Significance
Recently, a number of approaches have been developed to repurpose the CRISPR/Cas9 system as a sequence-specific transcriptional activator for gain-of-function experiments (CRISPR activators, or “CRISPRa”). While multiple CRISPRa strategies have been characterized in cell culture, little is known about their performance in vivo. We present an optimized strategy for generating a large-scale CRISPRa resource in Drosophila and show that this system has a high success rate and generates easily recognizable phenotypes in vivo. We describe a growing collection of transgenic fly lines to facilitate large-scale in vivo CRISPRa experiments.
Keywords: CRISPR/Cas9, Cas9-activators, gain-of-function, CRISPR mutagenesis, CRISPRa
Abstract
While several large-scale resources are available for in vivo loss-of-function studies in Drosophila, an analogous resource for overexpressing genes from their endogenous loci does not exist. We describe a strategy for generating such a resource using Cas9 transcriptional activators (CRISPRa). First, we compare a panel of CRISPRa approaches and demonstrate that, for in vivo studies, dCas9-VPR is the most optimal activator. Next, we demonstrate that this approach is scalable and has a high success rate, as >75% of the lines tested activate their target gene. We show that CRISPRa leads to physiologically relevant levels of target gene expression capable of generating strong gain-of-function (GOF) phenotypes in multiple tissues and thus serves as a useful platform for genetic screening. Based on the success of this CRISRPa approach, we are generating a genome-wide collection of flies expressing single-guide RNAs (sgRNAs) for CRISPRa. We also present a collection of more than 30 Gal4 > UAS:dCas9-VPR lines to aid in using these sgRNA lines for GOF studies in vivo.
Currently, it is possible to knock down nearly any gene in the Drosophila genome in any cell type with temporal control using several genome-wide UAS:RNAi resources (reviewed in ref. 1). A similar collection for gain-of-function (GOF) studies would be valuable, as GOF studies have led to many discoveries and serve as a powerful complement to loss-of-function (LOF) studies (reviewed in refs. 2 and 3). Several GOF collections exist, based either on expression of cDNAs using the Gal4-UAS system (4) or on randomly generated insertions of UAS sites in the genome (5, 6). However, there are drawbacks to both approaches. cDNA-based approaches are technically difficult to scale genome-wide and require a priori decisions about which isoform to express. In addition, for random UAS-insertion collections, the affected gene is not always easy to identify. Furthermore, when UAS is inserted into a transcription unit in the antisense orientation, antisense transcription can trigger RNAi, an issue estimated to affect up to one-third of existing lines (reviewed in ref. 3). Last, an issue that affects all Gal4-UAS–based GOF approaches is that this system typically induces extremely high levels of overexpression, which can affect the interpretation of such experiments.
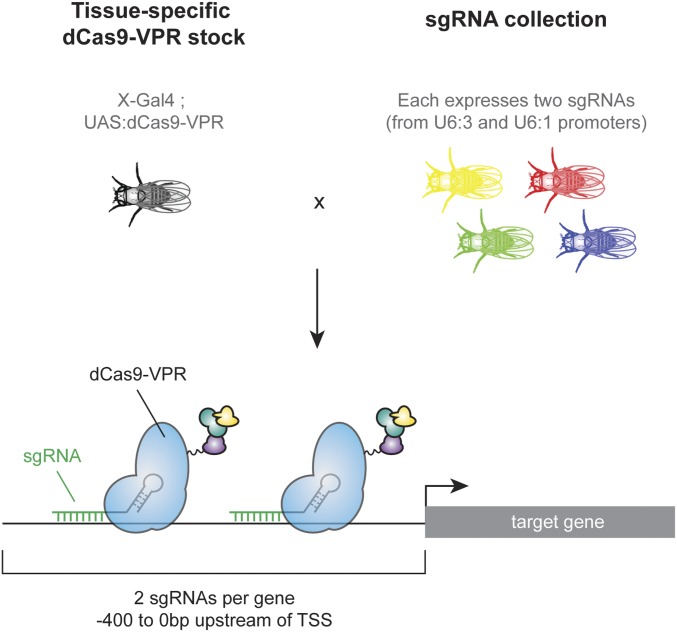
Cas9 activators, in which a catalytically dead Cas9 (“dCas9”) recruits transcriptional activation machinery to a DNA sequence upstream of a target gene’s transcriptional start site (TSS), can potentially overcome these obstacles. Cas9 activators appear to activate endogenous genes at near-physiological levels (7). In addition, the target specificity is conferred by 20-bp protospacer sequences within the single-guide RNA (sgRNA), such that production of reagents for CRISPR activators (CRISPRa) at genome-wide scale is feasible. For Drosophila, such a resource would consist of a collection of transgenic fly lines, each ubiquitously expressing sgRNAs targeting a region upstream of a given target gene, which could be crossed to flies expressing a Cas9 activator ubiquitously or in a tissue-specific manner (Fig. 1). However, the success rate of such a strategy in vivo is unknown, as only a single proof-of-principle experiment has been published using the dCas9-VPR system (8). In addition, a direct comparison of the seven currently available Cas9 activators recently showed that an alternative activator, Synergistic Activation Mediator (SAM) (9), outperforms dCas9-VPR in many cases in cell culture (10), prompting us to perform a similar comparison in vivo before generating a large resource. Here, we present an optimized strategy for generating a genome-wide collection of sgRNA lines for in vivo CRISPRa. We show that this strategy has a success rate of at least 75% and that easily recognizable phenotypes can be obtained in multiple tissues, thus serving as a useful strategy for GOF screening.
Fig. 1.
Schematic of a large-scale CRISPRa resource. Flies expressing tissue-specific Gal4 driving dCas9-VPR are crossed to a collection of flies constitutively expressing sgRNAs targeting upstream of the TSS of target genes. Tissue-specific activation of the target gene occurs in the offspring of the cross.
Results and Discussion
Comparative Analysis of CRISPRa Strategies in Vivo.
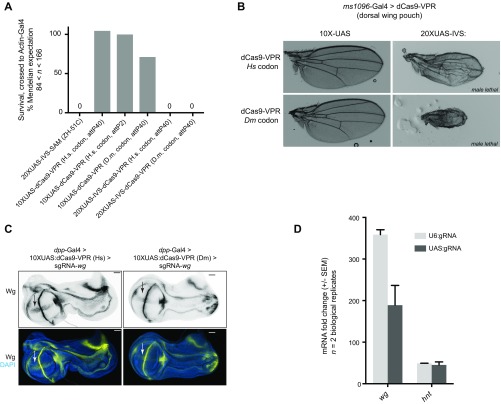
To establish a strategy for building a genome-wide GOF resource, we first optimized the protein component of the Cas9 activator for in vivo studies. The two most promising strategies for CRISPRa, dCas9-VPR and SAM, recruit transcriptional activation machinery using different mechanisms. In dCas9-VPR, dCas9 is directly fused to an optimized tripartite activator domain (VP64-p65-Rta, VPR) (7). In SAM, there are two protein components: a VP64 domain is fused to dCas9, and two additional activator domains, p65 and HSF, are recruited to the complex via MS2 stem loops in the sgRNA tail (9). Because it has been previously shown that SAM can outperform dCas9-VPR in Drosophila cells (10), we wished to compare the two methods in vivo. We created transgenic flies expressing the SAM component (MCP–p65–HSF) under UAS control. However, this UAS:SAM construct was 100% lethal when expressed ubiquitously (using actin-Gal4), in the absence of any sgRNA (Fig. S1A).
Fig. S1.
Optimization of expression level and codon use of dCas9-VPR and the sgRNA promoter. (A) Ubiquitous expression of 10XUAS-SAM, and three different 10XUAS:dCas9-VPR constructs using actin-Gal4 is not lethal, whereas expression of 20XUAS-IVS:dCas9-VPR is 100% lethal regardless of codon use. Dm, Drosophila melanogaster codon use; Hs, human codon use. (B) Expression of 20XUAS-IVS:dCas9-VPR constructs in the wing (using the ms1096-Gal4 line) was highly toxic to wing tissue in the female and caused male lethality. (C) Human codon-optimized 10UAS:dCas9-VPR led to higher levels of activation of the target gene wg than did Drosophila codon use, consistent with previous reports (8). (D) Expression of sgRNAs from UAS promoters (in the pCFD6 vector) did not lead to a higher expression than U6-driven sgRNAs (in the pCFD4 vector) in Drosophila S2R+ cells.
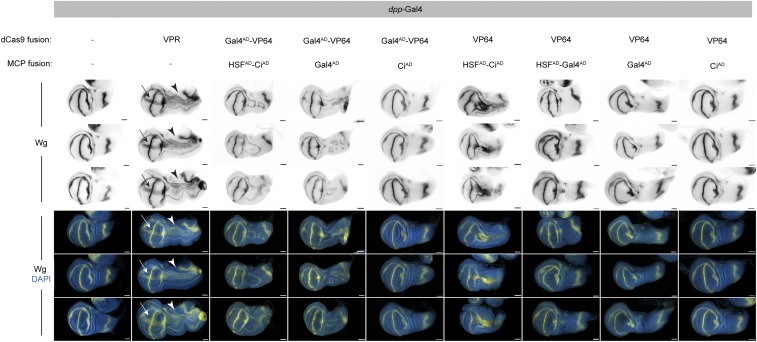
We next attempted to use the effect of recruiting additional domains via MS2 stem loops while overcoming the lethality of the SAM construct. To do so, we generated a series of seven “SAM-like” fly lines expressing a variety of activator domains known to function in flies (Gal4AD, HSFAD, VP64, and CiAD) fused to dCas9 and MCP in different configurations, all under UAS control, and we also created a modified sgRNA expression plasmid containing MS2 hairpins. To directly compare this panel of Cas9 activators in vivo, we used the dpp-Gal4 driver to express each SAM-like construct in an ectopic domain in the larval wing disc and crossed each of these lines to flies expressing validated sgRNAs targeting upstream of wingless (wg) (8). The level of CRISPRa can be assayed by immunostaining for Wg protein as well as by observing the extent of wing pouch duplication, a readout of Wg activity in this context (11). We found that dCas9–VPR unambiguously outperformed all seven SAM-like approaches (Fig. 2). Specifically, ectopic Wg expression levels in the wing pouch were far greater with dCas9–VPR than with any competing strategy (arrows in Fig. 2); accordingly, the extent of axis duplication was more extensive in dCas9–VPR than in any other strategy (arrowheads in Fig. 2). Thus, we decided to pursue dCas9–VPR for building a genome-wide CRISPRa resource.
Fig. 2.
dCas9-VPR outperforms SAM-like strategies. Each panel shows antibody detection of Wg following activation in the dpp-Gal4 domain (Fig. 3A) along the anterior–posterior boundary. Three representative discs are shown for each genotype. Successful CRISPRa results in detectable Wg signal (arrows) as well as altered patterning along the length of the disc (arrowheads). Identical protospacers were used in all cases; a standard sgRNA tail was used for dCas9-VPR, and an sgRNA containing two MS2 hairpins was used for all SAM-like crosses to recruit MCP-coupled activators. Anterior is up, dorsal is right. (Scale bars, 50 µm.)
To further optimize the dCas9–VPR construct, we tested whether expressing dCas9-VPR at a higher expression level or with Drosophila codon use could increase target gene activation levels. However, expression of dCas9-VPR at a higher level (with 20XUAS and the IVS translational enhancer, compared with 10XUAS) was lethal when expressed with actin-Gal4 and caused strong sgRNA-independent defects when expressed in the wing using ms1096-Gal4 (Fig. S1 A and B). Additionally, expression of a Drosophila codon-optimized 10XUAS-dCas9-VPR construct reduced function relative to the human codon-optimized construct (Fig. S1C). Together, our results demonstrate that the 10XUAS:dCas9-VPR construct (human codon optimized) outperforms all alternatives tested, including UAS:SAM, a panel of seven SAM-like constructs, and modifications of the expression level and codon use of the construct.
It has recently been reported that tissue-specific CRISPR can be improved in Drosophila by expressing sgRNAs under UAS control rather than from the commonly used U6:3 and U6:1 promoters (12). To test whether UAS-sgRNAs can also improve CRISPRa, we compared the activation of two target genes, wg and hindsight (hnt), in cell culture via qPCR. We found that UAS-sgRNAs did not lead to higher levels of activation and in fact dampened CRISPRa for wg (Fig. S1D).
Characterizing the Success Rate of dCas9-VPR.
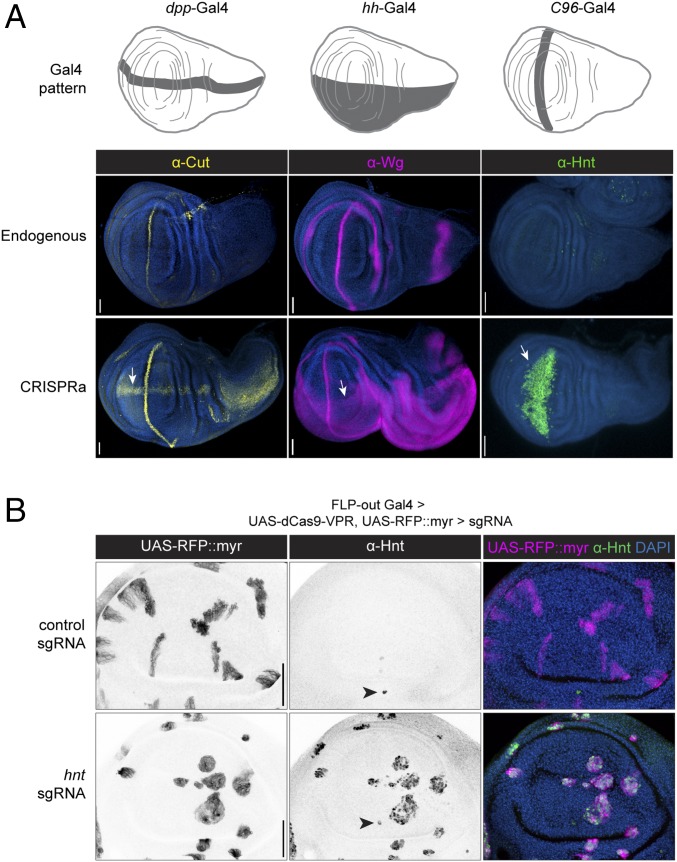
Having established that dCas9-VPR– and U6-driven sgRNAs outperform all variants tested, we next evaluated the success rate and versatility of this approach. Based on our previous observations, we generated transgenic flies expressing two sgRNAs per target gene, targeting a window between −400 and 0 bp upstream of the TSS (8). We tested three such sgRNA lines, each targeting a gene for which an antibody is available [cut, wg (8), and hnt] driven by three different Gal4 > dCas9-VPR lines. In all three cases, we observed the predicted ectopic expression for these genes (Fig. 3A), demonstrating that this technique is effective using different Gal4 drivers, and targeting different genes.
Fig. 3.
dCas9-VPR is effective and specific. (A) CRISPRa of three endogenous genes using different Gal4 drivers in the Drosophila wing disc, detected via antibody staining. Arrows indicate ectopic expression. Anterior is up. (B) FLP-out CRISPRa. Membrane-targeted RFP labels clones expressing dCas9-VPR in the presence of a control sgRNA (Upper Row) or sgRNA-hnt (Lower Row). Arrowheads indicate endogenous Hnt expression. Anterior is right. (Scale bars, 50 µm.)
A recent study of tissue-specific CRISPR has shown that leaky expression of Cas9 from a UAS promoter can lead to mutagenesis outside the desired Gal4 expression domain (12). To test whether the same problem affects CRISPRa, we generated a FLP-out Gal4 strain that, upon heat shock, creates RFP-labeled clones expressing UAS:dCas9-VPR. We crossed this line to sgRNA-hnt flies and examined FLP-out CRISPRa clones in the larval wing discs using an anti-Hnt antibody (Fig. 3B). We observed ectopic Hnt expression in 100% of RFP-marked clones and never observed ectopic Hnt outside the Gal4 domain (Fig. 3B), thus demonstrating that tissue-specific CRISPRa is both highly effective (all clones show activation of the target gene) and specific (no activation outside the Gal4 domain). This difference between CRISPR mutagenesis and CRISPRa is likely because CRISPR mutagenesis requires only a transient interaction between Cas9 and its target to permanently disrupt gene function, whereas CRISPRa presumably requires sustained interaction of dCas9-VPR and the TSS of its target gene.
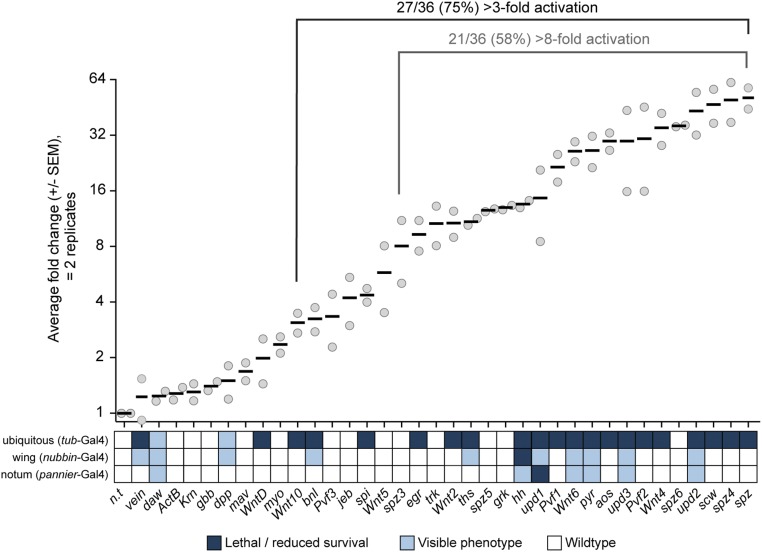
Generating a useful genome-wide collection requires that the large majority of the lines generated are functional. To estimate the proportion of sgRNA lines that function as predicted, we created a panel of transgenic fly stocks expressing sgRNAs targeting upstream of the TSS of 36 target genes, each of which encodes a secreted ligand for a highly conserved signaling pathway (Table S1). We crossed flies expressing dCas9-VPR ubiquitously (with tub-Gal4) to this panel and analyzed activation using qPCR (Table S2). To minimize lethality, we grew all the F1 of these crosses at 18 °C for 5 d to limit Gal4 activity and then transferred the larvae to 27 °C for 2 d. Seventy-five percent (27/36) of these sgRNA transgenes led to a greater than threefold increase in transcript levels when combined with dCas9-VPR, and 58% (21/36) caused a greater than eightfold increase, a success rate comparable to the proportion of transgenes in current RNAi stock collections that confer effective knockdown (Fig. 4) (13). We also note that several of the lines that do not appear up-regulated using tub-Gal4 when tested via qPCR do in fact produce specific phenotypes (see below), indicating that these success rates are in fact likely to be underestimated.
Table S1.
sgRNA lines used in this study
| Target name | FlyBase ID | sgRNA stock ID | sgRNA 1 | sgRNA 2 | Notes |
| spitz | FBgn0005672 | GS00182 | AGGGAGATGACACTATCGCGAGG | TTAAGTCTGTAATGTTTAGGTGG | sgRNA 2 is equidistant to CG10364 |
| gurken | FBgn0001137 | GS00045 | GAATGAAATCGATCGGTGAGCGG | ATAATAGTCGAAGAATCATATGG | |
| vein | FBgn0003984 | GS00144 | AGCATTCACAACGTATCTCACGG | GTGCCGAAGATATTCGAACACGG | |
| Keren | FBgn0052179 | GS00186 | ATGAAATTAGTTATTAACATCGG | TTAGGCTTATCGATTATATTCGG | sgRNA 2 is equidistant to CG7484 |
| argos | FBgn0004569 | GS00057 | CACTTCTCTGCCGGCTTCAACGG | ATTTGTTCCCAGTGCATGAGCGG | argos is within intron of CG33158 |
| PDGF- and VEGF-related factor 3 | FBgn0085407 | GS00022 | CAGTCGCGGCAGATGCGCATTGG | ACCAATCCGGCTGATCAGCCTGG | |
| PDGF- and VEGF-related factor 1 | FBgn0030964 | GS00032 | GCTATTGTTATTGGCGACTGTGG | AGCGCAATGCTGCGACAGCTGGG | |
| PDGF- and VEGF-related factor 2 | FBgn0031888 | GS00162 | TGTGGATCGCTTCTGTGAAACGG | TCTCTCACAGAGATTGGGAGAGG | |
| Trunk | FBgn0003751 | GS00062 | AGCCGAAAAAACTGCTCTGATGG | CACTCAGTAGTTGTAATACGAGG | sgRNA 2 is >400 bp upstream |
| Jelly belly | FBgn0086677 | GS00159 | GTTGAAAACAAGTGGTTGAGCGG | TCACTTTCTCTCATCCCGTTAGG | |
| branchless | FBgn0014135 | GS00107 | AGTCTCTGACTAGCACGCTTGGG | GCAAAAAGACCCTGTCGATGAGG | |
| thisbe | FBgn0033652 | GS00134 | AGGAAGAATCAGGGAAGAGGAGG | GGCGTGGCGGGTGGGCGGCGGGG | |
| pyramus | FBgn0033649 | GS00085 | CGGCGGGGGCGGTTCGAAGCGGG | CGCCGCCGACAATCCGAATGCGG | |
| unpaired 1 | FBgn0004956 | GS00169 | GCGAGCTGAGCGGCCTCTGCCGG | TGCCGACGAGCGGTACGCCATGG | |
| unpaired 2 | FBgn0030904 | GS00171 | CAGCAAGCGATTGTGATAGTTGG | TGCTGATGCTGATCGCTCCACGG | |
| unpaired 3 | FBgn0053542 | GS00129 | TGGAGTGGAGTGTTGTGGAGTGG | GGAGTTCAGTCAGTCTCCGCCGG | |
| decapentaplegic | FBgn0000490 | GS00163 | CGTCCAAAGCGGCCGAGGCAAGG | TCTGTGTTGACTGTAGACTGTGG | sgRNA 2 is >400 bp upstream, equidistant to CR44912 (ncRNA) |
| glass bottom boat | FBgn0024234 | GS00073 | TTGTTAAATTTATCATAGTCCGG | CCGATTCATTTTCCAAGTCGCGG | sgRNA1 is in 5′intron of YTR, sgRNA2 has U6 terminator |
| screw | FBgn0005590 | GS00080 | AGGTAGTTCAATTGCATCAGTGG | CCTGTACCTGACCAATGGGTTGG | screw is within intron of CG46244 and CG10443 |
| myoglianin | FBgn0026199 | GS00047 | ATCGGATACATACGGCTCACCGG | ATATCTTATATAGTTTCTTCGGG | |
| dawdle | FBgn0031461 | GS00051 | GCGGCCACAAAAGCCTGGAGCGG | ATGGGTGTGAGTATGGGCCTGGG | |
| maverick | FBgn0039914 | GS00018 | TGTTAAGTAAGCATTTGCTGTGG | ACACACCCACACATCTGCAGTGG | Equidistant to CR43009 (miRNA) |
| activin-β | FBgn0024913 | GS00028 | AAAAAGTCGGCAGATTCAAATGG | AGATAAATCTGTTAATGTTTAGG | |
| hedgehog | FBgn0004644 | GS00191 | TATGCCACTCGACGTTCGATCGG | TTCCACTTCCCTTGCGCATAAGG | |
| Wnt oncogene analog 2 | FBgn0004360 | GS00120 | AACACACGACCTCACTCACTTGG | GCGAGGTTCAAGTGGTTAAAAGG | |
| Wnt oncogene analog 4 | FBgn0010453 | GS00048 | CCGTGCCTGGCCGTCTTTGGTGG | ATATTGTGACTGCGATCAATTGG | |
| Wnt oncogene analog 5 | FBgn0010194 | GS00148 | AGTGAAAATGTGTATGGTGGTGG | GTTGCAGACGACGCAACGCGAGG | |
| Wnt oncogene analog 6 | FBgn0031902 | GS00049 | TGCTCTGCTGACGCTCTGTTTGG | ATCCGAGCGCTCAAAATCTGTGG | |
| wnt inhibitor of Dorsal aka Wnt8 | FBgn0038134 | GS00104 | CGCCCTGGCTTGGGATTTGCAGG | GAGCAGGCCCGAAATTCCCACGG | |
| Wnt oncogene analog 10 | FBgn0031903 | GS00034 | TTTAATTTGCATTTAGAGGCTGG | AGGACGAACGACGATGTCGATGG | |
| eiger | FBgn0033483 | GS00111 | GAGCAAGAGGGACAACGGATTGG | GCATCCTCGCCACGTGAGCGAGG | |
| spatzle | FBgn0003495 | GS00152 | GAGTCGAGTCGGCGGCCTCTGGG | TAAGCACTGAGCAATTGTTTGGG | |
| spatzle 3 | FBgn0031959 | GS00008 | TTTAAGGCGCACGGCGAGAACGG | AAGAGCATTTGCTCTTCGATTGG | |
| spatzle 4 | FBgn0032362 | GS00016 | CAAGTCCCACGGCAAACCGTCGG | AATGCACTGCAAGAAATTGGTGG | |
| spatzle 5 | FBgn0035379 | GS00130 | TGCCTCTTTTCCCGAGTTTGCGG | GGCAGAGAAAAGCAGAGCTTGGG | |
| spatzle 6 | FBgn0035056 | GS00088 | CTAGCCAGAGCATGAGAAAGTGG | CGGAAAGTATGCTTCTGTTTTGG | |
| QUAS (nontargeting control) | — | GS00089 | CTCGGGTAATCGCTTATCCTCGG | CGGATAAACAATTATCCTCACGG | |
| cut | FBgn0004198 | GS00041 | AAGCAACAACATAATGATAATGG | AGTCGTTTGTTGAATGGTGAGGG | |
| hindsight aka peb | FBgn0003053 | GS00052 | GAGAGAAGAGAGAAGCAGTCTGG | ATTTGAAACGAAGAATGAGAAGG | |
| scute | FBgn0004170 | GS00184 | TTCGAGTTCCCTACCTGTGCAGG | TTACAAACCTGATCAATAGATGG |
Table S2.
qPCR primers used in this study
| Target | 5′ primer | 3′ primer |
| spi | TGCGGTGAAGATAGCCGATC | TTCGCATCGCTGTCCCATAA |
| grk | GTCGCCGTCACAGATTGTTG | GATTGAGCAACTCAACCGCG |
| vn | GAACGCAGAGGTCACGAAGA | GAGCGCACTATTAGCTCGGA |
| Krn | CCGCTTTAATCGGCGCTTAC | ATCGGGAAGGTGACATTCGG |
| aos | TGCTGTTGGGTGAATTTCAGG | CGACTGGTCCAGATGATCCA |
| Pvf3 | TCGTGAAGAGCAGTAAGCATCG | AGGTGCAACTCAGTATGGTGG |
| Pvf1 | CTGTCCGTGTCCGCTGAG | CTCGCCGGACACATCGTAG |
| Pvf2 | GGTGGTCCACATCACGAGAG | CGACTTTGTCGCTGCATCTG |
| trk | CCAAAATTCTGGGACAGGCAT | AGATGATAGCTCTTCTCCTCGG |
| jeb | AAATCGAGTGTCCTACCGCC | CATCGCACAGCACATGATCG |
| bnl | AATGTCGCCCGCTGACAATA | TTGCTGATGGGCGTGTTACT |
| ths | CGTCCGCAACAACCATGAAG | CATTGCGCACATAGGTCAGC |
| pyr | GCAACGGATACCAAGTCCCA | TCTGGCTCGAACGCATTAGT |
| upd1 | CAGCGCACGTGAAATAGCAT | CGAGTCCTGAGGTAAGGGGA |
| upd2 | CGGAACATCACGATGAGCGAAT | TCGGCAGGAACTTGTACTCG |
| upd3 | AGCCGGAGCGGTAACAAAA | CGAGTAAGATCAGTGACCAGTTC |
| dpp | TGGCGACTTTTCAAACGATTGT | CAGCGGAATATGAGCGGCAA |
| gbb | CATCGACGAGAGCGACATCA | TAGTTGTCGTTGGGCACGTT |
| scw | TTTCCAACGAGGATCGACAGG | CCAAACTCAAATCCACTGGCA |
| myo | ATGCTGCGGTTGGAGAAAATA | CGTGACATATCGAGTTACACGG |
| daw | ATCCTTCGTCCGCATCCTAAG | CGGTTCCAGGTGTTTCAGC |
| mav | ACGAGGGCCAGGATCTAGG | CGAGTTTCTTGGAAGCCAACAT |
| Actβ | TGTGGTTGTAAAGTGCTGTTGC | TTGTGGAAATGACTTCCGGGA |
| hh | GGATTCGATTGGGTCTCCTAC | GGGAACTGATCGACGAATCT |
| Wnt2 | TTAGTGTCCAGCTTTACATCCGT | TCGCGGCACATATTTCGCT |
| Wnt4 | CGGGAACATGAACAGCACGAT | TCACCGAGGGTACACTCGATG |
| Wnt5 | ATATCTCGGCCTCAAGTCCCC | GTCATGTTCAGTGGTATGCTGG |
| Wnt6 | AGACGATTCGCCCGACTTTT | CTGGTCACATTGCACTCCCT |
| wntD | TTTGCCATCACATTCTTCATGGG | GGGTGTACTGGTAGTAGCTCA |
| Wnt10 | TGGAACTGTTCGTCGCTGAG | CAAACTGGAGGCATGCGGAT |
| egr | AGCTGATCCCCCTGGTTTTG | GCCAGATCGTTAGTGCGAGA |
| spz | GACACCTGGCAGTTAATTGTCA | CGAAGTCACAGGGTTGATCCG |
| spz3 | GCCGCAATCGGGTGGTTAT | GCTGCGTCTGCTGTACCTG |
| spz4 | CGCACCCAAATGGATAGGC | GCAATCGTCATTAGGATCAGCA |
| spz5 | GGAAAGACGTACTGCGAGCA | AAATCTTCCCAGGTTTCGTCC |
| spz6 | AAGTCTGCCGTGTCCGTTC | CGAGTACATTTTGTCCCAGCG |
| ct | TGAGGAGAACAAGGATGCGG | TTGTTGGCGCAATCATCGTC |
| hnt | ACATCCGGTGCCACAATTA | AGGGATGAAGCCGAGGATAGC |
| Rp49 (control) | ATCGGTTACGGATCGAACAA | GACAATCTCCTTGCGCTTCT |
| GAPDH (control) | CCAATGTCTCCGTTGTGGA | TCGGTGTAGCCCAGGATT |
Fig. 4.
sgRNA lines for CRISPRa have a high success rate. qPCR analysis of sgRNA lines crossed to flies expressing dCas9-VPR ubiquitously, shown on a log2 scale. Solid lines represent the mean of two biological replicates. Larvae in qPCR analyses were grown at 18 °C for 5 d to enhance survival and then were transferred to 27 °C for 2 d, whereas lethality was measured at 27 °C throughout development. Each gene is compared with a nontargeting sgRNA line (n.t.), and lethality/phenotypes are indicated on the heat-map below. See Methods for details of lethality/reduced survival.
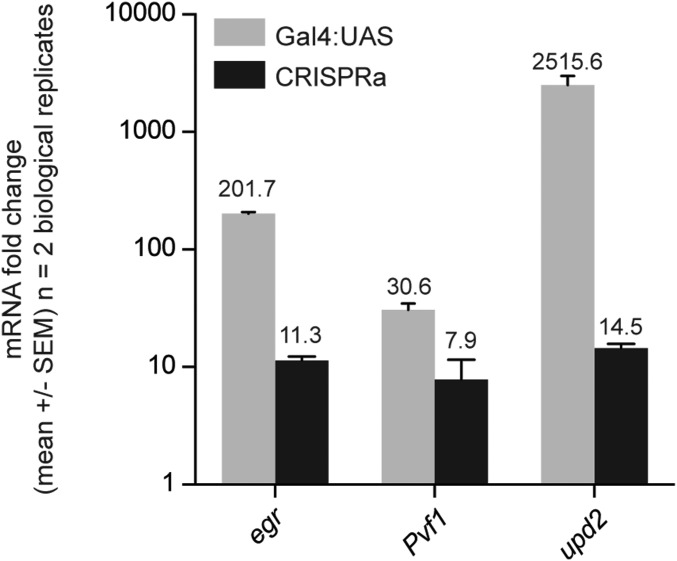
We next directly compared activation levels attained by CRISPRa versus Gal4 > UAS-cDNA for three genes using qPCR (in the wing using nubbin-Gal4) and found that, as predicted, CRISPRa consistently generated more modest levels of activation (Fig. S2). To address whether the level of activation induced by CRISPRa is biologically relevant in vivo, we scored phenotypes for these 36 genes using three different Gal4 lines: tubulin-Gal4 (ubiquitous), nubbin-Gal4 (wing), and pannier-Gal4 (notum). A nontargeting sgRNA had no effect on survival or morphology in any tissue tested (Figs. 4 and 5). For the 36 test lines, ubiquitous expression at 27 °C caused lethality for the majority of the genes, particularly those expressed at higher levels (Fig. 4).
Fig. S2.
Comparison of CRISPRa and Gal4-UAS-cDNA in vivo. Three target genes were expressed in the wing using the nubbin-Gal4 driver, and their activation level was measured using qPCR of dissected wing discs (∼30 wing discs per biological replicate).
Fig. 5.
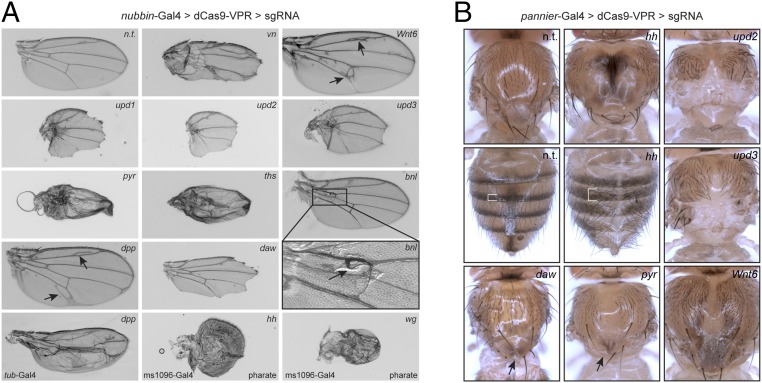
Phenotypes generated by in vivo CRISPRa. UAS:dCas9-VPR was driven in the wing by nubbin-Gal4 (A) or in the notum and dorsal cuticle by pannier-Gal4 (B). Additional Gal4s for select genes are shown in the bottom row of A; note that wg and hh depict wings from pharate adults, as these failed to eclose. Arrows and brackets indicate abnormal morphological features. See text for details.
We observed specific, predicted phenotypes for many genes (Fig. 5). CRISPRa of the EGFR ligand vein in the wing led to excess vein tissue (Fig. 5A) (14). Activation of each of the three JAK/STAT ligands upd1-3 led to nearly identical growth retardation in both wing (15) and notum (Fig. 5; note that upd1 CRISPRa in the notum was lethal). CRISPRa of the FGF ligands pyr and ths caused similar defects in wing growth and patterning, whereas activation of the remaining FGF, bnl, led to ectopic trachea at the base of the wing, as expected (Fig. 5A) (16). The TGF-β ligand dpp caused ectopic vein formation in the wing (Fig. 5A), to a lesser extent than seen with UAS:dpp (17), consistent with the minimal activation detected via qPCR for this line (Fig. 4). A stronger dpp phenotype was observed in the wing using tub-Gal4, confirming that the Gal4 expression level can influence phenotype strength (Fig. 5A). hh, which was lethal when overexpressed using nub-Gal4 (Fig. 4), caused overgrowth, bristle loss, and a pigmentation defect in the notum and abdomen. Using an additional wing Gal4, MS1096-Gal4, to drive hh CRISPRa led to severe patterning defects in pharate adult wings (Fig. 5A). Wnt6 caused overgrowth in the wing and notum as well as excess vein tissue in the wing (Fig. 5). wg, which was lethal when expressed with any of the three primary Gal4s tested, led to profound defects in the pharate adult wing when expressed using ms1096-Gal4 (Fig. 5A). Together, these results indicate that CRISPRa can activate physiologically relevant levels of transcription in vivo for a variety of genes and serves as a useful platform for screening.
We note that three GOF phenotypes reported based on UAS overexpression were not observed with CRISPRa. Specifically, CRISPRa of aos, egr, and scw did not lead to vein loss (18), tissue death (19, 20), or blistering/veination defects (21), respectively, even though qPCR analysis indicated these lines activate their target genes (Fig. 4). It is possible that these ligands require higher expression levels to trigger signaling. We also note an unexpected phenotype for the TGF-β ligand daw, which led to loss of the wing margin (Fig. 5). This is different from previous reports of no phenotype when driving UAS:daw in the wing (22, 23). The reason for this difference is unclear, but we note that it is unlikely to be an off-target effect, as the TSS of daw is >5 kb from the nearest neighboring TSS (Syt1, a synaptic vesicle protein), and no off-targets were predicted genome-wide in the vicinity of any TSS.
Gene-Flanking sgRNAs for LOF and GOF Studies.
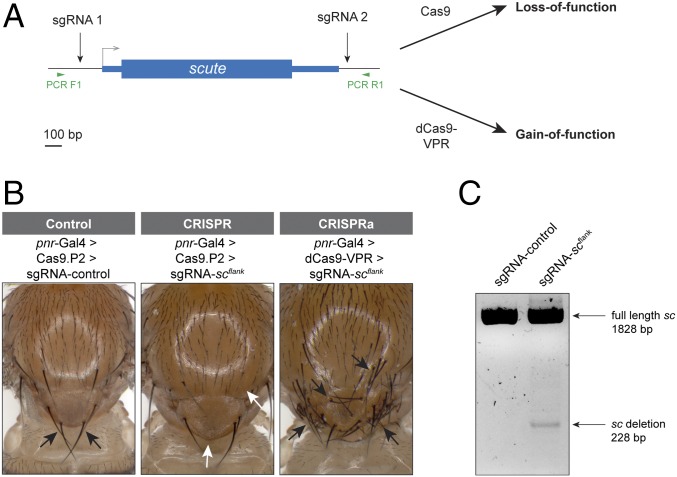
Next, we tested a strategy to generate sgRNA lines that could be used for both CRISPR mutagenesis and for CRISPRa. We hypothesized that flies expressing two sgRNAs, one upstream of the TSS and one downstream of the coding sequence (Fig. 6A), which we termed “sgRNAflank lines,” could serve such a dual function. When sgRNAflank lines are crossed to a Gal4 > UAS:dCas9-VPR fly stock, this should activate the target gene via CRISPRa, and when crossed to Gal4 > UAS:Cas9, this should generate LOF phenotypes by deleting the entire gene.
Fig. 6.
Dual-function sgRNAflank strategy for GOF and LOF. (A) Design strategy for sgRNA-scflank line. One sgRNA upstream of the TSS should activate CRISPRa, while the combination of both sgRNAs allows for deletion mutagenesis. (B) LOF and GOF phenotypes in the notum using sgRNA-scflank. sc LOF produces loss of scutellar bristles (white arrows) in 35% of flies, whereas sc GOF causes ectopic bristles (black arrows). (C) PCR analysis of dissected thoraces from pannier-Gal4 > UAS:Cas9.P2 > sgRNA-control or sgRNA-scflank flies using primers flanking the entire scute locus. Note that DNA was extracted from whole thoraxes, which include many wild-type cells that do not express pannier-Gal4.
As a proof of principle, we generated a sgRNAflank line targeting scute, a proneural transcription factor with well-characterized LOF and GOF phenotypes (Fig. 6A) (24). We crossed the sgRNA-scflank line to pannier-Gal4 > UAS:dCas9-VPR flies and observed strong GOF phenotype in the offspring: numerous ectopic bristles on the dorsal thorax, indicating successful CRISPRa (Fig. 6B). We then crossed sgRNA-scflank to pannier-Gal4 > UAS:Cas9.P2, which generated the predicted LOF phenotype of the loss of scutellar bristles (Fig. 6B). We confirmed the deletion of the scute locus via PCR (Fig. 6C). Thus, the sgRNAflank approach can work to generate both LOF and GOF functions from a single transgenic sgRNA line.
However, we note two caveats to this proof of principle: first, whereas the GOF phenotype was 100% penetrant, the LOF phenotype was observed in only 35% of F1 flies (n = 54). While this may reflect relatively low penetrance of the sc mutant phenotype, it also may be due to the fact that each sgRNA targets outside the sc coding sequence, and thus a LOF requires that both cuts be successful and induce a large deletion of the intervening sequence, potentially a relatively rare event. Second, this strategy relies on a single upstream sgRNA for CRISPRa. We have previously reported substantial variation in the success of individual sgRNAs for CRISPRa (8) and for this reason are building our large-scale CRISPRa resource with two sgRNAs per target gene to maximize the success rate. Nevertheless, the sgRNAflank approach is a promising strategy for generating dual-use lines for GOF and LOF.
Based on the high success rate of our CRISPRa experiments, we have initiated large-scale production of sgRNA fly stocks using the same platform that we previously used to generate a transgenic RNAi collection (25), with the aim of generating a genome-wide resource for GOF studies. At the time of writing, the collection includes over 250 lines. We hope that this resource will be useful for interrogating gene function via overexpression. GOF studies are an important complement to LOF studies and may be particularly useful for characterizing genes for which no LOF phenotype is apparent (2).
A number of studies have established that successful CRISPRa requires sgRNAs to be located within ∼500 bp of the target TSS (9, 26). This has implications for off-target effects: if the TSS of a target gene is within ∼500 bp of a neighboring TSS, there is likely to be off-target activation. In the Drosophila genome, we estimate that 3,509 of the 13,919 annotated protein coding genes (25%) are within 500 bp of another TSS, and 4,895 (35%) are within 1,000 bp. In such cases, it will be necessary to use a complementary approach to interrogate the function of each gene individually. In addition, for genes with multiple TSSs that are more than ∼400 bp apart, a decision about which isoform(s) to express will have to be made a priori.
To facilitate the use of CRISPRa in Drosophila, we have generated a collection of over 30 Drosophila stocks containing a variety of validated tissue-specific Gal4 lines driving UAS:dCas9-VPR, both with and without tubulin-Gal80ts for temporal control (Table S3). These lines, as well as information on the progress of our sgRNA collection, are described at https://fgr.hms.harvard.edu/fly-in-vivo-crispr-cas, and the stocks are available at fly.bio.indiana.edu/.
Table S3.
Gal4 + dCas9-VPR toolkit collection
| Line | Bloomington stock ID | Full genotype |
| UAS-dCas9-VPR (II) | 66561 | w; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; MKRS/TM6B, Tb[1] |
| UAS-dCas9-VPR (III) | 66562 | w; Kr[If-1[/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6b, Tb[1] |
| elav > dCas9-VPR | 67038 | P{w[+mW.hs]=GawB}elav[C155]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO |
| MS1096 > dCas9-VPR | 67039 | w[1118] P{w[+mW.hs]=GawB}Bx[MS1096]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO |
| pnr > dCas9-VPR | 67040 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; P{w[+mW.hs]=GawB}pnr[MD237]/TM6B, Tb[1] |
| dMef2 > dCas9-VPR | 67041 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; P{w[+mC]=GAL4-Mef2.R}R1/[TM6B, Tb[1]] |
| C96 > dCas9-VPR | 67042 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; P{w[+mW.hs]=GawB}bbg[C96]/TM6B, Tb[1] |
| Lpp > dCas9-VPR | 67043 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; P{Lpp-GAL4.B}/TM6B, Tb[1] |
| Mhc > dCas9-VPR | 67044 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; P{[ry+]MHC-82-Gal4}/TM6B, Tb[1] |
| dpp > dCas9-VPR | 67045 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; P{w[+mW.hs]=GAL4-dpp.blk1}/TM6B, Tb[1] |
| dpp > dCas9-VPR (SM5 balancer) | In progress | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40; P{w[+mW.hs]=GAL4-dpp.blk1}/SM5; TM6B Tb[1] |
| hh > dCas9-VPR | 67046 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; hh-Gal4/TM6B, Tb[1] |
| tubulin > dCas9-VPR | In progress | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; tub-Gal4/TM6B, Tb[1] |
| tubulin > dCas9-VPR (SM5 balancers) | 67048 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40; tub-Gal4/SM5; TM6B Tb[1] |
| en > dCas9-VPR + GFP | 67049 | w[*]; P{w[+mW.hs]=en2.4-GAL4}e16E, P{w[+mC]=UAS-2xEGFP}AH2/CyO; UASP{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6B, Tb[1] |
| spalt > dCas9-VPR | 67050 | w[*]; P{w[+mW.hs]=GawB}salm[LP39]/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6B, Tb[1] |
| blistered > dCas9-VPR | 67051 | w[*]; P{w[+mC]=bs-GAL4.Term}G1/CyO; UAS:P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6B, Tb[1] |
| nos > dCas9-VPR | 67052 | w[*]; P{w[+mC]=GAL4-nos.NGT}40/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6B, Tb[1] |
| twist > dCas9-VPR | 67053 | w[*]; P{w[+mC]=GAL4-twi.2xPE}1/CyO; P{w[+mC]=UAS-3xFLAG.dCas9.VPR}attP2 |
| esg > dCas9-VPR + GFP | 67054 | w[*]; esg-Gal4, P{ UAS-GFP}/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6B, Tb[1] |
| nubbin > dCas9-VPR | 67055 | w[*]; P{w[+mW.hs]=GawB}nubbin-AC-62/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6B, Tb[1] |
| nubbin > dCas9-VPR (SM5 balancers) | In progress | w[*]; P{w[+mW.hs]=GawB}nubbin-AC-62; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/SM5; TM6b Tb[1] |
| Myo1A > dCas9-VPR | 67057 | w[*]; P{GawB}Myo31DFNP0001/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6B, Tb[1] |
| DE-Gal4 > dCas9-VPR | In progress | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40; P{GawB}mirr[DE]/TM6b Tb[1] |
| GMR-Gal4 > dCas9-VPR | In progress | w[*]; “GMR-Gal4”/CyO; CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2/TM6B, Tb[1] |
| elav[ts] > dCas9-VPR | 67058 | P{w[+mW.hs]=GawB}elav[C155]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO |
| MS 1096[ts] > dCas9-VPR | 67059 | P{w[+mW.hs]=GawB}Bx[MS1096]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO |
| pnr[ts] > dCas9-VPR | 67060 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO; P{w[+mW.hs]=GawB}pnr[MD237]/TM6B, Tb[1]] |
| Lpp[ts] > dCas9-VPR | 67061 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO; P{Lpp-GAL4.B}/TM6B, Tb[1] |
| Delta[ts] > dCas9-VPR | In progress | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40/CyO; P{w[+mW.hs]=GawB}Dl[05151], P{w[+mC]=tubP-GAL80[ts]}[*]/TM6B, Tb[1] |
| Lpp[ts] > dCas9-VPR (SM5 balancer) | In progress | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]; P{Lpp-GAL4.B} SM5; TM6b Tb[1] |
| Mhc[ts] > dCas9-VPR | 67062 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO; P{[ry+]MHC-82-Gal4}/TM6B, Tb[1] |
| dMef2[ts] > dCas9-VPR | 67063 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO; P{w[+mC]=GAL4-Mef2.R}R1/(TM6B, Tb[1] |
| dMef2[ts] > dCas9-VPR (SM5 balancer) | In progress | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]; P{w[+mC]=GAL4-Mef2.R}R1/SM5; TM6b Tb[1] |
| C96[ts] > dCas9-VPR | 67064 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO; P{w[+mW.hs]=GawB}bbg[C96]/TM6B, Tb[1] |
| tub[ts] > dCas9-VPR | 67047 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO; tub-Gal4/TM6B, Tb[1] |
| tub[ts] > dCas9-VPR (SM5 balancer) | In progress | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]; tub-Gal4/SM5; TM6B, Tb[1] |
| dpp[ts] > dCas9-VPR | 67066 | w[*]; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp40, P{w[+mC]=tubP-GAL80[ts]}10]/CyO; P{w[+mW.hs]=GAL4-dpp.blk1}TM6B, Tb[1] |
| Myo1A[ts] > dCas9-VPR | 67067 | w[*]; P{GawB}Myo31DFNP0001/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2, P{w[+mC]=tubP-GAL80[ts]}2/TM6B, Tb[1] |
| en[ts] > dCas9-VPR + GFP | 67069 | w[*]; P{w[+mW.hs]=en2.4-GAL4}e16E, P{w[+mC]=UAS-2xEGFP}AH2/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2, P{w[+mC]=tubP-GAL80[ts]}2/TM6B, Tb[1] |
| spalt[ts ] > dCas9-VPR | 67070 | w[*]; P{w[+mW.hs]=GawB}salm[LP39]/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2, P{w[+mC]=tubP-GAL80[ts]}2/TM6B, Tb[1] |
| blistered[ts] > dCas9-VPR | 67071 | w[*]; P{w[+mC]=bs-GAL4.Term}G1/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2, P{w[+mC]=tubP-GAL80[ts]}2/TM6B, Tb[1] |
| esg[ts] > dCas9-VPR + GFP | 67072 | w[*]; esg-Gal4,P{UAS-GFP}/CyO; P{UAS-3XFLAG-dCas9-dCas9-VPR}attp2, P{w[+mC]=tubP-GAL80[ts]}2/TM6B, Tb[1] |
| FLP-out Gal4 > dCas9-VPR | NA (N.P. laboratory stock) | hsFlp; Ay-Gal4, UAS:RFP-myr; UAS:dCas9-VPR/SM6, TM6b |
Methods
Drosophila Strains and Rearing.
Except where mentioned, all experimental CRISPRa crosses were maintained at 27 °C to enhance Gal4 expression on standard Drosophila food supplemented with yeast powder or paste. We note that similar results were obtained at 25 °C in all cases tested. The Gal4 lines and UAS:dCas9 lines are described in detail in Table S3. The control nontargeting sgRNA line for all experiments expresses two sgRNAs targeting the QUAS sequence (27), which is not found in the Drosophila genome (Table S1).
sgRNAs.
All sgRNA sequences are listed in Table S1. sgRNAs (two per gene) were designed to target from 0 to 400 bp upstream of target genes (9, 26). sgRNAs were designed using the Drosophila Resource Screening Center Find CRISPRs v2 online tool (www.flyrnai.org/crispr/) (28) and were additionally screened for potential off-targets using an independent online tool, CRISPR Optimal Target Finder (tools.flycrispr.molbio.wisc.edu/targetFinder/index.php). Any sgRNAs found to have off-targets within 1 kb of any other TSS were redesigned. sgRNAs were cloned into pCFD4 (for U6) or pCFD6 (for UAS) as described (12, 29). To generate pCFD4-MS2, we modified pCFD4 to contain MS2 loops in the sgRNA tail (9) using Gibson assembly (30) (New England Biolabs) of GBlock fragments synthesized by Integrated DNA Technologies. For VPR versus SAM-like experiments, the same sgRNAs targeting upstream of wg were cloned into both pCFD4 (for VPR) and pCFD4-MS2 (for all SAM-like constructs) using a strategy analogous to pCFD4 cloning. All sgRNAs were integrated into the genome at the attP40 landing site (31) using PhiC31-mediated integration. We used y v nanos-integrase; attP40 (https://fgr.hms.harvard.edu/trip-toolbox) as the injection stock, and all stocks were balanced by backcrossing to y sc v; Gla, Bc/CyO.
Generation of CRISPRa Constructs and Stocks.
dCas9-VPR.
The 10XUAS-dCas9-VPR constructs (both human and Drosophila optimized) have been previously described (8). The human codon-optimized plasmid used to generate the stocks in this manuscript is available at Addgene (Addgene 78897), and fly lines have been submitted to the Bloomington Drosophila Stock Center. To generate high-expression constructs, both human and Drosophila dCas9-VPR sequences were cloned into pJFRC7 (32), which contains 20XUAS-IVS for enhanced expression, using Gibson assembly. An additional line with 10XUAS:dCas9-VPR (human codon-optimized) integrated into the attP2 landing site on the third chromosome (31) was generated so that second chromosome Gal4 lines could be used (Table S3). Lethality of these constructs in the absence of a sgRNA was assayed by crossing homozygous lines to yw; actin-Gal4/CyO and counting F1 compared with CyO siblings.
SAM.
The UAS:SAM construct (UAS:MCP-p65-HSF) was generated using Gibson assembly of PCR-amplified MCP-p65-HSF (Addgene 61426) and the pJFRC7 backbone (Addgene 26220). This construct was integrated into the ZH-51C attP site (33) on chromosome II (BestGene, Inc.) so that it could be recombined with the other SAM component, UAS:dCas9-VP64, which has been previously described (8), in the attP40 landing site. Lethality was assayed using actin-Gal4, as above.
SAM-like.
Two subclones (UAS:dCas9-XAD and UAS:MCP-XAD) were first generated in a VALIUM20 backbone (34) by restriction digest. dCas9 (D10A and H840A) was amplified from a published plasmid (35). MCP-XAD and the VP64 domains were synthesized by GENEWIZ. The transcriptional activation domains of Ci (CG2125) (36) and HSF (CG5748) (37) were cloned from cDNA of w1118 flies, and the Gal4 activation domain (38) was cloned from genomic DNA of act5C-Gal4/CyO flies. The lines were generated from these components using restriction digest cloning.
qPCR.
For qPCR, flies of the genotype w; UAS:dCas9-VPR; tub-Gal4/SM5, TM6B were crossed to homozygous sgRNA lines, and the F1 were maintained for 5 d at 18 °C to reduce Gal4 activity and thereby lethality. Larvae were then transferred to 27 °C for 48 h, and non-Tb larvae were used for qPCR. Three of the sgRNAs tested (Wnt6, pyr, and upd1) were fully lethal even at 18 °C; for these three lines, a tubGal80ts repressor was included to fully inhibit Gal4 for the first 5 d of growth at 18 °C. Three to six L3 larvae were flash-frozen on dry ice and homogenized in TRIzol (Thermo Fisher); then RNA was extracted following the manufacturer’s protocol. RNA was purified using either RNeasy (Qiagen) or Direct-zol (Zymo) kits, including a 15–20 min DNase treatment, and then was used as template for first-strand synthesis and qPCR as described (8), with the following modifications: all target genes were compared with the geometric mean of two reference genes (Rp49 and GAPDH), and an annealing temperature of 57 °C was used. Primers were designed using a precomputed database of Drosophila qPCR primers (39), and any that had efficiency outside ∼80–120% were redesigned either from the literature or using Primer3Plus (primer3plus.com/cgi-bin/dev/primer3plus.cgi). All experiments were conducted in biological duplicates, with technical triplicate qPCR reactions.
Phenotype Scoring.
To score for lethality following ubiquitous CRISPRa, each sgRNA line was crossed to w; UAS:dCas9-VPR; tub-Gal4/SM5, TM6B at 27 °C, and the number of F1 were compared with siblings possessing balancers. The following genes were 100% lethal: Pvf1, Pvf2, bnl, ths, upd, upd2, upd3, scw, hh, Wnt2, Wnt4, Wnt6, WntD, Wnt10, aos, and jeb. The following lines had highly reduced survival (<20% of Mendelian expectation): pyr, egr, spz, and spz4. For wing and notum screens, sgRNA lines were crossed to w; nubbin-Gal4; UAS:dCas9-VPR/SM5, TMBb or w; UAS:dCas9-VPR/CyO; pannier-Gal4/TM6B. Wings (10–20 of each sex per cross) were mounted in Permount (Fisher) and imaged using brightfield optics on a Zeiss Axioskop 2 microscope. Nota and abdomens were photographed using a Spot RT3 Color Slider Camera Model 25.4.x (Diagnostic Instruments, Inc.) mounted on a Nikon SMZ1500 dissecting stereo microscope. A z-stack was captured and compiled into a single image using Helicon Focus software. The images represent the sex displaying the strongest phenotype. For wings, females are shown for nontargeting (n.t.), Wnt6, dpp, and pyr; males are shown for vn, upd1, upd2, upd3, ths, daw, and bnl. For nota, females are shown for n.t., hh, and Wnt6; males are shown for upd2, upd3, daw, and pyr.
FLP-Out Gal4 CRISPRa.
Flies of the genotype hsFlp; actin-FRT-STOP-FRT-Gal4, UAS:RFP-myr; UAS-dCas9-VPR/SM6, TM6B were generated from stocks containing hsFlp on the X chromosome (40), Bloomington BL3953 and BL7118, and the UAS:dCas9-VPR in attP2 described above. This line was crossed to sgRNA lines for either QUAS (nontargeting control) or hnt. Eggs were laid over a 24-h period and 48 h later were heat-shocked for 10 min at 37 ° and then were returned to 27 °C until the wandering stage. Wing discs were dissected and stained for Hnt protein as described below. Antibody detection of RFP was unnecessary, as the native signal persisted through fixation and staining.
sgRNA-Scuteflank Experiments.
Male flies of the genotype y sc v; sgRNA-scuteflank or y sc v; sgRNA-QUAS (control) were crossed to females of the genotype w; UAS:Cas9.P2; pannier-Gal4/TM6B and w; UAS:dCas9-VPR; pannier-Gal4/TM6b. Progeny of both sexes were scored for loss of one or more scutellar bristles. Note that the female progeny of these crosses are heterozygous for sc, and thus the female progeny scored are heterozygous for sc. However, we did not observe the sc LOF phenotype in control crosses, confirming the recessive nature of this mutation.
For PCR analysis of sgRNA-scuteflank, nota from pannier-Gal4 > UAS:Cas9.P2 > control-sgRNA or sgRNA-scuteflank (n = 3 nota per genotype) were dissected and flash frozen on dry ice. Genomic DNA was extracted using standard techniques, and PCR was conducted using primers flanking the two sgRNAs (F: GAGTGTTTCTTTTGGACTGTCGAG, R: TTGTAAAACCAATGCAAAGCAACC) and visualized on a 1.5% agarose gel.
Antibody Staining.
L3 larval wing discs were dissected in PBS, fixed for 20–30 min in 4% paraformaldehyde, and then stained using standard protocols using the following antibodies: anti-Wg [4D4, Developmental Studies Hybridoma Bank (DSHB), 1:100], anti-Cut (2B10, DSHB, 1:10), and anti-Hnt (1G9-c; DSHB, 1:10). Anti-mouse secondary antibodies coupled to Alexa 488 or Alexa 555 were used at 1:250–1:400, and were imaged using a Zeiss LSM 780 confocal microscope. Maximum intensity projections are shown.
Cell Culture Analysis of pCFD6.
Experiments were conducted using Drosophila S2R+ cells from the Drosophila RNAi Screening Center. Plasmids encoding U6- or UAS-driven sgRNAs targeting wg and hnt (in pCFD4 or pCFD6, respectively) were cotransfected with pActin-Gal4 and UAS:dCas9-VPR, as described (8). RNA was extracted 3 d after transfection, and qPCR performed as described (8).
Acknowledgments
We thank F. Port and S. Bullock for sharing the UASt-sgRNA plasmid (pCFD6) prior to publication, H.-W. Tang for assistance generating FLP-out CRISPRa stocks, and R. Binari for expertise and assistance with fly work. This work was supported by NIH Grants R01GM084947 and R24OD021997 (to the N.P. laboratory). B.E.-C. received NIH funding under Ruth L. Kirschstein National Research Service Award F32GM113395 from the NIH General Medical Sciences Division. D.P.G. received funding from the Howard Hughes Medical Institute (HHMI) Exceptional Research Opportunities Program. V.R.F. received funding from the Brazil Scientific Mobility Program. S.E.M. is supported in part by NIH National Center Institute Cancer Center Support Grant 5 P30 CA06516 to the Dana Farber/Harvard Cancer Center. The J.-Q.N. laboratory was supported in part by National Key Technology Research and Development Program of the Ministry of Science and Technology of the People’s Republic of China Grants 2015BAI09B03; 2016YFE0113700, National Basic Research Program (973 Program) Grant 2013CB35102, and National Natural Science Foundation of China Grant 31571320. N.P. is an investigator of the HHMI.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707635114/-/DCSupplemental.
References
- 1.Mohr SE. RNAi screening in Drosophila cells and in vivo. Methods. 2014;68:82–88. doi: 10.1016/j.ymeth.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prelich G. Gene overexpression: Uses, mechanisms, and interpretation. Genetics. 2012;190:841–854. doi: 10.1534/genetics.111.136911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong J, Yedvobnick B. Targeted gain-of-function screening in Drosophila using GAL4-UAS and random transposon insertions. Genet Res. 2009;91:243–258. doi: 10.1017/S0016672309990152. [DOI] [PubMed] [Google Scholar]
- 4.Bischof J, et al. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. doi: 10.1242/dev.088757. [DOI] [PubMed] [Google Scholar]
- 5.Rørth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staudt N, et al. Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLoS Genet. 2005;1:e55. doi: 10.1371/journal.pgen.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S, Ewen-Campen B, Ni X, Housden BE, Perrimon N. In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics. 2015;201:433–442. doi: 10.1534/genetics.115.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavez A, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng M, Diaz-Benjumea FJ, Vincent JP, Wu J, Cohen SM. Specification of the wing by localized expression of wingless protein. Nature. 1996;381:316–318. doi: 10.1038/381316a0. [DOI] [PubMed] [Google Scholar]
- 12.Port F, Bullock SL. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods. 2016;13:852–854. doi: 10.1038/nmeth.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sopko R, et al. Combining genetic perturbations and proteomics to examine kinase-phosphatase networks in Drosophila embryos. Dev Cell. 2014;31:114–127. doi: 10.1016/j.devcel.2014.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wessells RJ, Grumbling G, Donaldson T, Wang SH, Simcox A. Tissue-specific regulation of vein/EGF receptor signaling in Drosophila. Dev Biol. 1999;216:243–259. doi: 10.1006/dbio.1999.9459. [DOI] [PubMed] [Google Scholar]
- 15.Hombría JC-G, Brown S, Häder S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288:420–433. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 16.Sato M, Kornberg TB. FGF is an essential mitogen and chemoattractant for the air sacs of the Drosophila tracheal system. Dev Cell. 2002;3:195–207. doi: 10.1016/s1534-5807(02)00202-2. [DOI] [PubMed] [Google Scholar]
- 17.de Celis JF. Expression and function of decapentaplegic and thick veins during the differentiation of the veins in the Drosophila wing. Development. 1997;124:1007–1018. doi: 10.1242/dev.124.5.1007. [DOI] [PubMed] [Google Scholar]
- 18.Schnepp B, et al. EGF domain swap converts a Drosophila EGF receptor activator into an inhibitor. Genes Dev. 1998;12:908–913. doi: 10.1101/gad.12.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol. 2002;12:1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- 20.Igaki T, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002;21:3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen M, Park S, Marqués G, Arora K. Interpretation of a BMP activity gradient in Drosophila embryos depends on synergistic signaling by two type I receptors, SAX and TKV. Cell. 1998;95:495–506. doi: 10.1016/s0092-8674(00)81617-7. [DOI] [PubMed] [Google Scholar]
- 22.Jensen PA, Zheng X, Lee T, O’Connor MB. The Drosophila activin-like ligand dawdle signals preferentially through one isoform of the Type-I receptor baboon. Mech Dev. 2009;126:950–957. doi: 10.1016/j.mod.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gesualdi SC, Haerry TE. Distinct signaling of Drosophila activin/TGF-β family members. Fly (Austin) 2007;1:212–221. doi: 10.4161/fly.5116. [DOI] [PubMed] [Google Scholar]
- 24.Campuzano S, Modolell J. Patterning of the Drosophila nervous system: The achaete-scute gene complex. Trends Genet. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- 25.Perkins LA, et al. The transgenic RNAi project at Harvard Medical School: Resources and validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: A repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Housden BE, et al. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci Signal. 2015;8:rs9. doi: 10.1126/scisignal.aab3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Port F, Chen H-M, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 31.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni J-Q, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren X, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA. 2013;110:19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogden SK, Ascano M, Jr, Stegman MA, Robbins DJ. Regulation of hedgehog signaling: A complex story. Biochem Pharmacol. 2004;67:805–814. doi: 10.1016/j.bcp.2004.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wisniewski J, Orosz A, Allada R, Wu C. The C-terminal region of Drosophila heat shock factor (HSF) contains a constitutively functional transactivation domain. Nucleic Acids Res. 1996;24:367–374. doi: 10.1093/nar/24.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riabinina O, et al. Improved and expanded Q-system reagents for genetic manipulations. Nat Methods. 2015;12:219–222. doi: 10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Y, et al. FlyPrimerBank: An online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 (Bethesda) 2013;3:1607–1616. doi: 10.1534/g3.113.007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang H-W, et al. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 2011;30:636–651. doi: 10.1038/emboj.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]