Significance
In the era of functional proteomics, a myriad of new interactions, notably those modification-dependent ones, are widely suggested by advanced proteomic approaches and bioinformatic analysis. Therefore, there exists an urgent need to develop a technology for high-throughput mapping and quantitative characterization of biomolecular binding events. This study achieved the immobilization and kinetic detection of various biomacromolecules (including modified peptides and modified nucleic acids) in high throughput through the 3D-carbene chip-based surface plasmon resonance imaging (SPRi) technology. Modified histone peptides and nucleic acids, which are key epigenetic marks, could be efficiently probed by this platform. We envision that the 3D-carbene SPRi technology described here will have wide appeal in profiling and discovering biological recognitions in and beyond epigenetics.
Keywords: SPR imaging, epigenetic interactions, histone modifications, nucleic acid modifications, 3D-carbene chip
Abstract
Chemical modifications on histones and DNA/RNA constitute a fundamental mechanism for epigenetic regulation. These modifications often function as docking marks to recruit or stabilize cognate “reader” proteins. So far, a platform for quantitative and high-throughput profiling of the epigenetic interactome is urgently needed but still lacking. Here, we report a 3D-carbene chip-based surface plasmon resonance imaging (SPRi) technology for this purpose. The 3D-carbene chip is suitable for immobilizing versatile biomolecules (e.g., peptides, antibody, DNA/RNA) and features low nonspecific binding, random yet function-retaining immobilization, and robustness for reuses. We systematically profiled binding kinetics of 1,000 histone “reader–mark” pairs on a single 3D-carbene chip and validated two recognition events by calorimetric and structural studies. Notably, a discovery on H3K4me3 recognition by the DNA mismatch repair protein MSH6 in Capsella rubella suggests a mechanism of H3K4me3-mediated DNA damage repair in plant.
Despite having identical genetic information, human cells differentiate into different identities during development. The mechanisms leading to differential transcriptional and developmental outcomes under the same genetic background are known as epigenetics. Posttranslational modification (PTM) on histones is a major epigenetic regulatory mechanism such that more than 30 types of PTMs occur on histones, with new ones still being discovered (1). The well-studied histone PTMs include methylation, acetylation, phosphorylation, and most recently discovered nonacetyl acylations (2). Histone PTMs regulate transcription either by directly affecting the structure/stability of single nucleosome and the high-order folding of chromatin, or by recruiting specific effector proteins (also called histone readers) recognizing them (3, 4).
The ever-expanding repertoire of histone readers are diverse in sequence and structure, and how they engage diversely modified chromatin is complicated, hard to predict, yet important for their functions. Often one type of reader domains can recognize different types of PTMs. For example, PHD finger proteins can recognize unmodified, methylated, or acylated lysine residue on histones (5, 6). Several YEATS domain proteins, originally identified as readers of lysine acetylation (Kac), in fact accommodate a wide range of lysine acylations with lysine crotonylation (Kcr) most preferred (7). These “mark–reader” interactions vary greatly in binding affinity, ranging from submicromolar to millimolar levels. To complicate the matter further, the recognition of marks by reader proteins is often either synergized or antagonized by other marks in close proximity (8). For instance, Spindlin1 recognizes histone H3 trimethylation of Lys-4 (H3K4me3) and asymmetrically dimethylation of Arg-8 (H3R8me2a) in concert, while phosphorylation of Ser-10 on H3 (H3S10ph) completely abrogates the binding of H3 Lys-9 trimethylation (H3K9me3) by HP1 protein (9, 10). BRDT protein binds neither histone H4 Lys-5 acetylation (H4K5ac) nor Lys-8 acetylation (H4K8ac) but engages H4 peptide bearing both marks with decent affinity (KD = 22 µM) (11). In sum, a more realistic and holistic understanding of how reader proteins engage chromatin necessitates screening known and potential readers against an exhaustive collection of histone PTMs and relevant combinations thereof, ideally in a high-throughput and quantitative manner. However, to the best of our knowledge, such a demand has not been addressed by the epigenetic field.
As an advanced optical-based quantitative detection method, surface plasmon resonance (SPR) sensing has been extended to the SPR microscopy or imaging (SPRi) technology for high-throughput probing of biomolecular interactions (12, 13). Despite some intrinsic disadvantages of SPR, such as influence of immobilization and surface fouling, SPRi stands out as a powerful detection tool as it probes analyte label-freely and measures binding kinetics in real time and in microarray format. The immobilization strategy and the spacing of ligands on the chip surface can influence the detecting efficiency of SPRi. Carbene-based cross-linking strategy, which rapidly immobilizes molecules in random orientation due to its high and diverse chemical reactivity (e.g., carbon insertion, Michael addition, nucleophilic reaction, etc.), is becoming an optimized immobilization method for microarray fabrications (14–16). Meanwhile, due to its high loading capacity for biomolecules and antifouling properties, 3D polymer surface with a designed structure often shows high signal-to-noise ratio in SPRi (17–19). Theoretically, a carbene-based 3D surface could immobilize biomolecules randomly in position and orientation to best retain the functionality and, in the meantime, has the advantage of high sensitivity and low nonspecific adsorption. However, binding detection of randomly immobilized biomolecules by the 3D-carbene SPRi technology has yet to be explored systematically.
Here, we report the quantitative and high-throughput profiling of epigenetic interactions by the 3D-carbene chip-based SPRi technology. We demonstrated that the 3D-carbene chip could efficiently immobilize various kinds of biomolecules such as peptide, protein, and DNA/RNA. The 3D-carbene chip surface features low nonspecific binding, random yet function-retaining immobilization of ligands, and robustness as well as stability. We immobilized 125 modified histone peptides on the chip and subjected the chip to interaction profiling of eight histone reader proteins. The positive and negative controls set in the experiments matched well with known results. We demonstrated that 3D-carbene–based SPRi technology was suitable to kinetically analyze epigenetic interactions in high throughput. We confirmed a strong interaction between TAF3–H3K4me3 and validated a unique weak interaction between TAF3–H3K4ac by isothermal titration calorimetry (ITC) and crystallography. Importantly, an epigenetic interaction between H3K4me3 and the tudor domain of mutS homolog 6 (MSH6) in Capsella rubella was discovered, suggesting a role of H3K4me3 in DNA mismatch repair in plant. Collectively, these results demonstrate that the 3D-carbene SPRi technology is suitable for kinetic and high-throughput analysis of the epigenetic “readership” and for interaction discovery.
Results
Design and Fabrication of the 3D-Carbene Chip.
The 3D-carbene SPRi technology was designed as in Fig. 1A. As the core element of the chip, a 3D-carbene surface was constructed on the SPRi gold substrate. Then an array of aqueous solutions of biomolecules (up to hundreds to thousands of spots) was printed on the surface and dried in dark. Afterward, the chip surface was radiated with 365-nm UV light, generating highly reactive carbene, which immobilizes the biomolecules in situ in random orientations. As a result, the binding sites of the immobilized ligands are presented in nearly all possible orientations. For real-time and high-throughput detection, the chip was assembled into the SPRi instrument, presenting a grayscale imaging of microarray. Then the binding protein was flowed over the chip surface and interacted with the immobilized ligands, resulting in the shifting of SPR angle. All binding signals were recorded as kinetic curves from which thermodynamic and kinetic values of the binding event were extracted with assistance of computer programs.
Fig. 1.
Surface design and characterization of 3D-carbene–based SPRi technology. (A) Scheme of high-throughput and label-free detection of biomolecular interactions on a 3D-carbene chip using SPRi instrument. The SPRi chip surface is grafted by brush-like polymer, which contains carbon backbones (blue), PEG branches (green), and diazirine-derived terminals (red). Biomolecules are randomly and covalently immobilized onto the terminals by carbene-based reaction under 365-nm UV light treatment. (B) Design and construction of 3D-polymer surface on gold substrate and conjugation of photo–cross-linker (PCL). (C) FT-IR characterization of surface construction and modification step by step. (D) XPS characterization of polymer/polymer-PCL surface by element analysis.
As shown in Fig. 1B, a 3D-carbene surface was fabricated over SPRi bare gold chip. Briefly, synthesized p(HEMA-co-PEGMA500) polymer (characterized by 1H NMR and GPC) was assembled on the bare gold surface (SI Appendix, Fig. S1). After carboxylation and EG3-OH blocking, the chip surface was activated by N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride crystalline (EDC)/N-hydroxysuccinimide (NHS) and coupled with photo–cross-linker (PCL). A detailed procedure of surface fabrication is described in SI Appendix. The whole process of the chemical modification on SPRi chip surface was recorded by a Fourier transform infrared spectroscopy (FT-IR) in grazing angle incidence mode (Fig. 1C). The characteristic peaks of C=O, O–H, C–O–C, C–H, and aromatic rings suggested a successful modification. In addition, the confirming of initial grafting and PCL coupling on the surface was also conducted by X-ray photoelectron spectroscopy (XPS) and atomic force microscopy (AFM), respectively (Fig. 1D and SI Appendix, Fig. S2 and Table S1). All of the characterization of the chip surface confirmed that the 3D-carbene surface was fabricated as we designed and could be used as a functional matrix for immobilizing biomolecules.
Immobilization of Biomacromolecules by 3D-Carbene Chip.
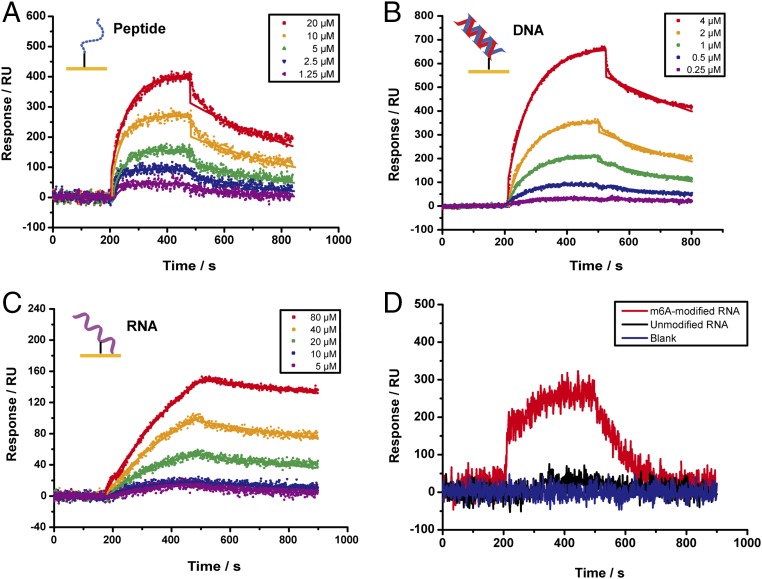
Carbene randomly inserts into C–H bonds that are ubiquitous in biomolecules, and in theory can immobilize peptide, protein, and DNA/RNA on 3D-carbene chip with full spectrum of orientations to best retain their respective functionality. To test our hypothesis, we immobilized histone H3 residues 1–15 with lysine 4 trimethylated [H3(1–15)K4me3] and determined its binding affinity to CHD1, a known histone reader protein specifically recognizing H3(1–15)K4me3 (20). H3(1–15)K4me3 showed significant signals on chip and the KD value determined by SPRi was 8.8 ± 0.5 µM (Fig. 2A). This result agrees reasonably with that determined by ITC (KD = 9.4 µM) (20). In addition, this affinity agrees well with that determined using the oriented immobilization strategy [KD = 8.2 ± 0.7 µM, C-terminal biotin labeled H3(1–15)K4me3 peptide] (SI Appendix, Fig. S3). The 3D-carbene chip can also immobilize stable proteins such as antibodies. We immobilized the rabbit IgG and flowed in goat anti-rabbit IgG, obtaining KD value of 8.4 ± 1.1 nM (SI Appendix, Fig. S4). These results demonstrated that the 3D-carbene chip could immobilize peptides/proteins and was suitable for characterizing modification-dependent interactions. Next, we immobilized an 18-bp double-stranded DNA (5′-CTTATGGAAAGCATGCTT-3′) on the 3D-carbene chip. A KD value of 3.1 ± 0.1 µM was determined for FOXP3, a protein recognizing this DNA sequence (21) (Fig. 2B). The mutant CHD1 (CHD1 W325A) and FOXP3 (FOXP3 H387A) lost the binding signal to their corresponding binding partners on this chip (SI Appendix, Fig. S5). We also immobilized a 16-nt RNA (5′-GACGACCGGACUGUCU-3′) and measured KD value of 17.0 ± 1.6 µM for the cognate binding protein HNRNPA2B1 (22) (Fig. 2C).
Fig. 2.
SPRi detection of epigenetic interactions by immobilizing peptide, DNA, and RNA on 3D-carbene chips. (A) SPRi detection of CHD1–peptides interactions with peptides immobilized on the surface. (B) SPRi detection of FOXP3–dsDNA interactions with dsDNA immobilized on the surface. (C) SPRi detection of HNRNPA2B1–RNA interactions with RNA immobilized on the surface. (D) Binding signals of 10 μM YTHDF1 to unmodified and m6A-modified RNA sequence by SPRi. All binding affinity values were determined according to three parallel experiments.
We also detected the signal of unmodified and m6A-modified RNA sequence to the m6A RNA reader YTHDF1 (23). We found that only m6A-modified RNA sequence had binding signal to 10 μM YTHDF1 (Fig. 2D). The binding affinity between YTHDF1 and m6A-modified RNA fitted by five gradients of protein concentration showed that the KD value was 3.6 ± 0.1 µM (SI Appendix, Fig. S6). This result demonstrated that the 3D-carbene SPRi technology had the ability to distinguish unmodified/m6A-modified RNA and discover readers of modified RNA.
In summary, modified peptide, antibody, double-stranded DNA, and single-stranded RNA can all be immobilized on the 3D-carbene chip without detectable activity loss. When coupled with SPRi technology, the 3D-carbene chip provides a convenient and versatile platform for profiling binding kinetics between these biomolecules in a high-throughput manner (see below).
Characterization of 3D-Carbene Chip.
After optimizations of the surface grating density, concentration of immobilized peptides, printing density, and regeneration conditions (SI Appendix, Figs. S7–S10), the 3D-carbene chip features low nonspecific binding, label-free detection, robustness, and stability. The 3D-carbene surface significantly reduced nonspecific binding compared with 2D-carbene surface (24) and bare gold surface. We immobilized H3(1–15)K4un and H3(1–15)K4me3 on both 3D-carbene chip, 2D-carbene chip, and bare gold chip. The binding signal to CHD1 showed that the background of 3D-carbene chip was much lower than that of 2D-carbene and bare gold chips (Fig. 3A and SI Appendix, Fig. S11). The high signal-to-noise ratio (61.3 of 3D-carbene chip versus 52.4 of 2D-carbene chip, −38.6 of bare gold chip) reduces false-positive results by reducing nonspecific binding to the surface (Fig. 3A and SI Appendix, Fig. S11 A–C). In addition, the Z′-factor of 3D-carbene chip (0.896) showed an overwhelming advantage compared with 2D-carbene (−0.479) and bare gold chips (−1.194), suggesting that the quality or performance of high-throughput analysis on 3D-carbene chip was ideal (SI Appendix, Fig. S11D).
Fig. 3.
The 3D-carbene chip features high signal-to-noise ratio (S/N), low nonspecific adsorption (NSA), label-free detection, and robustness. (A) S/N (Left) and NSA (Right) comparison among 3D-PCL, 2D-PCL, and bare gold surfaces by injecting 10 μM CHD1. (B) SPRi signals of 10 μM SGF29 binding to N-labeled, C-labeled, and unlabeled H3(1–15)K4me3 peptides. (C) SPRi signals of 10 μM CHD1 binding to H3(1–15)K4me3 peptides in 30 regeneration cycles. All error bars are obtained by calculating triplicates printed on the surface. CV, coefficient of variation.
Carbene immobilizes unlabeled peptides in random fashion, with the ensemble of peptide orientations best resembling that in solution. By contrast, the directional immobilization using labeled peptides risks epitope blocking without any precedent knowledge. For example, the histone reader protein SGF29 requires a free N terminus of histone H3 for binding, and thereby N-terminal biotin labeling of H3 peptide will disrupt this interaction (25) (SI Appendix, Fig. S12B). We immobilized unlabeled H3 and N-terminal and C-terminal biotin-labeled H3 peptides on the 3D-carbene chip. The binding signal showed that only unlabeled and C-terminally labeled H3 peptides were recognized by SGF29 (Fig. 3B and SI Appendix, Fig. S12A), highlighting the advantage of random immobilization.
To test the robustness and stability of 3D-carbene chip, we regenerated the chip immobilized with H3(1–15)K4me3 for 30 times. The binding signal of 10 µM CHD1 was consistent with no clear deterioration [coefficient of variation (CV) = 4.9%] during the 30 cycles of binding and regeneration (Fig. 3C and SI Appendix, Fig. S13), attesting to the robustness and stability of the 3D-carbene chip. In addition, parallel experiments using three different chips also showed consistent binding results (SI Appendix, Fig. S14).
Kinetic Profiling of Histone “Mark–Reader” Interactions.
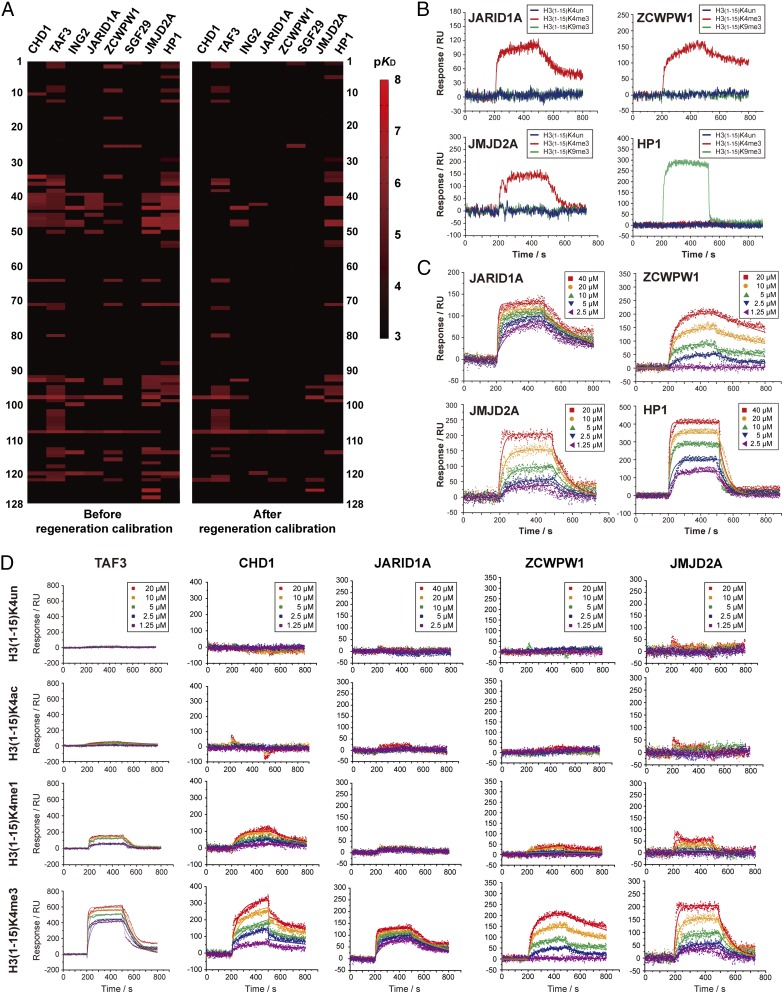
To demonstrate the potential of 3D-carbene–based SPRi technology in analyzing epigenetic interactions in high throughput, we immobilized 125 types of modified histone peptides and three types of negative controls in triplicates on the 3D-carbene chip (SI Appendix, Fig. S15 and Table S2) and tested eight histone reader proteins. This allowed us to analyze 1,000 pairs of interactions over a single chip. Each protein was injected using five gradients of concentration. The peptides are from different histones (histone H1, H2A, H2B, H3, and H4) of varied length (from 9 to 42 residues) and bear different modifications (methylation, acylation, and phosphorylation). The eight reader proteins include seven H3K4me3 readers (CHD1, TAF3, JMJD2A, SGF29, ZCWPW1, JARID1A, and ING2) and one H3K9me3 reader (HP1).
The heatmap for these 1,000 interaction pairs was generated based on the binding affinities (Fig. 4A, Left, and SI Appendix, Table S3). First, we checked the positive controls in this peptide array. The H3K4me3 readers binds to H3(1–15)K4me3 instead of H3(1–15)K4un peptides as expected (Fig. 4B). Second, the site specificity of H3K4me3 readers and H3K9me3 reader is also clear in the high-throughput result. All seven H3K4me3 readers recognize H3(1–15)K4me3 instead of H3(1–15)K9me3 peptides, while H3K9me3 reader HP1 engages H3(1–15)K9me3 instead of H3(1–15)K4me3 peptides (Fig. 4B). Third, the binding affinity determined by SPRi was in the same order of magnitude as that measured by ITC (Fig. 4C and SI Appendix, Table S4). These results demonstrate that the positive and negative controls can be clearly distinguished in this high-throughput experiment.
Fig. 4.
High-throughput kinetic analysis of 1,000 pairs of histone peptide–histone reader interaction. (A) Heatmaps of binding affinities between histone peptides and histone readers. The Left and Right panels are heatmaps before and after RC. The affinity values KD (in molar concentration) in various levels (10−3∼10−8 M) was transformed into −log10KD (3∼8) as pKD for comparison in the heatmap. The statistics of the heatmap is shown in SI Appendix, Table S3. (B) The SPRi curves of JARID1A, ZCWPW1, JMJD2A, and HP1. The binding curves to H3K4me3 are shown in red; the binding curves to H3K9me3 are shown in green; the binding curves to unmodified H3 are shown in blue. (C) The SPRi fitting curves of five gradients of protein concentration. All binding affinity values were determined according to three parallel experiments. (D) High-throughput kinetic SPRi curves of unmodified/modified H3(1–15)K4 peptides binding to TAF3, CHD1, JARID1A, ZCWPW1, and JMJD2A. All binding affinity values were determined according to three parallel experiments. The kinetic parameters of each binding pair are shown in SI Appendix, Table S6.
Another benefit of binding detection by SPRi is to monitor binding kinetics in real time. Unlike the fluorescent-chip–based detection, SPRi provides kinetic features to reflect binding specificity. We found that several long peptides (>25 residues), without methylation at H3K4 and H3K9, displayed nonspecific response signals likely due to over–cross-linking and surface fouling (SI Appendix, Table S5). To identify the false-positive hits, we defined the parameter of regeneration calibration (RC) value through analysis of SPRi kinetic curves. The RC value is defined as the 100 times the ratio of unregenerated signals (Δ1 shown in SI Appendix, Fig. S16A) to overall binding signals (Δ2 shown in SI Appendix, Fig. S16A). Empirically, the real positive result should have a RC value less than 10, which means that unregenerable signal contributes less than 10% to the overall signal. We chose H3(1–15)K4me3 and H3(1–34)K9crK27cr peptides as examples to analyze their respective signal response to CHD1. The result showed that the regenerable signal of H3(1–15)K4me3 peptide contributes to 99.8% of the overall signal while this value of H3(1–34)K9crK27cr peptide is only 53.3% (SI Appendix, Fig. S16A). This suggests that CHD1 binds specifically to H3(1–15)K4me3 but not H3(1–34)K9crK27cr, which is further confirmed by ITC studies (SI Appendix, Fig. S16B). The binding heatmap after RC is shown in Fig. 4A. In sum, the RC of SPRi kinetic curves efficiently helps to eliminate false-positive results.
The 3D-carbene SPRi platform makes high-throughput kinetic analysis of histone “mark–reader” pairs possible. We analyzed kinetic curves of four peptides [H3(1–15)K4un, H3(1–15)K4ac, H3(1–15)K4me1, and H3(1–15)K4me3] binding to five reader proteins (TAF3, CHD1, JARID1A, ZCWPW1, and JMJD2A) (Fig. 4D). For the same protein, we observed an increasing binding signal from H3(1–15)K4un to H3(1–15)K4me3. For TAF3, CHD1, ZCWPW1, and JMJD2A, H3(1–15)K4me3 showed 5.7-, 2.8-, 2.2-, and 2.5-fold stronger binding affinities to H3(1–15)K4me1 (SI Appendix, Table S6). This result demonstrated that carbene-based SPRi technology was suitable to compare binding affinities in high throughput. For the same peptide, such as H3(1–15)K4me3, we noticed that these five reader proteins exhibited different kinetic parameters. For example, the binding affinities of H3(1–15)K4me3 peptide were nearly the same to JARID1A (KD = 4.5 ± 1.0 μM) and JMJD2A (KD = 4.0 ± 1.1 μM). However, JARID1A showed a slower on and off pattern (ka = 196 M−1⋅s−1, kd = 8.83 × 10−4 s−1) while JMJD2A showed a faster on and off pattern (ka = 1,760 M−1⋅s−1, kd = 7.09 × 10−3 s−1) (SI Appendix, Table S6). This suggests that JMJD2A exhibits a more dynamic binding feature and might be more sensitive to the local protein concentration. Collectively, these results showed that SPRi could reflect different kinetic features even when the binding affinities are nearly the same.
Structural Verification of Histone–TAF3 Interactions.
Our high-throughput kinetic analysis showed that TAF3, a well-documented reader of H3K4me3, also recognized acetylated peptides (Fig. 5A). Specifically, TAF3 binds to H3K4ac peptide with an affinity of KD = 137 ± 13 µM measured by the SPRi experiment. To validate this weak interaction, we performed ITC, which revealed that KD between TAF3 and H3(1–15)K4ac peptide was 285 µM (Fig. 5B), consistent with our SPRi result. To further explore the molecular mechanism of this recognition, we solved the crystal structure of TAF3–H3K4ac complex (Fig. 5B and SI Appendix, Table S7) at 1.7 Å. The overall structure reveals that the H3K4ac peptide binds to the β3 surface (Fig. 5C). The H3K4ac peptide is induced to form an antiparallel β-sheet with the β3 strand of TAF3 (Fig. 5D, Upper). The H3R2 and H3Q5 residues form hydrogen bonds with the aspartic acid residues of TAF3 (Fig. 5D, Upper). The acetylated lysine residue inserts into an aromatic pocket formed by W867 and W890 (Fig. 5D, Upper). The amide group of H3K4ac stacks on the indole ring of W890 and is stabilized by amide–π interaction (Fig. 5D, Lower). H3K4ac has been reported to enrich at active gene promoters just upstream of H3K4me3 (26). Presumably, the observed “TAF3–H3K4ac” interaction might facilitate TAF3 searching for H3K4me3 at the transcription start site by priming the association of TAF3 with promoter chromatin. Collectively, the structural analysis establishes the molecular basis for H3K4ac recognition by TAF3 PHD finger. Weak interactions with high binding koff value are hard to detect by endpoint-based detection (e.g., fluorescence chip) because the analyte tends to be washed off. Our results suggest that the high-throughput detection by SPRi has the kinetic advantage to discover weak interactions in vitro.
Fig. 5.
Structural basis of TAF3–H3K4ac and TAF3–H3K4me3 interactions. (A) The SPRi fitting curves of five gradients of TAF3 concentrations to H3K4ac. The binding affinity was determined according to three parallel experiments. (B) The ITC curve of histone H3(1–15)K4ac peptide titrated into TAF3. (C) The overall crystal structure of TAF3–H3(1–15)K4ac complex. The TAF3 is shown in white; the histone H3 peptide is shown yellow. (D) The binding pocket of H3K4ac in TAF3. The key residues of TAF3 involved in H3K4ac peptide recognition are shown in the Upper. The amide–π stacking between Kac and W890 is shown in the Lower. (E) The ITC curve of histone H3(1–15)K4me3 peptide titrated into TAF3. (F) The overall crystal structure of TAF3–H3(1–15)K4me3 complex. The TAF3 is shown in white; the histone H3 peptide is shown in yellow. (G) The binding pocket and surface charge of H3K4me3 in TAF3. The N-terminal 15 residues of H3 are shown in the Lower. (H) The cation–π interaction between histone H3K4me3 and TAF3 W867/W890. (I) Histone H3A1 inserts into a shallow pocket in TAF3, highlighting the importance of label-free detection.
TAF3 is a core component of RNA polymerase II subunit TFIID, and its binding to H3K4me3 (a well-known transcription activation histone mark) facilitates the recruitment of RNA polymerase II to activated promoters (27). The SPRi analysis revealed that H3K4me3 bound to TAF3 with strong binding affinity (KD = 0.7 ± 0.1 µM; Fig. 4D and SI Appendix, Table S6). Our ITC analysis also confirmed a strong binding between TAF3 and H3(1–15)K4me3 with KD = 0.17 µM (Fig. 5E). Next, we solved the crystal structure of TAF3 in complex with H3(1–15)K4me3 at 1.3-Å resolution (Fig. 5F and SI Appendix, Table S7). The structure reveals that H3K4me3 peptide binds to the β3 surface of TAF3 PHD finger (Fig. 5F). Histone H3R2 inserts into a negative-charged pocket formed by TAF3 D885 and D888, while histone H3K4me3 inserts into an aromatic cage formed by TAF3 W867 and W890 (Fig. 5G). The two pockets are separated by the W890 residue, and this residue determines the site specificity of histone recognition. Histone H3R2 and H3K4 is separated by H3T3 while histone H3R8 and H3K9 has no linker residue (Fig. 5G, Lower). The W890-separated TAF3 pockets requires that a linker residue must exist between histone arginine and lysine residues. Our SPRi analysis also revealed that H3(1–15)K9me3 peptide did not bind to TAF3 (SI Appendix, Fig. S17). The recognition of K4me3 residues is achieved by cation–π interaction between trimethylated lysine residue and TAF3 W867/W890 (Fig. 5H). Notably, the free N terminus of H3A1 is registered at a shallow pocket in TAF3 (Fig. 5I); therefore, labeling on histone H3 N terminus will disrupt this interaction. Again, this highlights the advantage of label-free immobilization by 3D-carbene SPRi technology.
The Tudor Domain of crMSH6 Is an H3K4me3 Reader.
To demonstrate that 3D-carbene SPRi technology is suitable to discover epigenetic interactions, we studied a plant mismatch repair protein, MSH6 in Capsella rubella (crMSH6), which contains an N-terminal tudor domain of unclear histone binding activity (Fig. 6A). Interestingly, human MSH6 contains a PWWP domain instead of tudor at its N-terminal region, and it has been reported that this PWWP domain recognizes H3K36me3 to regulate DNA damage repair in human (28). We adopted the 3D-carbene SPRi technology to search for the target histone mark of crMSH6. Surprisingly, our SPRi experiment showed that crMSH6 binds to H3K4me3 with much stronger signal than that of H3K36me3 (Fig. 6B). SPRi curve fitting showed that crMSH6 binds to H3K4me3 at a KD value of 6.0 ± 0.2 μM (Fig. 6C), which is further verified by ITC experiments with a measured binding KD value of ∼4.4 μM (Fig. 6D). By contrast, the KD value between crMSH6 and H3K36me3 is >200 μM (Fig. 6D). This result suggests that, unlike human, plant may make use of H3K4me3 instead of H3K36me3 to facilitate DNA mismatch repair. The physiological importance of such an interaction remains to be explored in future functional studies.
Fig. 6.
The tudor domain of MSH6 in Capsella rubella is an H3K4me3 reader. (A) The domain architecture of MSH6 in Capsella rubella. The domain boundary is labeled as recorded in the server SMART (smart.embl-heidelberg.de/). (B) SPRi curves of 20 μM crMSH6 binding to H3 peptides modified at different sites. (C) The SPRi fitting curves of five gradients of crMSH6 concentration to H3(1–15)K4me3 peptide. The binding affinity was determined according to three parallel experiments. (D) The ITC curve of histone H3(1–15)un, H3K4(1–15)me3, H3.1(28–41)K36me3, and H3.3(28–41)K36me3 peptides titrated into crMSH6 protein.
Discussion
The 3D-carbene SPRi technology features label-independent and random yet function-retaining immobilization of ligands, label-free detection of biomolecules flowed-in, high signal-to-noise ratio, high-throughput and parallel comparison, real-time and kinetic detection, as well as robustness for reuses. It needs only 1–10 pmol of ligands for immobilization, much less demanding than other quantitative techniques (e.g., ITC typically requires 1–100 nmol of sample), and meanwhile is able to detect and compare hundreds to thousands binding events in one chip. Its high throughput and cost effectiveness render 3D-carbene SPRi attractive for studying biomolecular interactome in vitro and for discovering epigenetic interactions involving histones, DNA/RNA, or their modified forms. For interaction discovery, the 3D-carbene chip reduces false negatives due to the undermining effects of tagging and labeling ligands for immobilization, and is capable of identifying relatively weak interactions.
On the other side, the 3D-carbene SPRi technology also has its limitations. Although 3D-carbene chip offers superior random immobilization, it requires a “drying process” to minimize the carbene-blocking effect of water. In most cases, peptides, short oligos, and stable proteins like antibody could survive such a treatment. However, for proteins that are sensitive to water loss, the carbene-based immobilization strategy is usually not suitable. In such case, one has to adopt other water-compatible immobilization strategies for surface preparation. Due to the high density and high reactivity of the 3D-carbene surface, over–cross-linking, epitope blocking, and surface fouling could be other practical concerns. We found that long peptides (>25 residues) tend to give nonspecific response signals, alerting problems of over–cross-linking and surface fouling. As discussed previously, experimental parameter optimization during surface fabrication and SPRi screen as well as RC during data analysis could help to eliminate these false-positive results. Moreover, random immobilization will lead to inhomogeneous response of different anchor molecules, which may cause inaccuracy of curving fitting. However, since strongest binding event usually dominates the response signal, the 3D-carbene SPRi technology is still valid for binding detection and relatively accurate kinetic analysis. Currently, modular domain is most suitable for SPRi analysis. The application of full-length protein as analyte can still be challenging due to requirements of protein yield, purity, and stability, which awaits further improvements of the SPRi instrument to achieve higher sensitivity and lower analyte volume.
As evidenced by successful profiling of 1,000 pairs of histone “reader–mark” interactions, the 3D-carbene SPRi technology can be used to verify and quantify the myriad of interactions suggested by advanced proteomic approaches or bioinformatic analysis. Verified interactions can then be subjected to in-depth mechanistic and functional studies, such as cocrystal structural analysis. Besides the 3D-carbene SPRi platform, other fluorescence-based peptide or reader microarray technologies have also been successfully applied for profiling epigenetic targets (29, 30). Recently, DNA-barcoded designer nucleosome libraries (DNLs) have emerged as a unique technology that analyzes histone recognition and signaling, especially combinatorial histone modification readout at the nucleosome level (31). The kinetic and direct detection feature of the 3D-carbene SPRi platform complements well with other library-based methods for epigenetic discovery. For example, each single epigenetic interaction pair identified by 3D-carbene SPRi can be fed to the DNLs platform for complexity analysis at the nucleosome level.
With highly disordered regions, histone proteins epitomize a wide family of intrinsically disordered proteins (IDPs), which play pivotal roles in transcription regulation, signaling, and pathway control (32). Using histone peptides as exemplary IDPs, we demonstrated that 3D-carbene SPRi technology is suitable for systematic and kinetic characterization of interactions between modular domains and unstructured peptides in their native or modified states. Many important biological processes involve molecular recognition and physical binding among biomolecules. In the era of functional proteomics, we envision that the 3D-carbene SPRi technology will have wide appeal in profiling and discovering biorecognition events in and beyond the field of epigenetics.
Methods
Material.
Bis[2-(2-bromoisobutyryloxy)undecyl]disulfide (as initiator), copper(II) chloride (CuCl2), ascorbic acid (AA), 2,2′-bipyridyl (bpy), 2-hydroxyethyl methacrylate (HEMA) (Mr = 130), poly(ethylene glycol)monomethylether methacylate (PEGMA500) (Mr = 500), 4-(dimethylamino)pyridine (DMAP), succinic acid anhydride (SAA), EDC, NHS, and N,N′-diisopropylethylamine (DIPEA) were purchased from Sigma-Aldrich. HS-(CH2)11-EG3-OH (EG3-OH) and HS-(CH2)-EG3-COOH (EG3-COOH) were purchased from ProChimia Company. Ethanolamine (EA), dimethyl formamide (DMF), dichloromethane (DCM), dimethyl sulfoxide (DMSO), methanol (MeOH), ethanol (EtOH), and n-hexane were purchased from Sinopharm Chemical Reagent Beijing Company. [3-(Trifluoromethyl)-3H-diazirin-3-yl]benzoic acid was purchased from TCI. Rabbit IgG, goat anti-rabbit IgG, and BSA were purchased from Bioss.
The cDNA encoding the proteins ING2 (PHD finger domain; amino acids 161–173), HP1 (chromo domain; amino acids 39–74), and crMSH6 (tudor domain; amino acids 119–177) were cloned into the pGEX-6P1 vector (Novagen). The cDNA encoding the protein CHD1 (amino acids 270–443), SGF29 (amino acids 113–254), Foxp3 (amino acids 336–417), HNRNPA2B1 (full length), and JARID1A (amino acids 330–380) were cloned into the pRSFD vector (Novagen). TAF3 (amino acids 885–915), JMJD2A (amino acids 895–1,008), and ZCWPW1 (amino acids 246–307) were cloned into a modified pET28b vector (Novagen) with an N-terminal 10×His-SUMO tandem tag.
All histone peptides bearing different modifications were synthesized at SciLight Biotechnology. DNA and RNA oligos were synthesized at Genscript Biotechnology.
Protein Expression and Purification.
All proteins were expressed in the Escherichia coli BL21 (Novagen) and induced overnight by 0.2 mM isopropyl β-d-thiogalactoside at 16 °C in the LB medium. The collected cells were suspended in 100 mM NaCl and 20 mM Tris, pH 7.5. After cell lysis and centrifugation, the supernatant was applied to a HisTrap column (GE Healthcare) and GST column (GE Healthcare) according to the protein tag. After washing 5 column volumes with the suspension buffer, the GST-tagged proteins were eluted with the eluent buffer: 100 mM NaCl, 20 mM Tris, pH 7.5, 20 mM GSH. After washing 5 column volumes with the suspension buffer, the His-tagged and His-SUMO–tagged proteins were eluted with the eluent buffer: 100 mM NaCl, 20 mM Tris, pH 7.5, and 500 mM imidazole. GST-tagged proteins were digested overnight by the PreScission proteases. SUMO-tagged proteins were digested overnight by the ULP1. A HisTrap column was used to remove the cleaved 10×His-SUMO tag after removal of imidazole by desalting. All digested proteins were further purified by the HiTrap SP (GE Healthcare) cation-exchange column and a HiLoad 16/60 Superdex 75 (GE Healthcare) gel filtration column using AKTA Purifier 10 systems (GE Healthcare). All proteins were stored in 100 mM NaCl, 20 mM Tris, pH 7.5, at ∼10 mg/mL in an −80 °C freezer.
Synthesis of p(HEMA-co-PEGMA500).
In a typical experiment, polymerization solution was prepared by mixing the monomers HEMA (1.625 g) and PEGMA500 (6.25 g) and Bpy (32 mg) in 40 mL of 1:1 ddH2O/methanol (vol/vol). Then 2.5 mL of solution of CuCl2 (0.04 M) was added to reaction solution. After the solution was degassed by N2 for 30 min under stirring, 2.5 mL of AA (0.04 M) and 1 mL of EtOH solution of initiator (1 mM), and some more 1:1 ddH2O/methanol (vol/vol) was injected by syringe, forming a determined volume as 50 mL. The mixture was kept in a glove box with nitrogen atmosphere and the polymerization reacted for determined time (0.5, 1, 2, 4, 8, 16 h) at room temperature (25 °C). The polymerization was stopped when the bpy in solution was extracted by n-hexane, and methanol was removed by rotary evaporation. The aqueous solution was then purified by dialysis (MWCO, 8 kDa; Solarbio) for 48 h to remove the monomers and catalysts, followed with freeze-drying overnight. The lyophilized powder was redissolved into ethanol for chip surface modification. The polymer was characterized by 1H NMR (400 MHz, CDCl3, Avance 400; Bruker, SI Appendix, Fig. S1A) and GPC (1515; Waters, SI Appendix, Fig. S1B).
Fabrication of 3D-PCL and 2D-PCL Surfaces on Gold.
The procedure for preparing 3D-PCL polymer-coated gold chips is illustrated (Fig. 1B). Typically, a bare gold chip (Nanocapture; Plexera) was treated in a plasma cleaner (PDG-MG; Chengdu Mingheng Science and Technology Company), and then incubated in the synthesized polymer solution at room temperature for 1 h and then thoroughly rinsed with EtOH, ddH2O, and dried with N2 blowing before further treatment. The chip was then immersed in a DMF solution containing SAA (10 mg/mL) and DMAP (15 mg/mL) for 2 h to introduce carboxyl group onto the branches of the polymer brushes. After being thoroughly washed, the chip was further immersed in 1 mM EG3-OH (in EtOH) for 1 h to block the unreacted gold surface, and then cleaned by EtOH and ddH2O, and dried under nitrogen gas. Then, the carboxylated polymer on the chip surface was activated by EDC (0.4 M)/NHS (0.1 M) aqueous solution to form a semistable ester. After 15-min activation, the surface was immersed in the 10 mM amino-terminated photo–cross-linker (in DMF), which was synthesized by Kanoh’s method (14) for 4-h coupling. Finally, the chip was incubated in 1 M ethanolamine solution (in DMF) to block the unreacted carboxyl groups, washed by DMF, EtOH, and ddH2O, and dried by N2 gas stream. The chip was sealed and stored under −20 °C with argon atmosphere before further use.
To prepare a traditional 2D-PCL chip, a method reported was used for comparison in this research (24). Similar but not repeated, a gold-coated glass chip (Nanocapture; Plexera) was immersed in the ethanol solution containing 0.1 mM EG3-COOH and 0.9 mM EG3-OH for 16 h at room temperature to form a self-assembly monolayer. After being activated by EDC/NHS aqueous solution, the surface was coupled with photo–cross-linker for 4 h to obtain a photoreactive chip. Then the chip was rinsed successively with ethanol, water, and ethanol and dried under a nitrogen stream in UV-free room.
Surface Chemistry Characterization.
FT-IR spectra of modified surfaces was recorded on an FT-IR spectrometer using grazing angle specular reflectance accessory (Spectrum One; PerkinElmer) after each modification step, with 64 scans at a resolution of 4 cm−1 over a spectrum span of 4,000∼450 cm−1. An X-ray photoelectron spectrometer (ESCALAB 250Xi; Thermo Scientific) with a monochromatic AlK X-rays source (1,486.6 eV) was applied for characterizing the elemental composition of the surface, and ensuring the photo–cross-linker modification at a takeoff angle of 0° with a 0.78-mm2 spot size under a pressure of 3 × 10−9 mbar. An AFM (Bruker) was used to characterize the surface morphology and roughness before and after polymer coating in dry state and conducted under a tapping mode (silicon cantilever with nominal resonance frequency of 70 kHz and nominal spring constant 2 N/m).
Fabrication of Biomolecular Microarray on SPRi Chip.
Peptides (1 mM in ddH2O), DNAs/RNAs (100 μM in ddH2O), and antibodies (1 mg/mL in PBS buffer, pH 7.4) were prepared in 384 wells and spotted on 3D-PCL, 2D-PCL, and bare gold chips using an automated spotter (Qarray mini; Gentix) in a UV-free room with a humidity of 60%. The obtained PCL chips were dried in a vacuum chamber to evaporate the solvent, and the chips were then irradiated under a 365-nm UV cross-linker instrument (UVJLY-1; Beijing BINTA Instrument Technology Company). The irradiated energy on the surface amounted to 2.8 J/cm2. After UV treatment, the chips were rinsed in PBST (pH 7.4, 0.05% Tween 20), PBS, and ddH2O for 15 min each. For bare gold chip (as a reference), the chip was incubated in the fridge (4 °C) for 2 h right after printing. After carefully and thoroughly washed by PBST, PBS, and ddH2O, the chip was then incubated with 10 mg/mL BSA for another 2 h for blocking. Then the chip was repeatedly rinsed by PBST, PBS, and ddH2O to remove the physically adsorbed molecules. All of the chips after washing were dried under nitrogen gas for further use.
SPRi Analysis.
To measure the interaction between immobilized biomolecules and flowing proteins, an SPRi instrument (Kx5; Plexera) was used to monitor the whole procedure in real time. Briefly, a chip with well-prepared biomolecular microarray was assembled with a plastic flow cell for sample loading. The optical architecture and operation details of the PlexArray HT were described elsewhere previously (33). The protein samples were prepared at determined concentrations in TBS running buffer (20 mM Tris, 100 mM NaCl, pH 7.5) while a 10 mM glycine-HCl buffer (pH 2.0) was used as regeneration buffer. A typical binding curve was obtained by flowing protein sample at 2 μL/s for 300-s association and then flowing running buffer for 300-s dissociation, followed by 200-s regeneration buffer at 3 μL/s. To get a result of binding affinity, at least five gradient concentrations of flowing phase (proteins) were prepared and flowed in order. All of the binding signals were converted to standard refractive units (RU) by calibrating every spots with 1% glycerol (wt/vol) in running buffer with known refractive index change (1,200 RU). Binding data were collected and analyzed by a commercial SPRi analysis software (Plexera SPR Data Analysis Model; Plexera).
Isothermal Titration Calorimetry.
For ITC measurement, synthetic histone H3(1–15)K4me3 peptide and H3(1–34)K9crK27cr peptide (SciLight Biotechnology) and the protein CHD1 were extensively dialyzed against ITC buffer: 0.1 M NaCl and 20 mM Tris, pH 7.5. The titration was performed using a MicroCal iTC200 system (GE Healthcare) at 25 °C. Each ITC titration consisted of 17 successive injections with 0.4 μL for the first and 2.4 μL for the rest. H3 peptides at 1.0 mM were titrated into proteins at 0.1 mM. The resultant ITC curves were processed using Origin 7.0 software (OriginLab) according to the “One Set of Sites” fitting model. The titration condition of modified H3 peptides to TAF3 and crMSH6 was the same with CHD1.
X-Ray Crystallography.
Crystallization was performed by the sitting-drop vapor diffusion method under 18 °C by mixing equal volumes (1–2 μL) of protein and reservoir solution. For complex crystallization, TAF3 (2 mM) was first incubated with histone peptide (SciLight Biotechnology) at molar ratio of 1:1.5 for about 2 h. Crystals of TAF3–H3(1–15)K4ac and TAF3–H3(1–15)K4me3 complex were obtained under 0.03 M magnesium chloride, 0.03 M calcium chloride, 0.1 M Mes, 0.1 M imidazole, pH 6.5, 15% PEG MME 550, and 15% PEG 20K. Crystals were briefly soaked in a cryoprotectant composed of reservoir solution supplemented with 20% glycerol, and were flash frozen in liquid nitrogen for data collection at 100 K. Complex datasets were collected at beamline BL17U at the Shanghai Synchrotron Radiation Facility. All data were indexed, integrated, and merged using the HKL2000 software package (34). The complex structures were solved by molecular replacement using MOLREP (35). All structures were refined using PHENIX (36), with iterative manual model building using COOT (37). Model geometry was analyzed with PROCHECK (38). Detailed structural refinement statistics are shown in SI Appendix, Table S7. All structural figures were created using PyMOL (www.pymol.org/).
Supplementary Material
Acknowledgments
We thank the staff members at beamline BL17U of the Shanghai Synchrotron Radiation Facility and Dr. S. Fan at Tsinghua Center for Structural Biology for their assistance in data collection and the China National Center for Protein Sciences Beijing for providing facility support. We thank Dr. H. Xie in National Center for Nanoscience and Technology for her assistance in AFM experiment. This work was supported by Ministry of Science and Technology of China Grants 2016YFA0500700 and 2015CB910503, National Natural Science Foundation of China Grant 31470720, and the Tsinghua University Initiative Scientific Research Program (to H.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.L. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5WXG [TAF3PHD–H3(1–15)K4ac complex] and 5WXH [TAF3PHD–H3(1–15)K4me3 complex]).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704155114/-/DCSupplemental.
References
- 1.Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol. 2015;7:a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews FH, Strahl BD, Kutateladze TG. Insights into newly discovered marks and readers of epigenetic information. Nat Chem Biol. 2016;12:662–668. doi: 10.1038/nchembio.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Li H. Many keys to push: Diversifying the “readership” of plant homeodomain fingers. Acta Biochim Biophys Sin (Shanghai) 2012;44:28–39. doi: 10.1093/abbs/gmr117. [DOI] [PubMed] [Google Scholar]
- 6.Xiong X, et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat Chem Biol. 2016;12:1111–1118. doi: 10.1038/nchembio.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao D, Li Y, Xiong X, Chen Z, Li H. YEATS domain-A histone acylation reader in health and disease. J Mol Biol. 2017;429:1994–2002. doi: 10.1016/j.jmb.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X, et al. Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1. Genes Dev. 2014;28:622–636. doi: 10.1101/gad.233239.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 11.Morinière J, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 12.Scarano S, Scuffi C, Mascini M, Minunni M. Surface plasmon resonance imaging (SPRi)-based sensing: A new approach in signal sampling and management. Biosens Bioelectron. 2010;26:1380–1385. doi: 10.1016/j.bios.2010.07.056. [DOI] [PubMed] [Google Scholar]
- 13.Sendroiu IE, Gifford LK, Lupták A, Corn RM. Ultrasensitive DNA microarray biosensing via in situ RNA transcription-based amplification and nanoparticle-enhanced SPR imaging. J Am Chem Soc. 2011;133:4271–4273. doi: 10.1021/ja2005576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanoh N, et al. Immobilization of natural products on glass slides by using a photoaffinity reaction and the detection of protein-small-molecule interactions. Angew Chem Int Ed Engl. 2003;42:5584–5587. doi: 10.1002/anie.200352164. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, et al. Genetically encoded protein photocrosslinker with a transferable mass spectrometry-identifiable label. Nat Commun. 2016;7:12299. doi: 10.1038/ncomms12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang T, Li XM, Bao X, Fung YM, Li XD. Photo-lysine captures proteins that bind lysine post-translational modifications. Nat Chem Biol. 2016;12:70–72. doi: 10.1038/nchembio.1990. [DOI] [PubMed] [Google Scholar]
- 17.Krishnamoorthy M, Hakobyan S, Ramstedt M, Gautrot JE. Surface-initiated polymer brushes in the biomedical field: Applications in membrane science, biosensing, cell culture, regenerative medicine and antibacterial coatings. Chem Rev. 2014;114:10976–11026. doi: 10.1021/cr500252u. [DOI] [PubMed] [Google Scholar]
- 18.Huang CJ, Brault ND, Li Y, Yu Q, Jiang S. Controlled hierarchical architecture in surface-initiated zwitterionic polymer brushes with structurally regulated functionalities. Adv Mater. 2012;24:1834–1837. doi: 10.1002/adma.201104849. [DOI] [PubMed] [Google Scholar]
- 19.Jiang S, Cao Z. Ultralow-fouling, functionalizable, and hydrolyzable zwitterionic materials and their derivatives for biological applications. Adv Mater. 2010;22:920–932. doi: 10.1002/adma.200901407. [DOI] [PubMed] [Google Scholar]
- 20.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, et al. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res. 2015;43:1268–1282. doi: 10.1093/nar/gku1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alarcón CR, et al. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C, et al. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem. 2015;290:24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanoh N, et al. SPR imaging of photo-cross-linked small-molecule arrays on gold. Anal Chem. 2006;78:2226–2230. doi: 10.1021/ac051777j. [DOI] [PubMed] [Google Scholar]
- 25.Bian C, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30:2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guillemette B, et al. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011;7:e1001354. doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Li F, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bua DJ, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae N, et al. Developing Spindlin1 small-molecule inhibitors by using protein microarrays. Nat Chem Biol. 2017;13:750–756. doi: 10.1038/nchembio.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen UTT, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radivojac P, et al. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, et al. SPRi determination of inter-peptide interaction by using 3D supramolecular co-assembly polyrotaxane film. Biosens Bioelectron. 2015;66:338–344. doi: 10.1016/j.bios.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.