Significance
Pathogen recognition first occurs at the plasma membrane, where receptor-like kinases perceive pathogen-derived molecules and initiate immune responses. To abrogate this immune response, pathogens evolved effector proteins that act as virulence factors, often following delivery to the host cell. Plants evolved intracellular receptors, known as NOD-like receptors (NLRs), to detect effectors, thereby ensuring activation of effector-triggered immunity. However, despite their importance in immunity, the molecular mechanisms underlying effector recognition and subsequent immune activation by membrane-localized NLRs remain to be fully elucidated. Our analyses reveal the importance of and need for self-association and the coordinated interplay of specific domains and conserved residues for NLR activity. This could provide strategies for crop improvement, contributing to effective, environmentally friendly, and sustainable solutions for future agriculture.
Keywords: plant immunity, effector-triggered immunity, NLR receptor, oligomerization, Arabidopsis thaliana
Abstract
Plants evolved intracellular immune receptors that belong to the NOD-like receptor (NLR) family to recognize the presence of pathogen-derived effector proteins. NLRs possess an N-terminal Toll-like/IL-1 receptor (TIR) or a non-TIR domain [some of which contain coiled coils (CCs)], a central nucleotide-binding (NB-ARC) domain, and a C-terminal leucine-rich repeat (LRR). Activation of NLR proteins results in a rapid and high-amplitude immune response, eventually leading to host cell death at the infection site, the so-called hypersensitive response. Despite their important contribution to immunity, the exact mechanisms of NLR activation and signaling remain unknown and are likely heterogenous. We undertook a detailed structure-function analysis of the plasma membrane (PM)-localized CC NLR Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) using both stable transgenic Arabidopsis and transient expression in Nicotiana benthamiana. We report that immune signaling is induced only by activated full-length PM-localized RPM1. Our interaction analyses demonstrate the importance of a functional P-loop for in planta interaction of RPM1 with the small host protein RPM1-interacting protein 4 (RIN4), for constitutive preactivation and postactivation self-association of RPM1 and for proper PM localization. Our results reveal an additive effect of hydrophobic conserved residues in the CC domain for RPM1 function and RPM1 self-association and their necessity for RPM1–RIN4 interaction. Thus, our findings considerably extend our understanding of the mechanisms regulating NLR activation at, and signaling from, the PM.
During their life cycle, plants and animals encounter a variety of pathogens, such as viruses, bacteria, oomycetes, and fungi. Plant pathogens that breach the waxy cuticular layer atop the epidermis face an efficient plant immune system comprising two layers (1, 2). The first layer consists of plasma membrane (PM)-spanning pattern-recognition receptors (PRRs) that typically recognize and bind pathogen/microbe-associated molecular patterns (PAMPs/MAMPs) and initiate PAMP/MAMP-triggered immunity (PTI/MTI) (3, 4). PRR activation induces downstream signaling, including protein phosphorylation, changes in ion flux, production of reactive oxygen species, transcriptional reprogramming, vesicle transport for polarized delivery of newly synthesized antimicrobial compounds and pathogenesis-related proteins, as well as cell wall reinforcement (5). PTI/MTI is sufficient to prevent microbial colonization and growth in most cases; however, evolutionarily adapted pathogens have independently evolved large arsenals of effectors (virulence proteins), such as the type III secretion system (TTSS) effectors from bacterial pathogens or the RxLR family of effectors from oomycetes (6–8). Effectors are delivered by various means into the apoplast and/or the host cytoplasm, where they target and manipulate key pathways of the host cellular machinery to suppress PTI/MTI, thus leading to effector-triggered susceptibility (1, 9, 10).
To defend themselves against adapted pathogens, plants evolved a second layer of defense, referred to as effector-triggered immunity (ETI) (2). This branch of the plant innate immune system relies on the products of plant disease resistance (R) genes. Most R genes encode proteins of the NOD-like receptor (NLR) family (1). NLRs belong to a subclade of the AAA-ATPase superfamily and are molecular switches regulated via nucleotide binding and hydrolysis (11, 12). NLRs have a central nucleotide-binding site (NB-ARC) with homology to the animal immune receptors Apaf-1 and CED-4, C-terminal leucine-rich repeats (LRRs), and either a Toll-like/IL-1 receptor (TIR) domain or a non-TIR domain at their N termini. The latter contains a subclass containing N-terminal coiled-coil (CC) domains, subdividing NLRs into TNLs, CNLs, and others, respectively (13, 14). The conserved nucleotide-binding site is critical for NLR activation, and negative regulation of this domain, likely via intramolecular interactions, is required to limit ectopic activation, which can result in hyperimmune signaling and ectopic cell death (15, 16). The N-terminal CC or TIR domains are necessary, and in some cases sufficient, for cell death signaling (17–21).
The current model is that NLRs exist in an equilibrium between a “resting/off” ADP-bound state and an “active/on” ATP-bound state in which the “off” state is strongly favored (22, 23). NLRs can be activated either directly by binding an effector protein or indirectly by monitoring an effector-specific modification of a host target (or a host decoy of a true target) (24, 25). Effector recognition/binding presumably leads to the release of negative intramolecular regulation and conformational changes allowing the exchange of ADP for ATP, thereby tipping the balance toward the ATP-bound “on” state. Recent work on the flax TNLs L6 and L7 suggests that, at least in cases where NLRs directly bind effectors, the effector preferably binds to the ATP-bound “on” state of the receptor, stabilizing this state and preventing recycling to the “off” state (22). In this model, ATP hydrolysis drives a return to the “off” state, thus regulating the switch. The importance of nucleotide binding for NLR activity is reflected by loss-of-function and autoactivation phenotypes caused by mutations in the highly conserved Walker-A (or P-loop) motif (GxxxxGK[T/S]), the Walker-B motif (hhhDD/E), or the MHD motif (with a conserved histidine residue that can also be found in animal NLRs), important for nucleotide binding, ATP hydrolysis, and ADP binding, respectively (26–28). Effector-mediated activation of NLR proteins results in ETI, a rapid and high-amplitude activation of signaling pathways that largely overlaps with PTI/MTI (29, 30). ETI is usually associated with a rapid localized cell death at the infection site, the hypersensitive response (HR), which can inhibit further pathogen proliferation in some cases.
Recent studies of plant and animal NLR proteins support models in which self- and hetero-association of NLRs are key mechanisms of activation and signaling (13, 14, 31, 32) (SI Appendix, Table S1). The tobacco TIR-NLR (TNL) N self-associates only in the presence of its cognate effector, and this self-association is P-loop dependent (33). Many plant CC-NLRs (CNLs), i.e., Prf (tomato), RPS5 (Arabidopsis), and Rp-1D (maize), self-associate (15, 27, 34); however, data on the role of self-association or oligomerization for plant NLR proteins at both preactivation and postactivation steps are limited and sometimes conflicting. In the case of MLA10, Sr33, and Rx, self-association has been shown for the full-length receptors and for CC fragments (amino acids 1–160) that include a structurally important full fourth helix, but not for shorter CC fragments lacking the C-terminal end of this helix (18, 35, 36). These nondimerizing shorter CC fragments are monomeric in solution, as determined by size-exclusion chromatography- coupled multiangle light-scattering experiments and are biologically inactive, whereas the longer CC fragments (at least spanning amino acids 1–142) also form dimers and cause HR in transient expression in Nicotiana benthamiana. This suggests that at least the dimerization of the CC domains in these NLRs is required for activation.
In addition, the sites of NLR activation are heterogeneous. Some plant NLRs require coordinated nucleo-cytoplasmic trafficking to establish a full and adequate immune response, suggesting that NLRs can activate distinct signaling pathways in the cytoplasm and nucleus (35). In contrast, other NLRs require PM or endomembrane localization, and disruption of their proper localization severely affects or blocks function (26, 36, 37). Thus, various mechanisms of NLR activation may have evolved shaped by the constraints of coevolution among effectors, their host targets, and corresponding NLRs (24).
Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is a well-characterized PM-localized CNL from Arabidopsis. RPM1 perceives the P. syringae TTSS effectors AvrRpm1 and AvrB (26, 38–40). Once injected into the plant cell, these effectors are acylated and localized to the host PM, where they interact with the small host protein RPM1-interacting protein 4 (RIN4) and induce its phosphorylation, mediated by the receptor-like cytoplasmic kinase RIPK and closely related paralogs (41, 42). The phosphorylation of RIN4 at threonine residue 166 is necessary and sufficient for the activation of RIN4-associated RPM1 (42). RPM1 activation on the PM is necessary and sufficient for its function (26). Activated RPM1 leads to the HR and to growth restriction of Pseudomonas strains expressing AvrRpm1 or AvrB; however, the molecular mechanisms leading to RPM1 activation and downstream signaling are unknown, as are whether RPM1 self-associates before or after its activation, and whether this potential oligomerization is necessary for RPM1-mediated immune responses.
Therefore, we undertook a detailed structure-function analysis of RPM1 using both stable transgenic Arabidopsis and the heterologous N. benthamiana transient expression system. We found that immune signaling is induced only by the activated full-length PM-localized RPM1 protein. Furthermore, our results reveal that RPM1–RIN4 interaction, as well as RPM1 PM localization, is P-loop dependent. We also found an additive effect of conserved hydrophobic residues in the RPM1 CC domain for RPM1 function and self-association and their necessity for RPM1–RIN4 interaction. We characterized the self-association of RPM1 in both preactivation and postactivation states and found that self-association is P-loop dependent. Thus, our analyses considerably extend the sparse knowledge of the mechanisms regulating NLR activation at, and signaling from, the PM (15, 43).
Results
Immune Signaling Requires Full-Length RPM1.
Expression of just the N-terminal CC, CC-NB-ARC, or TIR domain of several NLRs is sufficient to initiate immune signaling, eventually leading to HR (19, 44–46). RPM1 activation and HR signaling can be reconstituted in N. benthamiana by coexpression of RPM1, RIN4, and AvrRpm1 (or AvrB), by coexpression of RPM1 and phosphomimetic RIN4T166D, or by expression of the autoactive MHD mutant RPM1D505V (26, 42).To analyze whether the CC or any other RPM1 domain can initiate a pathogen-independent HR in N. benthamiana, we generated nine RPM1 fragments including single domains, based on secondary structure predictions: CC-1, CC-2, CC-3, CC-4, CC-5, NB-ARC, LRR, CC-NB-ARC, and NB-ARC-LRR (SI Appendix, Figs. S1 and S2A). When transiently overexpressed in N. benthamiana, none of the individual domains or fragments of RPM1 induced HR-like cell death (SI Appendix, Fig. S2B). Coexpression of either epitope-tagged phosphomimetic RIN4T166D or wild-type RIN4 together with AvrRpm1 did not result in the activation of any RPM1 fragment (SI Appendix, Fig. S2C).
In addition, we were unable to reconstitute RPM1-induced cell death by co-overexpressing complementary domains in trans, i.e., CC, NB-ARC, and LRR (SI Appendix, Fig. S2C). This is different from the potato CNL RX or the tomato CNL Mi-1.2, in which in trans complementation of cell death induction by coexpression of individual domains has been demonstrated (16, 47). Introduction of the autoactivating MHD mutation D505V (26) into NB-ARC, CC-NB-ARC, or NB-ARC-LRR did not render any of these fragments autoactive. As a control for the negative effects of epitope tags on domains, we demonstrated that overexpression of identical but non–epitope-tagged RPM1 domains also did not lead to an HR (SI Appendix, Fig. S2D). All fragments accumulated to high levels, or to levels comparable to those of full-length RPM1 (SI Appendix, Fig. S2E).
RPM1 functions at the PM, where it interacts with RIN4. To analyze whether the loss of function of the different RPM1 domains tested could be due to loss of membrane localization, we conducted cell fractionation experiments with transiently expressed RPM1 domains. We found that all analyzed fragments localized to the membrane fraction, the CC, NB-ARC and CC-NB-ARC fragments more prominently than the LRR or NB-ARC-LRR fragments (SI Appendix, Fig. S3). These results suggest that the loss of HR induction is not explained simply by mislocalization of the fragments.
To test whether the absence of HR induction by the membrane-localized CC or CC-NB-ARC fragments in N. benthamiana is due to the heterologous expression system, we generated stable transgenic Arabidopsis plants expressing either of two C-terminal epitope-tagged RPM1 CCs (CC-2 and CC-4) or the CC-NB-ARC fragments under the control of either estradiol-inducible or the tobacco mosaic viral 35S promoters in rpm1-3 and rpm1-3 rps2-101c rin4 pRIN4::T7-RIN4 (T7-RIN4 r1r2r4) plants, respectively. We did not observe any morphological differences in the transgenic plants compared with the rpm1-3 mutant or T7-RIN4 r1r2r4 plants despite high expression levels from the 35S or the estradiol-inducible promoter after estradiol induction (SI Appendix, Fig. S2 F and G). This could be due to the presence of wild-type RIN4 suppressing any (auto) activity, as we previously demonstrated for RPM1D505V autoactivity (42). Therefore, we used two independent transgenic lines for each construct to determine whether the CC-2, CC-4, or CC-NB-ARC fragment could be activated by bacteria-delivered AvrRpm1. We found no HR in any of the infected transgenic plants, confirming the transient N. benthamiana results indicating that RPM1 fragments are insufficient to trigger the HR (SI Appendix, Fig. S2 F and G). Taken together, these data suggest that RPM1 function cannot be recapitulated by these fragments, supporting our contention that it is dependent on the integrity and cooperation of all domains.
In Planta RPM1–RIN4 Interaction Is Mediated by All RPM1 Domains.
RPM1 interaction with RIN4 at the PM is necessary to perceive effector-mediated phosphorylation on RIN4 Thr166 and is required to activate RPM1 (41, 42). Initial yeast two-hybrid experiments indicated that the first 176 amino acids of RPM1 (including the CC domain plus the linker region and a small part of the NB-ARC domain; SI Appendix, Fig. S1A) could interact weakly with RIN4 (40). To extend these observations, we measured the interaction of RPM1 domains with wild-type RIN4 and the phosphomimetic mutant RIN4T166D in planta. We performed coimmunoprecipitation (co-IP) analysis of transiently expressed RPM1 fragments and either RIN4 or RIN4T166D. We observed a strong interaction of wild-type RIN4 with the CC-1, NB-ARC, and NB-ARC-LRR fragments and only a very weak interaction with the other CC fragments, CC-NB-ARC, and LRR (SI Appendix, Fig. S4A). Interestingly, the CC fragments lost the ability to interact with RIN4T166D, whereas interactions of the NB-ARC, NB-ARC-LRR, and LRR with RIN4T166D were still observed (SI Appendix, Fig. S4B).
We next addressed whether RIN4 mutations that either abolish RIN4 Thr166 phosphorylation (T166A) or block RIN4–RPM1 interaction (F169A) (42) affect the association with the CC-1, NB-ARC, or LRR domain. We included the longer CC-2 fragment in our analysis, because this CC fragment ends exactly where the NB-ARC fragment starts (SI Appendix, Figs. S1 and S2A). This experiment confirmed the loss of interaction of RIN4T166D with CC-1 (SI Appendix, Fig. S4C) and the lack of interaction of any RIN4 allelic variant with the CC-2 fragment (SI Appendix, Fig. S4D). The NB-ARC and LRR fragments both interacted with RIN4 regardless of the phosphomimetic or phospho-dead status of Thr166 (SI Appendix, Fig. S4 E and F). As has been reported for full-length RPM1 (42), the interaction of NB-ARC and the LRR with RIN4 was lost with the RIN4F169A allele (SI Appendix, Fig. S4 E and F). These results indicate that off-state RPM1–RIN4 interaction is mediated by all RPM1 domains, and that the associations of the NB-ARC and LRR domains with RIN4 are not affected by the phosphomimetic or phospho-dead mutations T166D and T166A, respectively. In addition, phosphorylation of RIN4 Thr166 alters interactions with RPM1, leading to the loss of RIN4–CC interaction, potentially stimulating or stabilizing the activated state of RPM1.
RPM1 Self-Association Is Constitutive and P-Loop Dependent.
Self-association and/or dimerization of NLR proteins or their N-terminal domains has been demonstrated to be necessary for immune signaling in animals and plants (31, 32, 45, 48, 49) (SI Appendix, Table S1). Therefore, we asked whether RPM1 also self-associates, and whether such self-association is necessary for RPM1 function. Coexpression of differentially epitope-tagged RPM1 proteins in N. benthamiana followed by co-IP revealed self-association of the resting-state RPM1 protein in planta (Fig. 1A). To exclude the possibility that this self-association is due merely to transient overexpression in the heterologous system, we generated stable double-transgenic Arabidopsis lines by crossing a well-characterized C-terminally myc epitope-tagged RPM1 line (50) with a newly generated GFP epitope-tagged RPM1 line that fully complements the rpm1-3 mutant (SI Appendix, Fig. S5A). Both differentially epitope-tagged RPM1 proteins were expressed from the native RPM1 promoter. Co-IP experiments demonstrated self-association of resting-state RPM1 in Arabidopsis (Fig. 1B and SI Appendix, Fig. S5B), suggesting that RPM1 exists as at least a dimer in the preactivation state.
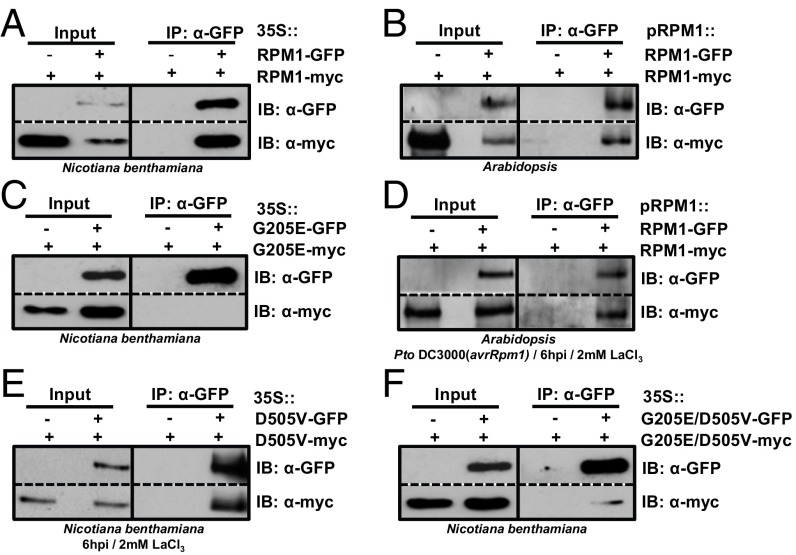
Fig. 1.
RPM1 constitutively self-associates preactivation and postactivation. (A) Self-association of 35S::RPM1-myc and 35S::RPM1-GFP. RPM1 constructs were transiently expressed in N. benthamiana, immunoprecipitated with anti-GFP magnetic beads, and then immunoblotted for both anti-myc and anti-GFP to assess input, immunoprecipitation, and co-IP. (B) Co-IP of native promoter-driven RPM1-myc and RPM1-GFP in transgenic Arabidopsis. Protein interaction was tested as described in A. (C) P-loop mutant RPM1G205E does not self-associate. Myc- and GFP-tagged 35S::RPM1G205E were transiently expressed in N. benthamiana and immunoprecipitated with anti-GFP magnetic beads. (D and E) Self-association of (D) effector-activated RPM1 in transgenic Arabidopsis (D) and of autoactive RPM1D505V in N. benthamiana (E). (D) Transgenic Arabidopsis, expressing both myc- and GFP-tagged RPM1 from the native RPM1 promoter, were hand-infiltrated with Pto DC3000 expressing avrRpm1 in 10 mM MgCl2 plus 2 mM LaCl3. Microsomal lysates were extracted at 6 h postinfection (hpi) and immunoprecipitated with anti-GFP beads. (E) Expression of C-terminally myc- and GFP-tagged RPM1D505V was induced with 20 μM estradiol plus 2 mM LaCl3 at 24 h postinfiltration in N. benthamiana leaves. Lysates were extracted at 6 h after estradiol induction (6 hpi) and immunoprecipitated with anti-GFP beads. (F) Self-association of RPM1G205E/D505V in N. benthamiana. Protein interaction was tested as described in A. IB, immunoblot.
We next analyzed whether this self-association is dependent on a functional P-loop, as has been demonstrated for the TNLs N and RPP1 (33, 45) (SI Appendix, Table S1). We transiently coexpressed differentially epitope-tagged P-loop loss-of-function mutant RPM1G205E (26) for co-IP analysis. Our results showed that a functional P-loop, and thus nucleotide binding, is required for resting-state RPM1 self-association (Fig. 1C).
We then tested whether effector-activated RPM1 or the autoactive MHD mutant RPM1D505V also self-associate. To prevent rapid degradation of active RPM1 (26, 50) and thus ensure sufficient protein for co-IP analysis, we blocked RPM1 degradation with lanthanum (LaCl3) treatment in these experiments. LaCl3 blocks the activity of divalent cation channels, mainly calcium channels, and also blocks RPM1-induced HR in Arabidopsis (51). We found that LaCl3 prevented the disappearance of activated RPM1 without altering effector (AvrRpm1)-induced posttranslational modification of RIN4 in our experiments, strongly suggesting that most, if not all, RPM1 precipitated in the presence of LaCl3 is in the activated state (SI Appendix, Fig. S6A). We also found that LaCl3 treatment in Arabidopsis did not affect bacterial growth of Pto DC3000 or growth restriction of Pto DC3000(avrRpm1) (SI Appendix, Fig. S6B), and thus that loss of HR is not due to a loss of bacterial fitness. LaCl3 also did not affect bacterial growth in rich medium (SI Appendix, Fig. S6C). Co-IP analysis of effector-activated myc- and GFP-tagged RPM1 in stable transgenic Arabidopsis and transiently coexpressed myc- and GFP-tagged autoactive RPM1D505V in N. benthamiana demonstrated self-association of active RPM1 in planta (Fig. 1 D and E). Taken together, these results suggest that RPM1 self-associates before and after activation, and that nucleotide binding (P-loop function) is important for resting-state RPM1 self-association.
We previously reported that the autoactivity of RPM1D505V is blocked by P-loop mutations in cis (RPM1G205E/D505V) (26). This prompted us to analyze whether RPM1G205E/D505V lost the ability to self-associate. To do so, we transiently coexpressed differentially epitope-tagged RPM1G205E/D505V in N. benthamiana and performed co-IP analysis. Similar to the P-loop mutant RPM1G205E, RPM1G205E/D505V lost self-association almost completely (Fig. 1F and SI Appendix, SI Materials and Methods). The loss-of-function phenotype of the cis double mutant (26) is likely due to a continual requirement for the P-loop in ATP binding and turnover (22, 23). Thus, we interpret our results as follows: RPM1 self-association depends on nucleotide binding, and a P-loop mutation alters NB-ARC domain structure or intramolecular domain interactions in such a way that both resting-state and active-state RPM1 self-association is inhibited or blocked.
RPM1 Plasma Membrane Localization Is P-Loop Dependent.
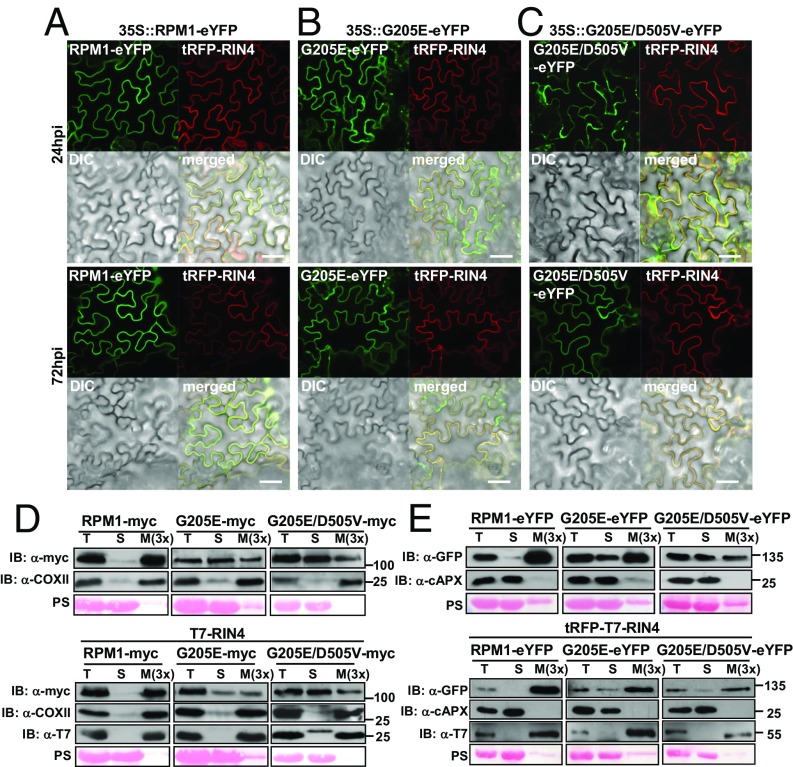
We previously reported that a functional P-loop is necessary for the proper localization of RPM1 to the microsomal fraction in cellular fractionation experiments (26) (SI Appendix, Fig. S5C). Thus, we examined the subcellular localization of RPM1, RPM1G205E, and the cis double-mutant RPM1G205E/D505V by confocal microscopy of C-terminally eYFP-tagged proteins transiently expressed in N. benthamiana. All three RPM1 proteins colocalized with tRFP-T7-tagged RIN4 at the PM (Fig. 2 A–C); however, we also observed a strong cytosolic fluorescence for RPM1G205E-eYFP and RPM1G205E/D505V-eYFP that almost disappeared at 72 h postinfiltration, likely due to degradation of the cytosolic (mis)localized proteins (Fig. 2 B and C). Cellular fractionation experiments in the presence or absence of coexpressed RIN4 confirmed the cytosolic localization of C-terminally myc- and eYFP-epitope tagged RPM1G205E and RPM1G205E/D505V in these experiments (26) (Fig. 2 D and E). Therefore, P-loop function is necessary for proper RPM1 PM localization, and the presence of Arabidopsis RIN4 is not sufficient to rescue the mislocalization of P-loop mutant RPM1G205E in N. benthamiana transient expression.
Fig. 2.
RPM1 PM localization requires a functional P-loop. (A –C) Live-cell imaging of RPM1-eYFP (A), RPM1G205E-eYFP (B), and RPM1G205E/D505V-eYFP (C) together with tRFP-T7-RIN4 illustrating PM localization of wild-type RPM1 and additional cytosolic localization of RPM1G205E and double-mutant RPM1G205E/D505V on transient expression in N. benthamiana. Leaves were imaged at 24 and 72 hpi. Note the reduced cytosolic fluorescence of RPM1G205E-eYFP and RPM1G205E/D505V-eYFP at 72 hpi. (Scale bar: 50 μM.) (D and E) Cell fractionation confirming cytosolic localization of RPM1G205E and RPM1G205E/D505V. (D) Agrobacteria containing either myc-tagged (D) or eYFP-tagged (E), 35S-driven RPM1 constructs were infiltrated either alone (Upper) or with pRIN4::T7-RIN4 (D) or pRIN4::tRFP-T7-RIN4 (E) (Lower) into N. benthamiana leaves. Tissue was harvested at 48 hpi for cell fractionation and immunoblotting with anti-myc (RPM1), anti-COXII (membrane marker), anti-APX (cytosol), and anti-T7 (RIN4) antibodies. Note that the band present in the anti-T7 blot in the soluble (S) fraction of the RPM1G205E/D505V-myc plus T7-RIN4 sample is nonspecific. Ponceau S (PS) staining served as loading control and marker for the cytosolic fraction. M(3×) indicates three times enrichment relative to T or S. IB, immunoblot; M, microsomal fraction; S, soluble; T, total extract.
RPM1 Function Is Affected by Mutations in Hydrophobic and Conserved Residues of the CC Domain.
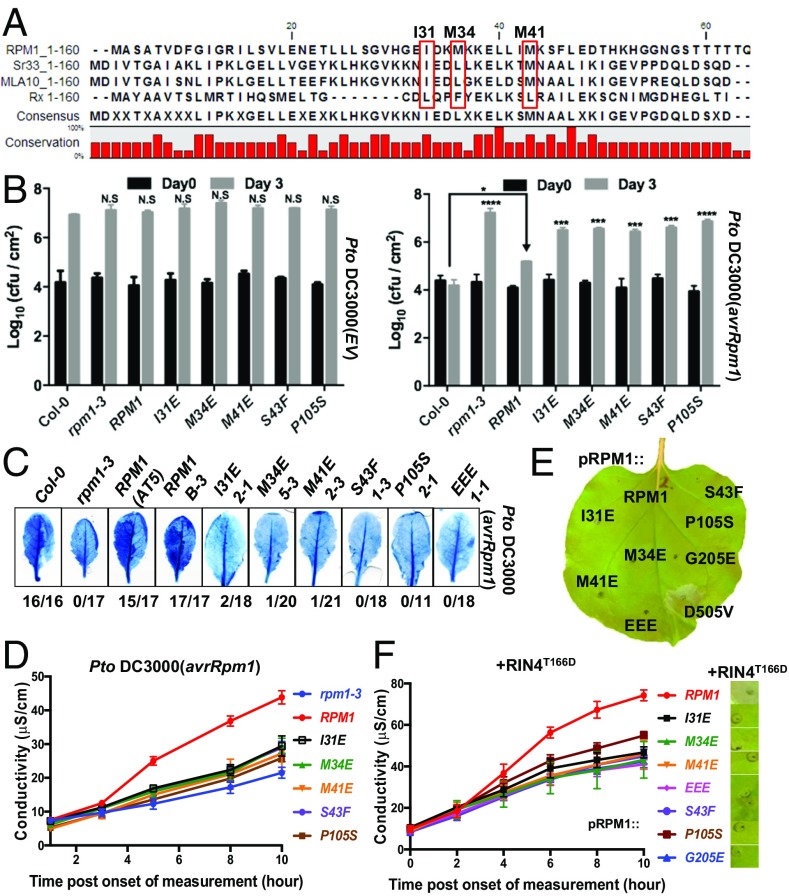
It was recently demonstrated that the solution structure of the wheat CNL Sr33 CC fragment differs significantly from that of the published crystal structure of a barley paralog, MLA10, and, rather surprisingly, resembles the structure of the CC fragment from the distantly related potato CNL Rx (52) solved in complex with its interacting protein RanGAP2 (53). Secondary structure prediction of an RPM1 CC fragment (amino acids 1–120) and homology modeling of this fragment onto the Sr33 CC domain NMR structure suggested that the RPM1 CC1–120 fragment also might adopt a four-helix bundle conformation (52) (SI Appendix, Figs. S7A and S8 A–D). Functional analyses demonstrated the importance of hydrophobic residues, located in the second helix of the four-helix bundle, in the CC domain for MLA10 function and CC dimerization (18). The hydrophobicity of these residues is conserved in RPM1, Sr33, and Rx (Fig. 3A), suggesting that these residues can be involved either in dimer formation or in holding together the monomeric four-helix bundle (52, 54). We wanted to know whether these conserved hydrophobic residues also play a role in RPM1 function. Our analyses also included two loss-of-function mutations in the CC domain that were isolated in a forward genetic screen to identify RPM1 mutations affecting the recognition of AvrRpm1 in Col-0 (55) (SI Appendix, Fig. S7A). These two mutations, S43F and P105S, map to the second helix and fourth helix, respectively, of the RPM1 CC domain defined by modeling onto the Sr33 CC solution structure with S43 and P105 likely surface-exposed (SI Appendix, Figs. S7A and S8 B and D).
Fig. 3.
Mutations in hydrophobic and conserved residues of the CC domain affect RPM1 function. (A) Alignment of amino acids 1–160 (only 1–62 shown) of RPM1, Sr33, MLA10, and Rx showing conservation of hydrophobic residues (highlighted; I31, M34, and M41) important for RPM1 function. (B) Loss of RPM1-induced bacterial growth restriction. Transgenic plants expressing the indicated RPM1 proteins were hand-infiltrated with Pto DC3000(EV) (Left) or Pto DC3000(avrRpm1) (Right). Error bars represent the SD among three samples. Asterisks indicate statistical significance calculated by one-way ANOVA. (C) Trypan blue staining of Arabidopsis leaves of indicated genotypes hand-infiltrated with Pto DC3000(avrRpm1) indicating loss of HR induction by RPM1 single mutants. The numbers below represent the ratio of total leaves infiltrated to leaves with clear HR and strong staining. (D) Quantitative measurement of cell death (conductivity) induced by wild-type RPM1 and RPM1 mutants on infiltration with Pto DC3000(avrRpm1) in transgenic Arabidopsis. (E) Cell death phenotype of eYFP-tagged RPM1 and indicated RPM1 alleles transiently expressed from the RPM1 promoter in N. benthamiana. (F) Mutations in the CC domain and the P-loop block the ability of RPM1 to induce HR when coexpressed with RIN4T166D. eYFP-tagged RPM1 proteins were expressed from the RPM1 promoter, T7-tagged RIN4T166D from the RIN4 promoter. Conductivity measurements and visual cell death in leaves expressing the indicated proteins are shown. EEE, RPM1I31/M34/M41E triple mutant; N.S, not significant. *P <0.05, ***P <0.001, ****P <0.0001.
We generated stable Arabidopsis transgenic plants for all five single mutants and infiltrated herbicide-resistant T2 plants with either Pto DC3000(EV) or Pto DC3000(avrRpm1) to measure bacterial growth restriction (Fig. 3B). None of the RPM1 alleles tested significantly restricted the growth of Pto DC3000(avrRpm1), and none affected Pto DC3000(EV) growth. Moreover, they all failed to induce HR on infiltration with a high inoculum of Pto DC3000(avrRpm1) (Fig. 3 C and D). Thus, all are loss-of- function alleles.
We also tested these mutant RPM1 proteins, as well as an RPM1I31/M34/M41E cis triple mutant, in our transient N. benthamiana reconstruction assays for their ability to induce cell death in response to coexpression of RIN4T166D. When transiently expressed from the weak RPM1 promoter, all accumulated, albeit to lower levels for the three hydrophobic residue alleles (SI Appendix, Fig. S7 I and J). None of the alleles was autoactive or able to be activated by RIN4T166D coexpression; all were loss of function (Fig. 3 E and F). However, transient overexpression from the 35S promoter allowed weak RIN4T166D-dependent RPM1 activation for the single mutations in the three hydrophobic residues (I31E, M34E, and M41E) and the conserved P105 (P105S) (SI Appendix, Fig. S7D). The RPM1S43F single mutant and the RPM1I31/M34/M41E triple mutant were not rescued by transient overexpression (SI Appendix, Fig. S7 D and F). Consistent with these findings, the single mutations I31E, M34E, M41E, and P105S altered effector-mediated activation of RPM1 only weakly following expression from the 35S promoter in transient overexpression conditions (SI Appendix, Fig. S7E), whereas neither the overexpressed RPM1I31/M34/M41E triple mutant nor RPM1S34F was activated by AvrRpm1 coexpression (SI Appendix, Fig. S7 E and F). This is also consistent with only the RPM1I31/M34/M41E triple mutant and RPM1S34F having a significant negative effect on the autoactivity of RPM1D505V in cis when transiently overexpressed (SI Appendix, Fig. S7 G and H). None of the tested mutants was autoactive when expressed from the 35S promoter (SI Appendix, Fig. S7 B and C). These results suggest weak effects of P105 and additive effects of the three hydrophobic residues I31, M34, and M41, as well as a strong effect for S43 on RPM1 activation in transient overexpression in N. benthamiana. We conclude that mutations in these hydrophobic and conserved residues represent at least partial loss-of-function alleles and putatively alter the structural requirements for RPM1 function or interdomain and intradomain interactions.
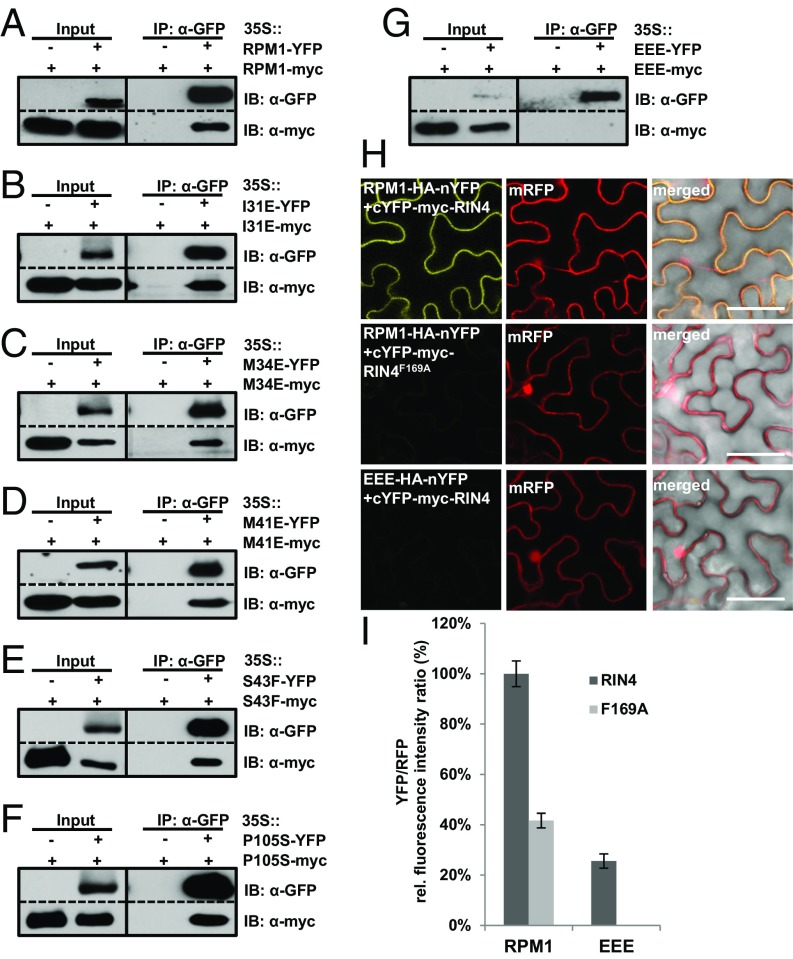
Loss-of-Function Mutations in Hydrophobic and Conserved Residues of the CC Domain Affect RPM1 Self-Association.
The loss-of-function phenotype of the RPM1 mutants that we tested prompted us to analyze whether these mutations also affect RPM1 self-association. We transiently coexpressed each single mutant and tested for self-association by co-IP. Each of these RPM1 alleles retained the ability to self-associate (Fig. 4 A–F). The proposed surface-exposed positions of S43 and P105 in the putative CC domain monomer model (SI Appendix, Fig. S8 B and D) suggests that these two residues could function in self-association; however, we did not observe any effect on self-association when these positions were mutated in the full-length RPM1 protein. Thus, they likely are involved in signaling or association with downstream interactors.
Fig. 4.
CC domain mutations affect full-length RPM1 self-association in an additive manner. (A–G) Co-IP of transiently expressed RPM1 indicating single and RPM1I31/M34/M41E triple mutants in N. benthamiana. RPM1 proteins were expressed from the 35S promoter. Total protein extract was immunoprecipitated with anti-GFP magnetic beads, and membranes were immunoblotted for anti-GFP or anti-myc to assess input, immunoprecipitation, and co-IP. (H) RPM1I31/M34/M41E loses the interaction with RIN4. In BiFC analyses of RPM1–RIN4 and RPM1I31/M34/M34E–RIN4 interactions in N. benthamiana, both RPM1 alleles and RIN4 were expressed from the 35S promoter from a single expression vector. RIN4F169A served as a control for loss of RPM1 interaction (42). (Scale bar: 50 mm.) (I) Quantification of the ratio of mean YFP fluorescence to RFP indicating the interaction of RPM1 with RIN4, but not with RIN4F169A, and the loss of interaction of RPM1I31/M34/M41E with RIN4. Data are mean ± SEM of 8–12 images selected at random over the surface of two leaf samples in each case. EEE, RPM1I31/M34/M41E triple mutant; IB, immunoblot; N.S, not significant.
Loss of RPM1 activity due to single mutations in any of the hydrophobic residues also is not caused by a mislocalization, given that all properly localized to the membrane in cell fractionation analysis (SI Appendix, Fig. S9A). To determine whether a combination of mutations of all three hydrophobic residues in cis might be necessary to completely block RPM1 self-association, we tested the strong loss-of-function RPM1I31/M34/M41E triple mutant in our co-IP analysis, and noted that it indeed lost self-association in planta (Fig. 4G). Interestingly, and in contrast to the loss of self-association mutants RPM1G205E and RPM1G205E/D505V, the PM localization of the RPM1I31/M34/M41E triple mutant was not affected (SI Appendix, Fig. S9 B and C). Thus, we anticipate that self-association per se is not sufficient for PM localization or function, but is required for signaling, potentially via interactions with RIN4, phosphorylated RIN4, or other signaling partners.
RPM1–RIN4 Interaction Is P-Loop Dependent and Affected by Loss-of-Function Mutations in the CC Domain.
To investigate whether these mutants express altered interactions with wild-type RIN4 or phosphomimetic RIN4T166D, we coexpressed each RPM1 single mutant or the RPM1I31/M34/M41E triple mutant together with wild-type RIN4 or with RIN4T166D in N. benthamiana and tested for interactions by co-IP. We also included the nonfunctional, mislocalized, and loss of self-association mutant RPM1G205E. All CC domain-localized single mutants retained the in planta interaction with both wild-type RIN4 and the phosphomimetic RIN4T166D (SI Appendix, Fig. S9 D and E). However, we consistently observed much weaker interactions of RIN4 and RIN4T166D with RPM1 alleles with mutations in any of the three hydrophobic residues I31, M34, and M41. This observation was supported by our finding that interaction of the triple-mutant RPM1I31/M34/M41E with wild-type RIN4 was lost compared with the single mutants (SI Appendix, Fig. S9D). We confirmed the loss of RPM1I31/M34/M41E–RIN4 interaction by bimolecular fluorescence complementation (BiFC) analysis in N. benthamiana (Fig. 4 H and I). The interaction of RPM1I31/M34/M41E with phosphomimetic RIN4T166D was also abolished (SI Appendix, Fig. S9E), suggesting that the integrity of the hydrophobic core of the RPM1 CC domain formed by residues I31, M34, and M41 is important not only for RPM1 self-association, but also for RPM1–RIN4 interaction both before and after activation.
We demonstrated that the P-loop mutant RPM1G205E lost interaction with both RIN4 and RIN4T166D (SI Appendix, Fig. S9 D and E), indicating that P-loop function is important for RIN4 interaction and supporting the notion that RPM1 self-association is crucial for RPM1–RIN4 interaction. To test whether restoration of strong PM localization of RPM1G205E could rescue the loss of RIN4 interaction, we tethered this mutant to the PM by adding the first 12 amino acids of calcineurin B-like protein 1 (CBL1) to the N terminus. This N-terminal fusion can target proteins to the PM due to myristoylation and palmitoylation of residues Glycine 2 and Cysteine 3, respectively (56). We previously demonstrated that CBL-tagged wild-type RPM1 is functional, but that functionality of RPM1G205E and RPM1G205E/D505V cannot be restored by forcing these mutants to the PM (26). We included the RPM1G205E/D505V cis double mutant in our experiment to also test whether the addition of the MHD mutation has any effect on RPM1–RIN4 interaction. As expected, we observed an interaction of the functional CBL-RPM1 wild-type protein with RIN4 (SI Appendix, Fig. S9G); however, the interaction of CBL-RPM1G205E and CBL-RPM1G205E/D505V with RIN4 was not restored, consistent with their loss of function, even when tethered to the PM (26) (SI Appendix, Fig. S9G).
All RPM1 Domains Contribute to Self-Association.
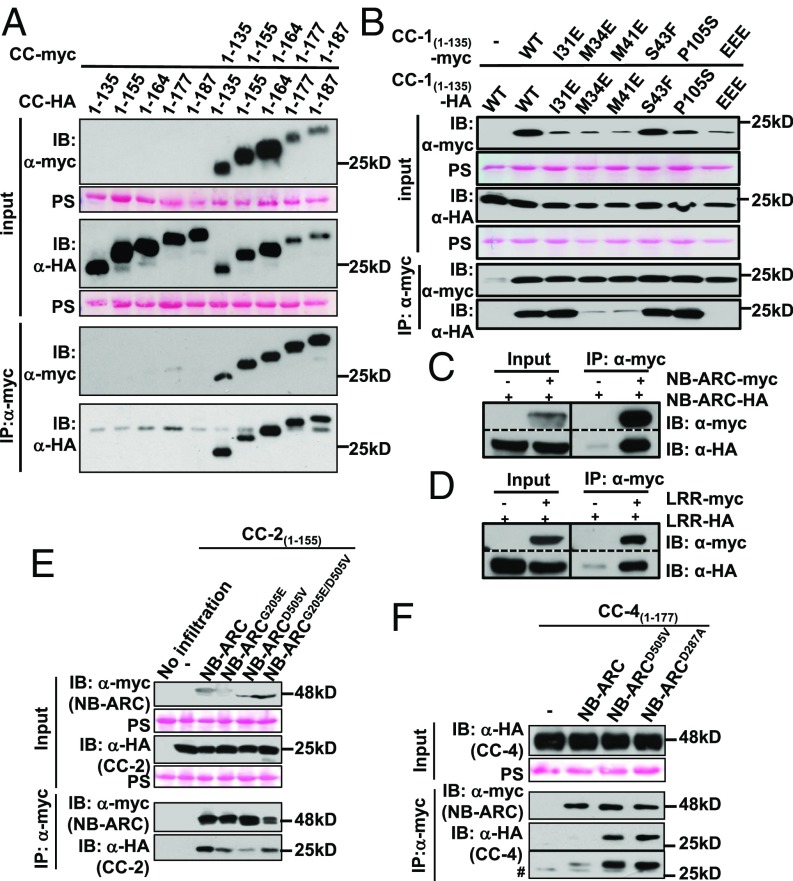
CC domain dimerization has been shown to be important for function of the barley CNL MLA10 (18). Recently, it was shown that only relatively longer, functional fragments of the MLA10, Sr33, and Sr50 CC domains dimerize in planta, whereas shorter, nonfunctional CC domain fragments that are disrupted in one alpha-helix presumed to be important for CC function do not (44, 52). Thus, we wanted to know whether the RPM1 CC domain also self-associates in planta. We coexpressed each of the five CC domain fragments with differential epitope tags in N. benthamiana and analyzed self-association by co-IP. In contrast to MLA10, Sr33, and Sr50, all tested RPM1 CC-domain fragments self-associated regardless of their length (Fig. 5A). The length of the self-associating RPM1 CC-1 fragment (amino acids 1–135) corresponds to the length of the short, nonfunctional, and non–self-associating Sr33 fragment (amino acids 1–130) (alignment in SI Appendix, Fig. S7A). However, the RPM1 CC-1 fragment (amino acids 1–135) includes the full predicted alpha-helical region possibly important for CC function (SI Appendix, Fig. S7A), as is the case for the larger and functional Sr33 CC fragment (amino acids 1–142) (44, 52). Therefore, all of our CC fragments should include the last alpha-helix, a structural characteristic important for MLA10 and Sr33 CC function. Thus, it seems likely that a complete four-helix bundle conformation, rather than a certain length, is important for CC self-association and function. We also confirmed self-association of the CC-2 fragment in BiFC analysis (SI Appendix, Fig. S10A), and demonstrated that the NB-ARC and LRR domains of RPM1 self-associate (Fig. 5 C and D). Thus, all domains of RPM1 can self-associate in planta and therefore might all contribute to self-association of full-length RPM1.
Fig. 5.
Intradomain and interdomain interactions contribute to RPM1 self-association. (A) In planta self-association of CC fragments of different lengths. All five CC fragments were transiently expressed in N. benthamiana leaves, and total protein extract was immunoprecipitated with anti-HA magnetic beads. Membranes were immunoblotted for anti-myc and anti-HA to assess input, immunoprecipitation, and co-IP. (B) CC domain mutations affect CC self-association in co-IP analysis of transiently expressed CC-1 fragments in N. benthamiana. Samples were processed as described in A. (C and D) Self-association of NB-ARC (C) and LRR (D) domains transiently expressed in N. benthamiana. Protein self-association was tested as described in A. (E) Mutations affecting nucleotide binding in the NB-ARC domain influence CC–NB-ARC interactions. CC-2 was transiently coexpressed with NB-ARC, NB-ARCG205E, NB-ARCD505V, and NB-ARCG205E/D505V to assess interaction by co-IP. Proteins were sampled as described in A. (F) NB-ARC alleles with mutations affecting ATP binding (D505V) and hydrolysis (D287) enhance CC-4–NB-ARC interactions. Proteins were transiently coexpressed in N. benthamiana to assess interaction by co-IP. The second immunoprecipitation panel, labeled with #, denotes longer exposure of the-HA (CC-4) blot. Proteins were sampled as described in A. Ponceau staining (PS) of the RuBisCO large subunit is shown as a protein-loading control for the input. EEE, RPM1I31/M34/M41E triple mutant; IB, immunoblot.
CC Dimerization Is Affected by Mutations in the Hydrophobic Residues of the CC Domain.
We next tested whether the CC domain mutations that render RPM1 inactive alter or inhibit the RPM1–RIN4 interaction and, at least as a triple mutant, abrogate full-length RPM1 self-association also affect self-association as isolated CC-1 fragments. We found that mutations in the hydrophobic residues M34 and M41 dramatically weakened CC-1 self-association, and, as for the full-length RPM1I13,M34,M41E, a combination of all three mutations completely disrupted CC-1 self-association (Fig. 5B). We also found a loss of self-association on BiFC analysis of the longer CC-2 fragment when all three residues were mutated (SI Appendix, Fig. S10 A and B). This finding indicates that alteration of these residues disrupts the intramolecular and intermolecular interactions of RPM1 important for function and thus could also affect possible interactions with downstream signaling components.
The RPM1 CC–NB-ARC Interaction Is Affected by Mutations in Residues Required for Nucleotide Binding.
Regulated intramolecular CC–NB-ARC interactions have been shown to be important for the function of some NLRs (27, 47). It is assumed that on activation, the intramolecular CC–NB-ARC interaction is altered in such a way that the signal-competent N-terminal domain is released from intramolecular repression, concomitant with, or as a result, of nucleotide exchange. We wanted to analyze whether the RPM1 CC domain interacts with the NB-ARC domain, and whether this interaction is altered on activation. Because none of the tested RPM1 fragments or domains was (auto)active in our in planta expression systems, we mimicked the activated state by introducing NB-ARC mutations generally known to affect nucleotide binding and hydrolysis in NLR proteins into our NB-ARC domain (28). We analyzed interactions of the CC-2 fragment (amino acids 1–155) with wild-type or mutated NB-ARC domains (amino acids 156–535) by co-IP of transiently overexpressed proteins in N. benthamiana. We observed a weakened interaction of CC-2 with either loss-of-function mimic NB-ARCG205E or autoactive mimic NB-ARCD505V mutant alleles (Fig. 5E). This was also the case in the interaction of the CC-2 with the P-loop MHD cis double-mutant NB-ARCG205E/D505V domain (Fig. 5E). Interestingly, when using the longer CC-4 fragment (amino acids 1–177), we observed an enhanced interaction with the NB-ARC domain harboring either of two autoactivating mutations in the MHD (D505V) or the Walker B (D287A) motifs (Fig. 5F). This suggests that the N-terminal amino acids (155–177) of the NB-ARC domain are responsible for a tighter CC-4–NB-ARC interaction on ATP binding (or the mimicking of ATP binding by MHD mutation).
Discussion
To successfully defend themselves against pathogens, animals and plants evolved efficient innate immune systems. In both kingdoms, proteins of the NLR family are responsible for the detection of effectors (and in animals MAMPs as well) and the subsequent induction of an appropriate immune response (1, 13, 14). In plants, the activation of NLR proteins is often accomplished by at least self-association of either the N-terminal (TIR or CC) domain or the full-length receptor (17, 18, 44, 46, 49, 52, 57, 58). Here we report that constitutive self-association of the PM-localized Arabidopsis CNL RPM1 is required for its function, and that all domains—the N-terminal CC, the central NB-ARC, and the C-terminal LRR domain—contribute to intermolecular interactions. Furthermore, our data support a model in which resting- and active-state RPM1 self-association and proper PM localization are P-loop dependent, and that conserved hydrophobic residues in the CC domain play important roles in RPM1 self-association, activation, and RIN4 interaction (Fig. 6).
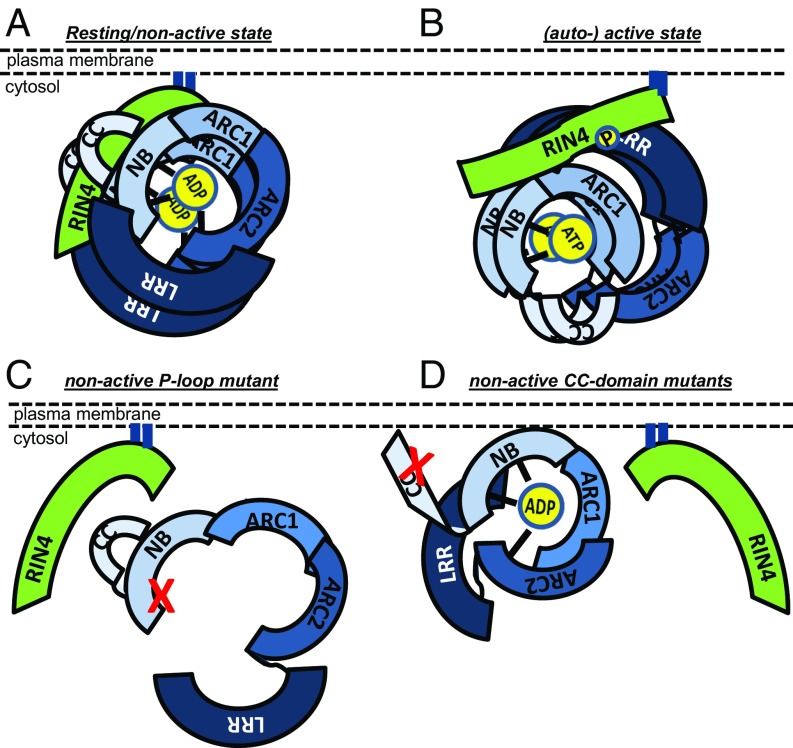
Fig. 6.
Model of RPM1 self-association and conformational switch on activation. (A) Resting/nonactive state: RPM1 interacts with RIN4 at the PM at least as a dimer in a closed conformation mediated by the NB-ARC, LRR, and CC domains. Intramolecular interactions between the CC and NB-ARC domains and intermolecular interactions mediated by all three domains contribute to RPM1 self-association. (B) Activated (and autoactive) RPM1 remains self-associated and in complex with RIN4. Intramolecular interactions between the CC and NB-ARC domains and interaction of the CC domain with phosphorylated RIN4 are altered and thus might allow for the recruitment of downstream signaling partners. (C) RPM1 resting-state self-association, full PM localization, and RIN4 interaction are dependent on a functional P-loop motif. P-loop mutation is indicated by the red “X” in the NB part of the NB-ARC domain. (D) Mutations in conserved and/or hydrophobic residues of the CC domain affect RPM1–RIN4 interaction, but not PM localization, presumably by altering intramolecular and intermolecular interactions important for activation and/or signaling. Mutations are indicated by a red “X” in the CC domain.
NLR-mediated immune signaling in animals and plants has been proposed to be dependent on their N-terminal domains, and in many cases (over)expression of this domain by itself is able to induce cell death signaling (18, 19, 45, 59). We were not able to induce cell death in either of our expression systems, transient in N. benthamiana or stable in transgenic Arabidopsis, regardless of the RPM1 domain, fragment, or combinations of domains or whether we used the phosphomimetic RIN4T166D to activate RPM1 (SI Appendix, Fig. S2). This suggests that tightly regulated cooperation of all of its domains is required for immune signaling initiated by RPM1. We previously demonstrated that RPM1 signals from the PM (26), and here we have extended this concept by showing that RPM1 membrane localization can be mediated by either the CC or the NB-ARC domain (SI Appendix, Fig. S3). Therefore, it is plausible to assume that the CC-NB-ARC domain drives RPM1 to the membrane, where RPM1 interacts with RIN4, which has a carboxyl-terminal acylation/prenylation site required for its localization and function (60), and potentially other components important for signaling. Interactions with putative signaling partners are very likely mediated by more than one or all RPM1 domains, and thus their integrity is necessary for signal induction. This idea is further supported by our finding that all RPM1 domains are involved in interaction with its guardee RIN4 in planta (SI Appendix, Fig. S4). This is unlike observations reported for other plant NLRs. In those cases, the N-terminal domains of the NLR mediate the interaction with their guardee, whereas the LRR domain is proposed to negatively regulate the NB-ARC and CC or TIR domains, thus keeping the NLR inactive (15, 47, 61–64). Alternatively, all domains of RPM1 are required for the CC domain to adopt the signal-competent state required for function. This model is consistent with the oligomerization observed on activation of animal NLRC4, where the N-terminal signaling domains appear to form a functional signaling platform only when the activated protein oligomerizes via its central NB-ARC, helical domain 1, and winged helix domain to form a functional inflammasome (31, 32). In this context, the functional capacity of any isolated N-terminal signaling domain would depend on its propensity to adopt and maintain a particular functional structure without contributions from any other domain of the full-length NLR. Once at the PM, RPM1 is in a stable association with RIN4, and this association together with intramolecular interactions keeps RPM1 in its “off” state. This “off”-state RPM1–RIN4 interaction depends on an intact P-loop and conserved hydrophobic residues in the RPM1 CC domain, which presumably are important for proper structural conformation of the CC domain and the intermolecular interactions necessary for RIN4 association. Upon effector-induced, host kinase-mediated phosphorylation of RIN4 Thr166, the intrinsically disordered RIN4 (65) might adopt a slightly different structure and contribute to RPM1 activation. This could drive RPM1 intramolecular rearrangements resulting in the loss of CC–RIN4 interaction, a weakened and/or changed interaction between the CC and NB-ARC domains, and eventually the exchange of ADP with ATP required for activation (Fig. 6B).
Complex intermolecular and intramolecular domain interactions have been hypothesized to be necessary for proper NLR activation, ADP-to-ATP exchange, and hydrolysis (11). It is widely accepted that the NB-ARC domain of plant NLRs adopts a more open conformation to allow for nucleotide exchange and to release the negative regulatory interactions with the N- or C-terminal domains on recognition of the appropriate pathogen signal (13). We noted a reduced, but not eliminated, interaction of the CC-2 fragment (amino acids 1–155) with RPM1 NB-ARC domains harboring mutations affecting nucleotide binding (Fig. 5E). Interestingly, when we used a slightly longer fragment of the CC domain (CC-4; amino acids 1–177) that overlapped with our NB-ARC domain by 22 amino acids, we observed a tighter interaction with the NB-ARC domain mutated in the MHD (ATP-binding) or Walker B (ATP hydrolysis) motif (Fig. 5F). Taken together, these findings indicate involvement of the linker region between CC and NB-ARC and of the N-terminal region of the NB-ARC domain—namely, the first part of the NB subdomain—in the intramolecular and also likely the intermolecular interactions (oligomerization) that accompany RPM1 activation. This is consistent with recovery of a loss-of-function RPM1 allele at residue G174 (55), which is extremely conserved in RPM1 orthologs as well as in most Arabidopsis CNLs (SI Appendix, Figs. S7A and S11).
Self-association/oligomerization of NLR proteins can occur either preactivation or postactivation, or can remain constant in both states (SI Appendix, Table S1). RPM1 self-association does not depend on activation status (Fig. 1); however, self-association is P-loop dependent. P-loop–dependent self-association on activation has been reported for the tobacco TNL N, Arabidopsis TNL RPP1Nd, and human NLRC4 proteins (31–33, 45, 46, 66). P-loop–dependent self-association is consistent with ATP binding being necessary for activation. Mutations in the MHD motif are thought to promote a conformational change, opening up the NB-ARC domain to allow for faster or easier exchange of ADP to ATP (67, 68). In addition, a mutation in the MHD motif of the flax TNL M exhibits higher affinity for ATP than for ADP, suggestive of a conformational change on ATP binding and/or activation (28). The RPM1G205E/D505V double mutant is not active and thus presumably does not bind ATP, as has been demonstrated for an equivalent cis double-mutant MK286L/D555V of the flax M NLR (23, 28) and thus also loses self-association. However, we speculate that activation of RPM1 leads to a conformational change that either restores or does not affect self-association, likely through the formation of a second, stable intramolecular interaction/oligomerization surface. Consistent with this hypothesis, the generation of an activation-dependent oligomerization surface also has been shown to be necessary for formation of the mammalian Prgj-NAIP2-NLRC4 inflammasome (31). It will be interesting to see whether a similar in cis double mutation in other NLRs also affects self-association and, in the case of inflammasome- or apoptosome-forming animal NLRs, function.
The recently published structural analysis of the CC fragments (amino acids 1–120) of wheat Sr33, rye Sr50, barley MLA10, and potato Rx NLRs suggests a shared and conserved monomeric four-helix bundle conformation of this domain (52). However, secondary structure predictions and functional analysis of a longer CC fragment (amino acids 1–160) of these CNLs indicated that the shorter CC fragment used to solve the respective structures lacks a C-terminal alpha-helix and is not functional; only longer CC fragments (consisting of at least amino acids 1–142) are autoactive (for cell death signaling in transient expression in N. benthamiana) and can self-associate in planta (44). Secondary structure predictions of the RPM1 CC domain indicate that all CC constructs used in this study should include the full C-terminal alpha-helix, likely explaining their observed self-association, yet none was autoactive (Fig. 5 and SI Appendix, Fig. S2). Hydrophobic residues, shown to be important for MLA10 function and dimerization, are highly conserved in RPM1 (18) (Fig. 3A). We have shown that these residues are also necessary for full activity of RPM1 and, in an additive manner, also for self-association of either the full-length protein or the CC domain. Loss of function of the single mutants RPM1I31E, RPM1M34E, and RPM1M41E is likely due to a weakened interaction of these mutants with RIN4 or RIN4T166D (SI Appendix, Fig. S9 D and E), resulting from loss of or altered CC folding (Fig. 5B). Their combination into the RPM1I31/M34/M41E cis triple mutant had an additive effect on RPM1 function. Taken together, these findings suggest that these residues are important for the structural conformation of the CC domain necessary for self-association, and thus affect intramolecular interactions important for RIN4 interaction and consequent RPM1 activation/signaling. We favor a model in which the CC and LRR domains interact with the “off”-state ADP-bound NB-ARC domain and both the CC and NB-ARC domains are necessary and sufficient for RPM1–RIN4 interaction at the PM. On effector-induced phosphorylation of RIN4 Thr166, the RIN4–CC domain association is released, and the CC–NB-ARC interaction eventually changes to allow the exchange of ADP by ATP, thus activating RPM1. Activated RPM1–RIN4 interactions would then be mediated by the NB-ARC and LRR domains (SI Appendix, Fig. S4). Phosphorylated RIN4 could stabilize such an activated RPM1 confirmation, as reflected by enhanced interaction of RPM1 with RIN4T166D (42).
We also analyzed the effect of two loss-of-function mutations, S43F and P105S, in the CC domain (55). Self-association of either the isolated CC domain or full-length RPM1 was not affected by either of these mutations. Thus, although S43 and P105 are required for RPM1 function, they are dispensable for resting-state self-association. However, our results show that transient co-overexpression of RIN4T166D does not activate RPM1S43F, but does activate RPM1P105S. The inability of RPM1S43F to be activated by RIN4T166D is not due to a loss of interaction, but could be explained by the structural requirements of S43 for RPM1 function or interdomain interactions that must occur subsequent to RIN4T166D binding. However, the finding that the S43F mutation also completely blocks D505V autoactivity in cis suggests an important function of S43 for RPM1 signaling as well as a structural role. Although RPM1P105S blocks effector-mediated RPM1 activation in Arabidopsis and activation by RIN4T166D in coexpression experiments in N. benthamiana when expressed from the RPM1 promoter, it does not do so in transient overexpression from the 35S promoter (SI Appendix, Fig. S7). In addition, this allele does not block autoactivity of RPM1D505V (26). A proline-to-serine substitution at P105 should have only a very minor effect on RPM1 structure, because serine residues may be relatively common within tight turns on protein surfaces and the serine side chain hydroxyl oxygen could form a hydrogen bond with the protein backbone, effectively mimicking a proline (www.russelllab.org/aas/Ser.html). Therefore, RPM1 function is not (or only very weakly) affected by P105S in transient overexpression in N. benthamiana, but this P-to-S substitution is sufficient to effectively interfere with RPM1 function in response to pathogen-delivered AvrRpm1 or AvrB in Arabidopsis. This is consistent with the fact that RPM1P105S expressed from the RPM1 promoter cannot complement the rpm1-3 mutant (Fig. 3).
Recent detailed biochemical and genetic analyses of different plant NLRs have elucidated NLR regulation, activation, and function. It is now generally accepted that plant NLRs, like their counterparts in animals, at least dimerize to initiate an appropriate immune response (14); however, we still lack evidence for plant NLR oligomerization upon activation into high-molecular signaling complexes that are functionally similar to animal apoptosomes or inflammasomes. Furthermore, as yet there is no published structure of a full-length NLR (neither plant nor animal NLR), and thus the exact molecular mechanism of activation via N-terminal domain function remains obscure. It will be interesting to explore whether RPM1 self-association after activation leads to the formation of high-molecular weight complexes and to determine the effect of release of the CC–RIN4 interaction on activation. Does the CC domain recruit downstream signaling components to the RPM1-RIN4 complex, or is it responsible for RPM1 oligomerization and the formation of a signaling hub? Or does the CC itself provide the necessary functions for disease resistance?
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis thaliana was grown in walk-in growth rooms maintained at 21 °C/18 °C (day/night) with a 9-h/15-h (day/night) cycle. Transgenic Arabidopsis lines were generated using a standard floral dip technique (69). N. benthamiana was grown in a walk-in growth room maintained at 26 °C/22 °C with a 12-h/12-h day/night cycle and a LGM550 professional LED grow light system (www.led-grow-master.com/). The following Arabidopsis genotypes were used: Col-0, rpm1-3 (39), pRIN4::T7-RIN4 rpm1-3 rps2-102c rin4 (42), pRPM1::RPM1-myc rpm1-3 (50), and pRPM1::RPM1-myc rpm1-3 Dex::AvrRpm1-HA (70). The RPM1-myc and RPM1-GFP double-transgenic Arabidopsis line was generated by crossing the pRPM1::RPM1-myc rpm1-3 line with a homozygous pRPM1::RPM1-GFP rpm1-3 line (this study).
Coimmunoprecipitation and Western Blot Analysis.
Frozen N. benthamiana leaf tissue (∼100–200 mg) was collected and ground in a mortar and pestle with liquid nitrogen and then resuspended in 2 mL of extraction buffer (50 mM Hepes-KOH pH 7.5, 50 mM NaCl, 10 mM EDTA pH 8.0, 0.5% Triton X-100, and 5 mM DTT with 1× plant protease inhibitor mixture; Sigma-Aldrich). Soluble supernatants were cleared by centrifugation at 10,600 × g for 5 min and at 20,800 × g for 15 min at 4 °C and then incubated for 2 h with end-over-end turning at 4 °C with 35 μL of α-myc–, α-GFP–, or α-HA–conjugated magnetic beads (Miltenyi Biotec). Samples were captured with MACS separation columns (Miltenyi Biotec), and washed three times with washing buffer (extraction buffer with 0.2% Triton X-100 and 150 mM NaCl). Bound proteins were eluted in 120 μL of elution buffer (50 mM Tris⋅HCl pH 6.8, 50 mM DTT, 1% SDS, 1 mM EDTA pH 8.0, 0.005% bromophenol blue, and 10% glycerol). Samples were resolved by electrophoresis on 8% (for RPM1), 12.5% or 15% (for RPM1 fragments), and 12.5% (for RIN4) SDS/PAGE gels, transferred to nitrocellulose membranes, and blotted with primary antibodies overnight at 4 °C in 5% nonfat dry milk diluted in TBS with 1% Tween. Primary and secondary antibody dilutions were as follows: α-Myc, 1:1,000 (Santa Cruz Biotechnology); α-HA, 1:1,000 (Roche); α-T7 HRP-conjugated, 1:10,000 (Novagen); α-GFP, 1:1,000 (Roche); and α-mouse HRP conjugated, 1:7,500 (R&D Systems). In two out of three experiments, a very weak self-association of RPM1G205E/D505V was observed, which in light of the overexpression in these co-IP experiments might not be biologically relevant.
LaCl3 Treatment.
LaCl3 was applied to N. benthamiana or Arabidopsis as described previously (51.). For this, 2 mM LaCl3 was injected into N. benthamiana leaves at 1 h before infiltration of Agrobacterium cell suspension expressing RPM1D505V-GFP or RPM1D505V-myc. To monitor the effects of LaCl3 on RPM1-mediated disease resistance, 2 mM LaCl3 was applied to Arabidopsis transgenic plants conditionally expressing AvrRpm1 by dexamethasone (Dex) treatment. AvrRpm1 was induced with 20 μM Dex at 1 h after LaCl3 application. A bacterial growth assay with Pto DC3000(EV) and Pto DC3000(avrRpm1) was performed by infiltrating 1 × 105 cfu/mL of bacterial inoculum in 10 mM MgCl2 with or without 2 mM LaCl3 into rosette leaves of 4- to 6-wk-old Col-0 plants. The same number of bacterial cells (1 × 105 cfu/mL) was grown for 3 h, equivalent to day 0 growth in the bacterial growth assay, at 28 °C in King’s B medium in the presence and absence of 2 mM LaCl3 to monitor the effects of LaCl3 on bacterial growth in medium. The effect of LaCl3 on RPM1 intracellular localization was also monitored by confocal microscopy of transiently expressed 35S::RPM1-eYFP and 35S::RPM1D505V-eYFP in N. benthamiana leaves, and no change was observed compared with mock treatment (data not shown).
Supplementary Material
Acknowledgments
We thank Lisa K. Wünsch and Paul McIntosh for technical assistance; Dr. Tony Perdue for advice and help with the confocal microscopy; Lisa K. Wünsch, Dr. Sarah Grant, Dr. Marc Nishimura, Dr. Li Yang, Dr. Gabriel Castrillo, Dr. Oliver Furzer, Dr. Freddy Monteiro, Dr. Daniel Slane, and the two reviewers for a critical reading of the manuscript and constructive comments for its improvement. This work was supported by the National Science Foundation (Grant IOS-1257373, to J.L.D.), the National Key Research and Development Program of China (Grant 2016YFD010060, to Z.G.), and the National Nature Science Foundation of China (Grant 31270315, to Z.G.). J.L.D. is an Investigator of the Howard Hughes Medical Institute, supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (Grant GBMF3030). F.E.K. was supported by a German Research Foundation Postdoctoral Fellowship (EL 734/1-1), and R.G.A. was supported by an NIH Ruth L. Kirschstein NRSA Fellowship (F32GM108226).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708288114/-/DCSupplemental.
References
- 1.Jones JDG, Vance RE, Dangl JL. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354:aaf6395. doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 4.Tang D, Wang G, Zhou J-M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell. 2017;29:618–637. doi: 10.1105/tpc.16.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 6.Anderson RG, Deb D, Fedkenheuer K, McDowell JM. Recent progress in RXLR effector research. Mol Plant Microbe Interact. 2015;28:1063–1072. doi: 10.1094/MPMI-01-15-0022-CR. [DOI] [PubMed] [Google Scholar]
- 7.Baltrus DA, et al. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011;7:e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asai S, Shirasu K. Plant cells under siege: Plant immune system versus pathogen effectors. Curr Opin Plant Biol. 2015;28:1–8. doi: 10.1016/j.pbi.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Dou D, Zhou J-M. Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe. 2012;12:484–495. doi: 10.1016/j.chom.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Weßling R, et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe. 2014;16:364–375. doi: 10.1016/j.chom.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonardi V, Cherkis K, Nishimura MT, Dangl JL. A new eye on NLR proteins: Focused on clarity or diffused by complexity? Curr Opin Immunol. 2012;24:41–50. doi: 10.1016/j.coi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takken FL, Albrecht M, Tameling WI. Resistance proteins: Molecular switches of plant defence. Curr Opin Plant Biol. 2006;9:383–390. doi: 10.1016/j.pbi.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B. Animal NLRs provide structural insights into plant NLR function. Ann Bot. 2017;119:827–702. doi: 10.1093/aob/mcw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duxbury Z, et al. Pathogen perception by NLRs in plants and animals: Parallel worlds. BioEssays. 2016;38:769–781. doi: 10.1002/bies.201600046. [DOI] [PubMed] [Google Scholar]
- 15.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Ooijen G, Mayr G, Albrecht M, Cornelissen BJC, Takken FLW. Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain. Mol Plant. 2008;1:401–410. doi: 10.1093/mp/ssn009. [DOI] [PubMed] [Google Scholar]
- 17.Bernoux M, et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9:200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maekawa T, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9:187–199. doi: 10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Collier SM, Hamel L-P, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact. 2011;24:918–931. doi: 10.1094/MPMI-03-11-0050. [DOI] [PubMed] [Google Scholar]
- 20.Swiderski MR, Birker D, Jones JDG. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact. 2009;22:157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- 21.Wang G-F, et al. Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog. 2015;11:e1004674. doi: 10.1371/journal.ppat.1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernoux M, et al. Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell. 2016;28:146–159. doi: 10.1105/tpc.15.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tameling WIL, et al. The tomato R gene products I-2 and MI-1 are functional ATP-binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Hoorn RAL, Kamoun S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Z, Chung EH, Eitas TK, Dangl JL. Plant intracellular innate immune receptor resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci USA. 2011;108:7619–7624, and erratum (2011) 108:8915. doi: 10.1073/pnas.1104410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G-F, et al. Correction: Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog. 2015;11:e1004830. doi: 10.1371/journal.ppat.1004830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams SJ, et al. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact. 2011;24:897–906. doi: 10.1094/MPMI-03-11-0052. [DOI] [PubMed] [Google Scholar]
- 29.Navarro L, et al. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135:1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao Y, et al. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Z, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350:399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350:404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutierrez JR, et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 35.Qi D, Innes RW. Recent advances in plant NLR structure, function, localization, and signaling. Front Immunol. 2013;4:348. doi: 10.3389/fimmu.2013.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelhardt S, et al. Relocalization of late blight resistance protein R3a to endosomal compartments is associated with effector recognition and required for the immune response. Plant Cell. 2012;24:5142–5158. doi: 10.1105/tpc.112.104992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takemoto D, et al. N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol Plant Microbe Interact. 2012;25:379–392. doi: 10.1094/MPMI-11-10-0272. [DOI] [PubMed] [Google Scholar]
- 38.Grant MR, et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 39.Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell. 1994;6:927–933. doi: 10.1105/tpc.6.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Elmore JM, Lin Z-JD, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9:137–146. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung E-H, et al. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi D, DeYoung BJ, Innes RW. Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 2012;158:1819–1832. doi: 10.1104/pp.112.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cesari S, et al. Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proc Natl Acad Sci USA. 2016;113:10204–10209. doi: 10.1073/pnas.1605483113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber KJ, Bentham A, Williams SJ, Kobe B, Staskawicz BJ. Multiple domain associations within the Arabidopsis immune receptor RPP1 regulate the activation of programmed cell death. PLoS Pathog. 2016;12:e1005769. doi: 10.1371/journal.ppat.1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, et al. Multiple functional self-association interfaces in plant TIR domains. Proc Natl Acad Sci USA. 2017;114:E2046–E2052. doi: 10.1073/pnas.1621248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moffett P, Farnham G, Peart J, Baulcombe DC. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu F, Cheng YT, Kapos P, Huang Y, Li X. P-loop-dependent NLR SNC1 can oligomerize and activate immunity in the nucleus. Mol Plant. 2014;7:1801–1804. doi: 10.1093/mp/ssu097. [DOI] [PubMed] [Google Scholar]
- 49.Nishimura MT, et al. TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc Natl Acad Sci USA. 2017;114:E2053–E2062. doi: 10.1073/pnas.1620973114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant M, et al. The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 2000;23:441–450. doi: 10.1046/j.1365-313x.2000.00804.x. [DOI] [PubMed] [Google Scholar]
- 52.Casey LW, et al. The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc Natl Acad Sci USA. 2016;113:12856–12861. doi: 10.1073/pnas.1609922113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hao W, Collier SM, Moffett P, Chai J. Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2) J Biol Chem. 2013;288:35868–35876. doi: 10.1074/jbc.M113.517417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Kasmi F, Nishimura MT. Structural insights into plant NLR immune receptor function. Proc Natl Acad Sci USA. 2016;113:12619–12621. doi: 10.1073/pnas.1615933113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tornero P, Chao RA, Luthin WN, Goff SA, Dangl JL. Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell. 2002;14:435–450. doi: 10.1105/tpc.010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batistic O, Sorek N, Schültke S, Yalovsky S, Kudla J. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell. 2008;20:1346–1362. doi: 10.1105/tpc.108.058123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams SJ, et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- 58.Césari S, et al. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014;33:1941–1959. doi: 10.15252/embj.201487923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanabe T, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim H-S, et al. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mucyn TS, et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rairdan GJ, Moffett P. Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell. 2006;18:2082–2093. doi: 10.1105/tpc.106.042747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bendahmane A, Farnham G, Moffett P, Baulcombe DC. Constitutive gain-of-function mutants in a nucleotide-binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002;32:195–204. doi: 10.1046/j.1365-313x.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- 64.Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132:449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun X, et al. The intrinsically disordered structural platform of the plant defence hub protein RPM1-interacting protein 4 provides insights into its mode of action in the host-pathogen interface and evolution of the nitrate-induced domain protein family. FEBS J. 2014;281:3955–3979. doi: 10.1111/febs.12937. [DOI] [PubMed] [Google Scholar]
- 66.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 67.van Ooijen G, et al. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot. 2008;59:1383–1397. doi: 10.1093/jxb/ern045. [DOI] [PubMed] [Google Scholar]
- 68.Tameling WIL, et al. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 2006;140:1233–1245. doi: 10.1104/pp.105.073510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 70.Serrano M, Hubert DA, Dangl JL, Schulze-Lefert P, Kombrink E. A chemical screen for suppressors of the avrRpm1-RPM1-dependent hypersensitive cell death response in Arabidopsis thaliana. Planta. 2010;231:1013–1023. doi: 10.1007/s00425-010-1105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.