Significance
The separation of daughter cells during cell division, or cytokinesis, is a process that requires contractile rings which develop tension using actin and myosin. Current models of contractile ring dynamics are based on quantitative data from two decades of research using the tractable fission yeast system. However, it is unknown how fission yeast’s essential myosin, Myo2, is regulated in the contractile ring. Here, we find that Myo2 does not assemble into minifilaments, consistent with its role in ring precursor nodes. Unphosphorylated Myo2 exhibits robust enzymatic and motor activity whereas phosphorylation of Myo2’s regulatory light chain reduces its affinity for actin. This reduction likely weakens the tension in the contractile ring, potentially to delay cytokinesis until segregation of chromosomes is complete.
Keywords: fission yeast, cytokinesis, myosin-II, phosphorylation
Abstract
Studies in fission yeast Schizosaccharomyces pombe have provided the basis for the most advanced models of the dynamics of the cytokinetic contractile ring. Myo2, a class-II myosin, is the major source of tension in the contractile ring, but how Myo2 is anchored and regulated to produce force is poorly understood. To enable more detailed biochemical/biophysical studies, Myo2 was expressed in the baculovirus/Sf9 insect cell system with its two native light chains, Rlc1 and Cdc4. Milligram yields of soluble, unphosphorylated Myo2 were obtained that exhibited high actin-activated ATPase activity and in vitro actin filament motility. The fission yeast specific chaperone Rng3 was thus not required for expression or activity. In contrast to nonmuscle myosins from animal cells that require phosphorylation of the regulatory light chain for activation, phosphorylation of Rlc1 markedly reduced the affinity of Myo2 for actin. Another unusual feature of Myo2 was that, unlike class-II myosins, which generally form bipolar filamentous structures, Myo2 showed no inclination to self-assemble at approximately physiological salt concentrations, as analyzed by sedimentation velocity ultracentrifugation. This lack of assembly supports the hypothesis that clusters of Myo2 depend on interactions at the cell cortex in structural units called nodes for force production during cytokinesis.
Animals, amoebas, and fungi undergo cytokinesis through the formation of a cleavage furrow, which is guided by tension generated by an actomyosin contractile ring. Relatively little is known about the physical interactions and juxtaposition of proteins in the contractile ring in animal cells (1, 2). However, pioneering classical and molecular genetics of the haploid fission yeast Schizosaccharomyces pombe have made it a well-established model organism for understanding how contractile rings assemble and undergo constriction. Elucidation of the inventory, timing, concentrations, and genetic interactions of cytokinesis proteins in the fission yeast contractile ring has led to computational models that account for many dynamic aspects of the contractile ring (2). To expand upon these models, better knowledge of the biophysical properties of the proteins involved is needed. How Myo2, the essential class-II myosin in fission yeast, is regulated and functions in the contractile ring represents a major gap in our understanding of the forces contributing to contractile ring dynamics.
Myo2 powers contractile ring assembly by the coalescence of precursor nodes that are modular units of the ring (3–5). The compaction of the ring has been proposed to occur by a “search, capture, pull, and release” mechanism, where Myo2, incorporated into nodes, pulls on actin filaments from neighboring nodes, thereby bringing the nodes together to form a contractile ring (6–8). The region that localizes Myo2 to nodes has been mapped to ∼150 to 300 aa at the carboxyl (C) terminus of Myo2 (9, 10). The binding partners of this region of Myo2 in the nodes are not established, but IQGAP Rng2 and anillin-like Mid1 have been implicated, based on localization dependency and immunoprecipitation experiments (4, 11). Unlike the two other myosins in the contractile ring (Myp2, an uncharacterized class-II myosin, and Myo51, an unusual single-headed class-V myosin) (12), Myo2 localization to the division site does not depend on actin filaments (11, 13, 14). Although Myo2’s interactions with other node proteins are unresolved at the molecular level, a plausible model for Myo2 function is that it forms clusters by being stably anchored to nodes.
Rng3, a Unc45/Cro1/She4 (UCS) family protein, has been suggested to activate Myo2 during cytokinesis (15). UCS proteins are chaperones that work with heat shock protein Hsp90 to fold newly synthesized myosin (16, 17). Consequently, Rng3 and the Hsp90 Swo1 are required for cytokinesis in fission yeast (18, 19). Rng3-defective, rng3-65 fission yeast strains exhibit failed contractile ring assembly at high temperature (37 °C), which is consistent with a loss of Myo2 function (18). GFP-tagged Rng3 is enriched ∼70-fold in the contractile ring in myo2-E1 mutant cells (18, 20). Yeast with the myo2-E1 allele harbor a mutation near the nucleotide binding pocket of the Myo2 motor domain, G345R, resulting in a total loss of motility and severely reduced ATPase (15, 21). Myo2-E1 protein is also more sensitive to limited proteolysis than WT (21), which suggests that unfolded Myo2-E1 recruits Rng3 to the ring. Additionally, the myo2-E1 rng3-65 double-mutant strains exhibit synthetically lethal cytokinesis defects (18). These findings are consistent with the role of fission yeast Rng3 as a conserved member of the UCS protein family that folds myosins. The hypothesis that Rng3 functions as an activator comes from the first study to isolate Myo2 from fission yeast, where it was observed that purified Myo2 did not bind actin filaments in motility assays unless Rng3 was added (15). Later studies, however, found that Rng3 did not affect ATPase rates, and enhanced Myo2 motility only at low myosin concentrations (22), but it was not clear why this occurs. Here, we offer an explanation for why Rng3 appeared to stimulate actin gliding in motility assays.
It is generally believed that Myo2 functions similarly to animal and amoeba class-II myosins. Class-II myosins are composed of two globular head domains that bind to actin and hydrolyze ATP, and a rod-like, α-helical coiled-coil tail that assembles the molecules into bipolar filaments. Molecules typically consist of two ∼1,500- to 2,000-aa heavy chains (HC), and two each of the essential light chains (ELC) and regulatory light chains (RLC). In animal cells, the nonmuscle myosins form minifilaments of ∼30 molecules and ∼300 nm in length (23) whereas the muscle thick filaments are 1.6 μm long and contain about 300 molecules. The ability of myosin-II to assemble is ancient and conserved in amoebozoans, such as Acanthamoeba castellanii and Dictyostelium discoideum, that form myosin filaments of variable size by unique rod–rod interactions (24, 25). In animal cells, smooth muscle and nonmuscle class-II myosins are activated by RLC phosphorylation. When the RLC is unphosphorylated, these myosins form inactive, folded 10S monomers that are autoinhibited by head–head interactions (26–30). Phosphorylation of the RLC is necessary for smooth and nonmuscle myosins to unfold into active 6S monomers that self-assemble into filaments at physiological salt conditions (23, 27). It is unclear how Myo2 is regulated by RLC phosphorylation, or whether it assembles into filaments, because the yields of Myo2 from fission yeast have been insufficient to fully explore these mechanisms in vitro. Our study takes advantage of the baculovirus/Sf9 insect cell expression system to obtain milligram yields of Myo2 for molecular characterization. Using recombinant Myo2, we examined the effects of RLC phosphorylation on enzymatic activity and motility, as well as the propensity for Myo2 to self-assemble, to address the question of how Myo2 produces force in the contractile ring.
Results
Myo2 Is Active Without Phosphorylation and Rng3.
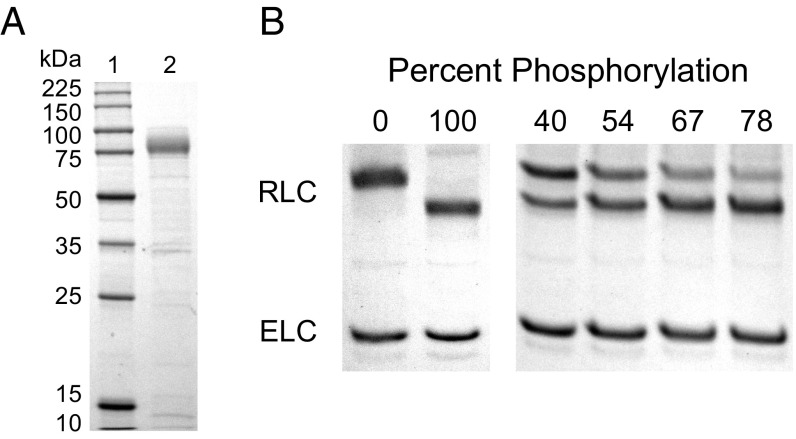
It was anticipated that Myo2 folding in the baculovirus/Sf9 insect cell system might require the coexpression of a species-specific chaperone, like certain unconventional myosins (31, 32), because animals and fission yeast are separated by 1 billion years of evolution (2). When Myo2 was coexpressed with its native light chains Cdc4 (ELC) and Rlc1 (RLC), it was not necessary to coexpress the native chaperone, Rng3, to obtain soluble Myo2 (Fig. 1A). Sedimentation velocity in the analytical ultracentrifuge showed symmetrical 7S peaks for two constructs (Fig. 1B), indicating that Myo2 is monodispersed. The 7S value corresponds to extended monomers, based on the well-documented animal and amoeba class-II myosins, which sediment at 6S to 8S as extended monomers. Phosphoprotein staining revealed that the RLC of the recombinant Myo2 was unphosphorylated (Fig. 1C). Despite lacking RLC phosphorylation, recombinant Myo2 exhibited ATPase activity with a Vmax of ∼2.3 s−1 (Fig. 1D), which is comparable with the rates (0.8 to 3.5 s−1) obtained with Myo2 purified from fission yeast in previous studies (15, 22, 33, 34). Unphosphorylated Myo2 was also capable of sliding actin filaments in motility assays, with average rates of 0.7 μm/s at 30 °C (Fig. 1E and Movie S1).
Fig. 1.
Unphosphorylated Myo2 has motor activity. (A) SDS gels of Myo2 heavy chain (HC) coexpressed with its native light chains, Rlc1 (RLC) and Cdc4 (ELC) in the baculovirus/insect cell expression system. Lane 1, molecular mass standards; lane 2, N-FLAG-Myo2; lane 3, Myo2-C-Biotin-C-FLAG; lane 4, N-FLAG-Myo2-C-Biotin. Asterisks (*) indicate breakdown products of the myosin tail. (B) Sedimentation velocity of Myo2 by analytical ultracentrifugation. Conditions were as follows: 20 °C, 10 mM imidazole, pH 7.0, 0.5 M KCl, 5 mM MgCl2, 1 mM EGTA, 1 mM DTT. OD, optical density; S, Svedberg. (C) SDS gel double-stained with phosphoprotein stain and then Coomassie to stain for total protein. The RLC of expressed N-FLAG-Myo2-C-Biotin has the same level of staining as the unphosphorylated smooth muscle myosin standard. (D) Rates of steady-state ATPase of Myo2 at various actin concentrations. Circles, squares, and triangles represent independent preparations of N-FLAG-Myo2-C-Biotin. Conditions were as follows: 30 °C, 10 mM imidazole, pH 7.0, 50 mM NaCl, 1 mM MgCl2, 1 mM ATP, and 2 mM DTT. (E) In vitro motility assays. (Left) A maximum projection image of a representative field (128 μm × 128 μm × 50 s) (Movie S1) showing trails of gliding actin filaments. (Right) Speeds of actin filament movement (n, number of moving filaments) from 11 experimental repeats with four independent preparations of Myo2 (all three tag variations). Error, ± SD.
In earlier work, when Myo2 was isolated from fission yeast, it was observed that Rng3 activated motility when the Myo2 concentration was low and no methylcellulose was present in the assay (15, 22). Because a minimum number of myosin heads are required for motility, a plausible explanation for activation is that Rng3, when bound nonspecifically to the nitrocellulose-coated coverslip, may have tethered actin down to the surface to bind myosin. We performed cosedimentation assays to assess whether Rng3 binds to actin in low salt, similar to the motility assay conditions. A small portion of Rng3 pelleted with the actin (Fig. 2A), indicating a weak interaction. We next examined whether Rng3 binds to actin in the motility assay. To assay relative actin binding, methylcellulose, a viscosity enhancer, was omitted to allow the actin to diffuse freely away from the coverslip surface (35). When Myo2 alone was applied to the chamber at 50 μg/mL, robust motility was observed, with an average speed of ∼0.7 μm/s; however, no filament binding or motility was seen when the Myo2 concentration was reduced to 16 μg/mL (Fig. 2B and Movie S2). Recapitulating the results of previous studies (15), the addition of Rng3 to the coverslip together with 16 μg/mL Myo2 restored motility (Fig. 2B and Movie S2). Furthermore, Rng3 by itself was observed to bind actin filaments near the coverslip surface (Fig. 2B and Movie S2). The addition of purified Rng3 to myosin specifically attached to a neutravidin-coated coverslip by a C-terminal biotin tag did not have a significant effect on the motility. Together, these data indicate that Rng3 binds only to actin and has no interaction with Myo2. Because we found Myo2 to be fully active (Figs. 1 D and E and 2B), the question remains whether Rng3′s interaction with actin is relevant in a cellular context.
Fig. 2.
Rng3 promotes motility in vitro by binding actin filaments nonspecifically. (A) SDS gel showing totals (T), supernatants (S), and pellets (P) from a cosedimentation assay in which 700 nM Rng3 and 75 μM actin were pelleted at 400,000 × g for 20 min in 25 mM imidazole, pH 7.4, 46 mM KCl, 23 mM NaCl, 1 mM EGTA, 4 mM MgCl2, and 2 mM DTT. Rng3 does not pellet in the absence of actin. (B) Panels showing maximum projections of representative fields (128 μm × 128 μm × 50 s) (Movie S2) and average speeds (blue) of motility in 50 mM KCl assay buffer performed in the absence of methylcellulose, to allow diffusion of actin away from the coverslip surface. Rng3 and Myo2 (N-FLAG-C-Biotin) were adhered nonspecifically to nitrocellulose-coated flow cells (15). Motility assay conditions were as follows: 30 °C, 25 mM imidazole, pH 7.4, 50 mM KCl, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT, and 1 mM ATP. Error, ± SD.
RLC Phosphorylation by PAK Lowers Actomyosin Affinity.
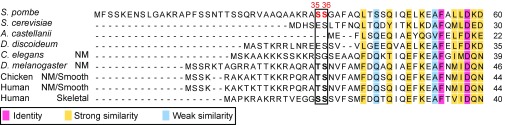
Studies in the fission yeast system by Loo and Balasubramanian (36) provided evidence both in vitro and in vivo that fission yeast p21/Cdc42-activated kinase (PAK), Pak1/Shk1/Orb2, can phosphorylate WT RLC but not RLCSer35,36Ala in which S35 and S36 were mutated to Ala. Serines 35 and 36 of fission yeast RLC are homologous to Thr-18 and Ser-19 of the smooth/nonmuscle RLC in mammals (Fig. 3). However, it is unknown how phosphorylation by PAK affects Myo2’s enzymatic and motor activity. To examine the effects of RLC phosphorylation on Myo2, we phosphorylated the RLC in vitro using purified fission yeast PAK expressed in Escherichia coli (Fig. 4A). Phosphorylation correlated with a single band shift on charge-separation gels (Fig. 4B), indicating that the expressed PAK phosphorylates a single serine.
Fig. 3.
The RLC of Myo2 has a homologous phosphorylation site. Multiple sequence alignment (N termini shown) using CLUSAL Omega (1.2.1). Sequence GenBank ID (GI) numbers are as follows: (S. pombe) 5824202, (S. cerevisiae) 151942958, (A. castellanii) 228753, (D. discoideum) 167833, (C. elegans) 351059042, (Gallus gallus) 45384118, (Homo sapiens) 205829213, 1025812257. The numbers above and to the right of the sequences are amino acid numbers, starting with the N-terminal Met as 1. The boxed area highlights the conserved phosphorylation sites, which are shown in red in S. pombe.
Fig. 4.
Fission yeast PAK phosphorylates one site on the RLC of Myo2. (A) SDS gel of (lane 1) molecular mass markers and (lane 2) purified recombinant fission yeast PAK. (B) Phosphorylated RLC migrates faster than unphosphorylated RLC on a 10% charge separation gel. (Right) The two bands, phosphorylated and unphosphorylated RLC, that occur when phosphorylation is incomplete.
Myo2 RLC phosphorylation was associated with an approximately twofold increase in actin-activated ATPase Vmax and an approximately fourfold increase in Km (Fig. 5A), which was significantly different (P < 0.0001) from unphosphorylated Myo2. The addition of tropomyosin (Tpm) to actin increased the Myo2 ATPase activity approximately twofold as shown previously (33, 34), but eliminated the effect of phosphorylation (P = 0.31) (Fig. 5A). Additionally, we examined the effects of phosphorylation on actin motility. To ensure equal availability of heads, Myo2 was attached to neutravidin-coated flow cells through the C-terminal biotin tag. Unphosphorylated Myo2 moved actin filaments at 0.74 ± 0.11 μm/s whereas phosphorylated Myo2 had a significantly lower average speed of 0.34 ± 0.18 μm/s (P < 0.0001) (Fig. 5B). Similar to the ATPase result, Tpm eliminated the effect of phosphorylation in the motility assay, which led to average speeds of ∼0.4 μm/s (P = 0.04).
Fig. 5.
Effects of RLC phosphorylation on ATPase activity and motility. (A) Steady-state ATPase of unphosphorylated (blue) or phosphorylated (red) Myo2 in the absence or presence of tropomyosin (Tpm). Circles and squares represent independent preparations of N-FLAG-Myo2-C-Biotin. Diamonds represent N-FLAG-Myo2. Conditions were as follows: 30 °C, 10 mM imidazole, pH 7.0, 50 mM NaCl, 1 mM MgCl2, 1 mM ATP, and 2 mM DTT. Tpm was added at a 2:1 Actin-Tpm molar ratio. (B) In vitro motility speeds of unphosphorylated or phosphorylated Myo2, in the absence or presence of Tpm. Speeds represent motility data from two experimental repeats conducted in parallel with two independent preparations of N-FLAG-Myo2-C-Biotin (n, number of moving filaments). To ensure that all heads were available for interaction with actin, Myo2 was attached to neutravidin-coated coverslips by a biotin tag at its C terminus. Conditions were as follows: 30 °C, 0.5% methylcellulose, 25 mM imidazole, pH 7.4, 50 mM KCl, 4 mM MgCl2, 1 mM EGTA, 1 mM ATP, and 10 mM DTT. When indicated, Tpm was added to a final concentration of 2 μM.
Tpm enhances actomyosin affinity in muscle, fission yeast, and other nonmuscle systems (33, 37, 38). To examine the effects of phosphorylation and Tpm on actomyosin affinity, we conducted a series of motility assays designed to reduce actin binding incrementally by removing methylcellulose and increasing the salt concentration. Without methylcellulose, phosphorylated Myo2 bound 25-fold fewer filaments per field (P = 0.01) on average compared with unphosphorylated Myo2 in 50 mM KCl (Fig. 6A and Movie S3). Tpm addition increased actin binding by Myo2 and resulted in no significant difference in the number of filaments bound between phosphorylated and unphosphorylated Myo2 (P = 0.3) (Fig. 6A and Movie S3). In 150 mM KCl, neither phosphorylated nor unphosphorylated Myo2 exhibited motility without methylcellulose (Fig. 6B and Movie S4). Tpm restored motility in 150 mM KCl for both phosphorylated and unphosphorylated Myo2; however, unphosphorylated Myo2 bound three- to fourfold more filaments (P < 0.0001) (Fig. 6B and Movie S4). Increasing the KCl to 200 mM reduced actin binding by phosphorylated Myo2 to nearly zero whereas unphosphorylated Myo2 still exhibited robust motility (Fig. 6C and Movie S5). These data show that phosphorylation of the RLC reduces the affinity of Myo2 for actin in the presence of Tpm at approximately physiological salt concentrations.
Fig. 6.
RLC phosphorylation reduces the number of Myo2 heads bound to actin filaments. Panels show maximum projections of representative fields (128 μm × 128 μm × 50 s) at different KCl concentrations (A–C). Phosphorylated Myo2 is shown on the Left (red) and unphosphorylated is shown on the Right (blue). Average speeds per condition are shown below. (Bar graphs) Numbers of motile filaments (n) identified by the analysis software (61) are shown as averages of multiple fields (N, number of fields). Myo2 was attached to neutravidin-coated coverslips through the C-terminal biotin tag. Conditions were as follows: 30 °C, 25 mM imidazole, pH 7.4, KCl as indicated, 4 mM MgCl2, 1 mM EGTA, 1 mM ATP, and 10 mM DTT. No methylcellulose was used to allow the diffusion of actin away from the surface. When indicated, Tpm was added to a final concentration of 2 μM. Data are from two independent preparations of Myo2 (N-FLAG-C-Biotin). Error, ± SD.
Myo2 Does Not Readily Assemble into Filaments.
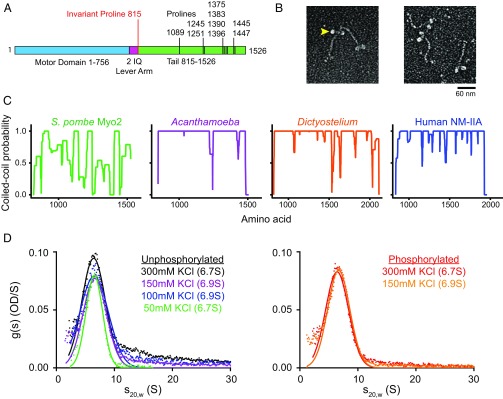
The rod-like tail domain is a canonical feature of class-II myosins that dictates self-assembly. Based on domain analysis, Myo2 is similar to other class-II myosins, except that the α-helical coiled-coil tail domain contains nine prolines distributed throughout its length (Fig. 7A). Despite the presence of prolines, which disrupt coiled coils, Myo2 has a rod-like tail and two heads (Fig. 7B). However, the coiled-coil probability distribution of the Myo2 tail is highly disrupted, compared with the tails of other class-II myosins that self-assemble (Fig. 7C) (39). The disrupted coiled coil of the rod is a unique feature of fungal class-II myosins that differentiates their functions and localizations in the cell (40–42). We examined whether Myo2 can self-assemble at various ionic strengths under conditions where other class-II myosins assemble, especially human nonmuscle myosin-II isoforms at 150 mM KCl (23) and Acanthamoeba myosin-II between 100 and 150 mM KCl (43). For all conditions tested by analytical ultracentrifugation, the fission yeast myosin formed a single 7S species (Fig. 7D), corresponding to a two-headed monomer. Myo2 was soluble even at 100 mM KCl; however, approximately half of the material precipitated at 50 mM KCl. Characterization of the precipitate by electron microscopy will be needed to determine whether Myo2 has the potential to form any bipolar filamentous structures. Despite Myo2’s partial insolubility at 50 mM KCl, we did not observe any faster sedimenting intermediate species (Fig. 7D). Importantly, fission yeast Myo2 is completely soluble at the approximately physiological ionic strength of 150 mM KCl, suggesting that Myo2 does not form filaments like class-II myosins that are expressed in animals and amoebas.
Fig. 7.
Myo2 does not have the propensity to assemble into filaments. (A) Diagram illustrating the domain structure of Myo2 and the high number of Pro residues interspersed throughout the tail. The invariant proline marks the junction between the lever arm and the tail. (B) Electron micrographs of platinum shadowed Myo2 (C-Biotin-C-FLAG) molecules showing that Myo2 has two heads and an ∼80-nm-long tail. Filled yellow arrowhead points to a motor domain. Image courtesy of Roger Craig. (C) Coiled-coil predictions using Paircoil2 analysis (62) comparing the tails of Myo2 and myosin-II from other species (A. castellanii, D. discoideum, and human nonmuscle myosin-IIA). Plots begin with the invariant proline and end at the final residue. (D) Sedimentation velocity of unphosphorylated or phosphorylated N-FLAG-Myo2 by analytical ultracentrifugation at varying KCl concentrations. A repeat analytical ultracentrifugation experiment with unphosphorylated Myo2 in 150 mM KCl gave a sedimentation coefficient of 7.0S. Conditions were as follows: 20 °C, 10 mM imidazole, pH 7.0, 5 mM MgCl2, 1 mM EGTA, 1 mM DTT (KCl as indicated). OD, optical density; S, Svedberg.
Discussion
To better understand the mechanism of cytokinesis, we investigated the function and regulation of Myo2, the myosin that is essential for cytokinesis in the well-established fission yeast model. Here, we examined the molecular properties of recombinant Myo2, which lacked RLC phosphorylation and did not require the yeast chaperone Rng3 for expression. Myo2 was fully active despite being unphosphorylated. Motility assays revealed that phosphorylation of the Myo2 RLC by p21/Cdc42-activated kinase (PAK) resulted in a significantly reduced affinity for actin and actin-Tpm. Moreover, hydrodynamic measurements indicated that Myo2 lacks self-assembly properties conserved with most class II myosins. Our findings underscore that Myo2 is an atypical class-II myosin in terms of molecular function and regulation, despite its conserved biological role during cytokinesis.
A Lack of Assembly Suggests Myo2 Clusters in Nodes.
Myo2 functions to make precursor nodes condense into a contractile ring that constricts during cytokinesis (Fig. 8A). Given that Myo2 expression levels remain constant throughout the cell cycle (20), how Myo2 is regulated during cytokinesis has been a basic question (15, 33). To maintain prolonged association with actin, a functional unit of Myo2 in the contractile ring is expected to consist of at least 10 molecules because Myo2, like other class-II myosins, spends only a small proportion of its ATPase cycle in strong-binding states: i.e., it has a low duty ratio. Duty ratio estimates (44) derived from our ATPase and motility data are consistent with Stark et al. (33), ranging from 4 to 10%, in the presence of Tpm. Without the ability to form oligomers, Myo2 therefore needs to be assembled into clusters through the association of the Myo2 tail with IQGAP (Rng2) and/or other proteins of the contractile ring nodes (Fig. 8B). Recent superresolution fluorescence microscopy of live fission yeast showed that the C terminus of the Myo2 tail is distributed with an average distance of ∼40 nm from the plasma membrane, which is similar to the distribution of Rng2 (45). Myo2 heads occupy a broader distribution than the tails, with an average distance of ∼100 nm away from the plasma membrane (45). This evidence suggests that Myo2 is arranged in polar clusters at the plasma membrane, unlike animal nonmuscle myosins that show a bipolar head-tail-head pattern by superresolution microscopy because of their assembly into bipolar minifilaments (46). Another difference is that the bipolar minifilaments in animal cells appear to expand into ordered contractile units in the contractile ring during cytokinesis (47). The in vivo superresolution light microscopy data revealing that Myo2 is localized to the plasma membrane in polarized clusters (45), together with our in vitro data showing that Myo2 lacks the propensity to self-assemble, are strong evidence that Myo2 is assembled through node binding rather than by rod–rod associations.
Fig. 8.
Myo2 activation by recruitment into nodes. (A) Diagram of fission yeast cytokinesis. RLC phosphorylation by PAK could delay contractile ring constriction (36) by reducing actomyosin affinity (this study). (B) Clustering by the node anchors Myo2 for force production in the contractile ring (45).
Substantial evidence suggests that the C-terminal 134 aa of Myo2 are a recruitment domain (9, 10), which might directly interact with IQGAP (4, 11). This domain begins at a junction in the rod where six prolines disrupt the α-helix (Fig. 7 A and B). A similar proline-rich recruitment domain occurs in the center of the rod of the budding yeast (Saccharomyces cerevisiae) class-II myosin ScMyo1, which relies on IQGAP (Iqg1) for localization during constriction (41). Like Myo2, ScMyo1 localizes before actin at the division site (the bud-neck). Early localization of ScMyo1 depends on an additional domain in the rod that binds to a septin-binding protein Bni5 (41) whereas fission yeast Myo2 depends on IQGAP during both assembly and constriction (4, 11). Thus, fungal class-II myosins seem to be adapted to associate with other proteins through the rod. A common theme in fungal cytokinesis is the formation of a cell wall-based septum on the extracellular interface of the cleavage furrow (Fig. 8). The clustering of myosin-II in protein complexes at the plasma membrane represents an efficient means for fungi to mechanically couple the contractile ring to the regulation and dynamics of transmembrane glucan synthases that build the cell wall (48, 49).
Myo2 Does Not Require Rng3 for Activity.
Our findings show that the chaperone Rng3 is not needed for Myo2 motor activity. Active recombinant Myo2 was obtained without coexpression of Rng3 (Fig. 1). Therefore, the insect homolog of Rng3, Unc45, must be sufficient to chaperone Myo2 into its native conformation. Our data suggest that Rng3 binds actin in the motility assay, which could be misinterpreted as increased actomyosin binding (Fig. 2B). Because we found that recombinant Myo2 is fully active, the tethering effect of Rng3 when bound nonspecifically to the coverslip could be an in vitro artifact. In general, the cellular role of UCS proteins as actin tethers has not been suggested in any other studies whereas there is substantial evidence for the involvement of UCS proteins in the unfolding response and protein turnover (50–52). UCS family proteins are widely known to be chaperones that assist in the folding of myosin (53). Evidence suggests that Rng3 is an essential chaperone for myosin in fission yeast (21) and is required for cytokinesis (18). The hypothesis that Rng3 activates motility stemmed, in part, from the observation that low concentrations (∼60 molecules) of Rng3 localize to the contractile ring in WT cells (15, 20). However, given the low abundance of Rng3 at the contractile ring, it is possible that a subpopulation of Myo2 can sometimes bring Rng3 along during the process of folding as Myo2 is incorporated into nodes since motor function is not required for the division site recruitment of Myo2 (9, 10, 18).
RLC Phosphorylation Decreases the Interaction of Myo2 with Actin in Force Generation.
We showed that Myo2 is active when the RLC is unphosphorylated, unlike vertebrate nonmuscle myosins (Fig. 1). Our results indicate that RLC phosphorylation leads to weaker actin binding, which is consistent with in vivo studies showing that RLC phosphorylation delays cytokinesis (36). Loo and Balasubramanian (36) reported that a nonphosphorylatable RLC (S35A/S36A) in fission yeast resulted in premature constriction, which sometimes led to aneuploidies. In fission yeast, the PAK kinase Pak1/Shk1/Orb2 colocalizes maximally with Myo2 in the contractile ring after coalescence and before constriction while the contractile ring is in the maturation (or dwell) phase (36). During the maturation phase, RLC phosphorylation by Pak1 may serve to slow node dynamics by reducing force production by Myo2, which permits sufficient time to allow chromosome segregation to occur before cytokinesis. In contrast to our findings, an earlier study (54) using Myo2 purified from fission yeast reported that the RLC (S35A/S36A) mutant had fourfold slower motility than the WT and phosphomimetic RLC (S35D/S36D) while ATPase rates were unaffected. The implication from this study was that phosphorylation enhanced the motility speed of Myo2, leading to increased rates of contractile ring constriction, in contradiction to the results we obtained with the native Ser in an unphosphorylated or phosphorylated state.
Tpm strongly enhances the affinity of Myo2 for actin, which reduced any effect of phosphorylation in motility and ATPase assays. Tpm has been shown to prolong the strong-binding states of budding yeast myosin-V, ScMyo2 (55), and has been suggested to do the same for fission yeast Myo2 (33). The unusually broad distribution of speeds in the motility assay for phosphorylated Myo2 in 50 mM KCl in the absence of Tpm (Fig. 5B) reflects the low affinity of phosphorylated Myo2 for actin, which leads to a range of submaximal speeds. This result is consistent with the low affinity of phosphorylated Myo2 for actin that was elucidated from experiments performed in the absence of methylcellulose, which allows weakly bound actin to diffuse away (Fig. 6A). The addition of Tpm enhances the affinity of Myo2 for actin, irrespective of the state of phosphorylation. Nonetheless, as the salt concentration was raised to more physiological levels (≥150 mM KCl), little or no binding of phosphorylated Myo2 occurred, even in the presence of tropomyosin, which is always bound to actin in the contractile ring. Unlike nonmuscle myosins in animal cells, where phosphorylation is required for activity, phosphorylation of Myo2 in fission yeast serves to down-regulate the interaction between myosin and actin. One unusual feature of fission yeast RLC is an extended N terminus that is distinct from RLCs of other species (Fig. 3) and may contribute to its unique regulatory properties. The structural basis by which RLC phosphorylation affects myosin function is not well-understood, but interactions between the RLC N terminus and the ELC may play a role (56). Although we have focused exclusively on the RLC in Myo2, other sites for posttranslational modification and interaction with binding partners may offer further clues to the mechanism of cytokinesis in fission yeast.
Materials and Methods
Expression and Purification of Myo2 and Rng3.
The coding sequence of the Myo2 heavy chain (HC) was cloned into the Sf9/baculovirus expression system vectors pAcSG2 (BD Biosciences) and pFastBac-1 (Thermo Fisher Scientific). The light chains (LCs), Cdc4 and Rlc1, were cloned into pAcUW51, a dual-promoter vector that drives expression of both LCs (BD Biosciences). The HC-coding sequence was linked at either the N or the C terminus by a FLAG tag (DYKDDDDK) to facilitate purification by affinity chromatography. Certain constructs additionally linked the HC-coding sequence to a C-terminal biotin tag (57) for attachment to streptavidin-coated surfaces. C-terminally FLAG-tagged Myo2 constructs copurified with a significant amount of tail breakdown product, which was identified by mass spectrometry (Fig. 1A). Sf9 cells were coinfected with recombinant baculovirus coding for the HC and LC constructs. After 72 h at 27 °C, the cells were harvested by centrifugation at 5,000 × g for 10 min at 4 °C. The cells were resuspended in ice-cold lysis buffer A [300 mM NaCl, 10 mM imidazole, pH 7.4, 5 mM MgCl2, and 1 mM ethylene glycol tetraacetic acid (EGTA)] supplemented with 2 mM DTT, 7% wt/vol sucrose, 0.5 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) (Fisher), 5 µg/mL leupeptin (Acros Organics), 0.5 mM phenylmethylsulfonyl fluoride (PMSF) (MP Biomedical), 0.5 mM Nα-tosyl-l-lysine chloromethyl ketone hydrochloride (TLCK) (Sigma-Aldrich), 0.4 mg/mL benzamidine (Sigma), and protease inhibitor mixture (P8340; Sigma), and lysed by sonication on an ice bath. Two mM MgATP was added, and cell debris was pelleted by centrifugation at 250,000 × g for 20 min at 4 °C. The clarified supernatant was batch-incubated with FLAG-affinity resin (Sigma) for 1 h at 4 °C. The resin was packed into a column and washed with lysis buffer before elution with 100 μg/mL FLAG peptide (Sigma) in lysis buffer A. The pooled elution fractions were concentrated by Amicon-Ultra filtration (EMD Millipore) and dialyzed at 4 °C against storage buffer [300 mM NaCl, 10 mM imidazole, pH 7.4, 1 mM EGTA, 1 mM NaN3, 50% glycerol (vol/vol)] with 2 mM DTT and 1 µg/mL leupeptin for storage at −20 °C. Rng3 with a C-terminal FLAG tag was expressed in the Sf9-baculovirus system and purified in the same way. Average yields of Myo2 and Rng3 were ∼1 mg per billion cells.
Purification of Other Proteins.
The p21/Cdc42-activated kinase (PAK), Pak1/Shk1/Orb2, coding sequence was cloned into the pET19 (EMD Millipore) vector upstream of a C-terminal FLAG tag sequence. Pak1 expression in Rosetta (DE3) E. coli (EMD Millipore) grown to OD600 0.8 to 1.0 in Luria-Bertani (LB) broth was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (Sigma) for 18 h at room temperature. After the induction, cells were pelleted by centrifugation at 5,000 × g for 20 min. Cells were lysed by sonication on ice in lysis buffer A supplemented with 2 mM DTT, 7% sucrose, 10 mM Na pyrophosphate, 10 mM NaF, 0.5 mM AEBSF (Fisher), 5 μg/mL leupeptin (Acros), 0.5 mM PMSF (MP), and 5 mM benzamidine (Sigma). Following lysis, the lysate was clarified, and FLAG-affinity chromatography was performed as outlined above. The full-length Pak1 used in this study was found to have a canonical N-terminal autoinhibitory domain (58), which partially suppressed its kinase activity.
Chicken skeletal actin was purified according to Spudich and Watt (59). To purify Tpm, bacterial expression construct pET3a (EMD Millipore) containing the coding sequence of fission yeast Cdc8 with N-terminal Ala-Ser (acetylation mimic) was transformed into BL21-DE3 E. coli (EMD Millipore). Transformed bacteria were grown in LB to OD600 0.8 to 1.0 and induced with 0.4 mM IPTG for 5 h at 25 °C. Cells were harvested by centrifugation and suspended in ice-cold lysis buffer B (20 mM imidazole, pH 7.5, 10 mM NaCl, and 2 mM EDTA) supplemented with 1 mM DTT, 1 μg/mL leupeptin, 0.5 mM PMSF, and 0.4 mg/mL benzamidine and lysed by sonication on ice. Cell debris was pelleted by centrifugation at 30,000 × g for 15 min at 4 °C. The supernatants were boiled for 5 min while stirring and cooled to 25 °C, and then denatured proteins were pelleted by centrifugation at 30,000 × g for 15 min at 4 °C. Tpm was precipitated by adjusting the pH of the supernatant to its isoelectric focusing point (pH 4.6), pelleted at 30,000 × g for 15 min at 4 °C, and dissolved by gentle agitation overnight at 4 °C in lysis buffer B supplemented with 1 mM DTT and 1 μg/mL leupeptin. The dissolved Tpm was dialyzed against Tpm buffer (50 mM NaCl, 10 mM imidazole, pH 7.5, and 1 mM DTT).
RLC Phosphorylation.
Myo2 was phosphorylated by adding a 0.3 molar ratio of PAK in kinase buffer (150 mM NaCl, 10 mM imidazole, pH 7.4, 5 mM MgCl2, 1 mM EGTA, 1 mM NaN3, and 2 mM DTT) with 5 mM MgATP, and incubating at 30 °C for 1 h. The control underwent the same treatment, but without Pak1. To evaluate phosphorylation levels, SDS/PAGE gels were stained with Pro Q Diamond Phosphoprotein Gel stain (Invitrogen) according to online protocol. Alternatively, samples were dissolved in 7.5 M urea and run on 40% glycerol gels (60) to resolve the RLC based on charge. The light chains of unphosphorylated smooth muscle myosin, or phosphorylated with myosin light chain kinase (MLCK) (60), were used as standards to determine the level of phosphorylation by Pro Q fluorescence intensity. Preliminary mass spectrometry analysis of the RLC detected phosphorylated serines 35 and 36; however, this analysis did not identify which site. For ATPase and motility assays, the RLC was 100% phosphorylated.
In Vitro Motility.
To remove ATP-insensitive heads, Myo2 was centrifuged for 20 min at 400,000 × g in the presence of twofold excess F-actin and 2 mM MgATP in myosin buffer (10 mM imidazole, pH 7.4, 0.3 M NaCl, 5 mM MgCl2, 1 mM EGTA, and 10 mM DTT). Nitrocellulose-coated flow cells were prepared with neutravidin by the sequential addition of 0.5 mg/mL biotinylated BSA, 1 mg/mL BSA, and 10 to 50 μg/mL neutravidin (Thermo Scientific) in buffer A (25 mM imidazole, pH 7.4, 0.15 M KCl, 4 mM MgCl2, 1 mM EGTA, and 10 mM DTT), each followed by a 1-min incubation and three washes of buffer A. Myo2 was then applied in myosin buffer at 50 μg/mL (unless otherwise specified) for 1 min. ATP-insensitive heads were further blocked by two additions of 1 μM vortexed F-actin in buffer B [25 mM imidazole, pH 7.4, 50 mM KCl, 4 mM MgCl2, 1 mM EGTA, and 10 mM DTT, 3 mg/mL glucose, 0.125 mg/mL glucose oxidase (Sigma-Aldrich), and 0.05 mg/mL catalase (Sigma-Aldrich)] over 1 min. Active heads were freed by three washes with buffer B containing 1 mM MgATP. The ATP was washed out with three more passes of buffer B, and then 10 nM F-actin labeled with 1.5-molar excess of rhodamine-phalloidin (Thermo Scientific) in buffer B was applied twice over 1 min. Buffer B with 0.5% methylcellulose and 1 mM MgATP was applied two times before imaging. Where indicated, methylcellulose was omitted in the final buffer. To decorate actin with tropomyosin, the AlaSer-Cdc8 acetyl mimic was added at 2 μM with the labeled actin stock, and 2 μM tropomyosin was added to the final buffer. For experiments where Myo2 and Rng3 were attached to the coverslip surface, instead of preparing the flow cells with neutravidin, combinations of Myo2 and Rng3 in myosin buffer were applied directly to the nitrocellulose-coated flow cells, and then 1 mg/mL BSA was used to block the remaining surface. In general, N-FLAG-Myo2-C-Biotin or C-Biotin-C-FLAG was attached to the coverslip with neutravidin to ensure that all heads are available for interaction with actin. N-FLAG-Myo2 had to be directly attached to nitrocellulose-coated coverslips.
Actin movement at 30 °C was observed using an inverted microscope (Zeiss Axiovert 10) equipped with a heated objective, epifluorescence, and a Rolera MGi Plus digital camera. A dedicated computer with the Nikon NIS Elements software package was used to control the microscope and record the image data. Tiff stacks were processed in ImageJ and analyzed using a filament tracking program described previously (61).
Actin-Activated ATPase Activity.
ATPase assays were performed in 10 mM imidazole, pH 7.0, 50 mM NaCl, 1 mM MgCl2, 1 mM Na azide, and 2 mM DTT in separate 1.5-mL Eppendorf tubes in a 30 °C water bath, while stirring. Myo2 was clarified by centrifugation for 20 min at 400,000 × g in myosin buffer and then diluted to twofold concentration (final concentration 25 μg/mL) in the assay buffer before being mixed 1:1 with skeletal actin at various concentrations. For Tpm-decoration, the actin stock was mixed 2:1 with AlaSer-Cdc8. Activity was initiated by the addition of 1 mM MgATP and quenched with SDS at 5-min intervals over 25 min. Inorganic phosphate concentration was determined colorimetrically (60).
Analytical Ultracentrifugation.
For assembly experiments, the N-flag Myo2 construct was dialyzed for 18 h against buffers containing 10 mM imidazole, pH 7.0, 5 mM MgCl2, 1 mM EGTA, and 1 mM DTT with varying concentrations of KCl. In an earlier experiment, the buffer was 10 mM potassium phosphate, pH 7.5, for the C-Biotin-FLAG construct (Fig. 1B). The addition of 0.2 mM ATP had no effect on the sedimentation of Myo2 in phosphate buffer at lower salt (150 mM KCl). After dialysis, Myo2 was clarified by centrifugation at 200,000 × g for 10 min at 4 °C. Recovery of protein was 70 to 80% of the concentration before dialysis. Precipitate was visible when Myo2 was dialyzed against buffer containing 50 mM KCl. This sample was clarified by centrifugation at 20,000 × g for 5 min at 4 °C, and ∼50% of the protein was recovered. A Beckman Optima XL-I analytical ultracentrifuge was used to perform sedimentation velocity runs at 40,000 rpm in an An-60Ti rotor (Beckman) at 20 °C. Sedimentation coefficients were calculated using DCDT+ (v.2.4.3) software by John Philo and were corrected for temperature and buffer composition using Sednterp software.
Graphing and Statistical Analysis.
Graphing and statistical analyses were done using GraphPad Prism 7.01. ATPase data were fit to the Michaelis–Menten equation and the best-fit values for Km and Vmax were compared using an Extra sum-of-squares F test. Motility speed and frequency data were fit to a Gaussian distribution function and compared using unpaired, two-tailed t tests. Welch’s correction was applied when datasets had significantly different variances, which were compared using F tests. Paircoil2 was used to predict the parallel coiled-coil fold from amino acid sequences. Paircoil2 calculates pairwise residue probabilities using the Paircoil algorithm and updated coiled-coil databases, resulting in fewer false-positive predictions compared with other coiled-coil prediction programs (62).
Supplementary Material
Acknowledgments
We thank members of the K.M.T. laboratory and Matthew Lord for early contributions to this project. We thank Kyoung Hwan Lee and Roger Craig (University of Massachusetts Medical School) for providing the platinum rotary-shadowed electron micrographs. This research was supported by National Institutes of Health Grants GM097193 (to S.L.) and GM078097 (to K.M.T.). Mass spectrometry protein identification of expressed protein bands was performed by the Vermont Genetics Network Core Facility, which is supported by National Institutes of Health Institutional Development Award P20GM103449.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703161114/-/DCSupplemental.
References
- 1.Cheffings TH, Burroughs NJ, Balasubramanian MK. Actomyosin ring formation and tension generation in eukaryotic cytokinesis. Curr Biol. 2016;26:R719–R737. doi: 10.1016/j.cub.2016.06.071. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD. The value of mechanistic biophysical information for systems-level understanding of complex biological processes such as cytokinesis. Biophys J. 2014;107:2499–2507. doi: 10.1016/j.bpj.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JQ, et al. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laplante C, et al. Three myosins contribute uniquely to the assembly and constriction of the fission yeast cytokinetic contractile ring. Curr Biol. 2015;25:1955–1965. doi: 10.1016/j.cub.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard TD, Wu JQ. Understanding cytokinesis: Lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 8.Lee IJ, Coffman VC, Wu JQ. Contractile-ring assembly in fission yeast cytokinesis: Recent advances and new perspectives. Cytoskeleton. 2012;69:751–763. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulvihill DP, Barretto C, Hyams JS. Localization of fission yeast type II myosin, Myo2, to the cytokinetic actin ring is regulated by phosphorylation of a C-terminal coiled-coil domain and requires a functional septation initiation network. Mol Biol Cell. 2001;12:4044–4053. doi: 10.1091/mbc.12.12.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol. 2004;165:685–695. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takaine M, Numata O, Nakano K. Fission yeast IQGAP maintains F-actin-independent localization of myosin-II in the contractile ring. Genes Cells. 2014;19:161–176. doi: 10.1111/gtc.12120. [DOI] [PubMed] [Google Scholar]
- 12.Tang Q, et al. A single-headed fission yeast myosin V transports actin in a tropomyosin-dependent manner. J Cell Biol. 2016;214:167–179. doi: 10.1083/jcb.201511102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaine M, Numata O, Nakano K. An actin-myosin-II interaction is involved in maintaining the contractile ring in fission yeast. J Cell Sci. 2015;128:2903–2918. doi: 10.1242/jcs.171264. [DOI] [PubMed] [Google Scholar]
- 14.Win TZ, Gachet Y, Mulvihill DP, May KM, Hyams JS. Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J Cell Sci. 2001;114:69–79. doi: 10.1242/jcs.114.1.69. [DOI] [PubMed] [Google Scholar]
- 15.Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni W, Odunuga OO. UCS proteins: Chaperones for myosin and co-chaperones for Hsp90. Subcell Biochem. 2015;78:133–152. doi: 10.1007/978-3-319-11731-7_7. [DOI] [PubMed] [Google Scholar]
- 17.Lee CF, Melkani GC, Bernstein SI. The UNC-45 myosin chaperone: From worms to flies to vertebrates. Int Rev Cell Mol Biol. 2014;313:103–144. doi: 10.1016/B978-0-12-800177-6.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong KC, Naqvi NI, Iino Y, Yamamoto M, Balasubramanian MK. Fission yeast Rng3p: An UCS-domain protein that mediates myosin II assembly during cytokinesis. J Cell Sci. 2000;113:2421–2432. doi: 10.1242/jcs.113.13.2421. [DOI] [PubMed] [Google Scholar]
- 19.Mishra M, D’souza VM, Chang KC, Huang Y, Balasubramanian MK. Hsp90 protein in fission yeast Swo1p and UCS protein Rng3p facilitate myosin II assembly and function. Eukaryot Cell. 2005;4:567–576. doi: 10.1128/EC.4.3.567-576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 21.Stark BC, James ML, Pollard LW, Sirotkin V, Lord M. UCS protein Rng3p is essential for myosin-II motor activity during cytokinesis in fission yeast. PLoS One. 2013;8:e79593. doi: 10.1371/journal.pone.0079593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lord M, Sladewski TE, Pollard TD. Yeast UCS proteins promote actomyosin interactions and limit myosin turnover in cells. Proc Natl Acad Sci USA. 2008;105:8014–8019. doi: 10.1073/pnas.0802874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Hong MS, Shu S, Yu S, Korn ED. Regulation of the filament structure and assembly of Acanthamoeba myosin II by phosphorylation of serines in the heavy-chain nonhelical tailpiece. Proc Natl Acad Sci USA. 2013;110:E33–E40. doi: 10.1073/pnas.1219727110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahajan RK, Pardee JD. Assembly mechanism of Dictyostelium myosin II: Regulation by K+, Mg2+, and actin filaments. Biochemistry. 1996;35:15504–15514. doi: 10.1021/bi9618981. [DOI] [PubMed] [Google Scholar]
- 26.Trybus KM, Huiatt TW, Lowey S. A bent monomeric conformation of myosin from smooth muscle. Proc Natl Acad Sci USA. 1982;79:6151–6155. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig R, Smith R, Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–439. doi: 10.1038/302436a0. [DOI] [PubMed] [Google Scholar]
- 28.Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J Cell Biol. 1999;147:1385–1390. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trybus KM, Lowey S. Conformational states of smooth muscle myosin: Effects of light chain phosphorylation and ionic strength. J Biol Chem. 1984;259:8564–8571. [PubMed] [Google Scholar]
- 30.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bookwalter CS, Kelsen A, Leung JM, Ward GE, Trybus KM. A Toxoplasma gondii class XIV myosin, expressed in Sf9 cells with a parasite co-chaperone, requires two light chains for fast motility. J Biol Chem. 2014;289:30832–30841. doi: 10.1074/jbc.M114.572453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird JE, et al. Chaperone-enhanced purification of unconventional myosin 15, a molecular motor specialized for stereocilia protein trafficking. Proc Natl Acad Sci USA. 2014;111:12390–12395. doi: 10.1073/pnas.1409459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell. 2010;21:989–1000. doi: 10.1091/mbc.E09-10-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clayton JE, Pollard LW, Murray GG, Lord M. Myosin motor isoforms direct specification of actomyosin function by tropomyosins. Cytoskeleton. 2015;72:131–145. doi: 10.1002/cm.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uyeda TQ, Kron SJ, Spudich JA. Myosin step size: Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J Mol Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 36.Loo TH, Balasubramanian M. Schizosaccharomyces pombe Pak-related protein, Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. J Cell Biol. 2008;183:785–793. doi: 10.1083/jcb.200806127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanBuren P, Palmiter KA, Warshaw DM. Tropomyosin directly modulates actomyosin mechanical performance at the level of a single actin filament. Proc Natl Acad Sci USA. 1999;96:12488–12493. doi: 10.1073/pnas.96.22.12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barua B, Nagy A, Sellers JR, Hitchcock-DeGregori SE. Regulation of nonmuscle myosin II by tropomyosin. Biochemistry. 2014;53:4015–4024. doi: 10.1021/bi500162z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May KM, Win TZ, Hyams JS. Yeast myosin II: A new subclass of unconventional conventional myosins? Cell Motil Cytoskeleton. 1998;39:195–200. doi: 10.1002/(SICI)1097-0169(1998)39:3<195::AID-CM2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Bezanilla M, Pollard TD. Myosin-II tails confer unique functions in Schizosaccharomyces pombe: Characterization of a novel myosin-II tail. Mol Biol Cell. 2000;11:79–91. doi: 10.1091/mbc.11.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang X, et al. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol. 2010;191:1333–1350. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lord M, Laves E, Pollard TD. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol Biol Cell. 2005;16:5346–5355. doi: 10.1091/mbc.E05-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinard JH, Stafford WF, Pollard TD. The mechanism of assembly of Acanthamoeba myosin-II minifilaments: Minifilaments assemble by three successive dimerization steps. J Cell Biol. 1989;109:1537–1547. doi: 10.1083/jcb.109.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huxley HE. Sliding filaments and molecular motile systems. J Biol Chem. 1990;265:8347–8350. [PubMed] [Google Scholar]
- 45.Laplante C, Huang F, Tebbs IR, Bewersdorf J, Pollard TD. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc Natl Acad Sci USA. 2016;113:E5876–E5885. doi: 10.1073/pnas.1608252113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beach JR, et al. Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fenix AM, et al. Expansion and concatenation of non-muscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol Biol Cell. 2016;27:1465–1478. doi: 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Z, et al. The contractile ring coordinates curvature-dependent septum assembly during fission yeast cytokinesis. Mol Biol Cell. 2015;26:78–90. doi: 10.1091/mbc.E14-10-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiyagarajan S, Munteanu EL, Arasada R, Pollard TD, O’Shaughnessy B. The fission yeast cytokinetic contractile ring regulates septum shape and closure. J Cell Sci. 2015;128:3672–3681. doi: 10.1242/jcs.166926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Etard C, Roostalu U, Strähle U. Shuttling of the chaperones Unc45b and Hsp90a between the A band and the Z line of the myofibril. J Cell Biol. 2008;180:1163–1175. doi: 10.1083/jcb.200709128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melkani GC, Lee CF, Cammarato A, Bernstein SI. Drosophila UNC-45 prevents heat-induced aggregation of skeletal muscle myosin and facilitates refolding of citrate synthase. Biochem Biophys Res Commun. 2010;396:317–322. doi: 10.1016/j.bbrc.2010.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melkani GC, Bodmer R, Ocorr K, Bernstein SI. The UNC-45 chaperone is critical for establishing myosin-based myofibrillar organization and cardiac contractility in the Drosophila heart model. PLoS One. 2011;6:e22579. doi: 10.1371/journal.pone.0022579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hutagalung AH, Landsverk ML, Price MG, Epstein HF. The UCS family of myosin chaperones. J Cell Sci. 2002;115:3983–3990. doi: 10.1242/jcs.00107. [DOI] [PubMed] [Google Scholar]
- 54.Sladewski TE, Previs MJ, Lord M. Regulation of fission yeast myosin-II function and contractile ring dynamics by regulatory light-chain and heavy-chain phosphorylation. Mol Biol Cell. 2009;20:3941–3952. doi: 10.1091/mbc.E09-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hodges AR, et al. Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Curr Biol. 2012;22:1410–1416. doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor KA, et al. Role of the essential light chain in the activation of smooth muscle myosin by regulatory light chain phosphorylation. J Struct Biol. 2014;185:375–382. doi: 10.1016/j.jsb.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cronan JE., Jr Biotination of proteins in vivo: A post-translational modification to label, purify, and study proteins. J Biol Chem. 1990;265:10327–10333. [PubMed] [Google Scholar]
- 58.Knaus UG, Bokoch GM. The p21Rac/Cdc42-activated kinases (PAKs) Int J Biochem Cell Biol. 1998;30:857–862. doi: 10.1016/s1357-2725(98)00059-4. [DOI] [PubMed] [Google Scholar]
- 59.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 60.Trybus KM. Biochemical studies of myosin. Methods. 2000;22:327–335. doi: 10.1006/meth.2000.1085. [DOI] [PubMed] [Google Scholar]
- 61.Kinose F, Wang SX, Kidambi US, Moncman CL, Winkelmann DA. Glycine 699 is pivotal for the motor activity of skeletal muscle myosin. J Cell Biol. 1996;134:895–909. doi: 10.1083/jcb.134.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: Improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.