Significance
Kindlin proteins play crucial roles in the integrin-signaling pathway by directly interacting with and activating integrins, which mediate the cell–extracellular matrix adhesion and signaling. Mutations of kindlins lead to diseases, such as Kindler syndrome, associated with skin blistering and atrophy; leukocyte adhesion deficiency; and cancers. However, the molecular basis underlying kindlin-mediated integrin activation remains to be determined. Here, we report the structural basis of the specific interaction between kindlins and integrins. Furthermore, we demonstrate that kindlins synergize integrin activation by forming a dimer, providing a model for understanding integrin signaling. Finally, we interpret disease-causing mutations found in kindlins at the atomic level, which can be useful for understanding and treating these diseases.
Keywords: kindlin, fermitin, FERMT2, Mig-2, integrin signaling
Abstract
Kindlins and talins are integrin-binding proteins that are critically involved in integrin activation, an essential process for many fundamental cellular activities including cell-matrix adhesion, migration, and proliferation. As FERM-domain–containing proteins, talins and kindlins, respectively, bind different regions of β-integrin cytoplasmic tails. However, compared with the extensively studied talin, little is known about how kindlins specifically interact with integrins and synergistically enhance their activation by talins. Here, we determined crystal structures of kindlin2 in the apo-form and the β1- and β3-integrin bound forms. The apo-structure shows an overall architecture distinct from talins. The complex structures reveal a unique integrin recognition mode of kindlins, which combines two binding motifs to provide specificity that is essential for integrin activation and signaling. Strikingly, our structures uncover an unexpected dimer formation of kindlins. Interrupting dimer formation impairs kindlin-mediated integrin activation. Collectively, the structural, biochemical, and cellular results provide mechanistic explanations that account for the effects of kindlins on integrin activation as well as for how kindlin mutations found in patients with Kindler syndrome and leukocyte-adhesion deficiency may impact integrin-mediated processes.
Integrins, composed of α- and β-subunits, are the major receptors mediating the cell–extracellular matrix (ECM) adhesion (1–3). By connecting specific ECM proteins and diverse cytoskeletal regulators, integrins mediate bidirectional transmembrane signaling (4, 5). Stable integrin–ECM interaction and subsequent signaling require integrin activation, which was reported to be mediated by talin, a 4.1-protein/ezrin/radixin/moesin (FERM) domain-containing protein (6). Recently, kindlins, another family of FERM-containing proteins, were found to play crucial roles in integrin activation and signaling (7–12).
The kindlin family consists of three members in vertebrates, kindlin1/2/3, each containing a FERM domain and a PH domain (Fig. 1A) (13). Compared with the typical FERM domain that consists of three lobes (F1, F2, and F3), kindlin-FERM contains an additional N-terminal F0 lobe. In kindlins, the F1 and F2 lobes are split by a largely unstructured insertion and the PH domain, respectively (Fig. 1A). Kindlins, although sharing high sequence similarity (SI Appendix, Fig. S1), show distinct tissue distributions and nonredundant functions. Kindlin1 is expressed mainly in epithelia, and nonfunctional kindlin1 mutations lead to Kindler syndrome, a congenital skin disease (14–16). Expression of kindlin3 is restricted to the hematopoietic system, and mutations in kindlin3 were found to associate with leukocyte-adhesion deficiency type III (LADIII) (17, 18). Kindlin2 is ubiquitously expressed, and loss of kindlin2 in mice leads to peri-implantation lethality (11). Kindlins are also involved in tumorigenesis and metastasis (19). The kindlin-associated diseases are due, at least in part, to impaired integrin activation, focal adhesion (FA) formation, and cell spreading (10, 12). However, due to the lack of structure information, the molecular mechanisms underlying kindlin-mediated integrin activation and signaling remain elusive. The current understanding of integrin recognition by kindlins largely relies on comparisons to the talin/integrin structure, despite the fact that kindlins and talins play distinct roles in integrin signaling.
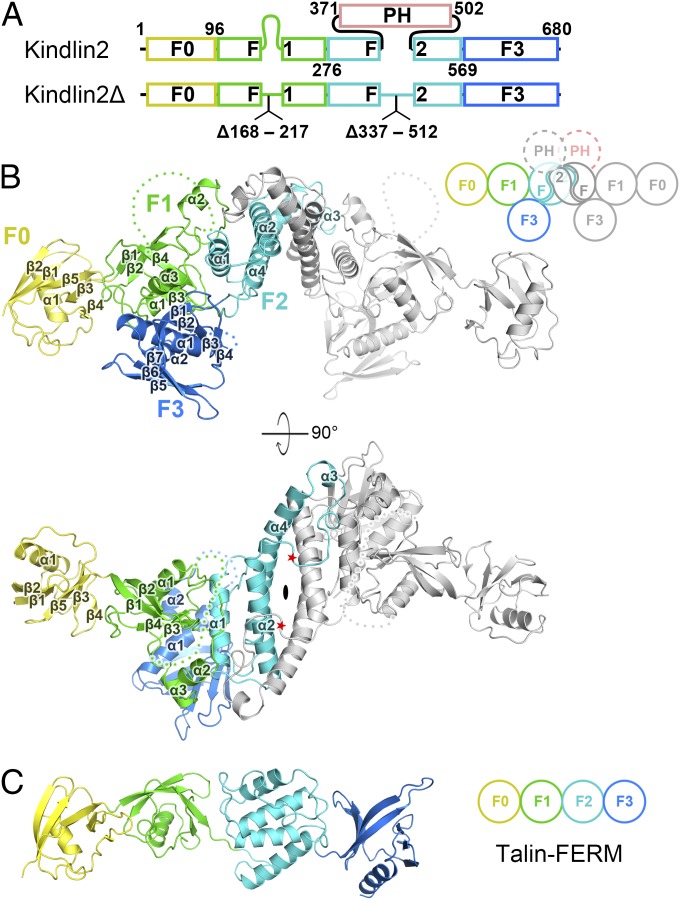
Fig. 1.
Overall structure of kindlin2. (A) The domain organizations of kindlin2. For structure determination, two regions were deleted as indicated in kindlin2Δ. The color coding of the regions is applied in all figures unless otherwise indicated. (B) Ribbon representation of the kindlin2Δ dimer structure. One protomer is colored follow the coding scheme in A, and the other identical protomer is colored in gray. The dimer is related by a twofold rotation axis, indicated by an ellipse. The disordered loops in the F1 and F3 lobes are indicated by hypothetical dotted lines. The PH domain deletion sites are indicated by red stars. The cartoon of the kindlin2 dimer schematically shows the interlobe interactions and the F2 domain-swapped dimer. (C) The talin-FERM structure (PDB ID code: 3IVF).
Talins activate integrins by binding to the cytoplasmic tail of β-integrin (β-tail) via its atypical FERM domain. The β-tail contains two conserved “NPxY” (“x” denotes any amino acids) sequence motifs. The membrane-proximal NPxY has been identified as the talin-binding site, and the membrane-distal NPxY specifically interacts with kindlins (8, 9). Through the interaction with the proximal NPxY motif and the membrane-proximal helix, talins release the inhibitory α/β-intersubunit interaction by changing the conformation of the integrin transmembrane region, eventually leading to increased binding affinities between integrins and ECM ligands (6, 20). Unlike talins, kindlins cannot directly alter the conformation of the integrin transmembrane helix and fail to activate integrin alone (21). Nevertheless, although it is widely accepted that kindlins and talins synergistically promote integrin activation (9, 11), the underlying mechanism is unclear.
To understand the molecular basis of kindlin-mediated integrin signaling, we solved crystal structures of kindlin2 in apo- and β-tail–bound forms. The complex structures reveal that beyond the canonical NPxY motif, the “TTV/STF” sequence between the proximal and distal NPxY motifs provides the binding specificity for kindlins. Disruption of the TTV-binding pocket of kindlin2 results in the loss of FA localization of kindlin2 and defects in integrin activation. Unexpectedly, we found that kindlin2 forms an F2 domain-swapped dimer in crystal and solution. The mutations limiting the dimerization hamper integrin activation. Our structural, biochemical, and cellular data suggest that the monomer–dimer transition is important for kindlin-mediated integrin activation. Additionally, our study provides structural insights into understanding the disease-causing mutations in kindlins.
Results
Overall Structure of Kindlin2.
We attempted to characterize the structure of kindlin2. However, extensive trials failed to yield any crystal, presumably due to high flexibility of the predicted unstructured region of the long inserted sequence (F1 insertion) in the F1 lobe and the inserted PH domain in the F2 lobe (Fig. 1A). By removing the two flexible regions (the truncated protein named hereafter kindlin2Δ), we successfully obtained high-quality crystals with diffraction up to 2.6 Å. The structure of kindlin2Δ was determined by preparing the Se-Met–labeled crystal (SI Appendix, Table S1).
Strikingly, the two kindlin2Δ molecules in one asymmetric unit form a dimer with a twofold rotation symmetry (Fig. 1B). The dimer is mediated mainly by the F2 lobe in a domain-swapped manner (see Kindlin2 Forms a Domain-Swapped Dimer via Its F2 Lobe for details). Each kindlin2Δ molecule is composed of four lobes, F0–F3, and adopts a compact fold via interlobe interactions (Fig. 1B). The F0 lobe keeps a similar fold with the isolated kindlin1-F0 and kindlin2-F0 (22, 23) and attaches to the F1 lobe using a similar interaction mode found in talin (Fig. 1C and SI Appendix, Fig. S2). The other three lobes, F1, F2, and F3, interact with each other to form the typical clover-like shape found in most FERM domains (SI Appendix, Fig. S3). Despite sharing structural similarity with kindlin2 in the respective lobes (SI Appendix, Fig. S4), the lobes in talin-FERM adopt a linear arrangement (24) due to the lack of interlobe interactions. In the kindlin2Δ structure, the interlobe interactions are enhanced by the F1 insertion. Although largely disordered, the long F1 insertion contains two short α-helices (α2F1 and α3F1) protruding from the F1-folding core (SI Appendix, Fig. S4). α2F1 is directly involved in F2-mediated dimerization, and α3F1 tightly packs with the F3 lobe (Fig. 1B). Interestingly, compared with other FERM domains, kindlin2 contains an additional α-helix (α2F3) at its C-terminal end, near the well-known target-binding region in other FERM domains (SI Appendix, Fig. S3), which likely contributes to creating a unique binding environment for target recognition. Given the high sequence similarity, the above structural features found in kindlin2 are very likely to be shared by other kindlins.
Molecular Details of the Kindlin2/β Integrin Interaction.
The β1-integrin contains two NPxY motifs in the membrane proximal tail (MPT) and the membrane distal tail (MDT) for the binding of talins and kindlins, respectively (Fig. 2 A and B). How can kindlins specifically recognize the NPxY motif in the MDT rather than the one in the MPT? To address this question, we set out to solve the structure of the kindlin/β-tail complex. Isothermal titration calorimetry (ITC)-based assays showed that both the full-length kindlin2 and kindlin2Δ bind to the β1-tail with a similar affinity (Kd of ∼20–30 μM) at a 1:1 ratio (Fig. 2 D and E). Because the binding is relatively weak, we fused the β1-MDT to the C terminus of kindlin2Δ to ensure a strict 1:1 stoichiometry in the complex. By using these strategies, we determined the kindlin-2Δ/β1-MDT complex structure (SI Appendix, Table S1).
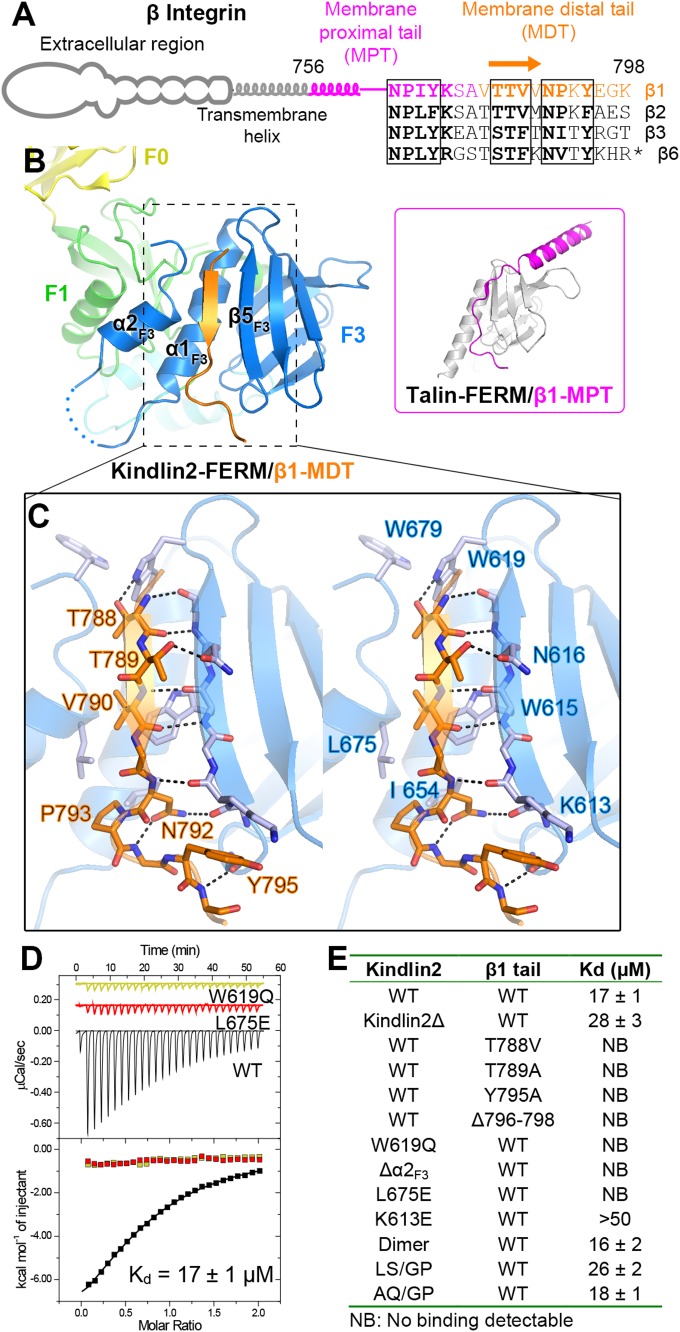
Fig. 2.
Structural and biochemical characterization of the kindlin2/β1-tail interaction. (A) The cartoon diagram of β-integrins. The sequences of the cytoplasmic tails from mouse β-integrins are aligned. The C-terminal 11 residues in the β6-tail are omitted from the alignment. The two NxPY motifs and the TTV motif are boxed. (B) The binding of the β1-MDT to kindlin2-F3. The complex structure of talin-FERM/β1-MPT (PDB ID: 3G9W) is shown in the Inset for comparison. (C) Molecular details of the β1-MDT/F3 interaction (stereoview). H-bonds are indicated by dashed lines. (D) ITC curves showing the interaction between the β1-tail and the kindlin2 proteins (wild-type as well as two TTV-binding deficient mutants). (E) The dissociation constants of the binding reactions of various forms of kindlin2 and the β1-tail (thioredoxin-tagged) derived from the ITC-based assays.
In the resulting complex, kindlin2 adopts a conformation essentially identical to the apo-form (the rmsd values of 0.7 Å for the overall structure and of 0.5 Å for the F3 lobe), indicating that the β1-MDT binding does not induce obvious conformational changes. The β1-MDT folds as a β-strand to bind with a groove (termed the αβ-groove, as formed mainly by α1F3 and β5F3) at the F3 lobe (Fig. 2B), which is a well-characterized binding region in FERM domains (25–30). The 792NPKY795 sequence in the β1-MDT adopts a turn-like shape in which N792β1 and P793β1 insert their side-chains into the αβ-groove, and Y795β1 closely packs with K613F3 (Fig. 2C). Mutating the interacting residues diminishes their interaction (8, 31, 32) (Fig. 2E). This NPxY-binding mode was also found in the talin-FERM/β1-MPT interactions (33, 34) (Fig. 2B and SI Appendix, Fig. S5), indicating that sequences other than NPKY are required for the β1-MDT's specific binding to kindlins.
Previous studies have suggested that several residues at the N-terminal to NPKY are involved in the kindlin/β-tail interaction (35). Consistently, the conserved 788TTV790 sequence in the β1-MDT binds with the F3 lobe in a sequence-dependent manner (Fig. 2 A and C). Specifically, T788β1 and T789β1, respectively, form H-bonds with W619F3 and N616F3, which are strictly conserved in kindlins but not in the corresponding position in talins (SI Appendix, Fig. S4B); although not previously reported as an interacting residue, V790β1 interacts with a hydrophobic patch in the αβ-groove. Interestingly, the hydrophobic patch is formed by residues from not only α1F3 and β5F3 but also the α2F3 helix, which is, to the best of our knowledge, found only in kindlins (Fig. 2B and SI Appendix, Fig. S3). Thus, W619F3 and N616F3 together with L675F3 in α2F3 provide a highly specific binding environment for the TTV motif. Because the T788β1-corresponding residue in the β1-MPT is an Asp (SI Appendix, Fig. S5C), it is unlikely to be accommodated by the hydrophobic αβ-groove. Consistent with our structural analysis, mutations in the TTV motif or the TTV-interacting site in kindlin2 disrupt the binding of kindlin2 to the β1-tail (Fig. 2 D and E). The interface residues are highly conserved in both kindlins and several β-integrin isoforms (Fig. 2A and SI Appendix, Fig. S1), indicating that the β-integrin binding mode found in the kindlin2Δ/β1-MDT structure is common to all kindlins. Consistently, the binding of kindlin1 to the β1-tail shows a similar affinity of ∼20 μM (SI Appendix, Fig. S6). Furthermore, by determining the kindlin2Δ/β3-MDT structure, we confirmed that the β3-tail binds to kindlin2 using the same mode as β1-MDT (SI Appendix, Fig. S5D).
In addition, although the very last three nonconserved residues (796EGK798) in the β1-tail were not well assigned (SI Appendix, Fig. S5), deletion of the EGK sequence abolishes the binding of integrins to kindlins in both our and others’ observations (Fig. 2E) (36, 37). Because the amide of E796β1 forms a main-chain/main-chain H-bond with the β4F3/β5F3 loop in kindlin2 (Fig. 2C), deletion of EGK eliminates the H-bond, thereby disrupting the interaction, which aligns with previous findings that the binding of the β1-tail to kindlin2 requires at least one residue to the C terminus of Y795β1 in a sequence-independent manner (36).
The Binding of Kindlin2 to the TTV Motif in β1-Integrin Is Required for Integrin Signaling.
Previously, we and others reported that kindlin2 is localized at FA and that the kindlin/integrin interaction is required for the FA localization of kindlin2 (7, 8, 35). Therefore, we used the localization of kindlin2 to FA as an assay to examine the TTV-binding pocket in kindlin2. As expected, kindlin2 localizes well to FAs in either wild-type or kindlin2 knockout HT1080 cells (Fig. 3A and SI Appendix, Fig. S7). In contrast, either the W619Q or L675E mutation, which disrupts the TTV-binding pocket, abolishes kindlin2 FA localization, suggesting that the TTV-mediated interaction is indispensable for kindlin2 to localize to FA. To further test whether the kindlin2 mutants affect cell function, we performed a cell-spreading assay using kindlin2 knockout cells. As expected, the reduced cell area of the knockout cells was largely recovered by expressing wild-type, but not TTV-binding deficient mutants (SI Appendix, Fig. S8).
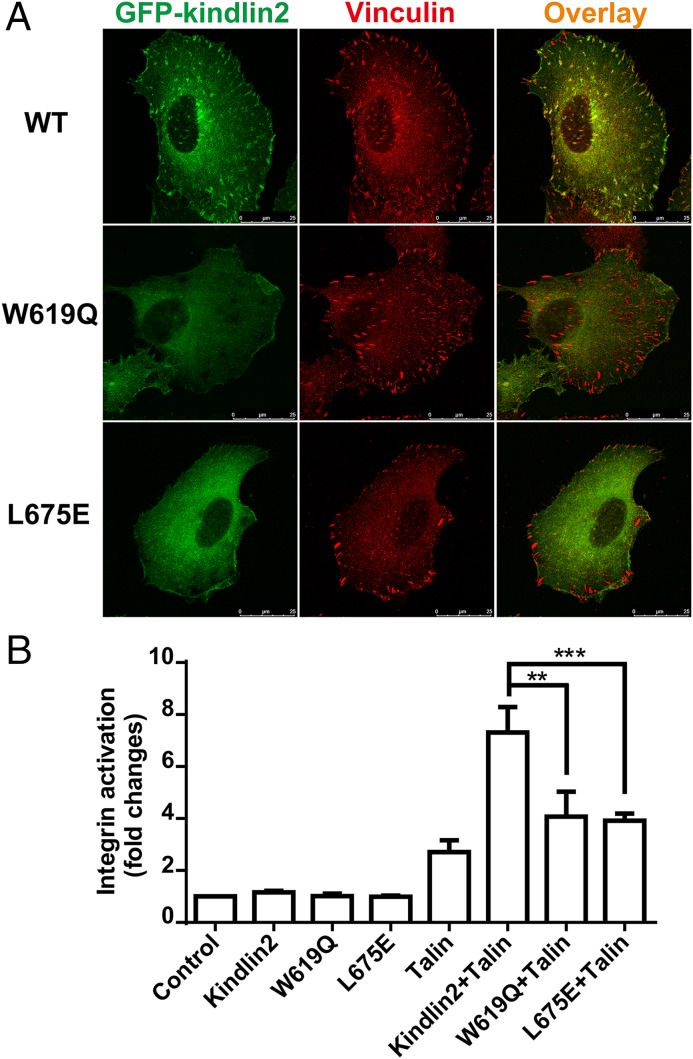
Fig. 3.
Functional characterization of the specific binding of kindlin2 to the β1-tail. (A) Transient expressed GFP-kindlin2 localized to FA indicated by vinculin in HT1080 cells. The W619Q or L675E mutations disrupt kindlin2’s localization to FA. (Scale bar, 25 μm.) (B) Individual GFP-tagged kindlin2 and its mutants, individual red fluorescence protein (RFP)-tagged talin-FERM, or a mixture of talin-FERM and various kindlin-2 were transiently transfected in αIIbβ3-CHO cells. Their effects on αIIbβ3-integrin activation were evaluated by PAC1 binding. Data were normalized to the control cells, and bars represent the means ± SD from four independent experiments. **P < 0.01; ***P < 0.001.
Because kindlins function as coactivators of integrin in the presence of talin-FERM (9, 11), we coexpressed talin-FERM with various kindlin2 constructs (the wild-type or the TTV-binding deficient mutants) in αIIbβ3 integrin-expressing CHO A5 cells and performed an integrin activation assay. Consistent with previous findings, wild-type kindlin2 synergized with talin to activate integrin (Fig. 3B). The two TTV-binding deficient mutants, however, showed little synergistic enhancement in integrin activation (Fig. 3B).
Taken together, the above structural, biochemical, and cellular characterizations clearly demonstrate that the binding of kindlin2 to the TTV motif of β1-integrin is crucial for kindlin-mediated integrin activation and signaling.
Kindlin2 Forms a Domain-Swapped Dimer via Its F2 Lobe.
Because we used the monomeric protein for crystallization, finding a dimeric kindlin2 structure in the crystals was unexpected. The symmetric dimer is mediated by the F2 lobe in a domain-swapped manner (Fig. 1B). Unlike talin-F2, the F2 lobe in the kindlin2 dimer adopts an extended, “open” conformation (SI Appendix, Fig. S4A). Instead of forming two separated helices as found in talin-F2, the corresponding part in kindlin2 becomes a long, continuous helix (α4F2). The rigidity of α4F2 prevents the region between α2F2 and α4F2 from forming an intramolecular interaction with the hydrophobic pocket formed by α1F2, α2F2, and α2F1, but promotes the intermolecular interaction with the same structural elements in the other molecule (Fig. 4A). Interestingly, although the dimeric conformation was preferred in most crystallization conditions that formed crystals, the monomeric structure was found under a few conditions. By further modifying the deletion boundary of the PH domain (residues 367–512), we obtained a structure of the monomeric kindlin2Δ′ (SI Appendix, Table S1). Although most parts remain the same as the dimer, the monomeric structure shows the “closed” conformation of the F2 lobe (Fig. 4A and SI Appendix, Fig. S9). Specifically, the α4F2 helix is bent into two helices, α4NF2 and α4CF2, resulting in a folding back of α3F2 and its preceding loop, which occupy the same hydrophobic pocket involved in dimerization (SI Appendix, Fig. S10). Therefore, the switch between the closed and open conformation of the F2 lobe controls the monomer–dimer exchange of kindlin2. In addition, the C-terminal part of α2F2 and the middle region of α4F2 are involved in forming another specific dimer interface (SI Appendix, Fig. S10), which may facilitate the monomer-to-dimer transition.
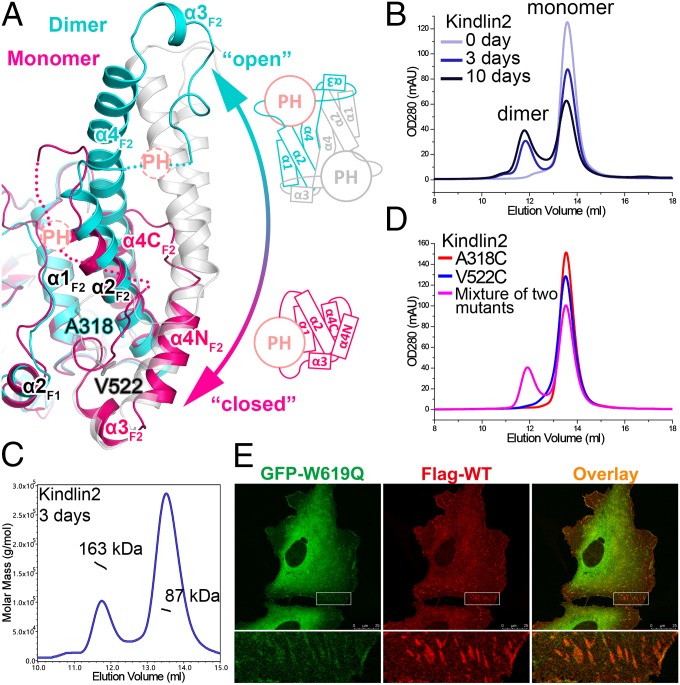
Fig. 4.
Structural basis of the monomer–dimer transition in kindlin2. (A) Structural comparison of the kindlin2 dimer and monomer. The insert PH domains and their connecting loops to the F2 lobes are indicated by dashed circles and lines, respectively. The open and closed conformations of the F2 lobe are also shown in the schematic views. The two residues, mutated to Cys, in D, are highlighted in the stick mode. (B) Analytical gel filtration analysis showing the dimer formation of kindlin2 in solution. The samples were prepared using the fresh-purified protein (0 d) or the same batch of protein placed at 4 °C for 3 d or 10 d. (C) The molecular weights of the dimer and monomer fractions were measured by using multiangle static light scattering. (D) Disulfide-bonding enhanced kindlin2 dimerization by structure-based mutation design. Before gel filtration analysis, the fresh-purified mutants and their mixture were kept at 4 °C overnight. (E) Cotransfection of the GFP-tagged W619Q mutant and the Flag-tagged wild-type protein in HT1080 cells. (Scale bar, 25 μm.)
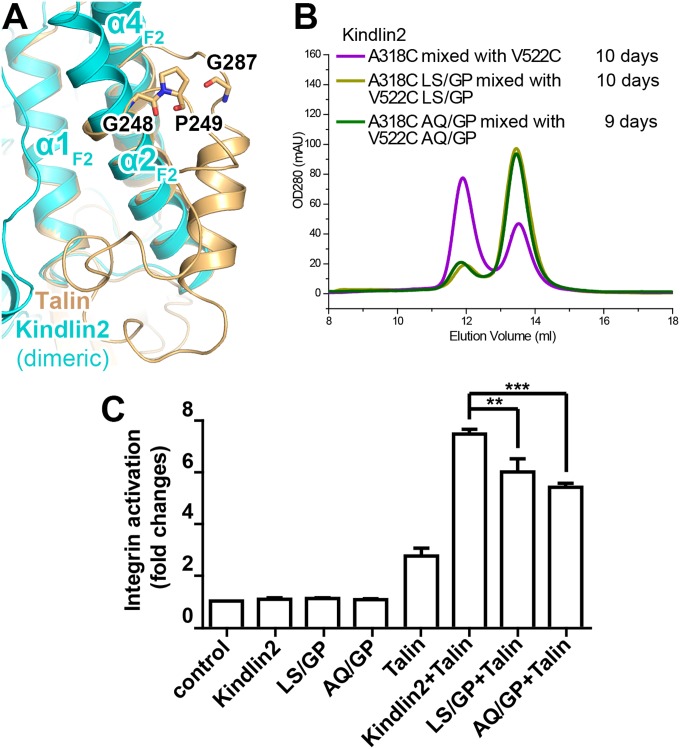
Next, we tried to confirm the dimer conformation in solution. The freshly purified kindlin2 protein is monomeric as shown by analytical gel filtration (Fig. 4B). Interestingly, the dimer was detected in a sample placed at 4 °C for several days, and the dimer population increased over time (Fig. 4 B and C). In comparison, no similar dimer formation was observed in talin-FERM protein placed at 4 °C (SI Appendix, Fig. S11A). To rule out the possibility that the dimer is formed nonspecifically by partially unfolded protein, we measured the binding of the dimer to β-integrin. The dimer showed essentially the same binding affinity as the monomer (Fig. 2E). Furthermore, we used a site-specific chemical cross-linking approach to probe the dimer state. In the kindlin2 dimer structure, V522 (in one protomer) and A318 (in another protomer) are close to each other in the dimer interface (Fig. 4A). If the dimer in solution adopts the same conformation found in the crystal structure, the respective substitution of V522 and A318 with Cys in the two protomers of the dimer would specifically promote the formation of a disulfide bond-mediated dimerization. Consistently, the dimerization process was greatly enhanced by mixing the V522C and A318C mutants, whereas neither the V522C nor the A318C mutant alone showed accelerated dimerization (Fig. 4D).
To investigate kindlin2 dimer formation in cells, we cotransfected GFP-tagged W619Q kindlin2, which shows abolished FA localizations, with Flag-tagged wild-type kindlin2. The dimerization of kindlin2 would presumably bring the W619Q mutant to FA by interacting with the wild-type protein. Indeed, we detected the partial FA localization of W619Q by cotransfecting it with wild-type kindlin2 (Fig. 4E).
Because kindlin family proteins are highly conserved (SI Appendix, Fig. S1), it would be interesting to know whether other kindlins also form dimers. By using analytical gel filtration, we observed the similar dimerization process of kindlin1 (SI Appendix, Fig. S11B). Consistent with this, the kindlin homolog in Caenorhabditis elegans, UNC112, was also shown to have the potential to form a dimer (38). Thus, the monomer–dimer transition is likely an evolutionarily conserved property of kindlins.
The Monomer–Dimer Transition Is Important for Kindlin-Mediated Integrin Activation.
To understand the functional roles of the dimer formation of kindlins, we set out to design mutations that disrupt the monomer–dimer transition in kindlin2. As talin-FERM keeps a monomeric state stably (SI Appendix, Fig. S11A), substitution of certain sequences from talins may lock kindlin2 into a monomer. Through structural and sequence comparison, we found that formation of the closed conformation of talin-F2 largely relies on two turns, formed by “248GP249” and “287GQ288,” respectively (Fig. 5A). The corresponding sequences in kindlin2 are “327LS328” and “547AQ548” (SI Appendix, Fig. S4B), which appear in turns much less often (39). To test whether the enhanced turn formation inhibits dimerization, we introduced either the LS/GP or the AQ/GP mutation into the kindlin2 Cys mutants. In agreement with our idea, the mixture of the V522C and A318C mutants carrying either the LS/GP or AQ/GP mutation showed diminished dimerization in solution (Fig. 5B), implying that both of the GP mutations disrupt the monomer-to-dimer transition.
Fig. 5.
Functional characterization of the mononer–dimer transition of kindlin2. (A) Structural comparison of the F2 lobes of talin and dimeric kindlin2. G248, P249, and G287 are likely to maintain the closed conformation of talin-F2 by stabilizing the two turns. (B) Analytical gel filtration analysis showing the impaired dimer formation by introduction of the GP mutations in kindlin2. (C) The two GP mutants show decreased integrin activation activity, compared with the wild-type kindlin2. Data were collected and analyzed using the same method as indicated in Fig. 3B. **P < 0.01; ***P < 0.001.
We next examined whether the two GP mutations affect the FA localization of kindlin2. Consistent with our ITC data showing that the GP mutants retained the ability to bind the β1-tail (Fig. 2E), the two GP mutants colocalized with vinculin, as did the wild-type kindlin2 (SI Appendix, Figs. S7 and S12). In contrast, the AQ/GP mutant failed to recruit W619Q-kindlin2 to FA (SI Appendix, Fig. S13), suggesting that dimer formation in cells can be specifically disrupted by the GP mutations. Next, we asked whether the GP mutants are able to mediate integrin activation upon FA recruitment. By using the integrin activation assay, we found that both GP mutants showed significantly decreased (∼20–30%) integrin activation (Fig. 5C). These data indicate that the monomer–dimer transition of kindlin2 is important for integrin activation.
Structural Implications of Kindlin Mutations in Diseases.
Defective mutations of kindlins lead to various human genetic diseases, such as Kindler syndrome and LADIII (40). More than 80 disease-causing mutations in kindlins have been identified (SI Appendix, Table S2). Most of these mutations are truncation or frame-shift mutations, thereby producing defective proteins (SI Appendix, Fig. S14A). Five missense mutations in kindlins have been reported (41–44). By fitting them in the kindlin2 structure, we analyzed potential effects on protein folding or target binding due to these mutations (SI Appendix, Fig. S14B). The R297G and W559R mutations found in Kindler syndrome likely disrupt the F2 folding as well as the F1/F2 interaction, whereas the Q595P mutation is likely to distort the β5F3 conformation and thereby disrupts the kindlin/integrin interaction. Consistent with our structural analysis, the corresponding mutations in kindlin2 either disrupt the folding or abolish the integrin binding (SI Appendix, Fig. S14 C and D). Thus, the genetic mutations found in kindlins are expected to alter their structures at different levels and thereby impair proper functions of kindlins.
Discussion
In this study, we have solved the crystal structures of kindlin2, a ubiquitously expressed member of the kindlin family, as well as the kindlin2/β-tails complex structures. The structures provide a major advancement in understanding the structural basis of the kindlin/integrin interaction as well as the genetic diseases associated with kindlin mutations.
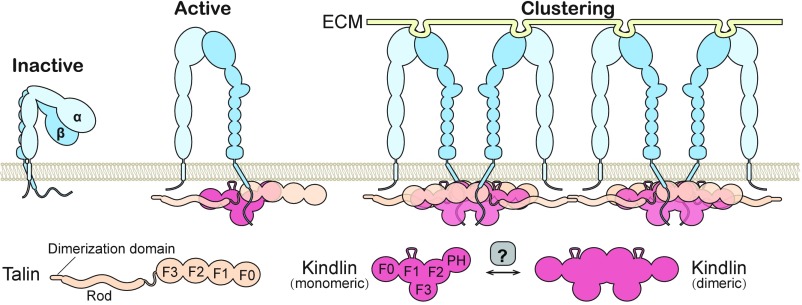
The monomer–dimer transition of kindlins described here provides insight into the mechanism of integrin signaling. Integrins form clusters upon full activation. There is evidence that kindlins activate integrins by promoting the clustering of talin-activated integrin and increasing integrin binding to multivalent ligands (45). The dimerization of kindlins provides a mechanistic explanation of how kindlins promote integrin clustering (Fig. 6). In this model, kindlins in either monomeric or dimeric forms are recruited to adhesion sites through interacting with the β-tails, whereas only dimeric kindlins can bridge talin-activated integrins to promote the clustering. Interestingly, talins are also dimerized antiparallelly, which is not mediated by the FERM domains but by the dimerization domain in their C terminus (46), which may further enhance integrin clustering (Fig. 6). In line with our model, disruption of the monomer–dimer transition of kindlin2 by the GP mutations led to impaired integrin activation and FA formation, but had little impact on the FA localization of kindlin2 (Fig. 5C and SI Appendix, Figs. S12 and S13). The in vitro dimerization process occurs spontaneously yet slowly (Fig. 4B). Therefore, in living cells, dimer formation needs to be accelerated locally for fast and efficient integrin signaling. The regulatory factors for the monomer–dimer transition remain to be identified. Because kindlins link integrin-mediated adhesion to the cytoskeleton via interacting with other actin cytoskeleton regulators (ILK, migfilin, paxillin, etc.) as well as actin (7, 47–49), it would be interesting to investigate whether dimeric kindlins impact these interactions or vice versa.
Fig. 6.
Model of kindlin-mediated integrin activation and clustering. The three states of integrin activation are shown. The binding of talins to the cytoplasmic tails of β-integrins promotes the extension of the extracellular domains of integrins. The activated integrins are further clustered and fully activated via multivalent ECM ligand binding as well as the interaction network mediated by talins and dimeric kindlins. The monomer–dimer transition of kindlins in vivo may require regulators (e.g., membrane environment through either increasing local concentration or altering the conformation of kindlins).
Materials and Methods
All proteins used in this study were expressed in Escherichia coli and purified by Ni2+-NTA affinity chromatography followed by size-exclusion chromatography. Crystals were obtained by the sitting drop vapor diffusion method at 16 °C. An extended description of the methods for protein preparation, crystallography, and biochemical and cellular assays is included in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Mingjie Zhang and Dr. Andrew Hutchins for their critical reading of our paper; the Life Science Research Center, Southern University of Science and Technology (SUSTech) for providing facilities; and the BL19U1 beamline at the National Center for Protein Sciences Shanghai and the BL17U beamline at Shanghai Synchrotron Radiation Facility for the X-ray beam time. This work was supported by National Natural Science Foundation of China Grants 31500621 (to C.Y.), 31570741 (to Z.W.), and 81430068 and 31471311 (to C.W.); by National Key R&D Program of China Grant 2016YFC1302100; National Institutes of Health Grant AR068950 (to C.W.); Natural Science Foundation of Guangdong Province Grant 2016A030312016 (to Z.W.); Shenzhen Science and Technology Innovation Commission Grants ZDSYS20140509142721429 and JCYJ20160331115658342 (to C.Y.), KQCX20140522150842929, JCYJ20150831142427959, and JCYJ20150331101823691 (to C.W. and Y.D.), and JCYJ20160229153100269 (to Z.W.); and funding from SUSTech. C.Y. and Z.W. are supported by the Recruitment Program of Global Youth Experts of China. Z.W. is a member of the Neural and Cognitive Sciences Research Center, SUSTech.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5XPY, 5XPZ, 5XQ0, and 5XQ1).
See Commentary on page 9234.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703064114/-/DCSupplemental.
References
- 1.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix and the cytoskeleton. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 2.Winograd-Katz SE, Fässler R, Geiger B, Legate KR. The integrin adhesome: From genes and proteins to human disease. Nat Rev Mol Cell Biol. 2014;15:273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 3.Iwamoto DV, Calderwood DA. Regulation of integrin-mediated adhesions. Curr Opin Cell Biol. 2015;36:41–47. doi: 10.1016/j.ceb.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 5.Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Tadokoro S, et al. Talin binding to integrin beta tails: A final common step in integrin activation. Science. 2003;302:103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 7.Tu Y, Wu S, Shi X, Chen K, Wu C. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 2003;113:37–47. doi: 10.1016/s0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 8.Shi X, et al. The MIG-2/integrin interaction strengthens cell-matrix adhesion and modulates cell motility. J Biol Chem. 2007;282:20455–20466. doi: 10.1074/jbc.M611680200. [DOI] [PubMed] [Google Scholar]
- 9.Ma YQ, Qin J, Wu C, Plow EF. Kindlin-2 (Mig-2): A co-activator of beta3 integrins. J Cell Biol. 2008;181:439–446. doi: 10.1083/jcb.200710196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moser M, Nieswandt B, Ussar S, Pozgajova M, Fässler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- 11.Montanez E, et al. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008;22:1325–1330. doi: 10.1101/gad.469408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ussar S, et al. Loss of Kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet. 2008;4:e1000289. doi: 10.1371/journal.pgen.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moser M, Legate KR, Zent R, Fässler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 14.Jobard F, et al. Identification of mutations in a new gene encoding a FERM family protein with a pleckstrin homology domain in Kindler syndrome. Hum Mol Genet. 2003;12:925–935. doi: 10.1093/hmg/ddg097. [DOI] [PubMed] [Google Scholar]
- 15.Lanschuetzer CM, et al. Characteristic immunohistochemical and ultrastructural findings indicate that Kindler’s syndrome is an apoptotic skin disorder. J Cutan Pathol. 2003;30:553–560. doi: 10.1034/j.1600-0560.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- 16.Siegel DH, et al. Loss of kindlin-1, a human homolog of the Caenorhabditis elegans actin-extracellular-matrix linker protein UNC-112, causes Kindler syndrome. Am J Hum Genet. 2003;73:174–187. doi: 10.1086/376609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mory A, et al. Kindlin-3: A new gene involved in the pathogenesis of LAD-III. Blood. 2008;112:2591. doi: 10.1182/blood-2008-06-163162. [DOI] [PubMed] [Google Scholar]
- 18.Kuijpers TW, et al. LAD-1/variant syndrome is caused by mutations in FERMT3. Blood. 2009;113:4740–4746. doi: 10.1182/blood-2008-10-182154. [DOI] [PubMed] [Google Scholar]
- 19.Rognoni E, Ruppert R, Fässler R. The kindlin family: Functions, signaling properties and implications for human disease. J Cell Sci. 2016;129:17–27. doi: 10.1242/jcs.161190. [DOI] [PubMed] [Google Scholar]
- 20.Wegener KL, et al. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 21.Bledzka K, et al. Spatial coordination of kindlin-2 with talin head domain in interaction with integrin β cytoplasmic tails. J Biol Chem. 2012;287:24585–24594. doi: 10.1074/jbc.M111.336743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goult BT, et al. The structure of the N-terminus of kindlin-1: A domain important for alphaiibbeta3 integrin activation. J Mol Biol. 2009;394:944–956. doi: 10.1016/j.jmb.2009.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perera HD, et al. Membrane binding of the N-terminal ubiquitin-like domain of kindlin-2 is crucial for its regulation of integrin activation. Structure. 2011;19:1664–1671. doi: 10.1016/j.str.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott PR, et al. The structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–1299. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 26.Hamada K, et al. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J. 2003;22:502–514. doi: 10.1093/emboj/cdg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Z, Yan J, Lu Q, Pan L, Zhang M. Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc Natl Acad Sci USA. 2011;108:3572–3577. doi: 10.1073/pnas.1016567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghai R, et al. Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proc Natl Acad Sci USA. 2013;110:E643–E652. doi: 10.1073/pnas.1216229110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Wei Z, Zhang J, Yang Z, Zhang M. Structural basis of the binding of Merlin FERM domain to the E3 ubiquitin ligase substrate adaptor DCAF1. J Biol Chem. 2014;289:14674–14681. doi: 10.1074/jbc.M114.551184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Z, Li Y, Ye F, Zhang M. Structural basis for the phosphorylation-regulated interaction between the cytoplasmic tail of cell polarity protein crumbs and the actin-binding protein moesin. J Biol Chem. 2015;290:11384–11392. doi: 10.1074/jbc.M115.643791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloeker S, et al. The Kindler syndrome protein is regulated by transforming growth factor-beta and involved in integrin-mediated adhesion. J Biol Chem. 2004;279:6824–6833. doi: 10.1074/jbc.M307978200. [DOI] [PubMed] [Google Scholar]
- 32.Bledzka K, et al. Tyrosine phosphorylation of integrin beta3 regulates kindlin-2 binding and integrin activation. J Biol Chem. 2010;285:30370–30374. doi: 10.1074/jbc.C110.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calderwood DA, et al. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J Biol Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 34.García-Alvarez B, et al. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 35.Harburger DS, Bouaouina M, Calderwood DA. Kindlin-1 and -2 directly bind the C-terminal region of beta integrin cytoplasmic tails and exert integrin-specific activation effects. J Biol Chem. 2009;284:11485–11497. doi: 10.1074/jbc.M809233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitzpatrick P, Shattil SJ, Ablooglu AJ. C-terminal COOH of integrin β1 is necessary for β1 association with the kindlin-2 adapter protein. J Biol Chem. 2014;289:11183–11193. doi: 10.1074/jbc.M113.535369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Z, et al. Interaction of kindlin-2 with integrin β3 promotes outside-in signaling responses by the αVβ3 vitronectin receptor. Blood. 2015;125:1995–2004. doi: 10.1182/blood-2014-09-603035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qadota H, Moerman DG, Benian GM. A molecular mechanism for the requirement of PAT-4 (integrin-linked kinase (ILK)) for the localization of UNC-112 (Kindlin) to integrin adhesion sites. J Biol Chem. 2012;287:28537–28551. doi: 10.1074/jbc.M112.354852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 40.Karaköse E, Schiller HB, Fässler R. The kindlins at a glance. J Cell Sci. 2010;123:2353–2356. doi: 10.1242/jcs.064600. [DOI] [PubMed] [Google Scholar]
- 41.Youssefian L, et al. The Kindler syndrome: A spectrum of FERMT1 mutations in Iranian families. J Invest Dermatol. 2015;135:1447–1450. doi: 10.1038/jid.2015.9. [DOI] [PubMed] [Google Scholar]
- 42.Has C, et al. Kindler syndrome: Extension of FERMT1 mutational spectrum and natural history. Hum Mutat. 2011;32:1204–1212. doi: 10.1002/humu.21576. [DOI] [PubMed] [Google Scholar]
- 43.McDowall A, et al. Two mutations in the KINDLIN3 gene of a new leukocyte adhesion deficiency III patient reveal distinct effects on leukocyte function in vitro. Blood. 2010;115:4834–4842. doi: 10.1182/blood-2009-08-238709. [DOI] [PubMed] [Google Scholar]
- 44.Suratannon N, et al. Adaptive immune defects in a patient with leukocyte adhesion deficiency type III with a novel mutation in FERMT3. Pediatr Allergy Immunol. 2016;27:214–217. doi: 10.1111/pai.12485. [DOI] [PubMed] [Google Scholar]
- 45.Ye F, et al. The mechanism of kindlin-mediated activation of integrin αIIbβ3. Curr Biol. 2013;23:2288–2295. doi: 10.1016/j.cub.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldmann WH, Bremer A, Häner M, Aebi U, Isenberg G. Native talin is a dumbbell-shaped homodimer when it interacts with actin. J Struct Biol. 1994;112:3–10. doi: 10.1006/jsbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- 47.Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- 48.Bledzka K, et al. Kindlin-2 directly binds actin and regulates integrin outside-in signaling. J Cell Biol. 2016;213:97–108. doi: 10.1083/jcb.201501006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Theodosiou M, et al. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 2016;5:e10130. doi: 10.7554/eLife.10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.