Significance
Nearly all proteins are posttranslationally modified, a phenomenon known to alter protein function. Recently, multiple posttranslational modifications (PTMs) have been documented to exist on the same proteins, revealing an additional level of complexity (named “PTM crosstalk”) that, due to its dynamic nature, is challenging to predict. Here, we propose a motif for PTM crosstalk between two of the most common PTMs: phosphorylation and O-GlcNAcylation. Through the use of a kinetic-based high-resolution mass spectrometry assay, we highlight specific residues that, when phosphorylated, hamper O-GlcNAcylation at nearby sites. In addition, we show that the Ser/Thr residues in one of the most common kinase motifs, PX(S/T)P, cannot be O-GlcNAcylated, demonstrating that reciprocal PTM crosstalk does not occur with Pro-directed kinases.
Keywords: O-GlcNAcylation, phosphorylation, crosstalk, signaling, regulation
Abstract
Proteins can be modified by multiple posttranslational modifications (PTMs), creating a PTM code that controls the function of proteins in space and time. Unraveling this complex PTM code is one of the great challenges in molecular biology. Here, using mass spectrometry-based assays, we focus on the most common PTMs—phosphorylation and O-GlcNAcylation—and investigate how they affect each other. We demonstrate two generic crosstalk mechanisms. First, we define a frequently occurring, very specific and stringent phosphorylation/O-GlcNAcylation interplay motif, (pSp/T)P(V/A/T)(gS/gT), whereby phosphorylation strongly inhibits O-GlcNAcylation. Strikingly, this stringent motif is substantially enriched in the human (phospho)proteome, allowing us to predict hundreds of putative O-GlcNAc transferase (OGT) substrates. A set of these we investigate further and show them to be decent substrates of OGT, exhibiting a negative feedback loop when phosphorylated at the P-3 site. Second, we demonstrate that reciprocal crosstalk does not occur at PX(S/T)P sites, i.e., at sites phosphorylated by proline-directed kinases, which represent 40% of all sites in the vertebrate phosphoproteomes.
Proteins are posttranslationally modified, yet the frequency at which these modifications occur and how they regulate protein function is largely unexplored. Over 450 different posttranslational modifications (PTMs) exist, having roles in cell signaling, regulation of gene expression, or protein–protein interactions (1). Many proteins are modified by multiple PTMs. These PTMs can affect each other in what is generally termed “PTM crosstalk,” which has become an intense subject of current research (2, 3). Examples of crosstalk entail the priming phosphorylation on a protein substrate by a single kinase that enables other kinases to phosphorylate sites in the neighborhood. One key example is the phosphorylation of Tau by the Cdk5 that leads to Tau hyperphosphorylation by GSK3β (4). Additionally, the interplay between ubiquitination and phosphorylation in the regulation of the EGF-mediated ERK signaling pathway represents a clear example of how two different PTMs can affect each other (5).
Another biologically intriguing PTM crosstalk is that between O-GlcNAcylation and phosphorylation, as these both target the same Ser/Thr amino acids (3, 6, 7). O-GlcNAcylation, whereby a β-N-acetylglucosamine (O-GlcNAc) is added to a protein by the enzyme O-β-(N-acetyl) glucosamine transferase (OGT), regulates proteins involved in cell division, metabolism, and cell signaling. Like phosphorylation, O-GlcNAcylation is a dynamic modification. Virtually every reported OGT substrate is also a phosphoprotein, making them all potentially affected by crosstalk (8). Initially, reciprocal interplay, whereby phosphorylation and O-GlcNAcylation occur on the same Ser/Thr site, was suggested as a major crosstalk mechanism (9). However, while reciprocal relationships between phosphorylation and O-GlcNAcylation have been suggested on a few proteins (10, 11), this “yin–yang” reciprocal model proved to be oversimplified. For instance, crosstalk can also occur whereby the O-GlcNAcylation/phosphorylation of a Ser/Thr residue is differentially affected by a phosphorylated/O-GlcNAcylated residue in close proximity. Experimental evidence for this crosstalk came from studies in which cellular stimuli were shown to increase both O-GlcNAcylation and phosphorylation levels simultaneously (6, 7).
Although, many studies have demonstrated that crosstalk occurs, studies that identify and colocalize both the phosphorylation and O-GlcNAcylation sites simultaneously on the same protein/peptide are sporadic. Thus, doubt still exists as to whether on a specific protein molecule O-GlcNAcylation directly affects phosphorylation or vice versa. Work by Trinidad et al. (12) suggested that phosphorylation and O-GlcNAcylation occur randomly with respect to each another and that negative crosstalk is likely not a common phenomenon. From that work it can be hypothesized that phosphorylation/O-GlcNAcylation crosstalk is not really widespread. Here, we address this hypothesis, assuming that new and better tools are needed to explore phosphorylation/O-GlcNAcylation crosstalk.
PTMs generically increase the mass of a substrate; therefore MS-based methods are ideal to investigate PTMs. Here, we use high-resolution MS to investigate the kinetics and substrate specificity of human OGT on an extensive library of substrate-mimicking peptides, whereby we addressed how such reactions are dependent on substrate sequence and phosphorylation state. We show that phosphorylation at specific sites on OGT substrates hampers O-GlcNAcylation, enabling us to define a phosphorylation/O-GlcNAcylation crosstalk motif based on kinetic data: (pST)P(TVA)(gST). This motif is enriched in the human proteome, strongly suggesting a positive evolutionary selection. Furthermore, our analysis revealed the critical role of a Pro at P+1 in this consensus motif in preventing O-GlcNAcylation, suggesting that reciprocal crosstalk is blocked when phosphorylation has been enforced by a Pro-directed kinase. As these phosphorylation events represent in frequency about 40% of all phosphorylation sites in human cells, this finding represents a significant step forward in predicting and evaluating PTM crosstalk.
Results
Phosphorylation at Specific Sites Hampers O-GlcNAcylation.
In an initial attempt to investigate whether phosphoproteins can be O-GlcNAcylated, we enriched phosphopeptides from A549 cells and O-GlcNAcylated them in vitro. As a positive control, we added a casein kinase II (CK2) peptide, a known, good substrate of OGT (13). The CK2 peptide became O-GlcNAcylated on only the predicted Ser347 with 100% occupancy. Intriguingly, although the majority of the phosphopeptides contained unoccupied Ser/Thr residues, only one of the thousands of phosphopeptides on which the phosphate and O-GlcNAc modifications could be mapped was found to be O-GlcNAcylated; this peptide originated from CRMP2 (SI Appendix, Fig. S1 and Table S1). Thus, we speculated that the primed phosphorylation might substantially hamper subsequent O-GlcNAcylation. To verify this, it is important to establish whether the presence of an O-GlcNAc moiety alters phosphopeptide enrichment. Thus, first a mixture of O-GlcNAc–modified and free unphosphorylated peptides were subjected to iron-immobilized metal ion affinity chromatography (Fe-IMAC) enrichment, and their relative abundances were compared before and in the flow through from the Fe-IMAC purification. The ratios of O-GlcNAc:free peptides remained constant, showing that O-GlcNAcylated peptides have no affinity for the Fe-IMAC resin (SI Appendix, Fig. S2A). Next, a mixture of O-GlcNAcylated and unmodified phosphopeptides was analyzed. The ratio of the intensities of the eluted O-GlcNAcylated:free phosphopeptides remained constant before and after elution from the Fe-IMAC column, confirming that the presence of an O-GlcNAc moiety on phosphopeptides does not affect their ability to bind and elute during phosphopeptide enrichment (SI Appendix, Fig. S2B). In additional support of our hypothesis, in vivo studies have shown that, upon treatment with kinase activators or phosphatase inhibitors, the levels of O-GlcNAcylation within cells decrease (14, 15).
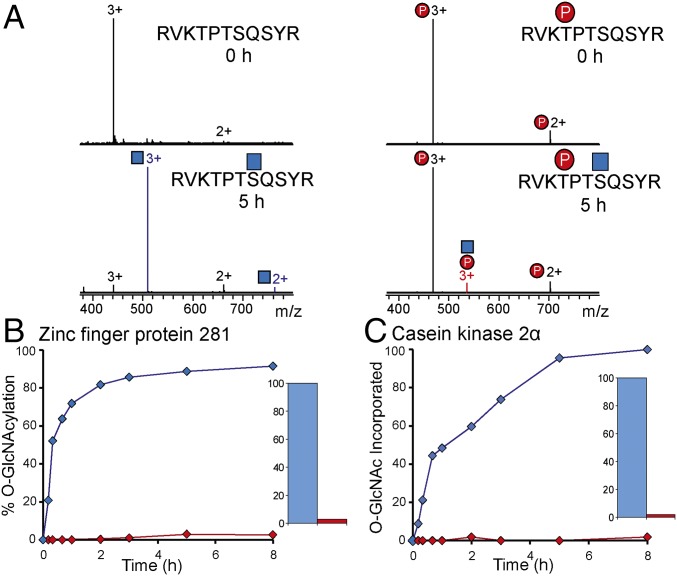
To further test our hypothesis, we synthesized two peptides corresponding to the zinc finger protein 281 (ZNF281res885–895) and CK2res343–365, respectively (SI Appendix, Table S2), as these earlier have been implicated in phosphorylation and O-GlcNAcylation crosstalk. Ser347 O-GlcNAcylation in CK2 was shown to be antagonistic to Thr344 phosphorylation, preventing CK2 from binding to Pin1, a process involved in enhancing CK2 stability (16). O-GlcNAcylation of ZNF281 at Ser891 also plays a key regulatory role that has been suggested to be involved in embryonic stem cell differentiation (17). Interestingly, phosphorylation on Thr888 in ZNF281 has been identified in large-scale phosphoproteomic datasets (18). However, whether direct interplay exists between these PTMs still remains to be verified. Here, we directly probed the effect of phosphorylation of Thr888 on ZNF281 on the rate of O-GlcNAcylation. Fig. 1 shows the mass spectra and the resulting O-GlcNAcylation kinetics of both the nonmodified and the phosphorylated ZNF281 peptide before and after incubation with OGT. Interestingly, less than 5% of the phosphorylated ZNF281 peptide was O-GlcNAcylated after 8 h compared with 90% of the free ZNF281 peptide (Fig. 1B), showing that phosphorylation at Thr888 hampers Ser891 O-GlcNAcylation. Hypothetically, this crosstalk observed could add an additional regulatory role to ZNF281 in stem cell differentiation, in addition to the previously suggested regulatory role of O-GlcNAcylation on Ser891 (17). To decipher whether this observed crosstalk is a more general phenomenon, an unmodified and phosphorylated CK2 peptide was also incubated with OGT, and the relative peptide O-GlcNAcylation levels were monitored (Fig. 1C). Consistent with work by Cole and coworkers (16), the rate of Ser347 O-GlcNAcylation was severely reduced by Thr344 phosphorylation (Fig. 1C).
Fig. 1.
Phosphorylation at P−3 hampers O-GlcNAcylation. (A) MS of free and phosphorylated ZNF281res885–895 upon incubation with OGT for 0 h (Top) and 5 h (Bottom). (B) Rates of O-GlcNAcylation of ZNF281res885–895 with (red trace) and without (blue trace) phosphorylation at P−3. (C) As in B but for CK2res343–365. The Insets show the O-GlcNAc incorporated after 8 h.
MS/MS analysis of the O-GlcNAcylated ZNF281- and CK2-derived peptide sequences revealed in both cases a single O-GlcNAcylation site (SI Appendix, Figs. S3 and S4). Interestingly, these phosphorylation sites that decreased the O-GlcNAcylation rate were located at P−3 with respect to the O-GlcNAcylation sites. Thus, we concluded that phosphorylation specifically at P−3 may reduce the rate of subsequent O-GlcNAcylation. To confirm that crosstalk primarily occurs when phosphorylation is present at P−3, a peptide from CRMP2 whose O-GlcNAcylation acceptor site is well established (19), was synthesized so that residues upstream and downstream of the O-GlcNAcylation site were either left unmodified or phosphorylated (SI Appendix, Table S2). The locations of the phosphorylation sites were chosen based on their previous detection in vivo (20). Interestingly, comparable O-GlcNAcylation levels were observed on Ser517 for the unmodified peptide and the peptide in which Ser522 had been phosphorylated (at P+5 relative to the O-GlcNAc site) (SI Appendix, Figs. S5 and S6). This reveals that phosphorylation downstream of the O-GlcNAcylation site does not substantially alter the rate of O-GlcNAcylation for CRMP2. In contrast, the quadruply phosphorylated CRMP2 peptide (Thr509, Thr514, Ser518, and Ser522) showed a dramatic decrease in O-GlcNAcylation kinetics with only 2% of this peptide being O-GlcNAcylated. Interestingly, these results on CRMP2 are in line with our aforementioned experiments, in which a large phosphopeptide library derived from A549 cells was O-GlcNAcylated. In these experiments, CRMP2 peptides were detected in which Thr509, Thr514, Ser518, and Ser522 were phosphorylated. However, upon O-GlcNAcylation of this phosphopeptide library, only one phosphopeptide became O-GlcNAcylated in which the phosphorylation site was at P+5 (Ser522) with respect to the O-GlcNAcylation site (Ser517) (SI Appendix, Fig. S1).
Since phosphorylation at P-3 seems to specifically constrain O-GlcNAcylation rates, we sought to investigate whether O-GlcNAcylation may also hamper protein phosphorylation. Therefore, we selected two in vitro O-GlcNAcylated peptide sequences (ZNF281res885–895 and CRMP2res507–525) and incubated them with a relevant kinase (Fig. 2). In neurons, phosphorylation of CRMP2 occurs initially at Ser522 by Cdk5, which primes CRMP2 for subsequent phosphorylation by GSK3β at Ser518 and Thr514 (21). Thus, the O-GlcNAcylated CRMP2 peptide was first incubated with Cdk5, followed by incubation with GSK3β. Interestingly, both the free and O-GlcNAcylated CRMP2 were phosphorylated on Ser522 with similar rates by Cdk5 (Fig. 2A and SI Appendix, Fig. S7); however, upon the addition of GSK3β, the presence of the O-GlcNAc moiety hampered phosphorylation, with only two phosphate groups incorporated on average compared with three for the unmodified CRMP2 (Fig. 2B). The O-GlcNAcylated ZNF281 peptide also hindered subsequent phosphorylation in assays in which we used six different protein kinases (Fig. 2C). Together, the data show that O-GlcNAcylation hampers phosphorylation, thus demonstrating that reciprocal crosstalk exists between O-GlcNAcylation and phosphorylation.
Fig. 2.
O-GlcNAcylation hampers phosphorylation on nearby Ser/Thr residues. (A) Normalized phosphate incorporation on the free (gray trace) and O-GlcNAcylated (blue trace) CRMP2res507–525 upon incubation with cdk5p25 kinase, ATP, and Mg2+. (B) The number of phosphorylation sites on the free (gray bar) and O-GlcNAcylated (blue bar) CRMP2 peptide (i.e., the phosphorylation stoichiometry) upon further incubation of the cdk5-phosphorylated CRMP2 peptides with the second kinase, GSK3β (t = 10 h). The error bars represent the SD from the mean. (C) Phosphorylation status of ZNF281res885-–895 upon incubation with the kinases PKA, ERK1, ERK2, cdk1cyclinA2, GSK3β, and cdk5p25. The normalized phosphate incorporation is shown for the free and O-GlcNAcylated ZNF281 peptide.
Phosphorylation/O-GlcNAcylation Crosstalk Sites Identified.
Very few data exist wherein O-GlcNAcylation and phosphorylation are detected simultaneously on the same protein molecule. Thus, we hypothesized that the phosphorylation/O-GlcNAcylation interplay reported here could represent a generic regulatory mechanism. We defined a stringent putative motif: (T/S)P(V/T/A)(S/T) (the O-GlcNAc and phospho sites are highlighted in bold and italic, respectively) (Fig. 3A). This motif is based on several sets of reported data, namely the O-GlcNAcylation motif extracted from the 26 best-known OGT substrates (22), the PX(S/T) O-GlcNAc motif described for HLA class I-bound peptides (23), the protein sequences O-GlcNAcylated in this study (SI Appendix, Table S2), and the finding reported in this paper that phosphorylation at P−3 hampers O-GlcNAcylation. Using bioinformatics analysis, we found that these sequences of four amino acids not only are very common in the human proteome (Fig. 3B) but also are significantly overrepresented (more than two times the number of occurrences), hinting at a positive evolutionary selection for either structural or functional reasons (Fig. 3B). Interestingly, more than 40% of these sequences have been deposited in the PhosphoSitePlus database in which 5,968 protein sequences had one or multiple Ser/Thr residues within the motif phosphorylated. Of all these sequences, 1,286 had a phosphorylated Ser/Thr residue at P−3. We next queried whether these 1,286 protein sequences might not only be substrates of kinases and OGT but also might display interplay in between phosphorylation and O-GlcNAcylation, whereby phosphorylation at P−3 in this consensus sequence should prevent O-GlcNAcylation. In support of our hypothesis, a few of these protein sequences have already been reported to be involved in phosphorylation/O-GlcNAcylation crosstalk [e.g., in the eIF4 that plays a role in mRNA translation (24) and the paired amphipathic helix protein Sin3a that acts cooperatively with OGT to repress transcription (25, 26)].
Fig. 3.
Predicting phosphorylation/O-GlcNAcylation crosstalk. (A) Motif for crosstalk with phosphorylation on P−3 and O-GlcNAc on 0. (B) The frequency of all four amino-acid combinations in the human proteome. The sequences matching the predicted crosstalk motif are in red. (C) The number of occurrences of each sequence from the crosstalk motif (red bars) compared with the number expected if this sequence were to occur at random (gray bars), taking into account that some amino acids are more abundant in the human proteome. The sequences that are also deposited in the PhosphoSitePlus database, i.e., those containing phosphorylation residues within or immediately surrounding the motif, are shown in orange. (D) O-GlcNAc incorporation after 24 h on nine bioinformatically predicted OGT substrates (Table 1) with and without phosphorylation at P−3.
To investigate whether crosstalk exists more widely for proteins exhibiting the motif (T/S)P(V/T/A)(S/T) defined here, two criteria must be satisfied. First, proteins containing these motifs must be OGT substrates. Second, phosphorylation at P−3 must decrease the extent of O-GlcNAcylation. Thus, nine substrate-mimicking peptides from the predicted proteins (Table 1 and SI Appendix, Table S3), none previouslyknown to be OGT substrates, were selected and individually incubated with OGT. The nine protein sequences were readily O-GlcNAcylated (Fig. 3C) with the O-GlcNAcylation sites matching precisely those predicted (Table 1 and SI Appendix, Figs. S8–S16 and Table S3). Next, the same nine peptides were synthesized with a phosphorylated Ser/Thr at P−3 (Table 1 and SI Appendix, Table S3) and were again incubated with OGT (Fig. 3C). In eight of the nine phosphopeptides, the O-GlcNAcylation became fully blocked. Only for the centromere protein V peptide was a minor amount of O-GlcNAcylation detected, albeit substantially less compared with its unphosphorylated peptide counterpart. Thus, since in all peptides analyzed, crosstalk was observed, we propose that all proteins containing the sequence motif (T/S)P(V/T/A)(S/T) could represent bona fide OGT substrates and be regulated by phosphorylation/O-GlcNAcylation crosstalk.
Table 1.
Pro and phosphorylation hamper promiscuous O-GlcNAcylation
| Peptide sequence | Predicted O-GlcNAc site | O-GlcNAc site | % O-GlcNAc incorporated |
| LSPATPTSEG | Thr377,Ser380 | Ser380 | 65.3 |
| LSPApTPTSEG | Thr377,Ser380 | Ser380 | 4.0 |
| LpSPATPTSEG | Thr377,Ser380 | Ser380 | 57.0 |
| SPGSTPTTPTSSQA | Thr437,Ser440 | Ser440 | 55.0 |
| SPGpSTPTTPTSSQA | Thr437,Ser440 | Ser440 | 45.2 |
| SPGSpTPTTPTSSQA | Thr437,Ser440 | Ser440 | 50.7 |
| SPGSTPTTPpTSSQA | Thr437,Ser440 | Ser440 | 0 |
| RKPVTVSPTTPTSPTEG | Thr508,Thr513, Ser516 | Thr508 | 9.1 |
ZNF687, Nup214, and dynein 1 light chain intermediate peptides contain multiple Ser/Thr residues but harbor only a single O-GlcNAc site.
Next we validated whether this crosstalk pertains in cellular systems. Using a specific O-GlcNAcase (OGA) inhibitor, GlcNAcstatin G, we substantially up-regulated the O-GlcNAcylation levels in human HeLa cells (SI Appendix, Fig. S17A) and queried whether this affected the extent of phosphorylation on peptides (following enrichment from HeLa cells using Fe-IMAC) harboring the (T/S)P(V/T/A)(S/T) sequence motif (Fig. 3A). Considering that reciprocal crosstalk exists between O-GlcNAcylation and phosphorylation (Fig. 2), our assumption was that if negative crosstalk occurs, whereby phosphorylation is at P−3 with respect to a putative O-GlcNAcylation site, the amount of phosphorylation will decrease. In the HeLa cells treated with the OGA inhibitor, 10 phosphorylated peptides were detected that harbored the (T/S)P(V/T/A)(S/T) sequence motif, all being down-regulated (SI Appendix, Fig. S17B), providing evidence that this negative crosstalk holds true in vivo for these substrates. Moreover, in vitro experiments showed nine of these down-regulated phosphopeptides were indeed OGT substrates (SI Appendix, Table S4). In addition, upon incubation of four of these peptides with the kinases cdk5 or p38α, decreased phosphate incorporation at P−3 was observed when compared with their O-GlcNAcylated (at P+3 with respect to the phosphorylation site) and unmodified counterparts (SI Appendix, Fig. S18).
Reciprocal Interplay Does Not Occur on Ser/Thr Sites Targeted by Pro-Directed Kinases.
Reciprocal phosphorylation/O-GlcNAcylation on the same site has been hypothesized to occur on various proteins; however, the frequency and biological relevance of this phosphorylation/O-GlcNAcylation competition in vivo remains unclear. Upon analysis of the 5,968 protein sequences containing our O-GlcNAcylation motif, (S/T)P(VTA)(S/T), 890 contained a phosphorylated Ser/Thr at the O-GlcNAc site followed by a Pro, i.e., putative substrates of Pro-directed kinases. To decipher whether reciprocal interplay occurs at Pro-directed kinase substrate sites, eight peptides were synthesized mimicking these putative substrates originating from phosphoproteins with diverse cellular functions (Table 1 and SI Appendix, Table S3). We hypothesized that if reciprocal interplay occurs at these sites, these protein sequences should also be good substrates of OGT, as they conform to our high-fidelity OGT substrate motif. Surprisingly, although all these eight peptides should undergo reciprocal interplay based on the computational prediction software Ying-O-Yang (27), none of them did, as no O-GlcNAcylation was observed (Table 1 and SI Appendix, Fig. S19). Thus, the presence of a Pro at P+1 has a dramatic obstructing effect on O-GlcNAcylation. To verify this preventative role of Pro on O-GlcNAcylation, a substrate-mimicking peptide from the PEST proteolytic signal-containing nuclear protein that is known to be a good substrate of OGT was synthesized. Two peptides were synthesized in which the naturally occurring alanine residue at P+1 was intentionally substituted with Pro (Fig. 3). Fig. 3A shows mass spectra after incubation with OGT for both the natural and Ala/Pro substitution. In contrast to the authentic protein sequence in which 60% was O-GlcNAc modified, no O-GlcNAcylation was observed when the +1 amino acid was substituted to Pro (Fig. 3A). Next, we chose a substrate-mimicking peptide in which the naturally occurring Pro residing at P+1 prevented O-GlcNAcylation and mutated this Pro to Ala (Fig. 4). No O-GlcNAcylation was observed with the natural protein sequence; however, O-GlcNAcylation was readily observed when the Pro was substituted for Ala. Electron transfer dissociation (ETD)-MS/MS confirmed that the O-GlcNAcylation site matched that predicted site, based on our strict O-GlcNAcylation motif (Figs. 3A and 4B). Therefore, our results show a strict negative preference for Pro at P+1 with respect to the O-GlcNAcylation site. From these data we conclude that reciprocal phosphorylation/O-GlcNAcylation interplay does not occur on sites targeted by Pro-directed kinases. This finding is of tremendous importance, since Pro-directed kinases represent in frequency about 40% of all human phosphorylation sites (28, 29).
Fig. 4.
Pro at P+1 obstructs O-GlcNAcylation. (A) MS of the PEST proteolytic signal-containing nuclear proteinres137–146 (Upper) and the same protein in which Ala at P+1 is substituted to Pro (Lower), upon incubation with OGT for 24 h. (B) Mass spectra of the DNA-binding protein RFX7res1023–1032 (Upper) and the same peptide sequence in which Pro at P+1 is substituted to an Ala (Bottom), upon incubation with OGT for 24 h. ETD-MS/MS confirmed the O-GlcNAc sites. Blue squares and asterisks indicate O-GlcNAc-containing and contaminant peaks, respectively. The percent O-GlcNAcylation detected is shown at right.
Using the same phosphoproteomics readout described above in the HeLa cells treated with the OGA inhibitor, we assumed that phosphopeptides harboring the (T/S)P(V/T/A)(S/T)P motif should not be affected. In the HeLa cells treated with the OGA inhibitor, 12 phosphopeptides containing the (S/T)P(V/A/T)(pS/pT)P motif were detected, all displaying no difference in phosphopeptide abundance (SI Appendix, Fig. S17C). Two of these detected phosphopeptides, protein FAM122B and cytoplasmic dynein 1 light intermediate chain 1, matched those made synthetically, (Tables 1 and 2 and SI Appendix, Table S3).
Table 2.
Peptides used to identify crosstalk motif
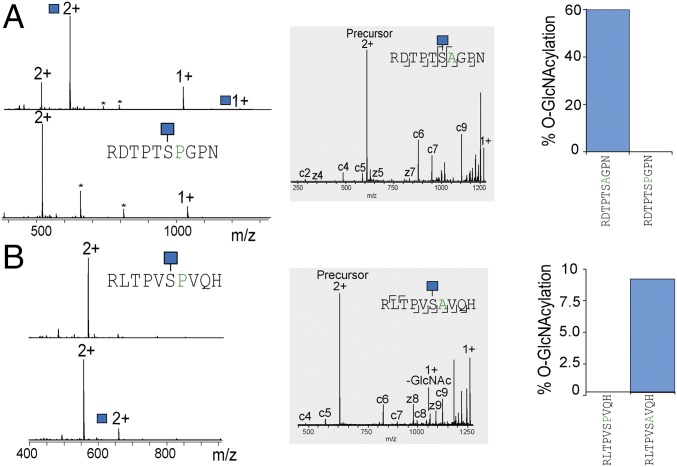
| Peptide sequence | Phospho site | Predicted O-GlcNAc site | O-GlcNAc confirmed |
| RSTTPTSSPF* | Thr2682 | Ser2685 | Yes |
| RDTPTSAGPN* | Thr139 | Ser142 | Yes |
| EKTPVSGSLK* | Thr1385 | Ser1388 | Yes |
| EKLTPTSKQL* | Thr678 | Ser681 | Yes |
| IPMTPTSSFV* | Thr392 | Ser395 | Yes |
| GPDTPVSADA* | Thr289 | Ser292 | Yes |
| LPTPTSAPTP* | Thr1191 | Ser1192 | Yes |
| TPATPTSSAS* | Thr101 | Ser104 | Yes |
| PEPSTPVSGQ* | Thr1226 | Ser1229 | Yes |
| RLTPVSPVQH† | Ser1028 | Ser1028 | ND |
| QKTPTSPLSP† | Ser1256 | Ser1256 | ND |
| SPRTPVSPVK† | Thr444,Ser447 | Ser447 | ND |
| DFTPVSPAPSPTR† | Ser115,Ser119 | Ser115 | ND |
| SRPSPGTPTSPSN† | Ser479,Thr482, Thr484,Ser485, Ser487 | Ser485 | ND |
| KSTWTPTSPG† | Ser684 | Ser684 | ND |
| LNPGTPVSPQ† | Ser2203 | Ser2203 | ND |
| YSPTSPSYSPTSPS† | x | x | ND |
ND, no O-GlcNAcylation detected; x, two heptad repeats from RNA polymerase II; thus this 14mer peptide represents any/all of these repeats.
Proteins with (S/T)PX(S/T) motif for which evidence exists that the first S/T can be phosphorylated. The predicted and observed O-GlcNAc sites are shown. The effect phosphorylation has on O-GlcNAcylation is shown in Fig. 3D.
Proteins that additionally have a Pro at P+1.
Thus, based on in vitro screens, both our hypotheses about crosstalk in (T/S)P(V/T/A)(S/T) and (T/S)P(V/T/A)(S/T)P motifs, seem to hold in human cells. Still further in vivo studies are required, as our assumptions are likely oversimplifying the genuine in vivo situation. Indeed, it is possible that O-GlcNAcylation levels could affect phosphorylation of these sites indirectly. In addition, we have not taken into account more regulatory levels of PTM crosstalk that occur in cells, such as the possible activation/deactivation of kinases by O-GlcNAcylation (30, 31).
Pro Prevents Promiscuous O-GlcNAcylation.
Ser/Thr-Pro–rich regions occur in proteins with high frequency. Most of these proteins are modified by a plethora of PTMs. Due to the broad specificity of protein kinases, multiple Ser/Thr residues within protein sequences can become phosphorylated. In sharp contrast, OGT has a much more restricted substrate sequence specificity (22); thus O-GlcNAcylation typically occurs at only one or a few Ser/Thr residues within these Ser/Thr-rich sequences. It has been suggested that there is little OGT substrate bias toward a specific amino acid at P+1 (22), in contrast to our finding that Pro at P+1 blocks O-GlcNAcylation. We investigated, therefore, whether Pro could act to guide OGT to specific Ser/Thr sites. Therefore, three Ser/Thr-rich peptides from the proteins ZNF687, Nup214, and cytoplasmic dynein 1 light intermediate chain 1 were synthesized (Table 2 and SI Appendix, Table S3). Interestingly, despite the fact that these contained multiple predicted O-GlcNAcylation sites (27), only one Ser/Thr on each peptide became O-GlcNAcylated (Table 2). In all three cases, the O-GlcNAcylation motif was PX(S/T) (the O-GlcNAc site is in bold, and “X” represents any amino acid) (SI Appendix, Figs. S20–S22). No O-GlcNAcylation occurred at the PX(S/T) motif when Pro was at P+1 with respect to the O-GlcNAcylation site. Thus, OGT shows a strict negative preference of Pro at P+1, preventing promiscuous O-GlcNAcylation in Ser/Thr-Pro–rich regions of proteins.
The Nup214 and ZNF687 sequences selected contain multiple reported phosphorylation sites in close proximity to the O-GlcNAcylation site observed. Thus, in addition to Pro preventing promiscuous O-GlcNAcylation, according to our phosphorylation/O-GlcNAcylation consensus motif, phosphorylation at specific sites may also prevent O-GlcNAcylation on these proteins. Therefore, ZNF687res373–382 was synthesized, in which we phosphorylated Ser374 and Thr377 (at P−6 and P−3, respectively, with respect to the O-GlcNAc site), and the percentage O-GlcNAcylation on the phosphopeptides was compared with that of the unmodified peptide (Table 2). The phosphosites were chosen based on their reported detection (32). Consistent with our previous data on CRMP2 (SI Appendix, Fig. S6), phosphorylation at P−3 dramatically reduced the rate of O-GlcNAcylation, whereas the P−6 phosphosite had little effect (Table 2 and SI Appendix, Fig. S23). Next, we synthesized peptides corresponding to Nup214res430–443 in which Ser433, Thr434, and Thr439 were phosphorylated (at P−7, P−6, and P−1, respectively, with respect to the O-GlcNAc site). Upon incubation with OGT, phosphorylation at either the P−6 or the P−7 site had no/very little effect on the O-GlcNAcylation rate, suggesting these are not involved in phosphorylation/O-GlcNAcylation crosstalk (Table 2 and SI Appendix, Fig. S24). In contrast, phosphorylation at P−1 prevented O-GlcNAcylation suggesting that, in addition to phosphorylation at P−3, phosphorylation at P−1 can also hamper the O-GlcNAcylation rate of Nup214 (Table 2 and SI Appendix, Fig. S24).
Data that underscore our finding that Pro at P+1 prevents promiscuous O-GlcNAcylation come from experiments performed on the microtubule-associated protein Tau (33). Based on prediction software (34) and the best known O-GlcNAcylation motif (22), O-GlcNAcylation is predicted to occur on Ser202, Thr205, and Ser208 in Tau (sequence R194SGYSSPGSPGTPGSR209) [i.e., all (S/T)PG(S/T) motifs; the O-GlcNAc site is highlighted in bold]. However, consistent with our work, O-GlcNAcylation was observed only on Ser208 (33), the only predicted O-GlcNAcylation site that does not contain a Pro at P+1. Thus, in addition to ZNF687 and Nup214, Pro can also guide OGT to specific sites on Tau, preventing O-GlcNAcylation on Ser202/Thr205.
Discussion
Previously, two distinctive mechanisms for crosstalk have been reported, namely one whereby O-GlcNAcylation is affected by the presence of a phosphate moiety on nearby Ser/Thr sites (or vice versa) and another whereby phosphorylation and O-GlcNAcylation occur on the same Ser/Thr residues and thus are mutually exclusive, known as “reciprocal interplay.” Here, we interrogate both these scenarios, introducing a MS-based kinetic assay to systematically monitor crosstalk, enabling rules for this PTM crosstalk to be defined.
To address the first mechanism, we show that in the most optimal OGT substrate sequence motif (S/T)P(V/T/A)(S/T), phosphorylation specifically at P−3 with respect to the O-GlcNAc site hinders or even blocks O-GlcNAcylation (Figs. 1 and 5). Phosphorylation on other sites, both up- and downstream of the OGT site, has a much smaller or even a negligible inhibiting effect. This dramatic effect of phosphorylation on P−3 can be partly explained upon examination of the OGT structure in which the CK2 peptide GSTPVSSA occupies the active site [Protein Data Bank (PDB) ID code 4GYY] (Fig. 5A). The P−3 residue is located immediately above the UDP cofactor; thus a phosphate at this position would cause steric hindrance, preventing the peptide from fitting tightly into the OGT active site. In support of this, native MS experiments on the TAB1 protein have shown differing rates of O-GlcNAcylation of Ser391 compared with Ser395, with the rate difference being attributed to the differing residues at P−3 (35). Taken together, our data not only confirm that O-GlcNAcylation is affected by the presence of a phosphate moiety on nearby Ser/Thr sites but also pinpoint this effect precisely to phosphorylation at P−3, a significant insight.
Fig. 5.
Crosstalk mechanisms. (A, Left) An optimal OGT substrate motif predicts a Ser/Thr near the P0 O-GlcNAc site. Phosphorylation at P−3 hampers O-GlcNAcylation. (Right) A structural basis for this observation can be postulated, looking at the OGT active site (gray surface) with UDP (white sticks) and the CK2 peptide substrate (green sticks) bound (PDB ID code 4GYY). CK2 residues are labeled with respect to the O-GlcNAcylation site. The small pocket size at P−3 suggests that the incorporation of a phosphate moiety here is sterically hindered. For clarity, the TPR domain on OGT is omitted. (B) Reciprocal crosstalk between phosphorylation/O-GlcNAcylation is not likely on Pro-directed phosphorylation sites.
Addressing the aforementioned reciprocal mechanism, our data reveal that reciprocal crosstalk cannot occur when the phosphorylation is carried out by a Pro-directed kinase (Fig. 5B and SI Appendix, Fig. S18), or in other words when the residue affected is followed by a Pro. Since in all vertebrates more than 40% of Ser/Thr residues that are phosphorylated are followed by Pro at P+1 (28), reciprocal phosphorylation/O-GlcNAcylation crosstalk, as suggested previously (12), is much more restricted and less frequent than initially proposed.
Finally, we provide insights into how OGT recognizes its substrates. We show that O-GlcNAcylation on our defined motif (S/T)P(V/T/A)(S/T) cannot occur when Pro is present at P+1 (Table 1 and SI Appendix, Fig. S1). Pro thus acts to guide OGT to specific Ser/Thr sites, preventing promiscuous O-GlcNAcylation on proteins containing Ser/Thr-Pro–rich sequences (Table 2 and SI Appendix, Fig. S23). Pro could be unfavorable for two reasons. First, it could prevent the hydrogen-bonding interactions needed for efficient enzyme-substrate recognition. Second, the presence of two Pro residues in the OGT substrate (at P−2 and P+1) could restrict its flexibility that is needed for the protein sequence to thread through the tetratrico peptide repeats (TPR) of OGT into its active site.
In summary, we define a very stringent crosstalk motif, (pSp/T)P(V/A/T)(gS/gT)(X-P), whereby phosphorylation at P−3 inhibits O-GlcNAcylation and “X-P” represents any amino acid except Pro. Since our defined motif is present in the consensus sequences for the kinases ERK1, ERK2, CKI, and GSK3β, this crosstalk likely has specific functional relevance in the protein substrates of these aforementioned protein kinases. All of the protein sequences queried here that contained this motif in the unphosphorylated form are in our assays good substrates of OGT, the modification of which is prevented upon phosphorylation at P−3. Thus, we propose that 1,048 protein sequences [1,286 total (pSp/T)P(V/A/T)(S/T) sequences minus 238 (S/T)P(V/A/T)(S/T)P sequences] in the PhosphoSitePlus database may undergo phosphorylation/O-GlcNAcylation crosstalk. This may even underrepresent the number of proteins that could undergo crosstalk at adjacent sites, as other motifs, such as (S/T)PG(S/T) in the case of Tau (33), and quite different sequences, such as res55-58 (LLPT) in the case of c-myc (11), can also participate in phosphorylation/O-GlcNAcylation crosstalk. In addition, a more global form of crosstalk also occurs whereby the enzymes involved in the crosstalk reported herein themselves contain PTMs that can alter their function (31, 36). For example, OGT has been reported to be activated by tyrosine phosphorylation (36), and CaMKII kinase has been reported to be activated by O-GlcNAcylation (31). Finally, we hypothesize that since the majority of O-GlcNAcylation substrates known to date occur either on intrinsically disordered regions of proteins (37) or cotranslationally before protein folding (38), the results described here will have implications for intact protein substrates, modulating their function in vivo.
Materials and Methods
O-GlcNAcylation Assays.
His-hOGT16–1036 was expressed in Sf9 insect cells and was purified using Ni affinity and size-exclusion chromatography. The synthetic peptides OGT and UDP-GlcNAc were incubated at 37 °C, 300 rpm, pH 8 in an Eppendorf Thermomixer. O-GlcNAcylation was quenched at various time points by dilution into 10% formic acid (FA).
Phosphorylation Assays.
Synthetic peptides, kinase, Mg, and ATP were incubated at 30 °C, 300 rpm in an Eppendorf Thermomixer. Aliquots were taken over time and quenched by the addition of an excess of EDTA and dilution into 10% FA.
Proteomics.
A549 and HeLa cells were lysed, reduced and alkylated, and digested with trypsin, and the resulting phosphopeptides were enriched using Fe-IMAC. For the O-GlcNAcylation assays with the A549 phosphopeptides, CK2res339–352 was added to the phosphopeptide library and subsequently incubated with OGT with a 50-fold excess of UDP-GlcNAc at 37 °C, for 24 h. To monitor crosstalk, the relative abundance of phosphopeptides enriched from WT and HeLa cells treated with O-GlcNAcstatin G were compared using label-free quantitation. Analysis was carried out on an Agilent 1290 UPLC system coupled to an Orbitrap Fusion Tribrid mass spectrometer. For more experimental details, see SI Appendix.
Supplementary Material
Acknowledgments
We thank Henk van den Toorn and Martin Fitzpatrick for the bioinformatics analysis. T. Celine Mulder provided A549 cells. OGT was produced by the Netherlands Cancer Institute protein facility. GlcNAcstatin G was kindly provided by the van Aalten laboratory (University of Dundee). This work was supported by Proteins At Work (Project 184.032.201), a program of the Netherlands Proteomics Centre financed by the Netherlands Organization for Scientific Research (NWO) as part of the National Roadmap Large-Scale Research Facilities of the Netherlands and by the Institute for Chemical Immunology, an NWO Gravitation Project funded by the Ministry of Education, Culture and Science of the Netherlands.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620529114/-/DCSupplemental.
References
- 1.Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: Nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip Rev Syst Biol Med. 2012;4:565–583. doi: 10.1002/wsbm.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: Crosstalk of posttranslational modifications. Proteomics. 2014;14:513–524. doi: 10.1002/pmic.201300344. [DOI] [PubMed] [Google Scholar]
- 3.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engmann O, Giese KP. Crosstalk between Cdk5 and GSK3β: Implications for Alzheimer’s disease. Front Mol Neurosci. 2009;2:2. doi: 10.3389/neuro.02.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen LK, Kolch W, Kholodenko BN. When ubiquitination meets phosphorylation: A systems biology perspective of EGFR/MAPK signalling. Cell Commun Signal. 2013;11:52. doi: 10.1186/1478-811X-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: Site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci USA. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Pandey A, Hart GW. Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol Cell Proteomics. 2007;6:1365–1379. doi: 10.1074/mcp.M600453-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 9.Hart GW, et al. O-linked N-acetylglucosamine: The “yin-yang” of Ser/Thr phosphorylation? Nuclear and cytoplasmic glycosylation. Adv Exp Med Biol. 1995;376:115–123. [PubMed] [Google Scholar]
- 10.Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: Post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001;276:10570–10575. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- 11.Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 12.Trinidad JC, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- 14.Griffith LS, Schmitz B. O-linked N-acetylglucosamine levels in cerebellar neurons respond reciprocally to pertubations of phosphorylation. Eur J Biochem. 1999;262:824–831. doi: 10.1046/j.1432-1327.1999.00439.x. [DOI] [PubMed] [Google Scholar]
- 15.Lefebvre T, et al. Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line. Biochim Biophys Acta. 1999;1472:71–81. doi: 10.1016/s0304-4165(99)00105-1. [DOI] [PubMed] [Google Scholar]
- 16.Tarrant MK, et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat Chem Biol. 2012;8:262–269. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers SA, Panning B, Burlingame AL. Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2011;108:9490–9495. doi: 10.1073/pnas.1019289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigbolt KTG, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 19.Lazarus MB, et al. Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat Chem Biol. 2012;8:966–968. doi: 10.1038/nchembio.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu Y, Hamajima N, Ihara Y. Neurofibrillary tangle-associated collapsin response mediator protein-2 (CRMP-2) is highly phosphorylated on Thr-509, Ser-518, and Ser-522. Biochemistry. 2000;39:4267–4275. doi: 10.1021/bi992323h. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimura T, et al. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Pathak S, et al. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat Struct Mol Biol. 2015;22:744–750. doi: 10.1038/nsmb.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marino F, et al. Extended O-GlcNAc on HLA class-I-bound peptides. J Am Chem Soc. 2015;137:10922–10925. doi: 10.1021/jacs.5b06586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gradi A, et al. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: Coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 26.Hahne H, Gholami AM, Kuster B. Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol Cell Proteomics. 2012;11:843–850. doi: 10.1074/mcp.M112.019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta R, Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 2002;7:310–322. [PubMed] [Google Scholar]
- 28.Giansanti P, et al. An augmented multiple-protease-based human phosphopeptide atlas. Cell Rep. 2015;11:1834–1843. doi: 10.1016/j.celrep.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 29.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dias WB, Cheung WD, Wang Z, Hart GW. Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification. J Biol Chem. 2009;284:21327–21337. doi: 10.1074/jbc.M109.007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erickson JR, et al. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013;502:372–376. doi: 10.1038/nature12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai C-F, et al. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun. 2015;6:6622. doi: 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smet-Nocca C, et al. Identification of O-GlcNAc sites within peptides of the Tau protein and their impact on phosphorylation. Mol Biosyst. 2011;7:1420–1429. doi: 10.1039/c0mb00337a. [DOI] [PubMed] [Google Scholar]
- 34.Steentoft C, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leney AC, Rafie K, van Aalten DMF, Heck AJR. Direct monitoring of protein O-GlcNAcylation by high-resolution native mass spectrometry. ACS Chem Biol. 2017 doi: 10.1021/acschembio.7b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whelan SA, Lane MD, Hart GW. Regulation of the O-linked β-N-acetylglucosamine transferase by insulin signaling. J Biol Chem. 2008;283:21411–21417. doi: 10.1074/jbc.M800677200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishikawa I, et al. Computational prediction of O-linked glycosylation sites that preferentially map on intrinsically disordered regions of extracellular proteins. Int J Mol Sci. 2010;11:4991–5008. doi: 10.3390/ijms11124991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, et al. O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat Chem Biol. 2015;11:319–325. doi: 10.1038/nchembio.1774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.