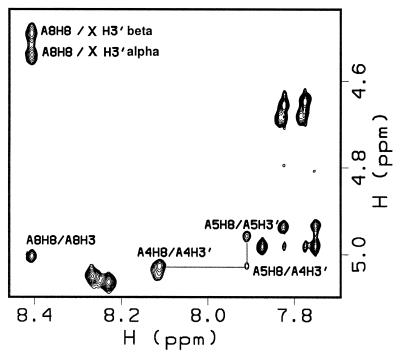
Abasic site conformation
Ten NOE constraints are assigned to each anomer. Four of these NOEs are unusual and hence structurally informative (Table 4). In B-form DNA the interaction between H3′ and the base proton 3′ to this sugar is generally weak (Fig. 4). However, the H3′ interaction with H8 of A8 is strong in both the α and β anomers. In B-form DNA the interaction between H2′ and H2″ and the base proton 3′ to this sugar is generally medium and strong, respectively. In both anomeric conformers of the abasic site the H2′ and H2″ protons showed very weak NOEs to H8 of A8 (data not shown). Finally, a weak NOE interaction was detected between H3′ of both abasic site anomers and the H3′ of G6. This information played a key role in defining a model structure for the two anomeric deoxyriboses. In the case of abasic site H3′ to A8 H8 the distance constraint was defined to vary between 2 and 3 Å, while in the case of H2′ and H2″ to A8 H8 it was 4.8–7 Å for the α anomer and 4.5–6.5 Å for the β anomer. These NOEs require sugar conformations for both anomers that deviate from standard B-form DNA.
Table 4. NOE interactions involving the α and β anomers of the abasic site (X) and their neighboring bases.
| NOE |
Strength of interaction |
| X H2′–A8 H8 | Weak |
| X H2″–A8 H8 | Weak |
| X H3′–A8 H8 | Strong |
| G6 H3′–X H3′ | Weak |
Figure 4.
H3′ to base proton region for 1, where X indicates the position of the abasic site, in a 200 ms NOESY experiment at 750 MHz. Strong NOEs are observed between the H3′ of the abasic site and its neighboring A8 H8. In B form DNA these NOEs are expected to be much weaker, as illustrated by the NOE between A5 H8 and A4 H3′.
Molecular dynamics
Ten separate structures were calculated for the α anomer and 20 structures for the β anomer of 1. A greater number of MD runs was performed for the β anomer because there was greater variation in the final structures. None of the final structures contain any NOE distance constraint violations >0.5 Å or dihedral angle constraint violations >5°. A stereo view of the overlay structures of the results for each anomer is seen in Figure 5. Seventy percent of the structures for each anomer are nearly identical, while the remainder of the solutions differ only in the location of the abasic site with respect to the helix. The results of the modeling runs for both anomers are summarized in Supplementary Material, Table S5.
Figure 5.
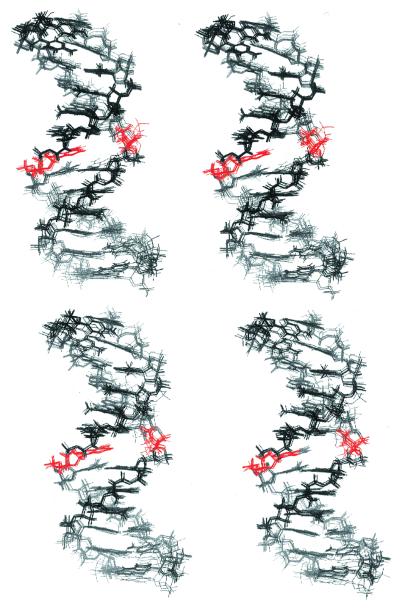
Stereo view of overlay structures of 10 α anomers (top) and 10 β anomers (bottom) of the abasic site-containing oligonucleotide. The abasic site and the base opposite the abasic site are indicated in red.
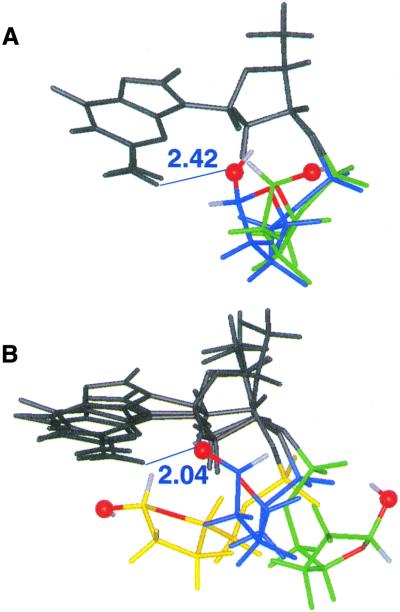
In the case of the α anomer all 10 structures have the abasic site deoxyribose flipped out of the helix ∼90° (Fig. 6A). The α anomer shows two of 10 structures with the abasic site O1′ in hydrogen bonding distance to the adjacent 2-amino group of G6 (Fig. 6A, blue), while the remaining eight structures are devoid of this putative interaction (Fig. 6A, green). For the β anomer, 14 of 20 structures have the abasic site flipped out of the helix ∼90° (Fig. 6B, blue). In these structures the abasic site O1′ is positioned to hydrogen bond with the 2-NH2 group of G6 in a conformation similar to the two observed for the α anomer (Fig. 6A and B, blue). These hydrogen bonds were not detected in our H2O experiments, suggesting that if they were present, they would have been in rapid exchange with the solvent. One of 20 structures of the β anomer shows the abasic deoxyribose flipped out of the helix ∼180° (Fig. 6B, green) and the remaining five structures show the abasic site as intrahelical (Fig. 6B, yellow). The five intrahelical structures show the abasic site O1′ within hydrogen bonding distance of the 6-NH2 group of the adenine opposite the lesion (A20, not shown). The intrahelical conformation of the abasic site deoxyribose for the β anomer most likely results from this putative hydrogen bonding stabilization between the deoxyribose and the opposite base, but it may not be representative of the conformation of the β anomer in solution.
Figure 6.
Representative conformations for the range of structures obtained by molecular modeling of both abasic site anomers. Possible hydrogen bonding is denoted by blue lines with putative hydrogen bonding distances given in Å. The abasic site O1′ (ball) and O4′ (stick) are shown in red, the H1′ and O1′H in white. (A) Two representative conformations of the α anomer (green, 8/10 solutions; blue, 2/10 solutions); (B) three representative conformations of the β anomer (5/20 solutions in yellow; 14/20 solutions in blue; 1/20 solutions in green).
Back-calculating structures
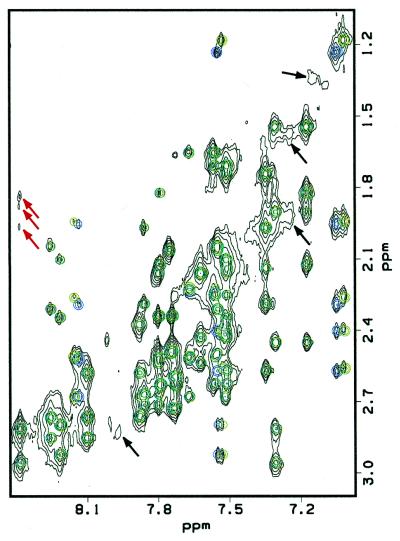
One method to obtain insight into the model that best describes the abasic site structures is to back-calculate the NMR data and to compare them with the experimental results. The 200 ms NOESY spectra for the α and β anomers were back-calculated using the IRMA protocol in InsightII. Ensembles of 10 structures in the major extrahelical conformation were back-calculated and showed good agreement to the experimental data (Fig. 7). Back-calculations of the 200 ms NOE spectrum of the β anomer of the abasic site in the intrahelical position do not agree with the experimental data. For example, they predict strong NOEs between the abasic site H1′ and G6 H1′ and G6 H4′, which were not observed experimentally.
Figure 7.
Back-calculation of a 200 ms NOESY spectrum (750 MHz). The experimental data is shown in black, the back-calculated spectrum of the α anomer is overlaid in blue and the back-calculated spectrum of the β anomer is overlaid in green. Back-calculations were performed on an average of 10 structures for both anomers. Black arrows indicate examples of NOEs belonging to a minor conformation in the experimental data which could not be assigned due to its low abundance in solution (∼5%). The weak NOEs of the abasic site (indicated by red arrows) can be seen in the back-calculated spectrum at a lower contour level.
DISCUSSION
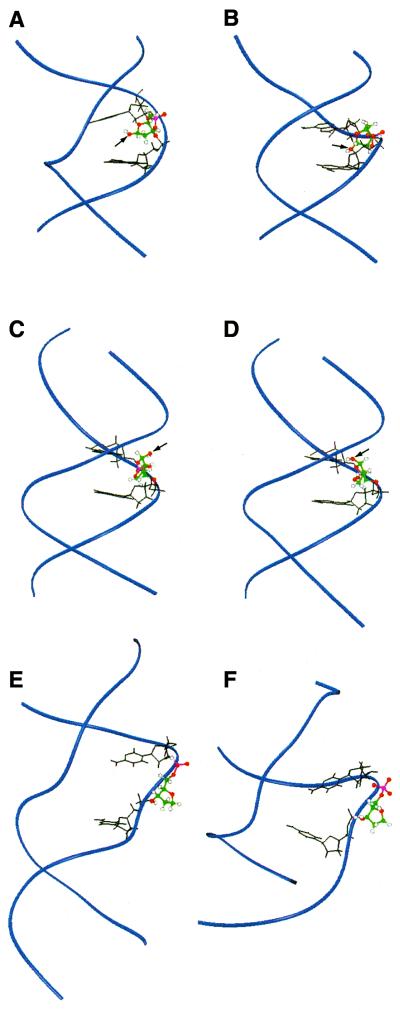
There are four reports in the literature of structural models of actual abasic sites in oligonucleotides with which to compare our results (29–32). We will focus on the work that has shed light on the importance of the base opposite the lesion and the sequence context of the lesion (29–31). Beger et al. (29) have investigated an abasic site (X) opposite adenine in the GAXAC sequence context (29,30) in which they observed a nearly equal population of α and β anomers (29,30). Their model structures showed that the β anomer of the deoxyribose was intrahelical and that the α anomer of the deoxyribose was somewhat more surface accessible but largely intrahelical. In their modeling protocol for both anomers a water molecule was included that formed a hydrogen bond to the abasic site hydroxyl group and to the amino group of the adenine opposite the abasic site (29). The distance between O1′ of the abasic site and the nearest proton of the 6-amino group of the adenine opposite this site was 1.7 Å for the α anomer and 4.6 Å for the β anomer. In our GXAC sequence of 1 our models suggest that both anomers have a similar degree of extrahelicity (Fig. 8C and D). The distance between O1′ of the abasic site and the nearest proton of the 6-amino group of the adenine opposite this site is 9.0 Å for the α anomer and 6.9 Å for the β anomer. Thus the neighboring bases flanking the abasic site influence its conformation with respect to the helix.
Figure 8.
Comparison between abasic site structures found in solution in naked duplex DNA and THF analog structures when co-crystallized with repair enzymes. For the NMR structures O1′ is indicated by an arrow. (A) Solution structures of the β anomer of the abasic site opposite dA (29); (B) α anomer of the abasic site opposite dA (29); (C) α anomer of the abasic site (our work); (D) β anomer of the abasic site (our work); (E) THF analog bound to APE1 (27); (F) THF analog bound to endonuclease IV (28). The DNA backbone is shown as blue ribbons. The basic site and analog are shown in color. Bases surrounding the abasic site are shown in black. Previous abasic site solution model structures are much more intrahelical than our model structures. The two enzyme-bound structures have distorted minor grooves and completely flipped-out abasic site analogs.
The importance of neighboring bases is further substantiated by studies of Wang et al. (31) on an abasic site opposite an A tract: d(CGCAAAAATGCG)·d(CGCATTXTTGCG). The abasic site disrupts the structure relative to the undamaged parent structure up to 4 bp in each direction of the lesion. Furthermore, the curvature of the intact oligonucleotide is absent in the corresponding abasic site-containing oligonucleotide. The influence of either anomer of the abasic site in our GXAC sequence extends only 1 or 2 bp in each direction.
In order to understand the influence of the base opposite the abasic site on structure, Beger et al. (29) examined GAXAC, described above, with a cytosine opposite X. Their modeling suggested that the abasic site was more surface accessible then when X was opposite adenine and further revealed that there were two different conformers of the β anomer and no α anomer (29). The absence of an α anomer is an unusual result and suggests that further studies are required. However, in this sequence context the A and C were both intrahelical, as we have observed in GXAC.
The predominant deoxyribose conformations of the abasic site in 1 have not been observed in solution for any of the other abasic site model structures and they are closest to conformations observed in crystal structures of abasic site analogs bound to repair enzymes (Fig. 8E and F). The crystal structures of both APE1 and E.coli endonuclease IV show THF analogs completely flipped out of the helix and into a specific binding pocket on the protein (27,28).
Structure-driven mutagenesis studies suggest that APE1 does not play an active role in flipping of the deoxyribose to its extrahelical conformation and that flexibility of the substrate is important in enzyme recognition (27). APE1 has also recently been shown to be processive over 200 bp or more (56). Successful scanning of the DNA is also likely to be related to its flexibility. Recent studies of diffusion coefficients by NMR demonstrate the flexibility of lesioned DNA relative to duplex (57). However, flexibility is not exclusively responsible for recognition of the lesioned duplex by APE1, as the enzyme binds a gapped duplex more tightly than a nicked duplex, and both would be expected to be flexible (58).
Despite increased flexibility (57), no significant bending in the helix in the region of the abasic site has been reported for any solution NMR structures (29–32). In fact, the abasic site in the dA tract discussed above eliminates the curvature of the intact duplex (31). These results contrast with the crystallographic data of THF-lesioned duplexes bound to repair enzymes (27,28). Bends in the helix away from the damage to varying extents have been observed with both APE1 and endonuclease IV.
The structure of APE1 bound to THF allowed Mol et al. (27) to suggest that the α anomer, the expected product of glycosylases, uniquely fits into the enzyme’s binding pocket. Our structural model is the only one to define the sugar pucker conformations accessible to an abasic site lesion. In our sequence context this conformation is C1′ exo for the β anomer. Our modeling studies using the APE1 coordinates reveal that β anomers with altered sugar puckers can also access the binding pocket. This is reasonable, since abasic sites generated non-enzymatically will be a 1:1 mixture of α and β anomers, both of which must be repaired.
Finally, evidence is mounting that recognition of abasic sites in the BER pathways depends on specific enzyme–enzyme interactions (59–61). While enzyme–enzyme interactions may play a role in recognition of abasic sites generated enzymatically, for example by glycosylases, abasic sites created non-enzymatically in vivo by hydrolysis are probably recognized directly by endonucleases. The possibility cannot be excluded, however, that abasic sites could be recognized by the glycosylases, which are known to bind product tightly. Thus understanding all the factors that contribute to recognition of the abasic site by the repair endonucleases remains an important goal. Further systematic studies of these lesions in different sequence contexts with different opposing bases and studies in larger pieces of DNA are essential.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Drs Phil Bolton, Vasilios Marathias, Dana Vanderwall and Helen Mao for helpful suggestions regarding molecular modeling. The authors also thank Dr Vouros and his laboratory for performing the electrospray mass spectrometry on our samples. This work was supported by the National Institutes of Health, grant no. GM34454. The NMR facility is supported by NIH grant RR-00995.
PDB accession nos 1G5D, 1G5E, 1GIZ, 1GJO
References
- 1.Sikic B.I., Rozencweig,M. and Carter,S.K. (eds) (1985) Bleomycin Chemotherapy. Academic Press, Orlando, FL.
- 2.Mir L.M., Tounekti,O. and Orlowski,S. (1996) Bleomycin: revival of an old drug. Gen. Pharmacol., 27, 745–748. [DOI] [PubMed] [Google Scholar]
- 3.Burger R.M. (1998) Cleavage of nucleic acids by bleomycin. Chem. Rev., 98, 1153–1169. [DOI] [PubMed] [Google Scholar]
- 4.Claussen C.A. and Long,E.C. (1999) Nucleic acid recognition by metal complexes of bleomycin. Chem. Rev., 99, 2797–2816. [DOI] [PubMed] [Google Scholar]
- 5.Povirk L.F. and Austin,M.J. (1991) Genotoxicity of bleomycin. Mutat. Res., 257, 127–143. [DOI] [PubMed] [Google Scholar]
- 6.Absalon M.J., Kozarich,J.W. and Stubbe,J. (1995) Sequence-specific double-strand cleavage of DNA by Fe bleomycin. 1. The detection of sequence-specific double-strand breaks using hairpin oligonucleotides. Biochemistry, 34, 2065–2075. [DOI] [PubMed] [Google Scholar]
- 7.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y.J., Kim,E.Y. and Demple,B. (1998) Excision of C-4′-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase beta. J. Biol. Chem., 273, 28837–28844. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhry M.A., Dedon,P.C., Wilson,D.M., Demple,B. and Weinfeld,M. (1999) Removal by human apurinic/apyrimidinic endonuclease 1 (Ape 1) and Escherichia coli exonuclease III of 3′-phosphoglycolates from DNA treated with neocarzinostatin, calicheamicin and gamma-radiation. Biochem. Pharmacol., 57, 531–538. [DOI] [PubMed] [Google Scholar]
- 10.Suh D., Wilson,D.M., and Povirk,L.F. (1997) 3′-Phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res., 25, 2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D.E. and Lindahl,T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholl I.D., Nealon,K. and Kenny,M.K. (1997) Reconstitution of human base excision repair with purified proteins. Biochemistry, 36, 7557–7566. [DOI] [PubMed] [Google Scholar]
- 13.Parikh S.S., Mol,C.D. and Tainer,J.A. (1997) Base excision repair enzyme family portrait: integrating the structure and chemistry of an entire DNA repair pathway. Structure, 5, 1543–1550. [DOI] [PubMed] [Google Scholar]
- 14.Mol C.D., Parikh,S.S., Putnam,C.D., Lo,T.P. and Tainer,J.A. (1999) DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys. Biomol. Struct., 28, 101–128. [DOI] [PubMed] [Google Scholar]
- 15.Slupphaug G., Mol,C.D., Kavli,B., Arvai,A.S., Krokan,H.E. and Tainer,J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- 16.Mol C.D., Arvai,A.S., Slupphaug,G., Kavli,B., Alseth,I., Krokan,H.E. and Tainer,J.A. (1995) Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell, 80, 869–878. [DOI] [PubMed] [Google Scholar]
- 17.Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,S. and Tainer,J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- 18.Lau A.Y., Scharer,O.D., Samson,L., Verdine,G.L. and Ellenberger,T. (1998) Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell, 95, 249–258. [DOI] [PubMed] [Google Scholar]
- 19.Loeb L.A. and Preston,B.D. (1986) Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet., 20, 201–230. [DOI] [PubMed] [Google Scholar]
- 20.Demple B. and Harrison,L. (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 21.Xanthoudakis S., Smeyne,R.J., Wallace,J.D. and Curran,T. (1996) The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA, 93, 8919–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winters T.A., Henner,W.D., Russell,P.S., McCullough,A. and Jorgensen,T.J. (1994) Removal of 3′-phosphoglycolate from DNA strand-break damage in an oligonucleotide substrate by recombinant human apurinic/apyrimidinic endonuclease 1. Nucleic Acids Res., 22, 1866–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manoharan M., Ransom,S.C., Mazumder,A., Gerlt,J.A., Wilde,J.A., Withka,J.A. and Bolton,P.H. (1988) The characterization of abasic sites in DNA heteroduplexes by site specific labeling with C-13. J. Am. Chem. Soc., 110, 1620–1622. [Google Scholar]
- 24.Manoharan M., Mazumder,A., Ransom,S.C., Gerlt,J.A. and Bolton,P.H. (1988) Mechanism of UV endonuclease-V cleavage of abasic sites in DNA determined by C-13 labeling. J. Am. Chem. Soc., 110, 2690–2691. [Google Scholar]
- 25.Mazumder A., Gerlt,J.A., Rabow,L., Absalon,M.J., Stubbe,J. and Bolton,P.H. (1989) UV endonuclease-V from bacteriophage-T4 catalyzes DNA strand cleavage at aldehydic abasic sites by a syn beta-elimination reaction. J. Am. Chem. Soc., 111, 8029–8030. [Google Scholar]
- 26.Mazumder A., Gerlt,J.A., Absalon,M.J., Stubbe,J., Cunningham,R.P., Withka,J. and Bolton,P.H. (1991) Stereochemical studies of the beta-elimination reactions at aldehydic abasic sites in DNA: endonuclease III from Escherichia coli, sodium hydroxide and Lys-Trp-Lys. Biochemistry, 30, 1119–1126. [DOI] [PubMed] [Google Scholar]
- 27.Mol C.D., Izumi,T., Mitra,S. and Tainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair and coordination. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- 28.Hosfield D.J., Guan,Y., Haas,B.J., Cunningham,R.P. and Tainer,J.A. (1999) Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell, 98, 397–408. [DOI] [PubMed] [Google Scholar]
- 29.Beger R.D. and Bolton,P.H. (1998) Structures of apurinic and apyrimidinic sites in duplex DNAs. J. Biol. Chem., 273, 15565–15573. [DOI] [PubMed] [Google Scholar]
- 30.Goljer I., Kumar,S. and Bolton,P.H. (1995) Refined solution structure of a DNA heteroduplex containing an aldehydic abasic site. J. Biol. Chem., 270, 22980–22987. [DOI] [PubMed] [Google Scholar]
- 31.Wang K.Y., Parker,S.A., Goljer,I. and Bolton,P.H. (1997) Solution structure of a duplex DNA with an abasic site in a dA tract. Biochemistry, 36, 11629–11639. [DOI] [PubMed] [Google Scholar]
- 32.Singh M.P., Hill,G.C., Peoc’h,D., Rayner,B., Imbach,J.L. and Lown,J.W. (1994) High-field NMR and restrained molecular modeling studies on a DNA heteroduplex containing a modified apurinic abasic site in the form of covalently linked 9-aminoellipticine. Biochemistry, 33, 10271–10285. [DOI] [PubMed] [Google Scholar]
- 33.Withka J.M., Wilde,J.A., Bolton,P.H., Mazumder,A. and Gerlt,J.A. (1991) Characterization of conformational features of DNA heteroduplexes containing aldehydic abasic sites. Biochemistry, 30, 9931–9940. [DOI] [PubMed] [Google Scholar]
- 34.Cuniasse P., Fazakerley,G.V., Guschlbauer,W., Kaplan,B.E. and Sowers,L.C. (1990) The abasic site as a challenge to DNA-polymerase—a nuclear-magnetic-resonance study of G, C and T opposite a model abasic site. J. Mol. Biol., 213, 303–314. [DOI] [PubMed] [Google Scholar]
- 35.Coppel Y., Berthet,N., Coulombeau,C., Garcia,J. and Lhomme,J. (1997) Solution conformation of an abasic DNA undecamer duplex d(CGCACXCACGC) × d(GCGTGTGTGCG): the unpaired thymine stacks inside the helix. Biochemistry, 36, 4817–4830. [DOI] [PubMed] [Google Scholar]
- 36.Cuniasse P., Sowers,L.C., Eritja,R., Kaplan,B., Goodman,M.F., Cognet,J.A., LeBret,M., Guschlbauer,W. and Fazakerley,G.V. (1987) An abasic site in DNA. Solution conformation determined by proton NMR and molecular mechanics calculations. Nucleic Acids Res., 15, 8003–8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalnik M.W., Chang,C.N., Grollman,A.P. and Patel,D.J. (1988) NMR studies of abasic sites in DNA duplexes: deoxyadenosine stacks into the helix opposite the cyclic analogue of 2-deoxyribose. Biochemistry, 27, 924–931. [DOI] [PubMed] [Google Scholar]
- 38.Cline S.D., Jones,W.R., Stone,M.P. and Osheroff,N. (1999) DNA abasic lesions in a different light: solution structure of an endogenous topoisomerase II poison. Biochemistry, 38, 15500–15507. [DOI] [PubMed] [Google Scholar]
- 39.Sklenar V., Piotto,M., Leppik,R. and Saudek,V. (1993) Gradient-tailored water suppression for H-1-N-15 experiments optimized to retain full sensitivity. J. Magn. Reson. Ser. A, 102, 241–245. [Google Scholar]
- 40.Bax A. and Lerner,L. (1988) Measurement of H-1-H-1 coupling-constants in DNA fragments by 2D NMR. J. Magn. Reson., 79, 429–438. [Google Scholar]
- 41.Sklenar V. and Bax,A. (1987) Measurement of H-1-P-31 NMR coupling-constants in double-stranded DNA fragments. J. Am. Chem. Soc., 109, 7525–7526. [Google Scholar]
- 42.Geen H. and Freeman,R. (1991) Band-selective radiofrequency pulses. J. Magn. Reson., 93, 93–141. [Google Scholar]
- 43.Lankhorst P.P., Haasnoot,C.A.G., Erkelens,C. and Altona,C. (1984) Nucleic-acid constituents. 36. C-13 NMR in conformational-analysis of nucleic-acid fragments. 2. A reparametrization of the Karplus equation for vicinal NMR coupling-constants in Ccop and Hcop fragments. J. Biomol. Struct. Dyn., 1, 1387–1405. [DOI] [PubMed] [Google Scholar]
- 44.Chary V.V.R., Rastogi,V.K. and Govil,G. (1993) An efficient 2D NMR technique HELCO for heteronuclear [31P-1H] long-range correlation. J. Magn. Reson. Ser. B, 102, 81–83. [Google Scholar]
- 45.Sklenar V., Miyashiro,H., Zon,G., Miles,H.T. and Bax,A. (1986) Assignment of the P-31 and H-1 resonances in oligonucleotides by two-dimensional NMR-spectroscopy. FEBS Lett., 208, 94–98. [DOI] [PubMed] [Google Scholar]
- 46.Markley J.L., Bax,A., Arata,Y., Hilbers,C.W., Kaptein,R., Sykes,B.D., Wright,P.E. and Wuthrich,K. (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Mol. Biol., 280, 933–952. [Reprinted from Pure Appl. Chem. (1998) 70, 117–142] [DOI] [PubMed] [Google Scholar]
- 47.van Wijk J., Huckriede,B.D., Ippel,J.H. and Altona,C. (1992) Furanose sugar conformations in DNA from NMR coupling constants. Methods Enzymol., 211, 286–306. [DOI] [PubMed] [Google Scholar]
- 48.Haasnoot C.A.G., de Leeuw,F.A.A.M. and Altona,C. (1980) The relationship between proton-proton NMR coupling constants and substituent electronegativities. Tetrahedron, 36, 2783–2796. [Google Scholar]
- 49.Haasnoot C.A.G., de Leeuw,F.A.A.M., de Leeuw,H.P.M. and Altona,C. (1981) The relationship between proton-proton NMR coupling constants and substituent electronegativities: II. Conformational analysis of the sugar ring in nucleosides and nucleotides in solution using a generalized Karplus equation. Org. Magn. Reson., 15, 43–52. [Google Scholar]
- 50.de Leeuw H.P.M., Haasnoot,C.A.G. and Altona,C. (1980) Empirical correlation between conformational parameters in β-D-furanoside fragments derived from a statistical survey of crystal structures of nucleic acid constituents: full description of nucleoside molecular geometries in terms of four parameters. Isr. J. Chem., 20, 108–126. [Google Scholar]
- 51.MacKerell A.D., Wiorkiewiczkuczera,J. and Karplus,M. (1995) An all-atom empirical energy function for the simulation of nucleic-acids. J. Am. Chem. Soc., 117, 11946–11975. [Google Scholar]
- 52.Boelens R., Koning,T.M.G. and Kaptein,R. (1988) Determination of biomolecular structures from proton-proton NOEs using a relaxation matrix approach. J. Mol. Struct., 173, 299–311. [Google Scholar]
- 53.Boelens R., Koning,T.M.G., Vandermarel,G.A., Vanboom,J.H. and Kaptein,R. (1989) Iterative procedure for structure determination from proton proton NOEs using a full relaxation matrix approach—application to a DNA octamer. J. Magn. Reson., 82, 290–308. [Google Scholar]
- 54.Wijmenga S.S., Mooren,M.W. and Hilbers,C.W. (1993) NMR of nucleic acids: from spectrum to structure. In Roberts,G.C.K. (ed.), NMR of Macromolecules: A Practical Approach. Oxford University Press, Oxford, UK, pp. 217–283.
- 55.Cortes S.J., Mega,T.L. and Van Etten,R.L. (1991) The 18-O isotope shift in 13-C nuclear magnetic resonance spectroscopy. 14. Kinetics of oxygen exchange at the anomeric carbon of D-ribose and D-2-deoxyribose. J. Org. Chem., 56, 943–947. [Google Scholar]
- 56.Carey D.C. and Strauss,P.R. (1999) Human apurinic/apyrimidinic endonuclease is processive. Biochemistry, 38, 16553–16560. [DOI] [PubMed] [Google Scholar]
- 57.Marathias V.M., Jerkovic,B. and Bolton,P.H. (1999) Damage increases the flexibility of duplex DNA. Nucleic Acids Res., 27, 1854–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masuda Y., Bennett,R.A. and Demple,B. (1998) Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J. Biol. Chem., 273, 30352–30359. [DOI] [PubMed] [Google Scholar]
- 59.Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waters T.R., Gallinari,P., Jiricny,J. and Swann,P.F. (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem., 274, 67–74. [DOI] [PubMed] [Google Scholar]
- 61.Bennett R.A., Wilson,D.M., Wong,D. and Demple,B. (1997) Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl Acad. Sci. USA, 94, 7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.