Significance
Craniosynostosis is a common congenital malformation resulting from premature fusion of the bones that comprise the cranial vault, requiring surgery in infancy to prevent adverse neurologic outcomes. Eighty-five percent of cases are non-syndromic and of unknown cause. By exome sequencing of families with non-syndromic midline craniosynostosis, we show that 5% of cases have de novo damaging mutations in negative regulators of the Wnt, bone morphogenetic protein (BMP), and Ras/ERK signaling pathways, developmental cascades that converge on common nuclear targets to promote bone formation. Another 5% have transmitted mutations in these pathways. Common variants near BMP2 show genetic interaction with these rare mutations. The results provide insight into pathophysiology and have immediate implications for the diagnosis and genetic counseling of families with craniosynostosis.
Keywords: de novo mutation, craniosynostosis, BMP signaling, Wnt signaling, Ras/ERK signaling
Abstract
Non-syndromic craniosynostosis (NSC) is a frequent congenital malformation in which one or more cranial sutures fuse prematurely. Mutations causing rare syndromic craniosynostoses in humans and engineered mouse models commonly increase signaling of the Wnt, bone morphogenetic protein (BMP), or Ras/ERK pathways, converging on shared nuclear targets that promote bone formation. In contrast, the genetics of NSC is largely unexplored. More than 95% of NSC is sporadic, suggesting a role for de novo mutations. Exome sequencing of 291 parent–offspring trios with midline NSC revealed 15 probands with heterozygous damaging de novo mutations in 12 negative regulators of Wnt, BMP, and Ras/ERK signaling (10.9-fold enrichment, P = 2.4 × 10−11). SMAD6 had 4 de novo and 14 transmitted mutations; no other gene had more than 1. Four familial NSC kindreds had mutations in genes previously implicated in syndromic disease. Collectively, these mutations contribute to 10% of probands. Mutations are predominantly loss-of-function, implicating haploinsufficiency as a frequent mechanism. A common risk variant near BMP2 increased the penetrance of SMAD6 mutations and was overtransmitted to patients with de novo mutations in other genes in these pathways, supporting a frequent two-locus pathogenesis. These findings implicate new genes in NSC and demonstrate related pathophysiology of common non-syndromic and rare syndromic craniosynostoses. These findings have implications for diagnosis, risk of recurrence, and risk of adverse neurodevelopmental outcomes. Finally, the use of pathways identified in rare syndromic disease to find genes accounting for non-syndromic cases may prove broadly relevant to understanding other congenital disorders featuring high locus heterogeneity.
Craniosynostosis affects 1 in 2,000 live births, constituting the second most-common craniofacial birth defect following orofacial clefts (1). Premature fusion of any cranial suture is treated surgically in infancy to prevent neurological impairment, with cases affecting the midline (sagittal and metopic) sutures accounting for more than half of all disease (2, 3). Fifteen percent of cases are syndromic, with extracranial malformations, while the remaining 85% are non-syndromic with isolated craniosynostosis (4).
Many genes have been implicated in human syndromic craniosynostoses, with at least 40 showing significant statistical support or having been identified in more than one study (5). Many genes have also been implicated by studies of mouse genetics (4, 6–9). These genes are involved in signaling pathways that converge on promotion of osteoblast differentiation and bone formation. The most prominent of these are the Ras/ERK, bone morphogenetic protein (BMP), and Wnt pathways, with ephrin, smoothened/hedgehog, STAT, and retinoic acid signaling pathways being implicated at lower frequency (5). Examples include gain-of-function (GOF) mutations in FGF receptors 1–3, which present with craniosynostosis of any or all sutures with variable hypertelorism, proptosis, midface abnormalities, and syndactyly, and loss-of-function (LOF) mutations in TGFBR1/2 that present with craniosynostosis in conjunction with severe cardiac/vascular abnormalities and other extracranial defects.
Whereas most syndromic craniosynostoses demonstrate patent Mendelian inheritance (5), the vast majority of non-syndromic cases are sporadic, suggesting a possible role for de novo mutation. To test this possibility, in a prior study of whole-exome sequencing of 191 probands with midline non-syndromic craniosynostosis (NSC), including 132 case-parent trios, we found a significant excess of damaging protein-altering de novo mutations, likely accounting for ∼11% of cases. One gene, SMAD6, surpassed genome-wide thresholds for significance, with three damaging de novo mutations and 10 rare independent transmitted LOF or damaging missense (D-mis) mutations (10). These SMAD6 mutations showed striking incomplete penetrance, which was explained by epistatic interaction with a previously identified (11) common risk allele near the BMP signaling ligand, BMP2 (10). No other gene had more than one damaging de novo mutation. In this prior study, the power to determine which other de novo mutations were pathogenic was limited by the study size and the apparent high degree of locus heterogeneity. We now report results of whole-exome sequencing of a much larger cohort of NSC families.
Results
Exome Sequencing of Midline NSC.
We studied a cohort of 384 probands with NSC affecting the sagittal (n = 237), metopic (n = 136), combined sagittal and metopic (n = 10), or combined sagittal and coronal (n = 1) sutures. These included 291 parent–offspring trios, 132 of which were previously reported (10). Whole-exome sequencing was performed as described in Methods. Of the targeted bases, 96.6% had 8 or more independent reads, and 88.6% had 20 or more (Table S1). In parallel, we analyzed de novo variants in parents and healthy siblings of 1,789 autism probands from the Simons simplex collection sequenced on the same platform. Variants in both cohorts were called using the same GATK pipeline, and de novo mutations were identified using TrioDeNovo (12); the impact of missense mutations was inferred using MetaSVM (Methods).
Table S1.
Exome metrics for all sequenced individuals (n = 966)
| Metric | Value |
| Read length, bp | 74 or 99 |
| No. of reads per sample, M | 66.1 |
| Percentage of targeted bases with at least eight independent reads, % | 96.6 |
| Percentage of targeted bases with more than 20 independent reads, % | 88.6 |
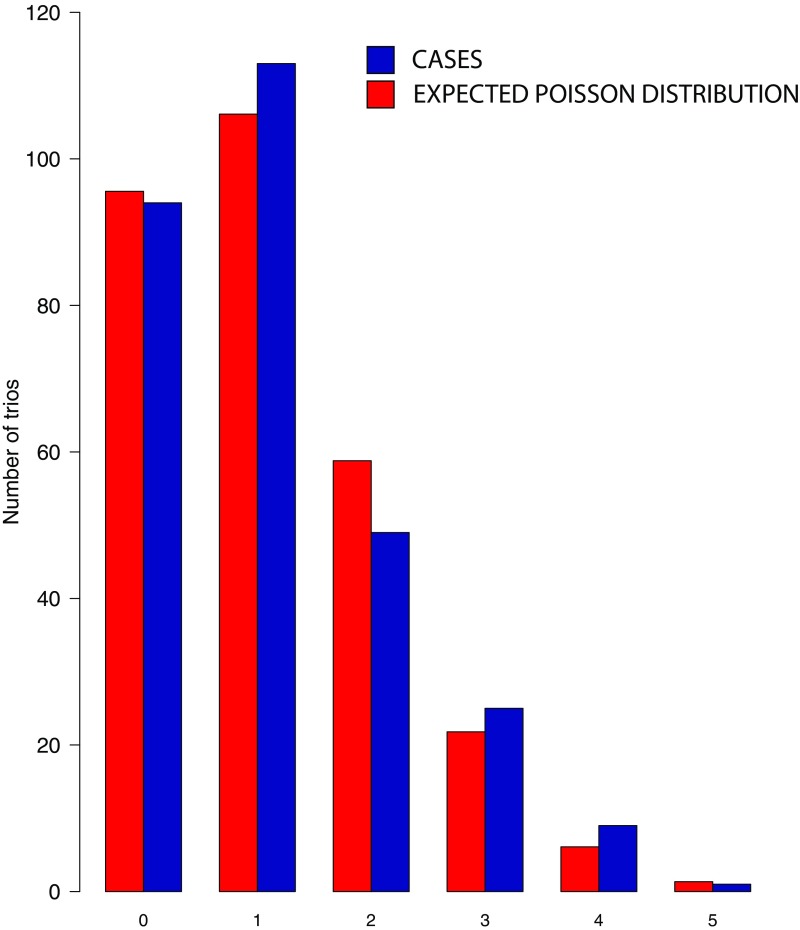
The results identified an average of 1.12 de novo mutations per proband, and closely approximated the expected Poisson distribution (Fig. S1). The observed mutation rate (1.69 × 10−8 per base pair) closely matched both expectation and prior experimental results (Table 1). The total burden of de novo mutations in controls was similar (Table 1).
Fig. S1.
Number of de novo mutations identified per proband in 291 craniosynostosis trios. The blue bars represent the observed distributions of de novo mutations identified per proband in craniosynostosis kindreds, and the red bars represents the expected number from the Poisson distribution. The observed distribution closely approximates expectation.
Table 1.
Burden of de novo mutations in 291 probands with midline craniosynostosis and 1,789 controls
| Mutation class | Cases, n = 291 | Controls, n = 1,789 | ||||||||||
| Observed | Expected | Enrichment | P | Observed | Expected | Enrichment | P | |||||
| n | Rate | n | Rate | n | Rate | n | Rate | |||||
| All genes | ||||||||||||
| Total | 325 | 1.12 | 325.6 | 1.12 | 1.00 | 0.51 | 1,830 | 1.02 | 1,999.5 | 1.12 | 0.92 | 1.00 |
| Synonymous | 61 | 0.21 | 92.3 | 0.32 | 0.66 | 1.00 | 484 | 0.27 | 567.6 | 0.32 | 0.85 | 1.00 |
| Protein altering | 264 | 0.91 | 232.9 | 0.80 | 1.13 | 0.024 | 1,346 | 0.79 | 1,431.9 | 0.80 | 0.94 | 0.99 |
| Total missense | 225 | 0.77 | 204.3 | 0.70 | 1.10 | 0.08 | 1,196 | 0.67 | 1,255.9 | 0.70 | 0.95 | 0.96 |
| T-mis | 175 | 0.60 | 166.0 | 0.57 | 1.05 | 0.28 | 974 | 0.54 | 1020.4 | 0.57 | 0.96 | 0.80 |
| D-mis | 50 | 0.17 | 38.3 | 0.13 | 1.31 | 0.040 | 222 | 0.12 | 235.5 | 0.13 | 0.94 | 0.82 |
| LOF | 39 | 0.13 | 28.6 | 0.10 | 1.36 | 0.037 | 150 | 0.08 | 175.9 | 0.10 | 0.85 | 0.98 |
| Damaging | 89 | 0.31 | 66.9 | 0.23 | 1.33 | 0.006 | 372 | 0.21 | 411.5 | 0.23 | 0.90 | 0.98 |
| Negative regulators of Ras/ERK, BMP, and Wnt signal transduction | ||||||||||||
| Total | 19 | 0.07 | 5.46 | 0.02 | 3.48 | 4.90 × 10−6 | 31 | 0.02 | 33.6 | 0.02 | 0.92 | 0.69 |
| Synonymous | 2 | 0.007 | 1.60 | 0.005 | 1.26 | 0.47 | 12 | 0.007 | 9.8 | 0.005 | 1.22 | 0.28 |
| Protein altering | 17 | 0.058 | 3.87 | 0.013 | 4.39 | 7.35 × 10−7 | 19 | 0.011 | 23.8 | 0.013 | 0.80 | 0.86 |
| Total missense | 9 | 0.031 | 3.42 | 0.012 | 2.63 | 8.56 × 10−3 | 17 | 0.010 | 21.0 | 0.012 | 0.81 | 0.84 |
| T-mis | 2 | 0.007 | 2.50 | 0.009 | 0.80 | 0.54 | 14 | 0.008 | 15.4 | 0.009 | 0.97 | 0.55 |
| D-mis | 7 | 0.024 | 0.92 | 0.003 | 7.65 | 4.82 × 10−5 | 3 | 0.002 | 5.6 | 0.003 | 0.53 | 0.92 |
| LOF | 8 | 0.027 | 0.45 | 0.002 | 17.6 | 3.04 × 10−8 | 2 | 0.001 | 2.8 | 0.002 | 0.72 | 0.77 |
| Damaging | 15 | 0.052 | 1.37 | 0.005 | 10.9 | 2.40 × 10−11 | 5 | 0.003 | 8.42 | 0.005 | 0.59 | 0.92 |
Damaging and tolerated missense called by MetaSVM (D-mis and T-mis, respectively). LOF denotes premature termination, frameshift, or splice-site mutation; damaging denotes LOF and D-mis mutations; n, number of de novo mutations; rate, number of de novo mutations per subject. P values represent the upper tail of the Poisson probability density function. Bold text indicates significant P values.
Significant Burden of Damaging de Novo Mutations.
The distribution of types of de novo coding mutations observed in probands was compared with that expected from the probability of mutation of each base in the coding region and flanking splice sites (13). Probands showed no significant enrichment of synonymous or missense mutations predicted to be tolerated; however, there was significant enrichment of inferred damaging missense (D-mis) and LOF mutations (Table 1). In contrast, no enrichment was observed in any class of coding region mutation in control subjects. From the observed excess of damaging de novo mutations in probands compared with expectation in this set, we infer that damaging protein-altering de novo mutations contribute to ∼8% of cases. SMAD6, with three different LOF and one D-mis de novo mutation, was the only gene with more than one damaging de novo mutation (probability of three or more de novo LOF mutations in SMAD6 by chance = 4.0 × 10−10) (Dataset S1).
Enrichment of Mutations in Negative Regulators of Wnt, BMP, and Ras/ERK Signaling.
As discussed above, mutations that cause rare syndromic forms of craniosynostosis in humans and in mouse cluster in developmental signaling pathways that converge on promotion of osteoblast differentiation and bone formation (5). These are encompassed by the Ras/ERK, Wnt, BMP, hedgehog, ephrin, STAT, and retinoic acid signaling pathways (5). We considered whether de novo mutations in these pathways are enriched in NSC. We anticipated that mutations would most frequently be LOF mutations in negative regulators of signaling rather than GOF in promoters of signaling, because of the generally much larger target size for mutations producing LOF rather than GOF. We first compared the observed and expected burden of damaging de novo mutations in all genes in these pathways (n = 963) as defined by Gene Ontology (GO) (Table S2). There were 19 damaging de novo mutations in cases compared with 4.7 expected from the Poisson distribution, a fourfold enrichment and highly statistically significant (P = 6.7 × 10−7) (Table S3). All 19 of these were confirmed as de novo mutations by Sanger sequencing. There was no enrichment of mutations in this gene set in controls, with 27 damaging mutations observed compared with 29.2 expected (enrichment = 0.92; P = 0.68).
Table S2.
GO terms comprising the curated gene set inclusive of known craniosynostosis pathways
| GO term | Term |
| GO:0030111 | Regulation of Wnt signaling pathway |
| GO:0030510 | Regulation of BMP signaling pathway |
| GO:0046578 | Regulation of Ras protein signal transduction |
| GO:0070372 | Regulation of ERK1 and ERK2 cascade |
| GO:1901187 | Regulation of ephrin signaling pathway |
| GO:0008589 | Regulation of smoothened signaling pathway |
| GO:1904892 | Regulation of STAT cascade |
| GO:0048385 | Regulation of retinoic acid receptor signaling pathway |
GO terms comprising the curated gene set used to assess for enrichment in known craniosynostosis pathways. Of note, the regulation of smoothened signaling pathway term encompasses regulators of hedgehog signaling, for which there is no independent GO term.
Table S3.
Burden of de novo mutations in 291 midline craniosynostosis trios in known craniosynostosis pathways (Wnt, BMP, Ras/ERK, ephrin, hedgehog, STAT, retinoic acid)
| All genes | Cases, n = 291 | Controls, n = 1,789 | ||||||||||
| Observed | Expected | Enrichment | P | Observed | Expected | Enrichment | P | |||||
| n | Rate | n | Rate | n | Rate | n | Rate | |||||
| Total | 36 | 0.12 | 20.3 | 0.07 | 1.78 | 0.001 | 113 | 0.06 | 124.6 | 0.07 | 0.91 | 0.86 |
| Synonymous | 07 | 0.02 | 5.9 | 0.03 | 1.19 | 0.37 | 34 | 0.02 | 36.0 | 0.03 | 0.94 | 0.65 |
| Protein altering | 29 | 0.10 | 14.4 | 0.05 | 2.01 | 4.7 × 10−4 | 79 | 0.04 | 88.6 | 0.05 | 0.89 | 0.86 |
| Total missense | 18 | 0.06 | 12.7 | 0.04 | 1.42 | 0.09 | 72 | 0.04 | 78.0 | 0.04 | 0.92 | 0.77 |
| T-mis | 010 | 0.03 | 9.7 | 0.03 | 1.03 | 0.48 | 52 | 0.03 | 59.4 | 0.03 | 0.88 | 0.85 |
| D-mis | 08 | 0.03 | 3.0 | 0.01 | 2.65 | 0.01 | 20 | 0.01 | 18.6 | 0.01 | 1.08 | 0.40 |
| LOF | 011 | 0.04 | 1.7 | 0.006 | 6.37 | 2.1 × 10−6 | 7 | 0.003 | 10.6 | 0.006 | 0.66 | 0.90 |
| Damaging | 019 | 0.07 | 4.75 | 0.02 | 4.00 | 6.7 × 10−7 | 27 | 0.02 | 29.2 | 0.02 | 0.92 | 0.68 |
Damaging and tolerated missense called by MetaSVM (D-mis and T-mis, respectively). LOF denotes premature termination, frameshift, or splice-site mutation; damaging denotes LOF + D-mis; n, number of de novo mutations in genes within this gene set in 291 subjects; rate, number of de novo mutations per subject. P values represent the upper tail of the Poisson probability density function. Bold text indicates significant P values.
Notably, all 19 of these mutations occurred in either the Wnt, BMP, or Ras/ERK pathway (4.8-fold enrichment, P = 4.8 × 10−8), again with no enrichment in controls (enrichment = 0.90; P = 0.72) (Table S4). We used GO annotation to partition these genes into positive regulators (n = 491) and negative regulators (n = 281) of Wnt, BMP, and Ras/ERK signal transduction, and assessed the burden of de novo mutation in each (Table S4). Fifteen of the damaging de novo mutations were in negative regulators of this pathway, compared with the 1.4 expected (Table 1), a highly significant result (P = 2.40 × 10−11) with extremely strong enrichment (10.9-fold). Among these 15 damaging de novo mutations, there were 8 LOF (17.6-fold enrichment), and 7 D-mis (7.7-fold enrichment); in contrast there were only 2 synonymous and 2 tolerated missense (T-mis) mutations (enrichment 1.0), demonstrating selective enrichment for damaging mutations (Tables 1 and 2). There was no significant enrichment of damaging de novo mutations in this gene set in controls (enrichment 0.6) (Table 1) or in positive regulators of these pathways (Table S4). Enrichment of damaging de novo mutations in negative regulators of these pathways remained highly significant after exclusion of mutations in the previously known gene SMAD6 (8.4-fold enrichment, P = 1.4 × 10−7) (Table S5). Moreover, negative regulators of each of the Wnt, BMP, and Ras/ERK pathways were individually enriched (Wnt enrichment 7.5-fold, P = 1.8 × 10−4; BMP enrichment 19-fold, P = 8.4 × 10−6; Ras/ERK enrichment 10.1-fold, P = 7.4 × 10−4) (Table S6). From the overall enrichment of these mutations, we infer that >90% contribute to NSC risk. We also infer that damaging de novo mutations in negative regulators of Wnt, BMP, or Ras/ERK signaling account for 4.7% of all midline NSC cases studied, and account for 62% of the total excess burden of damaging de novo mutations contributing to NSC in this cohort.
Table S4.
Burden of de novo mutations in 291 midline craniosynostosis trios in regulators of Wnt, BMP, and Ras/ERK signaling
| Regulators | Cases, n = 291 | Controls, n = 1,789 | ||||||||||
| Observed | Expected | Enrichment | P | Observed | Expected | Enrichment | P | |||||
| n | Rate | n | Rate | n | Rate | n | Rate | |||||
| All regulators of Wnt, BMP, and Ras-ERK signal transduction | ||||||||||||
| Total | 33 | 0.11 | 16.8 | 0.06 | 1.96 | 3.2 × 10−4 | 95 | 0.05 | 103.5 | 0.06 | 0.92 | 0.81 |
| Synonymous | 6 | 0.02 | 4.86 | 0.02 | 1.23 | 0.36 | 29 | 0.02 | 29.9 | 0.02 | 0.97 | 0.59 |
| Protein altering | 27 | 0.09 | 12.0 | 0.04 | 2.25 | 1.3 × 10−4 | 66 | 0.04 | 73.6 | 0.04 | 0.90 | 0.83 |
| Total missense | 16 | 0.05 | 10.5 | 0.04 | 1.52 | 0.07 | 60 | 0.03 | 64.8 | 0.04 | 0.93 | 0.74 |
| T-mis | 8 | 0.03 | 7.96 | 0.03 | 1.01 | 0.49 | 44 | 0.02 | 49.2 | 0.03 | 0.89 | 0.70 |
| D-mis | 8 | 0.03 | 2.54 | 0.01 | 3.15 | 0.005 | 16 | 0.01 | 15.6 | 0.01 | 1.02 | 0.50 |
| LOF | 11 | 0.04 | 1.43 | 0.005 | 7.66 | 3.6 × 10−7 | 6 | 0.003 | 8.83 | 0.005 | 0.68 | 0.87 |
| Damaging | 19 | 0.07 | 3.97 | 0.01 | 4.77 | 4.8 × 10−8 | 22 | 0.01 | 24.5 | 0.01 | 0.90 | 0.72 |
| Negative regulators of Wnt, BMP, and Ras-ERK signal transduction | ||||||||||||
| Total | 19 | 0.07 | 5.46 | 0.02 | 3.48 | 4.9 × 10−6 | 31 | 0.02 | 33.6 | 0.02 | 0.92 | 0.69 |
| Synonymous | 2 | 0.007 | 1.60 | 0.005 | 1.26 | 0.47 | 12 | 0.007 | 9.8 | 0.005 | 1.22 | 0.28 |
| Protein altering | 17 | 0.058 | 3.87 | 0.013 | 4.39 | 7.35 × 10−7 | 19 | 0.011 | 23.8 | 0.013 | 0.80 | 0.86 |
| Total missense | 9 | 0.031 | 3.42 | 0.012 | 2.63 | 8.56 × 10−3 | 17 | 0.010 | 21.0 | 0.012 | 0.81 | 0.84 |
| T-mis | 2 | 0.007 | 2.50 | 0.009 | 0.80 | 0.54 | 14 | 0.008 | 15.4 | 0.009 | 0.97 | 0.55 |
| D-mis | 7 | 0.024 | 0.92 | 0.003 | 7.65 | 4.82 × 10−5 | 3 | 0.002 | 5.6 | 0.003 | 0.53 | 0.92 |
| LOF | 8 | 0.027 | 0.45 | 0.002 | 17.6 | 3.04 × 10−8 | 2 | 0.001 | 2.8 | 0.002 | 0.72 | 0.77 |
| Damaging | 15 | 0.052 | 1.37 | 0.005 | 10.9 | 2.40 × 10−11 | 5 | 0.003 | 8.42 | 0.005 | 0.59 | 0.92 |
| Positive regulators of Wnt, BMP, and Ras-ERK signal transduction | ||||||||||||
| Total | 14 | 0.05 | 11.4 | 0.04 | 1.23 | 0.26 | 64 | 0.04 | 70.0 | 0.04 | 0.91 | 0.78 |
| Synonymous | 4 | 0.01 | 3.27 | 0.01 | 1.22 | 0.41 | 17 | 0.01 | 20.1 | 0.01 | 0.84 | 0.79 |
| Protein altering | 10 | 0.03 | 8.12 | 0.03 | 1.23 | 0.30 | 47 | 0.03 | 49.9 | 0.03 | 0.94 | 0.68 |
| Total missense | 7 | 0.02 | 7.14 | 0.02 | 0.98 | 0.57 | 43 | 0.02 | 43.9 | 0.02 | 0.98 | 0.57 |
| T-mis | 6 | 0.02 | 5.52 | 0.02 | 1.09 | 0.46 | 30 | 0.02 | 33.9 | 0.02 | 0.88 | 0.80 |
| D-mis | 1 | 0.003 | 1.62 | 0.006 | 0.61 | 0.80 | 13 | 0.007 | 10.0 | 0.006 | 1.30 | 0.21 |
| LOF | 3 | 0.01 | 0.98 | 0.003 | 3.05 | 0.07 | 4 | 0.002 | 6.04 | 0.003 | 0.66 | 0.85 |
| Damaging | 4 | 0.01 | 2.61 | 0.009 | 1.53 | 0.26 | 17 | 0.01 | 16.0 | 0.009 | 1.06 | 0.44 |
Damaging and tolerated missense called by MetaSVM (D-mis and T-mis, respectively). LOF denotes premature termination, frameshift, or splice-site mutation; damaging denotes LOF + D-mis; n, number of de novo mutations in 291 subjects or 1,789 controls; rate, number of de novo mutations per subject. P values represent the upper tail of the Poisson probability density function. Bold text indicates significant P values.
Table 2.
Damaging de novo mutations in negative regulators of Wnt, BMP, and Ras/ERK signal transduction in 291 midline NSC probands
| Craniosynostosis type | Gene | Mutation | ExAC frequency | pLI |
| Metopic | ARAP3 | IVS6+1delGT | Novel | 0.97 |
| Sagittal | AXIN1 | E322G | Novel | 0.25 |
| Sagittal | DVL3 | G327fs | Novel | 1.00 |
| Sagittal | MESP1 | E104* | 8.92 × 10−5 | 0 |
| Sagittal | NPHP4 | E453K | 0.0002 | 0 |
| Metopic | PSMC2 | R297G | Novel | 1.00 |
| Metopic | PSMC5 | R317W | Novel | 0.97 |
| Sagittal | RASAL2 | R571P | Novel | 0.23 |
| Sagittal+Metopic | SMAD6 | Q78fs | Novel | 0 |
| Metopic | SMAD6 | G88fs | Novel | 0 |
| Sagittal | SMAD6 | E374* | Novel | 0 |
| Metopic | SMAD6 | G390C | Novel | 0 |
| Metopic | SMURF1 | R468W | Novel | 1.00 |
| Sagittal | SPRY1 | Q6fs | Novel | 0 |
| Sagittal | SPRY4 | E160* | Novel | 0.11 |
pLI, measure of intolerance to LOF mutation. Most intolerant score is 1.0
Table S5.
Burden of de novo mutations in negative regulators of BMP/Wnt/Ras-ERK signaling in 277 midline NSC trios without SMAD6 mutations
| Class | Observed | Expected | Enrichment | P value | ||
| n | Rate | n | Rate | |||
| All mutations | 15 | 0.05 | 5.2 | 0.02 | 2.9 | 3.4 × 10−4 |
| Synonymous | 2 | 0.007 | 1.5 | 0.005 | 1.32 | 0.44 |
| Protein altering | 13 | 0.05 | 3.65 | 0.01 | 3.55 | 1.2 × 10−4 |
| Total missense | 8 | 0.03 | 3.25 | 0.01 | 2.46 | 0.02 |
| T-mis | 2 | 0.007 | 2.38 | 0.008 | 0.89 | 0.58 |
| D-mis | 6 | 0.02 | 0.87 | 0.003 | 6.89 | 2.9 × 10−4 |
| LOF | 5 | 0.02 | 0.43 | 0.002 | 11.54 | 8.9 × 10−5 |
| Damaging | 11 | 0.04 | 1.30 | 0.004 | 8.43 | 1.4 × 10−7 |
Damaging and tolerated missense called by MetaSVM (D-mis and T-mis, respectively). LOF denotes premature termination, frameshift, or splice site mutation; damaging denotes LOF + D-mis; n, number of de novo mutations in 277 subjects without a rare damaging de novo or transmitted SMAD6 mutation; rate, number of de novo mutations per subject. P values represent the upper tail of the Poisson probability density function. Bold text indicates significant P values.
Table S6.
Burden de novo mutations in negative regulators in separated Wnt, BMP, and Ras/ERK signaling pathways in 291 subjects with midline craniosynostosis
| Class | Observed | Expected | Enrichment | P value | ||
| n | Rate | n | Rate | |||
| Burden of de novo mutations in negative regulators of Wnt signaling | ||||||
| All mutations | 8 | 0.03 | 3.0 | 0.01 | 2.62 | 0.01 |
| Synonymous | 1 | 0.003 | 0.9 | 0.003 | 1.12 | 0.59 |
| Protein altering | 7 | 0.02 | 2.15 | 0.01 | 3.25 | 0.01 |
| Total missense | 5 | 0.02 | 1.9 | 0.01 | 2.63 | 0.04 |
| D-mis | 4 | 0.01 | 0.55 | 0.001 | 7.25 | 0.002 |
| LOF | 2 | 0.007 | 0.25 | 0.001 | 8.10 | 0.03 |
| Damaging | 6 | 0.02 | 0.80 | 0.002 | 7.50 | 1.8 × 10−4 |
| Burden of de novo mutations in negative regulators of BMP signaling | ||||||
| All mutations | 7 | 0.02 | 0.94 | 0.003 | 7.44 | 5.7 × 10−5 |
| Synonymous | 1 | 0.003 | 0.29 | 0.001 | 3.42 | 0.25 |
| Protein altering | 6 | 0.02 | 0.65 | 0.002 | 9.25 | 6.0 × 10−5 |
| Total missense | 3 | 0.01 | 0.58 | 0.002 | 5.16 | 0.02 |
| D-mis | 2 | 0.007 | 0.20 | 0.001 | 10.22 | 0.02 |
| LOF | 3 | 0.01 | 0.07 | 0.0002 | 44.6 | 4.8 × 10−5 |
| Damaging | 5 | 0.02 | 0.27 | 0.001 | 19.0 | 8.4 × 10−6 |
| Burden of de novo mutations in negative regulators of Ras/ERK signaling | ||||||
| All mutations | 4 | 0.01 | 1.81 | 0.006 | 2.21 | 0.11 |
| Synonymous | 0 | 0 | — | — | — | — |
| Protein altering | 4 | 0.01 | 1.30 | 0.004 | 3.09 | 0.04 |
| Total missense | 1 | 0.003 | 1.14 | 0.004 | 0.88 | 0.68 |
| D-mis | 1 | 0.003 | 0.24 | 0.001 | 4.24 | 0.20 |
| LOF | 3 | 0.01 | 0.16 | 0.0005 | 18.7 | 6.1 × 10−4 |
| Damaging | 4 | 0.01 | 0.40 | 0.001 | 10.1 | 7.4 × 10−4 |
Damaging and tolerated missense called by MetaSVM (D-mis and T-mis, respectively). LOF denotes premature termination, frameshift, or splice-site mutation; damaging denotes LOF + D-mis; n, number of de novo mutations in 291 subjects; rate, number of de novo mutations per subject. P values represent the upper tail of the Poisson probability density function. Bold text indicates significant P values.
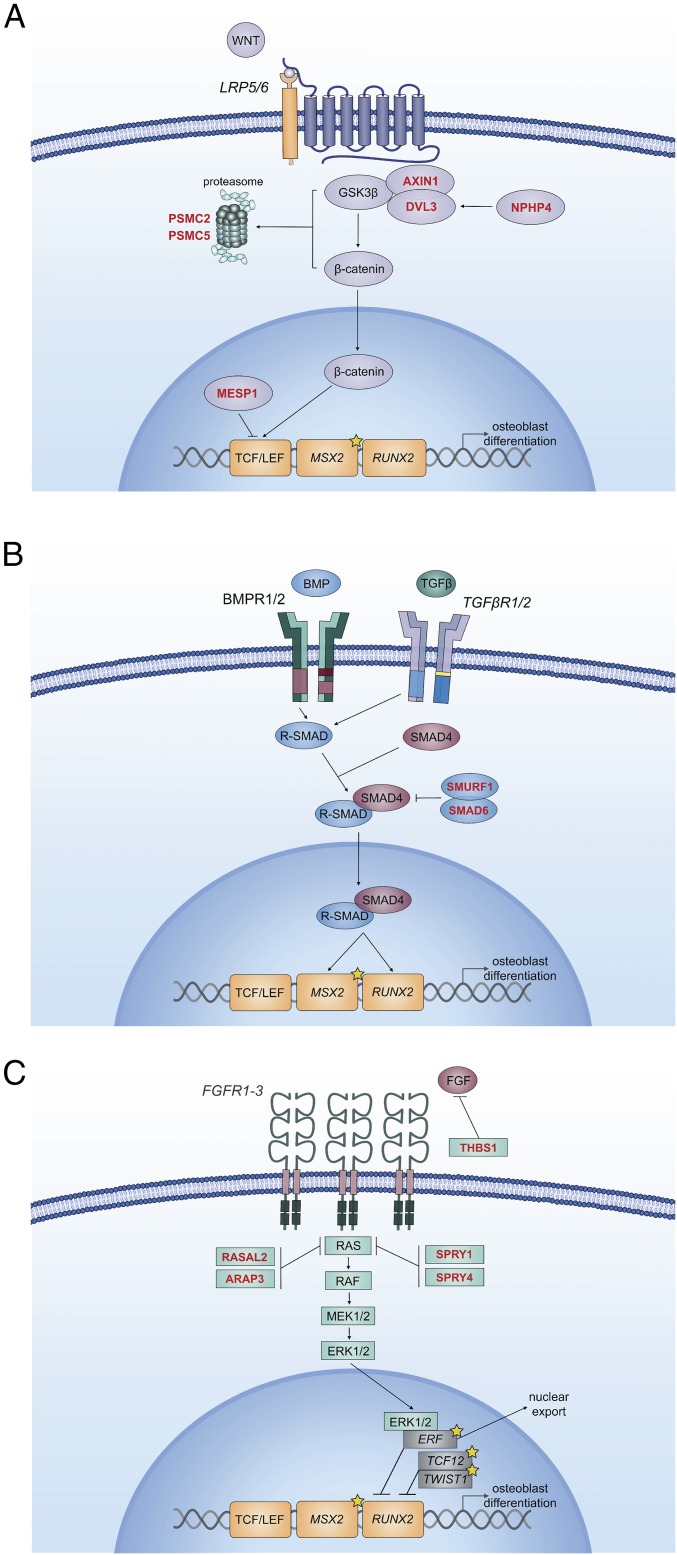
The finding that mutations in these pathways are highly enriched among negative regulators of Wnt, BMP, or Ras/ERK signaling is consistent with the known role of these signaling pathways in promoting osteoblast differentiation and premature bone formation (Fig. 1 and Table 2). In this cohort, damaging de novo mutations in negative regulators of BMP signaling included those in SMAD6 and its binding partner, SMURF1. SMAD6 is an inhibitory SMAD that inhibits nuclear translocation of phosphorylated receptor-SMADs, and SMURF1 is a ubiquitin ligase that ubiquitylates receptor-SMADs and activated BMP receptors, targeting them for proteasomal degradation. Mice with genetic deficiency for SMAD6 or SMURF1, or activation of upstream BMP-receptors each exhibit midline craniosynostosis phenotypes, supporting the human genetic evidence for these two genes (6, 14, 15).
Fig. 1.
De novo mutations in craniosynostosis probands identified in negative regulators of Wnt, BMP, and Ras-ERK signal transduction. Schematic of signaling in Wnt (A), BMP (B), and RAS/ERK (C) pathways are shown. All contribute to osteoblast differentiation and bone formation via common transcriptional targets. Genes in italics are mutated in syndromic craniosynostosis. Genes in red font are negative regulators of signaling that have damaging de novo mutations in probands. Genes noted with yellow stars are known syndromic genes found mutated in kindreds with midline NSC.
Damaging de novo mutations in negative regulators of Wnt signaling included those in AXIN1, MESP1, NPHP4, PSMC2, PSMC5, and DVL3. AXIN1 is part of the complex that constitutively degrades β-catenin, inhibiting Wnt signaling. MESP1 encodes a transcription factor that induces expression of DKKs, inhibitors of canonical Wnt signaling (16). Knockdown of NPHP4 results in accumulation of β-catenin, an effector of canonical Wnt signaling (17). DVL3 is part of a complex that recruits AXIN1 to the nuclear membrane upon Wnt activation of LRP5/6 signaling; however, DVL3 is also required for Wnt signal propagation (18, 19). PSMC2 and PSMC5 are components of the regulatory subunit of the 26S proteasome, and are known to bind ubiquitylated Wnt signaling substrates, promoting their degradation (20, 21). Thus, damaging mutations in these genes can plausibly increase signaling, contributing to NSC.
Damaging mutations in negative regulators of Ras/ERK signaling include those in SPRY1, SPRY4, RASAL2, and ARAP3. SPRY1 and SPRY4 are developmental regulators of EGF and FGF signaling that inhibit Ras activation. RASAL2 is also an inhibitor of the Ras-cAMP pathway. ARAP3 is an Arf and Rho GAP with a Ras-associating domain; however, its biological function is poorly characterized.
In addition to the above mutations there are several others of interest. Two probands with metopic NSC had de novo LOFs in chromatin modifiers (SUV420H1 and SMARCD2); de novo LOFs in these two genes, as well as other chromatin modifiers, have previously been implicated in the pathogenesis of other congenital disorders, including autism and congenital heart disease (22–24), and de novo LOFs in other chromatin modifiers, including ASXL1, KAT6A, and KMT2D have been reported in rare cases of syndromic craniosynostosis (5), suggesting that this gene set plays a role in a smaller fraction of NSC cases. Additional interesting mutations include a de novo D-mis variant in THBS1, a gene with very high expression in midline sutures that regulates the bioavailability of FGF signaling substrates at the plasma membrane (25), and a de novo D-mis variant in MAPK7, a nuclear effector of several RTK signaling pathways (Table 3).
Table 3.
Additional damaging de novo mutations of interest
| Craniosynostosis type | Gene | Mutation | ExAC frequency | pLI |
| Sagittal | ARHGEF18 | F1135fs | 8.70 × 10−6 | 0.91 |
| Sagittal | MACF1 | IVS89+1G > A | Novel | 1.00 |
| Sagittal | MAPK7 | R235W | 8.26 × 10−6 | 0.23 |
| Sagittal | NAA25 | F359fs | Novel | 1.00 |
| Metopic | SMARCD2 | R73* | Novel | 0.98 |
| Metopic | SUV420H1 | T97fs | Novel | 1.00 |
| Sagittal + Left Coronal | TCF12 | K287fs | Novel | 0.97 |
| Sagittal | THBS1 | R980C | 8.47 × 10−6 | 1.00 |
| Metopic | ZCCHC11 | E1275fs | Novel | 1.00 |
Transmitted Mutations in SMAD6 and Epistasis with a Common Risk Allele.
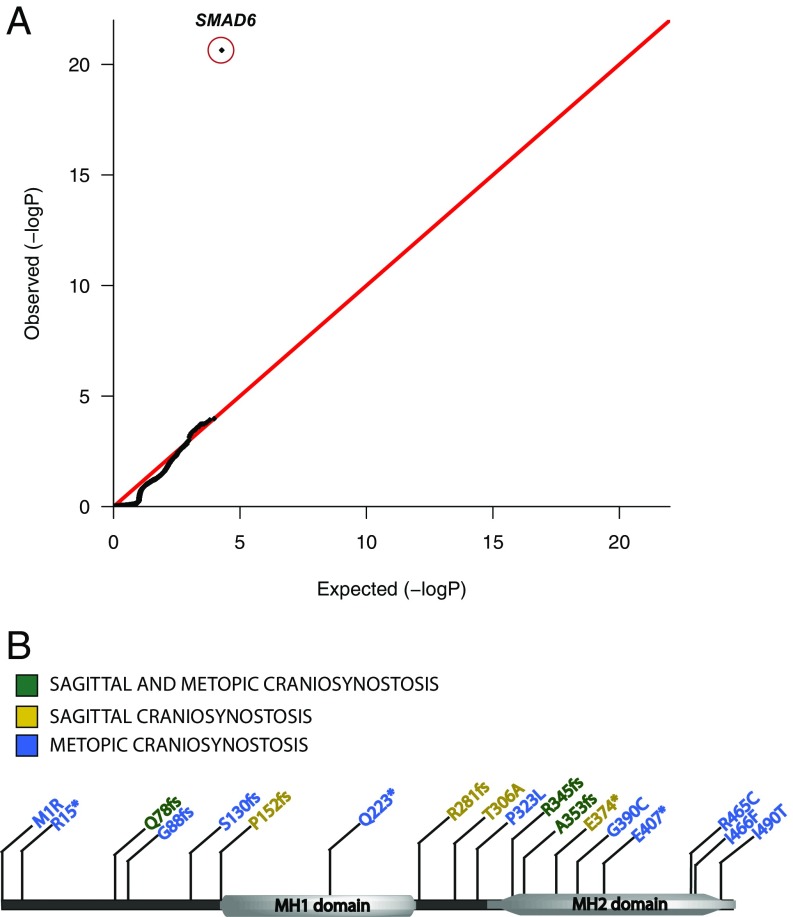
We previously reported a highly significant burden of de novo and rare [Exome Aggregation Consortium (ExAC) minor allele frequency (MAF) < 2 × 10−5] transmitted damaging alleles in SMAD6. Among 193 probands not previously reported, there were 5 novel SMAD6 damaging alleles, including 4 LOF and 1 D-mis; 4 of these were transmitted and 1 was de novo. This frequency of rare transmitted and de novo mutations was not expected by chance (P = 2.7 × 10−7 by binomial test). As before, all four transmitting parents had no sign of craniosynostosis or other congenital anomalies. These results strongly replicate our prior finding. In the combined set of 384 probands there were 18 independent damaging SMAD6 mutations (12 LOFs; LOF P = 2.3 × 10−21 by binomial test) (Fig. 2). In contrast, there were 0 LOFs among 3,337 parental controls and only 5 LOFs among ∼59,000 non-Finnish European alleles in ExAC. These SMAD6 LOFs are thus enriched more than 100-fold versus controls, and damaging SMAD6 alleles comprise 5% of all midline NSC: 8% of all metopic, 2% of all sagittal, and 30% of cases with both sagittal and metopic NSC.
Fig. 2.
Quantile-quantile plot of P values for LOF alleles in all protein-coding genes in 384 craniosynostosis probands. Rare (ExAC frequency < 2 × 10−5) LOF alleles were identified in probands. The probability of the observed number of variants in each gene occurring by chance was calculated from the total number of observed variants and the proportion of the coding length of the exome comprising each gene using the binomial test. (A) The observed distribution of P values matches the expected binomial distribution with the exception of SMAD6, in which 12 LOF alleles are observed compared with the expected 0.10. (120-fold enrichment; P = 2.28 × 10−21). (B) Distribution of all damaging mutations in SMAD6.
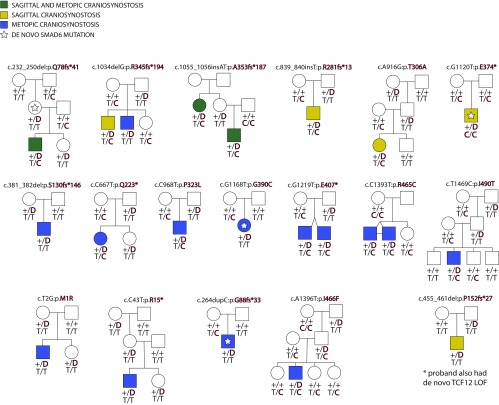
We previously demonstrated incomplete penetrance of SMAD6 mutations, and showed epistasis between these rare mutations and common risk alleles near BMP2 that were the strongest signal in a prior genome-wide association study (GWAS) of sagittal NSC (10, 11). Following addition herein of five new kindreds with a de novo or rare transmitted SMAD6 mutation, we reevaluated penetrance and interaction of SMAD6 and BMP2 alleles by association and parametric analysis of linkage under a two-locus model considering rare damaging alleles of SMAD6 and the common risk allele of SNP rs1884302. The overall estimate of penetrance of rare damaging SMAD6 alleles, excluding probands, is 17%. Among all SMAD6 mutation carriers, 24% of those without the common rs1884302 BMP2 risk allele had NSC, whereas 94% with the BMP2 risk allele had NSC (P = 1.4 × 10−5) (Table S7). There was also highly significant linkage of rare SMAD6 mutations and common BMP2 risk alleles under a parametric two-locus model [logarithm of the odds (LOD) score 8.2, odds >100 million:1 in favor of linkage], which is >1,400× more likely than the best single-locus model (Fig. S2 and Tables S8 and S9) (10).
Table S7.
Risk of craniosynostosis in SMAD6 mutation carriers in the presence or absence of a BMP2 risk allele
| SMAD6/BMP2 genotypes | Craniosynostosis (+) | Craniosynostosis (−) |
| SMAD6 (+) / BMP2 risk allele (+) | 15 | 1 |
| SMAD6 (+) / BMP2 risk allele (−) | 6 | 19 |
| SMAD6 (−) / BMP2 risk allele (+) | 0 | 24 |
All members of kindreds having a proband with SMAD6 mutation were included with exception of one individual with concurrent SMAD6 and TCF12 LOFs. SMAD6(+) indicates the presence of a rare heterozygous LOF or D-mis allele. The reported BMP2 risk allele is “C” at risk locus rs1884302, found within a gene desert ∼345-kb downstream of BMP2. P = 1.6 × 10−10 by the Freeman-Halton extension of Fisher’s exact test for 3 × 2 table analysis. Considering only the association of presence or absence of craniosynostosis in SMAD6 (+) subjects with or without BMP2 risk allele, P = 1.4 x 10−5 by Fisher’s exact test.
Fig. S2.
SMAD6 and BMP2 genotypes in probands with sagittal or metopic craniosynostosis. Pedigrees harboring de novo (denoted by stars within pedigree symbols) or rare transmitted variants in SMAD6. Filled and unfilled symbols denote individuals with and without craniosynostosis, respectively. The SMAD6 mutation identified in each kindred is noted above each pedigree. Below each symbol, genotypes are shown first for SMAD6 (with “D” denoting the damaging allele) and for rs1884302 risk locus downstream of BMP2, (with “T” conferring protection from and “C” conferring increased risk of sagittal craniosynostosis). Maximum LOD scores under two-locus and single-locus linkage models for each kindred are shown in Tables S8 and S9.
Table S8.
Parametric linkage of rare SMAD6 alleles to craniosynostosis under single locus and 2 locus models incorporating BMP2 risk alleles
| Genotype (SMAD6; BMP2) | Penetrance in two-locus model (SMAD6/BMP2) | Penetrance in single-locus model (SMAD6) |
| +,D; C,C | 1 | 0.12 |
| +,D; C,T | 0.85 | 0.12 |
| +,D; T,T | 0.07 | 0.12 |
| +,+; C,C | 0.0032 | 0.0032 |
| +,+; C,T | 0.0008 | 0.0008 |
| +,+; T,T | 0.0002 | 0.0002 |
Table S9.
Family specific LOD scores for each kindred under the two-locus and single-locus models
| Kindred | LOD score, two-locus model | LOD score, single-locus model |
| Q78fs*41 | 0.65 | 0.27 |
| R345fs*194 | 0.16 | 0.63 |
| A353fs*187 | 0.97 | 0.57 |
| R281fs*13 | 0.57 | 0.30 |
| T306A | 0.66 | 0.30 |
| E374* | NA | NA |
| S130fs*146 | 0.30 | 0.30 |
| Q223* | 0.65 | 0.27 |
| P323L | 0.57 | 0.30 |
| G390C | NA | NA |
| E407* | 1.13 | 0.60 |
| R465C | 0.65 | 0.32 |
| I490T | 0.99 | 0.35 |
| M1R | 0.28 | 0.27 |
| R15* | 0.27 | 0.24 |
| I466F | 0.36 | 0.35 |
| G88fs*33 | NA | NA |
| P152fs*27 | NA | NA |
| Total lod score | 8.22 | 5.07 |
| Odds ratio in favor of linkage | 1.7 x 108:1 | 1.2 x 105:1 |
Maximum LOD scores under the parametric models specified above are shown for each kindred harboring a SMAD6 mutation. Kindreds with de novo mutation in the proband and one in which there was co-occurrence of SMAD6 and TCF12 LOFs are not informative for segregation (NA). The LOD score for the two-locus model is >1400× more likely than the single-locus model under the optimized models.
No other individual gene approached genome-wide significance in analysis of LOF dominant alleles, and no gene had more than one rare recessive genotype. Among genes in these pathways with a damaging de novo mutation, AXIN1 had two rare (MAF < 2 × 10−5) heterozygous transmitted damaging mutations (one LOF, one D-mis) and RASAL2 had one. Interestingly, mice with genetic deficiency for the homolog AXIN2 have midline craniosynostosis (26).
Because of the role of the BMP2 risk allele at rs1884302 in modifying the phenotype of rare SMAD6 mutations, we considered whether it might play a similar role with rare damaging alleles in other genes in these pathways. We used the transmission disequilibrium test to determine whether the BMP2 risk allele was transmitted from heterozygous parents to affected offspring significantly more frequently than expected by chance in these 15 kindreds. Among 17 possible transmissions from heterozygous parents, the risk SNP was transmitted in 13 (P = 0.03). Considering only those kindreds with damaging de novo mutations in genes proposed to be more tolerant to heterozygous LOF mutation [measure of intolerance to LOF mutation (pLI) < 0.9; n = 10]—and perhaps in greater need of contribution from additional genetic or environmental factors to produce craniosynostosis—the risk SNP was transmitted in 10 of 11 possible transmissions (transmission disequilibrium test P = 0.007), including all 7 informative transmissions after exclusion of kindreds with de novo SMAD6 mutations (P = 0.008). Although the numbers of events are small, this result suggests that the BMP2 risk SNP might commonly interact with rare alleles in multiple pathways to produce midline craniosynostosis.
Variable Expressivity of Mutations in Syndromic Craniosynostosis Genes.
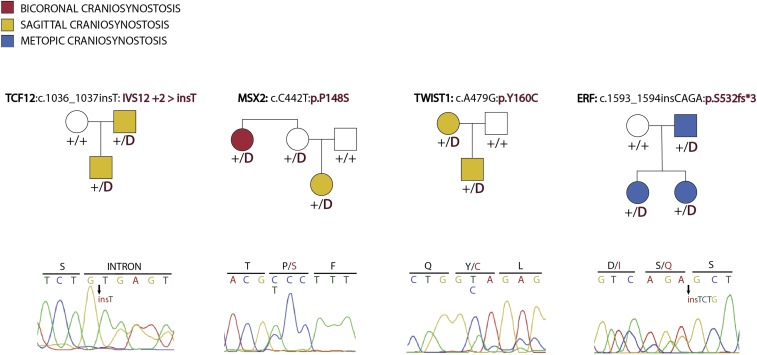
There were 49 probands with 1 or more additional relatives with craniosynostosis in our cohort, including 45 probands without SMAD6 mutations who had 49 affected first-degree relatives and 12 more distant relatives. Among these families, four had previously undescribed transmitted variants (absent in ExAC and other public databases) among 57 genes previously implicated in syndromic craniosynostoses (TWIST1, TCF12, ERF, and MSX2) (Table 4). By comparison, among 3,337 parental controls, there were only 9 damaging variants in 57 genes proposed to contribute to syndromic craniosynostosis (5) (P = 1.9 × 10−5 Fisher’s exact test). All four mutations found in probands were shared by all affected members of these kindreds, an event not likely to occur by chance (probability of cosegregation by chance 1 in 64). These findings make it highly likely that these novel variants are causally related to disease in these families.
Table 4.
Mutations in known syndromic genes in multiplex NSC kindreds
| Craniosynostosis type | Gene | Mutation | ExAC frequency | pLI |
| Metopic | ERF | S532fs | Novel | 0.81 |
| Sagittal, Coronal | MSX2 | P148S | Novel | 0.42 |
| Sagittal | TCF12 | IVS12+2 > insT | Novel | 0.97 |
| Sagittal | TWIST1 | Y160C | Novel | 0.18 |
The novel damaging variant in TWIST1 (p.Y160C) was identified in a parent and child who both had isolated sagittal craniosynostosis with no syndromic features. LOF mutations or missense mutations in the helix–loop–helix domain of TWIST1 cause Saethre-Chotzen syndrome, which almost always involves either uni- or bicoronal craniosynostosis in conjunction with any combination of ptosis, hypertelorism, strabismus, epicanthal folds, syndactyly, dental malocclusion, and mild-to-moderate learning disability (27). P.Y160 lies at the border of the helix–loop–helix domain (amino acids 108–159).
The TCF12 mutation (IVS12+2 > insT) was a “T” insertion that altered the splice donor site of intron 12 from GTGAG to GTTGAG. Multiple in silico splice predictors suggest that this mutant slice donor is less favorable than an alternative GTGAT sequence 29 bases upstream in exon 12 (Methods). This mutation was shared by a parent–offspring pair with isolated sagittal craniosynostosis. LOF mutations in TCF12, a transcription factor that heterodimerizes with TWIST1, cause a syndrome with considerable phenotypic overlap with Saethre-Chotzen syndrome, almost always involving the coronal sutures (28). To our knowledge, there are no previously described cases of isolated sagittal NSC in conjunction with TCF12 mutation.
The mutation in the transcription factor ERF (p.S532fs*3) was a novel frameshift and was shared among a proband, a sibling and their father who all had isolated metopic NSC with no syndromic features. LOF variants in ERF cause multisuture craniosynostosis with variable midface hypoplasia (29). To our knowledge, no cases of isolated metopic NSC have previously been described in conjunction with LOF mutation in ERF.
Finally, the mutation in the homeobox transcription factor MSX2 (p.P148S) was a novel missense variant found in a proband with sagittal NSC and an aunt with bicoronal NSC. Remarkably, this mutation introduces a different substitution at the same codon in the homeodomain as two different mutations previously reported (p.P148L and p.P148H) in two large kindreds with autosomal dominant multisuture craniosynostosis, brachydactyly, and neurologic features, including headache and seizure (Boston-type craniosynostosis) (30, 31). Functional studies of the previously identified missense variants demonstrated that they induced a GOF effect via increased affinity of MSX2 for its target DNA sequence (30, 32). Affected members of our kindred did not have brachydactyly or other syndromic features, and penetrance was incomplete, with an unaffected mother.
Given the atypical sutures involved and completely non-syndromic presentations in these four kindreds, these cases support variable expressivity of known disease loci (Fig. S3). These four genes are all nuclear effectors that modulate signaling downstream of Wnt, BMP, and Ras/ERK signaling, and strengthen the genetic evidence linking syndromic and NSC (Fig. 1).
Fig. S3.
Pedigrees of kindreds with atypical presentations resulting from mutations in known craniosynostosis genes. Four kindreds with dominant transmission of craniosynostosis and novel alleles in genes known to cause syndromic craniosynostosis. Filled and unfilled circles represent the presence or absence of craniosynostosis, with “D” representing the mutation specified above the pedigree, and “+” representing a wild-type allele. In each kindred, all affected individuals share the mutation found in the proband. Sanger sequence electropherograms demonstrating the mutation in each pedigree are shown. None of the affected individuals display the traits typical of the syndrome associated with each gene.
Discussion
These results demonstrate that de novo damaging mutations with large effect in negative regulators of Wnt, BMP, and Ras/ERK pathways play a frequent role in midline NSC. The evidence supporting this conclusion is extremely strong with highly significant P values along with very strong prior biological support of the role of these pathways in craniosynostosis (1, 5). These findings provide strong genetic evidence linking the biology underlying non-syndromic and syndromic forms of craniosynostosis. Among the 12 genes in these pathways with de novo damaging mutations, >90% are expected to be disease-related. The large fraction of mutations that are LOF implies haploinsufficiency as the mechanism of genetic effect, with half the normal gene dose insufficient to provide normal suppression of signaling in development. This suggests these mutations increase signaling, a known mechanism for promoting bone formation and premature suture closure. We presume that syndromic genes act in multiple developmental processes and tissues, whereas non-syndromic genes are either sufficiently expressed in other tissues to not be dose-limited or can be compensated.
Mutations in these three pathways account for 10% of probands in this cohort, with rare damaging de novo or transmitted mutations in SMAD6 found in 5%, damaging de novo mutations in 11 other negative regulators of these pathways in another 4%, and transmitted mutations in 4 genes previously implicated in syndromic craniosynostosis in 1%. De novo mutations in these pathways account for more than 60% of the excess of damaging de novo mutations seen in this cohort. GO analysis of pathway enrichment following removal of genes in these pathways identifies no additional pathways approaching significant enrichment.
These findings have implications for establishing genetic diagnosis and assessing risk of recurrent disease among NSC families. Among the 49 kindreds with familial disease, a likely genetic cause was identified in 16%, with 8% explained by rare damaging transmitted variants in SMAD6, and another 8% with transmitted mutations in genes previously implicated in syndromic disease. As more subjects are sequenced and individual genes are more firmly implicated, the finding of a likely causal de novo mutation in an affected child will provide reassurance to parents that recurrence is unlikely. Similarly, the finding of causal transmitted genotypes can be used for prospective counseling.
Our finding of mutations in genes that characteristically cause syndromic disease in probands with NSC demonstrates variable phenotypic expression of mutations. In the case of MSX2, the finding of a distinct phenotype from a different substitution at P148 in MSX2 strongly suggests an allelic effect; whether others are the result of stochastic factors, allele-specific effects, or genetic modifiers remains to be determined.
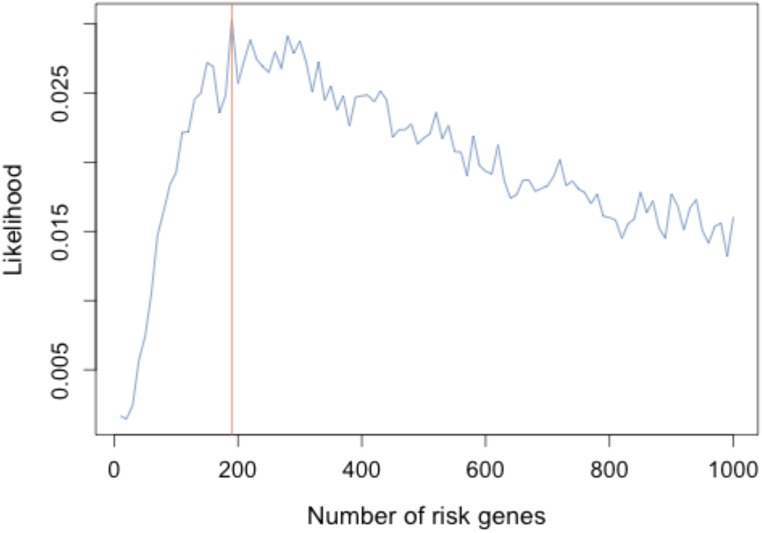
Like autism (24, 33–35) and congenital heart disease (22, 23), damaging mutations in many different genes contribute to NSC. From the observed excess of damaging de novo mutations and the number of genes mutated more than once, we estimate that the number of different genes that contribute to NSC is ∼190 (Fig. S4). Because only SMAD6 has more than one damaging de novo mutation, the confidence intervals of this estimate remain very large. This observation indicates that a large number of cases will need to be sequenced to approach acquisition of a complete set of midline craniosynostosis genes.
Fig. S4.
Estimation of the number of craniosynostosis risk genes by simulation. The likelihood of the model under the assumption of there being 0–1,000 risk genes is plotted, with maximum likelihood observed at 190 genes (Methods).
Although de novo mutations in these pathways impart very high relative risks of disease, the data provide significant evidence that the common risk allele near BMP2 modifies disease risk for many of these genes, as it does for SMAD6. While the results for these other genes are statistically significant, they are based on a small number of observations. Genes with high pLI (e.g., >0.95) are highly predictive of those in which heterozygous LOF mutations impair reproductive fitness and impart disease phenotypes. This is not generally expected for LOF mutations in genes with low pLI. Interestingly, six of the genes with heterozygous de novo LOF mutations among negative regulators of the Wnt, BMP, and Ras/ERK pathways have low pLI (all <0.25); probands with mutations in these genes are particularly enriched for the common BMP2 risk allele, consistent with these rare mutations being insufficient to consistently produce a disease phenotype without additional genetic contributions. This observation raises the broader question of whether other fitness-impairing traits associated with haploinsufficient mutations in genes with low pLI may also frequently require genetic or environmental modifiers to produce disease.
Whereas fusion of a cranial suture is the only physical manifestation of disease in NSC, more than one-third of all cases have subtle yet clinically recognizable neurocognitive deficits (36–38). The etiology of these deficits remains enigmatic to date. Precise regulation of BMP, Wnt, and Ras/ERK signaling is crucial to normal brain development (39–42), raising the possibility that genotype might influence neurocognitive outcome, as appears to be the case for congenital heart disease (22, 23). The results provide a means of stratifying non-syndromic patients to assess the extent to which specific genotypes contribute to neurocognitive outcome.
Finally, the genetic similarities and differences between syndromic and NSC are striking. Probands with craniosynostosis and distinctive extracranial phenotypes predictably have mutation in specific genes. In contrast, the genetic causes in probands with NSC are thus far explained in a small fraction of cases that are highly genetically heterogeneous, making their identification more challenging. Nevertheless, shared biological pathways provide genetic contributions to both rare syndromic and more common non-syndromic probands. Without the prior study of syndromic craniosynostosis, the pathway analysis that led to the current results would not have been as statistically powerful. This observation underscores the importance of solving all rare syndromic cases, as they commonly prove relevant to identification of pathways relevant to non-syndromic disease. These considerations also support the general use of rigorous pathway analysis in the study of non-syndromic diseases that are likely highly genetically heterogeneous.
Methods
Subjects and Samples.
Participants for this study were ascertained from the Yale Pediatric Craniofacial Clinic, the Pediatric Neurosurgery Clinic at the Medical University of Silesia, Poland, or by responding to an invitation posted on the Cranio Kids–Craniosynostosis Support and Craniosynostosis–Positional Plagiocephaly Support Facebook pages. All participating individuals or their parents provided written informed consent to participate in a study of the genetic cause of craniosynostosis in their family. Inclusion criteria included a diagnosis of sagittal or metopic craniosynostosis in the absence of known syndromic forms of disease by a craniofacial plastic surgeon or pediatric neurosurgeon. All probands had undergone reconstructive surgery. Participating family members provided buccal swab samples (Isohelix SK-2S buccal swabs) or saliva samples (DNA Genotek OGR-575), craniofacial phenotype data, medical records, operative reports, and imaging studies. The study protocol was approved by the Yale Human Investigation Committee Institutional Review Board.
Control trios were those sequenced from the Simons Foundation Autism Research Initiative Simplex Collection (24, 35, 43). Simplex families composed of two unaffected parents, one child with autism, and one unaffected sibling, underwent whole-exome sequencing, with 1,789 trios of unaffected family members serving as controls for this study.
Exome Sequencing and Analysis.
DNA was prepared from buccal swab or saliva samples according to the manufacturer’s protocol. Exome sequencing was performed by exon capture using the Roche MedExome, Roche V2, or IDT xGen capture reagent, followed by either 74 or 99 base paired-end sequencing on the Illumina HiSEq. 2000.
Sequence reads were aligned to the GRCh37/hg19 human reference genome using BWA-Mem. Local realignment and quality score recalibration were performed using the GATK pipeline, after which variants were called using the GATK Haplotype Caller. A Bayesian algorithm, TrioDeNovo, was used to call de novo mutations (12). VQSR “PASS” variants with ExAC allele frequency ≤ 10−3 sequenced to a depth of 8 or greater in the proband and 10 or greater in each parent with Phred-scaled genotype likelihood scores >30 and de novo quality scores [log10(Bayes factor)] >6 were considered. Independent aligned reads at variant positions were visualized in silico to remove false calls. All retained calls had de novo genotype quality scores of 100. Fifty de novo mutations were randomly selected for validation by bidirectional Sanger sequencing of the proband and both parents; 100% of these tests confirmed de novo mutation in the proband. All de novo variants contributing to significant results, specifically each of the damaging variants identified in the gene set for negative regulation of Wnt, BMP, and Ras/ERK signaling, were confirmed by Sanger sequencing. Transmitted variants were called as per above. All de novo and transmitted variants were annotated using ANNOVAR (44). Allele frequencies of identified variants were taken from the ExAC database. The impact of nonsynonymous variants was predicted using the MetaSVM rank score, with scores greater than 0.83357 serving as a threshold for predicting that the mutation was deleterious (MetaSVM “D”, D-mis) (45). Transmitted variants identified in known craniosynostosis genes (Table 4) were confirmed by Sanger sequencing.
Burden of de Novo Mutations.
Statistical analysis of the burden of de novo mutations in craniosynostosis cases and autism controls were performed in R using the denovolyzeR package, as previously described (10, 46). The expected number of de novo mutations in case and control cohorts across variant classes was calculated, and this value was compared with the observed number in each cohort using Poisson statistics (13). For gene-set enrichment analyses, only mutations observed or expected in genes within the specified gene set were included in each statistical test.
Contribution of de Novo Mutation to Craniosynostosis.
The number of cases with protein-altering de novo mutations genes was calculated. This number (n = 89), minus the number expected by chance (n = 66.9), is the number of mutations expected to contribute to craniosynostosis risk (n = 22.1). Dividing this number by the total number of trios (n = 291) provides the percentage of cases in which damaging de novo mutations genes are expected to cause craniosynostosis (7.6%).
Binomial Test.
The observed distribution of rare (ExAC frequency < 2 × 10−5) LOF alleles was compared with the expected distribution using the binomial test. The total number of LOF alleles in 384 probands was tabulated (n = 2,260), and the expected number for each gene was calculated from the proportion of the exome comprising the coding region of each gene, multiplied by the total number of LOF alleles identified in cases. Enrichment was calculated as the number of observed mutations divided by the expected number.
In Silico Splice-Site Prediction.
Splice-site predictors Human Splicing Finder (47) and Genie (48) were used to assess the impact of a single nucleotide insertion at the splice donor site of intron 12 of TCF12 that changed the donor site sequence from GTGAG to GTTGAG. Both programs predicted that the insertion resulted in a more favorable splice site 29 bases upstream in exon 12. Genie assigned a score of 0.55 to the wild-type canonical splice donor site; however, after mutation of the wild-type sequence to GTTGAG, the alternate splice donor 29 bases upstream was assigned a score of 0.52, while the GTTGAG sequence had a score <0.1. Using Human Splicing Finder, the wild-type donor-site sequence was deemed the most likely donor site with a MaxENT score of 7.0; however, after mutation of this sequence to GTTGAG, the alternative sequence 29 bases upstream became the most likely donor site with a MaxENT score of 5.99.
Estimating the Number of Risk Genes in Midline Craniosynostosis.
We followed a previously described method to estimate the number of genes in which damaging de novo mutations contribute risk to craniosynostosis (23). In summary, the number of observed damaging mutations in 291 craniosynostosis cases (K), the number of those genes mutated twice (R1), and number of those genes mutated at least three times (R2) was tabulated. We next estimated the fraction of damaging alleles in risk genes based on observed enrichment in cases compared with expectation [E = (M1 − M2)/M1, where M1 and M2 are the numbers of damaging mutations in cases and expectation, respectively]. The likelihood of risk gene number L(G) was simulated, with the number of damaging mutations (K) fixed at the observed number in cases. G risk genes were selected from all genes with equal probability, and the number of contributing damaging mutations in risk genes (C1) was established by sampling from a binomial distribution Binom(K,E). The number of noncontributing damaging mutations was set as C2 = K − C1. We then simulated C1 contributing damaging mutations by sampling with replacement from G genes and C2 noncontributing mutations from all genes using their background mutation rate as probability weights. In this process, 20,000 simulations were performed for every G from 1 to 1,000, and L(G) was set to be the proportion of simulations in which the number of genes with two damaging mutations was exactly R1 and the number of genes with more than two damaging mutations was exactly R2.
Supplementary Material
Acknowledgments
This project was supported by the Yale Center for Mendelian Genomics (NIH Grant M#UM1HG006504-05), the NIH Medical Scientist Training Program (NIH/National Institute of General Medical Sciences Grant T32GM007205), and the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: J.S. and R.P.L. were among 42 coauthors on a 2015 review article.
Data deposition: The sequences for case-parent trios reported in this paper have been deposited in the NCBI database of Genotypes and Phenotypes (dbGaP) (accession no. phs000744).
2Members of the Yale Center for Genome Analysis: Kaya Bilguvar, Shrikant Mane, Irina Tikhonova, Christopher Castaldi, and James Knight.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709255114/-/DCSupplemental.
Contributor Information
Collaborators: Kaya Bilguvar, Shrikant Mane, Irina Tikhonova, Christopher Castaldi, and James Knight
References
- 1.Heuzé Y, Holmes G, Peter I, Richtsmeier JT, Jabs EW. Closing the gap: Genetic and genomic continuum from syndromic to nonsyndromic craniosynostoses. Curr Genet Med Rep. 2014;2:135–145. doi: 10.1007/s40142-014-0042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenwood J, Flodman P, Osann K, Boyadjiev SA, Kimonis V. Familial incidence and associated symptoms in a population of individuals with nonsyndromic craniosynostosis. Genet Med. 2014;16:302–310. doi: 10.1038/gim.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slater BJ, et al. Cranial sutures: A brief review. Plast Reconstr Surg. 2008;121:170e–178e. doi: 10.1097/01.prs.0000304441.99483.97. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty K, Singh N, Richtsmeier JT. Understanding craniosynostosis as a growth disorder. Wiley Interdiscip Rev Dev Biol. 2016;5:429–459. doi: 10.1002/wdev.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Twigg SR, Wilkie AO. A genetic-pathophysiological framework for craniosynostosis. Am J Hum Genet. 2015;97:359–377. doi: 10.1016/j.ajhg.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komatsu Y, et al. Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J Bone Miner Res. 2013;28:1422–1433. doi: 10.1002/jbmr.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behr B, Longaker MT, Quarto N. Differential activation of canonical Wnt signaling determines cranial sutures fate: A novel mechanism for sagittal suture craniosynostosis. Dev Biol. 2010;344:922–940. doi: 10.1016/j.ydbio.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Behr B, Longaker MT, Quarto N. Absence of endochondral ossification and craniosynostosis in posterior frontal cranial sutures of Axin2(-/-) mice. PLoS One. 2013;8:e70240. doi: 10.1371/journal.pone.0070240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruyama T, et al. Rap1b is an effector of Axin2 regulating crosstalk of signaling pathways during skeletal development. J Bone Miner Res. 2017 doi: 10.1002/jbmr.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timberlake AT, et al. Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. eLife. 2016;5:5. doi: 10.7554/eLife.20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Justice CM, et al. A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nat Genet. 2012;44:1360–1364. doi: 10.1038/ng.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Q, et al. A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics. 2015;31:1375–1381. doi: 10.1093/bioinformatics/btu839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimazu J, Wei J, Karsenty G. Smurf1 inhibits osteoblast differentiation, bone formation, and glucose homeostasis through serine 148. Cell Reports. 2016;15:27–35. doi: 10.1016/j.celrep.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Retting KN. 2008. Smad proteins and the regulation of endochondral bone formation. PhD dissertation (University of California, Los Angeles)
- 16.David R, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 17.Burcklé C, et al. Control of the Wnt pathways by nephrocystin-4 is required for morphogenesis of the zebrafish pronephros. Hum Mol Genet. 2011;20:2611–2627. doi: 10.1093/hmg/ddr164. [DOI] [PubMed] [Google Scholar]
- 18.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HJ, et al. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 2006;20:1933–1945. doi: 10.1101/gad.1411206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li VS, et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homsy J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connerney J, et al. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- 26.Yu HM, et al. The role of Axin2 in calvarial morphogenesis and craniosynostosis. Development. 2005;132:1995–2005. doi: 10.1242/dev.01786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard TD, et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 28.Sharma VP, et al. 500 Whole-Genome Sequences (WGS500) Consortium Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat Genet. 2013;45:304–307. doi: 10.1038/ng.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Twigg SR, et al. Reduced dosage of ERF causes complex craniosynostosis in humans and mice and links ERK1/2 signaling to regulation of osteogenesis. Nat Genet. 2013;45:308–313. doi: 10.1038/ng.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jabs EW, et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 31.Florisson JM, et al. Boston type craniosynostosis: Report of a second mutation in MSX2. Am J Med Genet A. 2013;161A:2626–2633. doi: 10.1002/ajmg.a.36126. [DOI] [PubMed] [Google Scholar]
- 32.Liu YH, et al. Msx2 gene dosage influences the number of proliferative osteogenic cells in growth centers of the developing murine skull: A possible mechanism for MSX2-mediated craniosynostosis in humans. Dev Biol. 1999;205:260–274. doi: 10.1006/dbio.1998.9114. [DOI] [PubMed] [Google Scholar]
- 33.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Rubeis S, et al. DDD Study Homozygosity Mapping Collaborative for Autism UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magge SN, Westerveld M, Pruzinsky T, Persing JA. Long-term neuropsychological effects of sagittal craniosynostosis on child development. J Craniofac Surg. 2002;13:99–104. doi: 10.1097/00001665-200201000-00023. [DOI] [PubMed] [Google Scholar]
- 37.Shipster C, et al. Speech, language, and cognitive development in children with isolated sagittal synostosis. Dev Med Child Neurol. 2003;45:34–43. [PubMed] [Google Scholar]
- 38.Sidoti EJ, Jr, Marsh JL, Marty-Grames L, Noetzel MJ. Long-term studies of metopic synostosis: Frequency of cognitive impairment and behavioral disturbances. Plast Reconstr Surg. 1996;97:276–281. doi: 10.1097/00006534-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 39.Bier E, De Robertis EM. EMBRYO DEVELOPMENT. BMP gradients: A paradigm for morphogen-mediated developmental patterning. Science. 2015;348:aaa5838. doi: 10.1126/science.aaa5838. [DOI] [PubMed] [Google Scholar]
- 40.Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- 41.Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat Rev Neurosci. 2007;8:583–596. doi: 10.1038/nrn2189. [DOI] [PubMed] [Google Scholar]
- 42.Richtsmeier JT, Flaherty K. Hand in glove: Brain and skull in development and dysmorphogenesis. Acta Neuropathol. 2013;125:469–489. doi: 10.1007/s00401-013-1104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong C, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ware JS, Samocha KE, Homsy J, Daly MJ. Interpreting de novo variation in human disease using denovolyzeR. Curr Protoc Hum Genet. 2015;87:7.25.1–7.25.15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desmet FO, et al. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.