Significance
The growing size and sophistication of genome-wide association studies have led to the identification of variants which are clearly and reliably associated with obesity. A strong association between increased BMI and a region of human chromosome 2, near to the gene TMEM18, has been repeatedly demonstrated in children and adults. The function of TMEM18 in the control of appetitive behavior and body composition has been poorly characterized. In murine models, we show germline loss results in weight gain while adult onset hypothalamic overexpression results in weight loss, supporting the hypothesis that TMEM18 acting within the central nervous system can affect energy balance. We also report a structure and putative molecular function of TMEM18, challenging the current published model.
Keywords: TMEM18, GWAS, hypothalamus, obesity
Abstract
An intergenic region of human chromosome 2 (2p25.3) harbors genetic variants which are among those most strongly and reproducibly associated with obesity. The gene closest to these variants is TMEM18, although the molecular mechanisms mediating these effects remain entirely unknown. Tmem18 expression in the murine hypothalamic paraventricular nucleus (PVN) was altered by changes in nutritional state. Germline loss of Tmem18 in mice resulted in increased body weight, which was exacerbated by high fat diet and driven by increased food intake. Selective overexpression of Tmem18 in the PVN of wild-type mice reduced food intake and also increased energy expenditure. We provide evidence that TMEM18 has four, not three, transmembrane domains and that it physically interacts with key components of the nuclear pore complex. Our data support the hypothesis that TMEM18 itself, acting within the central nervous system, is a plausible mediator of the impact of adjacent genetic variation on human adiposity.
The understanding of human obesity has benefited greatly from advances in molecular genetics. In addition to the identification of many mechanistically illuminating, highly penetrant monogenic disorders, genome-wide association studies (GWAS) have identified multiple common genetic variants strongly associated with body mass index (BMI) (1–5). Many of these loci are within, or close to, genes which, to date, have not been recognized to encode proteins with a role in the control of energy homeostasis. Of note, these genes show a strong preponderance to being highly expressed in the central nervous system, a finding which is congruent with the fact that monogenic disorders leading to obesity largely exert their effects through a disruption of the central control of appetite and energy balance (6). Murine models have proven to be highly useful in bridging the gap between the identification of a variant as being associated with an adiposity phenotype and the understanding of how that variant actually influences energy balance. For example, the robust correlation between BMI and polymorphisms in the first intron of the human fat mass and obesity-associated (FTO) gene has been followed by loss and gain of function studies in genetically modified mice, supporting the notion that a number of neighboring genes including IRX3 (which is the gene most likely mediating the effect of the human SNP), RPGRIP1L, and FTO itself can all play a role in the control of energy balance and body composition (7–13). A strong association between increased BMI and a region of human chromosome 2, near to the gene TMEM18, has been repeatedly demonstrated in both adults and children (2, 14–18). Like the genes in the vicinity of FTO, TMEM18 had not been recognized as having a role in energy homeostasis before its identification by GWAS, and relatively little is known about its function, save that it is expressed in the brain and that it encodes a 140-aa protein reported to contain three transmembrane domain (19) which may act as a DNA-binding protein (20). We have therefore undertaken a range of studies to determine whether TMEM18 plays a role in the control of energy balance in mammals.
Results
Tmem18 Is Expressed within Hypothalamic Paraventricular Nucleus.
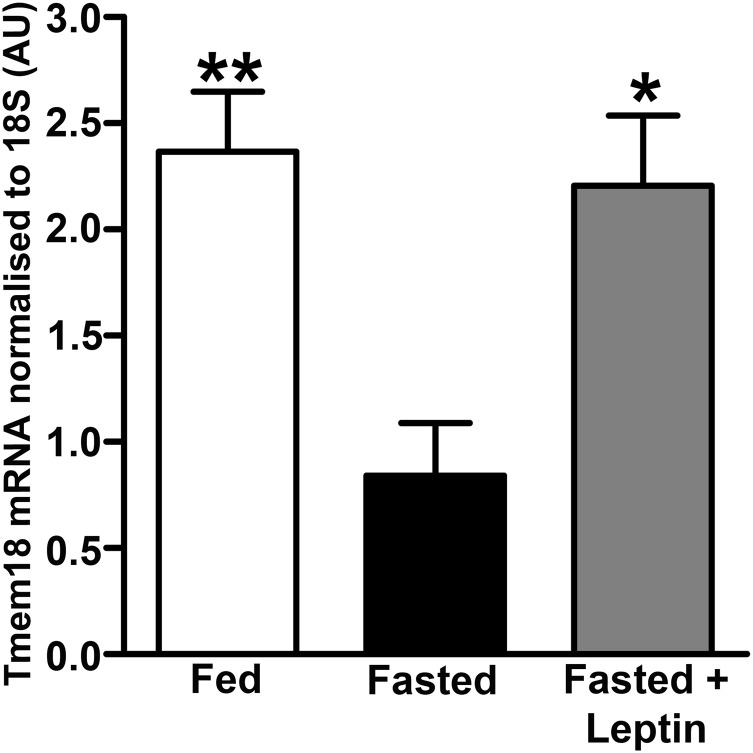
We initially examined the expression pattern of Tmem18 in a range of murine tissues and two mouse hypothalamic cell lines (N-46 and GT1-7). In keeping with previous reports (19), Tmem18 was widely expressed, with expression observed in several regions of the brain, in brown adipose tissue and in both hypothalamic cell lines (SI Appendix, Fig. S1). We undertook qPCR analysis on hypothalamic tissue acquired by laser capture microscopy (LCM) from three groups of wild-type mice (ad libitum fed, 48 h fasted and fasted for 48 h with leptin administered throughout the fast). Forty-eight-hour fasting decreased PVN Tmem18 expression by 70%, with leptin administration during the fast restoring expression back to fed levels (Fig. 1). To examine expression of Tmem18 in other hypothalamic nuclei and also to determine expression of the three genes vicinal to Tmem18 (Sh3yl1, Snth2, and Acp1), in a further set of wild-type animals (either ad libitum fed or fasted for 24 h), we undertook RNA-Seq analysis of LCM-acquired tissue from four hypothalamic nuclei (arcuate, ventral medial, paraventricular, and dorsal medial nuclei) (GEO accession no. GSE96627; SI Appendix, Fig. S2). Tmem18 expression was observed in all nuclei examined. (SI Appendix, Fig. S3). Of note, in this dataset of 24-h fasting vs. fed, none of the changes in expression of Tmem18, Sh3yl1, Snth2, or Acp1 reached statistical significance in any of the four nuclei studied (SI Appendix, Fig. S4).
Fig. 1.
Tmem18 expression within the hypothalamic paraventricular nucleus is nutritionally regulated. qRT-PCR analysis showing changes in Tmem18 gene expression in the PVN of wild-type mice that have been fed, fasted for 48 h, or fasted for 48 h with leptin administration. Data are expressed as mean ± SEM; *P < 0.05, **P < 0.01 vs. fasted mice.
Loss of Tmem18 Expression Results in an Increase in Body Mass.
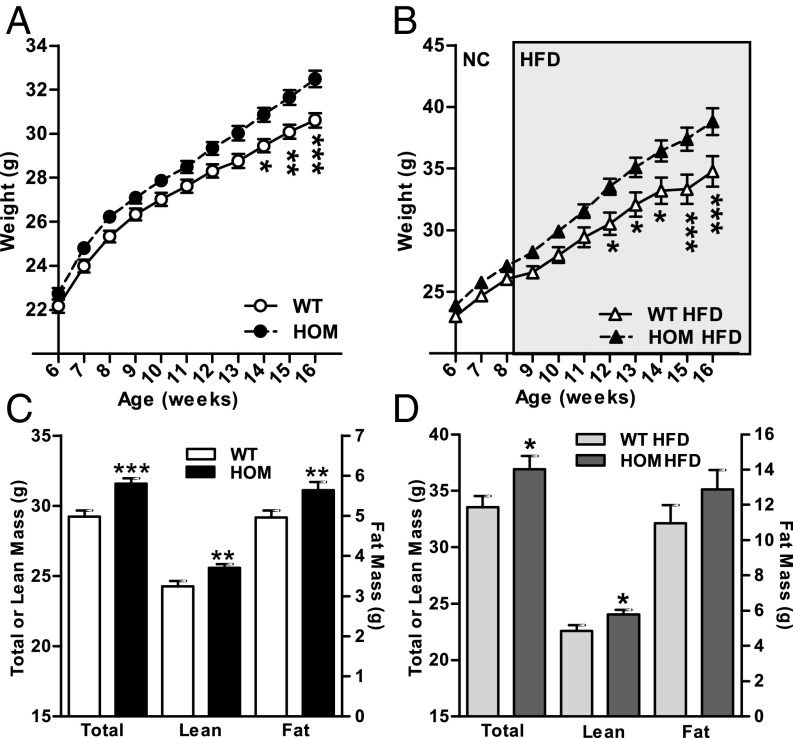
Knockout mice carrying mutant allele Tmem18tm1a(EUCOMM)Wtsi (abbreviated to “Tmem18tm1a”) were generated on a C57BL/6 genetic background as part of the European Conditional Mouse Mutagenesis Program (EUCOMM) (21). The introduction of the allele results in targeted disruption of exon 2 of Tmem18. qRT-PCR analysis revealed a very low level of residual Tmem18 transcript within the hypothalamus of homozygous mice (2.1% ±1.4), while heterozygous mice demonstrated a 50% decrease in transcript expression compared with Tmem18+/+ (49 ± 9 %) (SI Appendix, Fig. S5A). Tmem18 was the only transcript at that locus to be altered by introduction of the allele (SI Appendix, Fig. S5B), suggesting that there were no local “off target” effects. The mice were viable with expected homozygous mutant offspring born from heterozygous crosses. Female Tmem18tm1a homozygotes showed no differences in body weight or body composition compared with wild-type littermates (SI Appendix, Fig. S6 A, C, and E). Male Tmem18tm1a homozygotes, on normal chow, had significantly increased body weight by 14 wk of age (Fig. 2A) due to a significant increase in both fat and lean mass (Fig. 2C). At 16 wk, male homozygotes weighed on average 1.9 g more than wild-type littermates and had both increased gonadal white adipose tissue (gWAT) and brown adipose tissue (BAT) mass (SI Appendix, Fig. S7A).
Fig. 2.
Germline loss of Tmem18 results in increased body weight in male mice. Body weights of Tmem18 wt/wt (WT) and Tmem18 tm1a/tm1a (HOM) male mice on normal chow from weaning (WT, n = 23; HOM, n = 23) (A) or HFD (B) from 8 wk of age (WT, n = 15; HOM, n = 20). Body composition analyses showing total, lean, or fat mass of 14-wk old WT and HOM male mice on normal chow (WT, n = 16; HOM, n = 16) (C) or HFD from 8 wk of age (WT, n = 18; HOM, n = 20) (D). Data are expressed as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. WT mice.
Male Tmem18-Deficient Mice Fed an HFD Become More Obese Because of an Increase in Food Intake.
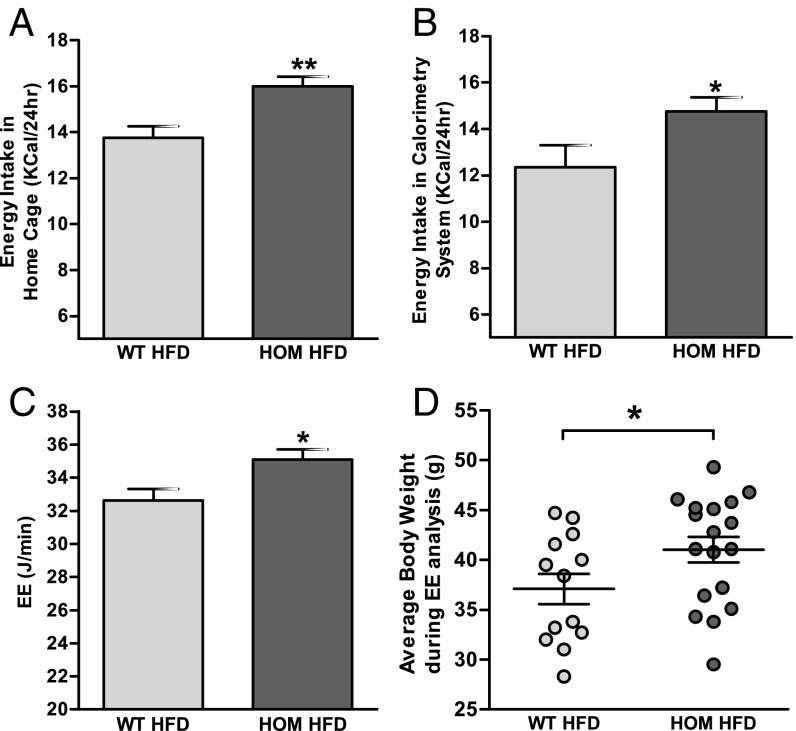
To determine the effects of a high fat diet on weight gain in Tmem18-deficient mice, WT and homozygous Tmem18tm1a mice were switched from normal chow to a 45% fat diet (HFD) at 8 wk of age. Female Tmem18tm1a homozygotes on a HFD showed no differences in body weight or body composition compared with wild-type littermates (SI Appendix, Fig. S6 B, D, and F). However, male Tmem18tm1a homozygotes on a HFD displayed significantly increased body weight by 12 wk of age (Fig. 2B) with an increase in both fat and lean mass (Fig. 2D). By 16 wk of age, male homozygotes on a HFD weighed ∼4.1 g heavier than wild-type littermates and had both increased gWAT and BAT (SI Appendix, Fig. S7B). Food intake in these mice was assessed on two occasions, first over a 10-d period at 15 wk of age while individually housed in home cages (Fig. 3A), and second over the 72-h period at 16 wk of age when mice were being analyzed for energy expenditure (Fig. 3B). On both occasions, Tmem18-deficient mice consumed significantly more energy than wild-type littermates. Interestingly, at the time of indirect calorimetry, Tmem18-deficient males on HFD had a significantly increased energy expenditure compared with wild-type littermates [analysis of covariance (ANCOVA) analysis with correction for body weight differences, *P = 0.015, Fig. 3 C and D]. Between genotypes, there was no difference in the expression of markers of thermogenesis in brown adipose tissue (SI Appendix, Fig. S8A) or in activity levels (SI Appendix, Fig. S8B). Thus, the increase in food intake (13.9%) in animals lacking Tmem18 was partially compensated for by a rise in energy expenditure (7.0%), resulting in only a modest increase in weight.
Fig. 3.
Effect of loss of expression of Tmem18 on energy homeostasis, activity levels, and food intake in male mice fed a HFD. Average 24-h energy intake of WT and homozygous male mice both measured in their home cage (A) and measured in the indirect calorimetry cage (B). (C) ANCOVA analysis of 24-h energy expenditure (EE) with body weight at time of analysis. (D) Average bodyweight of individual animals during 72-h analysis of energy expenditure. For all graphs, WT, n = 13; HOM, n = 18. Data are expressed as mean ± SEM; *P < 0.05, **P < 0.01 vs. WT mice.
Overexpression of Tmem18 Within the PVN Reduces Food Intake and Increases Energy Expenditure.
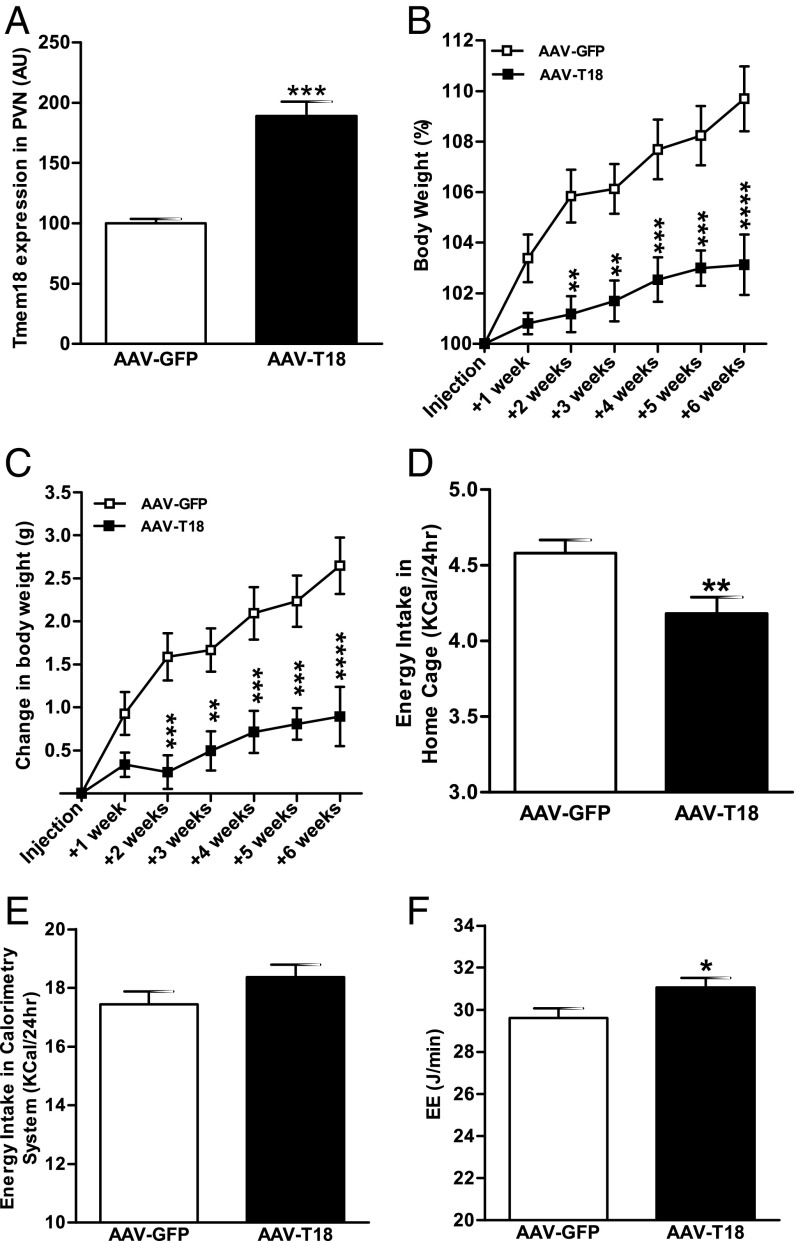
Having determined that Tmem18 expression in the PVN was altered by changes in nutritional state, we used adeno-associated viral vector (AAV-T18) to manipulate expression of Tmem18 within the PVN and study the subsequent effect on feeding behavior and body weight. Having confirmed targeting studies using AAV overexpressing GFP (SI Appendix, Fig. S9A), we then performed bilateral injections of an AAV-Tmem18-cDNA into the PVN of 12-wk-old C57/BL6 male mice. Compared with control mice, mice who received vectors expressing Tmem18 cDNA had a significant twofold increase in expression level (Fig. 4A). We found a significant difference in body weight gain at 2 wk after surgery and by 6 wk, mean weight gain in mice overexpressing Tmem18 was only 0.9 g compared with a 2.7 g in the control group (absolute weight gain, Fig. 4B; percentage weight gain, Fig. 4C). Analysis of food intake at 2 wk after surgery demonstrated AAV-T18–treated mice to have a significant reduction in food intake which was likely to have contributed to their reduction in weight gain (Fig. 4D). Interestingly, when reassessed at 6 wk, this relative hypophagia appeared to have resolved with no difference in energy intake between genotypes (Fig. 4E). However, at 6 wk after surgery, there was a significant difference in energy expenditure with AAV-T18–treated mice showing an increase compared with the control group (Fig. 4F; ANCOVA analysis with correction for body weight differences, *P = 0.03). Additionally, a separate group of mice (n = 6) overexpressing Tmem18 were placed on an HFD immediately after stereotactic surgery. Again, mice overexpressing Tmem18 gained significantly less weight (SI Appendix, Fig. S9D). There was a trend for a reduction in energy intake at 2 wk (SI Appendix, Fig. S9E), and at 6 wk, there was a significant difference in both total and fat mass between groups (SI Appendix, Fig. S9F).
Fig. 4.
Overexpression of Tmem18 within the PVN. (A) Change in Tmem18 gene expression within the PVN of mice 7 wk after bilateral injection of an adeno-associated vector (AAV-T18) (n = 10–11). Change in body weight of mice, measured weekly for 6 wk after bilateral PVN injections with either an AAV-T18 cDNA or AAV-GFP (n = 15 each group), expressed as grams (B) and as percentage of starting body weight (C). (D) Average 24-h energy intake of mice, measured in their home cage, 2 wk after surgery (n = 15 each group). (E) Average 24-h energy intake of mice, in the indirect calorimetry cage, 6 wk after surgery (n = 13 each group). (F) ANCOVA analysis of total EE, assessed over a 24-h period 6 wk after injection, with covariates evaluated at body weight = 29.41 g (n = 13 each group). Data are expressed as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001 vs. GFP-injected mice.
TMEM18 Contains Four, Not Three, Transmembrane Domains.
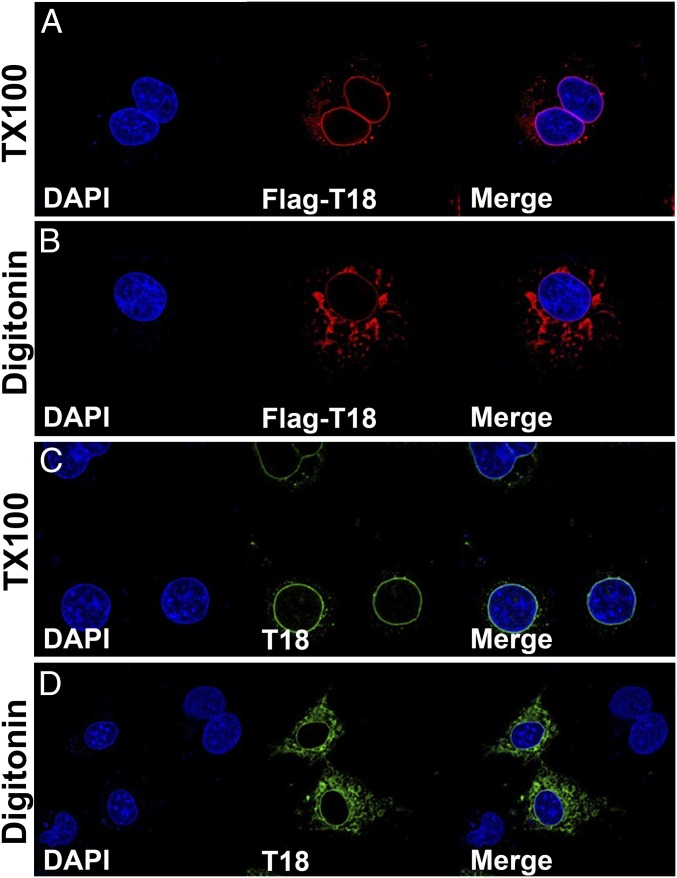
TMEM18 has been described to be a three-transmembrane protein that localizes to the nuclear membrane, courtesy of a nuclear localization signal, and reported to be involved in transcriptional repression (19, 20). To test this hypothesis, we first embarked on a deep phylogenetic analysis exploring sequence profile-to-profile homology (22) with MPI Bioinformatics Toolkit software (23). Database searches revealed remote but clear homology of TMEM18 to various ion channels. These included proteins from Pfam families (pfam.xfam.org) of fungal transient receptor potential ion channels (PF06011) and bacterial mechanosensitive ion channels (PF12794). Searches in metazoan proteomes yielded Unc-93 A homologs (mammalian gene UNC93A) annotated in Caenorhabditis elegans as an ion channel. The top hits (Toolkit parameter of probability of true positives approximately 80%) comprised a voltage-gated sodium channel from Caldalkalibacillus thermarum and an ion transport protein from Arcobacter butzleri (PDB ID codes 4BGN and 3RVY). Although their mutual phylogenetic distances are remote, they fold into a very similar structure and TMEM18 shares suggestive homology with their ion-transmitting domain. This region forms an oligomer consisting of two N- and C-terminal transmembrane helices pointing to the cytoplasm and two shorter inner helices fully embedded in the membrane. The conserved charged tip of their C-terminal helices is entirely outside the membrane and participates in the pore opening. It is highly probable that TMEM18 possesses a very similar topology and structure. Based on the homology to the C. thermarum channel, a tentative model can be proposed. SI Appendix, Fig. S10 shows the modeled membrane topology and a putative 3D structure under the assumption that TMEM18 forms a tetramer similar to the A. butzleri channel (24). As a nuclear membrane protein, it should expose both its termini to the cytoplasm and, therefore, would be comprised of four, rather than three, transmembrane domains. To test this experimentally, we transfected Cos cells with a N-terminal FLAG-tagged Tmem18 construct, then detected protein expression by using either a FLAG antibody or a TMEM18 antibody raised against C terminus amino acids 120–134. Protein expression was analyzed by using two different detergents, TX100 that permeabilizes all cellular membranes, and digitonin, that only permeabilizes the plasma membrane at a concentration of 40 μg/mL. Control antibodies (lamin B and calnexin) gave the expected results. Lamin B, which is localized to the inside of the nuclear membrane, could only be detected when TX100 was used to permeabilize both the plasma and nuclear membrane (SI Appendix, Fig. S11 A and B), whereas calnexin, which spans the nuclear membrane with the C terminus pointing into the cytoplasm, could be detected with both permeabilization reagents using a C-terminal antibody (SI Appendix, Fig. S11 C and D). Both the N terminus (FLAG antibody; Fig. 5 A and B) and C terminus (TMEM18 antibody; Fig. 5 C and D) of TMEM18 could be detected with digitonin permeabilization, indicating that both ends of the protein were located within the cytoplasm and confirming the bioinformatics analysis, which suggested that TMEM18 is a four-transmembrane protein.
Fig. 5.
Topology of the TMEM18 protein. Shown is the overexpression of N-terminal FLAG-tagged TMEM18 in COS cells treated with either TX-100 (permeabilizes both plasma and nuclear membrane) or digitonin (permeabilizes plasma membrane only). TMEM18 expression was detected with either a FLAG antibody (red, A and B) or an antibody to the C terminus of TMEM18 (green, C and D).
TMEM18 Is Unlikely To Directly Regulate Transcription.
It has been suggested that TMEM18 binds DNA and supresses transcription (20). To test this hypothesis, we performed RNA-Seq analysis of hypothalami from wild-type and Tmem18 knockout male mice (ENA accession no. PRJEB13884). Altogether, 27,691 (27,727 with nonzero total read count; 36 outliers per cooks Cutoff; ref. 25) annotated genes had reads mapped in at least one sample and were used for differential expression analysis (average number of uniquely mapped reads per sample: 32.4 × 10−6 ± 2.04 × 10−6 SD). Interestingly, after adjustment for multiple testing, only Tmem18 was significantly differentially expressed within the hypothalami of the two genotypes (padj = 3.02 × 10–22; SI Appendix, Fig. S12A). qRT-PCR analysis of Tmem18 and six additional genes with smallest (but not statistically significant) padj values confirmed the RNAseq data (SI Appendix, Fig. S12B), suggesting that TMEM18 is unlikely to be a global regulator of transcription as reported (20).
TMEM18 Interacts with Two Nuclear Pore Proteins, NDC1 and AAAS.
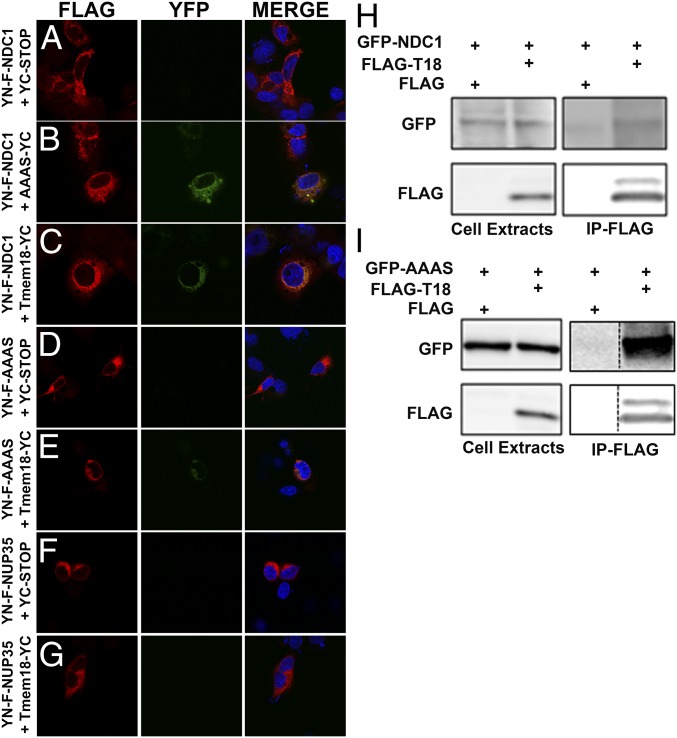
We used the methods of affinity purification and mass spectrometry (26) to identify TMEM18-interacting partners. Either FLAG-tagged TMEM18 or empty FLAG vector were overexpressed in HEK293 cells. Protein complexes were immunoprecipitated by using a FLAG antibody and mass spectrometry analysis performed to detect proteins bound to TMEM18. This process identified 116 proteins pulled down in control cells expressing the empty FLAG vector, 221 proteins common to both the control and experimental group, and 244 proteins unique to cells expressing FLAG-TMEM18 (see Dataset S1 for a list of all proteins identified as being pulled down by FLAG-TMEM18). Interestingly, three members of the nuclear pore complex, NDC1, AAAS, and NUP35/53 (27) showed high numbers of assigned spectra, indicating a high abundance of these proteins after pulldown. Therefore, biomolecular immunofluorescence complementation (BiFC) assays were used to confirm interactions between these proteins and TMEM18. Controls behaved as expected and showed that no YFP expression was seen if YN constructs were cotransfected with a YC-STOP plasmid (Fig. 6A). YN-FLAG-NDC1 and YC-AAAS served as an appropriate positive control (Fig. 6B) as these proteins have been reported to interact previously (28, 29). YFP expression could be detected in cells expressing YC-TMEM18 and either YN-FLAG-NDC1 or YN-FLAG-AAAS, but not YN-FLAG-NUP35 (Fig. 6 C–G). Positive BiFC protein–protein interactions were corroborated further by coimmunoprecipitation experiments using FLAG-TMEM18 and either GFP-tagged NDC1 (Fig. 6H) or GFP-tagged AAAS (Fig. 6I).
Fig. 6.
BiFC and co-IP confirmation of TMEM18 interaction with NDC1 and AAAS. (A–G) Physical interaction between TMEM18 and both NDC1 and AAAS was confirmed by BiFC. The N terminus of YFP was fused to FLAG-tagged NDC1 (YN-F-NDC1; C) or AAAS (YN-F-AAAS; E) or NUP35 (negative control, YN-F-NUP35; G), while the C terminus of YFP was fused to TMEM18 (TMEM18-Yc). FLAG expression was detected by using a FLAG antibody (red) while YFP signal is depicted in green. YN-F-NDC1 and AAAS-YC was used as a positive BiFC control (B). (H and I) Coimmunoprecipitation experiments using FLAG-tagged TMEM18 and GFP-tagged NDC1 (H) or AAAS (I) overexpressed in HEK cells. Dashed lines in the blot represents two different lanes in the same blot put together.
Discussion
Our data indicate that altering Tmem18 expression in mice, both globally and within the hypothalamus, can alter body weight. Male mice with a germline loss of Tmem18 have an increased body weight due to a significant increase in both fat and lean mass. This phenotype is more pronounced on an HFD, where weight gain is driven by hyperphagia. In contrast, overexpression of Tmem18 expression within the hypothalamic PVN can reduce food intake, increase energy expenditure, and reduce both total body and fat mass. The increased body weight phenotype of Tmem18 null mice was sexually dimorphic. Sex-specific differences in metabolic studies focusing on body weight are not uncommon and have been seen, for example, after embryonic TrkB inhibition or with disruption of GABA receptor signaling in proopiomelanocortin neurons (30, 31). In these reports and in our current study data, the mechanisms behind the sex-specific changes in body weight remain to be fully determined, but the differences in circulating gonadal derived hormones remain, of course, a potential contributor. Indeed, the relevance of estrogen in influencing the response to a dietary intervention was highlighted again in a study by Dakin et al. in which estradiol treatment to male mice fed a high-fat, high-sugar “obesogenic” diet prevented increases in adipose tissue mass (32).
Male mice on an HFD gained weight because of increased energy intake. Of note, data from indirect calorimetry also showed Tmem18 null mice to have a ∼10% increase in energy expenditure compared with wild-type mice, but as food intake was increased by ∼30%, the dominant drive was to caloric excess and weight gain. This could be considered an example of “diet induced thermogenesis,” a state seen as an effect of a change to a higher calorie diet in which mice increase energy expenditure and food intake simultaneously (33). The potential biological purpose and the tissues relevant to this phenomenon continue to be disputed (34). There is more agreement that the ambient temperature of a study can have a strongly qualitative effect on the outcome of metabolic studies (35). Our studies were conducted under “standard” animal house conditions that could be considered a chronic thermal stress to mice. Future studies of energy expenditure in Tmem18-deficient mice living at thermoneutrality may be helpful in further understanding the role of this molecule in metabolic control.
Several groups have previously shown Tmem18 to be highly expressed within the hypothalamus, although not all have reported it to be nutritionally regulated (19, 36, 37). Our data also indicate that Tmem18 is expressed within a number of hypothalamic regions. We chose to focus on the PVN, an anatomical region enriched with neuronal populations involved in appetitive behavior and energy expenditure (38) and one in which we saw nutritional regulation. The character of Tmem18-expressing neurons within the PVN remain to be determined. Further, the nature of the different perturbations (germline loss in Tmem18tm1a vs. delivery of AAV via stereotactic injection) is likely to mean that the PVN neuronal population which lost Tmem18 expression in the null mice may not be the same population that gained overexpression in the stereotactic-driven AAV study. We were unfortunately unsuccessful in our attempts to use siRNA and Cre to selectively knock down and/or delete Tmem18 in the PVN despite promising preliminary studies in vitro. In the future, transgenic lines with specific anatomical Cre drivers (e.g., Sim1-Cre, Agrp-Cre) may be helpful to further delineate the role of TMEM18 in both hypothalamic nuclei and specific neuronal populations.
We report interesting data on the potential function of TMEM18. Previous reports have indicated that TMEM18 is highly conserved, has no family members, localizes at the nuclear membrane, and that the C terminus of the protein may be involved in the regulation of transcription. While the localization of TMEM18 is not disputed, we propose that a role for TMEM18 in transcriptional regulation is unlikely given our RNA-Seq data showing that the hypothalamic transcriptome of Tmem18tm1a mice is not significantly different to that of wild-type littermates. Previous bioinformatic analysis had predicted a three-transmembrane structure (19, 39). However, our cellular experiments reveal that TMEM18 is in fact comprised of four transmembrane segments, indicating that it is unlikely that the C terminus is involved in binding DNA as earlier reported (20). Instead, we hypothesize that TMEM18 may be involved in the transport of molecules across the nuclear envelope. Pull-down experiments, corroborated by BiFC and IP studies, have identified two binding partners for TMEM18–NDC1 and AAAS/ALADIN. These proteins are 2 of the 30 NUPs that make up the nuclear pore complex (NPC) (40). As NDC1 and AAAS have been reported to interact with one another (29), it is possible that TMEM18 is not interacting directly with both proteins. By enabling transport of molecules across the nuclear envelope, NUPs are involved in many fundamental cellular processes, consequently abnormal expression/function of these NUPs has been linked to various human diseases such as cancer, cardiovascular disorders, autoimmune disease, and neurological defects (41). Further investigation into the interaction of TMEM18 with members of the NPC will hopefully shed new light on the molecular function of TMEM18 and its role in obesity.
The association of single nucleotide polymorphisms (SNPs) near human TMEM18 with obesity was first reported by Willer et al. (2) and has since been identified by multiple GWAS (reviewed in ref. 6). Although these loci have taken on the moniker of “near to TMEM18,” to date it has remained unclear whether these variants have any influence on the regulation of TMEM18 expression or function, or indeed if TMEM18 has a direct role in energy homeostasis. Efforts to advance understanding from association to biologically relevant mechanisms have combined multiple techniques and evidence from several model platforms. For example, when investigating obesity-associated noncoding sequences within FTO, Smemo used chromatin conformation capture techniques (CCST) to show a region of FTO directly interacts with, and forms part of the regulatory landscape of, the homeobox gene IRX3 (10). They also reported that relevant SNPs were associated with expression of IRX3, but not FTO, in human brains. In a more recent study of obesity-associated FTO variants, Claussnitzer combined chromatin capture technology, expression quantitative trait loci (eQTL) data, and a powerful bioinformatics approach to investigate a putative role for FTO adipocyte biology (42). Thus far, our attempts to interrogate the region near TMEM18 through the examination of various gene expression databases shows no eQTL data relevant to these SNPs. Further, our data are insufficient to confirm or refute a role for other genes vicinal to TMEM18 as candidates of the GWAS association. A brain tissue-specific approach may prove more fruitful but unfortunately RNA expression data specifically from human hypothalamus linked to genotype is not a currently publicly available resource. The studies we have described have strengthened the candidacy of TMEM18 as, at least in part, the mediator of the association between genetic variation in this region of chromosome 2 and human adiposity, one of the strongest associations of a common variant with human obesity. Our work has also provided information about the topology and binding partners of this enigmatic protein associated with the nuclear pore complex.
Methods
Detailed study methods are provided in SI Appendix, SI Methods.
Animals.
All procedures were carried out in accordance with guidelines of the United Kingdom Home Office. Animals were kept under controlled temperature (22 °C) and a 12-h light, 12-h dark schedule (lights on 700–1900).
AAV Vectors.
The cDNA of murine Tmem18 was cloned into an AAV backbone plasmid under the control of the CMV promoter. AAV vectors were generated by helper virus-free transfection of HEK293 cells.
Stereotactic Surgery.
Mice were stereotaxically injected with AAV while under isoflurane induced anesthesia (coordinates: 1.0 mm caudal to bregma, ±0.25 mm lateral to the midline, 5.0 mm below the surface of the skull).
Metabolic Phenotyping.
Body composition was determined by using Lunar PIXImus2 mouse densitometer (General Electric Medical System). Energy expenditure was determined by using indirect calorimetry in a custom-built monitoring system (Ideas Studio).
Laser Capture Microdissection.
Coronal sections of 20-μm thickness were prepared on a cryostat, mounted on RNase-free membrane-coated glass slides with laser microdissection performed by using a P.A.L.M. MicrolaserSystem (P.A.L.M. Microlaser Technologies).
Pulldown and Mass Spec Analysis.
For protein overexpression studies, transient transfection in HEK293 cells was performed by using a CalPhos kit according to the manufacturer’s protocol. Forty-eight hours after transfection, cells were harvested and Flag immunoprecipitations (IPs) were performed from the resulting lysates.
Statistical Analysis.
All values are expressed as mean ± SEM. Statistical analysis was performed by using Graph Pad Prism software (GraphPad Prism) or SPSS (IBM). For the analysis of food intake and body weight over time, two-way repeated measures ANOVA, with a Bonferroni post hoc test, was used with time and treatment as variables for comparison. For energy expenditure (EE) ANCOVA was performed to assess body weight/EE interactions with body weight as a covariate, genotype, or AAV treatment as a fixed factor and EE as a dependent variable. Multiple linear regression analysis was carried out with no selection criteria. Differential expression in LCM tissue analysis was calculated by using edgeR, with correction for multiple comparisons using the Benjamini–Hochberg procedure. Significant differences were designated as P < 0.05.
Supplementary Material
Acknowledgments
We thank Helen Westby, Will Gee, and Elizabeth Wynn for technical assistance and Satish Patel and Koini Lim for assistance with the BiFC protocol. R.L., Y.-C.L.T., D.R., G.S.H.Y., S.O.R., and A.P.C. are funded by Medical Research Council (MRC) Metabolic Disease Unit Grant MRC_MC_UU_12012/1, and animal work was carried out with the assistance of MRC Disease Model Core of the Wellcome Trust MRC Institute of Metabolic Sciences Grant MRC_MC_UU_12012/5 and Wellcome Trust Strategic Award 100574/Z/12/Z. F.B. is the recipient of an award from the Institució Catalana de Recerca i Estudis Avançats (ICREA) Academia, Generalitat de Catalunya, Spain. Vector generation and production were funded by Ministerio de Economía y Competitividad, Spain, Grant SAF 2014-54866-R. C.D. and D.W.L. were supported by the Wellcome Trust Grant WT098051, and C.D. was supported by Wellcome Trust PhD Programme for Clinicians Grant 100679/Z/12/Z.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE96627), and the European Nucleotide Archive, www.ebi.ac.uk/ena (accession no. PRJEB13884).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707310114/-/DCSupplemental.
References
- 1.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willer CJ, et al. Wellcome Trust Case Control Consortium Genetic Investigation of ANthropometric Traits Consortium Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speliotes EK, et al. MAGIC Procardis Consortium Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorleifsson G, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 5.Meyre D, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 6.Loos RJ. Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab. 2012;26:211–226. doi: 10.1016/j.beem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Church C, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer J, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 9.McMurray F, et al. Adult onset global loss of the fto gene alters body composition and metabolism in the mouse. PLoS Genet. 2013;9:e1003166. doi: 10.1371/journal.pgen.1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stratigopoulos G, et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J Clin Invest. 2016;126:1897–1910. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tung YC, et al. Hypothalamic-specific manipulation of Fto, the ortholog of the human obesity gene FTO, affects food intake in rats. PLoS One. 2010;5:e8771. doi: 10.1371/journal.pone.0008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stratigopoulos G, et al. Hypomorphism for RPGRIP1L, a ciliary gene vicinal to the FTO locus, causes increased adiposity in mice. Cell Metab. 2014;19:767–779. doi: 10.1016/j.cmet.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felix JF, et al. Bone Mineral Density in Childhood Study (BMDCS) Early Genetics and Lifecourse Epidemiology (EAGLE) consortium Early Growth Genetics (EGG) Consortium Bone Mineral Density in Childhood Study BMDCS Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet. 2016;25:389–403. doi: 10.1093/hmg/ddv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei YF, et al. Meta-analysis of genome-wide association data identifies novel susceptibility loci for obesity. Hum Mol Genet. 2014;23:820–830. doi: 10.1093/hmg/ddt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherag A, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paternoster L, et al. Genome-wide population-based association study of extremely overweight young adults–the GOYA study. PLoS One. 2011;6:e24303. doi: 10.1371/journal.pone.0024303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, et al. Role of BMI-associated loci identified in GWAS meta-analyses in the context of common childhood obesity in European Americans. Obesity (Silver Spring) 2011;19:2436–2439. doi: 10.1038/oby.2011.237. [DOI] [PubMed] [Google Scholar]
- 19.Almén MS, et al. The obesity gene, TMEM18, is of ancient origin, found in majority of neuronal cells in all major brain regions and associated with obesity in severely obese children. BMC Med Genet. 2010;11:58. doi: 10.1186/1471-2350-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurvansuu JM, Goldman A. Obesity risk gene TMEM18 encodes a sequence-specific DNA-binding protein. PLoS One. 2011;6:e25317. doi: 10.1371/journal.pone.0025317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 23.Alva V, Nam SZ, Söding J, Lupas AN. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016;44:W410–W415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai CJ, et al. Two alternative conformations of a voltage-gated sodium channel. J Mol Biol. 2013;425:4074–4088. doi: 10.1016/j.jmb.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 27.Kabachinski G, Schwartz TU. The nuclear pore complex–structure and function at a glance. J Cell Sci. 2015;128:423–429. doi: 10.1242/jcs.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kind B, Koehler K, Lorenz M, Huebner A. The nuclear pore complex protein ALADIN is anchored via NDC1 but not via POM121 and GP210 in the nuclear envelope. Biochem Biophys Res Commun. 2009;390:205–210. doi: 10.1016/j.bbrc.2009.09.080. [DOI] [PubMed] [Google Scholar]
- 29.Yamazumi Y, et al. The transmembrane nucleoporin NDC1 is required for targeting of ALADIN to nuclear pore complexes. Biochem Biophys Res Commun. 2009;389:100–104. doi: 10.1016/j.bbrc.2009.08.096. [DOI] [PubMed] [Google Scholar]
- 30.Byerly MS, Swanson RD, Wong GW, Blackshaw S. Stage-specific inhibition of TrkB activity leads to long-lasting and sexually dimorphic effects on body weight and hypothalamic gene expression. PLoS One. 2013;8:e80781. doi: 10.1371/journal.pone.0080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito Y, et al. GABA type B receptor signaling in proopiomelanocortin neurons protects against obesity, insulin resistance, and hypothalamic inflammation in male mice on a high-fat diet. J Neurosci. 2013;33:17166–17173. doi: 10.1523/JNEUROSCI.0897-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dakin RS, Walker BR, Seckl JR, Hadoke PW, Drake AJ. Estrogens protect male mice from obesity complications and influence glucocorticoid metabolism. Int J Obes. 2015;39:1539–1547. doi: 10.1038/ijo.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281:31–35. doi: 10.1038/281031a0. [DOI] [PubMed] [Google Scholar]
- 34.Kozak LP. Brown fat and the myth of diet-induced thermogenesis. Cell Metab. 2010;11:263–267. doi: 10.1016/j.cmet.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 36.Yoganathan P, Karunakaran S, Ho MM, Clee SM. Nutritional regulation of genome-wide association obesity genes in a tissue-dependent manner. Nutr Metab (Lond) 2012;9:65. doi: 10.1186/1743-7075-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid PM, et al. Expression of fourteen novel obesity-related genes in Zucker diabetic fatty rats. Cardiovasc Diabetol. 2012;11:48. doi: 10.1186/1475-2840-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowley MA, et al. Integration of NPY, AGRP, and melanocortin signals in the hypothalamic paraventricular nucleus: Evidence of a cellular basis for the adipostat. Neuron. 1999;24:155–163. doi: 10.1016/s0896-6273(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 39.Jurvansuu J, et al. Transmembrane protein 18 enhances the tropism of neural stem cells for glioma cells. Cancer Res. 2008;68:4614–4622. doi: 10.1158/0008-5472.CAN-07-5291. [DOI] [PubMed] [Google Scholar]
- 40.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 41.Nofrini V, Di Giacomo D, Mecucci C. Nucleoporin genes in human diseases. Eur J Hum Genet. 2016;24:1388–1395. doi: 10.1038/ejhg.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.